Abstract
The timing of major life-history events, such as migration and moult, is set by endogenous circadian and circannual clocks, that have been well characterized at the molecular level. Conversely, the genetic sources of variation in phenology and in other behavioral traits have been sparsely addressed. It has been proposed that inter-individual variability in the timing of seasonal events may arise from allelic polymorphism at phenological candidate genes involved in the signaling cascade of the endogenous clocks. In this study of a long-distance migratory passerine bird, the willow warbler Phylloscopus trochilus, we investigated whether allelic variation at 5 polymorphic loci of 4 candidate genes (Adcyap1, Clock, Creb1, and Npas2), predicted 2 major components of the annual schedule, namely timing of spring migration across the central Mediterranean sea and moult speed, the latter gauged from ptilochronological analyses of tail feathers moulted in the African winter quarters. We identified a novel Clock gene locus (Clock region 3) showing polyQ polymorphism, which was however not significantly associated with any phenotypic trait. Npas2 allele size predicted male (but not female) spring migration date, with males bearing longer alleles migrating significantly earlier than those bearing shorter alleles. Creb1 allele size significantly predicted male (but not female) moult speed, longer alleles being associated with faster moult. All other genotype–phenotype associations were statistically non-significant. These findings provide new evidence for a role of candidate genes in modulating the phenology of different circannual activities in long-distance migratory birds, and for the occurrence of sex-specific candidate gene effects.
Keywords: Adcyap1, avian migration, candidate genes, clock, phenology, ptilochronology
The annual schedule of migratory birds is controlled by an endogenous program, which is synchronized with seasonal changes primarily by daily changes in photoperiod (e.g., Gwinner 1986; Gwinner 2003; Sharp 2005; Pulido 2007; Visser et al. 2010). The endogenous clock that modulates circadian and circannual rhythmicity has been extensively studied in several organisms, from prokaryotes to vertebrates, and the genes controlling such mechanisms have been well characterized (Bell-Pedersen et al. 2005). Conversely, the genetic basis of phenotypic variation in the timing of seasonal events is poorly understood, and only a few candidate genes have been rather firmly linked to phenological variability in wild organisms (e.g., the Clock gene; Liedvogel et al. 2009; Caprioli et al. 2012; Saino et al. 2015a).
It has been suggested that differences in the timing of life-history events among individuals could arise from polymorphism at genes involved in the signaling cascade of the endogenous clock (Visser et al. 2010). Studies of among-individual and among-population phenological variability in vertebrate species have mainly focused on length polymorphism at 4 candidate genes, namely Adcyap1, Clock, Creb1, and Npas2 (e.g., Liedvogel et al. 2009; O'Malley et al. 2010, Caprioli et al. 2012; Chakarov et al. 2013; Bourret and Garant 2015). Clock and its paralog Npas2 show a polymorphic polyglutamine (polyQ) repeat sequence in their exonic regions (Fidler and Gwinner 2003; Steinmeyer et al. 2009). Short tandem repeat sequences at 3′-UTR have been detected in Creb1, a transcription factor involved in the light-induced clock entrainment (Gau et al. 2002; Tischkau et al. 2003), and Adcyap1, encoding for PACAP, a neurotransmitter with several biological functions related to the circadian and circannual rhythmicity (Simonneaux et al. 1993; Hannibal et al. 1997; Nagy and Csernus 2007; Racz et al. 2008; Schwartz and Andrews 2013).
In birds, allele size variation at Clock and Npas2 has been linked with differences in the timing of breeding among individuals, longer alleles being associated with delayed reproduction and in some species with shorter incubation periods (Liedvogel et al. 2009; Caprioli et al. 2012, Bourret and Garant 2015; but see Liedvogel and Sheldon 2010; Dor et al. 2012). Moreover, timing of migration (Bazzi et al. 2015; Saino et al. 2015a) and of complete annual moult (Saino et al. 2013) was delayed among individuals bearing longer Clock alleles in some long-distance migratory bird species. Polymorphism at Adcyap1 and Creb1 genes was found to be associated with juvenile dispersal behavior in buzzards Buteo buteo: individuals dispersing earlier carried longer Adcyap1 alleles and shorter Creb1 alleles than those dispersing later (Chakarov et al. 2013). Furthermore, Creb1 allele size was related to incubation duration in male tree swallows Tachycineta bicolor, though in combination with spring temperatures only (Bourret and Garant 2015). Finally, Adcyap1 allele size was associated with laying date in female tree swallows; however, the direction of the association varied with latitude, being negative at lower latitudes but becoming positive at higher latitudes (Bourret and Garant 2015). Although other studies did not report any significant association between candidate genes and phenology (e.g., Liedvogel and Sheldon 2010; Dor et al. 2012), there is evidence that polymorphism at some of such genes is associated with other behavioral traits that may be indirectly linked to circannual rhythms and/or photoperiodic response, such as migratory restlessness and migration distance (Mueller et al. 2011; Peterson et al. 2013; Bazzi et al. 2016). Moreover, a latitudinal cline in the frequency of alleles of different length has been reported for Clock and Adcyap1 in a few species (e.g., 1 out of 2 species in Johnsen et al. 2007; Bazzi et al. 2016; but see Kuhn et al. 2013): allele size of both candidate genes increased northwards, hinting at a possible role of polymorphism in the adaptation to different photoperiodic regimes or to the timing of breeding season, that is delayed and shorter at higher latitudes (Gwinner 1986; Berthold 1996; Johnsen et al. 2007; Bazzi et al. 2016; Bazzi et al. in press).
Taken together, there is evidence that polymorphism at candidate genes may underlie variability in the timing of life-history events through the whole life-cycle of birds and at different life stages, but the general picture is still patchy. In this study of the willow warbler Phylloscopus trochilus, we aimed at assessing whether length polymorphism at 5 polymorphic loci of 4 candidate genes (the previously studied Adcyap1, Clock, Creb1, and Npas2 genes and a newly identified polymorphic region of Clock gene; see Materials and Methods) predicted the timing of spring migration and the speed of winter moult, as assessed by measuring the growth rate of tail feathers by means of ptilochronological techniques (Grubb 2006). We assumed that a larger feather growth rate (FGR) corresponds to a faster moult (De la Hera et al. 2011). The willow warbler, a small (ca. 10 g) trans-Saharan migratory passerine that breeds in Eurasia at medium-high latitudes and overwinters in sub-Saharan Africa, is among the few species performing 2 complete annual moults, one of which occurs during winter, while the birds are in Africa (Underhill et al. 1992). Birds leave for spring migration in late February–March and reach the breeding grounds in mid-March to late May (Cramp 1998), and were sampled during spring migration across the central Mediterranean sea. According to previous studies of candidate gene–phenotype associations conducted on other migratory species (see above), we expected migration date to be delayed among birds with longer Clock and Npas2 alleles. Conversely, due to the variable genotype–phenotype associations reported in previous studies, we had no clear predictions on the allele size-phenology or FGR association for the other candidate genes.
