Abstract
Background:
Sudden death of a newborn is a rare entity, which may be caused by genetic cardiac arrhythmias. Among these diseases, Long QT syndrome is the most prevalent arrhythmia in neonates, but other diseases such as Brugada syndrome, Short QT syndrome and Catecholaminergic Polymorphic Ventricular Tachycardia also cause sudden death in infants. All these entities are charac-terized by well-known alterations in the electrocardiogram and the first symptom of the disease may be an unexpected death. Despite the low prevalence of these diseases, the performance of an electro-cardiogram in the first hours or days after birth could help identify these electrical disruptions and adopt preventive measures. In recent years, there has been an important impulse by some experts in the scientific community towards the initiation of a newborn electrocardiogram-screening program, for the detection of these electrocardiographic abnormalities. In addition, the use of genetic analysis in neonates could identify the cause of these heart alterations. Identification of relatives carrying the ge-netic alteration associated with the disease allows adoption of measures to prevent lethal episodes.
Conclusion:
Recent technological advances enable a comprehensive genetic screening of a large number of genes in a cost-effective way. However, the interpretation of genetic data and its translation into clinical practice are the main challenges for cardiologists and geneticists. However, there is im-portant controversy as to the clinical value, and cost-effectiveness of the use of electrocardiogram as well as of genetic testing to detect these cases. Our review focuses on these current matters of argue.
Keywords: Electrocardiogram, neonates, arrhythmia, sudden cardiac death, long QT syndrome, genetics
1. INTRODUCTION
A sudden death (SD) is a non-traumatic, unexpected fatal event occurring within 1 hour of the onset of symptoms in an apparently healthy subject. The definition applies when the victim was in good health 24 hours before the event if death is not witnessed. It can be caused by a wide range of conditions [1]. Cardiac alterations are the most prevalent origin of SD, referred to as Sudden Cardiac Death (SCD). This term is used when a congenital, or acquired, potentially fatal cardiac condition was known to be present during life; or autopsy has a cardiac or vascular anomaly as the probable cause of the event; or no obvious extra-cardiac causes have been by post-mortem examination and therefore an arrhythmic event is a likely cause of death. In young people under 35 years old, the main cause of SCD is primary inherited electrical diseases [2]. When post-mortem examination is performed and both autopsy and toxicology investigations are inconclusive, and the heart is structurally normal at gross and histological examination, and non-cardiac aetiologies are excluded, in adults is called Sudden Arrhythmic Death Syndrome (SADS) [3], as well as Sudden Infant Death Syndrome (SIDS) in infants [4]. In addition, clinical assessment and genetic analysis identify an arrhythmogenic disease in 19-30% of families, reinforcing the comprehensive analysis of paediatric family members in SADS [5]. In SIDS cases, molecular autopsy identifies a lower burden of ion channel disease compared with SADS and sporadic genetic disease as a cause of sudden death may be more frequent [6].
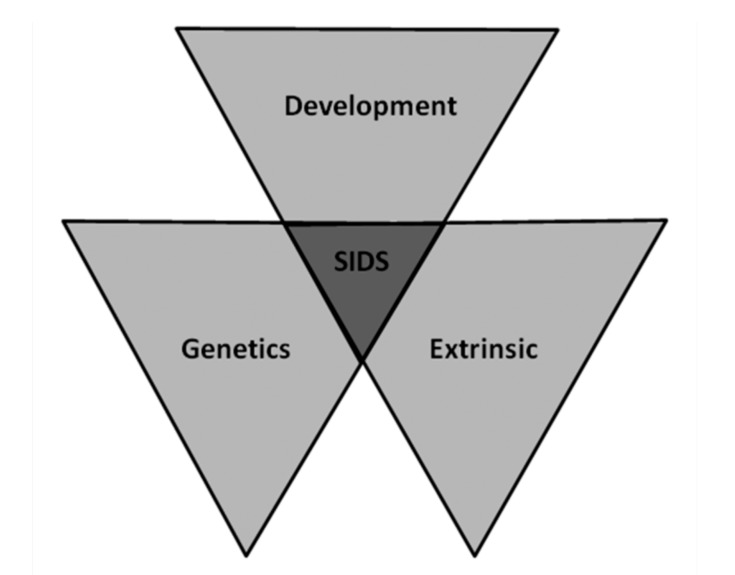
SIDS is the leading cause of death in the first year of life, more frequently in infants between 2 and 4 months old and decrease thereafter [7]. Boys are more likely to die from SIDS (ratio of 3:2), and racial and ethnic disparities exist in its incidence [8]. Different mechanisms have been proposed to be responsible for SIDS although decisive pathogenic substrates or mechanisms triggering an infant’s sudden demise remain unclear Fig. (1) (triple-risk model) [9]. Arrhythmogenic origin has been suggested as one of the reasons after no conclusive comprehensive autopsy [10]. A lethal event can be the first manifestation of the disease and molecular autopsy may unravel the cause of death in the neonates [11]. These malignant arrhythmogenic entities are commonly called cardiac channelopathies, causing a percentage of SIDS cases up to 15% [12, 13].
Fig. (1).
Triple risk factors for SIDS.
