Abstract
Porcine circovirus 2 (PCV-2) is a virus characterized by a high evolutionary rate, promoting the potential emergence of different genotypes and strains. Despite the likely relevance in the emergence of new PCV-2 variants, the subtle evolutionary patterns of PCV-2 at the individual-host level or over short transmission chains are still largely unknown. This study aimed to analyze the within-host genetic variability of PCV-2 subpopulations to unravel the forces driving PCV-2 evolution. A longitudinal weekly sampling was conducted on individual animals located in three farms after the first PCV-2 detection. The analysis of polymorphisms evaluated throughout the full PCV-2 genome demonstrated the presence of several single nucleotide polymorphisms (SNPs) especially in the genome region encoding for the capsid gene. The global haplotype reconstruction allowed inferring the virus transmission network over time, suggesting a relevant within-farm circulation. Evidences of co-infection and recombination involving multiple PCV-2 genotypes were found after mixing with pigs originating from other sources. The present study demonstrates the remarkable within-host genetic variability of PCV-2 quasispecies, suggesting the role of the natural selection induced by the host immune response in driving PCV-2 evolution. Moreover, the effect of pig management in multiple genotype coinfections occurrence and recombination likelihood was demonstrated.
Introduction
Porcine circovirus 2 (PCV-2) is a non-enveloped single-stranded DNA virus with a small circular genome of 1766–1768 bp1. The PCV-2 genome consists in at least four major open reading frames (ORFs) encoding the replicase protein (ORF1), the viral capsid protein (ORF2) and proteins with either apoptotic (ORF3) or anti-apoptotic capacities (ORF4)2,3. PCV-2 belongs to the genus Circovirus from the Circoviridae family, together with two other porcine circoviruses, PCV-14 and PCV-35. Although its small size, PCV-2 is of great importance in the global swine industry economy, since it is the primary causative agent of porcine circovirus diseases (PCVD)6, especially PCV-2 systemic disease (PCV-2-SD, formerly known as postweaning multisystemic wasting syndrome), which causes severe losses in intensive pig production worldwide7. PCV-2 has a complex epidemiology, featured by an effective among-animal transmission due to the different shedding routes and the long viral persistence in infected animals8.
Four main genotypes have been determined based on the sequence analysis of the whole genome and cap gene, with PCV-2a, PCV-2b, PCV-2c and PCV-2d being globally distributed9. More recently, two additional genotypes have been proposed10,11. ORF2 is particularly relevant because of not only a higher genetic heterogeneity, facilitating viral classification and molecular epidemiology studies, but also for its relevance in the host immune response12–14. The genetic variability of this virus has been demonstrated to be remarkable, with the highest evolutionary rate among DNA viruses15. Several mutations, particularly in the ORF2 region have been reported worldwide with different outcomes both in vivo and in vitro, with recombinant strains being also relevant in the distribution of different genotypes12. Moreover, during the last decades, novel variants of PCV-2 have been identified16,17, increasing the interest in monitoring the possibility of emergence of new strains and the role of these circulating strains in PCV-2 evolution. The evolutionary pathways of this virus have shown to be particularly relevant regarding the clinical manifestations occurring through time12. Despite the likely relevance in the emergence of new PCV-2 variants, the subtle evolutionary patterns of PCV-2 at individual-host level or over short transmission chains are still largely unknown. The aim of this study was to analyze the genetic variability of PCV-2 subpopulations at individual pig level in different farms and unravel the forces driving PCV-2 evolution in vivo.
Results
A total of 20 piglets from each of three different Spanish farms analyzed (farms A-C) were sampled and checked weekly to detect their first evidence of PCV-2 infection in life. Once detected, 5 PCV-2-positive animals per farm were sampled weekly for 4 weeks post infection (wpi) and the full PCV-2 genome was sequenced using next generation technology. A summary of PCV-2 positive animal sampling and sequencing results is reported in Supplementary Fig. S1. Briefly, a mean coverage greater than approximately 300 reads was obtained for all the pigs included in the study, with the only exception of pig 737 at week 8 (farm C) and 776 at week 11 (farm A).