Materials and Methods
Field methods
Willow warblers were sampled at Ventotene (40°48′N–13°25′E), a small island located in the central Mediterranean Sea, ca. 50 km off the Italian coast, during the period 22 March–27 May 2013; this sampling period encompassed the entire spring migration of the study species at Ventotene (Spina et al. 1993; Messineo et al. 2001; Saino et al. 2010). Birds were trapped using mist-nets following standard capture protocols and individually marked with metal rings (Spina et al. 1993; Saino et al. 2010). We used the length of the primary feather number 8 (according to the centrifugal numeration of primaries), that is, the third outermost primary feather, as a highly accurate estimate of wing length (Jenni and Winkler 1989) (wing length hereafter). Wing length and tarsus length were recorded to the nearest 0.5 and 0.1 mm using a pin ruler and a dial caliper, respectively. Wing length and tail length (but not tarsus length) can be used as rough proxies of breeding destination among willow warblers breeding in Fennoscandia (Bensch et al. 1999): both wing and tail length show a strong increase with breeding latitude (r2 > 0.58). Since willow warblers migrating through the central Mediterranean are directed mostly toward Fennoscandia (Jonzén et al. 2006a, 2006b), we used wing length and tail feather length (see Ptilochronological analyses; wing and tail feather length were strongly positively correlated in our sample of birds: males, r = 0.83; females, r = 0.87) as rough proxies of breeding latitude. Birds usually rest on Ventotene for a few hours before resuming their travel toward breeding quarters (Goymann et al. 2010; Tenan and Spina 2010). We considered only first capture dates (i.e., we excluded recaptures of birds previously ringed at the study site during the same season). We assumed that the distribution of first capture dates (expressed in Julian dates, with January 1 = day 1) reflects the phenology of species’s timing of spring migration at Ventotene (see Saino et al. 2010, 2015a).
We aimed at sampling ca. 100 individuals, evenly distributed along the whole spring migration season. According to the number of willow warblers captured during the previous years (2006–2011, ca. 800 birds/year), we sampled 1 every 8 captured individuals (see Saino et al. 2010, 2015a). For each individual we collected a small blood (ca. 10–30 µL, collected in heparinized capillary tubes and stored at −20°C) or feather (3–4 undertail coverts, stored in 99% ethanol at room temperature) sample as a source of DNA. Moreover, the fourth outermost rectrix (hereafter R4) was collected and stored in individual bags for ptilochronological analyses. The total sample size was 124 individuals.
Ptilochronological analyses
Moult speed was indirectly assessed by measuring growth bar width (GBW) on R4 (see De la Hera et al. 2009). A single feather growth bar consists of 1 light band and 1 dark band, which correspond to the portion of the feather grown during a single night–day cycle (Brodin 1993). Wider growth bars reflect faster feather growth (Grubb 2006). Although moult speed depends on the number of feathers growing simultaneously, as well as on the individual FGR, it has been shown that individuals with high FGRs moult many feathers at the same time (Bensch and Grahn 1993). Hence, we can assume that GBW roughly reflects moult speed of all feather tracts (De la Hera et al. 2011; see also Saino et al. 2012). We measured the width of 6 bars, 3 on either side of a point located at two-third of feather length [modified from Grubb (2006) and De la Hera et al. (2009) according to the number of growth bars clearly recognizable on willow warblers’ R4]. The total width of bars was measured with a digital caliper (to the nearest 0.01 mm) on the dorsal surface of the vane. GBW was expressed as the total width of bars/6. In order to avoid any bias, all measures were taken by the same observer (SP). Repeatability of GBW, as assessed on a sample of feathers measured twice, was very high (n = 20, r = 0.96, P < 0.001).
After measuring GBW, feathers were taped to tracing paper across the shaft, and scanned; tail feather length (to the nearest 0.01 mm) was measured on the resulting images using the “segmented line” tool of ImageJ 1.46r software (rsbweb.nih.gov) (Saino et al. 2015b). Individuals whose feather tips were broken, for which feather length could not be measured, were excluded from moult speed analyses. We obtained GWB from 118 individuals.
Genetic analyses
Genomic DNA was extracted from blood samples by means of alkaline lysis of 6 µL of blood in 100 µL of a 50 mM NaOH solution at 100°C for 20 min. Extracted DNA was quantified using a spectrophotometer and diluted to a final concentration of 50–100 ng/µL. Genomic DNA from feathers was extracted using a commercial kit (5 PRIME, ArchivePure DNA purification kit, Hilden, Deutschland). The procedure is described in detail in Saino et al. (2015a).
Willow warblers are sexually size dimorphic (males are larger than females) but sexually monochromatic (Cramp 1998), and sex cannot be determined in the field. Hence, sex was determined using CHD1 primers (for DNA extracted from blood samples; details on primers and PCR amplification in Saino et al. 2015a). As PCR amplification performed on DNA extracted from feathers with CHD1 primers did not produce reliable results, we designed a new set of primers on Passer montanus CHD gene (Sequence ID in GenBank: gb|GU370350.1|): PassexF 5′-GAGAAACTGTGCAAAACAGG-3′ and PassexR 5′-GAGTCACTATCAGATCCAGARTATC-3′. PCR amplification were performed in a final volume of 15 µL, with 6 µL DNA solution, 1× PCR buffer (Promega), 1.5 mM of Mg2+, 0.3 µL of each primer (stock 10 mM), 1.5 µL of dNTPs (stock 2 mM), and 1 U Taq DNA polymerase (Promega). PCR amplification profile was as follows: 95°C for 3 min, 35 cycles at 95°C for 45 s, 55°C for 45 s and 72°C for 50 s, and further extension at 72°C for 5 min. PCR products were then separated on 2.5% agarose gel and visualized after ethidium bromide staining. All 124 sampled individuals were sexed (64 males and 60 females).
We assessed polymorphism at Adcyap1, Creb1, and Npas2 genes and at 2 polymorphic Clock gene regions [region 1 (r1) and region 3 (r3); Clock r1 was the locus investigated by Johnsen et al. (2007) and by subsequent studies on Clock gene polymorphism, while Clock r3 was a newly identified polymorphic region; see below] by means of PCR amplification followed by fragment analysis. Primers for Adcyap1 PCR amplifications were taken from Saino et al. (2015a), whereas Clock r1 primers are described in Caprioli et al. (2012). Finally, Creb1 and Npas2 primers correspond to those described in Steinmeyer et al. (2009) [with the slight modifications proposed by Bourret and Garant (2015) for the Creb1 gene].
The Clock r3 locus was identified by aligning all Clock avian gene sequences available in GenBank (55 genomic sequences retrieved in November 2015) and searching for polymorphic regions that vary in number of glutamine residues among species. We identified a predicted exonic region containing a variable number of glutamine-coding triplets (3–9) located at ca. 200 bp from Clock r1 toward the NH2 terminus of the protein. Specific Clock r3 primers (Clock r3.F 5′-TCTGCTGCTTTCCCACTACA-3′ and Clock r3.R 5′-ATCAGTCATCTTGTCAGTTCTGTG-3′) were designed ex novo.