2. CHANNELOPATHIES
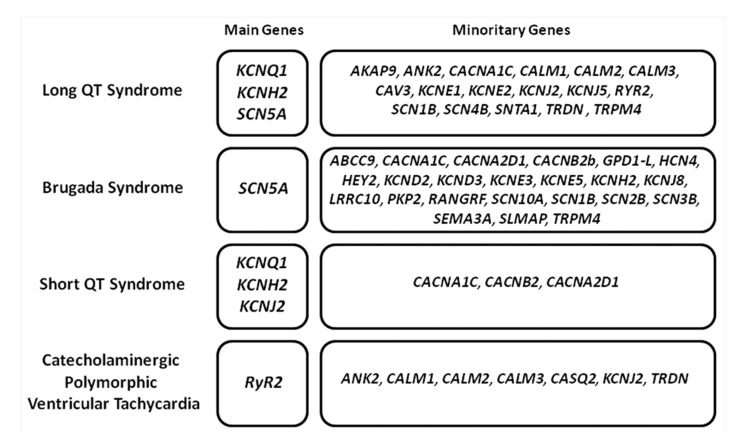
Electrical heart disruption may be due to pathogenic alterations in genes encoding for ion channels, their subunits or associated proteins that play a key role in the function of the channel. These genetic alterations lead to an arrhythmogenic substrate in a structurally normal heart [14]. Cardiac channelopathies may be clinically identified by the presence of characteristic abnormalities in the Electrocardiogram (ECG) [15]. However, the distinctive ECG patterns that characterize these disorders may be masked due to incomplete penetrance and variable expressivity, hallmarks of inherited arrhythmogenic disorders. This group mainly includes the Long QT Syndrome (LQTS), Short QT Syndrome (SQTS), Brugada Syndrome (BrS), and Catecholaminergic Polymorphic Ventricular Tachycardia (CPVT). Currently, hundreds of pathogenic or potentially pathogenic variants have been identified in nearly 40 genes, which encode mainly sodium, potassium and calcium ion channels Fig. (2) [16]. Concerning genetic analysis in SIDS cases, contradictory results have been published so far, mainly due to reduced cohorts and a different number of genes analyzed. Therefore, recent studies suggest that pathogenic variants in a cardiac ion-channel-related gene may be the putative cause of death up to 15% of SIDS cases [17, 18]. It is well established that de novo genetic variants were more frequent than inherited gene variants [8]. In addition, all data conclude that LQTS is the main responsible arrhythmogenic entity of lethal episodes in SIDS population.
Fig. (2).
Main arrhythmogenic entities associated with SIDS. Distribution of genes associated with each arrhythmogenic syndrome, showing both main and minority genes.
2.1. Long QT Syndrome
The LQTS is an inherited arrhythmogenic disease characterized by delayed ventricular repolarization resulting as a QT prolongation on the 12-lead ECG, always in a structurally normal heart Fig. (3). Nowadays, LQTS is diagnosed with either QTc ≥480 ms in repeated 12-lead ECGs or LQTS risk score >3. Diagnosis of LQTS should be also considered in the presence of a QTc ≥460 ms in repeated 12-lead ECGs in patients with an unexplained syncopal episode in the absence of secondary causes for QT prolongation [1]. Finally, LQTS is diagnosed in the presence of a confirmed pathogenic LQTS mutation, irrespective of the QT duration, despite establishing a definitely conclusive pathogenic role of a genetic variant remains a current matter of discussion. The phenotype can range from asymptomatic (10%-35% of patients present with a normal QT interval when measured on a resting 12-lead ECG) to ventricular tachyarrhythmias (torsade de pointes), and even SCD [19]. The prevalence is estimated from 1:2000 to 1:5000 due to the heterogeneity of the disease [20, 21]. To date, more than 600 pathogenic variants (Human Genome Mutation Database, HGMD) have been identified in 20 genes following mainly an autosomal dominant pattern of inheritance. Few recessive cases have also been reported and usually associated with severe phenotypes. All these genes together are responsible for 80%–85% of all LQT cases being KCNQ1 (LQT type 1) (30%-35%), KCNH2 (LQT type 2) (25%-30%), and SCN5A (LQT type 3) (5%-10%), three major genes associated with the disease [22]. Therefore, part of LQT cases continue without a genetic diagnosis and the real prevalence of these LQT-related genes remain to be clarified [14]. Current clinical guidelines recommend only the screening of the main genes focused on a cost-effective way [1]. In addition, genetic testing of relatives is also recommended to identify carriers at risk. In SIDS population, the LQT type 3 is considered the most lethal entity as the proportion of pathogenic variants in sodium channel is ten times higher in comparison to older population suffering of LQTS [23]. Usually, SIDS occurs during the night, at rest, accordingly to pathophysiological mechanisms related to sodium channels [24, 25]. Treatment with beta-blockers has been demonstrated effective, with mortality below 3% among treated patients carrying pathogenic variants in the SCN5A gene (LQTS type 3) [26], despite ICD should be considered in severe cases at risk of SCD. Therefore, early identification and adoption of preventive therapeutic measures help to avoid lethal events in pediatric population.
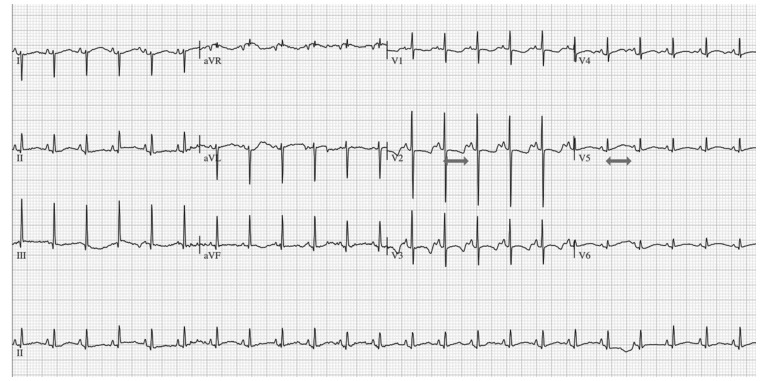
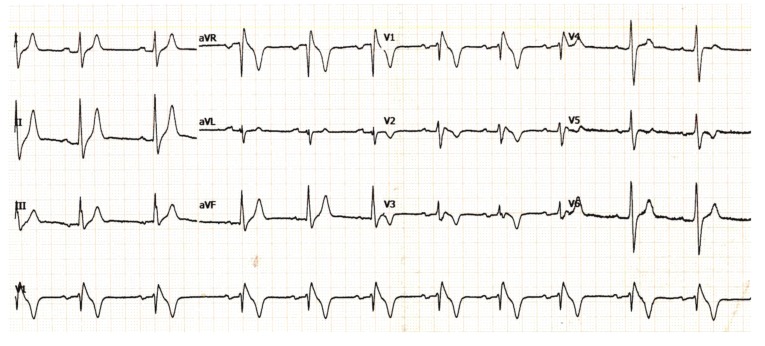
Fig. (3).