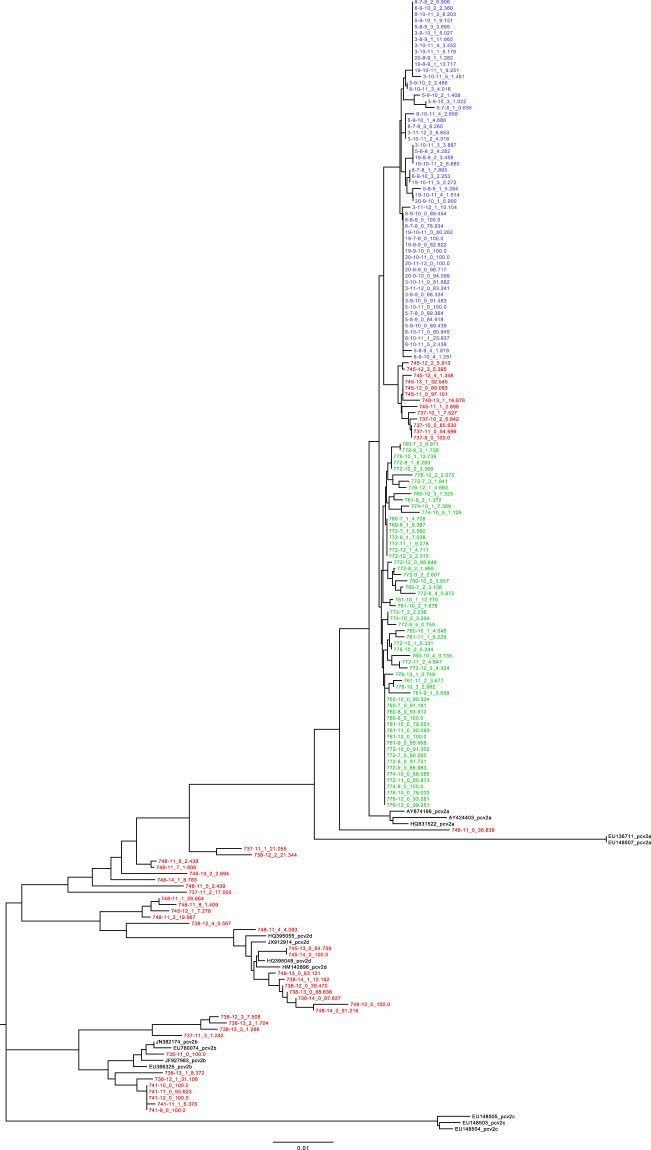
The study of viral global haplotypes performed at individual animal level (Fig. 1) revealed the presence of remarkably different scenarios among the farms. Particularly, all strains sampled from both farm A and B formed two distinct farm-specific clades belonging to the PCV-2a genotype. In farm C, PCV-2b genotype was the only one detected until 11 weeks of age. At that age, all pigs were relocated within fattening facilities remaining exclusively in contact with pigs from the same origin except for farm C where they were mixed with animals from different herds. After this event, the scenario became much more complex in farm C and strains belonging to different genotypes (PCV-2a, PCV-2b and PCV-2d) were identified in different animals at different time points. Additionally, several co-infections with multiple genotypes were observed and both tree topology and recombination analysis suggested the presence of several recombination events. Based on these results, the experiment was further split into two studies. Farms A and B were selected to evaluate the pattern of PCV-2 genetic variability within and between animals over time (Study 1). The presence of multiple genotypes impeded this analysis in farm C since most of the variability would be attributable to the long term evolution separating the PCV-2 genotypes. At the same time, this genetic distance facilitates the differentiation of different strains despite the unavoidable limits inherent to NGS data analysis. Consequently, farm C was selected to investigate the within-farm spreading pattern of PCV-2 and its consequences (Study 2).
Figure 1.
Maximum likelihood phylogenetic tree reconstructed using the ORF2 gene of estimated haplotypes and the reference sequences proposed by Franzo et al., 2015. Individual haplotypes have been named using the following scheme: AnimalID-SamplingWeek_HaplotypeNumber_HaplotypePrevalence. Farm A, B and C have been color-coded in green, blue and red, respectively.
Study 1: Analysis of PCV-2 genetic variability over time
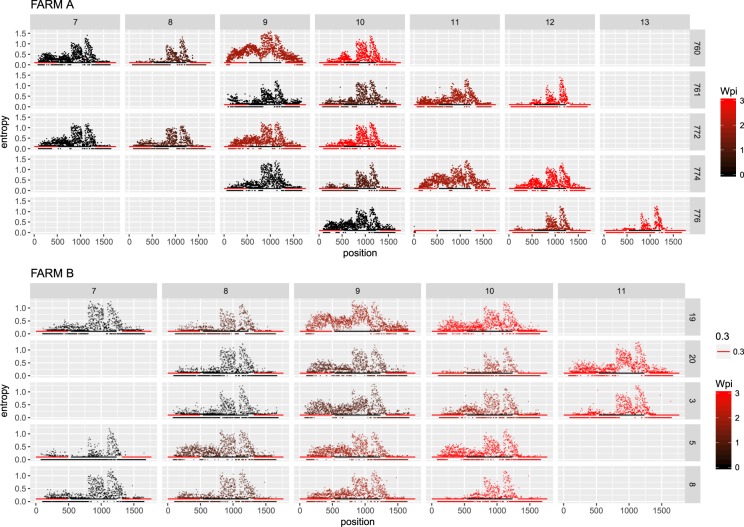
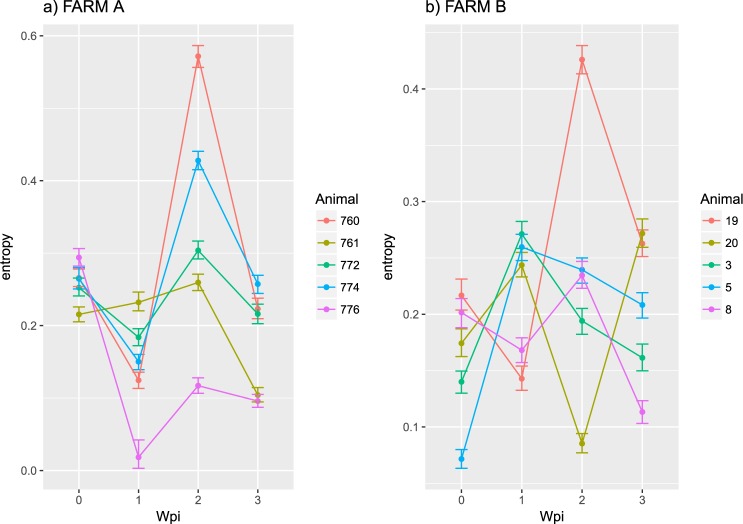
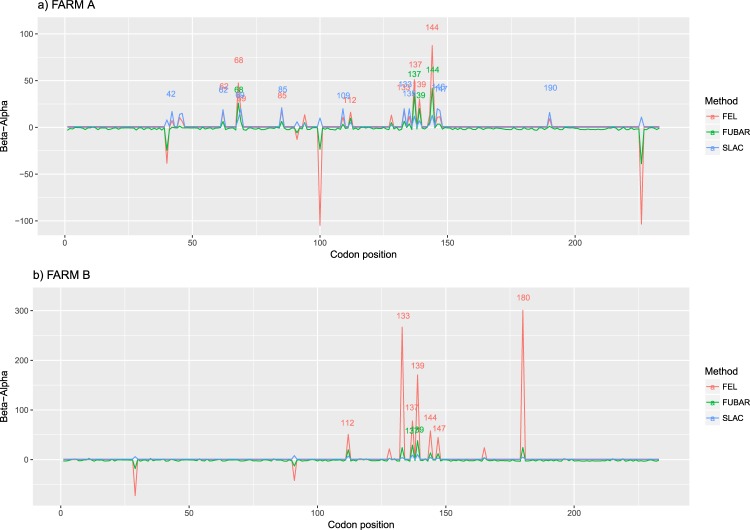
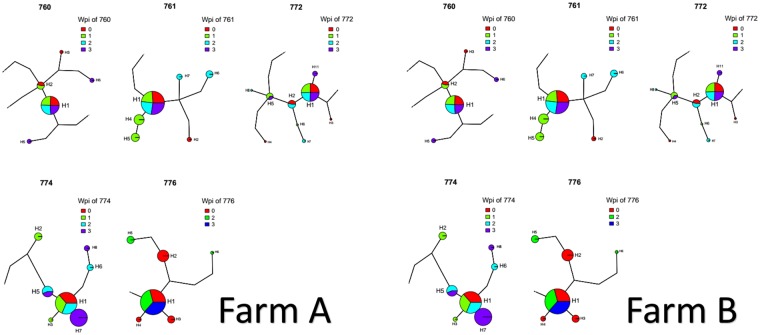
Entropy was calculated throughout the whole genome to obtain a statistical summary representing the genetic variability at each genome position. The overall pattern analysis evidenced higher variability in the ORF2 region compared to the rest of the genome, irrespective of the farm, animal and sampling date, as depicted in Fig. 2. A comparable scenario was observed when nucleotide frequencies were considered. Despite the more stringent single nucleotide variation (SNV) acceptance criteria, several subpopulations were detected both in farm A and B (Supplementary Fig. S2). Although some SNV were identified in the ORF1 region, most of them affected the ORF2. The average entropy displayed different trends when analyzed in the two farms under study. In farm A, there was a fluctuating pattern in overall variability, which showed a decreased tendency at 1 wpi, increased at the following sampling week and, finally, decreased by the end of the study (Fig. 3a). However, the scenario was much more complex in farm B, where entropy patterns over time differed remarkably among animals and a consistent trend could hardly be