PCR amplification was performed using a commercial kit (Qiagen, Multiplex PCR Kit) in a final volume of 25 µL with 12.5 µL 2× QIAGEN Multiplex PCR Master Mix, 2.5 µL 10× primer mix (0.5 µL of each primer) (final concentration 0.2 µM), 2 µL RNase-free water (for genomic DNA extracted from blood only), 5 µL 5× Q-Solution and 3 µL of DNA solution (5 µL for DNA extracted from feather samples). PCR amplification profile was: 95°C for 15 min, 35 cycles at 94°C for 30 s, 56°C for 90 s, 72°C for 60 s, and a final extension at 60°C for 30 min. PCR products were labeled with 6-FAM (Clock r1 and Creb1), HEX (Clock r3 and Npas2), or TAMRA (Adcyap1). Polymorphism at candidate genes was determined using fragment analysis (Macrogen Inc., Seoul, Republic of Korea) (see Caprioli et al. 2012; Bazzi et al. 2015; Saino et al. 2015a). The sample size of individuals genotyped for each locus is shown in Table 1.
Table 1.
List of candidate genes, sample size (n), number of alleles observed at each locus (K), range of allele length (size range, in bp), mean allele size (in bp, with associated standard error, in brackets), and observed heterozygosity (Ho)
| Candidate gene | n | K | Size range | Mean allele size (SE) | Ho |
|---|---|---|---|---|---|
| Adcyap1 | 112 | 10 | 160–176 | 170.21 (0.22) | 0.83 |
| Clock r1 | 121 | 5 | 114–126 | 120.03 (0.14) | 0.47 |
| Clock r3 | 97 | 2 | 108–111 | 108.29 (0.06) | 0.15 |
| Creb1 | 92 | 4 | 271–277 | 274.01 (0.10) | 0.50 |
| Npas2 | 93 | 5 | 166–178 | 172.37 (0.11) | 0.38 |
Statistical analyses
We tested for deviations from Hardy–Weinberg equilibrium (HWE) for the 5 loci using the Markov chain method (Guo and Thompson 1992) implemented in GENEPOP (dememorization = 1000, batches = 100, iterations per batch = 1000). We quantified the extent of genetic differentiation between the sexes at the 5 loci separately as well as for the combination of all loci by estimating FST between males and females using Fstat 2.9.3 software (Goudet 2001).
To investigate the association between candidate genes allele size (mean of the long and short allele, mean allele size hereafter) and migration date, while controlling for variation in migration timing due to sex, we ran a linear model of migration date (1 = January 1) as a function of sex (0 = females, 1 = males) and Adcyap1, Clock r1, Clock r3, Creb1, and Npas2 allele size as covariates. Within individuals, the mean allele sizes of the different microsatellites were not significantly correlated (|r| always < 0.12). Hence, the simultaneous inclusion of the mean allele size of all loci in a single model is feasible, and aims at testing the phenotypic associations of each locus while controlling for any concomitant effect of the other loci. Since any possible association between candidate genes’ allele size and migration date may arise from a latitudinal cline of allele size, we included wing length as a further covariate, representing a rough proxy of breeding latitude (wing length increases with latitude across Europe in several passerine bird species besides the willow willow warbler; Cramp 1998; Bensch et al. 1999; Peiró 2003; Evans et al. 2009; Tarka et al. 2010).
We then tested whether candidate genes’ mean allele size predicted GBW. Since GBW and tail feather length were strongly correlated (r = 0.51, P < 0.001), to control for the effect of tail feather length on GBW we computed the residuals of a linear regression of GBW on tail feather length (FGR). Then, similarly to migration date, we ran a linear model of FGR as a function of sex and Adcyap1, Clock r1, Clock r3, Creb1, and Npas2 mean allele size, while including wing length as a further covariate.
Both for migration date and FGR, we tested for sex-specific phenotypic effects of candidate genes by including in the models all the 2-way interactions between each locus and sex. Statistically significant interaction terms were retained in final models and were interpreted by checking sex-specific slopes of genotype–phenotype associations. We relied on 81 individuals genotyped at all loci for migration date, and 78 for FGR.
Finally, we tested for associations between morphology (wing and tail feather length) and mean allele sizes of candidate genes by running linear models of wing or tail feather length as a function of the mean size of the alleles (all loci included simultaneously). To account for marked sex differences in morphology [males are significantly larger than females; see Cramp (1998) and Results], these models were run separately for each sex.
All linear models were also run by including the long (instead of the mean) allele sizes of all loci as predictors: this was done because previous studies highlighted a possible dominance of the longer alleles in shaping phenology and other phenotypic traits of migratory birds for Clock and for other candidate genes (see Liedvogel et al. 2009; Saino et al. 2015a; Bazzi et al. 2016). Within individuals, the long allele sizes of the different microsatellites were not significantly correlated (|r| always < 0.10): hence, the simultaneous inclusion of the long allele sizes of all loci in a single model was feasible.
Results
Migration phenology and morphology
The willow warbler is a highly protandrous species, with mean migration date of males [99.7 (11.0 SD), n = 64] being much earlier than that of females [117.0 (12.5 SD), n = 60; t122 = 8.26, P < 0.001; see also Saino et al. 2010]. Males were significantly larger than females for all biometrics [wing length, males: 53.3 mm (1.9 SD), females: 49.6 mm (1.9 SD); tail feather length, males: 56.5 mm (1.8 SD), females: 52.5 (2.2 SD); tarsus length, males: 19.7 mm (0.7 SD), females: 18.6 mm (0.6 SD); all t > 9.39, all P < 0.001] (see also Cramp 1998).
Wing and tail feather length of males did not significantly vary with migration date [wing length, estimate: −0.022 (0.022 SE) mm/day, t62 = 0.99, P = 0.36; tail feather length, estimate: −0.001 (0.021 SE) mm/day, t62 = 0.03, P = 0.98]. On the other hand, wing and tail length of females significantly declined with migration date [wing length, estimate: −0.049 (0.019 SE) mm/day, t58 = 2.60, P = 0.012; tail feather length, estimate: −0.057 (0.022) mm/day, t58 = 2.61, P = 0.011]. Tarsus length did not significantly vary with migration date in both sexes (males, estimate: −0.012 (0.008 SE) mm/day, t61 = 1.60, P = 0.12; females, estimate: −0.009 (0.006 SE) mm/day, t58 = 1.51, P = 0.14).
Candidate genes variation
We successfully genotyped 93–112 individuals, depending on locus (Table 1). Polymorphism broadly varied among candidate genes: the Clock r3 locus showed very low variability, with 2 alleles only, 1 of which (108 bp) had an allelic frequency of 90.2% (Table 1). On the other hand, the Adcyap1 locus was highly variable (Table 1). The other candidate genes showed intermediate levels of observed heterozygosity (Table 1). The Creb1 locus significantly deviated from the HWE (P < 0.001), while this was not the case for the other loci (P > 0.21). Allele frequencies of males and females were similar for all loci, as indicated by the small FST values (Adcyap1: FST = 0.001, P = 0.20; Clock r1: FST = −0.002, P = 0.75; Clock r3: FST = −0.009, P = 0.70; Creb1: FST = 0.015, P = 0.30; Npas2: FST = −0.005, P = 0.70; all loci pooled: FST = 0.008, P = 0.25).