Electrocardiogram showing Long QT syndrome in a neonate. Both arrows show a prolonged QT interval in two different leads.
2.2. Short QT Syndrome
This lethal arrhythmogenic disease was reported in 2000 [27]. It is characterized by a reduced duration of cardiac repolarization (QTc<330 ms), which constitutes the substrate for the development of life-threatening arrhythmias. In the ECG, a high sharp T wave and a short interval between the peak and the end of the T wave can also be identified Fig. (4). Clinical manifestations range from lack of symptoms to syncope and even SCD. In the young population less than 40 years old, the probability of a first cardiac arrest is >40% [28]. Given the small size of the populations reported so far, the high lethality may partially reflect a reporting bias related to the under-detection of SQTS in asymptomatic patients, especially young population [29]. Regarding genetics, few pathogenic alterations have been identified in different genes (KCNQ1, KCNJ2, KCNH2, CACNA1C, CACNB2, CACNA2D1, and SLC4A3), following an autosomal dominant pattern of inheritance [30]. A comprehensive genetic analysis identifies the genetic origin in nearly 40% of families. Concerning SIDS population, only one report including three families with SQTS described two male patients who died suddenly (at 8 and 3 months), carrying a pathogenic variant in KCNH2 [31]. A negative autopsy of a neonate who died suddenly could be explained by this arrhythmogenic entity if the family is diagnosed with this lethal disease or if a de novo pathogenic variant is identified in a SQTS-associated gene.
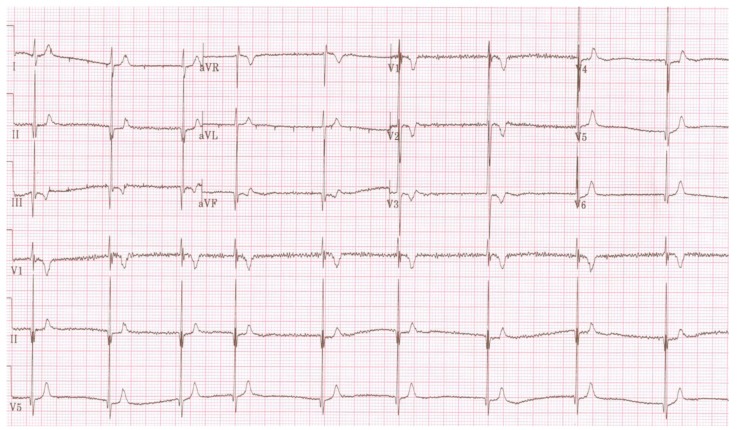
Fig. (4).
Electrocardiogram showing Short QT syndrome in a child.
2.3. Brugada Syndrome
This rare arrhythmogenic entity is characterized by an ECG pattern consisting of coved-type ST-segment elevation in atypical right-bundle branch block in leads V1 to V3 (often referred to as type-1 Brugada ECG pattern) Fig. (5). It supposes an increased risk for SCD resulting from episodes of polymorphic ventricular tachyarrhythmias [32]. Brugada syndrome is estimated to be responsible for up to 20% of SCD in patients with structurally normal hearts. It involves mainly young male adults (about 40 years old), and syncope usually occurs at rest [33] Patients with BrS usually remain asymptomatic and modulating factors such as fever, exercise or drugs (www.brugadadrugs.org), may play a major role in the dynamic nature of the ECG. The ECG pattern can be baseline or intermittent, and it can be unmasked during a drug test (class IC sodium channel-blockers) [34]. Currently, 25 genes have been associated to the disease (ABCC9, CACNA1C, CACNA2D1, CACNB2b, FGF12, GPD1-L, HCN4, HEY2, KCND2, KCND3, KCNE3, KCNE5, KCNH2, KCNJ8, LRRC10, PKP2, RANGRF, SCN10A, SCN1B, SCN2B, SCN3B, SCN5A, SEMA3A, SLMAP, and TRPM4) but a comprehensive genetic analysis only identify the genetic alteration in 35% of all diagnosed cases [35]. Approximately 30% of patients with BrS carry a pathogenic alteration in the SCN5A gene [36]. The prevalence of BrS in children remain unclear due to few cases have been published so far. BrS exist in children but becomes clinically unmasked with increasing age. Concerning the association of SIDS and BrS, a study reported several family members suffering of BrS (including two SIDS of male infants) carrying a pathogenic variant in the SCN5A gene [37]. Despite the risk of ventricular arrhythmias is generally low in children, these circumstances might be especially pertinent to SIDS. A negative autopsy of a neonate who died during night could be explained by this arrhythmogenic entity.
Fig. (5).
Electrocardiogram showing Brugada syndrome in an infant.
2.4. Catecholaminergic Polymorphic Ventricular Tachycardia
It is an inheritable disorder characterized by a normal ECG at rest (occasionally with bradycardia and U waves), and triggered exclusively by adrenergic stimulus (mainly exertion, extreme stress or emotion) leading to bidirectional and polymorphic VT in a normal heart [38]. The estimated prevalence is 1 in 10,000 [1]. The clinical manifestations of CPVT usually occur in the first decade of life and are prompted by physical activity or emotional stress [39]. CPVT shows a high mortality rate occurring mainly in children and adolescents and it is increasingly recognized as a cause of SADS in young individuals, predominantly males [40]. To date, more than 200 pathogenic alterations have been identified in eight genes (ANK2, CALM1, CALM2, CALM3, CASQ2, KCNJ2, RyR2, and TRDN) being RyR2 the main gene responsible of nearly 50% of all cases. A comprehensive genetic analysis explains around 60% of CPVT cases [22]. CPVT diagnosis can be difficult especially in children because patients with CPVT have a normal ECG, therefore an exercise stress test that elicits atrial and ventricular arrhythmias (bidirectional or polymorphic VT) is recommended [1]. Concerning SIDS, few cases have been reported showing pathogenic variants (mainly de novo) [41, 42]. Because of adrenergic triggers are main inductors of CPVT, this entity is a potential cause of death in neonates.
