Significance
Muscle wasting and weakness can be observed under either physiological or pathological conditions, which are partly due to an imbalance between autophagy (“self-eating”) and apoptosis (“self-killing”). How microRNAs coordinate autophagy and apoptosis in the metabolic regulation of cell death remains largely unknown. This work identifies miR-378 as a critical component of metabolic checkpoints, which integrates metabolic information into an adaptive response to reduce the propensity of myocytes to undergo apoptosis by enhancing autophagy and suppressing apoptosis via directly targeting phosphoinositide-dependent protein kinase 1 and Caspase 9, respectively. Our study highlights a crucial role of miR-378 in maintaining normal muscle homeostasis by orchestrating autophagy and apoptosis processes and provides a potential therapeutic target to treat myopathies.
Keywords: miR-378, skeletal muscle, autophagy, apoptosis
Abstract
The metabolic regulation of cell death is sophisticated. A growing body of evidence suggests the existence of multiple metabolic checkpoints that dictate cell fate in response to metabolic fluctuations. However, whether microRNAs (miRNAs) are able to respond to metabolic stress, reset the threshold of cell death, and attempt to reestablish homeostasis is largely unknown. Here, we show that miR-378/378* KO mice cannot maintain normal muscle weight and have poor running performance, which is accompanied by impaired autophagy, accumulation of abnormal mitochondria, and excessive apoptosis in skeletal muscle, whereas miR-378 overexpression is able to enhance autophagy and repress apoptosis in skeletal muscle of mice. Our in vitro data show that metabolic stress-responsive miR-378 promotes autophagy and inhibits apoptosis in a cell-autonomous manner. Mechanistically, miR-378 promotes autophagy initiation through the mammalian target of rapamycin (mTOR)/unc-51-like autophagy activating kinase 1 (ULK1) pathway and sustains autophagy via Forkhead box class O (FoxO)-mediated transcriptional reinforcement by targeting phosphoinositide-dependent protein kinase 1 (PDK1). Meanwhile, miR-378 suppresses intrinsic apoptosis initiation directly through targeting an initiator caspase—Caspase 9. Thus, we propose that miR-378 is a critical component of metabolic checkpoints, which integrates metabolic information into an adaptive response to reduce the propensity of myocytes to undergo apoptosis by enhancing the autophagic process and blocking apoptotic initiation. Lastly, our data suggest that inflammation-induced down-regulation of miR-378 might contribute to the pathogenesis of muscle dystrophy.
A growing body of evidence suggests a critical involvement of macroautophagy (hereafter referred to as autophagy) as a metabolic checkpoint that dictates cell fate in response to metabolic fluctuations (1). Autophagy (“self-eating”) is a dynamic homeostasis process stimulated on stress, in which cytoplasmic portions and organelles are engulfed, degraded, and recycled to support cellular metabolism (2). Proper autophagy maintains homeostasis, whereas uncontrolled autophagy is implicated in pathological progression (3, 4). As another self-destructive process, apoptosis (“self-killing”) determines the turnover of entire cells and is also crucial for tissue homeostasis and pathogenesis (2). The coordination of autophagy and apoptosis after metabolic stress is complex and highly context dependent. The overall design of the cross-talk between autophagy and apoptosis is to reduce the propensity of cells to undergo apoptosis (2). In general, autophagy tends to be antiapoptotic by raising the threshold of stress required to induce apoptosis (5). Although our knowledge has greatly expanded recently, the mechanisms that determine the basal levels of autophagy and the threshold for apoptosis are not fully understood (6).
As the most abundant tissue in human body, skeletal muscle not only functions to produce force and motion but also, serves as a source of amino acids to be utilized for energy production on dwindling nutrient resources (7). Muscle wasting and weakness can be observed under either physiological or pathological conditions, which are speculated to be partly due to an imbalance between autophagy and apoptosis (8–10). Of note, inhibition of autophagy has been linked to muscle atrophy in both mouse models and human, whereas aberrantly enhanced apoptosis has been reported in muscle atrophy due to immobilization or diseases (4, 7, 9, 10). In addition, excessive apoptosis, inhibition of autophagy, and accumulation of dysfunctional organelles have been reported in muscles from patients with muscle dystrophy as well as in mouse models of muscle dystrophy (11–14). Although autophagy and apoptosis have been extensively studied in skeletal muscle, the coordination and interplay between autophagy and apoptosis on metabolic stress or after muscle damage are complex and are not fully understood (8). Key apoptotic and autophagic regulators, especially those involved in the pathogenesis of myopathies, remain to be identified.
MicroRNAs (miRNAs) are endogenous small noncoding RNAs that regulate target gene expression posttranscriptionally (15, 16). The discovery of miRNAs has provided new insights into the mechanisms that control multiple cellular processes, including autophagy and apoptosis. However, whether miRNAs are able to respond to metabolic stress, reset the threshold of cell death, and attempt to reestablish homeostasis is largely unknown. miR-378 and miR-378* are located in the first intron of its host gene peroxisome proliferator-activated receptor gamma coactivator-1β (PGC1β). Functions of miR-378 and miR-378* have been implicated in mitochondrial metabolism, systemic energy homeostasis, and classic brown adipose tissue-specific expansion as well as cancer metabolism (17–19). We previously established fasting-responsive hepatic miR-378 as a critical regulator of hepatic insulin signaling (20). Here, our study reveals that miR-378 is a vital coordinator of autophagy and apoptosis in maintaining normal muscle homeostasis.
Results
miR-378/378* KO Mice Have Decreased Muscle Weight.
In this study, we found the highest expression of both miR-378 and PGC1β in skeletal muscle compared with other tissues (Fig. 1A and SI Appendix, Fig. S1A). Interestingly, the miR-378 levels in skeletal muscle were increased significantly on fasting (Fig. 1B), suggesting a functional involvement of miR-378 in physiological adaptations to metabolic stress in skeletal muscle. We then took advantage of our previously generated miR-378/378* KO mice to test this hypothesis (20). Compared with WT mice, we did not observe obvious changes in body weight in KO mice (SI Appendix, Fig. S1B); instead, we noticed a significant decrease in ratios of slow-twitch soleus (SOL), mixed gastrocnemius (GAS), and fast-twitch tibialis anterior (TA) muscle weights to body weight as well as a significant smaller myofiber size in KO mice compared with WT mice (Fig. 1 C and D and SI Appendix, Fig. S1C). Accordingly, KO mice ran for a significantly shorter distance on the treadmill than their WT littermates (Fig. 1E). These observations suggest that the skeletal muscle of KO mice might undergo atrophy. Consistently, the protein levels of muscle ring finger protein 1 (MURF) (21), a marker of atrophy process, were markedly increased in the GAS muscle of KO mice compared with WT mice (SI Appendix, Fig. S1D). Together, these data indicate that miR-378/378* is required for maintaining normal muscle mass.