Candidate genes, timing of migration, and morphology
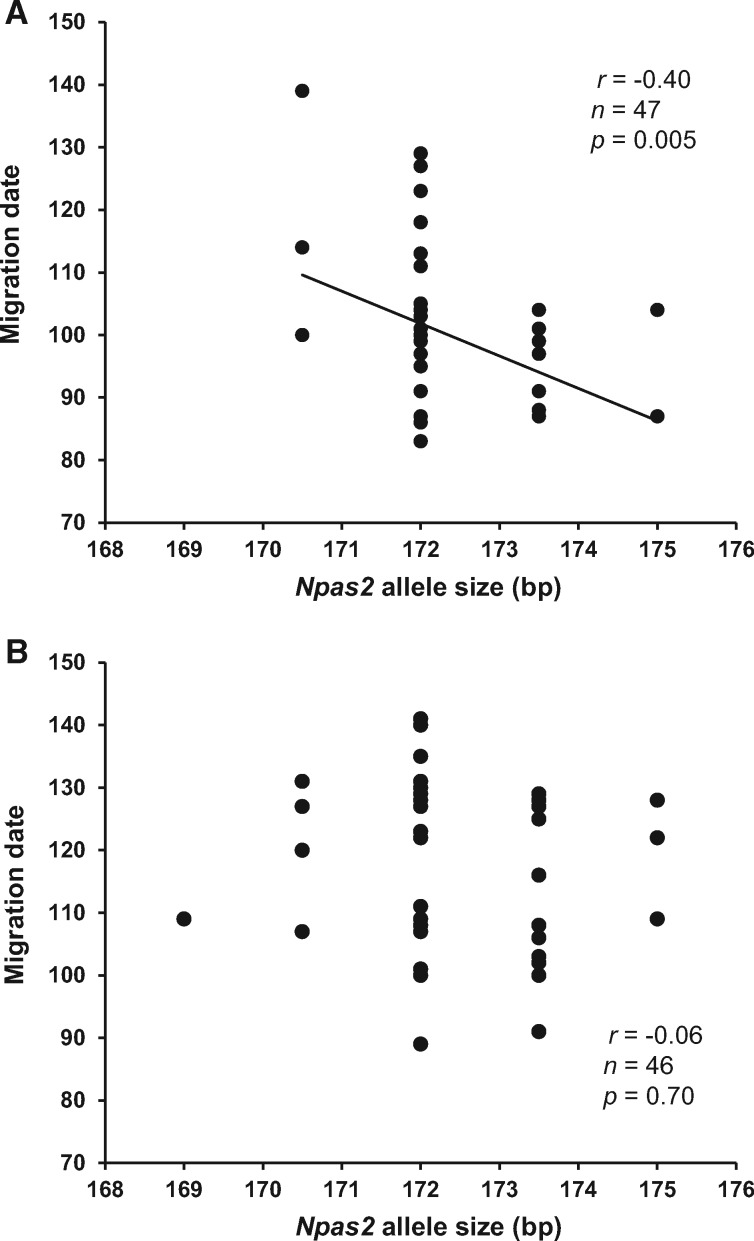
The linear model analysis showed that the mean allele size of the different loci did not significantly affect migration date, with the exception of Npas2, that significantly predicted migration date in a different way according to sex (Npas2 × sex interaction, Table 2): male birds bearing longer Npas2 alleles had a significantly earlier migration date, whereas this was not the case for females (Table 2, Figure 1). A similar linear model run using long allele sizes showed no statistically significant sex-specific effect of any locus (all P > 0.10), and no significant association between long allele size of any locus and migration date (model with interactions removed, all P > 0.16; details not shown for brevity).
Table 2.
Linear model of the effect of candidate genes’ mean allele size (5 loci) on migration date (1 = January 1)
| Variable | Estimate (SE) | df | F | P |
|---|---|---|---|---|
| Sex | —a | 1, 72 | 15.49 | <0.001b |
| Wing length | −1.718 (0.672) | 1, 72 | 6.54 | 0.017 |
| Adcyap1 | 0.502 (0.627) | 1, 72 | 0.64 | 0.43 |
| Clock r1 | −1.529 (0.899) | 1, 72 | 2.90 | 0.09 |
| Clock r3 | −0.171 (2.149) | 1, 72 | 0.01 | 0.94 |
| Creb1 | 1.043 (1.853) | 1, 72 | 0.32 | 0.58 |
| Npas2 | — | 1, 72 | 4.45 | 0.038 |
| Npas2 × sex | —c | 1, 72 | 6.09 | 0.016 |
Notes: Estimates for covariates included in retained interaction terms are not shown because they are not meaningful: details about these effects are shown in the table footnotes.
Estimated means (SE) at mean values of covariates: males, 116.1 (2.2); females, 102.2 (2.2).
Test statistics of estimated means at mean values of the covariates.
Model-derived estimate (SE): males, −5.714 (1.947), P = 0.004; females, 0.517 (1.560), P = 0.74.
Figure 1.
Migration date (1 = January 1) in relation to Npas2 mean allele size in (A) male and (B) female willow warblers. The line represents simple linear regression with a statistically significant (P < 0.05) slope. The correlation coefficient (Pearson’s r) is also shown.
Wing and tail feather length did not significantly covary with the allele size (both mean and long) in linear models of morphology in relation to allele sizes of all loci (models run separately for each sex; 4 linear models; all P > 0.07).