3. MOLECULAR AUTOPSY
Post-mortem genetic analysis may help to identify the cause of deaths due to an arrhythmogenic cardiac disease but also allow the identification of relatives who carry the same inherited genetic disorder and, consequently, are at risk of SCD [3]. All relatives of a SD victim with a no conclusive cause of death may undergo a clinical assessment by a multidisciplinary team including cardiologists, forensic pathologists and geneticists because unraveling the cause of death is extremely complicated, mainly when the victim is an infant [43, 44]. Therefore, as genetic analysis is nowadays widely accepted as a diagnostic tool, current guidelines recommend performing the molecular autopsy as part of the comprehensive medico-legal investigation in SD cases without conclusive cause of death, especially in pediatric population [45, 46]. However, molecular autopsy is not performed in most part of forensic centers mainly due to lack of economic resources or legal restrictions involved with the sampling and storage of DNA [47]. Collection of blood and/or suitable tissue for molecular autopsy is recommended in SIDS cases, because post-mortem genetic testing focused on arrhythmia syndromes can be useful [48].
An additional crucial point is which genes should be analyzed. Nowadays, more than 40 genes have been associated with cardiac channelopathies. However, current guidelines recommend performing a genetic analysis of only the main channelopathies-associated genes (KCNQ1, KCNH2, SCN5A for LQTS, SCN5A for BrS, RyR2, CASQ2 for CPVT, and KCNQ1, KCNJ2, KCNH2 for SQTS), focused on cost-effective approach [1]. Focused on SIDS cases, most part of pathogenic variants identified so far are located in the SCN5A gene, associated with LQTS and BrS, despite punctual variants have been reported in genes encoding potassium channels [8]. Curiously, in recent years additional variants have been identified in genes encoding structural genes. These reports analyzed SIDS cases (showing completely normal hearts at post-mortem examination) and identified rare variants in genes associated with hypertrophic cardiomyopathy, dilated cardiomyopathy, arrhythmogenic cardiomyopathy, and valvular cardiac disease [42, 49]. All variants were predicted as possibly pathogenic or probably pathogenic by in silico analysis [50]; no other criteria were used in order to classify the potential pathogenicity of these variants, mainly family segregation [51]. Despite these limitations, data suggest that subtle forms of cardiomyopathy might predispose to SIDS, even in the absence of an overt phenotype at autopsy [52].
Recent developments in massively parallel genetic sequencing (Next Generation Sequencing -NGS-) allow a cost-effective analysis of several genes in a reduced time [53]. NGS can detect simple genetic variants, insertions/deletions (indels) of single nucleotide polymorphisms (SNPs) and variations in the number of copies (Copy Number Variations, CNVs). These technologies have been used with success in cardiac genetics [45, 54], such as studies for the detection of pathogenic variants associated with LQTS [55, 56] as well as in SIDS cases [25, 51, 57]. The utilization of NGS technology allows a comprehensive genetic analysis, identifying several rare variants in practically all samples analyzed. The main current challenge in genetic area is the interpretation of variants identified as well as clinical translation. Hence, only up to 10% of rare variants previously classified as pathogenic play a conclusive deleterious role in SIDS after a suitable carefully classification [13, 48]. These results entail that most parts of identified rare variants remain of unknown/ambiguous significance. The identification of variants of uncertain significance leaves clinicians with inconclusive data that does not help either the diagnosis or the adoption of potential therapeutic measures for treatment and prevention of malignant arrhythmias. Therefore, translation into clinical practice should be performed carefully. Guidelines focusing on this point have been recently published by American College of Medical Genetics/Association for Molecular Pathology (ACMG/AMP) [58, 59] suggesting functional analysis (in vitro, in silico, in vivo), family history information, segregation of genetic variant and genotype-phenotype correlations as crucial items to elucidate a definite role of each variant identified [60]. It is very important to remark that genetic testing is just one more tool that complements the diagnosis; it should not be used as the only factor to confirm or exclude a clinical diagnosis.
4. ELECTROCARDIOGRAPHIC ASSESSMENT
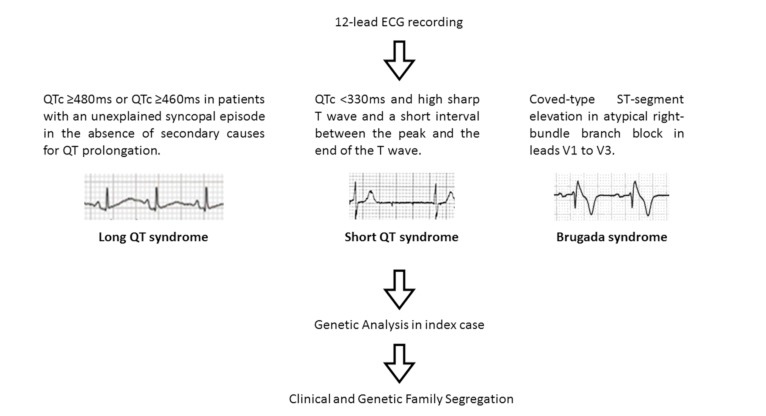
In 1951 Ziegler published electrocardiographic alterations in infants [61]. Most part of these alterations concerns to the physiologic adaption of heart and it is well-accepted that during the first days after birth, the ECG shows changes in relation to these adaptations. Normal values, as well as ranges of ECG amplitudes and interval durations in each state, have been published [62]. In addition, structural and electric heart defects that are not associated with normal evolution are also identified [63]. Hence, several reports have advocated for the inclusion of an ECG in neonatal screening, with the argument that the abnormal findings may carry a lethal outcome, and that early identification may potentially prevent SCD [25]. However, due to the lack of conclusive studies, it remains as a controversial area and, each pediatrician and cardiologist implement [64] or not [65] the neonatal ECG screening Fig. (6).
Fig. (6).
Algorithm for identification and genetic analysis.