Fig. 1.
miR-378/378* KO mice exhibit multiple abnormalities in skeletal muscle. (A) qPCR analysis of relative miR-378 levels in different tissues [brain (Bra), epididymal fat (Ep), heart (He), kidney (Kid), liver (Liv), lung (Lun), skeletal muscle (Mus), pancreas (Pan), spleen (Spl), and testis (Tes)] from C57BL/6J mice as indicated. (B) qPCR analysis of relative miR-378 levels in the TA muscle of fed and fasted mice. (C) Relative muscle weight (MW), including SOL, GAS, and TA weight, to body weight (BW) ratio of miR-378/378* KO and WT mice (n = 11–12). (D) Immunofluorescence staining of Caveolin (Left) and average muscle fiber area (Right) in the GAS muscle of KO and WT mice. (E) Maximal treadmill running distance for KO and WT mice (n = 5–6). (F) EM images of autophagosomes (arrowheads) and abnormal mitochondria in the GAS muscle of WT and KO mice, respectively. The areas indicated by boxes in Upper are shown at higher magnification in Lower. (Scale bars: F, Upper, 2 μm; F, Lower, 500 nm.) (G) qPCR analysis of the mRNA levels of LC3b, ATG12, ATG4b, Garbarapl 1, BNIP3, Beclin, and VPS34 in the GAS muscle of KO and WT mice. (H) Western blots of LC3 (LC3-I and LC3-II) as indicated by arrows in the GAS muscle of KO and WT mice. (I) Western blots of LC3 (LC3-I and LC3-II) as indicated by arrows in the GAS muscle of fed or fasted KO and WT mice. (J) Western blots of PARP in the GAS muscle of KO and WT mice. Cleaved PARP proteins are indicated by an arrow. Means ± SEM are shown. *P < 0.05; **P < 0.01; ***P < 0.001.
miR-378/378* KO Mice Display Defective Autophagy and Excessive Apoptosis in Muscle.
We then observed an accumulation of abnormal swollen mitochondria and significantly fewer autophagosomes in the GAS muscle of KO mice, suggesting possibly impaired autophagy in KO mice (Fig. 1F and SI Appendix, Fig. S1 E and F). Consistently, mRNA expression of autophagy-related genes, including LC3b, ATG12, ATG4b, Gabarapl 1, BNIP3, Beclin, and VPS34, was significantly decreased in the GAS muscle of KO mice compared with WT mice (Fig. 1G). Similar patterns were observed in the SOL muscle, although not all of the changes reached statistical significance (SI Appendix, Fig. S1G). Accordingly, the expression levels of microtubule-associated protein 1 light chain 3 (LC3) (SI Appendix, Fig. S1H) and lipidated LC3 (LC3-II) (Fig. 1H) were both markedly reduced in the GAS muscle of KO mice compared with WT mice. Additionally, we noticed an elevation of p62 protein levels in the GAS muscle of KO mice compared with WT mice (SI Appendix, Fig. S1I). As the accumulation of p62 protein is always used as a readout for autophagy impairment (22), these results consistently indicate a defective autophagy in the absence of miR-378/378*. Since it is known that autophagy is required for maintaining muscle mass and that inhibition of autophagy leads to accumulation of abnormal and dysfunctional mitochondria (7), our data indicate that the defective autophagy might contribute to the muscle loss and the accumulation of abnormal mitochondria in KO mice.
In addition to being responsible for the disposal of damaged organelles, autophagy supplies nutrients for cell survival under poor environmental conditions. We next tested whether autophagy was affected in KO mice on metabolic stress after fasting. As expected, we found that loss of miR-378/378* could remarkably decrease the levels of lipidated LC3 and increase the protein levels of p62 in the GAS muscle of mice in either the fed state or the fasted state (Fig. 1I and SI Appendix, Fig. S1 J and K). These results indicate that miR-378/378* is critical for not only constitutive basal autophagy but also, fasting-induced adaptive autophagy.
Given that activation of the apoptotic program is always coupled to the suppression of autophagy (2), we examined the apoptosis in the skeletal muscle of KO mice with impaired autophagy. We found that the levels of cleaved poly-ADP ribose polymerase (PARP) were increased in the GAS muscle of KO mice compared with WT mice (Fig. 1J), suggesting increased apoptosis in KO mice. Consistently, we found an increased leaking of cytochrome c from mitochondria in the GAS muscle of KO mice compared with WT mice (SI Appendix, Fig. S1L) (23). Collectively, our results indicate that the defective autophagy, accumulation of abnormal mitochondria, and excessive apoptosis might contribute to the muscle wasting and poor running performance in KO mice.
miR-378 Promotes Skeletal Muscle Autophagy in a Cell-Autonomous Manner.
To further explore the physiological role of miR-378/378* in regulating autophagy in muscle cells, we next investigated the expression of miR-378 and miR-378* in response to nutrient deprivation. Increased expression levels of miR-378 and miR-378* were observed within 2 and 4 h after C2C12 myotubes were exposed to either glucose-free DMEM (No Glu) or amino acid-free Krebs-Ringer Buffer (KRB) medium (Fig. 2A and SI Appendix, Fig. S2A), suggesting that miR-378/378* might be implicated in the cellular adaptation to dwindling nutrient resources. Notably, the mRNA levels of host gene PGC1β increased within 2 h; however, they dropped back to or below the starting level after 4 h on nutrient deprivation (SI Appendix, Fig. S2B).
Fig. 2.
miR-378 promotes autophagy in muscle cells. (A) qPCR analysis of relative miR-378 levels in C2C12 myotubes cultured in (Left) No Glu or (Right) KRB for indicated time periods. (B) Western blots of LC3 (LC3-I and LC3-II) as indicated by arrows in Ad-378–infected C2C12 myotubes cultured in (Left) No Glu or (Right) KRB as indicated. (C) Western blots of LC3 (LC3-I and LC3-II) as indicated by arrows in C2C12 myotubes transfected with miR-378 mimics (miR-378) and control oligos (Control) or Ant-378 and antagomir control (Ant-Ctrl) as indicated. (D) Immunofluorescence staining of LC3 (red) in C2C12 myoblasts transfected with miR-378 mimics (miR-378), control oligos (Control), Ant-378, or Ant-Ctrl as indicated. (E) Immunofluorescence images of LC3 in Ad-mCherry-GFP-LC3–infected C2C12 myoblasts transfected with miR-378 mimics (miR-378) or control oligos (Control; Left) and Ant-378 or Ant-Ctrl (Right) as indicated. (F) Western blots of p62 in C2C12 myotubes transfected with miR-378 mimics (miR-378), control oligos (Control), Ant-378, or Ant-Ctrl as indicated. (G) Western blots of p-mTOR, mTOR, p-ULK1 (Ser757), ULK1, p-FoxO [p-FoxO1 (Thr24) and p-FoxO3 (Thr32)], FoxO1, and FoxO3 in C2C12 myotubes transfected with miR-378 mimics (miR-378), control oligos (Control), Ant-378, or Ant-Ctrl as indicated. (H) Western blots of LC3 (LC3-I and LC3-II) as indicated by arrows in Ad-378–infected C2C12 myotubes in the presence or absence of Baf A1 (Upper) or CQ (Lower) as indicated. Means ± SEM are shown. **P < 0.01; ***P < 0.001.