Candidate genes and moult
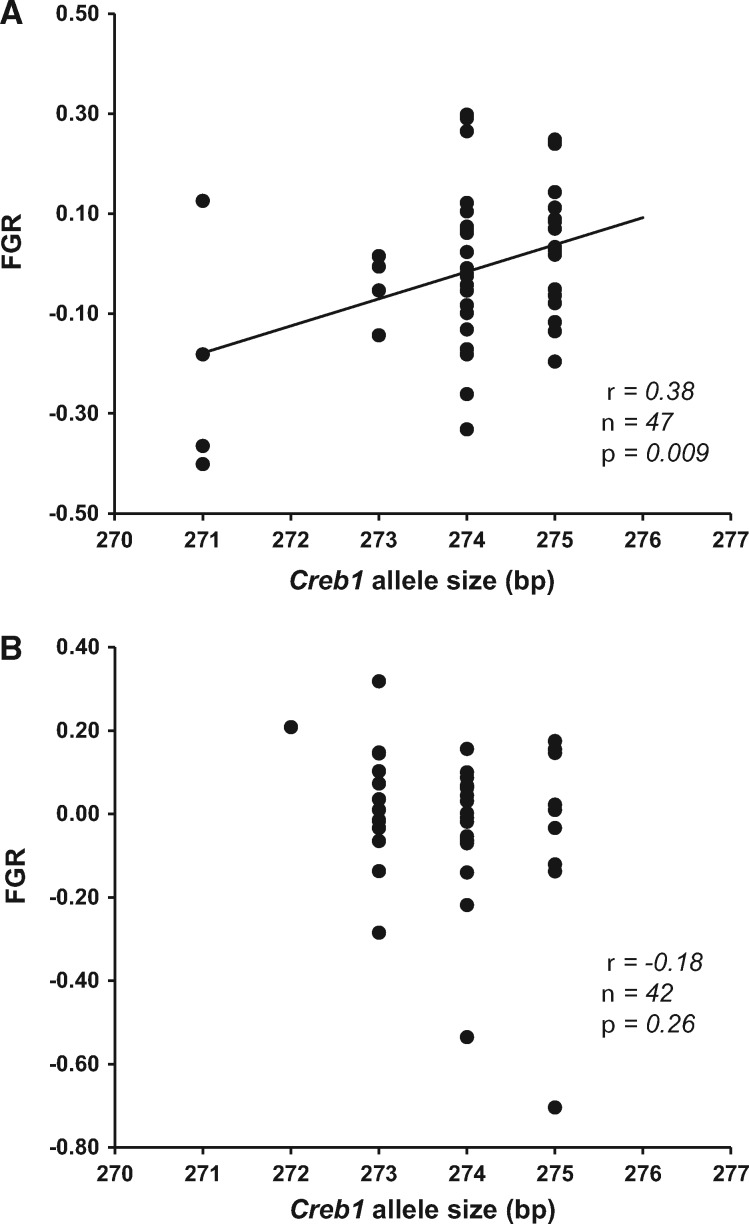
FGR did not significantly differ between the sexes [males: −0.01 (0.18 SD), n = 60; females: 0.01 (0.18 SD), n = 58; t116 = 0.76; P = 0.44]. The full linear model including all interaction terms revealed that 2 loci, Clock r1 and Creb1, showed a marginally non-significant (P = 0.06 in both cases) tendency for sex-specific effects on FGR (details not shown), while the phenotypic effects of the other loci were far from significance (all P values > 0.26, details not shown for brevity). We thus decided to retain these 2 interactions in the final model (Table 3): in both cases, genotype–FGR associations were statistically significant for 1 sex but not for the other (footnotes to Table 3). To investigate this further, we increased sample size (78–88 individuals; different loci had different sample sizes, see Table 1) by running an additional model where we removed data for all loci that had a non-significant effect on FGR (i.e., Adcyap1, Clock r3, and Npas1; see Table 3). The larger sample size yielded a statistically significant Creb1 × sex interaction (F1,81 = 7.98, P = 0.006), while the Clock r1 × sex interaction was not significant (F1,81 = 3.02, P = 0.09). Inspection of sex-specific slopes from this model indicated that no slope was statistically different from 0 for Clock r1 (both P > 0.12), whereas a significant positive effect of Creb1 on male (but not female) FGR [males: 0.056 (0.022 SE), P = 0.012; females: −0.056 (0.033), P = 0.10] emerged. Analyses run on data from all individuals genotyped for Creb1 (n = 89; Figure 2) and Clock r1 (n = 115) confirmed the robustness of this last model (details not shown). Hence, we conclude that our data support a statistically significant sex-specific genotype–FGR association for Creb1 but not for the other loci, longer Creb1 alleles being associated with faster feather growth in males but not in females. Models run using long allele sizes did not highlight any significant genotype–phenotype association (details not shown). However, a model including data for all birds genotyped for Creb1, together with sex and wing length, confirmed a sex-specific effect of long Creb1 (Creb1 × sex, F1,84 = 4.30, P = 0.041).
Table 3.
Linear model of the effect of candidate genes’ mean allele size (5 loci) on FGR (residuals of a regression of GBW on feather length; see Materials and Methods)
| Variable | Estimate (SE) | df | F | P |
|---|---|---|---|---|
| Sex | —a | 1, 68 | 0.13 | 0.72b |
| Wing length | 0.001 (0.010) | 1, 68 | 0.02 | 0.90 |
| Adcyap1 | 0.006 (0.007) | 1, 68 | 0.32 | 0.58 |
| Clock r1 | — | 1, 68 | 0.32 | 0.57 |
| Clock r3 | 0.001 (0.027) | 1, 68 | 0.16 | 0.70 |
| Creb1 | — | 1, 68 | 0.73 | 0.40 |
| Npas2 | 0.011 (0.017) | 1, 68 | 0.68 | 0.41 |
| Clock r1 × sex | —c | 1, 68 | 4.40 | 0.040 |
| Creb1 × sex | —d | 1, 68 | 3.27 | 0.075 |
Notes: Estimates for covariates included in retained interaction terms are not shown because they are not meaningful: details about these effects are shown in the table footnotes.
Estimated means (SE) at mean values of covariates: males, −0.003 (0.031); females, −0.021 (0.032).
Test statistics of estimated means at mean values of the covariates.
Model-derived estimate (SE): males, −0.019 (0.021), P = 0.36; females, 0.034 (0.015), P = 0.028.
Model-derived estimate (SE): males, 0.026 (0.043), P = 0.62; females, −0.073 (0.035), P = 0.038.
Figure 2.
FGR (residuals of a regression of GBW on feather length; see Materials and Methods) versus Creb1 mean allele size in (A) male and (B) female willow warblers. High FGR values are assumed to reflect faster moult. The line represents simple linear regression with a statistically significant (P < 0.05) slope. The correlation coefficient (Pearson’s r) is also shown (the result for females was similar after removing the 2 extreme data points with FGR < −0.40; details not show for brevity).
Discussion
We investigated whether allelic variation at 5 candidate genes’ loci (Adcyap1, Clock r1, Clock r3, Creb1, and Npas2) predicted the timing of 2 important life-history activities, timing of spring migration across the central Mediterranean sea, and speed of tail feather moult in the African winter quarters, in the long-distance migrating willow warbler. Allelic variation broadly differed between the 5 loci, ranging from the low values of observed heterozygosity shown by the novel Clock r3 locus to the high variability of Adcyap1. The Clock r3 locus, a newly identified region of the Clock gene showing polyQ polymorphism (see Materials and Methods), had in fact 2 alleles only, and a very low variability (Ho was equal to 0.15; Table 1). Although polymorphism at this region was not associated with any phenotypic trait, suggesting that its phenotypic associations are weak, future studies testing Clock–phenotype associations in avian species might consider genotyping this region besides the well-studied Clock r1.
We highlighted a novel association between Creb1 allele size and FGR, a proxy of overall moult speed, faster feather growth being associated with longer Creb1 alleles in male (but not female) willow warblers. Npas2 allele size was associated with migration date in male (but not female) willow warblers, but the relationship was opposite to our expectations based on previous research, with individuals bearing shorter Npas2 alleles migrating later through the study site compared with those bearing longer alleles. Moreover, we observed that early migrating individuals, especially females, had longer wings, suggesting that birds from northern populations migrate earlier across the central Mediterranean than those from southern populations.
