Abstract
There is no reliable cure for chronic Hepatitis B Virus (HBV). In this issue of Structure, Eren et al. (2018) show how antibody-derived proteins bind different forms of the HBV capsid protein, blocking assembly. This interaction may also affect downstream signaling. These antibody-derived molecules mark a new strategy that may ultimately contribute to a cure.
HBV is an endemic disease. It is prevalent in Asia and Africa, with around 257 million people suffering from chronic hepatitis B (CHB). HBV contributes to more than 800,000 deaths annually, according to the World Health Organization, mainly from liver failure, cirrhosis, and hepatocellular carcinoma. There is a high incidence of HIV-HBV coinfection that introduces new challenges to patient care for both diseases.
Despite the outsize footprint of HBV on human health, its genome has only 3,200 base pairs encoding four overlapping open reading frames: core protein, surface antigen, polymerase, and X protein (a factor regulating transcription and persistence of viral DNA) (Venkatakrishnan and Zlotnick, 2016). The virion has a lipid envelope studded with surface protein. This envelope surrounds an icosahedral capsid composed of 120 core protein dimers; the capsid surrounds the viral genome and a copy of polymerase. In an infection, core protein assembles into empty capsids or specifically packages one copy of the pregenomic RNA and a copy of polymerase. Once packaged, the polymerase reverse transcribes the linear RNA to the circular, partially double-stranded DNA of the mature virus. The mature virus can go on to acquire an envelope or get transported back to the nucleus. In the nucleus, the DNA is released, repaired by host enzymes, decorated with nucleosomes, and serves as the template for viral RNA.
There are two products of the core gene, HBV core antigen (HBcAg) and HBV e antigen (HBeAg), initiated from different, in-frame start codons and resulting in two unique homo-dimeric proteins with different structures and functions. Both proteins have the same 149 residue assembly domain (Venkatakrishnan and Zlotnick, 2016). HBcAg retains a highly basic C-terminal domain. The HBcAg accounts for capsid assembly and genome packaging to form the nucleocapsid core, where it affects intracellular trafficking and reverse transcription. It is also associated with nuclear viral DNA. HBeAg starts at the earlier methionine, resulting in an N-terminal signal sequence that is removed at the ER and leaving behind a 10-residue propeptide (DiMattia et al., 2013). About 70%–85% of HBeAg is secreted, losing all or most of the C-terminal basic domain. Secreted HBeAg acts as an immune modulator suppressing inflammatory and non-inflammatory T cells (Milich et al., 1990). The HBeAg that is retained in cytoplasm can assemble into decoy capsids and also modulate the innate immune response.
Direct-acting antivirals have been designed to target specific steps of the viral life cycle. Nucleotide analogs and reverse transcriptase inhibitors suppress viremia and improve liver health, but almost never lead to viral clearance even after years (Levrero et al., 2018). Myrcludex B, a peptide based on the HBV surface protein, is a competitive inhibitor for HBV binding to its cellular receptor (Lempp and Urban, 2014). siRNA targeting viral transcripts has also shown encouraging results (Wooddell et al., 2017). Core protein assembly modulators (CpAMs) act by promoting capsid assembly, leading to empty capsids and/or misdirected assembly products (Schlicksup et al., 2018, Venkatakrishnan and Zlotnick, 2016). Preliminary clinical studies have shown that all of these approaches can suppress virus production, but longer trials will be required to see if these new strategies can lead to or contribute to a cure.
In this issue of Structure, Wingfield and colleagues (Eren et al., 2018) present a new approach to fight CHB: targeting HBeAg and dimeric HBcAg. They present three crystal structures of HBeAg and HBcAg dimers in complex with a Fab fragment or an scFV that binds with sub-nanomolar affinity (Zhuang et al., 2017). The HBeAg and HBcAg monomers are very similar: an all-helix protein where long helices 3 and 4 form the intradimer interface and helix 5, along with its following loop and extended structure, has the potential to form the interdimer contacts that support capsid assembly (Figure 1A). The dimer interfaces of HBcAg and HBeAg are amazingly different (Figure 1B). In HBcAg, the dimer interface is parallel and forms a spike that extends from the capsid surface; in all known HBcAg structures, the base of the spike is structurally highly conserved (Venkatakrishnan and Zlotnick, 2016). In HBeAg, the dimer interface is antiparallel, held in place by a disulfide from helix 3 to the propeptide (DiMattia et al., 2013). The HBeAg structure from Eren et al. (2018) has a monomer-monomer angle that is substantially different from the angle in the previously determined HBeAg structure (DiMattia et al., 2013) (Figure 1B). Given the structural changes observed in HBeAg and HBcAg structures and in molecular dynamics simulations (Venkatakrishnan and Zlotnick, 2016, Hadden et al., 2018), both are flexible proteins and binding to antibodies can alter their conformations and activities.