identified (Fig. 3b). In both farms, individual animals showed peculiar patterns in viral genetic heterogeneity, with some pigs evidencing major fluctuations in entropy values over time while an essentially constant variability was detected in others through the whole study period. Despite these differences, some similarities were observed in the analysis of selective pressures acting on the Cap gene of those strains. Particularly, an area under diversifying selection was observed approximately between AA 130 and 150 in sequences from both farms, while some evidences of a certain selective diversifying selection were observed to act on other protein regions only for sequences obtained from farm A (Fig. 4). Nevertheless, it must be stressed that only for some specific positions the deviation from neutral selection was statistically significant. Particularly, the MEME method, able to detect episodic diversifying selection, detected evidences of positive selection in AA position 137 (p-value 0.08) in farm A and at codons 133 (p-value 0.10) and 180 (p-value 0.08) in farm B. A summary of codons detected under diversifying selection using other methods is reported in Table 1 and depicted in the protein 3D-structure in Supplementary Fig. S3 (see also Supplementary Video). Similarly, the haplotype network analysis performed in the ORF2 gene demonstrated a fairly repeatable dynamics among different farms and animals, characterized by the presence of a major haplotype persisting for the whole period of time (except for animal 774 at 3 wpi) and minor ones emerging over time. Some of those haplotypes were detected only once, while others were able to persist for longer periods of time, as shown in Fig. 5. No evidences of a relevant correlation between humoral immune response and PCV-2 variability were apparent either globally or in sub-genomic regions (Supplementary Figs S4 and S5).
Figure 2.
Entropy value at each genome position are reported for farm A (top) and farm B (bottom). Data for different animal and week of age have been reported separately. Week post infection has been color coded. The ORF1 and ORF2 have been respectively represented as red and black line in the lower part of each graph.
Figure 3.
Mean entropy (point) and 95 confidence intervals (error-bars; calculated using bootstrap) are reported for each animal at different wpi.
Figure 4.
Estimate of selective pressures acting on the Cap gene. Normalized dN–dS is displayed for each codon position. Results of SLAC, FUBAR and FEL method have been color-coded.
Table 1.
Sites under diversifying selection in farm A (upper) and farm B (lower).
| POSITION | FEL | FUBAR | SLAC | |||
|---|---|---|---|---|---|---|
| beta-alpha | p-value | beta-alpha | Pos. Prob. | beta-alpha | p-value | |
| 68 | 47.485 | 0.187 | 26.094 | 0.955 | 26.007 | 0.297 |
| 112 | 16.192 | 0.275 | 9.704 | 0.904 | 15.625 | 0.444 |
| 137 | 50.997 | 0.057 | 34.919 | 0.988 | 46.875 | 0.087 |
| 139 | 30.033 | 0.238 | 20.113 | 0.951 | 19.558 | 0.509 |
| 144 | 87.469 | 0.111 | 41.912 | 0.977 | 54.687 | 0.303 |
| 112 | 50.779 | 0.261 | 19.922 | 0.935 | 41.509 | 0.444 |
| 133 | 266.901 | 0.077 | 24.066 | 0.904 | 20.754 | 0.666 |
| 137 | 77.874 | 0.169 | 29.263 | 0.966 | 62.264 | 0.087 |
| 139 | 170.577 | 0.214 | 38.084 | 0.962 | 63.036 | 0.509 |
| 180 | 300.844 | 0.055 | 24.432 | 0.911 | 37.907 | 1 |
Figure 5.
