Abstract
Aim
To analyze the distribution of SLC6A4 gene polymorphisms in Crohn’s disease (CD) patients and their association with the disease.
Methods
We evaluated the presence/absence of promoter (5-HTTLPR, rs25531) and intron 2 (STin2 VNTR) polymorphic variants of SLC6A4 gene in a retrospective case-control study including 192 CD patients and 157 healthy controls (HC). Genotyping was performed by polymerase chain reaction. The association of polymorphisms with CD and its clinical subtypes was analyzed using χ2 and Fisher exact test, binary logistic regression, and haplotype analysis.
Results
CD patients and healthy controls had similar sex (88 [45.8%] vs 84 [53.5%] women, respectively; P = 0.154) and age (41.3 ± 12.8 years vs 41.7 ± 8.8 years, respectively, P = 0.091) distribution. Significant differences were observed in the STin2 genotype and allele distribution between CD patients and healthy controls (P = 0.003 and P = 0.002, respectively) and between the corresponding female subgroups (P = 0.004 and P = 0.007, respectively), with a significant negative association of biallelic ss (STin2.9 and Stin2.10) STin2 genotype with CD (P = 0.013, age- and sex-adjusted odds ratio [OR] 0.5, 95% confidence interval [CI] 0.29-0.86; women: P = 0.006, age-adjusted OR 0.32, 95% CI 0.14-0.72) and a significantly higher S-STin2.12 (5-HTTLPR/rs25531: S-STin2: STin2.12) haplotype distribution in CD patients (P = 0.004, OR 1.62, 95% CI 1.16-2.26). There was no significant association between 5-HTTLRP and rs25531 genotype or allele frequencies and CD and between any SLC6A4 polymorphic loci with clinical CD subtypes.
Conclusion
STin2 VNTR polymorphism of SLC6A4 gene may contribute to CD pathogenesis.
Inflammatory bowel disease (IBD), with its constituent clinical phenotypes, Crohn’s disease (CD) and ulcerative colitis (UC), represents a major relapsing gastrointestinal (GI) disorder, with a combined incidence of 2-20 per 100 000 individuals in the developed countries (1-4). IBD susceptibility is influenced by a variety of factors, including genetic polymorphism, GI motility, stress response, visceral hypersensitivity, abnormal immune response, and its reaction to enteromicrobial pathogens (1-4).
Neuroimmunological interactions are also important because various pro- and anti-inflammatory cytokines may affect neuronal activity and the release of neurotransmitters influencing the activity of immuno-effector cells in the GI tract. The best characterized among them is serotonin (5-HT), 90% of which is produced and secreted by intestinal enterochromaffin cells (5-8). Alterations in 5-HT biosynthesis, quantity, release, or clearance are important for both sensory signal transduction in GI motility and the development of visceral hypersensitivity (5-8). 5-HT is also a chemotactic molecule and may promote lymphocyte activation and secretion of pro-inflammatory cytokines (5-8). Therefore, upon its release and binding to targeted receptors, 5-HT action must be rapidly terminated. This is maintained by the action of serotonin re-uptake transporter (SERT), expressed by serotonergic neurons and the mucosal enterocytes (5-8).
Animal models and studies on human cell lines and tissue indicate increased 5-HT availability and reduced SERT expression in the inflamed colon, accompanied with an increased expression of inflammatory genes, thus supporting the idea that the loss of SERT gene (SLC6A4) transcription can either cause intestinal inflammation or result from it (8,9). SLC6A4 gene polymorphisms have also been linked with its translation and expression (10,11).
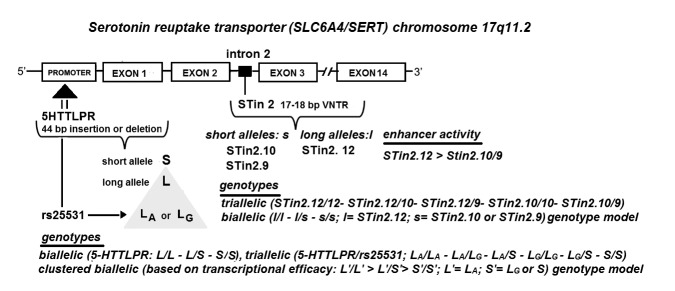
The most extensively studied among them are 5-HTTLPR, rs25531 (A/G) single nucleotide (SNP), and STin2 VNTR (variable number of tandem repeats) polymorphic regions found in the promoter (5-HTTLPR and rs25531) and intron 2 region (STin2 VNTR) of SLC6A4 gene (Figure 1) (12-19).
Figure 1.