The significant associations reported in this study should however be interpreted cautiously because the results may be affected, among others, by: 1) age-related variation in migration date and FGR (age cannot be assessed in spring because the species performs a complete winter moult, Jenni and Winkler 1994); 2) unknown origin/destination of populations migrating through Ventotene (wing and tail feather length are only rough proxies of geographic origin; see Materials and Methods); and 3) the fact that FGR is only a rough proxy of overall moult speed (De la Hera et al. 2011).
Notwithstanding the possible confounds listed above, sampling birds during spring migration allowed us to try to make inferences about proxies of the speed of the complete winter moult by means of ptilochronological analyses of tail feathers (Grubb 2006; De la Hera et al. 2011). Studying proxies of moult speed in relation to candidate genes polymorphism could improve our understanding of the genetic regulation of annual scheduling. Moult requires considerable amounts of resources, and overlap between moult and other circannual activities is largely avoided by most species (Jenni and Winkler 1994; Hemborg and Lundberg 1998). Hence, in winter moulting species, such as the willow warbler, moult speed may constrain the timing of spring migration (Hedenström et al. 2007; Møller et al. 2011). Indeed, comparative studies of trans-Saharan migrants with different moult strategies showed that species performing a complete moult during wintering migrate later than those moulting in Europe before autumn migration (Rubolini et al. 2005).
We had no a priori expectation on the possible effect of candidate genes allele size on proxies of moult speed, since the single previous study investigating the relationship between genotype and moult phenology focused on the Clock gene only, highlighting that individual barn swallows Hirundo rustica bearing a rare long Clock variant (Q7/Q8) had a delayed moult of wing feathers compared with the other genotypes (Saino et al. 2013). Moreover, Chakarov et al. (2013) found that longer Creb1 alleles were associated with delayed juvenile dispersal in buzzards. Hence, the Creb1 allele size–moult speed association we detected may arise from a delayed onset of plumage moult among individuals bearing longer Creb1 alleles. A delayed timing of moult might constrain its duration, leading to faster feather growth, as demonstrated in small migratory passerines experimentally subjected to shorter moult periods by altering photoperiod (e.g., Dawson et al. 2000; Hall and Fransson 2000). Alternatively, we might speculate that Creb1 allele size directly affected moult speed through its involvement in the melanin synthesis pathway [see e.g., Kondo and Hearing (2011) for mammals], but the specific mechanism linking Creb1 allele size variation to melanin synthesis is unknown.
The delayed migration of males bearing shorter Npas2 alleles was opposite to expectations. According to the few studies investigating the association between Npas2 gene polymorphism and phenology (Chakarov et al. 2013; Bourret and Garant 2015) and the hypothesis that Npas2 could overtake Clock gene functions, representing an alternative or additional source of adaptive polyQ variation for the regulation of timing of seasonal events (Debruyne 2008; Steinmeyer et al. 2009), we expected Npas2 allele size to increase with migration date.
A possible explanation for this findings is that different willow warbler populations that have diverged for Npas2 migrate through the study site at different times. The negative association between Npas2 and migration date could thus originate because of geographic differentiation in Npas2. This possibility is corroborated by the rather unusual migration pattern of this species at Ventotene, whereby wing length decreased in the course of the spring migration season. Wing length generally increases with latitude across Europe in several passerine species (including the willow warbler; Bensch et al. 1999) and northern populations usually migrate later than southern ones (see e.g., Cramp 1998; Rubolini et al. 2005; Conklin et al. 2010), while the opposite was apparently the case in this study. The willow warbler may not be an exception, as similar results emerged for 2 other long-distance migratory passerines sampled at the same study site (Luscinia megarhyonchos and Ficedula hypoleuca; Saino et al. 2015a).
However, wing length did not covary with Npas2 mean or long allele size in either sex, and the statistically significant relationship between Npas2 genotype and male migration date was obtained when controlling for wing length (see Results), which should at least partly account for intraspecific variation in the latitude of breeding.
Hence, the explanation for a negative association between Npas2 mean allele size and migration date remains elusive. Clearly, these findings suggest that candidate gene–phenotype associations may be complex and broadly vary among species and populations (e.g., Peterson et al. 2013; Bourret and Garant 2015).
Our results showed that Npas2 and Creb1 genes had sex-specific phenotypic effects. Sex-specific effects of candidate genes have been previously highlighted for different life-history events by several studies (Caprioli et al. 2012; Bourret and Garant 2015; Saino et al. 2015a; Bazzi et al. 2016). Sex-specific effects may originate because of sex-specific selective pressures on timing of life-history events. For instance, in proterandrous migratory species, males are subjected to stronger selective pressures for early arrival at the breeding grounds than females (e.g., Morbey and Ydenberg 2001; Spottiswoode et al. 2006; Newton 2008; Reudink et al. 2009, Spottiswoode and Saino 2010). Proximately, sex-specific genotype–phenotype associations may arise because of sex-specific genetic architecture. For instance, the autosomal genome is shared by both sexes, but gene expression and regulation is often sexually dimorphic, leading to genotype–sex interactions in genotype–phenotype association studies (review in Ellegren and Parsch 2007; Ober et al. 2008). An alternative possibility is that males and females migrating at Ventotene originated from different breeding populations and that the observed sex-specific genotype–phenotype associations may instead originate because of population-specific candidate gene effects. However, the lack of genetic differentiation at candidate genes between the sexes (both for single loci and for the combination of the 5 loci) argues against this possibility.
To conclude, our study provides novel insights into avian migratory phenotype–genotype associations for a broad set of candidate genes’ loci. Our findings suggest that different candidate genes may contribute to regulating different life-history events in a sex-specific fashion, and that candidate gene polymorphism underlies among-individuals variation in phenology throughout the annual cycle. Intriguingly, the association between Creb1, a candidate gene which constitutes a key element for the light entrainment of the endogenous clock, and a proxy for moult speed, a life-history event that occurs at equatorial latitudes, may suggest that daylength plays a role in the synchronization of circadian and circannual rhythms of birds even where daily changes in photoperiod are small.
We thank M. Caprioli and C.D. Possenti for assistance during laboratory and field work; A. Galimberti for the support with statistical analyses; and 4 anonymous reviewers for constructive criticism on a previous draft. We thank the Riserva Naturale Isole di Ventotene e Santo Stefano for the logistic support and the field assistants and ringers that helped collecting the data. Results from the Progetto Piccole Isole (INFS-ISPRA): paper no. 55.
