Significance
Delivery remains a significant challenge for robust implementation of CRISPR/Cas9. We report an efficient CRISPR/Cas9 delivery system comprising PEGylated nanoparticles based on the α-helical polypeptide PPABLG. Assisted by the high membrane-penetrating ability of the polypeptide, P-HNPs achieved efficient cellular internalization and endosomal escape. The CRISPR/Cas9 delivery system could reach 47.3% gene editing in cells, 35% gene deletion in vivo, and HeLa tumor growth suppression >71%, demonstrating an advantage over the existing conventional polycationic transfection reagents. Efficient also in knock-in and gene activation, the reported CRISPR/Cas9 delivery system serves to advance gene editing in vitro and in vivo.
Keywords: genome editing, CRISPR/Cas9, helical polypeptide, nanomedicine, cell-penetrating peptide
Abstract
Effective and safe delivery of the CRISPR/Cas9 gene-editing elements remains a challenge. Here we report the development of PEGylated nanoparticles (named P-HNPs) based on the cationic α-helical polypeptide poly(γ-4-((2-(piperidin-1-yl)ethyl)aminomethyl)benzyl-l-glutamate) for the delivery of Cas9 expression plasmid and sgRNA to various cell types and gene-editing scenarios. The cell-penetrating α-helical polypeptide enhanced cellular uptake and promoted escape of pCas9 and/or sgRNA from the endosome and transport into the nucleus. The colloidally stable P-HNPs achieved a Cas9 transfection efficiency up to 60% and sgRNA uptake efficiency of 67.4%, representing an improvement over existing polycation-based gene delivery systems. After performing single or multiplex gene editing with an efficiency up to 47.3% in vitro, we demonstrated that P-HNPs delivering Cas9 plasmid/sgRNA targeting the polo-like kinase 1 (Plk1) gene achieved 35% gene deletion in HeLa tumor tissue to reduce the Plk1 protein level by 66.7%, thereby suppressing the tumor growth by >71% and prolonging the animal survival rate to 60% within 60 days. Capable of delivering Cas9 plasmids to various cell types to achieve multiplex gene knock-out, gene knock-in, and gene activation in vitro and in vivo, the P-HNP system offers a versatile gene-editing platform for biological research and therapeutic applications.
The clustered regularly interspaced short palindromic repeats, CRISPR-associated protein 9 (CRISPR/Cas9) system has emerged as one of the most exciting tools for gene editing in recent years (1). Compared with other systems that recognize target sequences by programmable protein–DNA interactions (2), the CRISPR/Cas9 system relies simply on base-pairing between the single-guide RNA (sgRNA) and the target DNA. With a minimal requirement in target design and straightforward construction of sgRNAs, it facilitates widespread application in either fundamental research or therapeutic settings. However, effective delivery of active CRISPR/Cas9 remains a challenge (3).
Two critical components, Cas9 nuclease and sgRNA, are essential for CRISPR/Cas9 activity. To enable effective gene editing, the Cas9 and sgRNA expression cassettes are frequently encoded within one plasmid for ease of codelivery. However, this approach increases the plasmid size, which hinders effective transfection. Moreover, in multiplexed genome editing, the cloning effort for “all-in-one” vector construction can be burdensome and challenging for optimization, for example, in screening different sgRNAs to realize multiplex gene editing after vector construction. Alternatively, the delivery of Cas9 expression plasmids and sgRNAs could be separated with either sgRNA expression plasmids or in vitro-synthesized sgRNAs to provide flexibility.
Until now, most of the CRISPR/Cas9 delivery systems had relied on physical or viral approaches. Physical methods, including electroporation and microinjection, afford high transfection efficiency but may suffer from low cell viability and cell-specific efficiency; physical methods are also difficult for in vivo application (4, 5). Similarly, viral vectors achieve good performance on delivery of CRISPR/Cas9, but are limited by restricted packaging capacity (6) or unwanted genetic mutations and immunogenicity (7), posing concerns about safety in clinical translation. In comparison, nonviral delivery methods via nanoparticles have the potential to overcome many of these limitations, particularly with respect to safety, large packaging capacity, and in vivo application (7).
Efficient delivery of Cas9 plasmids and synthesized sgRNAs by nanoparticles requires proficient navigation through the extracellular and intracellular barriers. Membrane-disruptive and endosomolytic materials are potent facilitators. Moreover, the timing of intracellular release of Cas9 expression plasmids and sgRNAs is also critical. And there is another consideration: nanocarriers optimal for Cas9 expression plasmid delivery may not be suitable for sgRNAs, as Cas9 expression plasmids are long double-stranded DNA (9.3 kb) with high charge density while sgRNAs are 100-nt single-strand RNAs with lower charge density. Thus, nanocarriers independently optimized for the delivery of Cas9 expression plasmids and sgRNAs would be desirable.