Nowadays, it is suggested that nearly 10% of cases of SIDS may actually be caused by LQTS [66]. However, few studies have been published assessing the value of neonatal QTc screening and reporting a prolonged QTc between 2.5% and 50% of neonates; this wide range depends on QT correction formula to be used in ECG, the value of QTc prolongation and method of ECG acquisition [67, 68]. Hence, exist a controversy regarding the choice of the QT correction formula to be used in ECG in neonates for the screening of QT interval. Recently, an exhaustive revision was published concerning this point, concluding that Bazett’s correction provides an effective HR independent QT correction and accurately identifies the neonates affected by LQTS with a cut-off value of 460ms. It can be used with confidence to make recommendations for diagnostic or therapeutic decisions in neonates. However, Fridericia’s or Framingham correction methods could also be employed, albeit using different cut-off values [69]. Regarding QT interval measurements, the reading error, which occurs among experts, is not negligible. Practically all published studies state that at least two independent cardiologists should measure the value in order to avoid QTc discrepancies. Hence, in neonates, if the QTc interval is >450ms, the expected rate of false positive is 4.5/1,000 meanwhile if the QTc interval is >460ms, the expected rate decrease to 1/1.000 [68]. Current guidelines establish a QTc of >470ms as potentially positive but specified that repetitive measures should be performed at different times in order to avoid false positive values [1]. It is stated that the appropriate period for ECG recording is at 3-4 weeks of life, when the QT pattern has stabilized and in a period before the most common window for neonatal death [68]. Concerning the method of acquisition of ECG, current recording systems are equipped with interpretation software to facilitate the work of cardiologists, following recommendations by the American Heart Association, the American College of Cardiology, and the Heart Rhythm Society [70]. However, the gold standard for an accurate ECG interpretation is the expert in the field, the pediatric cardiologist. Recently, Murphy et al performed a randomized crossover study of methods of acquiring ECG in stable infants [71, 72]. The authors identified that connecting the ECG leads to the machine before applying the leads to the neonate resulted in quicker heart rate acquisition and proposed that this method should be used when acquiring an ECG in neonates. In 2015, a review focused on available evidence on the cost-effectiveness of genetic and ECG testing strategies for the diagnosis of LQTS in neonates was published [73]. The authors concluded that ECG screening in neonates is cost-effective in high-risk groups but no strong data are available regarding global screening.
In recent years, there has been an impulse in the medical community to introduce the ECG into routine neonatal screening [64] despite an equal steady impulse has been performed in the opposite way. This ECG testing is crucial in order to identify QT alterations but also other electric alterations that could be related to any arrhythmogenic syndrome and, in consequence, at risk of SCD [74]. Therefore, up to 20% of neonatal ECGs obtained in the acute setting may have clinically significant abnormal findings and an early identification, as well as proper interpretation, allow the adoption of personalized measures avoiding potential lethal episodes [75]. Despite published data, not everybody agrees though, and it has become an area of active discussion and health policy deliberation, due to concerns about the economic, clinical, social and even psychological implications of having a borderline QTc or any ECG alteration without a conclusive diagnosis [65]. Regarding genetic testing, it remains as not cost-effective due to the high economic cost and lack of genetic services in all hospitals, especially in underprivileged countries. Thus, genetic testing is at present only recommended in clear familial diseases, as well as in highly suspected symptomatic cases.
CONCLUSION
Performing an ECG in the neonatal population is an action that may identify malignant arrhythmias. The first manifestation of the disease could be the death, therefore, early identification is crucial to adopt preventive antiarrhythmic therapies. New genetic technologies allow a comprehensive analysis of the main genes associated with arrhythmogenic diseases. In SIDS cases, molecular autopsy identifies genetic alterations associated with cardiac channelopathies up to 15% of cases, mainly due to LQTS. As these are inherited diseases, relatives can also benefit from a clinical and genetic analysis. The identification of genetic carriers, despite remaining asymptomatic, allow the adoption of personalized preventive measures in order to reduce the risk of SCD. Despite all these facts, the cost-effectiveness of ECG screening and genetic analysis remains controversial, especially due low incidence of malignant arrhythmias in the neonatal population.
ACKNOWLEDGEMENTS
This work was supported by Fundació La Marato TV3 (358/U/2015), Fundació Daniel Bravo Andreu, and Obra Social ‘La Caixa’. The CIBERCV is an initiative of the Instituto de Salud Carlos III (ISCIII), Spanish Ministry of Economy and Competitiveness.
Consent for Publication
Not applicable.
CONFLICT OF INTEREST
All authors declare no conflict of interest to disclose, financial or otherwise.
REFERENCES