Haplotype network drawn for each animal (animal ID is reported above the graph). The size of the circle is proportional to the prevalence of the haplotype while the different week post infection have been color coded. The five animals from farm A are represented on the left while the ones from farm B are represented on the right.
Study 2. Spreading pattern of PCV-2 and its consequences
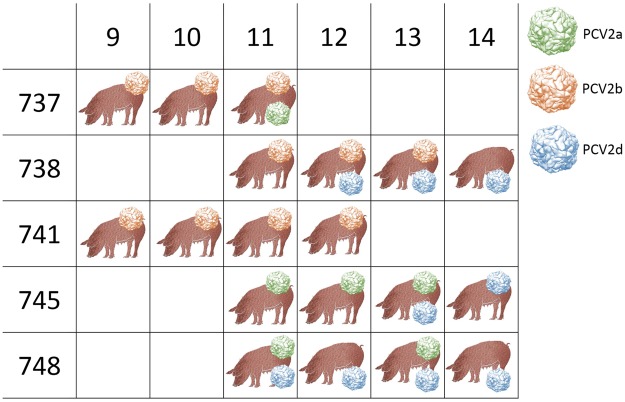
The analysis of viral circulation within the herd showed a remarkable variation in the farm C epidemiological scenario after the mixing with animals from different origins. While two animals were already infected with PCV-2b in the origin farm, the other became positive with PCV-2a or PCV-2d after the mixing (Fig. 6). In 4 out of 5 animals, co-infections were detected, persisting for more than two weeks in 3 out of 4 pigs (Fig. 6). The co-infection presence was further confirmed by the SNV analysis using LoFreq, which showed an extremely high SNV frequency in some animals (Supplementary Fig. S2). In all co-infection events where PCV-2d was identified, it persisted longer than other genotypes, suggesting a better fitness of this genotype or, at least, of the particular strain. Because PCV-2d infection occurred after the other genotypes in 2 out of 3 pigs, a still incomplete clearance by the host immune system cannot be excluded. Additionally, a strong recombination signal was detected in the haplotype alignment and RDP4 analysis identified 18 recombinant haplotypes emerging after animal mixing (Table 2).
Figure 6.
Summary of genotypes (color-coded) detected in different animals at different weeks of age. All considered animals originated from farm C.
Table 2.
Detected recombinant strains and the genotype of the minor and major parental strains are shown together with the potential recombination breakpoints.
| Identified Recombinant | Beginning breakpoint | Ending breakpoint | Minor parent | Major parent | Recombination event |
|---|---|---|---|---|---|
| 745-13_2 | 386 | 681 | PCV2a | PCV2b | Event 1 |
| 748-11_6 | |||||
| 748-11_7 | |||||
| 748-11_5 | 314 | 699 | PCV2a | PCV2b | Event 2 |
| 737-11_1 | |||||
| 738-12_2 | |||||
| 748-14_1 | |||||
| 748-11_0 | 79 | 500 | PCV2b | PCV2d | Event 3 |
| 738-12_4 | 441 | 702 | PCV2b | PCV2d | Event4 |
| 737-11_2 | |||||
| 745-12_1 | |||||
| 748-11_1 | |||||
| 748-11_2 | |||||
| 748-11_8 | |||||
| 738-13_2 | 81 | 454 | PCV2b | PCV2d | Event 5 |
| 737-11_3 | |||||
| 738-12_3 | |||||
| 738-13_3 |
Discussion
PCV-2 is characterized by a high substitution and recombination rate18, which has led to the remarkable genetic heterogeneity demonstrated worldwide9. Even if overall features of PCV-2 variability have been investigated at epidemiological level, the fine mechanism acting at individual level and their overall impact are still obscure.
The present study, evaluating the within animal viral variability, found a remarkable difference in terms of entropy between the Rep and Cap regions. Such a marked heterogeneity cannot be realistically explained either by a differential viral replication fidelity or by variable NGS error rates. Although the presence of both PCR and sequencing errors together with the complexity in differentiating them from true variations still represents one of the main challenge in the use of NGS for viral subpopulation estimation19,20, the consistency of the results obtained from different animals and farms in this study, as well as their biological plausibility, supports the observations described herein. Therefore, a combination of selective constraints acting on proteins involved in viral replication and the presence of multiple ORFs in different orientation and reading frames1,12 could limit the plasticity of the Rep coding region by causing most of the mutations to be deleterious and rapidly purged by natural selection. On the other hand, the Cap gene could benefit from a higher genetic plasticity because of the immunological pressures favouring the emergence of immune response evading variants. Capsid entropy demonstrated variable patterns among individuals. Interestingly, several animals showed a fluctuating trend, which was already described for another persisting swine infection like porcine reproductive and respiratory syndrome virus (PRRSV) and ascribed to the interaction with the host immune response21. Nevertheless, several exceptions were observed among individual animals and, more generally, an overall different pattern was found between farms A and B. Therefore, the observed scenario is likely the result of a complex network of interactions whose outcome depends on peculiar host and viral strain features as well as environmental conditions not measured here. This high background noise surely hampers the understanding of the underling biological process, and larger studies will be required. Similarly, the individual variability can probably explain the absence of any clear relationship between viral heterogeneity and host immune response. Additionally, the global IgM and IgG titers represent a quite raw measure to investigate the fine interactions between a swarm of viral variants and a population of epitope-specific antibodies and immune system cells. Analogous conclusions can be drawn regarding the evaluation of virus neutralization titers, since different epitopes, antibody classes and lymphocyte clones are likely involved in the phenomena. Nevertheless, despite the limited animal number and the apparent individual peculiarities, several common patterns could be identified.