Recently, cell-penetrating peptides (CPPs) (8), lipid-based nanoparticles (9–12), DNA nanoclews (13), gold nanoparticles (14, 15), and “core-shell” artificial viruses (16) have been applied to Cas9/gRNA delivery. The CPPs, based on materials for membrane destabilization, such as HIV-TAT, GALA, and oligoarginine, have been commonly incorporated into delivery vectors for membrane destabilization to increase uptake, promote endosomal escape, and improve overall transfection efficiency (17). We hypothesize that using CPP alone as the delivery vector may be even more potent; a single-component delivery vector is also advantageous for eventual translation. However, for Cas9 expression plasmid delivery, the conventional CPPs either are too small or lack an adequate cationic charge density to function as stand-alone gene vectors. Additionally, after complexing with the nucleic acids, the overall positive charge of CPPs may be neutralized and the membrane-active domains of the short oligopeptide would be partially shielded, thus reducing their membrane activities as well as transfection efficiencies (17, 18). As such, CPPs often play only a supportive role to improve the delivery efficiencies of existing systems (17).
We previously reported the synthesis of water-soluble, α-helical polypeptides bearing a cationic side-chain terminus (19) and demonstrated the helicity-associated membrane activity that rendered these polypeptides effective gene vectors (20, 21). This type of helical polypeptide poly(γ-4-((2-(piperidin-1-yl)ethyl)aminomethyl)benzyl-l-glutamate) (PPABLG) can bind and condense both plasmid DNA and short siRNA while retaining the helical structure to provide high membrane-penetrating ability for enhanced cellular internalization and endosomal escape. PPABLG can also maintain stable helical structure against changes of environmental conditions, such as pH, ionic strength, temperature, and the presence of denaturing reagents (19), making it particularly attractive for in vivo application. More importantly, PPABLG features high stability against proteases, a distinct advantage over the majority of CPPs that are prone to hydrolytic degradation in vivo (20, 22). The desired enzymatic stability of PPABLG is mainly due to its nonnatural amino acid sequences that cannot be recognized by proteases and the long hydrophobic side chains that hinder the access of proteases (19). Our previous data have also shown that this kind of helical polypeptide has superior cell-membrane permeability over HIV-TAT and Arg9 by up to two orders of magnitude and thus affords excellent DNA and siRNA delivery efficiencies in various mammalian cells (23, 24). After complexing siRNA, the PPABLG could still maintain high membrane activities (23, 24), a desired property that short oligopeptides may not possess. PPABLG is a cationic polypeptide that could serve the dual function of an efficient gene carrier and a cell-membrane–penetrating agent. We thus hypothesized that the helical PPABLG, with high positive charge, could complex Cas9 expression plasmids and sgRNAs to form nano-complexes (referred to as helical polypeptide nanoparticles, or HNPs). To enhance the extracellular stability and the potential for in vivo application, the HNPs were PEGylated (referred to as P-HNPs) through incorporation of PEG-Polythymine40 (PEG-T40) in the formulation. We first evaluated this delivery system for multiplex gene editing in vitro (Fig. 1), followed by studying gene disruption in vivo.
Fig. 1.
Schematic illustration showing the formation of P-HNPs and the intracellular activity of Cas9 expression plasmid/sgRNA in performing genome editing or gene activation.
Results and Discussion
PPABLG Synthesis and Nanoparticle Formulation.
The cationic, α-helical polypeptide PPABLG was synthesized and characterized via ring-opening polymerization of N-carboxyanhydride monomers and subsequent side-chain functionalization through ozonolysis and reductive amination (SI Appendix, Fig. S1) (20).
HNPs containing only the Cas9 expression plasmid (px165) were formulated by adding the PPABLG solution into the plasmid solution at various weight ratios (PPABLG:px165 = 1:1–15:1). The size of the polyplexes decreased from 230 nm to around 100 nm as the PPABLG/px165 ratio (wt/wt) increased from one to five (SI Appendix, Fig. S2A), and the zeta potential increased from 1 to above 20 correspondingly. The polydispersity index (PDI) value of HNPs at all PPABLG/px165 ratios was less than 0.2, indicating relatively high uniformity.
Delivery of Cas9 Expression Plasmids into Various Primary and Immortalized Cells.
The plasmid that fuses the reporter gene GFP to the Cas9 expression cassette [pSpCas9(BB)-2A-GFP and abbreviated as px458] was used to facilitate the detection of Cas9 expression in the transfected cells. As shown in SI Appendix, Fig. S2B, HNPs formed by PPABLG:px458 (wt/wt = 30:1) demonstrated the best transfection efficiency (∼48%) in HEK293T cells, which was comparable to the transfection efficiency of the pEGFP plasmid with dramatically smaller size (4.7 kb) (25). However, such HNPs caused ∼30% cell death (SI Appendix, Fig. S2C). Moreover, HNPs were colloidally unstable in serum-containing medium (SI Appendix, Fig. S2D), which would compromise their in vivo performance. To address such an issue, we next developed P-HNPs to decrease the cytotoxicity and enhance the stability in serum (Fig. 1). Particularly, PEG-T40 was synthesized and added to the plasmid solution at various weight ratios before it was mixed with PPABLG. We varied the molecular weight of PEG from 2 to 5 kDa, and the PEG-T40 content in P-HNPs from 0.66% (PPABLG:px458:PEG-T40 = 30:1:0.2) to 3.22% (PPABLG:px458:PEG-T40 = 30:1:1). Addition of PEG-T40 did not change the size and zeta potential of the nanoparticles compared with HNPs (SI Appendix, Fig. S3 A and B), and the obtained P-HNPs with 3.22% PEG2k-T40 showed desired colloidal stability in 10% FBS-containing medium, as evidenced by inappreciable alteration of the particle (SI Appendix, Fig. S2D). As shown in SI Appendix, Fig. S3 C and D, P-HNPs modified with PEG2k-T40 at the relative content of 3.22% showed the best GFP-positive percentage and GFP mean fluorescence intensity compared with polyethylenimine (PEI)/px458 treatment or HNPs treatment. Cells treated by P-HNPs with PEG5k-T40 (1.61% PEG-T40 content) also showed a high GFP-positive percentage, but were twofold lower on the GFP mean fluorescence intensity compared with P-HNPs modified with PEG2k-T40 at the relative content of 3.22%.