- 1.Priori S.G., Blomstrom-Lundqvist C. 2015 European Society of Cardiology Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death summarized by co-chairs. Eur. Heart J. 2015;36:2757–2759. doi: 10.1093/eurheartj/ehv445. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez-Calvo M.S., Brion M., Allegue C., Concheiro L., Carracedo A. Molecular genetics of sudden cardiac death. Forensic Sci. Int. 2008;182:1–12. doi: 10.1016/j.forsciint.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Behr E.R., Dalageorgou C., Christiansen M., et al. Sudden arrhythmic death syndrome: familial evaluation identifies inheritable heart disease in the majority of families. Eur. Heart J. 2008;29:1670–1680. doi: 10.1093/eurheartj/ehn219. [DOI] [PubMed] [Google Scholar]
- 4.Raju H., Behr E.R. Unexplained sudden death, focussing on genetics and family phenotyping. Curr. Opin. Cardiol. 2013;28:19–25. doi: 10.1097/HCO.0b013e32835b0a9e. [DOI] [PubMed] [Google Scholar]
- 5.Giudici V., Spanaki A., Hendry J., et al. Sudden arrhythmic death syndrome: Diagnostic yield of comprehensive clinical evaluation of pediatric first-degree relatives. Pacing Clin. Electrophysiol. 2014;37(12):1681–1685. doi: 10.1111/pace.12479. [DOI] [PubMed] [Google Scholar]
- 6.Krous H.F., Beckwith J.B., Byard R.W., et al. Sudden infant death syndrome and unclassified sudden infant deaths: A definitional and diagnostic approach. Pediatrics. 2004;114:234–238. doi: 10.1542/peds.114.1.234. [DOI] [PubMed] [Google Scholar]
- 7.Moon R.Y., Fu L. Sudden infant death syndrome: An update. Pediatr. Rev. 2012;33:314–320. doi: 10.1542/pir.33-7-314. [DOI] [PubMed] [Google Scholar]
- 8.Baruteau A.E., Tester D.J., Kapplinger J.D., Ackerman M.J., Behr E.R. Sudden infant death syndrome and inherited cardiac conditions. Nat. Rev. Cardiol. 2017;14:715–726. doi: 10.1038/nrcardio.2017.129. [DOI] [PubMed] [Google Scholar]
- 9.Filiano J.J., Kinney H.C. A perspective on neuropathologic findings in victims of the sudden infant death syndrome: The triple-risk model. Biol. Neonate. 1994;65:194–197. doi: 10.1159/000244052. [DOI] [PubMed] [Google Scholar]
- 10.Goldwater P.N. A perspective on SIDS pathogenesis. the hypotheses: Plausibility and evidence. BMC Med. 2011;9:64. doi: 10.1186/1741-7015-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campuzano O., Allegue C., Partemi S., Iglesias A., Oliva A., Brugada R. Negative autopsy and sudden cardiac death. Int. J. Legal Med. 2014;128:599–606. doi: 10.1007/s00414-014-0966-4. [DOI] [PubMed] [Google Scholar]
- 12.Van Niekerk C., Van Deventer B.S., du Toit-Prinsloo L. Long QT syndrome and sudden unexpected infant death. J. Clin. Pathol. 2017;70:808–813. doi: 10.1136/jclinpath-2016-204199. [DOI] [PubMed] [Google Scholar]
- 13.Tester D.J., Wong L.C.H., Chanana P., et al. Cardiac genetic predisposition in sudden infant death syndrome. J. Am. Coll. Cardiol. 2018;71:1217–1227. doi: 10.1016/j.jacc.2018.01.030. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Falgueras A., Sarquella-Brugada G., Brugada J., Brugada R., Campuzano O. Cardiac channelopathies and sudden death: Recent clinical and genetic advances. Biology (Basel) 2017;6(1):E7. doi: 10.3390/biology6010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schimpf R., Veltmann C., Wolpert C., Borggrefe M. Channelopathies: Brugada syndrome, long QT syndrome, short QT syndrome, and CPVT. Herz. 2009;34:281–288. doi: 10.1007/s00059-009-3238-1. [DOI] [PubMed] [Google Scholar]
- 16.Campuzano O., Sarquella-Brugada G., Brugada R., Brugada J. Genetics of channelopathies associated with sudden cardiac death. Glob. Cardiol. Sci. Pract. 2015;2015:39. doi: 10.5339/gcsp.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilders R. Cardiac ion channelopathies and the sudden infant death syndrome. ISRN Cardiol. 2012;2012:846171. doi: 10.5402/2012/846171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis A.M., Glengarry J., Skinner J.R. Sudden infant death: QT or not QT? That is no longer the question. Circ Arrhythm Electrophysiol. 2016;9:e003859. doi: 10.1161/CIRCEP.115.003859. [DOI] [PubMed] [Google Scholar]
- 19.Crotti L., Celano G., Dagradi F., Schwartz P.J. Congenital long QT syndrome. Orphanet J. Rare Dis. 2008;3:18. doi: 10.1186/1750-1172-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz P.J., Ackerman M.J. The long QT syndrome: A transatlantic clinical approach to diagnosis and therapy. Eur. Heart J. 2013;34:3109–3116. doi: 10.1093/eurheartj/eht089. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz P.J., Crotti L., Insolia R. Long-QT syndrome: From genetics to management. Circ Arrhythm Electrophysiol. 2012;5:868–877. doi: 10.1161/CIRCEP.111.962019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campuzano O., Sarquella-Brugada G., Cesar S., et al. Genetics of inherited arrhythmias in pediatrics. Curr. Opin. Pediatr. 2015;27:665–674. doi: 10.1097/MOP.0000000000000272. [DOI] [PubMed] [Google Scholar]
- 23.Arnestad M., Crotti L., Rognum T.O., et al. Prevalence of long-QT syndrome gene variants in sudden infant death syndrome. Circulation. 2007;115:361–367. doi: 10.1161/CIRCULATIONAHA.106.658021. [DOI] [PubMed] [Google Scholar]
- 24.Stattin E.L., Westin I.M., Cederquist K., et al. Genetic screening in sudden cardiac death in the young can save future lives. Int. J. Legal Med. 2016;130:59–66. doi: 10.1007/s00414-015-1237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glengarry J.M., Crawford J., Morrow P.L., Stables S.R., Love D.R., Skinner J.R. Long QT molecular autopsy in sudden infant death syndrome. Arch. Dis. Child. 2014;99:635–640. doi: 10.1136/archdischild-2013-305331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilde A.A., Moss A.J., Kaufman E.S., et al. Clinical aspects of type 3 long-QT syndrome: An international multicenter study. Circulation. 2016;134:872–882. doi: 10.1161/CIRCULATIONAHA.116.021823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gussak I., Brugada P., Brugada J., et al. Idiopathic short QT interval: A new clinical syndrome? Cardiology. 2000;94:99–102. doi: 10.1159/000047299. [DOI] [PubMed] [Google Scholar]