Figure 1. Dimeric Forms of the HBV Core Gene Products.
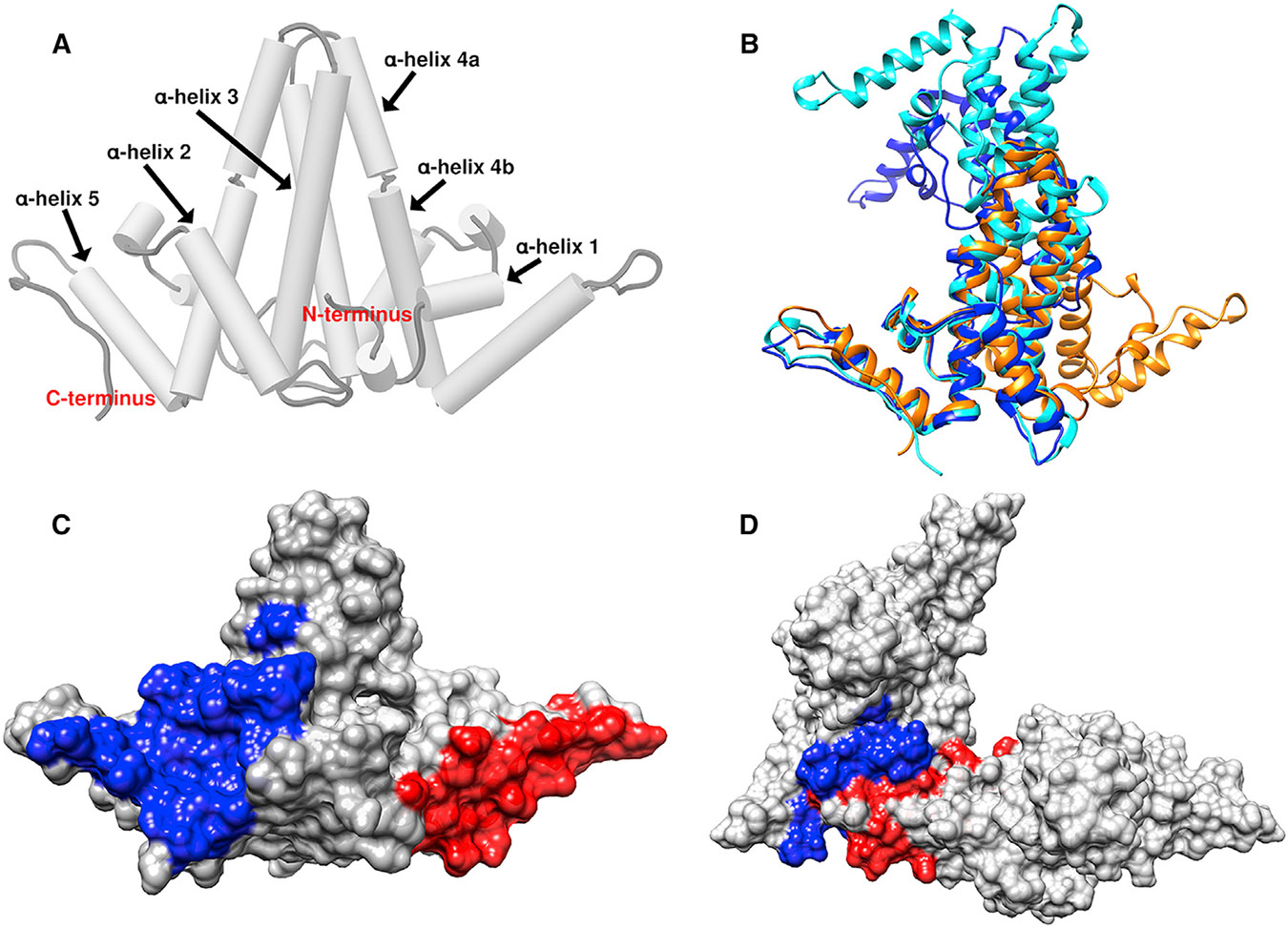
(A) The capsid (HBcAg) conformation has a spike at the intradimer interface formed by helices 3 and 4 from both monomers. Helix 5 and the following sequence form the interdimer interface.