The existence of sensu stricto quasispecies in vivo is still highly debated; however, the presence of a complex network of viral subpopulations or quasispecies, evolving within the same individual over time is demonstrated in the present study for the first time for PCV-2. The analysis of the Cap region showed the presence of a major haplotype, typically persisting during the whole study duration, together with other haplotypes that showed variable prevalence and persistence. Comparable results were obtained by the analysis of SNV over time. Therefore, the described scenario is suggestive of the existence of a stable reservoir population from which different evolutionary paths do occur. Even if PCV-2 is recognized as a rapidly evolving virus18, the variability herein observed is quite surprising since several mutations were directed towards amino-acidic changes (i.e. leading to phenotypic modifications). In fact, the analysis of selective pressures demonstrated that a neutral to positive selection was primarily acting on the viral capsid. Although the number of animals limited the study power and expose to errors due to random fluctuations, several evidences pose in favor of the results’ robustness. Particularly, several factors were consistently observed on the two farms including similar dN/dS ratios and statistically significant sites. Additionally, all sites detected to be under significant diversifying selection were exposed on the viral surface or, at least, on the capsomer surface, which is compatible with an immune derived immune selection. Therefore, the picture described here enforces the pivotal role of immune derived selective pressure in shaping the within-host PCV-2 diversity. However, the present results are in contrast of other studies demonstrated a purifying selection significantly acting on PCV-2 capsid12.
Different biological processes could explain this discrepancy. Firstly, the study duration could have been too short for the removal of variants carrying deleterious mutations, leading to an excess of non-synonymous mutations. Secondly, non-synonymous mutations useful in evading individual pig immune responses could be over specific and negatively affecting the transmission or population level fitness. Interestingly, a similar gap between individual host and population level evolutionary rate has been reported for other viruses22,23.
Although a high within-host heterogeneity does not automatically entail a high overall viral fitness, it can represent an effective strategy for within host persistence. At the same time the forces favoring intra-animal diversification can lead to the genesis of some variants that are able to succeed and spread on a broader scale24. At least two positions in the capsid (i.e. 68 and 133) proven to be under diversifying selection in the present study, were also reported to be under this kind of selection in other studies investigating PCV-2 evolution12 and vaccine escape25. Additionally, mutations inside the capsid region at position 133, have been shown to affect cross-reactivity to monoclonal antibodies in experimental studies26. Based on these results, the reduction of PCV-2 circulation and persistence, achievable by proper biosecurity measures and vaccination application, appears of major importance since it can constrain the evolutionary potential of this virus and the consequent emergence of variants with different virulence or antigenic/immunological features.
The analysis of viral sub-populations in farm C demonstrated the impact of animal management in herd PCV-2 epidemiology. After animal mixing, the relatively homogeneous PCV-2b population was replaced by a mixed genotype circulation with several co-infection events. The most likely explanation is that animals from different sources were infected with different strains which were rapidly spread among animals. This is clinically relevant for several reasons. At first, co-infections with multiple genotypes have been proposed as a risk factor for clinical signs development27 and even when no overt clinical signs were observed as in this study, a negative effect on productive performances is still likely28. Co-infection is also a prerequisite for recombination to occur. Several recombinant strains were identified in the present study, all originating after animal mixing. The occurrence of in silico recombinants cannot be excluded and represents one of the still unsolved-limits of global haplotype reconstruction in NGS data analysis29. However, the virtual simulation made with mixed reads belonging to different genotypes, strongly supports the implemented pipeline ability to accurately reconstruct PCV-2 genotypes minimize in silico recombination. The estimated haplotype frequency mirrored the actual ones, set during artificial read generation independently of the selected reference strain genotype, although deviations were observed only when low frequency variants (<5%) were simulated; in this case extremely low frequency haplotypes (<1%) were sometimes missed. Additionally, closely related recombinant sequences were detected in different animals (i.e. independent NGS runs and data analysis), supporting the actual existence of the recombinant forms rather than technical errors, and suggesting a certain fitness of these strains which were able to persist and infect other pigs. Recombination events have been frequently described in different countries and recombinant clusters have been reported to display a fitness and distribution comparable to the “classical” genotypes12, representing a potential threat for pig farming. Therefore, demonstrating the high within-host recombination frequency, the present study stresses once more the relevance of adequate control measures to reduce opportunities of recombination and its consequences.
The present study illustrates the remarkable genetic variability of PCV-2 at the individual pig level, particularly in the Cap gene, likely driven by the host immune response. Moreover, this study also points out how farm management (animal flow, mixing of animals) played a major role in the occurrence of multiple genotype co-infections and likelihood of recombination.
Methods
Sample collection
Three different farms from the area of Catalonia (Spain) were screened for this study. Sera samples from 20 piglets starting from 3 weeks of age (weaning age), were collected weekly. At 11 weeks of age, all animals from farms A and B were moved to fattening units, remaining in contact only with pigs originating from the same farm in the case of farms A and B. In contrast, animals coming from the third farm (farm C), were mixed with other fattening pigs originating from different herds from the same swine integrator. From 3 weeks of age onwards, serum samples were tested for PCV-2 detection using conventional PCR as previously described30. Quantification of PCV-2 DNA in serum was done as described31 and IgM and IgG titers were estimated from the sixth week onwards using a commercial Ingezim Circo IgG ELISA kit as recommended (Inmunologia y Genetica Aplicada). A total of 5 infected animals from each farm were selected due to both positive detection of PCV-2 in sera and low maternally derived antibody levels. Longitudinal sampling during four weeks after the first positive PCV-2 detection was performed for each piglet. All methods were carried out in accordance with relevant guidelines and regulations. The present study was approved by the Ethics Committee for Animal Experimentation from the Universitat Autònoma de Barcelona and the Animal Experimentation Commission from the local government (Dpt. de Medi Ambient i Habitatge from the Generalitat de Catalunya; Reference 5796).
