Abstract
MDCK cells are widely used to study the differential targeting of membrane transporters to apical and basolateral membrane but its canine origin limited the commercial tools available for the analysis of protein trafficking machinery. Because apical and basolateral membranes are only found in differentiated epithelial cells, genes critical for differential targeting may be specifically up-regulated upon MDCK cell differentiation. To search for these genes, a cross-species screening strategy was used. We first analyzed the human microarray data for protein trafficking-related genes that were up-regulated in colon carcinoma Caco2 cells upon differentiation. The results of mouse 44K gene expression microarray analysis were then used to extract additional candidate genes that showed higher expression in normal colon epithelium compared to primary embryonic fibroblasts. Finally, NCBI genomic sequence information was used to design RT-PCR primers for 13 candidate and 10 negative control genes and used to analyze MDCK cells at 2, 13 and 17 days after seeding. To determine whether the gene up-regulation was specific in epithelial differentiation, we also performed RT-PCR on rat non-differentiating intestinal IEC-6 cells and mouse C2C12 cells, a differentiating myoblast model. Of the 13 candidate genes, 3 genes, SDCBP2, KIF12, KIF27, met all criteria of specific up-regulation in differentiated MDCK cells. In addition, KIF13A showed up-regulation in differentiated MDCK and C2C12 cells but not in IEC-6 cells cultured for the same duration. The functions of these genes need to be analyzed in the future. This cross-species screening strategy may be useful for other non-human, non-rodent cell models.
Keywords: MDCK cells, Differentiation, Epithelium, Kinesin, RAB, SDCBP2
1. Introduction
Epithelial cells are responsible for the material exchange between multi-cellular organisms and the environment [1]. For example, membrane transporters in the intestine and kidney epithelial cells are in charge of the unidirectional nutrient absorption/reabsorption as well as the foreign chemical and waste secretion [2], [3], [4], [5]. To accomplish these specialized functions, differentiated epithelial cells have two plasma membrane domains: an apical membrane facing the luminal environment and a basolateral membrane facing the interior of the organism [2]. Many membrane transporters are targeted differentially to these two domains upon the formation of tight junction [6], [7], [8], [9], [10]. Although peptide signals for the differential targeting have been studied extensively [7], [11], only limited information is available on the trafficking machinery. Because luminal and basolateral bifurcation is only present in differentiated epithelial cells, it is likely that epithelial cell type will have a unique set of gene expression for the differential targeting. Furthermore, the genes involved in the differential targeting may also increase their expression during epithelial differentiation, either in a sustained or transient way that may start at early or late stages of the differentiation of the epithelia.
The cell line Madin–Darby canine kidney (MDCK) is the most widely used epithelial model [12], [13]. The strengths of this model include its origination from normal kidney and its spontaneous differentiation [14]. MDCK cells develop apical/basolateral polarity and tight junctions [15], [16], [17], structure that constitutes the frontier between the two plasma membrane domains as well as a fence that avoids the lateral diffusion of components of one membrane domain to the other [18], [19]. Although it is ideally suited to examine the machinery that targets transporters to apical and basolateral membrane, limited endogenous gene expression information is available for MDCK cells. The canine origin of the cell line has limited the availability of commercial tools. For example, the latest Affymetrix Canine array was based on 2005 Canis lupus familiaris genome information. In addition, the presence of canine gene transcripts was mostly predicted without experimental validation.
This study demonstrated the use of available non-canine gene expression information to predict genes that are up-regulated during the differentiation of canine epithelial cells but not during long-term culturing of non-differentiating epithelial cells or during the differentiation of muscle cells. One focus was on gene families that are known to play roles in the protein targeting such as GTP-binding member RAS oncogene family (RAB) [20], [21] and kinesin gene family (KIF) [22], [23]. The cross-species prediction has reasonable accuracy as expected from evolution conservation. Three genes, SDCBP2, KIF12 and KIF27, meet the criteria. Our results also indicate the expression of 16 previously predicted canine genes in MDCK cells.
2. Materials and methods
2.1. Materials
All cell culture reagents were from Invitrogen Corp. (Carlsbad, CA) except characterized fetal bovine serum purchased from Hyclone, USA (Logan, UT). All other chemicals used were of reagent grade. Primers used for RT-PCR were custom-synthesized by Eurofins Genomics (Huntsville, AL).
2.2. Gene expression microarray analysis
The workflow of the entire study is shown in Fig. 1. Two-color gene expression microarray data are available for human colon carcinoma cell line, Caco2 [24], [25] (NCBI GEO DataSets Accession: GSE7442). We compared the level of gene expression between pre-confluent Caco-2 cells (day 1 and 2 post-seeding) (n = 4 total) and differentiated Caco-2 cells (day 14 and 21 post-seeding) (n = 3 for each time point). Results from two sets of mouse single-color gene expression arrays (4 × 44K, Agilent Technologies) described in our previous publications were also compared: primary mouse embryonic fibroblasts (MEF) (representing non-epithelial cell type) (n = 6) [26], and mouse proximal (ascending) colon epithelium (n = 9) [27]. The two sets of mouse array were prepared, processed and analyzed by the same facility [26], [27]. Some genes were in multiple features within the array and our cross-array comparisons were all made between signals from matching features. Non-normalized array signal intensities were used for the comparison.
Fig. 1.
Study design. For the Caco2 analysis, 4 arrays of preconfluent Caco2 cells were compared to 6 arrays of differentiated Caco2 cells. For the mouse tissue analysis, 6 mouse primary embryonic fibroblast arrays were compared to 9 mouse proximal colon epithelium arrays. For all culture cells, cells at D2 after seeding (n = 3) were compared to D13 (n = 3) and D17 (n = 3) after seeding. MDCK and C2C12 cells were well differentiated at D13 and D17 after seeding.