We found that adenoviral miR-378/378* (Ad-378) infection led to an increase in lipidated LC3 levels in C2C12 myotubes cultured in either complete cell culture medium or two different types of nutrient-deficient (No Glu or KRB) medium as indicated, suggesting that Ad-378 infection could enhance both basal and adaptive autophagy in myocytes (Fig. 2B). Moreover, overexpression of miR-378 by transfection of specific mimics for miR-378 increased the lipidation of LC3, whereas inhibition of miR-378 by transfection of specific antagomir for miR-378 (Ant-378) decreased the lipidation of LC3 in both C2C12 myotubes (Fig. 2C and SI Appendix, Fig. S2C) and L6 myotubes (SI Appendix, Fig. S2 D and E). Transfection of either miR-378* mimics or miR-378* inhibitor had no effect on lipidated LC3 levels in C2C12 myotubes (SI Appendix, Fig. S2F), indicating that miR-378, but not miR-378*, contributed to the proautophagic effect of Ad-378 in muscle cells. Together, these results suggest that miR-378 promotes autophagy in skeletal muscle in a cell-autonomous manner.
Consistent with above results, we found that miR-378 overexpression increased the number of LC3 puncta, while miR-378 inhibition decreased the number of LC3 puncta in either C2C12 or L6 myoblasts (Fig. 2D and SI Appendix, Fig. S2 G and H) (22). When Ad-mCherry-GFP-LC3 was used to track LC3 expression in autophagosomes and autolysosomes, we observed an increase of both yellow LC3 puncta (representing autophagosomes) (Fig. 2E, Left and SI Appendix, Fig. S2I) and red puncta (representing autolysosomes) (SI Appendix, Fig. S2J) in C2C12 myoblasts overexpressing miR-378, suggesting that miR-378 is able to promote autophagosomes formation and enhance autophagy flux. Results obtained from Ant-378 treatment (Fig. 2E, Right and SI Appendix, Fig. S2K) as well as p62 immunoblots (Fig. 2F) (24) also supported the above conclusion.
To further explore the role of miR-378 in autophagy, key regulators of autophagy were examined. Mammalian target of rapamycin (mTOR) complex 1 is a dominant regulator of autophagy induction in skeletal muscle (25), which inhibits autophagosome formation by phosphorylation of unc-51-like autophagy activating kinase 1 (ULK1) (3, 26). Consistently, we found that phosphorylated mTOR (p-mTOR) and p-ULK1 levels were reduced by miR-378 overexpression and elevated by miR-378 inhibition in C2C12 myotubes (Fig. 2G). Forkhead box class O (FoxO) family members are highly conserved transcription factors. Among them, FoxO1 and FoxO3 have been shown to control the expression of many autophagy-related genes in muscle cells (27, 28). We found that FoxO1 and FoxO3 protein levels were elevated by miR-378 overexpression and reduced by miR-378 inhibition in C2C12 myotubes (Fig. 2G). Accordingly, p-FoxO [p-FoxO1 (Thr24) and p-FoxO3 (Thr32)] levels were decreased by miR-378 overexpression and increased by miR-378 inhibition in C2C12 myotubes (Fig. 2G). These results indicate that miR-378 not only promotes the autophagy initiation through the mTOR/ULK1 pathway but also, sustains the autophagy via FoxO-mediated transcriptional reinforcement.
As either an excessive autophagy induction or a reduced exhaustion of autophagic vesicles could contribute to the accumulation of lipidated LC3 and LC3-positive autophagosomes, to discriminate between these two possibilities and further assess the role of miR-378 on autophagic process, lipidated LC3 was examined in C2C12 myotubes treated with bafilomycin A1 (Baf A1), which is able to trap newly formed autophagosomes by inhibiting the fusion between autophagosomes and lysosomes. We found that the levels of lipidated LC3 were elevated by Baf A1 treatment, which could be further increased by Ad-378 infection (Fig. 2H, Upper). Similar results were obtained when chloroquine (CQ) was used to inhibit both fusion of autophagosome with lysosome and lysosomal protein degradation (Fig. 2H, Lower). These results suggest that the increased LC3 lipidation by miR-378 overexpression was at least partly caused by enhanced autophagy induction. Consistent with our observation in myocytes, miR-378 overexpression increased the number of LC3 puncta in HeLa cells (SI Appendix, Fig. S2L). Moreover, the number of LC3 puncta was increased after Baf A1 treatment, which could be further increased by miR-378 overexpression, further supporting that miR-378 might promote autophagy flux (SI Appendix, Fig. S2L).
Given that the up-regulation of miR-378 after fasting (Fig. 1B) or nutrient deprivation (Fig. 2A) was accompanied by an increase in FoxO1 expression (SI Appendix, Fig. S2 M and N) and LC3 lipidation (Fig. 2B and SI Appendix, Fig. S2M), we speculated that FoxO1 might regulate miR-378 expression on metabolic stress. Indeed, we found that FoxO1 could regulate miR-378 expression at the transcriptional level in muscle cells (SI Appendix, Fig. S2 O–R), indicating that miR-378 and FoxO1 may form a positive feedback loop to sustain autophagy on metabolic stress.
PDK1 Is a Direct Target Gene of miR-378 in Skeletal Muscle.
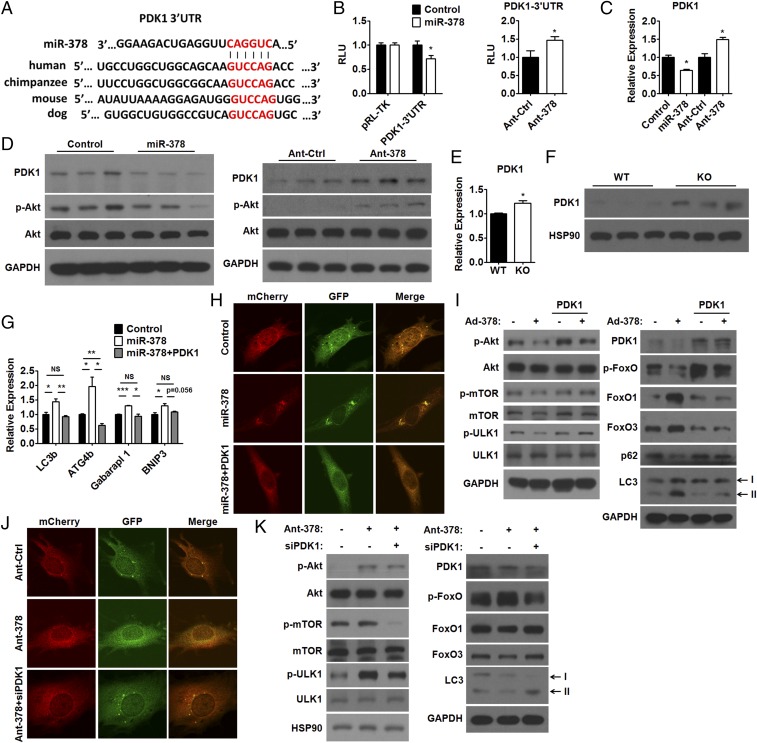
We next screened predicted target genes for miR-378 in silico to look for autophagy-associated putative target genes. Since the regulation of mTOR and FoxOs by serine/threonine kinase Akt is well established, it is generally assumed that alteration in Akt signaling would have a great impact on autophagy. Intriguingly, we found that phosphoinositide-dependent protein kinase 1 (PDK1), which activates Akt by phosphorylating Akt at Thr-308, contains a highly conserved putative miR-378 binding site in its 3′UTR (Fig. 3A). Luciferase reporter assay with the 3′UTR of PDK1 (PDK1-3′UTR) containing putative or mutated miR-378 recognition element further validated that PDK1 is a direct target gene of miR-378 in muscle cells (Fig. 3B and SI Appendix, Fig. S3A). Moreover, the finding that PDK1 expression was regulated on overexpression or inhibition of miR-378 in C2C12 cells further supported the above conclusion (Fig. 3 C and D and SI Appendix, Fig. S3B). Consistent with the expression pattern of PDK1, p-Akt (Thr-308) levels were decreased by miR-378 overexpression or increased by miR-378 inhibition in C2C12 myotubes (Fig. 3D and SI Appendix, Fig. S3C). Similar results were obtained in L6 myotubes (SI Appendix, Fig. S3 D–F). The regulation of PDK1 by miR-378 was further confirmed by the observation that the PDK1 expression was increased in the GAS muscle of KO mice (Fig. 3 E and F and SI Appendix, Fig. S3G).