References
- Bazzi G, Ambrosini R, Caprioli M, Costanzo A, Liechti F. et al. , 2015. Clock gene polymorphism and scheduling of migration: a geolocator study of the barn swallow Hirundo rustica. Sci Rep 5:12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi G, Galimberti A, Hays QR, Bruni I, Cecere JG. et al. , 2016. Adcyap1 polymorphism covaries with breeding latitude in a Nearctic migratory songbird, the Wilson’s warbler Cardellina pusilla. Ecol Evol 6:3226–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi G, Cecere JGC, Caprioli M, Gatti E, Gianfranceschi L. et al. , in press. Clock gene polymorphism, migratory behaviour and geographic distribution: a comparative study of trans-Saharan migratory birds. Molecular Ecology. [DOI] [PubMed]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE. et al. , 2005. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 6:544–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensch S, Andersson T, Åkesson S, 1999. Morphological and molecular variation across a migratory divide in willow warblers, Phylloscopus trochilus. Evolution 53:1925–1935. [DOI] [PubMed] [Google Scholar]
- Bensch S, Grahn M, 1993. A new method for estimating individual speed of molt. Condor 95:305–315. [Google Scholar]
- Berthold P, 1996. Control of Bird Migration. Berlin, Germany: Springer Science & Business Media. [Google Scholar]
- Bourret A, Garant D, 2015. Candidate gene–environment interactions and their relationships with timing of breeding in a wild bird population. Ecol Evol 5:3628–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin A, 1993. Radio-ptilochronology tracing radioactively labelled food in feathers. Ornis Scand 24:167–173. [Google Scholar]
- Caprioli M, Ambrosini R, Boncoraglio G, Gatti E, Romano A. et al. , 2012. Clock gene variation is associated with breeding phenology and maybe under directional selection in the migratory barn swallow. PLoS ONE 7:e35140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakarov N, Jonker RM, Boerner M, Hoffman JI, Kruger O, 2013. Variation at phenological candidate genes correlates with timing of dispersal and plumage morph in a sedentary bird of prey. Mol Ecol 22:5430–5440. [DOI] [PubMed] [Google Scholar]
- Conklin JR, Battley PF, Potter MA, Fox JW, 2010. Breeding latitude drives individual schedules in a trans-hemispheric migrant bird. Nat Commun 1:67.. [DOI] [PubMed] [Google Scholar]
- Cramp S, 1998. The Complete Birds of the Western Palearctic on CD-ROM. Oxford: Oxford University Press. [Google Scholar]
- Dawson A, Hinsley SA, Ferns PN, Bonser RH, Eccleston L, 2000. Rate of moult affects feather quality: a mechanism linking current reproductive effort to future survival. Proc R Soc B 267:2093–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Hera I, Pérez-Tris J, Tellería JL, 2009. Migratory behaviour affects the trade-off between feather growth rate and feather quality in a passerine bird. Biol J Linn Soc 97:98–105. [Google Scholar]
- De la Hera I, Schaper SV, Díaz JA, Pérez-Tris J, Bensch S. et al. , 2011. How much variation in the molt duration of passerines can be explained by the growth rate of tail feathers? Auk 128:321–329. [Google Scholar]
- Debruyne JP, 2008. Oscillating perceptions: the ups and downs of the CLOCK protein in the mouse circadian system. J Genet 87:437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dor R, Cooper CB, Lovette IJ, Massoni V, Bulit F. et al. , 2012. Clock gene variation in Tachycineta swallows. Ecol Evol 2:95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, Parsch J, 2007. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet 8:689–698. [DOI] [PubMed] [Google Scholar]
- Evans KL, Gaston KJ, Sharp SP, McGowan A, Hatchwell BJ, 2009. The effect of urbanisation on avian morphology and latitudinal gradients in body size. Oikos 118:251–259. [Google Scholar]
- Fidler AE, Gwinner E, 2003. Comparative analysis of avian BMAL1 and CLOCK protein sequences: a search for features associated with owl nocturnal behaviour. Comp Biochem Physiol B 136:861–874. [DOI] [PubMed] [Google Scholar]
- Gau D, Lemberger T, Von Gall C, Kretz O, Le Minh N. et al. , 2002. Phosphorylation of CREB Ser142 regulates light-induced phase shifts of the circadian clock. Neuron 34:245–253. [DOI] [PubMed] [Google Scholar]
- Goudet J, 2001. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Department of Ecology and Evolution, University of Lausanne, Switzerland.
- Goymann W, Spina F, Ferri A, Fusani L, 2010. Body fat influences departure from stopover sites in migratory birds: evidence from whole-island telemetry. Biol Lett 6:478–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb TC, 2006. Ptilochronology: Feather Time and the Biology of Birds .New York: Oxford University Press. [Google Scholar]
- Guo SW, Thompson EA, 1992. A Monte Carlo method for combined segregation and linkage analysis. Am J Hum Genet 51:1111–1126. [PMC free article] [PubMed] [Google Scholar]
- Gwinner E, 1986. Circannual Rhythms. Berlin: Springer-Verlag. [Google Scholar]
- Gwinner E, 2003. Circannual rhythms in birds. Curr Opin Neurobiol 13:770–778. [DOI] [PubMed] [Google Scholar]
- Hall KSS, Fransson T, 2000. Lesser Whitethroats under time-constraint moult more rapidly and grow shorter wing feathers. J Avian Biol 31:583–587. [Google Scholar]
- Hannibal J, Ding JM, Chen D, Fahrenkrug J, Larsen PJ. et al. , 1997. Pituitary adenylate cyclase-activating peptide (PACAP) in the retinohypothalamic tract: a potential daytime regulator of the biological clock. J Neurosci 17:2637–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedenström A, Barta Z, Helm B, Houston AI, Mcnamara JM. et al. , 2007. Migration speed and scheduling of annual events by migrating birds in relation to climate change. Clim Res 35:79–91. [Google Scholar]
- Hemborg C, Lundberg A, 1998. Costs of overlapping reproduction and moult in passerine birds: an experiment with the pied flycatcher. Behav Ecol Sociobiol 43:19–23. [Google Scholar]
- Jenni L, Winkler RR, 1989. The feather-length of small passerines: a measurement for wing-length in live birds and museum skins. Bird Study 36:1–15. [Google Scholar]
- Jenni L, Winkler RR, 1994. Moult and Ageing of European Passerines. London: Academic Press. [Google Scholar]
- Johnsen A, Fidler AE, Kuhn S, Carter KL, Hoffmann A. et al. , 2007. Avian Clock gene polymorphism: evidence for a latitudinal cline in allele frequencies. Mol Ecol 16:4867–4880. [DOI] [PubMed] [Google Scholar]
- Jonzén N, Linden A, Ergon T, Knudsen E, Vik JO. et al. , 2006a. Rapid advance of spring arrival dates in long-distance migratory birds. Science 312:1959–1961. [DOI] [PubMed] [Google Scholar]
- Jonzén N, Piacentini D, Andersson A, Montemaggiori A, Stervander M. et al. , 2006b. The timing of spring migration in trans-Saharan migrants: a comparison between Ottenby, Sweden and Capri, Italy. Ornis Svecica 16:27–33. [Google Scholar]