DNA extraction and sequencing
DNA was extracted from these samples using the QiAmp MiniElute Virus Spin Kit (QIAGEN) and the full PCV-2 genome was amplified using the primer PCV2 SacF TCCGCGGGCTGGCTGAACTTTTGA and PCV2 SacR CCCGCGGAAATTTCTGACAAACGT as described elsewhere32. Amplicons were purified using the QIAquick PCR purification kit, eluted in 30 µL Tris buffer and both quality and quantity of nucleic acids were evaluated on a BioDrop DUO (BioDrop Ltd). Barcoded libraries were individually constructed for each animal and sampling time at Servei de Genomica Autonomous University of Barcelona using the Nextera® XT DNA Sample Preparation Kit (Illumina®, San Diego, CA) and the MiSeqTM Illumina platform (pair-end 2 × 250 bp) following the manufacturer instructions (MS-102-2003 MiSeq® Reagent Kit v2, 500 cycle).
Data analysis
Read quality of obtained FASTQ files was visually inspected using PRINTSEQ33. Reads that fulfilled the following criteria were filtered out, shorter length than 30 nt, average quality lower than 20 or with more than one base with a quality lower than 10. Additionally, adapters and barcodes were removed and trimming of poor quality bases, poly-A/T and poly-N at the 5′ and 3′ tails were performed using the same software.
Overlapping paired reads were merged using mothur34 and aligned to the PCV-2 reference genome (NC_005148) using Bowtie235. Samtools36 was used to convert, sort and remove duplicates from the obtained SAM files. Remaining reads were extracted and converted to FASTA format using the same software. Finally, sample specific coverage and consensus sequence were obtained using QUASR37.
Subpopulation study
The analysis of viral subpopulation was conducted at two levels: single nucleotide variation (SNV and entropy calculation) and global haplotype. To limit the bias introduced by the PCR and sequencing error in viral subpopulation estimation, an error correction approach assuming a Poisson distribution of errors parameterized differently in homopolymeric and non-homopolymeric regions was applied in the obtained FASTA sequences38 and the corrected read sets were used for further analysis. The number of variations, their coverage and the entropy statistic for each genome position was calculated using the QuRe software38. Each position-specific SNV and its frequency was identified with the highly specific LoFreq method39, also implementing a Poisson–binomial distribution model and accounting for base-call quality value as well as other sources of uncertainty to identify true variants from sequencing errors. In order to achieve only highly reliable SNV, the significance level was set to p < 0.001. Additionally, global haplotypes and their prevalence were reconstructed using the heuristic algorithm implemented in QuRe, which matches multinomial distributions of distinct viral variants overlapping across the genome division38. For each sample, the full genome consensus sequence previously obtained was used as reference. The QuRe software was selected because it was proven to provide a good compromise between sensitivity and false positivity in haplotype reconstruction under experimental conditions29. However, to diminish the impact/likelihood of in silico recombination and because of the higher phylogenetic signal only the Cap gene was used in the analyses based on global haplotype reconstruction. Additionally, in silico simulations were performed to evaluate the accuracy of haplotype reconstruction. Paired reads with features (i.e. read length and error rate) comparable with the experimental setting were simulated using the wgsim tool of Samtools36. To evaluate the co-infection detection capability, fastq files were generated based on PCV2a, PCV2b and PCV2d genotype reference sequences and merged in different proportions. A similar approach was used to evaluate haplotype reconstruction performances. Particularly, closely related variants were assumed as reference and used to generate simulated reads which were then mixed in variable proportions. All obtained simulated dataset were analyzed with the same pipeline used for the experimental samples and selecting a PCV2a genome as reference.
Analysis of sub-population evolution and epidemiological relationship
Evidences of recombination between specific haplotypes identified in each farm were assessed using the GARD40 method implemented in HyPhy41 and the Phi test implemented in Splittree442. When recombination was detected, a further characterization was performed using RDP443, adjusting methods’ settings based on dataset features. Individual haplotypes were genotyped by comparing their ORF2 sequence with the reference dataset proposed by Franzo et al.9. Sequences were aligned at amino-acid level and then back translated to nucleotide sequences using the MAFFT44 method implemented in TranslatorX45. Phylogenetic tree was reconstructed using the Maximum likelihood method implemented in PhyML46 selecting as substitution model the one with the lowest Bayesian Information Critera, calculated using Jmodeltest47. Pig specific haplotype networks were reconstructed using the parsimnet function (which find one of the most parsimonious network within a 95% probability of parsimony as defined in Templeton et al.48 implemented in the haplotypes R library. The within sample haplotype prevalence was accounted in the analysis. The presence of selective forces acting on the Cap gene of the identified variants was evaluated on farm-specific alignments using the MEME49, FUBAR50, SLAC and FEL51 methods implemented in HyPhy41. Less stringent significance value (p-value < 0.1) of posterior probability (>0.9) were accepted considering the limited sequence number.
Relationship between genetic variability and Ab
Potential interaction between host antibody response and viral evolution was globally evaluated by comparing the average entropy with antibody levels at different wpi. Considering the specificity of host immune response toward specific regions of viral proteins, a more focused analysis was performed using a sliding window approach: the windows and step sizes were set to 40 and 20 nucleotides, respectively. The relationship between antibody titer and genetic variability was assess by calculating the correlation between mean entropy in the specific windows region and the antibody titer (IgM and IgG). Also, all sera samples were tested for their ability to neutralize PCV-2 by an in vitro neutralization assay. The virus neutralization assay was performed as described previously52. Data obtained from all animals and wpi were used in the correlation calculation.