2.3. Cell culture and RNA extraction
Three cell lines for RT-PCR were used based on their origination and their biological properties. None of the cell lines were originated from tumors. Canine MDCK cells were used as the model for differentiating epithelial cells [12], [13], [14] while rat intestinal IEC-6 cells as the model of non-differentiating epithelial cells [28]. Mouse skeletal muscle C2C12 cells was chosen as the model of differentiating cells from a different embryonic layer [29]. All cells were cultured following our previous publication in 6-well plates [6], [30], [31]. Day 2, 13 and 17 were chosen for gene expression analysis based on our past studies with MDCK cells. MDCK cells were sub-confluent at 2 days after seeding at 1 × 104 cells/cm2. Domes in the monolayer, a late sign of tight junction and cell polarity development, started to appear at 4 days after seeding. We observed polarized distribution of membrane transporters in MDCK cells between 13 days and 17 days after seeding [6], [7], [11], [32]. For comparison, IEC-6 cells and C2C12 cells were also cultured and harvested at 2 days, 13 days and 17 days after seeding. By adjusting the seeding density to 2 × 103 cells/cm2 for C2C12 cells and 2 × 104 cells/cm2 for IEC-6 cells, all three cell lines were sub-confluent at 2 days after seeding and reached 90–100% confluence at around 4 days after seeding. Different from MDCK cells, IEC-6 cells did not show any dome formation even at 17 days after seeding. The fusion of C2C12 myoblasts started at 4 days after seeding, which led to the formation of well-organized multinucleated myofibrils by Day 13 and 17 [29]. For all cell lines, RNA extraction was performed following the manufacturer's protocol (MasterPure RNA purification kit, Epicentre, Madison, WI, USA). All extracted RNA samples had OD260 to OD280 ratios greater than 1.9 and 28S rRNA bands (stained by ethidium bromide) nearly twice as strong as those of 18S rRNA. RNA concentrations in samples were determined by OD260. Protein concentrations in similarly cultured cells were also determined using a modified Lowry assay [33]. Triplicate samples at each time points were from three independent wells of cells.
2.4. Known tissue presence, canine genome annotation, and primer design for RT-PCR
The UniGene database was used to obtain the information on the presence of these genes of interest in kidney and intestinal tissues across several mammalian species. The database also provided information on the status of each gene, predicted or known expression, in the canine genome. Canine RT-PCR primers were designed based on CanFam3.1 assembly (build released 2012) for 23 genes and the control GAPDH (Table 1). The known or predicted mRNA sequences of selected genes in the C. lupus familiaris genome were first obtained through UniGene in NCBI website. BLAST was then applied to identify all known and predicted transcript variants. Cross-species BLAST was also used to determine the likely transcripts in the C. lupus familiaris genome. The common exons among the transcript variants of each gene were used for the primer design with Primer-BLAST. PCR primer pairs were all designed to amplify sequences across several intron–exon junctions, and the PCR products ranged from 300 to 1000 bp in sizes (Table 1). To perform RT-PCR on rat IEC-6 cells and mouse C2C12 cells, additional primers were designed from the genomes of Rattus norvegicus and Mus musculus (Table 1). The rodent primers were all from the same region of the genes as the MDCK primers (Table 1).
Table 1.
Epithelial tissue expression and canine genome status of genes studied and their RT-PCR primers used for the semi-quantitative gene expression analysis in MDCK, IEC-6 and C2C12 cells.
| Genes | Intestine and kidneya | Canine genomeb | Primers for MDCK (forward and reverse, 5′–3′)b | Product (bp) | Primers for IEC-6 and C2C12 (forward & reverse, 5′–3′)c | Product (bp) |
|---|---|---|---|---|---|---|
| Primer pair for RT-PCR normalization gene | ||||||
| GAPDH | yes | yes | AGTCCATCTCCATCTTCCAG CGTCACGCCACATCTTCC |
381 | ATCCCATCACCATCTTCCAG GCCATCACGCCACAGTTTCC |
393 |
| Primer pairs for differentiation-related genes (identified through human microarray) | ||||||
| C2CD2L | yes | predicted | CACCTGGGAAGTGAGTTGG CGTTGGAAAGGTGGCTCTCC |
622-5 | ||
| EXOC2 | yes | predicted | TCCGCTTGGTATTGAGATTG TTTCCGTTCCATCCGCTTC |
296-9 | GCGAATCCTCTTGGCATTG CTTTTTCAGTTCCATCTGCTTC |
307 |
| MYLK | yes | predicted | CTCCAGTGTCAGGTGTCGTC GCTGCCATTCTCACTGGTC |
531 | ||
| MYO7B | yes | predicted | CGACATCCACTTCAACCCC CCTGGTGACAAACTCCCCTC |
502 | ACCATCCGCAATGACAACTC GGACAGGGATTGTGTGTTTG |
523 |
| SDCBP2 | yes | predicted | TCTGGAGGACCTGAAGATGG CAGCCCCACAAGAGACG |
403 | TCTGGAGGACCTGAAGATGG GCAAACCCACAAGGGATGC |
419-31 |
| Primer pairs for chosen KIF genes (identified through mouse microarray) | ||||||