(B) A comparison of HBcAg from capsid (orange, 1QGT), the older HBeAg structure (blue, 3V6Z), and the new HBeAg dimer (cyan, 6CVK). The three structures are aligned based on one monomer; the other halves of the dimers are in substantially different positions.
(C) An HBcAg dimer with a molecular surface highlighting the epitopes bound by FAB e21 (red) and scFV e13 (blue) (see table S1 in Eren et al. (2018)). Both epitopes include helix 5 and are distal to the major epitope of HBcAg, the tip of the spike.
(D) Two HBcAg dimers, extracted from a capsid, interact via contacts with helix 5. These interactions occlude e13 and e21 epitopes.
The two novel antibody-derived proteins presented in this paper, scFv e13 and Fab e21, have extremely high affinity (in the picomolar range) for the dimer states of HBcAg and HBeAg. The epitopes for these two antibodies do not overlap and are completely separate from the major antigenic site of HBcAg at the tip of the spike (Venkatakrishnan and Zlotnick, 2016). scFv e13 primarily interacts with α helix 2, α helix 4b, and α helix 5, whereas the Fab e21 mainly binds to α helix 1 and α helix 5 of both proteins. These epitopes overlap the sites of interdimer interaction (Figures 1C and 1D).
Thus, e13 and e21 led Eren et al. (2018) to propose a new antiviral strategy: trapping HBcAg and HBeAg in the dimeric state. This interaction blocks assembly in vitro and will presumably have the same effect in vivo, preventing the formation of real and “decoy” capsids. Interestingly the two molecules do not disrupt intact capsids. The authors speculate that bound antibody will also affect other intracellular activities of HBcAg and HBeAg dimers. Also, extracellularly, bound e13 or e21 may attenuate immunomodulatory activity of secreted HBeAg.
Though the target is the same protein, if scFV e13 and FAB e21 are assembly antagonists, then CpAMs would be agonists that stimulate over-assembly of capsid and effectively deplete dimer concentration. The two approaches are not mutually exclusive. Eren et al. (2018) have enriched the landscape of possible combination therapies for HBV by identifying two antibodies with special binding sites and strong binding affinity that target multiple viral processes.
REFERENCES
- DiMattia MA, Watts NR, Stahl SJ, Grimes JM, Steven AC, Stuart DI, and Wingfield PT (2013). Antigenic switching of hepatitis B virus by alternative dimerization of the capsid protein. Structure 21, 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren E, Watts NR, Dearborn AD, Palmer IW, Kaufman JD, Steven AC, and Wingfield PT (2018). Structures of hepatitis b virus core- and e-antigen immune complexes suggest multi-point inhibition. Structure 26, this issue, 1314–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadden JA, Perilla JR, Schlicksup CJ, Venkatakrishnan B, Zlotnick A, and Schulten K (2018). All-atom molecular dynamics of the HBV capsid reveals insights into biological function and cryo-EM resolution limits. eLife 7, e32478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempp FA, and Urban S (2014). Inhibitors of hepatitis B virus attachment and entry. Intervirology 57, 151–157. [DOI] [PubMed] [Google Scholar]
- Levrero M, Subic M, Villeret F, and Zoulim F (2018). Perspectives and limitations for nucleo(t) side analogs in future HBV therapies. Curr. Opin. Virol 30, 80–89. [DOI] [PubMed] [Google Scholar]
- Milich DR, Jones JE, Hughes JL, Price J, Raney AK, and McLachlan A (1990). Is a function of the secreted hepatitis B e antigen to induce immunologic tolerance in utero? Proc. Natl. Acad. Sci. USA 87, 6599–6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicksup CJ, Wang JC, Francis S, Venkatakrishnan B, Turner WW, VanNieuwenhze M, and Zlotnick A (2018). Hepatitis B virus core protein allosteric modulators can distort and disrupt intact capsids. eLife 7, e31473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan B, and Zlotnick A (2016). The structural biology of hepatitis b virus: form and function. Annu. Rev. Virol 3, 429–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooddell CI, Yuen MF, Chan HL, Gish RG, Locarnini SA, Chavez D, Ferrari C, Given BD, Hamilton J, Kanner SB, et al. (2017). RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci. Transl. Med 9, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Watts NR, Palmer IW, Kaufman JD, Dearborn AD, Trenbeath JL, Eren E, Steven AC, Rader C, and Wingfield PT (2017). Chimeric rabbit/human Fab antibodies against the hepatitis Be-antigen and their potential applications in assays, characterization, and therapy. J. Biol. Chem 292, 16760–16772. [DOI] [PMC free article] [PubMed] [Google Scholar]