Electronic supplementary material
Acknowledgements
This work was supported by the Boehringer Ingelheim European Animal Health PCV-2 award (2014). We also would like to thank the Italian Society of Swine Pathology and Farming (SIPAS) for supporting the Short Term Scientific Missions of Giovanni Franzo.
Author Contributions
J.S. and T.K. conceived and planned the experiments. A.L. performed the experiments. F.C.F. and G.F. processed the experimental data, performed the analysis, drafted the manuscript and designed the figures. J.S. and T.K. discussed the results and contributed to the final manuscript.
Data Availability
The entire sequence dataset is available in the NCBI database, BioProject PRJNA450261 (available at http://www.ncbi.nlm.nih.gov/bioproject/450261).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Florencia Correa-Fiz and Giovanni Franzo contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33849-2.
References
- 1.Lv QZ, Guo KK, Zhang YM. Current understanding of genomic DNA of porcine circovirus type 2. Virus Genes. 2014;49:1–10. doi: 10.1007/s11262-014-1099-z. [DOI] [PubMed] [Google Scholar]
- 2.Juhan NM, LeRoith T, Opriessnig T, Meng XJ. The open reading frame 3 (ORF3) of porcine circovirus type 2 (PCV2) is dispensable for virus infection but evidence of reduced pathogenicity is limited in pigs infected by an ORF3-null PCV2 mutant. Virus Res. 2010;147:60–66. doi: 10.1016/j.virusres.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Gao Z, et al. ORF4-protein deficient PCV2 mutants enhance virus-induced apoptosis and show differential expression of mRNAs in vitro. Virus Res. 2014;183:56–62. doi: 10.1016/j.virusres.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 4.Rosario K, et al. Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch. Virol. 2017;162:1447–1463. doi: 10.1007/s00705-017-3247-y. [DOI] [PubMed] [Google Scholar]
- 5.Palinski R, et al. A Novel Porcine Circovirus Distantly Related to Known Circoviruses Is Associated with Porcine Dermatitis and Nephropathy Syndrome and Reproductive Failure. J. Virol. 2017;91:e01879–16. doi: 10.1128/JVI.01879-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segalés J, Allan GM, Domingo M. Porcine circovirus diseases. Anim. Heal. Res. Rev. 2005;6:119–142. doi: 10.1079/AHR2005106. [DOI] [PubMed] [Google Scholar]
- 7.Allan GM, Ellis JA. Porcine Circoviruses: A Review. J. Vet. Diagnostic Investig. 2000;12:3–14. doi: 10.1177/104063870001200102. [DOI] [PubMed] [Google Scholar]
- 8.Franzo G, et al. International trades, local spread and viral evolution: The case of porcine circovirus type 2 (PCV2) strains heterogeneity in Italy. Infect. Genet. Evol. 2015;32:409–415. doi: 10.1016/j.meegid.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Franzo G, et al. Revisiting the taxonomical classification of Porcine Circovirus type 2 (PCV2): still a real challenge. Virol. J. 2015;12:131. doi: 10.1186/s12985-015-0361-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harmon KM, et al. Whole-Genome Sequences of Novel Porcine Circovirus Type 2 Viruses Detected in Swine from Mexico and the United States. Genome Announc. 2015;3:e01315. doi: 10.1128/genomeA.01315-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bao F, et al. Retrospective study of porcine circovirus type 2 infection reveals a novel genotype PCV2f. Transbound. Emerg. Dis. 2018;65:432–440. doi: 10.1111/tbed.12721. [DOI] [PubMed] [Google Scholar]
- 12.Franzo G, Cortey M, Segalés J, Hughes J, Drigo M. Phylodynamic analysis of porcine circovirus type 2 reveals global waves of emerging genotypes and the circulation of recombinant forms. Mol. Phylogenet. Evol. 2016;100:269–280. doi: 10.1016/j.ympev.2016.04.028. [DOI] [PubMed] [Google Scholar]
- 13.Mahé D, et al. Differential recognition of ORF2 protein from type 1 and type 2 porcine circoviruses and identification of immunorelevant epitopes. J. Gen. Virol. 2000;81:1815–1824. doi: 10.1099/0022-1317-81-7-1815. [DOI] [PubMed] [Google Scholar]
- 14.Darwich L, Mateu E. Immunology of porcine circovirus type 2 (PCV2) Virus Res. 2012;164:61–67. doi: 10.1016/j.virusres.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Karuppannan Anbu, Opriessnig Tanja. Porcine Circovirus Type 2 (PCV2) Vaccines in the Context of Current Molecular Epidemiology. Viruses. 2017;9(5):99. doi: 10.3390/v9050099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo L, et al. A porcine circovirus type 2 (PCV2) mutant with 234 amino acids in Capsid protein showed more virulence in vivo, compared with classical PCV2a/b strain. PLoS One. 2012;7:e41463. doi: 10.1371/journal.pone.0041463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Opriessnig T, et al. Mutant USA strain of porcine circovirus type 2 (mPCV2) exhibits similar virulence to the classical PCV2a and PCV2b strains in caesarean-derived, colostrum-deprived pigs. J. Gen. Virol. 2014;95:2495–2503. doi: 10.1099/vir.0.066423-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Firth C, Charleston MA, Duffy S, Shapiro B, Holmes EC. Insights into the evolutionary history of an emerging livestock pathogen: porcine circovirus 2. J. Virol. 2009;83:12813–21. doi: 10.1128/JVI.01719-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCrone JT, Lauring AS. Measurements of Intrahost Viral Diversity Are Extremely Sensitive to Systematic Errors in Variant Calling. J. Virol. 2016;90:6884–95. doi: 10.1128/JVI.00667-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Posada-Cespedes S, Seifert D. Recent advances in inferring viral diversity from high-throughput sequencing data. Virus Res. 2017;239:17–32. doi: 10.1016/j.virusres.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 21.Chen N, Trible BR, Kerrigan MA, Tian K, Rowland RRR. ORF5 of porcine reproductive and respiratory syndrome virus (PRRSV) is a target of diversifying selection as infection progresses from acute infection to virus rebound. Infect. Genet. Evol. 2016;40:167–175. doi: 10.1016/j.meegid.