| KIF1A | yes | yes | CCCCTTGATGTCCGAGTGTC GAGTAGAGCGTGTCGGTGAG |
780 | ATCAAAGATGGCGTCACCAG TCGTGTCTCTCGGTCTTTGG |
804 |
| KIF5A | yes | predicted | TGCGGGGAGACAAGTTCATC GCACCCGATTCTTGTCCTCA |
417 | TGCGGGGAGACAAGTTCATC CTTGACAAACGGCACCCG |
428 |
| KIF5B | yes | predicted | GCAAGCAGAAAACGATGCC GTTGCTCTCAGTTTGTGTGC |
427 | ||
| KIF12 | yes | predicted | GGAAGACCTACACCCTGACC GACTCCCAAACTCCACCACC |
276 | TACACTCTGACTGGACCTCC CTGTTCCACATAGAAGCCTC |
423 |
| KIF13A | yes | predicted | AAGTGATGCGAGTGACAGGG CCAGCACAGCGTTTCTTTCC |
969 | GGCGGAGGATGACTCTTC CACTGCTCCACCAGCCTG |
858 |
| KIF21A | yes | predicted | CCGAAGGAGAACCACCACTC CAGGAAATGGGTTGATTATGCCC |
524 | ||
| KIF27 | yes | predicted | CACTTTGTGGACTTGGCTG GTTAGTTCTTCGCTCAGTTG |
923-38 | CACTTTGTGGACTTGGCTG TGAAATCCTGTCTTGATGGG |
1084 |
| KIFAP3 | yes | predicted | TTTCCACCAAGCCACAAGAG GGAAATGAATGCTGCCCAAC |
411 | GCCACAAGAGATGTCATAATC GGAAATGAGTGCTGTCCAACIEC−6 AAATGCGTGTTGCCCCACC2C12 |
399-401 |
| Primer pairs for chosen RAB genes (identified through mouse microarray) | ||||||
| RAB1A | yes | yes | GCGACTCTGGGGTTGGAAAG TTGCTCCAGGACCCATTCGC |
489 | ||
| RAB2A | yes | yes | GGTTAGAAGATGCCCGCCAG GTGGCATTAGTAGCAGCGTG |
292 | ||
| RAB5A | yes | yes | AGCAACAAGACCCAACGGG AACACTGACTCCTGGTTGGC |
623 | AGCAACAAGACCCAACGGG AACACTGACTCCTGGTTGGC |
623 |
| RAB8A | yes | yes | TGGTCAAGAACGGTTTCGG TGGCGAGAGTGAAAAATGCG | 305 | ||
| RAB10 | yes | yes | CCAGGAGCGATTTCACACC TTGCTCTTCCAGCCTGTCAC |
393 | ||
| RAB15 | yes | predicted | GAAGACCATAGAGGTAGATG TCAGGTTTGCCCTCATCC |
462 | ||
| RAB19 | yes | predicted | GAACACGATTGGGGTGGAC CATCAGCGTGAAGACCTCG |
385 | ||
| RAB25 | yes | predicted | ATGGGGAACAGAACCGAGG TGGAGGTCACTCTTGTTGCC |
389 | ||
| RAB27A | yes | yes | GGCAAGAGAGGTTTCGTAGC TCCCCTTCTCCTTTGCTTC |
423 | GGCAAGAGAGGTTTCGTAGC ACCCCTTCTCCTTCTCCTC |
423 |
| RAB27B | yes | predicted | ACCTCACCAGTCAACAGAGC TCTCTGCTGGTTTTTCCCC |
342 | ACCTCACCAGTCAACAGAGC TCTCTGCTGGTTTTTCCCC |
342 |
The expression in the intestine and kidney is confirmed by experimental evidences from a variety of mammalian species listed in the UniGene database.
Genes belong to the “yes” category if the presence in canine genome has been experimentally validated. Otherwise, primer design was based on the predicted transcripts in the canine genome.
IEC6 and C2C12 primers were designed based on validated Rattus norvegicus or Mus musculus genes.
2.5. Semi-quantitative PCR analysis
cDNA was all synthesized using oligo(dT)15 primer (#C1101, Promega Corp., Madison, WI, USA) and Omniscript Reverse Transcription Kit (#205111, Qiagen Corp., Hilden, Germany) following the manufacturer's instruction. All RT reactions were carried out with 2 μg total RNA/20 μl reaction. PCR reactions were performed with GoTaq G2 Hot Start Colorless Master Mix (#M7432, Promega Corp) following the manufacturer's instruction. All PCR reactions were carried out with 3 μl properly diluted cDNA/25 μl reaction. For all PCR reactions, the following conditions were used: lid temperature 105 °C; initial denaturing: 95 °C, 5 min; each cycle, denaturing: 95 °C, 1 min, annealing: 57 °C, 1 min, extension: 68 °C, 2 min; final extension: 68 °C, 15 min. PCR products and molecular weight markers were visualized in the ethidium bromide-containing 2% agarose gels using Gel Doc EZ Imager (Bio-Rad) and quantified by ImageJ64. All PCR reactions yielded bands at the expected size. Within each cell type, RT reactions of all RNA samples were carried out at the same time using the same batch of master mix. Three independent RNA samples per time point yielded three independent cDNA samples. A single batch of cDNA was serial diluted and used for the PCR reactions of all genes. Preliminary studies were carried out for each gene to determine the cDNA dilution and PCR cycle number that would yield non-saturating concentrations of PCR bands for samples from all three time points of culturing. The folds of cDNA dilution and cycle numbers of PCR reaction were shown in the Supplemental Table 1. For the final quantitative comparison, all nine PCR reactions of a gene in a cell line (from three time points with three independent cDNA samples per time point) were carried out in the same PCR run. These nine PCR products were visualized in the same gel.
2.6. Statistical analysis
For Caco-2 array analyses, GEO2R at the NCBI website was used to compare the expression level in subconfluent cells with that in cells at 14 days after seeding; and to compare the expression level in subconfluent cells with that in cells at 21 days after seeding. For the mouse array analyses, Student's t test was used to compare the expression in MEF and colon epithelium. For RT-PCR, One-way ANOVA was performed to determine the effect of culture days (2, 13, and 17 days) using Bonferroni/Dunn test for the post hoc multiple comparison. A significant effect was defined as p < 0.01 in all tests.