Location of SLC6A4 gene polymorphic regions and their genotype combinations examined in this study. SLC6A4 gene is affected by the insertion/deletion variation of the promoter region 5-HTTLPR, resulting in two alleles, long L and small S. This promoter region also functionally couples with the common rs25531 (A/G) single nucleotide polymorphism (SNP), resulting in alleles LA and LG. LA allele is associated with the higher transcriptional activity, whereas the LG allele exhibits lower serotonin uptake and the transcriptional activity equivalent to S allele of the 5-HTTLPR polymorphic region. Transcriptional activity of SLC6A4 gene is further modulated by the enhancer activity of STin2 polymorphic region, 17–18 bp long variable number of tandem repeats (VNTR), found in intron 2. The STin2 allelic variants were identified as 10-repeat and 12-repeat alleles that have been identified in all ethnicities, and the less common 9-repeat allele found only in individuals of European or African descent. Thus, promoter polymorphic region may be presented in biallelic, triallelic, and clustered biallelic genotype model while STin2 VNTR region may be presented in triallelic and biallelic genotype forms. Rare allele forms (eg, XL or STin2.7) not encountered in this study, are not presented.
Sikander et al (20) has reported a significant association of 5-HTTLPR polymorphism with UC and microscopic colitis (MC). However, the association between 5-HTTLPR and CD has never been determined. The same was true for other SLC6A4 polymorphic regions in any form of IBD (11,20). Therefore, the aim of this study was to compare genotype and allele frequencies of 5-HTTLPR, rs25531and STin2 VNTR polymorphic regions in CD patients and healthy control participants and investigate their association with CD. Given the crucial role of SERT protein in the regulation of intestinal 5-HT availability, we hypothesized that the SLC6A4 polymorphism may contribute to the genetic predisposition for the development of CD.
PATIENTS AND METHODS
Patients
This retrospective case-control study included all 192 consecutive CD patients diagnosed between December 2012 and 2016 at the University Hospital Center Zagreb (Table 1). CD diagnosis was established according to standard endoscopic, radiological, and histopathological criteria (21). The healthy control group consisted of 157 participants with no family history of IBD or irritable bowel disease. They were selected from 217 completely healthy voluntary participants (hospital employees or their acquaintances) and age- and sex-matched to CD patient group. Genetically related voluntary control participants and participants who were incompatible according to age (outside the age range of CD group members) were excluded from further analysis. All participants signed the informed consent for study participation and data publication. The study protocol was approved by the Ethics Committee of the University Hospital Center Zagreb (Approval number 04/31-JG; contract number 108-1081874-1917, start date January 1, 2007).
Table 1.
Clinical characteristics of patients with Crohn’s disease (CD) analyzed in the study
| No. (%) of CD patients |
|||
|---|---|---|---|
| Characteristics | total | women | men |
| Age at diagnosis (years) | |||
| ≤16 | 21 (10.93) | 5 (23.8) | 16 (76.2) |
| 17-41 | 153 (79.69) | 76 (49.7) | 77 (50.3) |
| >41 | 18 (9.38) | 7 (38.9) | 11 (61.1) |
| Disease location | |||
| ileum | 60 (31.3) | 29 (48.3) | 31 (51.7) |
| colon | 34 (17.7) | 14 (41.2) | 20 (58.8) |
| ileocolon | 91 (47.4) | 41 (45.1) | 50 (54.9) |
| upper gastrointestinal (UGI) tract | 29 (15.1) | 17 (58.6) | 12 (41.4) |
| no UGI location | 157 (81.8) | 68 (43.3) | 89 (56.7) |
| Disease type | |||
| inflammatory | 89 (46.4) | 40 (44.9) | 49 (55.1) |
| stricturing | 64 (33.3) | 31(48.4) | 33 (51.6) |
| penetrating | 39 (20.3) | 17(43.6) | 22 (56.4) |
| Perianal disease | |||
| yes | 63 (32.8) | 24 (38.1) | 39 (61.9) |
| no | 129 (67.2) | 64(49.6) | 65 (50.4) |
| Extra-intestinal manifestation | |||
| yes | 43 (22.4) | 23 (53.5) | 20 (44.7) |
| no | 134 (69.8) | 57 (42.54) | 77 (57.46) |
| Family history | |||
| yes | 17 (8.85) | 8 (47.1) | 9 (52.9) |
| no | 175 (91.15) | 80 (45.71) | 95 (54.29) |
| Surgery | |||
| yes | 105 (54.69) | 45 (42.9) | 60 (57.1) |
| no | 87 (45.31) | 43 (49.4) | 44 (50.6) |
DNA extraction and genotyping
Blood samples were collected by venipuncture in EDTA BD Vacutainer® blood collection tubes (Becton, Dickinson and Company, Franklin Lakes, NJ, USA), and genomic DNA was isolated from peripheral blood leukocytes using the salting out procedure (22). Study participants were genotyped for both promoter and intron 2 polymorphisms of the SLC6A4 as previously described (22-24). Biallelic 5-HTTLRP variants were discriminated with some modifications as previously described (22-24). The following primers were used: forward 5′-ATG CCA GCA CCT AAC CCC TAA TGT-3′, reverse 5′-GGA CCG CAA GGT GGG CGG GA-3′. The 5-HTTLPR polymorphic region was subdivided into functional variants (S, LA, LG) by rs25531 (A/G) SNP using a TaqMan qPCR procedure according to Hu et al (25). Amplification was performed in a 7500 qPCR System under the following conditions: at 50°C for 2 minutes, at 95°C for 15 seconds, and at 62.5°C for 1.5 minutes. Genotype sequencing/capillary electrophoresis (ABI PRISM 3100 Genetic Analyzer, TermoFisher Scientific, Waltham, MA, USA) was performed to acquire controls. The STin2 VNTR polymorphism was discriminated using the method described by Ito et al (26). For the PCR reaction, the following primer pair was used: forward 5′-GTCAGTATCACAGGCTGCGAG-3′, reverse 5′-TGTTCCTAGTCTTACGCCAGTG-3′.