We also coated the PPABLG/px458 HNP complex with PEG2k-T40 by adding PEG2k-T40 after forming the PPABLG/px458 HNPs complex [named P-HNP(coating)]. P-HNP(coating) made at the ratio of 30:1:1 (PPABLG:px458:PEG2k-T40) has a similar size (87 nm) and zeta potential (20.8 mV) with P-HNPs made by mixing the PEG2k-T40 with the plasmid before adding the PPABLG. However, P-HNP(coating) reduced the transfection efficiency to 5% (SI Appendix, Fig. S4), likely caused by the masking action of PEG-T40 in blocking the cellular penetration ability of PPABLG. For all subsequent studies, we therefore settled on the formulation of mixing PEG2k-T40 with the plasmid before adding PPABLG at the ratio of 30:1:1 (PPABLG:px458:PEG2k-T40).
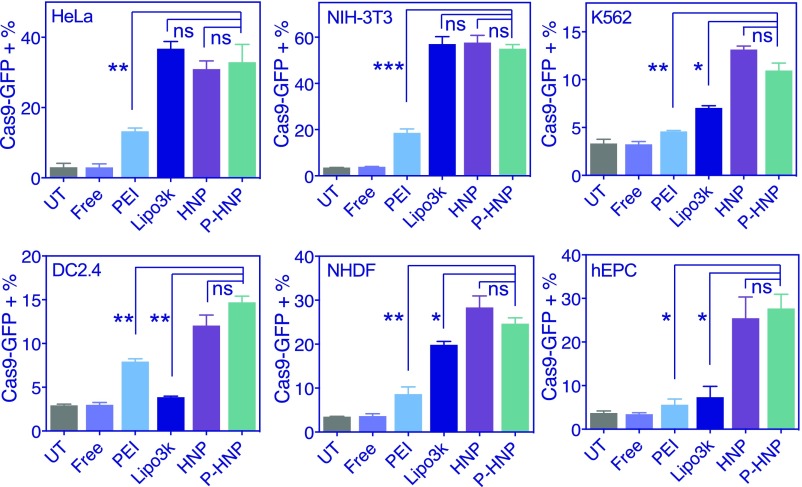
To evaluate the broad applicability of P-HNPs as Cas9 expression plasmid delivery vehicles, we performed plasmid transfection experiments in a variety of cell types (Fig. 2). In cancer cell lines, including HeLa and K562 cells, P-HNPs showed 32.9 and 11.0% transfection efficiency, respectively. In other immortalized cell lines, such as mouse fibroblast cells NIH 3T3 and dendritic cells DC2.4, P-HNP transfection resulted in 55.0 and 14.7% of Cas9-GFP expression, respectively. In primary cells of normal human dermal fibroblasts (NHDF) and human umbilical cord blood-derived endothelial progenitor cells (hEPCs), P-HNPs showed 24.6 and 27.7% transfection efficiency, respectively. P-HNPs for px458 plasmid transfection in all these six cell lines yielded no significant difference with HNP-based px458 plasmid transfection. Compared with Lipofectamine 3000, the most potent commercially available in vitro transfection reagent, P-HNPs performed better in the K562 cells, DC2.4 cells, NHDF cells, and hEPCs. Noteworthy is that P-HNPs lead to 1.9- to 4.9-fold higher transfection efficiency than PEI/px458 treatment, which is the most commonly used polycation gene transfection reagent (Fig. 2).
Fig. 2.
Transfection of Cas9-GFP plasmid px458 in a variety of cells. The HNPs were formed by PPABLG/px458 at the wt/wt ratio of 30:1. The P-HNPs were formed by PPABLG/px458/PEG2k-T40 at the wt/wt/wt ratio of 30:1:1. Free, treatment with naked plasmids; UT, untreated group.
Cellular Internalization and Endosomal Escape of P-HNPs.