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 22.Duchêne S, Holmes EC, Ho SYW. Analyses of evolutionary dynamics in viruses are hindered by a time-dependent bias in rate estimates. Proceedings. Biol. Sci. 2014;281:20140732. doi: 10.1098/rspb.2014.0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pybus OG, Fraser C, Rambaut A. Evolutionary analysis of the dynamics of viral infectious disease. Phil. Trans. R. Soc. B. 2013;368:20120193. doi: 10.1098/rstb.2012.0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franzo Giovanni, Tucciarone Claudia Maria, Cecchinato Mattia, Drigo Michele. Reconciling individual and population levels of porcine reproductive and respiratory syndrome virus evolution. Virologica Sinica. 2017;32(4):342–345. doi: 10.1007/s12250-017-3981-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franzo G, Tucciarone CM, Cecchinato M, Drigo M. Porcine circovirus type 2 (PCV2) evolution before and after the vaccination introduction: A large scale epidemiological study. Sci. Rep. 2016;6:39458. doi: 10.1038/srep39458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saha D, et al. Antigenic subtyping and epitopes’ competition analysis of porcine circovirus type 2 using monoclonal antibodies. Vet. Microbiol. 2012;157:13–22. doi: 10.1016/j.vetmic.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 27.Harding JCS, Ellis JA, McIntosh KA, Krakowka S. Dual heterologous porcine circovirus genogroup 2a/2b infection induces severe disease in germ-free pigs. Vet. Microbiol. 2010;145:209–219. doi: 10.1016/j.vetmic.2010.03.026. [DOI] [PubMed] [Google Scholar]
- 28.Alarcon P, Rushton J, Wieland B. Cost of post-weaning multi-systemic wasting syndrome and porcine circovirus type-2 subclinical infection in England - An economic disease model. Prev. Vet. Med. 2013;110:88–102. doi: 10.1016/j.prevetmed.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prosperi MCF, et al. Empirical validation of viral quasispecies assembly algorithms: state-of-the-art and challenges. Sci. Rep. 2013;3:2837. doi: 10.1038/srep02837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quintana J, et al. Experimental inoculation of porcine circoviruses type 1 (PCV1) and type 2 (PCV2) in rabbits and mice. Vet. Res. 2002;33:229–237. doi: 10.1051/vetres:2002011. [DOI] [PubMed] [Google Scholar]
- 31.Olvera A, Sibila M, Calsamiglia M, Segalés J, Domingo M. Comparison of porcine circovirus type 2 load in serum quantified by a real time PCR in postweaning multisystemic wasting syndrome and porcine dermatitis and nephropathy syndrome naturally affected pigs. J. Virol. Methods. 2004;117:75–80. doi: 10.1016/j.jviromet.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Kekarainen T, Gonzalez A, Llorens A, Segalés J. Genetic variability of porcine circovirus 2 in vaccinating and non-vaccinating commercial farms. J. Gen. Virol. 2014;95:1734–1742. doi: 10.1099/vir.0.065318-0. [DOI] [PubMed] [Google Scholar]
- 33.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schloss PD, et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, et al. The Sequence Alignment/Map format ans SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watson SJ, et al. Viral population analysis and minority-variant detection using short read next-generation sequencing. Philos. Trans. R. Soc. London.Series B, Biol. Sci. 2013;368:20120205. doi: 10.1098/rstb.2012.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prosperi MCF, Salemi M. QuRe: Software for viral quasispecies reconstruction from next-generation sequencing data. Bioinformatics. 2012;28:132–133. doi: 10.1093/bioinformatics/btr627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilm A, et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40:11189–201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW. GARD: A genetic algorithm for recombination detection. Bioinformatics. 2006;22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- 41.Kosakovsky Pond SL, Frost S, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 42.Huson, D. H. & Bryant, D. Estimating phylogenetic trees and networks using SplitsTree 4. Bioinforma. LNCS IEEE/ACM Trans. Comput. Biol. Bioinforma. J. Classif. LNBI Bioinforma. 2452 (2002).
- 43.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:1–5. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Standley K. MAFFT multiple sequence alignment software version 7: improvements in performance and usability (outlines version 7) Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abascal F, Zardoya R, Telford MJ. TranslatorX: Multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38:W7–W13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 47.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772–772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 1992;132:619–33. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murrell B, et al. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012;8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murrell B, et al. FUBAR: A fast, unconstrained bayesian AppRoximation for inferring selection. Mol. Biol. Evol. 2013;30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kosakovsky Pond SL, Frost SD. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 52.Fort M, Olvera A, Sibila M, Segalés J, Mateu E. Detection of neutralizing antibodies in postweaning multisystemic wasting syndrome (PMWS)-affected and non-PMWS-affected pigs. Vet. Microbiol. 2007;125:244–255. doi: 10.1016/j.vetmic.2007.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The entire sequence dataset is available in the NCBI database, BioProject PRJNA450261 (available at http://www.ncbi.nlm.nih.gov/bioproject/450261).