3. Results and discussion
3.1. Prediction based on microarrays of Caco-2 cells and mouse MEF and colon
For the sodium-dependent vitamin C transporter (SVCT) family, SVCT1 was targeted to the apical membrane while SVCT2 was targeted to basolateral membrane in both differentiated Caco-2 cells and MDCK cells [6], [7]. Thus, intestine and kidney epithelial cells likely have similar membrane protein targeting machinery. Based on this hypothesis, we decided to use existing intestinal microarrays to identify the protein targeting machinery in MDCK cells, the non-tumor epithelial cell model that will eventually allow functional analysis of the targeting machinery.
From the Caco-2 microarrays, we first identified 135 genes that showed significant differentiation-related up-regulation in Day 14 and Day 21. These 135 genes included genes that had not been named as well as redundant detection of the same genes in different parts of the array. After sorting to remove the unknowns and redundant ones, we found that 80 known genes showed differentiation-associated up-regulation (Supplemental Table 2). Based on the functional information in the literature [34], [35], [36], [37], [38], five of the 80 genes, C2 domain-containing protein 2-like (C2CD2L), Exocyst complex component 2 (EXOC2), Myosin Light Chain Kinase (MYLK), Myosin VIIB (MYO7B), and Syndecan binding protein 2 (SDCBP2), may participate in the membrane protein targeting. These five genes were also shown to express in the normal intestine and kidney tissues (Table 1, first data column). We then compared the expression levels of these five genes between mouse colon epithelium and MEF in the microarray analysis. The genes that are involved in the epithelial membrane protein targeting would expect to have higher expression in the colon epithelium than in the fibroblasts. Indeed, three genes, MYLK, MYO7B, and SDCBP2, showed significantly higher expression in colon epithelium compared to in MEF (Table 2, comparing the first and second data columns).
Table 2.
Using differentiating and non-differentiating cell lines for RT-PCR analysis of genes that showed significantly higher expression levels in colon epithelium compared to fibroblasts.a
| Genes | Array expressionb (Unnormalized signal) |
Expression level determined by RT-PCRc (Normalized by the expression level of Gapdh) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MEFb (n = 6) | Colonb (n = 9) | MDCK cellsc (n = 3 each day) |
IEC-6 cellsc (n = 3 each day) |
C2C12 cellsc (n = 3 each day) |
|||||||
| Day 2 | Day 13 | Day 17 | Day 2 | Day 13 | Day 17 | Day 2 | Day 13 | Day 17 | |||
| MYLK | 3380 | 45531 | 4.10 | 2.90 | 2.19 | ||||||
| MYO7B | 47 | 7709 | 0.35A | 1.84B | 5.47C | 0.23A | 1.23B | 1.24B | 0.10A | 0.45B | 1.14C |
| SDCBP2 | 396 | 64199 | NDA | 5.53B | 1.89A | ND | ND | ND | 0.63 | 0.60 | 0.54 |
| KIF5A | 623 | 2578 | 2.42A | 6.98B | 6.43B | 0.68A | 0.98B | 1.06B | 0.66A | 0.74A | 1.22B |
| KIF12 | 18 | 1785 | 3.66A | 6.78B | 6.27B | 1.12 | 0.93 | 0.95 | ND | ND | ND |
| KIF13A | 208 | 548 | 2.75A | 7.49B | 6.77B | 1.23 | 1.10 | 1.10 | 0.17A | 0.98B | 0.85B |
| KIF21A | 231 | 790 | 3.74A | 5.97B | 3.54A | ||||||
| KIF27 | 9 | 154 | 1.51A | 5.53B | 5.26B | ND | ND | ND | ND | ND | ND |
| RAB15 | 4250 | 54009 | 5.60 | 7.13 | 6.06 | ||||||
| RAB19 | 171 | 5795 | 5.10 | 5.12 | 4.42 | ||||||
| RAB25 | 23 | 13342 | 6.93 | 6.50 | 4.98 | ||||||
| RAB27A | 260 | 1981 | 7.48A | 5.14B | 2.91C | 1.40A | 0.69B | 0.88B | 0.83A | 0.42B | 0.67C |
| RAB27B | 86 | 2781 | 1.42A | 4.70B | 3.20C | 1.17A | 0.89B | 0.79B | 0.49A | 1.02B | 0.66C |
A,B,CDifferent superscript capital letters indicate significantly different from each other within the same cell type at p < 0.01. ND: not detectable.
Data of gene expression array and RT-PCR analysis shown in this table are means of groups. Three genes with expression levels in bold fonts meet all proposed criteria as candidate machinery genes in epithelial apical and basolateral protein targeting.
These gene expression microarrays used cDNA prepared from mouse embryonic fibroblasts (MEF) or from colon epithelium (Colon). The differences between two groups are significant at p < 0.01.
These gene expression analyses used cDNA of MDCK cells (canine kidney epithelium-origin) at Day 2 after seeding (prior to differentiation) and Day 13 and 17 after seeding (differentiated); IEC-6 cells (rat intestinal epithelium-origin but non-differentiating) after the same interval of culturing as other two cell lines; C2C12 cells (mouse skeletal muscle-origin) at Day 2 after seeding (prior to differentiation) and Day 13 and 17 after seeding (differentiated).