- Kondo T, Hearing VJ, 2011. Update on the regulation of mammalian melanocyte function and skin pigmentation. Expert Rev Dermatol 6:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn K, Schwenk K, Both C, Canal D, Johansson US. et al. , 2013. Differentiation in neutral genes and a candidate gene in the pied flycatcher: using biological archives to track global climate change. Ecol Evol 3:4799–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedvogel M, Sheldon BC, 2010. Low variability and absence of phenotypic correlates of Clock gene variation in a great tit Parus major population. J Avian Biol 41:543–550. [Google Scholar]
- Liedvogel M, Szulkin M, Knowles SC, Wood MJ, Sheldon BC, 2009. Phenotypic correlates of Clock gene variation in a wild blue tit population: evidence for a role in seasonal timing of reproduction. Mol Ecol 18:2444–2456. [DOI] [PubMed] [Google Scholar]
- Messineo A, Grattarola A, Spina F, 2001. Dieci anni di Progetto Piccole Isole. Biol Conserv Fauna 106:1–244. [Google Scholar]
- Møller AP, Nuttall R, Piper SE, Szép T, Vickers EJ, 2011. Migration, moult and climate change in barn swallows Hirundo rustica in South Africa. Clim Res 47:201–205. [Google Scholar]
- Morbey YE, Ydenberg RC, 2001. Protandrous arrival timing to breeding areas: a review. Ecol Lett 4:663–673. [Google Scholar]
- Mueller JC, Pulido F, Kempenaers B, 2011. Identification of a gene associated with avian migratory behaviour. Proc R Soc B 278:2848–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy AD, Csernus VJ, 2007. The role of PACAP in the control of circadian expression of clock genes in the chicken pineal gland. Peptides 28:1767–1774. [DOI] [PubMed] [Google Scholar]
- Newton I, 2008. The Migration Ecology of Birds. London: Academic Press. [Google Scholar]
- O'Malley KG, Ford MJ, Hard JJ, 2010. Clock polymorphism in Pacific salmon: evidence for variable selection along a latitudinal gradient. Proc R Soc B 277:3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober C, Loisel DA, Gilad Y, 2008. Sex-specific genetic architecture of human disease. Nat Rev Genet 9:911–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiró G, 2003. Intraspecific variation in the wing shape of the long-distance migrant reed warbler Acrocephalus scirpaceus: effects of age and distance of migration. Ardeola 50:31–37. [Google Scholar]
- Peterson MP, Abolins-Abols M, Atwell JW, Rice RJ, Mila B. et al. , 2013. Variation in candidate genes CLOCK and ADCYAP1 does not consistently predict differences in migratory behavior in the songbird genus Junco. F1000Research 2:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido F, 2007. The genetics and evolution of Avian migration. Bioscience 57:165–174. [Google Scholar]
- Racz B, Horvath G, Faluhelyi N, Nagy AD, Tamas A. et al. , 2008. Effects of PACAP on the circadian changes of signaling pathways in chicken pinealocytes. J Neurosci 36:220–226. [DOI] [PubMed] [Google Scholar]
- Reudink MW, Marra PP, Kyser TK, Boag PT, Langin KM. et al. , 2009. Non-breeding season events influence sexual selection in a long-distance migratory bird. Proc R Soc B 276:1619–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubolini D, Spina F, Saino N, 2005. Correlates of timing of spring migration in birds: a comparative study of trans-Saharan migrants. Biol J Linn Soc 85:199–210. [Google Scholar]
- Saino N, Rubolini D, Serra L, Caprioli M, Morganti M. et al. , 2010. Sex-related variation in migration phenology in relation to sexual dimorphism: a test of competing hypotheses for the evolution of protandry. J Evol Biol 23:2054–2065. [DOI] [PubMed] [Google Scholar]
- Saino N, Romano M, Caprioli M, Ambrosini R, Rubolini D. et al. , 2012. A ptilochronological study of carry‐over effects of conditions during wintering on breeding performance in the barn swallow Hirundo rustica. J Avian Biol 43:513–524. [Google Scholar]
- Saino N, Romano M, Caprioli M, Fasola M, Lardelli R. et al. , 2013. Timing of molt of barn swallows is delayed in a rare Clock genotype. PeerJ 1:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saino N, Bazzi G, Gatti E, Caprioli M, Cecere JG. et al. , 2015a. Polymorphism at the Clock gene predicts phenology of long-distance migration in birds. Mol Ecol 24:1758–1773. [DOI] [PubMed] [Google Scholar]
- Saino N, Romano M, Romano A, Rubolini D, Ambrosini R. et al. , 2015b. White tail spots in breeding barn swallows Hirundo rustica signal body condition during winter moult. Ibis 157:722–730. [Google Scholar]
- Schwartz C, Andrews MT, 2013. Circannual transitions in gene expression: lessons from seasonal adaptations In: Wassarman PM, editor. Current topics in developmental biology .Elsevier, Oxford (UK): Academic Press, 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PJ, 2005. Photoperiodic regulation of seasonal breeding in birds. Ann N Y Acad Sci 1040:189–199. [DOI] [PubMed] [Google Scholar]
- Simonneaux V, Ouichou A, Pévet P, 1993. Pituitary adenylate cyclase-activating polypeptide (PACAP) stimulates melatonin synthesis from rat pineal gland. Brain Res 603:148–152. [DOI] [PubMed] [Google Scholar]
- Spina F, Massi A, Montemaggiori A, Baccetti N, 1993. Spring migration across central Mediterranean: general results from the “Progetto Piccole Isole”. Vogelwarte 37:1–94. [Google Scholar]
- Spottiswoode C, Saino N, 2010. Sexual selection and climate change In: Møller AP, Fiedler W, Berthold P, editors. Effects of Climate Change in Birds. Oxford: Oxford University Press, 169–189. [Google Scholar]
- Spottiswoode CN, Tøttrup AP, Coppack T, 2006. Sexual selection predicts advancement of avian spring migration in response to climate change. Proc R Soc B 273:3023–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmeyer C, Mueller JC, Kempenaers B, 2009. Search for informative polymorphisms in candidate genes: clock genes and circadian behaviour in blue tits. Genetica 136:109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarka M, Åkesson S, Beraldi D, Hernández-Sánchez J, Hasselquist D. et al. , 2010. A strong quantitative trait locus for wing length on chromosome 2 in a wild population of great reed warblers. Proc R Soc B 277:2361–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenan S, Spina F, 2010. Timing and condition-related effects on recapture probability, mass change and stopover length of spring migrating songbirds on a small mediterranean island. Ardeola 57:121–132. [Google Scholar]
- Tischkau SA, Mitchell JW, Tyan SH, Buchanan GF, Gillette MU, 2003. Ca2+/cAMP response element-binding protein (CREB)-dependent activation of Per1 is required for light-induced signaling in the suprachiasmatic nucleus circadian clock. J Biol Chem 278:718–723. [DOI] [PubMed] [Google Scholar]
- Underhill LG, Prys-Jones RP, Dowsett RJ, Herroelen P, Johnson DN. et al. , 1992. The biannual primary moult of willow warblers Phylloscopus trochilus in Europe and Africa. Ibis 134:286–297. [Google Scholar]
- Visser ME, Caro SP, Van Oers K, Schaper SV, Helm B, 2010. Phenology, seasonal timing and circannual rhythms: towards a unified framework. Phil Trans R Soc Lond B 365:3113–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]