To investigate the ability of P-HNPs to overcome cellular barriers, we assessed the cellular uptake of P-HNPs by delivering YOYO-1–labeled Cas9 expression plasmid px165. SI Appendix, Fig. S5A, reveals that P-HNPs had a comparable cellular uptake level with HNPs but outperformed PEI, which is consistent with the px458 plasmid transfection results (SI Appendix, Fig. S3 C and D). The cellular uptake for P-HNPs was inhibited by 36.1% at 4 °C, suggesting that most of the complexes enter the cell via energy-independent nonendocytosis pathways (SI Appendix, Fig. S5B). To investigate if the membrane permeability of PPABLG was well-maintained after forming P-HNPs, we monitored the cellular uptake of FITC–Tris, a hydrophilic membrane-impermeable fluorescent dye, in the presence of HNPs and P-HNPs (20). As shown in SI Appendix, Fig. S5C, P-HNPs promoted FITC–Tris uptake by ∼300-ng/mg protein as well as the HNP-induced FITC–Tris uptake, suggesting that the PEGylation did not abrogate the cell-membrane–penetrating ability of P-HNP. The cellular uptake was also partially inhibited by chlorpromazine and wortmannin (SI Appendix, Fig. S5B), demonstrating that the clathrin-mediated pathway and macropinocytosis were involved in the endocytosis of P-HNPs. Additionally, the significant inhibition by mβCD and genistein implied that the internalization of P-HNPs was closely associated with caveolae-mediated uptake, which is beneficial for plasmid release in the cytoplasm via the bypass of lysosome formation and more direct translocation to the Golgi or endoplasmic reticulum to facilitate nuclear transport (26). Confocal laser scanning microscopy observation confirmed the endosomal escape of the P-HNPs. As shown in SI Appendix, Fig. S5D, in cells treated with P-HNPs as well as with HNPs, YOYO-1–labeled px165 was distributed in the whole cytoplasm and showed low colocalization with Lysotracker Red-stained endosomes. The YOYO-1–labeled px165 was also localized to the DAPI-stained nuclei in the HNP- and P-HNP–treated groups, validating that Cas9 expression plasmids could be transferred into cell nuclei to enable the Cas9 gene transcription (SI Appendix, Fig. S5D).
Delivery of Cas9 Plasmids and sgRNAs for in Vitro Gene Disruption and Endogenous Gene Editing.
After establishing the efficiency of P-HNPs in delivering Cas9 plasmids, we also confirmed their suitability for delivering sgRNA (SI Appendix, Fig. S5E). The treatment by P-HNPs (30:1:1) loaded with FITC-labeled sgRNAs showed the best cellular uptake efficiency at 67.4% (SI Appendix, Fig. S5E) and strongly outperformed PEI/sgRNA complexes (9.8%) by sevenfold and Lipofectamine 3000 (33.5%) by twofold.
The U2OS.EGFP cell line, which constitutively expresses a destabilized EGFP, was used as a model for the Cas9-induced reporter gene disruption assay. P-HNPs codelivering two plasmids expressing Cas9 and EGFP-targeting sgRNAs at the ratio of 2:1 (wt/wt for Cas9:sgRNA plasmids) produced the highest EGFP absence efficiency (29.0%) and outperformed PEI/plasmid gene disruption efficiency (7.9%) (Fig. 3A). No reduction in EGFP-positive cells was observed in control groups containing naked plasmids or Cas9 plasmid alone.
Fig. 3.
In vitro gene disruption. (A) EGFP disruption assay of Cas9 expression plasmid and sgRNA expression plasmid delivery by HNPs or P-HNPs in U2OS.EGFP reporter cells. The ratios in the figure are weight ratios for Cas9 expression plasmid:sgRNA expression plasmid. (B and C) EGFP disruption assay of Cas9 expression plasmid and in vitro-synthesized sgRNA delivery in U2OS.EGFP reporter cells. Short dash under the bars in B indicates the group treated by only Cas9 expression plasmids. (D–F) GCD assay of indels in the EGFP gene in U2OS.EGFP cells (D), the AAVS1 gene in HEK293T cells (E), and the HPRT1 gene in HEK293T cells (F). “Multi” is the group treated by Cas9 expression plasmid delivery and subsequent codelivery of AAVS1- and HPRT1-targeting sgRNAs. Arrows indicate the cutting products. Free, treatment with naked nucleic acids; UT, untreated group.