Fig. 3.
PDK1 mediates the effect of miR-378 on autophagy in skeletal muscle. (A) Sequence alignment of miR-378 and the PDK1-3′UTR from various species. (B) Luciferase assay showing the effect of miR-378 mimics (miR-378) transfection (Left) or Ant-378 transfection (Right) on the activity of the reporter containing the PDK1-3′UTR with a putative miR-378 recognition element in C2C12 myoblasts. (C and D) qPCR analysis of relative PDK1 mRNA levels (C) and Western blots (D) of PDK1 and p-Akt (Thr-308) in C2C12 myotubes transfected with miR-378 mimics (miR-378), control oligos (Control), Ant-378, or antagomir control (Ant-Ctrl) as indicated. (E and F) qPCR analysis of relative PDK1 mRNA levels (E) and Western blots of PDK1 (F) in the GAS muscle of KO and WT mice. (G) qPCR analysis of the mRNA levels of LC3b, ATG4b, Gabarapl 1, and BNIP3 in C2C12 myotubes transfected with miR-378 mimics (miR-378) or control oligos (Control) with or without PDK1 expression plasmids as indicated. (H) Immunofluorescence images of LC3 in Ad-mCherry-GFP-LC3–infected C2C12 myoblasts transfected with control oligos (Control), miR-378 mimics (miR-378) alone, or miR-378 mimics together with PDK1 expression plasmids. (I) Western blots of PDK1, p-Akt (Thr-308), Akt, p-mTOR, mTOR, p-ULK1 (Ser757), ULK1, p-FoxO [p-FoxO1 (Thr24) and p-FoxO3 (Thr32)], FoxO1, FoxO3, p62, and LC3 in Ad-378–infected C2C12 myotubes transfected with or without PDK1 expression plasmids as indicated. (J) Immunofluorescence images of LC3 in Ad-mCherry-GFP-LC3–infected C2C12 myoblasts transfected with Ant-Ctrl, Ant-378 alone, or Ant-378 together with siRNA specific for PDK1 (siPDK1). (K) Western blots of PDK1, p-Akt (Thr-308), Akt, p-mTOR, mTOR, p-ULK1 (Ser757), ULK1, p-FoxO [p-FoxO1 (Thr24) and p-FoxO3 (Thr32)], FoxO1, FoxO3, and LC3 in C2C12 myotubes transfected with Ant-378 and/or siPDK1 as indicated. Means ± SEM are shown. NS, not significant; RLU, relative luciferase units. *P < 0.05; **P < 0.01; ***P < 0.001.
In agreement with the notion that autophagic response preferentially occurs in fast-switch muscle, we found that the miR-378 levels were higher, while PDK1 expression levels were lower in the GAS muscle than those in SOL muscle (SI Appendix, Fig. S3 H and I). The positive correlation between the levels of miR-378 and lipidated LC3 and the negative correlation between the levels of miR-378 and p62 suggest that miR-378 might be implicated in the determination of basal levels of autophagy in GAS and SOL muscles (SI Appendix, Fig. S3 H and I). Additionally, we found that the PDK1 mRNA levels were decreased in C2C12 myotubes after glucose or amino acids starvation for 4 h (SI Appendix, Fig. S3J), suggesting that the increased levels of miR-378 after nutrient deprivation might contribute to the down-regulation of PDK1.
To further evaluate whether PDK1 mediates the effect of miR-378 on autophagic process, we performed a series of rescue experiments in C2C12 myotubes. First, PDK1 overexpression diminished the effect of miR-378 overexpression on the expression of autophagy-related genes and LC3 puncta formation (Fig. 3 G and H and SI Appendix, Fig. S3 K and L). Second, PDK1 overexpression attenuated the effect of miR-378 overexpression on p-Akt (Thr-308), p-mTOR, p-ULK1, p-FoxO [p-FoxO1 (Thr24) and p-FoxO3 (Thr32)], FoxO1, FoxO3, p62, and lipidated LC3 levels (Fig. 3I). Third, inhibition of PDK1 attenuated the effect of Ant-378 on LC3 puncta formation (Fig. 3J and SI Appendix, Fig. S3M) and protein levels of p-Akt (Thr-308), p-mTOR, p-ULK1, p-FoxO [p-FoxO1 (Thr24) and p-FoxO3 (Thr32)], FoxO1, FoxO3, and lipidated LC3 in C2C12 myotubes (Fig. 3K). Fourth, overexpression of the constitutively active form of FoxO3 attenuated the effect of Ant-378 on lipidated LC3 levels and LC3 puncta formation (SI Appendix, Fig. S3 N and O) (28). Fifth, the effect of Ant-378 on lipidated LC3 levels and Akt signaling was abrogated after ablation of PI3K signaling by LY294002 (SI Appendix, Fig. S3P). These results collectively show that PDK1 mediates the proautophagic effect of miR-378 through Akt signaling.
Caspase 9 Is a Direct Target Gene of miR-378 in Skeletal Muscle.