The 80 up-regulated genes in differentiated Caco-2 cells did not include any family members of KIF or RAB, important proteins in protein targeting/trafficking [20], [21], [22], [23]. When the colon and MEF arrays were compared, out of total 35 KIF members in the mouse array, we identified five KIF members, KIF5A, KIF12, KIF13A, KIF21A and KIF27, which showed significantly higher expression in the colon epithelium (Table 2, comparing the first and second data columns). Of the 53 RAB members in the mouse array, we identified five, RAB15, RAB19, RAB25, RAB27A and RAB27B, that showed significantly higher expression in colon compared to in MEF (Table 2, comparing the first and second data columns). All these KIF and RAB genes were also found to express in normal intestine and kidney tissues (Table 1, first data column). These mouse array studies were originally designed to determine the treatment effects of nutrients [26], [27] and none of the genes that we examined were affected by these nutrient treatments. This is consistent with the expected role of these genes as part of the general protein trafficking machinery.
3.2. RT-PCR analysis of MDCK gene expression
Altogether, we experimentally examined the expression of 23 genes in MDCK cells plus the control GAPDH. The 23 genes include the 13 candidate genes (Table 2) as well as 10 negative control genes (Supplemental Table 3). They were all expressed in the intestine and kidney tissues (Table 1, the first data column). While the candidate genes showed higher expression in the colon epithelium compared to the fibroblasts, the negative control genes had either similar expression or lower expression in colon epithelium when compared to MEF. Two previous MDCK studies used canine arrays for 5573 and 16,145 genes, respectively, to examine the effects of hepatocyte growth factors [39] and to determine the expression of membrane transporters and cytochome P450 enzymes [40]. This study is the first systemic analysis of endogenously expressed RAB and KIF family members in MDCK cells during differentiation.
Prior to our RT-PCR, only 7 of the 23 genes were experimentally validated in canine cells (Table 1, the second data column), the expression of the rest 16 genes is now supported by our data. Some RT-PCR bands from our MDCK analysis were also extracted and sequenced and the sequencing results all matched with the prediction. A representative gel for MDCK gene expression analysis is shown in Fig. 2A. The expression of GAPDH was not different from Day 2, 13 to 17, and hence the PCR band intensities of GAPDH were used for the normalization. The normalized gene expression levels are shown in Table 2 and the Supplemental Table 3 (the third to fifth data columns). Of the three genes that we initially identified through Caco-2 array analysis and showed higher expression in colon, two, MYO7B and SDCBP2, showed increased expression in differentiated MDCK cells.
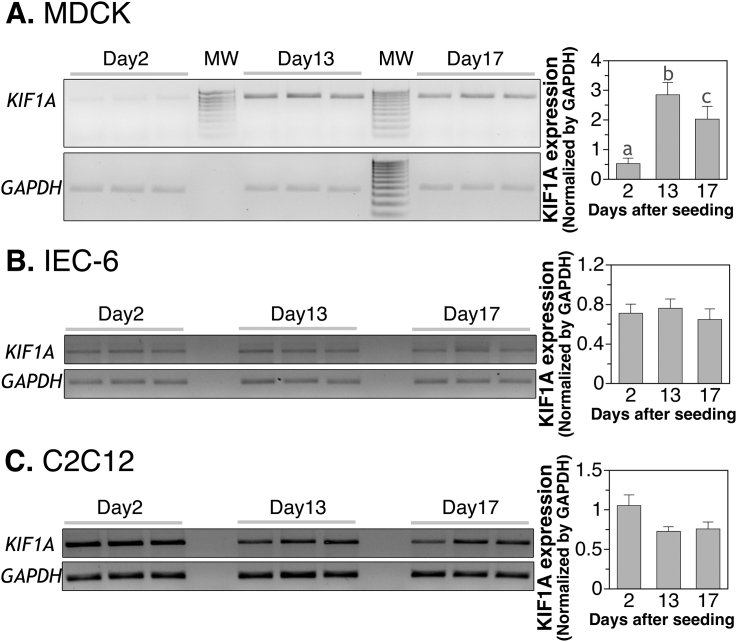
Fig. 2.
Semi-quantitative RT-PCR analysis of KIF1A and GAPDH expression in (A) MDCK, (B) IEC-6 and (C) C2C12 cells at 2, 13 and 17 days after seeding. MW: molecular weight markers from 0.1 to 1 kB a,b,cSignificantly different from each other by ANOVA and post-hoc with p < 0.01.
All five KIF members, KIF5A, KIF12, KIF13A, KIF21A and KIF27, that had higher expression in colon also showed increased expression in Day 13 differentiated MDCK cells (Table 2, the third to fourth data columns). However, KIF21A showed a decreased expression upon further culturing. At Day 17, the level became similar to that of the Day 2 (Table 2, the third and fifth data columns). This drop was not observed for the other 4 KIF genes in Table 2. Of the five RAB members that had higher expression in colon, only RAB27B showed more expression in differentiated MDCK cells. Other RAB members showed no change and RAB27A even had decreased expression upon differentiation (Table 2, the third to fifth data columns). Of the 10 negative control genes in Supplemental Table 3, EXOC2, KIF1A, KIFAP3 and RAB5A had increased expression in Day 13 MDCK cells but the expression dropped at Day 17 for EXOC2 and KIF1A (Supplemental Table 2, the third to fifth data columns).
3.3. Comparison of MDCK, IEC-6 and C2C12 cells
Of the genes in MDCK cells that showed expression increases from the subconfluent state (Day 2) to Day 13 and 17 after seeding, we wanted to distinguish expression changes that were specific to epithelial differentiation from those changes resulting from an increase in the culture duration. To this end, these MDCK differentiation-related genes were also analyzed in non-differentiating intestinal IEC-6 cells and myobalst C2C12 cells (Table 2, data column 6-11). These three cell lines grew comparably under our experimental condition. MDCK cells had total protein content per well (n = 3–6 independent wells, mean ± SD) of 0.228 ± 0.009, 1.20 ± 0.04, and 1.11 ± 0.02 mg at 2 days, 13 days and 17 days after seeding. At the same three time points, IEC-6 cells had 0.114 ± 0.002, 0.772 ± 0.025 and 1.27 ± 0.05 mg protein per well and C2C12 cells had 0.025 ± 0.002, 1.76 ± 0.04 and 2.09 ± 0.09 mg protein per well.