EGFP deletion was also observed when the sgRNAs were delivered in the synthetic nucleic acid format by H-HNPs instead of via transfection (SI Appendix, Fig. S6A). However, this only resulted in a weakened EGFP gene disruption efficiency (19.2%). Similar finding was reported by Miller et al. (27) when they used zwitterionic amino lipid-based nanoparticles to deliver Cas9 expression mRNAs and synthetic sgRNAs. We hypothesize that the presence of Cas9 and sgRNA in the nuclei needs to be synchronized for maximal gene editing. Observing a peak Cas9 protein at ∼24 h posttransfection in our other experiments, we delivered the synthetic sgRNAs 24 h after the Cas9 plasmid transfection and did observe a higher EGFP disruption efficiency at 35.7% (Fig. 3 B and C). The lower performance of the HNP group (24.9%) and the PEI treatment group (8.2%) appeared to be related to the poorer cellular uptake of the sgRNA nanoparticles (SI Appendix, Fig. S5E). We also compared the EGFP disruption efficiency of P-HNPs with the commercial formulations Lipofectamine 2000, Lipofectamine 3000, and Lipofectamine CRISPRMAX (SI Appendix, Fig. S6B). Although the Lipofectamine CRISPRMAX could induce a higher gene disruption (51.2%) compared with P-HNPs (36.5%), the delivery of Cas9 proteins may face concerns such as affordability, purity of the Cas9 protein, and nonspecific nuclease action-mediated toxicity, especially humoral immunity to Cas9 (28). In direct comparison with Lipofectamine 2000 and 3000 in Cas9 plasmid and sgRNA delivery, P-HNPs outperformed Lipofectamine 2000 and were comparable to Lipofectamine 3000 in U2OS.EGFP cells. To verify that the EGFP disruption arose from insertion-deletion (indel) events, we applied the genomic cleavage detection (GCD) assay to quantify the frequency of indels at the target EGFP locus (Fig. 3D). We observed 39.7, 22.5, and 6.2% indels for the P-HNP, HNP, and PEI treatment, respectively, which supported the results of flow cytometry analysis (Fig. 3 B and C). The indel frequencies at EGFP-target sites in cells treated by P-HNPs were also assayed by DNA sequencing (SI Appendix, Fig. S6C). The sequencing of 21 clones showed 8 clones with mutations near the target site, confirming that the gene disruption was mediated by the CRISPR/Cas9 system. The EGFP disruption efficiency of the P-HNP delivery system was comparable with the self-assembled DNA Nanoclews for the Cas9 protein/sgRNA complex in the same cell line (13).
A major concern in using the CRISPR/Cas9 for genome engineering is the off-target editing. Using the GCD assay to detect the indels at two off-target sites for the EGFP-targeting sgRNA, we observed a lower off-target cleavage (<2.8%) in cells treated by sequentially delivering the Cas9 expression plasmid and the synthetic sgRNAs (SI Appendix, Fig. S6D), compared with lipids or short CPP-mediated Cas9 plasmid delivery if differences in the targeted gene and sequence were neglected (8, 29).
After validating the reporter gene disruption in a model cell line, we evaluated the genome editing of endogenous genes in HEK293T cells. Noting the potential biomedical relevance of editing the adeno-associated virus integration site 1 (AAVS1) gene (30) and the hypoxanthine phosphoribosyltransferase 1 (HPRT1) gene (31), we evaluated the transfection of P-HNPs loaded with Cas9 expression plasmids and sgRNA targeting AAVS1 or HPRT1. The disruption efficiencies of 42.5% for the AAVS1 gene or 46.0% for the HPRT1 gene (Fig. 3 E and F), respectively, was higher than the published value using short CPP (28% for the AAVS1 gene) (8), but lower than Lipofectamine 3000 (63% for the AAVS1 gene and 64% for the HPRT1 gene) (29).
We also explored the potential of multiplex gene editing based on P-HNPs. We used P-HNPs to codeliver sgRNAs targeting AAVS1 and HPRT1 at the weight ratio 1:1 at 24 h after cells were treated by Cas9 expression plasmids, and detected the indels formed at the AAVS1 and HPRT1 targeting site, respectively (Fig. 3 E and F). It yielded 41.9% indels for the AAVS1 gene and 47.3% indels for the HPRT1 gene. Single delivery of sgRNAs targeting AAVS1 or HPRT1 would not cause discernible off-target editing to the other (Fig. 3 E and F), indicating the high specificity for genome editing based on P-HNP delivery of CRISPR/Cas9.
Delivery of Cas9 Plasmids and sgRNA for in Vitro Gene Recombination.
CRISPR/Cas9 is a versatile editing tool not only for gene disruption but also for gene correction, insertion, and activation, offering an immense potential for biological and therapeutic applications. To investigate whether the P-HNP system could attain gene insertion by homology-directed repair (HDR), we first delivered the Cas9 expression plasmid, followed by the delivery of AAVS1 targeting sgRNA and the HDR donor template 24 h later using the same P-HNP carrier (Fig. 4A). The GFP and puromycin expression sequences were involved in the HDR donor template. Three days after transfection, the HDR events were verified by PCR using a forward primer recognizing the AAVS1 genome and a reverse primer binding to the puromycin gene in the donor template (Fig. 4A and SI Appendix, Table S1). PCR assay demonstrated that only cells treated with the delivery of Cas9 expression plasmids, sgRNAs, and HDR donor templates exhibited HDR events (Fig. 4B). Cells treated with nanoparticles delivering sgRNA and donor only, without Cas9 expression plasmid, did not show any PCR product. This result demonstrated that the increased HDR events were initiated by CRISPR/Cas9 cleavage, not by traditional HDR events. Although both the transfection of PEI complexes and HNPs showed HDR events, the PCR products of P-HNP transfection were more intense. Several clones with strong GFP expression were formed after selection with puromycin for 2 wk (Fig. 4C). The HDR event was also confirmed by Sanger sequencing of the selected clone’s genome and detection of the expected DNA bases at the genome-donor boundary (Fig. 4D).
Fig. 4.