The observation that apoptosis was increased in the skeletal muscle of KO mice prompted us to test whether miR-378/378* simultaneously regulates both autophagy and apoptosis. Indeed, one of the initiator caspases Caspase 9 (CASP9) (29), which controls intrinsic apoptotic initiation, was predicted as a putative target gene of miR-378. The putative miR-378 binding site in the 3′UTR of CASP9 (CASP9-3′UTR) is highly conserved (Fig. 4A). Luciferase reporter assay with the CASP9-3′UTR containing putative or mutated miR-378 recognition element validated that CASP9 is a direct target gene of miR-378 in muscle cells (Fig. 4B and SI Appendix, Fig. S4A). The finding that CASP9 expression could be regulated by miR-378 overexpression or inhibition in C2C12 (Fig. 4 C and D and SI Appendix, Fig. S4B) and L6 myotubes (SI Appendix, Fig. S4 C and D) further substantiated the above conclusion. Additionally, we noticed a dynamic change of CASP9 mRNA expression on nutrient deprivation (SI Appendix, Fig. S4E), which could partly be explained by miR-378 modulation.
Fig. 4.
CASP9 is a direct target gene of miR-378 in skeletal muscle. (A) Sequence alignment of miR-378 and the CASP9-3′UTR from various species. (B) Luciferase assay showing the effect of miR-378 mimics (miR-378) transfection (Left) or Ant-378 transfection (Right) on the activity of CASP9-3′UTR with the putative miR-378 binding site in C2C12 myoblasts. RLU, relative luciferase units. (C and D) qPCR analysis of relative CASP9 mRNA levels (C) and Western blots (D) of CASP9 in C2C12 myotubes transfected with miR-378 mimics (miR-378), control oligos (Control), Ant-378, or antagomir control (Ant-Ctrl) as indicated. (E) Immunofluorescence staining of cleaved CASP3 (red) in C2C12 myoblasts transfected with miR-378 mimics (miR-378), control oligos (Control), Ant-378, or Ant-Ctrl as indicated. (F) Western blots of PARP in C2C12 myotubes transfected with miR-378 mimics (miR-378), control oligos (Control), Ant-378, or Ant-Ctrl as indicated. (G–I) qPCR analysis of relative CASP9 mRNA levels (G), Western blots of CASP9 and cleaved CASP9 (H), and relative CASP9 activity (I) in the GAS muscle of KO and WT mice. (J) Western blots of CASP3 and cleaved CASP3 in the GAS muscle of KO and WT mice. (K) Maximal treadmill running distance for KO mice treated with or without z-VAD (n = 6). (L) Western blots of p62, LC3, PARP, CASP3, and cleaved CASP3 in the GAS muscle of KO mice treated with or without z-VAD as indicated. LC3-I, LC3-II, and cleaved PARP proteins are indicated by arrows. Means ± SEM are shown. *P < 0.05; **P < 0.01.
To test whether the regulation of CASP9 by miR-378 would affect apoptosis, we investigated the cleavage of Caspase 3 (CASP3) and PARP on overexpression or inhibition of miR-378 in myotubes cultured in amino acid-free KRB medium. Studies in both C2C12 and L6 myotubes concluded that miR-378 could regulate apoptosis in myotubes on nutrient deprivation (Fig. 4 E and F and SI Appendix, Fig. S4 F and G). To be noted, neither miR-378* mimics nor miR-378* inhibitor had effects on the cleavage of PARP in C2C12 myotubes (SI Appendix, Fig. S4H). The effect of miR-378 on apoptosis was further confirmed by TUNEL assay in either C2C12 or L6 myocytes (SI Appendix, Fig. S4 I and J) and Annexin V assay in C2C12 myocytes (SI Appendix, Fig. S4K).
Importantly, the regulation of CASP9 by miR-378 was further confirmed by the observation that the expression levels of CASP9 were increased in the GAS muscle of KO mice compared with WT mice (Fig. 4 G and H and SI Appendix, Fig. S4L). We also noticed an increase in cleaved PARP levels (Fig. 1J), cleaved CASP9 levels (Fig. 4H and SI Appendix, Fig. S4L), CASP9 activity (Fig. 4I), cleaved CASP3 levels, and CASP3 activity (Fig. 4J and SI Appendix, Fig. S4 M and N) in KO mice compared with WT animals. Together, all of these data suggest an antiapoptotic role of miR-378 in skeletal muscle, which could be mediated by targeting CASP9.
Given that the poor running performance in KO mice was accompanied by excessive apoptosis in skeletal muscle, we speculated that block of apoptosis might improve the athletic performance in these mice. Indeed, inhibition of apoptosis by z-VAD-FMK (z-VAD), a pancaspase inhibitor, significantly increased the maximal exercise capacity of KO mice (Fig. 4K). Interestingly, z-VAD treatment not only reduced the levels of cleaved CASP3 and cleaved PARP but also, increased the autophagy as evident from decreased levels of p62, increased levels of lipidated LC3, and enhanced LC3 staining in the GAS muscle of KO mice (Fig. 4L and SI Appendix, Fig. S4O). Consistent with the notion that the cleavage of Beclin 1 (BECN1) by CASP3 is able to inactivate autophagy and promote apoptosis (30), we found decreased levels of cleaved BECN1 in both C2C12 myotubes (SI Appendix, Fig. S4P) and GAS muscle of KO mice on z-VAD treatment (SI Appendix, Fig. S4Q).
To see whether miR-378 can regulate apoptosis through other mechanisms, we checked the effect of miR-378 on CASP3 and suppression of tumorigenicity 7-like (ST7L), which previously have been implicated in miR-378–regulated apoptosis (31–34). Our results showed that both miR-378 overexpression and inhibition had no effect on the protein levels of both CASP3 and ST7L in C2C12 myotubes (SI Appendix, Fig. S4 R–U). Notably, the CASP3 protein levels were decreased rather than increased in the GAS muscle of KO mice compared with WT mice (Fig. 4J and SI Appendix, Fig. S4M, Right). These results suggest that CASP3 and ST7L are not direct targets for miR-378 in the regulation of apoptosis by miR-378 in skeletal muscle.
Overexpression of miR-378 Induces Autophagy and Represses Apoptosis in Skeletal Muscle.
To further confirm the regulatory role of miR-378 on autophagy and apoptosis in vivo, C57BL/6J mice were treated with agomir-378. In vivo administration of agomir-378 led to a significant increase in miR-378 levels in the GAS and TA muscles (Fig. 5A) and a concurrent decrease in both mRNA and protein levels of PDK1 and CASP9 (Fig. 5 B and C and SI Appendix, Fig. S5A). Moreover, agomir-378 treatment led to decreased levels of cleaved CASP9, cleaved CASP3, and cleaved PARP along with increased levels of lipidated LC3 (Fig. 5 C–E and SI Appendix, Fig. S5 B–E), indicating enhanced autophagy and inhibited apoptosis in skeletal muscle. Accordingly, the mRNA expression of autophagy-related genes tended to be increased in the GAS muscle of mice injected with agomir-378 (Fig. 5F and SI Appendix, Fig. S5F). Consistent with the finding in KO mice (Fig. 1I and SI Appendix, Fig. S1 I and J), agomir-378 treatment led to an increase in LC3 lipidation and a decrease in p62 protein levels in GAS muscle of mice under both fed and fasted conditions (Fig. 5G), further supporting the notion that miR-378 controls both constitutive basal autophagy and fasting-induced adaptive autophagy. Together, our results establish miR-378 as a proautophagic and antiapoptotic factor in the skeletal muscle of mice.