- 28.Mazzanti A., Kanthan A., Monteforte N., et al. Novel insight into the natural history of short QT syndrome. J. Am. Coll. Cardiol. 2014;63:1300–1308. doi: 10.1016/j.jacc.2013.09.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pereira R., Campuzano O., Sarquella-Brugada G., et al. Short QT syndrome in pediatrics. Clin. Res. Cardiol. 2017;106(6):393–400. doi: 10.1007/s00392-017-1094-1. [DOI] [PubMed] [Google Scholar]
- 30.Behere S.P., Weindling S.N. Inherited arrhythmias: The cardiac channelopathies. Ann. Pediatr. Cardiol. 2015;8:210–220. doi: 10.4103/0974-2069.164695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brugada R., Hong K., Dumaine R., et al. Sudden death associated with short-QT syndrome linked to mutations in HERG. Circulation. 2004;109:30–35. doi: 10.1161/01.CIR.0000109482.92774.3A. [DOI] [PubMed] [Google Scholar]
- 32.Brugada P., Brugada J. Right bundle branch block, persistent ST segment elevation and sudden cardiac death: A distinct clinical and electrocardiographic syndrome. A multicenter report. J. Am. Coll. Cardiol. 1992;20:1391–1396. doi: 10.1016/0735-1097(92)90253-j. [DOI] [PubMed] [Google Scholar]
- 33.Gehi A.K., Duong T.D., Metz L.D., Gomes J.A., Mehta D. Risk stratification of individuals with the Brugada electrocardiogram: A meta-analysis. J. Cardiovasc. Electrophysiol. 2006;17:577–583. doi: 10.1111/j.1540-8167.2006.00455.x. [DOI] [PubMed] [Google Scholar]
- 34.Benito B., Brugada R., Brugada J., Brugada P. Brugada syndrome. Prog. Cardiovasc. Dis. 2008;51:1–22. doi: 10.1016/j.pcad.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Brugada R., Campuzano O., Sarquella-Brugada G., Brugada J., Brugada P. Brugada syndrome. Methodist DeBakey Cardiovasc. J. 2014;10:25–28. doi: 10.14797/mdcj-10-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarquella-Brugada G., Campuzano O., Arbelo E., Brugada J., Brugada R. Brugada syndrome: Clinical and genetic findings. Genet. Med. 2016;18:3–12. doi: 10.1038/gim.2015.35. [DOI] [PubMed] [Google Scholar]
- 37.Priori S.G., Napolitano C., Giordano U., Collisani G., Memmi M. Brugada syndrome and sudden cardiac death in children. Lancet. 2000;355:808–809. doi: 10.1016/S0140-6736(99)05277-0. [DOI] [PubMed] [Google Scholar]
- 38.Refaat M.M., Hassanieh S., Scheinman M. Catecholaminergic polymorphic ventricular tachycardia. Card. Electrophysiol. Clin. 2016;8:233–237. doi: 10.1016/j.ccep.2015.10.035. [DOI] [PubMed] [Google Scholar]
- 39.Priori S.G., Napolitano C., Memmi M., et al. Clinical and molecular characterization of patients with catecholaminergic polymorphic ventricular tachycardia. Circulation. 2002;106:69–74. doi: 10.1161/01.cir.0000020013.73106.d8. [DOI] [PubMed] [Google Scholar]
- 40.Ylanen K., Poutanen T., Hiippala A., Swan H., Korppi M. Catecholaminergic polymorphic ventricular tachycardia. Eur. J. Pediatr. 2010;169:535–542. doi: 10.1007/s00431-010-1154-2. [DOI] [PubMed] [Google Scholar]
- 41.Tester D.J., Dura M., Carturan E., et al. A mechanism for Sudden Infant Death Syndrome (SIDS): Stress-induced leak via ryanodine receptors. Heart Rhythm. 2007;4:733–739. doi: 10.1016/j.hrthm.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neubauer J., Lecca M.R., Russo G., et al. Post-mortem whole-exome analysis in a large sudden infant death syndrome cohort with a focus on cardiovascular and metabolic genetic diseases. Eur. J. Hum. Genet. 2017;25:404–409. doi: 10.1038/ejhg.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klaver E.C., Versluijs G.M., Wilders R. Cardiac ion channel mutations in the sudden infant death syndrome. Int. J. Cardiol. 2011;152:162–170. doi: 10.1016/j.ijcard.2010.12.051. [DOI] [PubMed] [Google Scholar]
- 44.Dettmeyer R.B., Kandolf R. Cardiomyopathies--misdiagnosed as Sudden Infant Death Syndrome (SIDS). Forensic Sci. Int. 2010;194:e21–e24. doi: 10.1016/j.forsciint.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 45.Semsarian C., Bagnall R.D. Sudden cardiac death in children and young adults. N. Engl. J. Med. 2016;375:1301–1302. doi: 10.1056/NEJMc1609620. [DOI] [PubMed] [Google Scholar]
- 46.Semsarian C., Ingles J. Molecular autopsy in victims of inherited arrhythmias. J. Arrhythm. 2016;32:359–365. doi: 10.1016/j.joa.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michaud K., Mangin P., Elger B.S. Genetic analysis of sudden cardiac death victims: a survey of current forensic autopsy practices. Int. J. Legal Med. 2011;125:359–366. doi: 10.1007/s00414-010-0474-0. [DOI] [PubMed] [Google Scholar]
- 48.Ackerman M.J., Priori S.G., Willems S., et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: This document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Europace. 2011;13:1077–1109. doi: 10.1093/europace/eur245. [DOI] [PubMed] [Google Scholar]
- 49.Hertz C.L., Christiansen S.L., Larsen M.K., et al. Genetic investigations of sudden unexpected deaths in infancy using next-generation sequencing of 100 genes associated with cardiac diseases. Eur. J. Hum. Genet. 2016;24:817–822. doi: 10.1038/ejhg.2015.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brion M., Allegue C., Santori M., et al. Sarcomeric gene mutations in sudden infant death syndrome (SIDS). Forensic Sci. Int. 2012;219:278–281. doi: 10.1016/j.forsciint.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 51.Campuzano O., Allegue C., Sarquella-Brugada G., et al. The role of clinical, genetic and segregation evaluation in sudden infant death. Forensic Sci. Int. 2014;242:9–15. doi: 10.1016/j.forsciint.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 52.Sarquella-Brugada G., Campuzano O., Cesar S., et al. Sudden infant death syndrome caused by cardiac arrhythmias: Only a matter of genes encoding ion channels? Int. J. Legal Med. 2016;130:415–420. doi: 10.1007/s00414-016-1330-7. [DOI] [PubMed] [Google Scholar]