Representative gels for IEC-6 and C2C12 gene expression analysis are shown in Fig. 2B and C. The expression of GAPDH was not different from day 2 to day 17 in IEC-6 and C2C12 cells, and thus was also used for the normalization. Of the candidate genes analyzed that showed increased expression in differentiated MDCK cells, three genes, SDCBP2, KIF12 and KIF27 appeared to have up-regulation specific for MDCK cells (Table 2, bold). SDCBP2 and KIF27 were not detected in IEC-6 cells while KIF12 and KIF27 were not detected in C2C12 cells. Some of the negative control genes in Supplemental Table 3 such as EXOC2, KIF1A, and KIFAP3 also showed up-regulation specific for MDCK cells. Different from SDCBP2, KIF12 and KIF27, EXOC2, KIF1A, and KIFAP3 did show detectable expression in both IEC-6 cells and C2C12 cells.
3.4. Other genes
We also considered the possible importance of other genes in differential targeting by systemically comparing the results of mouse colon epithelium and embryonic fibroblast gene expression arrays as we did in Step 1 (Fig. 1). Of the 44K features that we compared, 6282 features showed significant differences between colon epithelium and embryonic fibroblasts in the expression levels. Of them, 1572 known genes including those that we already analyzed showed significantly higher expression in colon epithelium than fibroblasts at p < 0.01. After further examination of the known function and localization of the 1572 genes, we found some additional genes that may merit further analysis as shown in the Supplemental Table 4.
3.5. Significance and alternative explanations of discovery
Overall, our discovery adds new information to the limited knowledge on these protein trafficking-related genes. Among genes that we found specific changes in expression upon epithelial differentiation, SDCBP2 [38] proteins were previously found to play trafficking roles in MDCK cells. KIF12 was implicated in the polycystic kidney disease [41], [42]. KIF27 knockout led to congenital hydrocephalus [43] and this is the first study analyzing the expression in epithelial differentiation. Our observation also provides new insight into KIF5A, a gene with increased expression upon prolonged culturing of all three cell lines (Table 2). KIF5A was not previously examined for epithelial cells but was a known neuronal motor [44] involving in hereditary spastic paraplegia [45]. In addition, KIFAP3 was previously found to be part of the mitotic apparatus [46] and thus explains its higher expression in MEF than in colon (Supplemental Table 3, first data group).
Embryonic fibroblasts and colon epithelium have large functional and morphological differences. For example, of the 1572 known genes that showed higher expression in colon epithelium, many of them are transporters and metabolic enzymes. It is possible that SDCBP2, KIF12 and KIF27 that we identified here only contribute to the morphological but not functional differences between fibroblasts and epithelium. The analyses we performed, both microarray and RT-PCR, examined the mRNA levels. The gene expression results need to be confirmed by protein level measurement. Unfortunately, very few commercial antibodies have been validated for canine proteins. Direct functional analysis such as gene knock-out or knock-down may be the only way to verify the proposed functional role in polarized protein targeting.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgement
We thank UB Confocal Microscopy and Flow Cytometry core facility for giving access to the Gel Doc EZ Imager for the PCR band quantification. Dr. Christine E. Campbell (University at Buffalo)'s assistance in the sequencing of PCR bands to confirm the RT-PCR products is greatly appreciated.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.biopen.2017.04.002.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Marchiando A., Graham W., Turner J. Epithelial barriers in homeostasis and disease. Ann. Rev. Pathol. 2010;5:119–144. doi: 10.1146/annurev.pathol.4.110807.092135. [DOI] [PubMed] [Google Scholar]
- 2.Ullrich K., Frömter E., Murer H. Principles of epithelial transport in the kidney and intestines. Klin. Wochenschr. 1979;57:977–991. doi: 10.1007/BF01479983. [DOI] [PubMed] [Google Scholar]
- 3.Verrey F., Ristic Z., Romeo E., Ramadan T., Makrides V., Dave M., Wagner C., Camargo S. Novel renal amino acid transporters. Annu. Rev. Physiol. 2005;67:557–572. doi: 10.1146/annurev.physiol.67.031103.153949. [DOI] [PubMed] [Google Scholar]
- 4.Takano M., Yumoto R., Murakami T. Expression and function of efflux drug transporters in the intestine. Pharmacol. Ther. 2006;109:137–161. doi: 10.1016/j.pharmthera.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Regårdh C. Factors contributing to variability in drug pharmacokinetics. IV. Renal excretion. J. Clin. Hosp. Pharm. 1985;10:337–349. doi: 10.1111/j.1365-2710.1985.tb00932.x. [DOI] [PubMed] [Google Scholar]
- 6.Boyer J.C., Campbell C.E., Sigurdson W.J., Kuo S.-M. Polarized localization of vitamin C transporters, SVCT1 and SVCT2, in epithelial cells. Biochem. Biophys. Res. Commun. 2005;334:150–156. doi: 10.1016/j.bbrc.2005.06.069. [DOI] [PubMed] [Google Scholar]
