Abstract
Spatially resolved single-cell RNA sequencing (scRNAseq) is a powerful approach to infer connections between a cell’s identity and its position within a tissue. We recently combined scRNAseq with spatially-mapped landmark genes to infer the expression zonation of hepatocytes. However, determining zonation of small cells with low mRNA content or without highly expressed landmark genes, remains challenging. Here, we present paired-cell sequencing, whereby mRNA from pairs of attached cells are sequenced and gene expression from one cell type is used to infer the pairs’ tissue coordinates. We apply the method to pairs of hepatocytes and liver endothelial cells (LECs). Using the spatial information from hepatocytes, we reconstruct LEC zonation and extract a landmark gene panel that we use to spatially map LEC scRNAseq data. Our approach reveals expression of both Wnt ligands and the Dkk3 Wnt antagonist in distinct pericentral LEC sub-populations. This approach can be used to reconstruct spatial expression maps of non-parenchymal cells in other tissues.
An outstanding challenge in biology is to characterize the cell types that make up mammalian tissues1–3. Since the coordinates of a cell within a tissue are a critical determinant of its molecular identity, approaches for spatial transcriptomics are necessary to resolve the connection between location and function4–13. In the mammalian liver, hepatocytes and diverse non-parenchymal cells (NPCs) operate in repeating hexagonal-shaped anatomical units termed lobules (Fig. 1a). Each lobule is comprised of a central vein, radial sinusoidal networks, and portal nodes that consist of arteries, veins and bile ducts. The lobule blood vessels, which are in direct contact with hepatocytes, are lined with liver endothelial cells (LECs) and contain diverse resident and circulating immune cells. Blood emanates from the portal nodes and flows towards draining central veins, creating gradients of oxygen, nutrients and hormones14,15. In addition, morphogens such as Wnt and Rspo3 secreted by central vein LECs create an inverse polarizing field16–18. The graded lobule microenvironment gives rise to spatial division of labour among hepatocytes residing at different radial coordinates9,19–21. Whether the liver NPCs exhibit similar spatial division of labour is unknown.
Figure 1. Strategy for paired-cell reconstruction of liver LEC zonation.
a) A diagram of the liver lobule. Blow up represents a typical porto-central sinusoidal unit. A typical unit consists of 10-15 hepatocytes and was coarse-grained into 8 or 4 concentric layers when analyzing paired cells and single cells respectively. b) Paired-cell RNA sequencing utilizes the hepatocyte zonation to determine tissue localization and single cell RNAseq of LECs to extract LEC-specific genes (1). LEC zonation is obtained by averaging expression of LEC genes in the spatially-localized pairs. This dataset is also used to extract LEC landmark genes for localizing single LECs (2).
LECs make up about 50% of the tissue’s NPCs22 and carry critical functions - they form the building blocks of the blood vessels, clear endotoxins, bacteria and other compounds, regulate host immune responses to pathogens, present antigens and secrete morphogens that shape hepatocyte gene expression22–24. Several studies have identified morphological differences in LECs located at different lobule radial coordinates, including the amounts and sizes of LECs fenestrae and of the cells themselves14. However, we lack a comprehensive picture of LEC spatial diversity in terms of their gene expression signatures.
We have recently applied spatially resolved single cell transcriptomics to reconstruct the zonation patterns of all hepatocyte genes9. We used Massively parallel single cell RNA-Seq (MARS-Seq25) to sequence thousands of hepatocytes, and constructed a concise panel of zonated hepatocyte landmark genes, quantified with single molecule fluorescence in-situ hybridization (smFISH) to retrospectively map the hepatocytes back to their original radial lobule layers. A weighted-average of the hepatocytes’ expression in each layer yielded the expression profiles of all hepatocyte genes. Uncovering similar zonation patterns of LECs is much more challenging. Since LECs are small cells, and scRNAseq techniques sparsely sample only a small fraction of the total cellular mRNA, transcripts of most genes will not be present in individual LECs. Consequently, a suitable landmark gene panel would need to be large. Since we lack prior knowledge regarding the zonation patterns of more than a handful of LEC genes, new approaches need to be developed to reveal such a suitable landmark gene panel.
To overcome these limitations we developed paired-cell sequencing (pcRNAseq), that profiles gene expression of hepatocytes and adjacent LECs that were attached to them in the tissue and determines their localization within tissues using the expression of hepatocyte landmark genes. In this way, the spatial zonation patterns of LEC genes can be resolved with high spatial resolution (Fig. 1).
Results
A cell atlas of liver NPCs
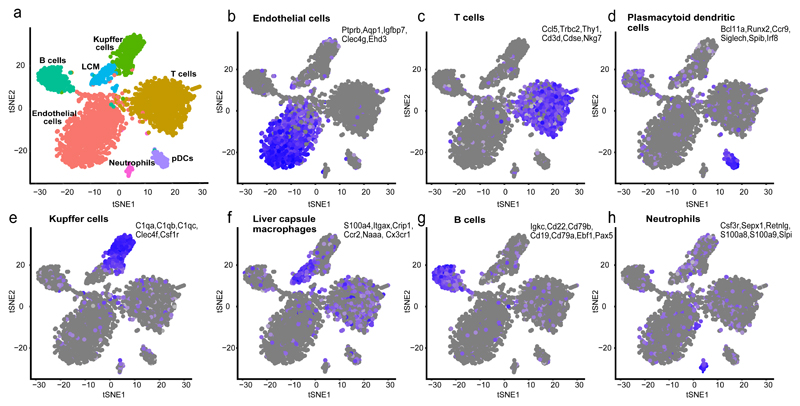
To analyze the identities of NPCs we sorted single perfused mouse liver cells labeled with the pan-immune surface marker CD45 and cells labeled with the endothelial surface marker CD31. We next used MARS-Seq25 to measure the global gene expression of 3,151 sorted single cells (Fig. 2, Supplementary Fig.1, Supplementary Data 1,2). The cells clustered into 7 groups (Fig. 2a) that were enriched for markers of endothelial cells (Fig. 2b), T cells, (Fig. 2c), Plasmacytoid Dendritic cells (pDCs26, Fig. 2d), Kupffer cells (Fig. 2e), liver capsule macrophages (LCM27, Fig. 2f), B cells (Fig. 2g) and neutrophils (Fig. 2h).
Figure 2. Single cell RNAseq reveals the expression signatures of liver non-parenchymal cells.
a) tSNE map colored by the identified seven clusters, consisting of endothelial cells (b, 1,203 cells), T cells (c, 958 cells), plasmacytoid dendritic cells (pDCs, d, 119 cells), Kupffer cells (e, 340 cells), liver capsule macrophages (LCMs, f, 164 cells), B cells (g, 307 cells) and Neutrophils (h, 60 cells). Blue is high expression, gray is low expression in b)-h).
We used a database of known ligand-receptor pairs28,29 to identify potential interactions between these liver sub-populations (Supplementary Fig. 2, Supplementary Data 3). This analysis highlighted LECs as communication hubs that interact with all other liver cell types (Supplementary Fig. 2a), as expected based on direct physical contacts with hepatocytes on one side and immune cells on the other. Notable ligand receptor pairs included the LEC ligand Csf1 and its Kupffer cell receptor Csf1r, the Kupffer cell ligand C1qa and its LEC receptor Cd93, the LEC-hepatocyte pairs Rspo3-Lgr4 and Wnt2-Fzd8 (Supplementary Fig. 2b, Supplementary Data 3), the LEC-LEC juxtacrine signaling pairs Dll4-Notch1 and Efnb2-Ephb4 and the hepatocyte complement C4a and its Kupffer cell receptor C3ar1 (Supplementary Fig. 2c).