- 53.Faita F., Vecoli C., Foffa I., Andreassi M.G. Next generation sequencing in cardiovascular diseases. World J. Cardiol. 2012;4:288–295. doi: 10.4330/wjc.v4.i10.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanchez O., Campuzano O., Fernandez-Falgueras A., et al. Natural and undetermined sudden death: Value of post-mortem genetic investigation. PLoS One. 2016;11:e0167358. doi: 10.1371/journal.pone.0167358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Campuzano O., Sarquella-Brugada G., Mademont-Soler I., et al. Identification of genetic alterations, as causative genetic defects in long qt syndrome, using next generation sequencing technology. PLoS One. 2014;9:e114894. doi: 10.1371/journal.pone.0114894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams V.S., Cresswell C.J., Ruspi G., et al. Multiplex ligation-dependent probe amplification copy number variant analysis in patients with acquired long QT syndrome. Europace. 2015;17:635–641. doi: 10.1093/europace/euu288. [DOI] [PubMed] [Google Scholar]
- 57.Priest J.R., Ceresnak S.R., Dewey F.E., et al. Molecular diagnosis of long QT syndrome at 10 days of life by rapid whole genome sequencing. Heart Rhythm. 2014;11:1707–1713. doi: 10.1016/j.hrthm.2014.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amendola L.M., Jarvik G.P., Leo M.C., et al. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the clinical sequencing exploratory research consortium. Am. J. Hum. Genet. 2016;98:1067–1076. doi: 10.1016/j.ajhg.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Campuzano O., Allegue C., Fernandez A., Iglesias A., Brugada R. Determining the pathogenicity of genetic variants associated with cardiac channelopathies. Sci. Rep. 2015;5:7953. doi: 10.1038/srep07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ziegler R.F. Characteristics of the unipolar precordial electrocardiogram in normal infants. Circulation. 1951;3:438–443. doi: 10.1161/01.cir.3.3.438. [DOI] [PubMed] [Google Scholar]
- 62.Rijnbeek P.R., Witsenburg M., Schrama E., Hess J., Kors J.A. New normal limits for the paediatric electrocardiogram. Eur. Heart J. 2001;22:702–711. doi: 10.1053/euhj.2000.2399. [DOI] [PubMed] [Google Scholar]
- 63.Brockmeier K., Nazal R., Sreeram N. The electrocardiogram of the neonate and infant. J. Electrocardiol. 2016;49:814–816. doi: 10.1016/j.jelectrocard.2016.07.027. [DOI] [PubMed] [Google Scholar]
- 64.Saul J.P., Schwartz P.J., Ackerman M.J., Triedman J.K. Rationale and objectives for ECG screening in infancy. Heart Rhythm. 2014;11:2316–2321. doi: 10.1016/j.hrthm.2014.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skinner J.R., Van Hare G.F. Routine ECG screening in infancy and early childhood should not be performed. Heart Rhythm. 2014;11:2322–2327. doi: 10.1016/j.hrthm.2014.09.046. [DOI] [PubMed] [Google Scholar]
- 66.Schwartz P.J., Priori S.G., Bloise R., et al. Molecular diagnosis in a child with sudden infant death syndrome. Lancet. 2001;358:1342–1343. doi: 10.1016/S0140-6736(01)06450-9. [DOI] [PubMed] [Google Scholar]
- 67.Schwartz P.J., Stramba-Badiale M., Segantini A., et al. Prolongation of the QT interval and the sudden infant death syndrome. N. Engl. J. Med. 1998;338:1709–1714. doi: 10.1056/NEJM199806113382401. [DOI] [PubMed] [Google Scholar]
- 68.Schwartz P.J., Stramba-Badiale M., Crotti L., et al. Prevalence of the congenital long-QT syndrome. Circulation. 2009;120:1761–1767. doi: 10.1161/CIRCULATIONAHA.109.863209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stramba-Badiale M., Karnad D.R., Goulene K.M., et al. For neonatal ECG screening there is no reason to relinquish old Bazett’s correction. Eur. Heart J. 2018;39(31):2888–2895. doi: 10.1093/eurheartj/ehy284. [DOI] [PubMed] [Google Scholar]
- 70.Kligfield P., Gettes L.S., Bailey J.J., et al. Recommendations for the standardization and interpretation of the electrocardiogram: Part I: The electrocardiogram and its technology a scientific statement from the American Heart Association Electrocardiography and Arrhythmias Committee, Council on Clinical Cardiology; the American College of Cardiology Foundation; and the Heart Rhythm Society endorsed by the International Society for Computerized Electrocardiology. J. Am. Coll. Cardiol. 2007;49:1109–1127. doi: 10.1016/j.jacc.2007.01.024. [DOI] [PubMed] [Google Scholar]
- 71.Murphy M.C., Angelis L., Fitzgerald E., McCarthy L.K. ODonnell CPF. Randomised crossover study comparing speed of heart rate display in newborns using ECG plus pulse oximeter versus pulse oximeter alone. Arch. Dis. Child. Fetal Neonatal Ed. 2017;102:F464–F5. doi: 10.1136/archdischild-2017-313149. [DOI] [PubMed] [Google Scholar]
- 72.Murphy M.C., De Angelis L., McCollum D., McCarthy L.K., O’Donnell C.P. A randomised cross-over study of methods of acquiring ECG heart rate in newborns. Arch. Dis. Child. Fetal Neonatal Ed. 2017;102:F369–F70. doi: 10.1136/archdischild-2017-312866. [DOI] [PubMed] [Google Scholar]
- 73.Gonzalez F.M., Veneziano M.A., Puggina A., Boccia S. A systematic review on the cost-effectiveness of genetic and electrocardiogram testing for long QT syndrome in infants and young adults. Value Health. 2015;18:700–708. doi: 10.1016/j.jval.2015.03.1788. [DOI] [PubMed] [Google Scholar]
- 74.McCorquodale A., Poulton R., Hendry J., et al. High prevalence of early repolarization in the paediatric relatives of sudden arrhythmic death syndrome victims and in normal controls. Europace. 2017;19:1385–1391. doi: 10.1093/europace/euw248. [DOI] [PubMed] [Google Scholar]
- 75.Chan T.C., Sharieff G.Q., Brady W.J. Electrocardiographic manifestations: Pediatric ECG. J. Emerg. Med. 2008;35:421–430. doi: 10.1016/j.jemermed.2007.09.039. [DOI] [PubMed] [Google Scholar]