Fig. 5.
Overexpression of miR-378 induces autophagy and represses apoptosis in skeletal muscle. (A) qPCR analysis of relative miR-378 levels in the GAS and TA muscles of mice treated with agomir-378 or control. (B) qPCR analysis of relative mRNA levels of PDK1 and CASP9 in the GAS muscle of mice treated with agomir-378 or control. (C–E) Western blots of CASP9, cleaved CASP9, PDK1, and LC3 (C); CASP3 and cleaved CASP3 (D); and PARP (E) in the GAS muscle of mice treated with agomir-378 or control. LC3-I, LC3-II, and cleaved PARP proteins are indicated by arrows. (F) qPCR analysis of LC3b, ATG12, BNIP3, BNIP3l, Cathepsin L, Atrogin 1, VPS34, and Beclin in the GAS muscle of mice treated with agomir-378 or control. (G) Western blots of p62 and LC3 (LC3-I and LC3-II) in the GAS muscle of fed or fasted mice treated with agomir-378 or control as indicated. Means ± SEM are shown. *P < 0.05; **P < 0.01.
miR-378 Is Implicated in the Pathogenesis of Muscle Dystrophy.
We also examined miR-378 expression in the muscle biopsy samples obtained from patients with Duchenne muscular dystrophy (DMD) or the less severe Becker muscular dystrophy (BMD), and we found that the miR-378 levels were markedly decreased in BMD patients and were further decreased in DMD patients, suggesting that miR-378 levels were negatively correlated with the disease severity (SI Appendix, Fig. S6A). In contrast, expression levels of the miR-378 targets—PDK1 and CASP9—were significantly increased in DMD subjects (SI Appendix, Fig. S6 A–C). In agreement with the notion that muscular dystrophy exhibits defective autophagy and excessive apoptosis (35, 36), we found that the levels of lipidated LC3 were decreased while the levels of cleaved PARP were increased in muscle biopsy specimens from DMD patients compared with those from normal subjects (SI Appendix, Fig. S6 D and E). Similar results were obtained in muscle tissues obtained from patients with congenital muscle dystrophy carrying mutations in collagen VI (11) (SI Appendix, Fig. S6F). Chronic inflammation represents a hallmark feature of DMD pathogenesis (37). We found significantly reduced miR-378 expression in C2C12 myotubes treated with inflammatory cytokines (e.g., TNFα and IFNγ) (SI Appendix, Fig. S6G), indicating that inflammation-induced down-regulation of proautophagic and antiapoptotic miR-378 contributes to the pathogenesis of muscle dystrophy and highlighting a potential therapeutic target to treat myopathies.
Taken together, our data establish miR-378 as a common upstream modulator of autophagy and apoptosis in skeletal muscle. On metabolic stress or under inflammatory condition, miR-378 coordinates autophagy and intrinsic apoptosis through targeting PDK1 and CASP9, respectively (Fig. 6).
Fig. 6.
Schematic diagram of the working model of miR-378 in the coordination of autophagy and apoptosis in skeletal muscle. mTORC1, mammalian target of rapamycin complex 1.
Discussion
Understanding the survival strategies used by cells is of particular importance. Growing evidence suggests that metabolism and cell death are deeply intertwined at multiple levels. In most cases, metabolic checkpoints generate an adaptive response in an attempt to reestablish the homeostasis and prevent the cell from undergoing cell death on metabolic stress (1). Identifying components of metabolic checkpoints will help us to not only understand the metabolic regulation of cell death but also, find pharmacological approaches to treat pathologies involving unwarranted cell death. In this study, we found that skeletal muscle miR-378 is able to respond to metabolic stress and raise the threshold for cell death initiation by enhancing autophagic process and blocking intrinsic apoptosis directly (Fig. 6). Our results establish metabolic stress-responsive miR-378 as a critical modulator that coordinates both autophagy and apoptosis, adding miR-378 to the growing list of components of metabolic checkpoints.
In addition to supplying cellular energy, mitochondria are also a source of proapoptotic factors (38). Therefore, the ability to withstand mitochondrial damage is especially critical for the survival of cells. It is known that autophagy is the only known means to remove defective mitochondria, which often occurs after damage or stress (7, 39). Here, we discovered a mechanism engaged by muscle cells that highly restricts cell death and ensures cell survival. Our results suggest that the levels of miR-378 in myocytes determine the threshold for cell death initiation, since miR-378 is able to not only promote autophagy to facilitate the disposal of dysfunctional or damaged mitochondria through targeting PDK1 but also, inhibit mitochondria-mediated intrinsic apoptosis by targeting CASP9. As the expression of miR-378 in skeletal muscle was the highest among the tissues examined (Fig. 1A), the high levels of miR-378 in myocytes should be of great importance for myocytes to restrict apoptosis and preserve cell viability. Failure to maintain the high levels of miR-378 in skeletal muscle would lead to the increased vulnerability to cell death observed in muscle dystrophy (SI Appendix, Fig. S6 A and F).
PDK1 belongs to the AGC serine/threonine kinase family, which is crucial for the activation of Akt and many other AGC kinases by phosphorylating these kinases at their activation loops (40). The regulation of Akt by PDK1 on PI3K activation has been extensively studied. PDK1 is recruited to the cell membrane by PI3K-generated PIP3 and activates AKT by phosphorylation at Thr308 (40). PDK1 itself is believed to be constitutively active. The mechanisms involved in the regulation of PDK1 comprise autophosphorylation, subcelluar localization, regulator interaction, and conformation change (41). In this study, we found that PDK1 expression can be regulated by miR-378 at the posttranscriptional level in skeletal muscle. Our discovery adds another important layer of regulation for PDK1, which sheds light on how PI3K/Akt signaling is dynamically and precisely controlled on metabolic stress.
CASP9, as an initiator caspase in intrinsic caspase cascade, is sensitive to activation signals. Indeed, unlike the executioner caspases, CASP9 is not activated by cleavage but by dimerization, which avoids adventitious activation (29). However, whether other mechanisms for the regulation of CASP9 exist remains unknown. Here, we provide evidence that CASP9 can be regulated by miR-378 at the posttranscriptional level. On metabolic stress, the up-regulation of miR-378 in myocytes strengthens the threshold for intrinsic apoptosis by suppressing CASP9 expression. In contrast, inhibition or loss of miR-378 leads to an increase in CASP9 levels in skeletal muscle, thereby increasing the propensity of cells to die. Thus, based on our findings, we establish miR-378 as a gatekeeper of the intrinsic apoptotic pathway in skeletal muscle, which also provides insight into the mechanisms underlying the metabolic control of intrinsic apoptosis.
Inhibition of autophagy not only exacerbates muscle loss during denervation and fasting but also, is considered to be involved in the pathogenesis of muscle degeneration and weakness in many myopathies (e.g., DMD) (13, 14). In line with the current knowledge, we found decreased lipidation of LC3, indicative of impaired autophagy, in muscle biopsy specimens from DMD patients compared with normal subjects (SI Appendix, Fig. S6D). Interestingly, the miR-378 levels were markedly decreased while PDK1 expression was significantly increased in DMD patients compared with normal subjects (SI Appendix, Fig. S6 A and B). Furthermore, both TNFα and IFNγ could down-regulate miR-378 in muscle cells, suggesting that inflammation-induced down-regulation of miR-378 might play a role in the pathogenesis of DMD. Thus, our findings offer strategies to treat muscle pathologies.