Paired cell sequencing of hepatocytes and attached LECs
To test whether LEC genes exhibit spatial zonation in their expression along the radial lobule axis we examined the expression of LEC genes that were previously shown to be zonated – the pericentral Rspo3, Wnt2 and Wnt9b16–18. The UMI levels of these genes were too low to enable robust spatial inference of the single sequenced cells (less than 200 of the 1,203 LECs had more than a single UMI of any of these genes). The total numbers of UMIs in LECs were about 23-fold lower than in single hepatocytes sequenced with the same technology (a median of 470 total UMIs in LECs vs. a median of 10,710 total UMIs in hepatocytes). Since these few hundred UMIs were spread over several thousands of LEC-expressed genes we concluded that robust spatial inference with a small panel of landmark genes would be infeasible. We therefore sought to isolate pairs of hepatocytes and directly adjacent LECs, with the aim of identifying their location by the expression of highly abundant hepatocyte landmark genes.
We dissociated liver tissue with Collagenase D, an enzyme that we found to be less efficient in tissue dissociation compared to enzymes such as Liberase9 (Supplementary Fig. 3a-c). We used FACS to isolate hepatocytes by gating their size and enriched for cells that also expressed CD31 (Fig. 3a-c, Supplementary Fig. 3). This sorting strategy enriched for hepatocyte-LEC pairs (Fig. 3d-f, Supplementary Fig. 3d, 69%). We performed MARS-Seq on the isolated pairs and computationally filtered out wells which did not contain both hepatocyte and endothelial cell markers (Online Methods) resulting in a dataset of 4,602 paired-cells (Supplementary Data 4).
Figure 3. Sorting strategy to isolate pairs of attached hepatocytes and LECs.
a-c) FACS gates used in the isolation. a) FSC-A and SSC-A are used to select hepatocytes based on size. b) FSC-W is used to gate out clusters of hepatocytes. c) CD31 and CD45 fluorescense is used to select pairs that contain a LEC. Numbers in a-c) represent the percent of gated cells. d) Imagestream example of a sorted pair. BF – bright field. Hoechst staining in blue, CD31 in magenta. The experiment was repeated 3 independed times with similar results. e) Expression of hepatocyte pericentral genes (Cyp2e1, Oat, Gulo, Lect2, Cyp1a2) is enriched in the Rspo3+\Wnt9b+ pairs (p=3.3e-29, two-sided Wilcoxon rank-sum test) and depleted in the Efnb2+\Dll4+ pairs (p=6.45e-13). Expression of hepatocyte periportal genes (Cyp2f2, Sds, Hsd17b13, Aldh1b1, Alb, Cps1, Arg1, Pck1) is enriched in the Efnb2+\Dll4+ pairs (p=9.35e-9) and depleted in the Rspo3+\Wnt9b+ pairs (p=1.5e-29, n=4602 paired-cells). Expression is in units of fraction of total cellular UMIs. Box plot elements: center line, median; box limits, first to third quartile (Q1 to Q3); whiskers, extend to the most extreme data point within 1.5× the interquartile range (IQR) from the box; circles, data points. f) Reconstructed zonation profiles of hepatocyte genes based on the pcRNAseq data overlap profiles validated with smFISH 9. Patches are standard errors of the means. smFISH plots were based on n=10 lobules from 2 mice, pcRNAseq plots were based on n=4602 paired-cells.
To demonstrate that the hepatocyte-LEC pairs sequenced were directly adjacent within the tissue, rather than coming into contact during the cell isolation procedure, we selected genes that have been previously shown to be zonated and that were expressed in LECs and not in hepatocytes (Supplementary Data 2) - the pericentral Rspo3 and Wnt9b16–18 and the periportal Dll4 and Efnb224. Pairs that had transcript reads for either Rspo3 or Wnt9b had significantly higher expression of pericentral hepatocyte genes and lower expression of periportal genes. Conversely, pairs that had transcript reads for either Efnb2 or Dll4 had significantly higher expression of periportal hepatocyte genes and lower expression of pericentral genes (Fig. 3e). In addition, we sorted single CD31+ endothelial cells from a mouse constitutively expressing EGFP and hepatocytes from a mouse constitutively expressing DsRed and incubated them together. Upon sorting the mixture we observed <0.15% CD31+CD45- pairs (0 cells in paired-cell optimized sorting gate out of 713 analyzed cells vs. 1,626 in the same paired-cell gate out of 87,872 analyzed cells in Figure 3c, Fisher test p=3.72*10-6) (Supplementary Fig. 3h,i). These experiments demonstrate that the sequenced hepatocyte-LEC pairs were directly adjacent in the tissue.
To infer the radial lobule coordinates of the sequenced pairs of cells we assigned each pair a scaled coordinate denoted η, which was based on the ratio of the summed expression of 21 pericentral and 30 periportal hepatocyte landmark genes (Supplementary Fig. 4a,b). These zonated landmark genes were selected based on their high expression levels and low inter-mouse variability (Online Methods). Pairs of cells that originated in the pericentral layers had a small value of η, whereas pairs from the periportal layers had a large η value (Supplementary Fig. 4c). We used our previous spatial reconstruction of hepatocytes9 to map each η value to the concentric radial lobule layers (Supplementary Fig. 4d, Online Methods). Finally, to obtain the average expression of each gene in each lobule layer we summed the expression of all pairs, weighted by their probability to belong to that layer, based on their η values (Supplementary Fig. 4d, Online Methods). This algorithm yielded the zonation table of 33,856 genes (Supplementary Data 5).
Our zonation inference algorithm recapitulated the previous global zonation patterns of hepatocyte genes (Supplementary Fig. 5). Moreover, the reconstructed zonation profiles of hepatocyte genes in the paired-cell data overlapped zonation profiles that we have validated using smFISH9 (Fig. 3f).
Global zonation of LEC gene expression
We next turned our attention to LEC genes. We focused on genes that had a relative expression that was at least 23-fold higher in single LECs (Fig. 2) compared to single hepatocytes9 (Supplementary Data 2), so that most of the transcripts in the sequenced pairs would have originated from LECs (Online Methods). To avoid the erroneous inference of a gene’s zonation based on its zonation in hepatocytes we also excluded genes that were zonated in single hepatocytes9 (Online Methods). We further removed genes that were highly expressed in the single immune cells (Fig. 2) and selected genes with a maximal zonation level higher than 10−6 of the total cellular transcripts, resulting in 1,303 LEC-specific genes. Our analysis revealed profound spatial heterogeneity of LECs, with around 35% of these genes (475) significantly zonated (Fig. 4a). We validated the predicted zonation patterns of 12 genes using smFISH (Fig. 4b,c, Online Methods).
Figure 4. Global zonation of LEC genes.
a) Reconstructed zonation profiles of the 475 significantly zonated LEC genes. Genes are sorted by the centers of mass of their zonation profiles and normalized by their maximum. Plots on the left show examples of pericentral (Wnt9b, Rspo3), non-monotonic (Pcdhgc5, Ecm1) and periportal (Ltbp4, Efnb2) profiles. Patches are standard errors of the means, plots were based on n=4602 paired-cells. b) smFISH image of the zonated gene Dll4. LEC marker Aqp1 in magenta, the periportal hepatocyte marker Acly in green and Dll4 in red. CV – central vein, PN – portal node. Blue – DAPI stained nuclei, gray – phalloidin stained membranes. Insets show a pericentral LEC (magenta outline), a mid-lobule LEC (yellow outline) and a periportal LEC (green outline). Red dots, transcripts of the Dll4, green dots, transcripts of Acly. Large orange blobs outside of LECs are lipofuscin – non-specific fluorescence common in liver tissue. Red outlines and white arrowheads mark LECs, green outlines mark hepatocytes. Scale bar is 20μm in the scan and 5μm in the insets. c) Validations of the zonation profiles using smFISH for 12 zonated LEC genes. Blue curves are the inferred zonation profile based on pcRNAseq, patches are s.e.m. Red curves are the average zonation profiles based on the smFISH of the sinusoidal LECs, patches are s.e.m. KruskalWallis p-values confirming the significance between the pericentral and periportal endothelial cells are shown at each plot title. Box plot elements: center line, median; box limits, first to third quartile (Q1 to Q3) of the smFISH expression; Gray patches in each plot mark the pericentral layers (CV, left) and periportal layers (PN, right). As opposed to the pcRNAseq, within these layers smFISH enabled distinguishing between LECs that line the central and portal vessels (the extreme boxes) and the sinusoidal LECs. N=30 cells for each layer from 2 mice were quantified for every gene.