- 7.Varma S., Sobey K., Campbell C.E., Kuo S.-M. Hierarchal contribution of N- and C-terminal sequences to the differential localization of homologous sodium-dependent vitamin C transporters, SVCT1 and SVCT2, in epithelial cells. Biochemistry. 2009;48:2969–2980. doi: 10.1021/bi802294v. [DOI] [PubMed] [Google Scholar]
- 8.Fletcher S., Rappoport J. Tight junction regulation through vesicle trafficking: bringing cells together. Biochem. Soc. Trans. 2014;42:195–200. doi: 10.1042/BST20130162. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez-Boulan E., Nelson W. Morphogenesis of the polarized epithelial cell phenotype. Science. 1989;245:718–725. doi: 10.1126/science.2672330. [DOI] [PubMed] [Google Scholar]
- 10.Cereijido M., Contreras R., Shoshani L. Cell adhesion, polarity, and epithelia in the dawn of metazoans. Physiol. Rev. 2004;84:1229–1262. doi: 10.1152/physrev.00001.2004. [DOI] [PubMed] [Google Scholar]
- 11.Kuo S., Wang L., Yu S., Campbell C., Valiyaparambil S., Rance M., Blumenthal K. The N-terminal basolateral targeting signal unlikely acts alone in the differential trafficking of membrane transporters in MDCK cells. Biochemistry. 2013;52:5103–5116. doi: 10.1021/bi4005914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lever J. Expression of differentiated functions in kidney epithelial cell lines. Min. Electrolyte Metab. 1986;12:14–19. [PubMed] [Google Scholar]
- 13.Simmons N. Cultured monolayers of MDCK cells: a novel model system for the study of epithelial development and function. Gen. Pharmacol. 1982;13:287–291. doi: 10.1016/0306-3623(82)90047-7. [DOI] [PubMed] [Google Scholar]
- 14.Rindler M., Chuman L., Shaffer L., Saier M.J. Retention of differentiated properties in an established dog kidney epithelial cell line (MDCK) J. Cell Biol. 1979;81:635–648. doi: 10.1083/jcb.81.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cereijido M., Robbins E., Dolan W., Rotunno C., Sabatini D. Polarized monolayers formed by epithelial cells on a permeable and translucent support. J. Cell Biol. 1978;77:853–880. doi: 10.1083/jcb.77.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cereijido M., Stefani E., Palomo A. Occluding junctions in a cultured transporting epithelium: structural and functional heterogeneity. J. Membr. Biol. 1980;53:19–32. doi: 10.1007/BF01871169. [DOI] [PubMed] [Google Scholar]
- 17.Cereijido M., Ehrenfeld J., Meza I., Martínez-Palomo A. Structural and functional membrane polarity in cultured monolayers of MDCK cells. J. Membr. Biol. 1980;52:147–159. doi: 10.1007/BF01869120. [DOI] [PubMed] [Google Scholar]
- 18.Farquhar M., Palade G. Junctional complexes in various epithelia. J. Cell Biol. 1963;17:375–412. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Meer G., Simons K. The function of tight junctions in maintaining differences in lipid composition between the apical and the basolateral cell surface domains of MDCK cells. EMBO J. 1986;5:1455–1464. doi: 10.1002/j.1460-2075.1986.tb04382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jordens I., Marsman M., Kuijl C., Neefjes J. Rab proteins, connecting transport and vesicle fusion. Traffic. 2005;6:1070–1077. doi: 10.1111/j.1600-0854.2005.00336.x. [DOI] [PubMed] [Google Scholar]
- 21.Pfeffer S. Structural clues to Rab GTPase functional diversity. J. Biol. Chem. 2005;280:15485–15488. doi: 10.1074/jbc.R500003200. [DOI] [PubMed] [Google Scholar]
- 22.Schroer T., Schnapp B., Reese T., Sheetz M. The role of kinesin and other soluble factors in organelle movement along microtubules. J. Cell Biol. 1988;107:1785–1792. doi: 10.1083/jcb.107.5.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirokawa N., Noda Y., Tanaka Y., Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 24.Pinto M., Robine-Leon S., Appay M.D., Kedinger M., Triadou N., Dussaulx E., Lacroix B., Simon-Assmann P., Haffen K., Fogh J., Zweibaum A. Enterocyte-like differentiation and polarization of the human colon carcinoma cell line, Caco-2 in culture. Bio. Cell. 1983;47:323–330. [Google Scholar]
- 25.Sääf A., Halbleib J., Chen X., Yuen S., Leung S., Nelson W., Brown P. Parallels between global transcriptional programs of polarizing Caco-2 intestinal epithelial cells in vitro and gene expression programs in normal colon and colon cancer. Mol. Biol. Cell. 2007;18:4245–4260. doi: 10.1091/mbc.E07-04-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo S.-M., Burl L.R., Hu Z. Cellular phenotype-dependent and -independent effects of vitamin C on the renewal and gene expression of mouse embryonic fibroblasts. PLoS One. 2012;7:e32957. doi: 10.1371/journal.pone.0032957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuo S.-M., Chan W.-C., Hu Z. Wild-type and IL10-Null mice have differential colonic epithelial gene expression responses to dietary supplementation with synbiotic bifidobacterium animalis subspecies lactis and inulin. J. Nutr. 2014;144:245–251. doi: 10.3945/jn.113.185249. [DOI] [PubMed] [Google Scholar]