In vitro gene insertion and gene activation. (A) Donor architecture for HDR of the AAVS1 gene. The arrows show the locations of PCR primers for detecting successful HDR events. (B) PCR assay to detect HDR events 3 d after transfection. (C) Microscopic images of two GFP+ clones after treatment by P-HNPs loaded with Cas9 expression plasmids and the synthetic sgRNAs and HDR donor after selection with puromycin for 2 wk. (Scale bar, 100 μm.) (D) Sanger sequencing of the HDR events in the selected GFP+ clones in C. (E) The transfection efficiency of P-HNPs for pMax-VP64dCas9-BFPVP64 plasmid by detecting the BFP-positive cells. (F) The qRT-PCR evaluation of relative Myod1 mRNA level to detect the Myod1 activation. The results were shown as the ratio relative to the mRNA expression of the untreated cells. Free, treatment with naked nucleic acids; UT, untreated group.
Delivery of dCas9 Plasmids and sgRNA for in Vitro Gene Activation.
The CRISPR/Cas9 technology can also be applied to activate gene expression by introduction of point mutations into the Cas9 nuclease domain to generate a deactivated Cas9 (dCas9) and then by fusing a transactivator such as VP64 to dCas9 (32, 33). We tested the delivery efficiency of the pMax-VP64dCas9-BFPVP64 plasmid of PEI, HNP, and P-HNP. The design of pMax-VP64dCas9-BFPVP64 is such that the dCas9 is fused to two transactivation VP64 domains for higher activation and a BFP protein for ease of detection and optimization (32, 33). P-HNP transfection on NIH 3T3 cells resulted in 41.5% BFP expression, with no significant difference compared with HNP transfection but 2.6-fold higher than PEI transfection (Fig. 4E). This is consistent with the data for the px458 plasmid transfection on NIH 3T3 cells. We next targeted the Myod1 gene, a critical factor that, when activated, can induce the reprogramming of mouse embryonic fibroblasts to skeletal myocytes (33). By delivery of pMax-VP64dCas9-BFPVP64 plasmids and Myod1-targeted sgRNA, P-HNPs efficiently increased the Myod1 mRNA level 5.4-fold after a single transfection and 8.4-fold after three sequential transfections (Fig. 4F). After three sequential transfections, P-HNPs showed a twofold higher level of Myod1 activation compared with PEI-based transfection.
Delivery of Cas9 Plasmids and sgRNA for in Vivo Gene Disruption and Tumor Growth Suppression.
There is often a disconnect between the in vitro and in vivo performance of nanomedicines. To evaluate if the promising P-HNP delivery system is also applicable for gene editing in vivo, we tested the in-tumor gene-editing feasibility of P-HNPs in A549.GFP tumor-bearing mice. Two days after the last injection, many A549.GFP cells near the injection site showed loss or decrease of GFP fluorescence (SI Appendix, Fig. S7A). In contrast, the tumors in the PBS-treated mice or in the mice treated with P-HNPs delivering Cas9 expression plasmids alone did not show any loss of GFP signal. The GFP gene disruption was confirmed by indel detection at the target gene site in the lysed tumor tissue (SI Appendix, Fig. S7B). Treatment by P-HNPs loaded with Cas9 expression plasmids and with synthesized GFP targeting sgRNAs caused 29.7% of the indels at the target site.
After validating the capability of P-HNPs in reporter GFP gene disruption in tumors, we further investigated the gene editing of the survival gene, polo-like kinase 1 (Plk1), for anticancer therapy. Plk1 is a highly conserved serine–threonine kinase that promotes cell division, and overexpression of Plk1 is found in various tumors (34). Inhibition or depletion of Plk1 expression leads to cell-cycle arrest, apoptosis, and “mitotic catastrophe” in cancer cells, which provides a promising modality for anticancer therapy. Delivery of Cas9 expression plasmids and synthesized Plk1-targeting sgRNAs (denoted as P-HNPPCas9+sgPlk1) to HeLa cells caused obvious indels at the target site in HeLa cells in vitro (Fig. 5A). In comparison, P-HNPs encapsulating only the Cas9 expression plasmid (denoted as P-HNPPCas9) showed no cleavage of Plk1. As a result of Cas9/sgRNA-mediated Plk1 disruption, the P-HNPPCas9+sgPlk1 inhibited the proliferation of HeLa cells by 43.2% and caused cell apoptosis by 46.2%, which was significantly higher than P-HNPPCas9 (Fig. 5B and SI Appendix, Fig. S7C). Next, we evaluated the tumor inhibition efficacy of P-HNPs in HeLa xenograft tumor-bearing mice. P-HNPPCas9+sgPlk1 achieved significant tumor inhibition in comparison with PBS, free Cas9 plasmid/free sgPlk1, and P-HNPPCas9, leading to a tumor suppression of >71% (Fig. 5C). Accordingly, P-HNPPCas9+sgPlk1 notably improved the survival rate of mice to 60% (Fig. 5D). The body weight of mice did not change noticeably compared with the control group, suggesting low toxicity of P-HNPs (SI Appendix, Fig. S7D). In consistence with the antitumor efficacy, P-HNPPCas9+sgPlk1 led to remarkable cancer cell necrosis in hematoxylin and eosin (H&E)-stained tumor tissues and afforded the highest cell remission level in tumor tissue sections immune-stained with EdU-594 (SI Appendix, Fig. S7E). To further confirm the CRISPR/Cas9-mediated in vivo gene editing of Plk1, the indel frequency at the Plk1 locus in tumor tissues following P-HNPPCas9+sgPlk1 treatment was measured by deep sequencing. Among the sequenced 20 clones, 7 clones had mutations near the target site, including two single nucleic acid insertions, one G → A single nucleic acid mutation, and four large deletions (SI Appendix, Fig. S8). The final genome-editing efficiency in the Plk1 locus mediated by P-HNPPCas9+sgPlk1 was calculated to be 35%. Western blot analysis further confirmed that the manipulation in the Plk1 genome caused a 67% decrease of the Plk1 protein expression level (Fig. 5E), supporting the finding that P-HNPPCas9+sgPlk1 inhibited the tumor growth by depletion of the Plk1 gene in tumor tissues. While these results suggest the applicability of P-HNPs for local in vivo CRISPR/Cas9 gene editing toward anticancer therapy, its potential for systemic application remains a subject of ongoing investigation. Work is in progress to modify the synthesized sgRNAs for higher stability, to understand the biodistribution via different administration routes, and to evaluate the targeting ability of this delivery system for various gene-editing applications in vivo.