Statistical analysis
The normality of data distribution was analyzed by Shapiro-Wilk test. Data are presented as frequencies or means with standard deviations (±SD). The groups were compared using t test or Mann-Whitney test for parametric and non-parametric continuous variables, respectively, and Pearson χ2 test for categorical variables. Conformity of genotype distributions to Hardy–Weinberg equilibrium was assessed using goodness-of-fit χ2 test. The difference in allele frequency was determined by using either χ2 or Fisher exact test as appropriate. Crude and adjusted odds ratios (OR) (controlling for age and sex) and the corresponding 95% confidence intervals (CI) determined by binary logistic regression were also used to test the association between CD and genotype variants of both promoter and intron 2 SLC6A4 gene polymorphisms. Association analysis based on patient sex, or clinical CD subtypes such as age at diagnosis, localization, and disease behavior were also performed. Considering that LG allele has the same transcriptional activity as the S allele, the triallelic 5-HTTLPR-rs25531 genotypes were also coded in a clustered biallelic form (L'L', L'S' and S'S'; Figure 1). Likewise, the triallelic STin2 VNTR polymorphism was also coded in a biallelic (ll - ls - ss; Figure 1) genotype form. All biallelic genotype forms were analyzed under the codominant, dominant, and recessive model. Construction of haplotypes, linkage disequilibrium (LD), and haplotype association analysis were calculated by permutation test using the online SHEsisPlus software (27).
The power analysis (to compute a priori sample size and the post-hoc power of the study) was performed using G*Power software (version 3.1.9.2 for Windows) (28). For the analysis of 5-HTTLRP genotype frequencies with a χ2 goodness-of-fit test with the small effective size (w = 0.2, df = 2); power (1-β) = 0.80, and α = 0.017 (Bonferroni corrected P value), the required total sample size was 313 and actual sample size was 349. Post-hoc computed achieved power (1-β) of analysis was 0.085 (w = 0.2), 0.997 (w = 0.3; medium effective size), 1.00 (w = 0.5; large effective size) for genotype distribution, and 0.80 for allele frequency analysis (w = 0.2 - 0.5; df = 1). For the a priori analysis of STin2 VNTR genotype frequencies with a χ2 goodness-of-fit test with the small effective size (w = 0.23, df = 4); power (1-β) = 0.80 and α = 0.008 (Bonferroni corrected P value), the required total sample size was 329, and the post-hoc achieved power (1-β) of analysis was 0.83-1.00 for genotype distribution and 0.99-1.00 (w = 0.2 - 0.5; df = 2) for allele frequency analysis. Furthermore, with a power of 0.80 we were able to detect an effect size of 0.223 for a significant difference in STin2 VNTR genotype frequencies. The two-tailed P < 0.05 was considered significant and corrected according to Bonferroni procedure (the corrected level of significance is: Pc = 0.05/N; N - number of independent tests). All reported P values were uncorrected unless stated otherwise. Statistical analysis was performed using SPSS Statistics software trial version (IBM Corp., Armonk, NY, USA), unless stated otherwise.
RESULTS
Population characteristics
There were 88 (45.8%) women and 104 (54.2%) men in the CD group, and 84 (53.5%) women and 73 (46.5%) men in the control group (Pearson χ2 = 2.033, P = 0.154). The groups were also comparable according to age (mean±SD: 41.3 ± 12.8 years vs 41.7 ± 8.7 years, respectively; Mann-Whitney test, P = 0.091).