- 28.Quaroni A., Wands J., Trelstad R., Isselbacher K. Epithelioid cell cultures from rat small intestine. Characterization by morphologic and immunologic criteria. J. Cell Biol. 1979;80:248–265. doi: 10.1083/jcb.80.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMahon D., Anderson P., Nassar R., Bunting J., Saba Z., Oakeley A., Malouf N. C2C12 cells: biophysical, biochemical, and immunocytochemical properties. Am. J. Physiol. 1994;266:C1795–C1802. doi: 10.1152/ajpcell.1994.266.6.C1795. [DOI] [PubMed] [Google Scholar]
- 30.Kuo S.-M., Morehouse H.F., Lin C.-P. Effect of antiproliferative flavonoids on ascorbic acid accumulation in human colon adenocarcinoma cells. Cancer Lett. 1997;116:131–137. doi: 10.1016/s0304-3835(97)00183-3. [DOI] [PubMed] [Google Scholar]
- 31.Kuo S.-M., Tan D., Boyer J.C. Cellular vitamin C accumulation in the presence of copper. Biol. Trace Elem. Res. 2004;100:125–136. doi: 10.1385/BTER:100:2:125. [DOI] [PubMed] [Google Scholar]
- 32.Varma S., Campbell C.E., Kuo S.-M. Functional role of conserved transmembrane segment 1 residues in human sodium-dependent vitamin C transporters. Biochemistry. 2008;47:2952–2960. doi: 10.1021/bi701666q. [DOI] [PubMed] [Google Scholar]
- 33.Peterson G.L. Determination of total protein. Methods Enzymol. 1983;91:95–119. doi: 10.1016/s0076-6879(83)91014-5. [DOI] [PubMed] [Google Scholar]
- 34.Pottekat A., Becker S., Spencer K., Yates J.R., Manning G., Itkin-Ansari P., Balch W. Insulin biosynthetic interaction network component, TMEM24, facilitates insulin reserve pool release. Cell Rep. 2013;4:921–930. doi: 10.1016/j.celrep.2013.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hazelett C., Yeaman C. Sec5 and Exo84 mediate distinct aspects of RalA-dependent cell polarization. PLoS One. 2012;7:e39602. doi: 10.1371/journal.pone.0039602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Usatyuk P., Singleton P., Pendyala S., Kalari S., He D., Gorshkova I., Camp S., Moitra J., Dudek S., Garcia J., Natarajan V. Novel role for non-muscle myosin light chain kinase (MLCK) in hyperoxia-induced recruitment of cytoskeletal proteins, NADPH oxidase activation, and reactive oxygen species generation in lung endothelium. J. Biol. Chem. 2012;287:9360–9375. doi: 10.1074/jbc.M111.294546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crawley S., Shifrin D.J., Grega-Larson N., McConnell R., Benesh A., Mao S., Zheng Y., Zheng Q., Nam K., Millis B., Kachar B., Tyska M. Intestinal brush border assembly driven by protocadherin-based intermicrovillar adhesion. Cell. 2014;157:433–446. doi: 10.1016/j.cell.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fialka I., Steinlein P., Ahorn H., Böck G., Burbelo P., Haberfellner M., Lottspeich F., Paiha K., Pasquali C., Huber L. Identification of syntenin as a protein of the apical early endocytic compartment in Madin-Darby canine kidney cells. J. Biol. Chem. 1999;274:26233–26239. doi: 10.1074/jbc.274.37.26233. [DOI] [PubMed] [Google Scholar]
- 39.Balkovetz D., Gerrard E.J., Li S., Johnson D., Lee J., Tobias J., Rogers K., Snyder R., Lipschutz J. Gene expression alterations during HGF-induced dedifferentiation of a renal tubular epithelial cell line (MDCK) using a novel canine DNA microarray. Am. J. Physiol. Ren. Physiol. 2004;286:F702–F710. doi: 10.1152/ajprenal.00270.2003. [DOI] [PubMed] [Google Scholar]
- 40.Quan Y., Jin Y., Faria T., Tilford C., He A., Wall D., Smith R., Vig B. Expression profile of drug and nutrient absorption related genes in madin-darby canine kidney (MDCK) cells grown under differentiation conditions. Pharmaceutics. 2012;4:314–333. doi: 10.3390/pharmaceutics4020314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gong Y., Ma Z., Patel V., Fischer E., Hiesberger T., Pontoglio M., Igarashi P. HNF-1beta regulates transcription of the PKD modifier gene Kif12. J. Am. Soc. Nephrol. 2009;20:41–47. doi: 10.1681/ASN.2008020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mrug M., Zhou J., Yang C., Aronow B., Cui X., Schoeb T., Siegal G., Yoder B., Guay-Woodford L. Genetic and informatic analyses implicate Kif12 as a candidate gene within the Mpkd2 tocus that modulates renal cystic disease severity in the Cys1cpk mouse. PLoS One. 2015;10:e0135678. doi: 10.1371/journal.pone.0135678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogel P., Read R., Hansen G., Payne B., Small D., Sands A., Zambrowicz B. Congenital hydrocephalus in genetically engineered mice. Vet. Pathol. 2012;49:166–181. doi: 10.1177/0300985811415708. [DOI] [PubMed] [Google Scholar]
- 44.Macioce P., Gambara G., Bernassola M., Gaddini L., Torreri P., Macchia G., Ramoni C., Ceccarini M., Petrucci T. Beta-dystrobrevin interacts directly with kinesin heavy chain in brain. J. Cell Sci. 2003;116:4847–4856. doi: 10.1242/jcs.00805. [DOI] [PubMed] [Google Scholar]
- 45.Fichera M., Lo Giudice M., Falco M., Sturnio M., Amata S., Calabrese O., Bigoni S., Calzolari E., Neri M. Evidence of kinesin heavy chain (KIF5A) involvement in pure hereditary spastic paraplegia. Neurology. 2004;63:1108–1110. doi: 10.1212/01.wnl.0000138731.60693.d2. [DOI] [PubMed] [Google Scholar]
- 46.Haraguchi K., Hayashi T., Jimbo T., Yamamoto T., Akiyama T. Role of the kinesin-2 family protein, KIF3, during mitosis. J. Biol. Chem. 2006;281:4094–4099. doi: 10.1074/jbc.M507028200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.