Zonated LEC genes included the pericentral ligands Wnt2 and Wnt9b and the Rspo3 ligand (Fig. 4a,c). Dll4 (Fig. 4b,c) which was previously shown to be enriched in arterial endothelial cells30,31 was periportally zonated (Fig. 4c), as were EphrinB232 and Cldn533. Other genes such as Ecm1, Lyve1 and Ccnd1 exhibited a non-monotonic zonation pattern, with the highest expression in the mid-lobule layers (Fig. 4).
Molecular signature of pericentral LECs
The pericentral LECs have been recently shown to be essential for maintaining hepatocyte zonation, through the specific secretion of Wnt ligands and Rspo316–18. Our reconstructed LEC zonation profiles revealed a molecular signature for this pericentral niche, including, in addition to Rspo3, Wnt2 and Wnt9b, thrombomodulin (Thbd), Cdh13, Fabp4 and Kit (Fig. 4c, Fig. 5a). The smFISH validations revealed a bimodal expression pattern for these pericentral genes - Rspo3, Thbd and Cdh13 were highly expressed in both the pericentral LECs that line the sinusoidal channels, as well as in those that line the central vein (Fig. 4c, Fig. 5b), whereas Wnt2 and Kit were expressed to a higher level in the pericentral sinusoidal LECs with lower levels in the LECs that line the central vein (Fig. 4c, Fig. 5c,d). Other genes such as Bmp2 and Stab1 also showed specific repression in the pericentral LECs that line the central vein (Fig. 4c). Notably, we identified expression of the Wnt antagonist Dkk3 in a sub-population of pericentral LECs (Fig. 5a,d). This expression was anti-correlated with Wnt2 in the LECs that line the central vein (Fig. 5e, R=-0.37, p=3.9e-4).
Figure 5. Expression signature of pericentral LECs.
a) Zonation profiles of representative pericentral genes, selected out of the 60 zonated LEC genes with the highest ratio of expression between pericentral layer 1 and pericentral layer 8. Patches are s.e.m. plots were based on n=4602 paired-cells. b) Rspo3 (green dots) is highly expressed both in LECs that line the central vein (CV, arrows), and in sinusoidal pericentral LECs (arrowheads). c) Kit (green dots) is repressed in LECs that line the central vein (CV, arrows), and up-regulated in sinusoidal pericentral LECs (arrowheads). Red dots are mRNAs of the LEC marker Aqp1, yellow blobs are hepatocyte lipofuscins that fluoresce in both red and green channels. Scale bar is 10μm. d) Dkk3 (green dots) is expressed in a subset of LECs that line the central vein (green arrow), distinct from cells that express Wnt2 (red dots, cell marked by red arrow). Scale bar is 5μm. In (b-d) micrographs are representative of 10 lobules and two mice exhibiting similar results. e) Expression of Dkk3 and Wnt2 is anticorrelated in LECs that line the central vein, Spearman R=-0.37, p=3.9e-4 (n=92 cells).
Spatial reconstruction of single LECs
A limitation of pcRNAseq for spatial reconstruction is the inability to infer LEC zonation for genes that are also abundantly expressed in hepatocytes. To overcome this we used pcRNAseq to extract a panel of landmark genes that could be used to retrospectively localize the single sequenced LECs along the lobule radial axis (Fig. 6a,b). Since individual LECs had very low levels of UMIs due to their small size, reliable inference required a large panel of 70 pericentral and 70 periportal LEC genes. As with the paired-cells, we assigned each sequenced LEC a scaled coordinate η that was based on the summed expression of these landmark genes (Online Methods). This coordinate correlated with the cells’ distances from the central vein. We classified the single LECs into four spatially-stratified populations, based on the value of η (Online Methods). The lower spatial resolution of four radial layers, compared to the eight layers obtained with the pcRNAseq reconstruction was required in order to obtain sufficient transcript representation for each layer. In addition, reliable reconstruction was only possible for 2,145 genes with sufficiently high expression, again due to the sparseness of single LEC gene expression (Online Methods).
Figure 6. Spatial reconstruction of single LECs using landmark genes obtained from pcRNAseq.
a-b) pcRNAseq-based zonation profiles of the panel of LEC landmark genes used to localize cells in the scRNAseq data. a) 70 pericentral LEC genes. b) 70 periportal LEC genes. Profiles were scaled between 0 and 1. c) Overlap of zonation profiles based on scRNAseq (blue) n=1163 single-cells and pcRNAseq (red) n=4602 paired-cells. Patches are s.e.m. Zonation profiles were normalized to their means across all layers (pcRNAseq profiles contain 8 layers whereas scRNAseq profiles contain 4). d) Ctsl (green dots) is highly expressed in hepatocytes (white dashed outline) and also expressed in the adjacent LECs (arrows). micrographs are representative of 10 lobules and two mice exhibiting similar results. e) scRNAseq spatial reconstruction reveals zonated expression of Ctsl in LECs, with a reduced expression level in pericentral LECs (1) compared to mid-lobule LECs (2). Box plot elements: center line, median; box limits, first to third quartile (Q1 to Q3) of the smFISH expression, horizonatal lines are medians. Quantification based on n= 30 cells from each layer from 2 mice. Gray patches mark the pericentral layers (CV, left) and periportal layers (PN, right). Blue is the scRNAseq-based zonation profile. Patches are s.e.m. Panels on the left show representative images. Blue is DAPI nuclear stain. Scale bar is 5μm in d)-e).
Our reconstructed profiles (Supplementary Data 6) overlapped the ones obtained with pcRNAseq for the highly zonated LEC genes (Fig. 6c). Importantly, however, this approach uncovered new zonated LEC profiles that included genes that were also expressed in hepatocytes. Examples included the Notch target Hes1 and Ctsl (Supplementary Data 6, Fig. 6d,e).
Spatial sorting of LECs
In light of the global spatial heterogeneity of LECs we sought to identify zonated surface markers for prospective isolation of bulk spatially-resolved LEC populations. Indeed, our analysis revealed several zonated transcription factors34 (Supplementary Fig. 6a) and surface markers (Supplementary Fig. 6b). Kit, encoding the CD117 surface marker, exhibited a zonated profile with a gradual decline in expression from the central vein torwards the portal node (Fig. 4c, Fig. 5a). We used an anti-CD117 antibody to sort CD31+CD45- LECs according to their CD117 levels. qPCR measurements of these bulk spatially-sorted populations validate the identities of these spatially resolved LECs (Supplementary Fig. 6 c-d). This sorting approach can therefore yield massive amounts of LECs from distinct lobule layers, which can be used for future interrogation of their genome, epigenome, proteome and other cellular features that cannot currently be robustly measured at the single cell level.