The effect of PGC1β on autophagy and atrophy has been reported in skeletal muscle; however, the results are controversial and inconclusive. An early report showed that PGC1β could prevent the induction of autophagy and atrophy in muscle cells (42). However, a recent study suggested that long-term PGC1β overexpression could lead to apoptosis, autophagy, and atrophy in the skeletal muscle of mice (43). Additionally, it has been suggested that PGC1β overexpression decreases apoptosis in breast cancer cells (44). In our study, we noticed a dynamic change of either miR-378 or PGC1β mRNA expression on metabolic stress (Fig. 2A and SI Appendix, Fig. S2B). The expression of both miR-378 and PGC1β was increased within 2 h after nutrient depletion in C2C12 myotubes. The increase in miR-378 expression could be observed even after 4 h of nutrient deprivation. In contrast, the PGC1β mRNA levels dropped back to the original levels or even lower than the control group 4 h after nutrient deprivation. These results indicate that miR-378 and PGC1β might work dynamically and coordinately in the cellular adaptation to dwindling nutrient resources. We speculate that the combinational effect of miR-378 and PGC1β on the metabolic regulation of cell death in skeletal muscle is complicated and requires additional study.
In this study, KO mice exhibited muscle atrophy as evident from decreased muscle weight and myofiber size and increased protein levels of MURF, an atrogene transcriptionally regulated by FoxOs (Fig. 1 D and C and SI Appendix, Fig. S1D) (28). Given that FoxO levels were decreased due to Akt activation after miR-378 inhibition in myotubes (Figs. 2G and 3K), one would expect down-regulation rather than up-regulation of MURF in miR-378 null muscles. This unexpected finding suggests that miR-378–deficient muscles underwent atrophy regardless of FoxO activity. Notably, although mTOR has been appreciated as a positive regulator of protein synthesis in muscle, sustained activation of mTOR complex 1 by deletion of tuberous sclerosis complex 1 led to atrophy, which was attributed mainly to inability to induce autophagy due to ULK1 inactivation (25, 45). The concomitant muscle atrophy observed in autophagy-defective miR-378 KO mice suggests that similar mechanisms may account for the atrophy in miR-378 KO mice. We also speculate that, similar to those suggested for Atg7 null muscles (7), compensatory up-regulation of proteasomal function and excessive apoptosis due to autophagy inhibition might contribute, at least partially, to muscle atrophy in miR-378 KO mice. Nevertheless, we cannot rule out that other miR-378 target genes might also contribute to the atrophy phenotype of KO mice.
Taken together, we discovered previously undescribed essential roles of miR-378 in metabolic regulation of cell death. miR-378 is able to act as a component of metabolic checkpoints and raise the threshold for cell death initiation by coordinating autophagy and apoptosis through targeting two critical nodes, PDK1 and CASP9. On the basis of this discovered dual autophagy–apoptosis regulatory potential of miR-378, we propose that miR-378 may represent a potential therapeutic target to treat myopathies involving unwarranted cell death.
Materials and Methods
In Vivo Study.
All experimental procedures and protocols were approved by the Institutional Review Board of the Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (permit no. 2015-AN-12). Mice were randomly assigned to each group; however, blinding was not possible. Mice with similar age or from the same litters had the priority of use. We estimated the sample size by using an online program for animal study. miR-378/378* KO mice were generated as described before (20). Male KO mice aged 8–14 wk old and their WT littermates were maintained at a temperature of 23 °C ± 3 °C and a humidity of 35 ± 5% under a 12-h dark/light cycle in a specific pathogen-free animal facility. Muscle specimens from patients with BMD or DMD and patients with congenital muscle dystrophy due to mutations in collagen VI were obtained surgically after obtaining their written informed consent at Children’s Hospital of Fudan University. Muscle specimens were immediately frozen in liquid nitrogen and stored at −80 °C until additional analysis. All of the procedures were reviewed and approved by the Ethics Committee of Children’s Hospital of Fudan University.
Immunostaining and EM.
Cells were seeded into 12-well plates, transfected with RNA oligonucleotides, and infected with Ad-mCherry-GFP-LC3 (Hanbio Biotech). After 48 h, the cells were fixed with cold methanol and permeabilized with 1% Triton X-100 in PBS for 15 min. Cell images were taken using a microscope (BX61; Olympus). Regarding tissue immunohistochemical staining, skeletal muscle or cells were fixed and processed by the paraffin-embedded method. Rabbit monoclonal antibodies against LC3 (Cell Signaling), cleaved CASP3 (Cell Signaling), and Caveolin (Cell Signaling) were used. The sections were counterstained with hematoxylin. For EM, mice were killed, and skeletal muscle was rapidly fixed in 2.5% glutaraldehyde and 0.1 M cacodylate buffer. LC3 puncta quantification was determined by ImageJ (version 1.61; NIH).
Muscle Cross-Sectional Area Measurement.
After immunostaining of skeletal muscle against Caveolin, digital images were captured at room temperature using a microscope (BX61; Olympus), a cooled charge-coupled device camera (QICAM Fast; QImaging), and the software package Q-Capture (version 2.9.11; QImaging) with a UPlan-Apochromat 10×/0.40 N.A. (Olympus) objective lens. Ten fields were used for calculating the cross-sectional area of muscle fibers using ImageJ software (version 1.61; NIH).
Statistical Analysis.
All experiments were performed at least three times, and representative data are shown. Data were presented as means ± SE. Student’s t test was performed to assess whether the means of two groups are statistically significant from each other (P < 0.05). GraphPad Prism 5.0 (GraphPad Software) was applied to all statistical analysis.
Mice phenotyping and treatment, plasmids, RNA oligonucleotides, cell culture, transfection, luciferase assay, real-time PCR and Western blot analysis, TUNEL assay, Annexin V assay, analysis of CASP activity, and ChIP assay are described in SI Appendix, SI Materials and Methods. The information on primers used for real-time PCR, cloning, and ChIP assay are provided in SI Appendix, Tables S1 and S2. Ad-378 was prepared as described previously (20, 46). The protein band density was determined by using ImageJ software (version 1.61; NIH). Uncropped images of Western blots are presented in SI Appendix, Fig. S7.
Supplementary Material
Acknowledgments
This work was supported by Ministry of Science and Technology of China Grants 2016YFA0500102 and 2016YFC1304905; National Natural Science Foundation of China Grants 31371189, 31525012, 31600954, 81471016, 31871195, and 81870541; Chinese Academy of Sciences Grants ZDBS-SSW-DQC-02 and ZDRW-ZS-2017-1; Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDA12040324; and Fundamental Research Funds for the Central Universities Grants JUSRP51708A and JUSRP11842.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.M.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803377115/-/DCSupplemental.