Fig. 5.
In vivo gene disruption and tumor growth suppression. (A) Indels of the Plk1 gene in HeLa cells treated with P-HNPs containing px165 only (P-HNPPCas9) or P-HNPs containing px165 and Plk1-targeting sgRNAs (P-HNPPCas9+sgPlk1). (B) Viability of HeLa cells at 72 h after transfection with P-HNPPCas9 or P-HNPPCas9+sgPlk1. Free, cells treated with free px165 and free Plk1-targeting sgRNAs; UT, untreated cells. (C) The inhibition of tumor growth in HeLa xenograft tumor-bearing mice after intratumoral injection of P-HNPPCas9 or P-HNPPCas9+sgPlk1 (n = 7). Black and red arrows indicate the injection day of P-HNPPCas9 or P-HNPs containing Plk1-targeting sgRNAs (P-HNPsgPlk1), respectively. The injection dose was 20 μg for PCas9 or sgPlk1. (D) Survival of HeLa xenograft mice in C. Euthanasia was performed when tumor volumes reached 1,000 mm3. (E) Expression level of Plk1 protein in tumor tissues in C, analyzed at 48 h post last injection.
Conclusions.
In this work, we reported the synthesis and application of a PEGylated nanoparticle, P-HNP, based on the cationic α-helical polypeptide PPABLG and Cas9 expression plasmids/sgRNAs for genome editing in mammalian cells and in tumor tissue. The P-HNPs showed high membrane-penetrating ability to facilitate cell uptake, escape from the endosome, and translocate into the nucleus for CRISPR/Cas9-based genome editing. The P-HNPs efficiently delivered Cas9 plasmid or sgRNA, as a complex or separately, to various cell types including tumor cells, fibroblasts, dendritic cells, and human endothelial progenitor cells to achieve multiplex gene knockout, demonstrating an advantage over the existing polycation-based gene vectors. P-HNPs could also mediate gene knock-in and gene activation. In vivo studies further indicated the efficiency of this nonviral gene-editing system for topical anticancer therapy. Given the potency of PPABLG for systemic siRNA delivery in vivo, this work suggests the broad potential of P-HNPs for nonviral gene editing.
Materials and Methods
Detailed materials and methods are provided in SI Appendix, including the synthesis of PPABLG and PEG-T40, preparation and characterization of HNPs and P-HNPs, and delivery of plasmids and sgRNAs in vitro and in vivo.
Supplementary Material
Acknowledgments
We thank Prof. J. Keith Joung (Massachusetts General Hospital) for providing the U2OS.EGFP cell line and Prof. Jun Wang and Prof. Yucai Wang (University of Science and Technology of China) for providing the A549.GFP cell line. Funding support was received from the NIH (Grants HL109442, AI096305, GM110494, and UH3 TR000505); National Natural Science Foundation of China (Grants 51573123 and 51722305); the Ministry of Science and Technology of China (Grant 2016YFA0201200); Guangdong Innovative and Entrepreneurial Research Team Program (Grant 2013S086); and Global Research Laboratory Program (Korean NSF GRL; 2015032163).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712963115/-/DCSupplemental.