Discussion
An outstanding challenge in spatial transcriptomics is the expression mapping of small non-parenchymal tissue cells. Here, we introduced paired-cell sequencing, a method that extracts spatial information from attached adjacent parenchymal cells. We used this approach to uncover a high degree of spatial heterogeneity of LECs, with around 35% of LEC genes zonated. We also inferred the molecular signature of pericentral LECs, an important liver niche that secretes key morphogens such as Wnts and Rspo3, and identified an expression of the Wnt antagonist Dkk3 in a sub-population of the LECs that line the central vein that was distinct from the Wnt-producing cells (Fig. 5d,e). Expression of both Wnt ligands and Dkk antagonists has been demonstrated in niche cells of the intestine and hair-follicle35–37. Our findings highlight a potential balancing effect of these positive and negative regulators of liver Wnt signaling by pericentral LECs.
To extend the zonation profiles to genes that are also expressed in hepatocytes we extracted a large panel of LEC landmark genes from the pcRNASeq data and used it to localize the single LECs along the lobule radial axis. This revealed zonation profiles for genes that were masked by the hepatocyte expression in the paired-cell data. While more genes could be examined with this approach, the spatial resolution of this reconstruction was lower (four layers vs. eight layers for the pcRNAseq data). Technologies that provide more sequenced cells38,39 along with higher capture rate40 could facilitate a higher spatial resolution and statistical power for identifying additional zonated lowly-expressed genes. Thus, our study demonstrated the utility of pcRNAseq not only for inferring zonation profiles of NPC-specific genes, but also as a method for unbiased detection of a large panel of NPC landmark genes, to be used in single-cell based spatially resolved transcriptomics.
While our reconstruction had high spatial resolution, it did not capture the entire complexity of LECs, due to limitations related to the parenchymal cells used for spatial inference. Endothelial cells are comprised of arterial, venous and sinusoidal cells22,41. A hepatocyte at a given radial lobule layer is often spatially adjacent to more than one type of LEC. For example, sorted pericentral hepatocytes can either carry an attached pericentral LEC that lines the radial sinusoids or one that lines the central vein (Fig. 5b,c). Thus our layer 1 LEC expression is an average of these distinct pools of cells. Indeed, using smFISH measurements we found that some pericentral LEC genes such as Rspo3 and Cdh13 were high in the LECs that line the central vein, whereas others such as Wnt2 and Kit were more highly expressed in the pericentral sinusoidal LECs. Moreover, LECs at the same tissue location can exhibit distinct sub-populations and additional variability, as exemplified in the pericentral LECs that express either Dkk3 or Wnt2. The fact that the spatial structure of LECs did not naturally emerge from the single cell data (Figure 2b) may also indicate that LEC identity is defined by a complex overlay of different factors, where lobule coordinate is only one of which. Our spatial blueprint should thus serve as a basis for exploring these additional layers of variability within individual layers.
Modelling hepatocyte functions ex-vivo has been a challenging field due to the difficulties of emulating the complex liver microenvironment42. For example, the expression of the pericentrally-zonated xenobiotic metabolism enzymes is often lost within several hours after extraction of hepatocytes, but partially retrieved by co-culturing hepatocytes with LECs42. Our identification of zonated LEC surface markers such as CD117 (Supplementary Fig. 6) could be used to obtain co-cultures of hepatocytes and spatially stratified LECs, which could potentially be even more efficient in reconstructing zonated hepatocyte functions of interest ex-vivo. Such spatial sorting could also be used to explore other cellular features of spatially-stratified LEC populations, such as histone modifications, DNA methylations and protein content.
PcRNAseq can be generically applied to other tissues and cell types. In the liver, natural candidates for similar reconstructions are hepatic stellate cells, which are physically adjacent to hepatocytes within the space of Disse43 (Supplementary Fig. 7e, Supplementary Data 7). Enterocytes, the most abundant epithelial cells in the intestine, have recently been shown to exhibit global expression gradients along the crypt-villus axis44. Several intestinal cell types of key importance, such as enteroendocrine cells, goblet cells, tuft cells and intraepithelial lymphocytes are interleaved and physically attached to enterocytes within the epithelial sheet45,46 (Supplementary Fig. 7a-d). PcRNAseq could be readily used to uncover the spatial expression gradients of these cells. Other examples of relevant cell pairs include neurons and astrocytes, glia and microglia in the brain, a tissue in which neurons exhibit broad zonation47,48, as well as peripheral neurons and diverse cells such as hepatocytes, enterocytes and adipose cells49 (Supplementary Data 7). PcRNAseq can also be used to explore the interactions between diverse tumor cell populations and their adjacent stromal neighbors, such as cancer associated fibroblasts or immune cells (Supplementary Fig. 7f 50). In such samples, the ‘landmark genes’ expressed in the cancer cells could either intrinsically encode spatial landmarks such as core vs. periphery of the tumor or adjacency to blood vessels, or alternatively clonally or transcriptionally distinct populations that do not have a clear spatial correlate.
For each tissue and for each pair of cell types, pcRNAseq would require optimization of the dissociation protocols, to enable isolation of pairs rather than single cells or clumps of more than two cells (Supplementary Fig. 3). Combining the approach with DNA stains such as Hoechst could facilitate pair-enrichment in cases where FACS cell size properties are insufficient. Moreover, cell-type-specific surface markers should be used for the enrichment of the relevant cell types. In summary, pcRNASeq opens avenues for detailed spatial characterization of cells in diverse tissues.
Online Methods
Mice and tissues
All animal studies were approved by the Institutional Animal Care and Use Committee of WIS. C57bl6 male mice age 6-12 weeks were housed under reverse phase cycle, and fasted for 2 hours starting at 8AM (note that the change from 5 hours fasting in9 to 2 hours fasting did not changed the spatial reconstruction (Supplementary Fig. 5a)). C57BL/6-Tg(CAG-EGFP)51 and C57BL/6-Actb-DsRed.T352 mice were used for the dual color FACS (Supplementary Fig. 3h,i). All mice were anesthetized with an intraperitoneal injection of a ketamine (100 mg/kg) and xylazine (10 mg/kg) mixture. For smFISH, liver tissues were harvested and fixed in 4% paraformaldehyde for 3 hours, incubated overnight with 30% sucrose in 4% paraformaldehyde and then embedded in OCT. 7 µm cryosections were used for hybridization. Mouse liver cells for RNAseq were extracted from five mice (3 for pcRNASeq and 2 for scRNASeq). All smFISH quantifications were perfromed on at least 2 mice. smFISH images in figure 5 were done on ad libitum mice.
Antibodies used in this study
The following antibodies were used for cell isolation: CD31 (APC 102510, PE-Cy7 102418),CD45 (APC-Cy7 103116, PE-Cy7 103114), CD3 (PE 100205), CD19 (PE 152407). CD117 (Kit, APC 105815) All antibodies were purchased from Biolegend.
Hybridization and imaging
Probe library constructions, hybridization procedures and imaging conditions were previously described53,54. SmFISH probe libraries (Supplementary Data 8) were coupled to Cy5 or Alexa594. Endothelial cells were detected by Aqp1 staining. To detect cell borders alexa fluor 488 conjugated phalloidin (Rhenium A12379) was added to the GLOX buffer wash54. Portal nodes were identified morphologically using the DAPI channel based on bile ductile, central vein was identified using smFISH for Glul in TMR, included in all hybridizations. For zonation validation profiles, images were taken as scans extending from the portal node to the central vein. Endothelial cells were classified into 7 layers as follows: cells that were in contact with Glul+ cells and that lined the central vein were classified as layer 1, cells that were in contact with Glul+ cells but resided in the sinusoidal channels were classified as layer 2, cells that surrounded the portal vessels at a distance of up to 1 hepatocyte were classified as layer 6, cells that lined the inside of the vessels in the portal node were classified as layer 7, sinusoidal cells in the mid-lobule area were assigned layers 3-5 using equal distances from layer 2 to layer 6. To validate the predicted zonation we selected 15 genes that were significantly zonated and had an average expression in the scRNAseq data of LECs of more than 10-4 of the cellular UMIs - Rspo3, Cdh13, Thbd, Wnt2, Kit, Bmp2, Bmp4, Ccnd1, Lyve1, Stab1, Dll4, Zeb2, Efnb2, Ltbp4 and Klf4. smFISH did not yield a signal for Bmp4, Zeb2 and Klf4, whereas the remaining 12 genes are presented in Figure 4c. The results in Figure 4c were based on at least 30 cells from each layer and from 2 mice. Quantification of smFISH data was done using ImageM54. Dots were counted in the first 5μm of the Z-stack, and divided by the segmented cell volume to obtain the mRNA concentration per cell.