References
- 1.Green DR, Galluzzi L, Kroemer G. Cell biology. Metabolic control of cell death. Science. 2014;345:1250256. doi: 10.1126/science.1250256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: The interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wirawan E, Vanden Berghe T, Lippens S, Agostinis P, Vandenabeele P. Autophagy: For better or for worse. Cell Res. 2012;22:43–61. doi: 10.1038/cr.2011.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roos WP, Thomas AD, Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer. 2016;16:20–33. doi: 10.1038/nrc.2015.2. [DOI] [PubMed] [Google Scholar]
- 6.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 7.Masiero E, et al. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Dirks AJ, Leeuwenburgh C. The role of apoptosis in age-related skeletal muscle atrophy. Sports Med. 2005;35:473–483. doi: 10.2165/00007256-200535060-00002. [DOI] [PubMed] [Google Scholar]
- 9.Dupont-Versteegden EE. Apoptosis in muscle atrophy: Relevance to sarcopenia. Exp Gerontol. 2005;40:473–481. doi: 10.1016/j.exger.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Jackman RW, Kandarian SC. The molecular basis of skeletal muscle atrophy. Am J Physiol Cell Physiol. 2004;287:C834–C843. doi: 10.1152/ajpcell.00579.2003. [DOI] [PubMed] [Google Scholar]
- 11.Grumati P, et al. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat Med. 2010;16:1313–1320. doi: 10.1038/nm.2247. [DOI] [PubMed] [Google Scholar]
- 12.Irwin WA, et al. Mitochondrial dysfunction and apoptosis in myopathic mice with collagen VI deficiency. Nat Genet. 2003;35:367–371. doi: 10.1038/ng1270. [DOI] [PubMed] [Google Scholar]
- 13.De Palma C, et al. Autophagy as a new therapeutic target in Duchenne muscular dystrophy. Cell Death Dis. 2012;3:e418. doi: 10.1038/cddis.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spitali P, et al. Autophagy is impaired in the tibialis anterior of dystrophin null mice. PLoS Curr. 2013;5:ecurrents.md.e1226cefa851a2f079bbc406c0a21e80. doi: 10.1371/currents.md.e1226cefa851a2f079bbc406c0a21e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang D, et al. miR-182 regulates metabolic homeostasis by modulating glucose utilization in muscle. Cell Rep. 2016;16:757–768. doi: 10.1016/j.celrep.2016.06.040. [DOI] [PubMed] [Google Scholar]
- 17.Pan D, et al. MicroRNA-378 controls classical brown fat expansion to counteract obesity. Nat Commun. 2014;5:4725. doi: 10.1038/ncomms5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eichner LJ, et al. miR-378(*) mediates metabolic shift in breast cancer cells via the PGC-1β/ERRγ transcriptional pathway. Cell Metab. 2010;12:352–361. doi: 10.1016/j.cmet.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Carrer M, et al. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc Natl Acad Sci USA. 2012;109:15330–15335. doi: 10.1073/pnas.1207605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu W, et al. Hepatic miR-378 targets p110α and controls glucose and lipid homeostasis by modulating hepatic insulin signalling. Nat Commun. 2014;5:5684. doi: 10.1038/ncomms6684. [DOI] [PubMed] [Google Scholar]
- 21.Bodine SC, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 22.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li P, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 24.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 25.Castets P, et al. Sustained activation of mTORC1 in skeletal muscle inhibits constitutive and starvation-induced autophagy and causes a severe, late-onset myopathy. Cell Metab. 2013;17:731–744. doi: 10.1016/j.cmet.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Sandri M, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riedl SJ, Salvesen GS. The apoptosome: Signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8:405–413. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Y, et al. Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein Cell. 2010;1:468–477. doi: 10.1007/s13238-010-0048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, et al. MicroRNA-378 protects against intestinal ischemia/reperfusion injury via a mechanism involving the inhibition of intestinal mucosal cell apoptosis. Cell Death Dis. 2017;8:e3127. doi: 10.1038/cddis.2017.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang N, et al. MicroRNA-378 alleviates cerebral ischemic injury by negatively regulating apoptosis executioner caspase-3. Int J Mol Sci. 2016;17:E1427. doi: 10.3390/ijms17091427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S, Yang F, Wang M, Cao W, Yang Z. miR-378 functions as an onco-miRNA by targeting the ST7L/Wnt/β-catenin pathway in cervical cancer. Int J Mol Med. 2017;40:1047–1056. doi: 10.3892/ijmm.2017.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang J, et al. Overexpression of microRNA-378 attenuates ischemia-induced apoptosis by inhibiting caspase-3 expression in cardiac myocytes. Apoptosis. 2012;17:410–423. doi: 10.1007/s10495-011-0683-0. [DOI] [PubMed] [Google Scholar]
- 35.Bargiela A, et al. Increased autophagy and apoptosis contribute to muscle atrophy in a myotonic dystrophy type 1 Drosophila model. Dis Model Mech. 2015;8:679–690. doi: 10.1242/dmm.018127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pauly M, et al. AMPK activation stimulates autophagy and ameliorates muscular dystrophy in the mdx mouse diaphragm. Am J Pathol. 2012;181:583–592. doi: 10.1016/j.ajpath.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 37.Rosenberg AS, et al. Immune-mediated pathology in Duchenne muscular dystrophy. Sci Transl Med. 2015;7:299rv4. doi: 10.1126/scitranslmed.aaa7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Green DR, Kroemer G. The pathophysiology of mitochondrial cell death. Science. 2004;305:626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 39.Nakahira K, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manning BD, Toker A. AKT/PKB signaling: Navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calleja V, et al. Acute regulation of PDK1 by a complex interplay of molecular switches. Biochem Soc Trans. 2014;42:1435–1440. doi: 10.1042/BST20140222. [DOI] [PubMed] [Google Scholar]
- 42.Brault JJ, Jespersen JG, Goldberg AL. Peroxisome proliferator-activated receptor gamma coactivator 1alpha or 1beta overexpression inhibits muscle protein degradation, induction of ubiquitin ligases, and disuse atrophy. J Biol Chem. 2010;285:19460–19471. doi: 10.1074/jbc.M110.113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sopariwala DH, et al. Long-term PGC1β overexpression leads to apoptosis, autophagy and muscle wasting. Sci Rep. 2017;7:10237. doi: 10.1038/s41598-017-10238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L, et al. Apoptosis induced by PGC-1β in breast cancer cells is mediated by the mTOR pathway. Oncol Rep. 2013;30:1631–1638. doi: 10.3892/or.2013.2628. [DOI] [PubMed] [Google Scholar]
- 45.Yoon MS. mTOR as a key regulator in maintaining skeletal muscle mass. Front Physiol. 2017;8:788. doi: 10.3389/fphys.2017.00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song Y, et al. Ligand-dependent corepressor acts as a novel corepressor of thyroid hormone receptor and represses hepatic lipogenesis in mice. J Hepatol. 2012;56:248–254. doi: 10.1016/j.jhep.2011.07.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.