References
- 1.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox DBT, Platt RJ, Zhang F. Therapeutic genome editing: Prospects and challenges. Nat Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang HX, et al. CRISPR/Cas9-based genome editing for disease modeling and therapy: Challenges and opportunities for nonviral delivery. Chem Rev. 2017;117:9874–9906. doi: 10.1021/acs.chemrev.6b00799. [DOI] [PubMed] [Google Scholar]
- 4.Wells DJ. Gene therapy progress and prospects: Electroporation and other physical methods. Gene Ther. 2004;11:1363–1369. doi: 10.1038/sj.gt.3302337. [DOI] [PubMed] [Google Scholar]
- 5.Maurisse R, et al. Comparative transfection of DNA into primary and transformed mammalian cells from different lineages. BMC Biotechnol. 2010;10:9. doi: 10.1186/1472-6750-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Z, Yang H, Colosi P. Effect of genome size on AAV vector packaging. Mol Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin H, et al. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 8.Ramakrishna S, et al. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014;24:1020–1027. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuris JA, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin H, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M, et al. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc Natl Acad Sci USA. 2016;113:2868–2873. doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo Y-L, et al. Macrophage-specific in vivo gene editing using cationic lipid-assisted polymeric nanoparticles. ACS Nano. 2018;12:994–1005. doi: 10.1021/acsnano.7b07874. [DOI] [PubMed] [Google Scholar]
- 13.Sun W, et al. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew Chem Int Ed Engl. 2015;54:12029–12033. doi: 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang P, et al. Genome editing for cancer therapy: Delivery of Cas9 protein/sgRNA plasmid via a gold nanocluster/lipid core-shell nanocarrier. Adv Sci (Weinh) 2017;4:1700175. doi: 10.1002/advs.201700175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mout R, et al. Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing. ACS Nano. 2017;11:2452–2458. doi: 10.1021/acsnano.6b07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, et al. Artificial virus delivers CRISPR-Cas9 system for genome editing of cells in mice. ACS Nano. 2017;11:95–111. doi: 10.1021/acsnano.6b04261. [DOI] [PubMed] [Google Scholar]
- 17.Komin A, Russell LM, Hristova KA, Searson PC. Peptide-based strategies for enhanced cell uptake, transcellular transport, and circulation: Mechanisms and challenges. Adv Drug Deliv Rev. 2017;110–111:52–64. doi: 10.1016/j.addr.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Meade BR, Dowdy SF. Exogenous siRNA delivery using peptide transduction domains/cell penetrating peptides. Adv Drug Deliv Rev. 2007;59:134–140. doi: 10.1016/j.addr.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Lu H, et al. Ionic polypeptides with unusual helical stability. Nat Commun. 2011;2:206. doi: 10.1038/ncomms1209. [DOI] [PubMed] [Google Scholar]
- 20.Gabrielson NP, et al. Reactive and bioactive cationic α-helical polypeptide template for nonviral gene delivery. Angew Chem Int Ed Engl. 2012;51:1143–1147. doi: 10.1002/anie.201104262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin L, et al. Supramolecular self-assembled nanoparticles mediate oral delivery of therapeutic TNF-α siRNA against systemic inflammation. Angew Chem Int Ed Engl. 2013;52:5757–5761. doi: 10.1002/anie.201209991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin L, et al. Non-viral gene delivery via membrane-penetrating, mannose-targeting supramolecular self-assembled nanocomplexes. Adv Mater. 2013;25:3063–3070. doi: 10.1002/adma.201205088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He H, et al. Suppression of hepatic inflammation via systemic siRNA delivery by membrane-disruptive and endosomolytic helical polypeptide hybrid nanoparticles. ACS Nano. 2016;10:1859–1870. doi: 10.1021/acsnano.5b05470. [DOI] [PubMed] [Google Scholar]
- 24.Zheng N, et al. Manipulating the membrane penetration mechanism of helical polypeptides via aromatic modification for efficient gene delivery. Acta Biomater. 2017;58:146–157. doi: 10.1016/j.actbio.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Zheng N, et al. Maximizing gene delivery efficiencies of cationic helical polypeptides via balanced membrane penetration and cellular targeting. Biomaterials. 2014;35:1302–1314. doi: 10.1016/j.biomaterials.2013.09.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iversen TG, Skotland T, Sandvig K. Endocytosis and intracellular transport of nanoparticles: Present knowledge and need for future studies. Nano Today. 2011;6:176–185. [Google Scholar]
- 27.Miller JB, et al. Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew Chem Int Ed Engl. 2017;56:1059–1063. doi: 10.1002/anie.201610209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charlesworth CT, et al. Identification of pre-existing adaptive immunity to Cas9 proteins in humans. bioRxiv. 2018 doi: 10.1101/243345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang X, et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 30.Smith JR, et al. Robust, persistent transgene expression in human embryonic stem cells is achieved with AAVS1-targeted integration. Stem Cells. 2008;26:496–504. doi: 10.1634/stemcells.2007-0039. [DOI] [PubMed] [Google Scholar]
- 31.Gibbs RA, Nguyen PN, McBride LJ, Koepf SM, Caskey CT. Identification of mutations leading to the Lesch-Nyhan syndrome by automated direct DNA sequencing of in vitro amplified cDNA. Proc Natl Acad Sci USA. 1989;86:1919–1923. doi: 10.1073/pnas.86.6.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez-Pinera P, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chakraborty S, et al. A CRISPR/Cas9-based system for reprogramming cell lineage specification. Stem Cell Reports. 2014;3:940–947. doi: 10.1016/j.stemcr.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eckerdt F, Yuan J, Strebhardt K. Polo-like kinases and oncogenesis. Oncogene. 2005;24:267–276. doi: 10.1038/sj.onc.1208273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.