Hepatocytes isolation
Mouse liver cells were isolated by a modification of the two-step collagenase perfusion method of Seglen55 from 2 hours fasted, 6-7 weeks old male C57bl6 mice for the pcRNAseq (3 mice) or 3-4 months for the scRNAseq (2 mice). Digestion step was performed with Collagenase D (Sigma 11088858001) for the pcRNAseq or Liberase Blendzyme 3 recombinant collagenase (Roche Diagnostics) for the scRNAseq according to the manufacturer’s instruction. Isolated cells were stained and taken directly to sorting. We found that Collagenase D was superior to Liberase for isolating pairs, since it is less efficient in tissue dissociation.
Multispectral imaging flow cytometry analysis (ImageStream)
Cells were imaged using a multispectral Imaging Cytometer (ImageStreamX mark II imaging flow-cytometer; Amnis Corp, Part of EMD Millipore, Seattle, WA,). At least 5x103 cells were collected from each sample and data were analyzed using the manufacturer’s image analysis software (IDEAS 6.2; Amnis Corp). Images were compensated for fluorescent dye overlap by using single-stain controls. Cells were gated for focused cells, using the Gradient RMS feature, as previously described (George et al., 2006). Cropped cells were further eliminated by plotting the cell area of the bright field image against the Centroid X feature (the number of pixels in the horizontal axis from the left corner of the image to the center of the cell mask). Only cells negative for PI staining were included in the analysis. Cells were then gated for single cells or doublets, using the area and aspect ratio features of the bright-field channel. Additional singles were further gated for area of the bright field and the aspect ratio of the Hoechst staining (normalized for intensity), and added to the ‘single’ population. CD31 positive cells were gated according to the intensity (total fluorescence within the image) and Max pixel (the highest intensity pixel within the image) values of the CD31 staining. We found that 69% of the cells were hepatocyte-LEC pairs, 23% were single hepatocytes, 8% had more than one hepatocyte attached to a single LEC (Supplementary Fig. 3d). We computationally selected only pairs that had both hepatocytes and endothelial cells markers (below). While pairs that had more than a single hepatocyte (i.e. triplets of two hepatocytes and one LEC) could not be omitted computationally, they most probably originated from hepatocytes that were attached in the tissue. Since the hepatocyte gene expression is used to localize the pairs, such triplets should not introduce a significant bias.
Single-cell and paired-cell sorting
Paired Cells were sorted with SORP-FACSAriaII machine using a 130 μm nozzle, single NPCs were sorted using 100 μm nozzle. Dead cells were excluded on the basis of 500 ng/ml Dapi incorporation. To enrich for hepatocytes a #1.5 ND filter and was used, whereas a #1 ND filter was used to enrich for NPCs. To enrich for hepatocyte-LEC pairs, cells were gated according to size that match the hepatocytes distribution by FSC-A and SSC-A. The cells were next gated by FSC-W to remove clusters of hepatocytes, and also gated for CD45 negative CD31 positive staining to enrich for pairs of hepatocyte-endothelial cells. We selected the top 2% of cells according to CD31 staining, to ensure the inclusion of LECs. Cells were sorted into 384-well cell capture plates containing 2 μl of lysis solution and barcoded poly(T) reverse-transcription (RT) primers for single-cell RNA-seq25. Barcoded single cell capture plates were prepared with a Bravo automated liquid handling platform (Agilent) as described previously25. Four empty wells were kept in each 384-well plate as a no-cell control during data analysis. To enrich for pericentral pairs, which are a minority due to the hexagonal lobule geometry, in 4 out of the 17 384-well plates of pairs sorted we collected pairs that were also positive for CD73, a hepatocyte-specific surface marker that we previously found to be pericentrally zonated9. Immediately after sorting, each plate was spun down to ensure cell immersion into the lysis solution, snap frozen on dry ice and stored at - 80°C until processed.
Spatial sorting
Livers were perfused with Liberase Blendzyme 3 recombinant collagenase (Roche Diagnostics). Cells were enriched for NPCs by 3 minute of centrifugation at 30g. supernatant was centrifuged at 300g for 5 minute. Cell pellet was treated with red blood cell lysis buffer (Sigma R7757) according to the manufacturer’s instruction. Isolated cells were stained and taken to sorting. Cells were gated for CD31 positive, CD45 negative population to enrich for LECs. CD117-positive LECs (relative to the fluorophore isotype control) were divided to 4 equal populations according to fluorescent levels where highet expression represented the most pericentral population (Supplementary Fig. 6). Cells were sorted into lysis buffer.
qPCR quantification
RNA was isolated from cell lysates by Dynabeads mRNA DIRECT Micro Kit (Invitrogen 61021) according to the manufacturer’s instruction. RNA was revese transcribe with Maxima H minus RT enzyme (Thermo scientific EP0753) and subjected to qPCR with fast SYBR green on selected genes (Supplementary Data 9).
Massively Parallel Single Cell RNA-Seq (MARS-Seq) library preparation
Single cell libraries were prepared, as described in25. Briefly, mRNA from cells sorted into MARS-Seq capture plates were barcoded and converted into cDNA and pooled using an automated pipeline. The pooled sample was then linearly amplified by T7 in vitro transcription and the resulting RNA was fragmented and converted into sequencing ready library by tagging the samples with pool barcodes and Illumina sequences during ligation, reverse transcription and PCR. Each pool of cells was tested for library quality and concentration was assessed as described in25. Mapping of single-cell reads to mouse reference genome (mm9) was done using HISAT version 0.1.6-beta and reads with multiple mapping positions were excluded. Reads were associated with genes if they were mapped to an exon defined by a reference set obtained from the UCSC genome browser. Exons of different genes that share genomic position on the same strand were considered as a single gene with concatenated gene symbol. Corrected read counts were evaluated based on unique molecular identifiers (UMI) as described in25.
scRNAseq data processing
For each single cell and for each gene we first subtracted the estimated background expression. Background was calculated for each 384-well plate separately, as the mean gene expression in the four empty wells and in ‘drop-out’ wells that had low signal. These drop-out wells were defined as wells with both a number of expressed genes that was lower than the minimal number of expressed genes in the empty wells and a total UMI count that was smaller than the maximal total UMI count in the empty wells. After subtraction, negative values were set to zero. Next, cells with total UMI counts lower than 200 or higher than 6,000 were removed.
We used Seurat v2.0.1 package in R56 to visualize and cluster the single cell RNAseq data (Fig. 2). Gene expression measurements (UMIs per gene) were normalized for each cell by the summed UMI, multiplied by a scale factor 10,000, and then log-transformed. To avoid undesired sources of variation in gene expression, we used Seurat to regress out cell-cell variation driven by batch, total number of UMIs, and mouse identity. For detection of variable genes we set a bottom cutoff of 0.2 and a top cutoff of 3.5 on the regressed log-transformed average gene expression, as well as a bottom cutoff of 0.6 on the dispersion. Cell clustering was based on PCA dimensionality reduction using the first 15 PCs, and a resolution value of 0.5.
We used cell type-specific markers to interpret the resulting 7 clusters, based on literature search and the Immunological Genome project database57. Ptprb, Igfbp7, Clec4g, Aqp122 and Ehd3 were highly expressed in the endothelial cell cluster, C1qa, C1qb, C1qc, Clec4f and Csf1r in the Kupffer cell cluster, Ccl5, Trbc2, Cd3e, Cd3d, Nkg7 and Thy1 in the T cell cluster, Igkc, Cd22, Cd79b, Cd19, Cd79a, Ebf1 and Pax5 in the B cell cluster, S100a9, S100a8, Csf3r, Slpi, Sepx1 and Retnlg in the neutrophil cluster, Bcl11a, Runx2, Ccr9, Siglech, Spib, and Irf8 in the Plasmacytoid Dendritic cell (pDC) cluster26 and Cd11c (Itgax) and Cx3cr1 in the liver capsular macrophage cluster27.
Ligand-receptor analysis
To examine the interactions between liver cell types we used Seurat v2.0.1 R package to obtain the clustering and tSNE coordinates of a combined dataset, that included the single cells sequenced in this study as well as the single cell sequencing data of 9. Data in Seurat was regressed on UMI counts and mice (however, hepatocytes were regressed in a pooled manner). Data was normalized and clustered as described in the previous section for the single NPC data, with the exception that the resolution value here was 0.35. This analysis resulted in 8 clusters representing endothelial cells, hepatocytes, kupffer cells, T cells, B cells, pDCs, LCMs and neutrophils.
Ligand-receptor murine pairs were extracted from Graeber et al.28 (708 unique ligands and 691 unique receptors). For each gene g and each cluster c we calculated the average expression We next computed a Z-score, representing the enrichment of each ligand and receptor in each cluster:
| [1] |
Where the means and were taken over the 8 cluster values. We next defined an interaction score as:
| [2] |
Were, is the ligand Z-score for cluster c, and is the receptor Z-score for cluster c. This resulted in 6689 interactions annotated by ligand, receptor, source cluster and target cluster (Supplementary Data 3, all pairs sheet). We next considered only interactions for which both the average ligand expression in the source cluster, and the average receptor expression in the target cluster were above 0.1 and for which the fraction of positive cells for the ligand/receptor were higher than 2% in the source/target cluster respectively (note that expression values here are in units of Seurat’s log-transformed normalized data). In addition we removed pairs in which either the ligand/receptor were expressed in less than 10 cells in the source/target cluster. This resulted in 2895 interactions (Supplementary Data 3, expression-filtered sheet). To highlight cluster-specific interactions we reported all interactions with ligand/receptor Z-score above 1.5 and Zinteraction > 2 (Supplementary Data 3, cluster-specific sheet). Visualization of representative pairs was done with Matlab (Supplementary Fig. 2). Supplementary Fig.2a shows the network of cluster interactions, visualized with Cytoscape.
Processing of paired-cell data
For the CD31+ hepatocyte pairs, we used the same background subtraction method used for the single NPCs. We selected cells with total number of UMIs between 2,500 and 50,000, and at least 500 expressed genes. For each well, we normalized the expression by the total UMIs in that well, so that the expression was in units of fraction of total UMIs to obtain the expression matrix D ∈ ℝ#genes,#cells. To ensure that each pair includes an hepatocyte we removed pairs with an expression less than 0.01 for Alb9. To ensure that each pair contains an endothelial cell, we excluded wells for which the summed expression of the endothelial markers: Ptprb, Igfbp7, Clec4g, Aqp1 and Ehd3 was lower than 5 ⋅ 10−5. Additionally we excluded wells for which the summed expression of the Kupffer cell markers C1qa, C1qb, C1qc, Clec4f and Csf1r was higher than 0.0005. These threshold values were chosen as the 80% values in the respective clusters in the single NPC dataset, after correcting for the 23-fold dilution of NPC transcripts that occurs when they are mixed with the large hepatocytes. This procedure resulted in 4,602 paired cells that were retained for further analysis.
Selection of hepatocyte landmark genes for pcRNAseq reconstruction
We selected a large panel of hepatocyte landmark genes for inferring the original lobule coordinates of the pairs based on our previous study of hepatocyte zonation9. We chose zonated genes with a maximum expression level of at least 2 ⋅ 10−4 across layers, with a ratio between the maximum and minimum zonation values that exceeded 1.5. We excluded genes that were previously found to vary in a circadian manner (JTK q-value lower than 0.0158, to reduce mouse-to-mouse variability in the expression of the landmark genes. We also removed genes that had an average expression that was at least 2-fold higher in the LEC scRNAseq data compared to pcRNAseq of hepatocytes9. Given the 23-fold dilution of LEC transcripts in the pairs, this ensured that any LEC contamination among the landmark genes would not be higher than 10%. The pericentral signature genes were selected from the remaining gene list, as genes with maximum expression in the most pericentral layer and minimum in most periportal layer, as well as center of mass lower than layer 4.5 (21 genes, Supplementary Fig. 4a). The periportal genes were selected as genes with maximum expression in the most periportal layer and minimum in most central layer, as well as center of mass higher than 5 (30 genes, Supplementary Fig. 4b). While this panel did not include some classic periportal genes such as Pck1, Cps1 and Arg1, reconstruction is essentially unchanged when including these landmark genes (data not shown).
Zonation reconstruction algorithm
To reconstruct the zonation profiles from the pcRNAseq data we used the combined expression of the landmark gene (LM) panel. Each cell i was assigned a coordinate ηi, which reflected its location along the radial lobule axis, where ηi = 0 was the most pericentral coordinate and ηi = 1 the most periportal coordinate. We normalized the expression of every landmark gene by the maximum across all cells, to avoid giving excessive weight to highly expressed hepatocyte genes. This resulted in a normalized expression matrix E ∈ ℝ#LMgenes,#cells.
For each cell i we divided the summed expression of the portal LM genes (pLM), by the summed expression of the central (cLM) and portal LM genes to yield a number xi which we normalized between 0-1 to obtain ηi.
| [3] |
| [4] |
To map η values to lobule layers, we estimated the η of the single hepatocytes form Bahar-Halpern at el.9. We combined lobule layers 8-9 to yield 8 layers since there were only few cells in layer 9. We fitted a Gamma distribution to the histogram of η values for each layer L∈[1 8] (Supplementary Fig. 4d). We used this Gamma distribution to compute the probability that each pair ηi belongs to each lobule layer L, P(η = ηi |layeri = L).
According to Bayes’ law:
| [5] |
We assumed a uniform prior P(layeri = L) rather than a prior that incorporates the hexagonal geometry of the lobule9 since we used CD73 in some of the sorted plates to enrich for pericentral pairs.
| [6] |
We thus calculated a matrix of probabilities M ∈ ℝ#cells,#layers such that:
| [7] |
To transform M into a weight matrix W ∈ ℝ#cells,#layers, we divided each value by the sum of its column The final zonation matrix Z ∈ ℝ#genes,#layers was obtained by multiplying the weight matrix by the expression matrix Z = D × W. As in the expression matrix D the units of the zonation matrix Z are fraction of total cellular UMIs, however here they represent the average over all cells in the layer.
We used 500 bootstrap iterations to obtain standard errors for the mean zonation profiles. To assign zonation significance we extracted a summary statistic for each gene as the difference between the maximum and minimum values of the mean-normalized profile and compared it to the summary statistics in 1,000 datasets in which the cells’ η was randomly reshuffled. We calculated Z-scores for the summary statistic of each gene and used the normal distribution to obtain p-values. We used Benjamini-Hochberg multiple hypothesis correction to assign a q-value for each profile.
Determining the set of genes expressed uniquely in endothelial cells
Our paired-cell sequencing yields UMI counts that may originate from either a hepatocyte or its paired endothelial cell. Since around 50% of the genes expressed in hepatocytes exhibit zonation9 we first excluded genes that were found to be zonated in hepatocytes9, as well as genes with a maximal zonation level lower than 10−6 in the zonation matrix Z. For each gene we next calculated the mean over all cells of the expression (in fraction of total UMIs) in single hepatocytes9 and in every NPC cluster in the scRNAseq data. We removed genes for which the mean expression in one of the non-endothelial cell types was higher than five-fold the mean in the endothelial scRNAseq cluster. When computing hepatocyte mean expression in the data of 9 we removed cells with a summed expression higher than 5 ⋅ 10−4 of endothelial markers (Ptprb, Igfbp7, Clec4g, Aqp1, Ehd3) or Kupffer cell markers (C1qa, C1qb, C1qc, Clec4f, Csf1r). We computed the ratio between the median total number of UMIs in single hepatocytes (TH = 10,710) and in single endothelial cells (TE = 470, This higher mRNA content in hepatocytes is compatible with their much larger size (Fig. 3d). We selected genes with endothelial expression that was 23-fold higher than the hepatocyte expression, and for which the fraction of cells with non-zero expression was higher in endothelial cells than in hepatocytes. This resulted in 1,303 endothelial genes. Genes with zonation q-value lower than 0.2 were considered significantly zonated, resulting in 475 zonated endothelial genes.
Zonation Reconstruction of single LECs
To spatially reconstruct the scRNAseq LEC data we established a panel of 140 LEC landmark genes, extracted from the pcRNAseq data. We chose among the 475 zonated LEC genes the ones with maximal zonation values above 2 ⋅ 10−6. We sorted the zonation profiles by their center of mass, and chose the portal and central landmark genes as the top and bottom 70 genes. We used equations [3-4] to compute η for every cell over this LEC landmark panel and partitioned the cells into 4 layers (0 ≤ η ≤ 0.25, 0.25 < η ≤ 0.5, 0.5 < η ≤ 0.75, 0.75 < η ≤ 1). By averaging the expression of cells in each group we established the spatial expression patterns of all genes. We removed 40 of the 1,203 sequenced LECs that had no expression of any of the 140 landmark genes. Due to the sparseness of the scRNAseq data we focused on 2,158 genes with a mean expression higher than 10−4. Additionally, we removed 11 genes with mean expression higher than 0.01 in the single hepatocytes, assuming that expression of these genes in LEC could be contamination from hepatocyte mRNA. Significance of zonation was computed using Kruskal-Wallis test followed by Benjamini-Hochberg multiple hypothesis correction.
Statistics and reproducibility
No statistical method was used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to animal allocation during experiments and outcome assessment. All replicates were technical replicates performed in independed experiments, data is represented as s.e.m. In figure 3e, two-sided Wilcoxon rank-sum test was used. In figure 4c and supplementary figure 6d p-values were calculated using KruskalWallis method. In figure 5e and supplementary figure 5b Spearman correlation test was performed. In supplementary figure 3i two-sided Fisher test was performed. A life sciences reporting summary is available
Supplementary Material
Ed Summary.
The expression profile and tissue coordinates of liver endothelial cells are determined using a paired-cell sequencing approach that extracts spatial information from attached hepatocytes.
Acknowledgements
We thank M. Kolesnikov and S. Jung for the C57BL/6-Actb-DsRed.T3 mice and D. Jaitin for the C57BL/6-Tg(CAG-EGFP) mice. S.I. is supported by the Henry Chanoch Krenter Institute for Biomedical Imaging and Genomics, The Leir Charitable Foundations, Richard Jakubskind Laboratory of Systems Biology, Cymerman-Jakubskind Prize, The Lord Sieff of Brimpton Memorial Fund, the I-CORE program of the Planning and Budgeting Committee and the Israel Science Foundation (grants 1902/12 and 1796/12), the Israel Science Foundation grant No. 1486/16, the EMBO Young Investigator Program and the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement number 335122, the Bert L. and N. Kuggie Vallee Foundation and the Howard Hughes Medical Institute (HHMI) international research scholar award. S.I. is the incumbent of the Philip Harris and Gerald Ronson Career Development Chair.
Footnotes
Code availability
Code 1 - ZONATION_pcRNAseq-MATLAB code that loads expression data for pairs of hepatocytes and endothelial cells, localizes each pair along the lobule axis according to hepatocyte landmark genes and outputs a table of zonated expression and a list of zonated endothelial genes.
Data availability
Data generated in this study have been deposited in Gene Expression Omnibus with the accession code: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108561).
Data referenced in9 is available in GEO with the accession code: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE84498. Data referenced in Supplementary Fig. 2 is available in doi:10.1038/ng755. Data referenced in Supplementary Fig. 6a is available in doi:10.1038/nature13182 (Supplementary Table 8).
Author Contribution
K.B.H., C.M., B.T., A.E., A.E.M., E.E.M., Z.P. performed experiments. R.S., S.I., H.M., A.G. and E.D. performed data analysis. I.A. contributed to project design. S.I. supervised the study. S.I., K.B.H. and R.S. wrote the paper. All authors discussed the results and commented on the manuscript. K.B.H. and R.S. contributed equally to the study.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Regev A, et al. The Human Cell Atlas. bioRxiv. 2017 doi: 10.1101/121202. 121202. [DOI] [Google Scholar]
- 2.Transcriptomic characterization of 20 organs and tissues from mouse at single cell resolution creates a Tabula Muris. [Accessed: 27th December 2017];bioRxiv. Available at: https://www.biorxiv.org/content/early/2017/12/20/237446.
- 3.Han X, et al. Mapping the Mouse Cell Atlas by Microwell-Seq. Cell. 2018;172:1091–1107.e17. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Crosetto N, Bienko M, van Oudenaarden A. Spatially resolved transcriptomics and beyond. Nat Rev Genet. 2015;16:57–66. doi: 10.1038/nrg3832. [DOI] [PubMed] [Google Scholar]
- 5.Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The Technology and Biology of Single-Cell RNA Sequencing. Molecular cell. 2015;58:610–620. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotech. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Achim K, et al. High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nat Biotech. 2015;33:503–509. doi: 10.1038/nbt.3209. [DOI] [PubMed] [Google Scholar]
- 8.Karaiskos N, et al. The Drosophila embryo at single-cell transcriptome resolution. Science. 2017;358:194–199. doi: 10.1126/science.aan3235. [DOI] [PubMed] [Google Scholar]
- 9.Bahar Halpern K, et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542:352–356. doi: 10.1038/nature21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ståhl PL, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 11.Lein E, Borm LE, Linnarsson S. The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science. 2017;358:64–69. doi: 10.1126/science.aan6827. [DOI] [PubMed] [Google Scholar]
- 12.Lee JH. De Novo Gene Expression Reconstruction in Space. Trends in Molecular Medicine. 2017;23:583–593. doi: 10.1016/j.molmed.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moor AE, Itzkovitz S. Spatial transcriptomics: paving the way for tissue-level systems biology. Curr Opin Biotechnol. 2017;46:126–133. doi: 10.1016/j.copbio.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Jungermann K, Keitzmann T. Zonation of Parenchymal and Nonparenchymal Metabolism in Liver. Annual Review of Nutrition. 1996;16:179–203. doi: 10.1146/annurev.nu.16.070196.001143. [DOI] [PubMed] [Google Scholar]
- 15.Gebhardt R. Metabolic zonation of the liver: regulation and implications for liver function. Pharmacol Ther. 1992;53:275–354. doi: 10.1016/0163-7258(92)90055-5. [DOI] [PubMed] [Google Scholar]
- 16.Wang B, Zhao L, Fish M, Logan CY, Nusse R. Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature. 2015;524:180–185. doi: 10.1038/nature14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donovan A, et al. The RSPO–LGR4/5–ZNRF3/RNF43 module controls liver zonation and size. Nature Cell Biology. 2016;18:467. doi: 10.1038/ncb3337. [DOI] [PubMed] [Google Scholar]
- 18.Rocha AS, et al. The Angiocrine Factor Rspondin3 Is a Key Determinant of Liver Zonation. Cell Reports. 2015;13:1757–1764. doi: 10.1016/j.celrep.2015.10.049. [DOI] [PubMed] [Google Scholar]
- 19.Braeuning A, et al. Differential gene expression in periportal and perivenous mouse hepatocytes. FEBS J. 2006;273:5051–5061. doi: 10.1111/j.1742-4658.2006.05503.x. [DOI] [PubMed] [Google Scholar]
- 20.Gebhardt R, Matz-Soja M. Liver zonation: Novel aspects of its regulation and its impact on homeostasis. World J Gastroenterol. 2014;20:8491–8504. doi: 10.3748/wjg.v20.i26.8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colnot S, Perret C. Liver Zonation. In: Monga SPS, editor. Molecular Pathology of Liver Diseases. Springer; US: 2011. pp. 7–16. [Google Scholar]
- 22.Aird WC. Phenotypic heterogeneity of the endothelium: II. Representative vascular beds. Circ Res. 2007;100:174–190. doi: 10.1161/01.RES.0000255690.03436.ae. [DOI] [PubMed] [Google Scholar]
- 23.Strauss O, Phillips A, Ruggiero K, Bartlett A, Dunbar PR. Immunofluorescence identifies distinct subsets of endothelial cells in the human liver. Sci Rep. 2017;7 doi: 10.1038/srep44356. 44356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rafii S, Butler JM, Ding B-S. Angiocrine functions of organ-specific endothelial cells. Nature. 2016;529:316. doi: 10.1038/nature17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaitin DA, et al. Massively Parallel Single-Cell RNA-Seq for Marker-Free Decomposition of Tissues into Cell Types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chistiakov DA, Orekhov AN, Sobenin IA, Bobryshev YV. Plasmacytoid dendritic cells: development, functions, and role in atherosclerotic inflammation. Front Physiol. 2014;5 doi: 10.3389/fphys.2014.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sierro F, et al. A Liver Capsular Network of Monocyte-Derived Macrophages Restricts Hepatic Dissemination of Intraperitoneal Bacteria by Neutrophil Recruitment. Immunity. 2017;47:374–388.e6. doi: 10.1016/j.immuni.2017.07.018. [DOI] [PubMed] [Google Scholar]
- 28.Graeber TG, Eisenberg D. Bioinformatic identification of potential autocrine signaling loops in cancers from gene expression profiles. Nat Genet. 2001;29:295–300. doi: 10.1038/ng755. [DOI] [PubMed] [Google Scholar]
- 29.Zhou JX, Taramelli R, Pedrini E, Knijnenburg T, Huang S. Extracting Intercellular Signaling Network Of Cancer Tissues Using Ligand-Receptor Expression Patterns From Whole-Tumor And Single-Cell Transcriptomes. bioRxiv. 2017 doi: 10.1101/127167. 127167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shutter JR, et al. Dll4, a novel Notch ligand expressed in arterial endothelium. Genes Dev. 2000;14:1313–1318. [PMC free article] [PubMed] [Google Scholar]
- 31.Hellström M, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 32.Adams RH, Klein R. Eph Receptors and Ephrin Ligands: Essential Mediators of Vascular Development. Trends in Cardiovascular Medicine. 2000;10:183–188. doi: 10.1016/s1050-1738(00)00046-3. [DOI] [PubMed] [Google Scholar]
- 33.Rahner C, Mitic LL, Anderson JM. Heterogeneity in expression and subcellular localization of claudins 2, 3, 4, and 5 in the rat liver, pancreas, and gut. Gastroenterology. 2001;120:411–422. doi: 10.1053/gast.2001.21736. [DOI] [PubMed] [Google Scholar]
- 34.FANTOM Consortium and the RIKEN PMI and CLST (DGT) et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore KA, Lemischka IR. Stem cells and their niches. Science. 2006;311:1880–1885. doi: 10.1126/science.1110542. [DOI] [PubMed] [Google Scholar]
- 36.Gregorieff A, et al. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129:626–38. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 37.Sick S, Reinker S, Timmer J, Schlake T. WNT and DKK determine hair follicle spacing through a reaction-diffusion mechanism. Science. 2006;314:1447–1450. doi: 10.1126/science.1130088. [DOI] [PubMed] [Google Scholar]
- 38.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klein AM, et al. Droplet Barcoding for Single-Cell Transcriptomics Applied to Embryonic Stem Cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.mcSCRB-seq: sensitive and powerful single-cell RNA sequencing. [Accessed: 27th December 2017];bioRxiv. Available at: https://www.biorxiv.org/content/early/2017/10/18/188367.
- 41.Vanlandewijck M, et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018;54:475. doi: 10.1038/nature25739. [DOI] [PubMed] [Google Scholar]
- 42.Godoy P, et al. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol. 2013;87:1315–1530. doi: 10.1007/s00204-013-1078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol. 2011;6:425–456. doi: 10.1146/annurev-pathol-011110-130246. [DOI] [PubMed] [Google Scholar]
- 44.Moor AE, et al. Spatial reconstruction of single enterocytes uncovers broad zonation along the intestinal villus axis. bioRxiv. 2018 doi: 10.1101/261529. [DOI] [PubMed] [Google Scholar]
- 45.Olivares-Villagómez D, Van Kaer L. Intestinal Intraepithelial Lymphocytes: Sentinels of the Mucosal Barrier. Trends in Immunology. doi: 10.1016/j.it.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boisset J-C, et al. Mapping the physical network of cellular interactions. Nat Methods. 2018 doi: 10.1038/s41592-018-0009-z. [DOI] [PubMed] [Google Scholar]
- 47.Shah S, Lubeck E, Zhou W, Cai L. seqFISH Accurately Detects Transcripts in Single Cells and Reveals Robust Spatial Organization in the Hippocampus. Neuron. 2017;94:752–758.e1. doi: 10.1016/j.neuron.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 48.Codeluppi S, et al. Spatial organization of the somatosensory cortex revealed by cyclic smFISH. bioRxiv. 2018 doi: 10.1101/276097. [DOI] [PubMed] [Google Scholar]
- 49.Zeng W, et al. Sympathetic neuro-adipose connections mediate leptin-driven lipolysis. Cell. 2015;163:84–94. doi: 10.1016/j.cell.2015.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhowmick NA, Moses HL. Tumor–stroma interactions. Current Opinion in Genetics & Development. 2005;15:97–101. doi: 10.1016/j.gde.2004.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 52.Vintersten K, et al. Mouse in red: red fluorescent protein expression in mouse ES cells, embryos, and adult animals. Genesis. 2004;40:241–246. doi: 10.1002/gene.20095. [DOI] [PubMed] [Google Scholar]
- 53.Bahar Halpern K, et al. Bursty Gene Expression in the Intact Mammalian Liver. Molecular Cell. 2015;58:147–156. doi: 10.1016/j.molcel.2015.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lyubimova A, et al. Single-molecule mRNA detection and counting in mammalian tissue. Nat Protocols. 2013;8:1743–1758. doi: 10.1038/nprot.2013.109. [DOI] [PubMed] [Google Scholar]
- 55.Seglen PO. Preparation of rat liver cells. 3. Enzymatic requirements for tissue dispersion. Exp Cell Res. 1973;82:391–398. doi: 10.1016/0014-4827(73)90357-1. [DOI] [PubMed] [Google Scholar]
- 56.Butler A, Satija R. Integrated analysis of single cell transcriptomic data across conditions, technologies, and species. bioRxiv. 2017 doi: 10.1101/164889. 164889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shay T, Kang J. Immunological Genome project and systems immunology. Trends Immunol. 2013;34:602–609. doi: 10.1016/j.it.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.