Association analysis
According to the Hardy-Weinberg equilibrium, there was no deviation in the 5-HTTLPR, STin2, and rs25531 genotype distribution in CD group (5-HTTLPR: χ2 = 0.557, df = 1, P = 0.756; rs25531: χ2 = 1.151, df = 3, P = 0.949; STin2: χ2 = 0.585, df = 3, P = 0.964) and healthy control group (5-HTTLPR: χ2 = 0.156, df = 1, P = 0.924; rs25531: χ2 = 2.358, df = 3, P = 0.797; STin2 VNTR: χ2 = 7.224, df = 3, P = 0.124). Furthermore, allele and genotype frequencies of all polymorphic loci in the control group corresponded well to the previously published data for Croatian and other European populations (18,29-31).
CD group and control group showed a different distribution of STin2 VNTR allele frequencies, which remained significant after Bonferroni correction (Table 2). This was also the case for women with CD (χ2 = 9.850, df = 2, P = 0.007) and their healthy female controls (data not shown). In addition, STin2.12 allele showed a positive and STin2.9 allele a negative association with CD both in the overall (Table 2) patient sample and in the subgroup of female patients (data not shown). However, the association remained significant only for the rare STin2.9 allele in the overall study sample and the subgroup of female patients after Bonferroni correction (Table 2). No significant association following Bonferroni correction was found between STin2 VNTR allele frequency and clinical CD subtypes (data not shown).
Table 2.
5-HTTLPR, STin2 VNTR and rs25531 SNP allele frequency distribution in patients with Crohn’s disease and healthy controls*
| Polymorphism | No. (%) of participants |
Pearson χ2 |
Individual comparisons |
||||
|---|---|---|---|---|---|---|---|
| CD | controls | χ2 | P‡ | χ2 | P‡ | OR (95% CI)§ | |
| 5-HTTLRP | |||||||
| L | 231 (60.2) | 208 (66.2) | 2.74 | 0.098 | 2.74 | 0.098 | 0.77 (0.56-1.05) |
| S | 153 (39.8) | 106 (33.8) | 2.74 | 0.098 | 1.30 (0.95-1.77) | ||
| rs25531 | |||||||
| LA | 212 (55.2) | 185 (58.9) | 3.76 | 0.152 | 0.97 | 0.325 | 0.86 (0.64-1.16) |
| LG | 19 (5.0) | 23 (7.3) | 1.73 | 0.188 | 0.66 (0.35-1.23) | ||
| S | 153 (39.8) | 106 (33.8) | 2.74 | 0.097 | 1.30 (0.95-1.77) | ||
| STin2 VNTR | |||||||
| 12 | 242 (63.0) | 174 (55.4) | 12.03 | 0.002† | 4.15 | 0.042 | 1.37 (1.01-1.86) |
| 10 | 141 (36.7) | 130 (41.4) | 1.59 | 0.207 | 0.82 (0.61-1.12) | ||
| 9 | 1 (0.3) | 10 (3.2) | 0.002 | 0.08 (0.01-0.62) | |||
*CD – Crohn’s disease; 5-HTTLPR – serotonin reuptake transporter (SLC6A4) length polymorphic region; STin2 VNTR – variable number of tandem repeats (VNTR) found in intron 2 of SLC6A4 gene; rs25531 – single nucleotide (SNP) polymorphism found in the background of longer 5-HTTLPR allele variant; L – long 5-HTTLPR allele; S – short 5-HTTLPR allele; LA – long 5-HTTLPR allele associated with the higher transcriptional activity; LG – long 5-HTTLPR allele exhibiting lower serotonin uptake and transcriptional activity equivalent to the short (S) allele of the 5-HTTLPR polymorphic region; 12, 10, 9 – STin2 VNTR alleles; OR – odds ratio; CI – confidence interval.
†Bonferroni correction Pc = 0.05/3 = 0.017.
‡Bonferroni non-adjusted P values.
§Logistic regression adjusted for age and sex.
There was no significant association in 5-HTTLRP and rs25531 allele frequency between the CD and control group or between the corresponding sex subgroups (data not shown). The results remained the same after the analysis for clinical CD subtypes (data not shown). Nevertheless, we did observe a positive, although non-significant, association of S allele of 5-HTTLRP and rs25531 polymorphism with CD (Table 2).
Pearson χ2 test showed significant differences in STin2 genotype distribution (Table 3) between CD patients and healthy controls (P = 0.003) and between the corresponding female (χ2 = 15.326, df = 4, P = 0.004) subgroups (data not shown). This was even more pronounced when carriers of the rare STin2 genotype forms (STin2 12/9 + STin2 10/9; Fisher exact test for CD vs controls; P = 0.002) were grouped together (overall CD: χ2 = 15.720, df = 3, P = 0.001; female subgroup: χ2 = 15.326, df = 3, P = 0.002). Furthermore, STin2 12/12 and Stin2 12/10 genotypes were more common, while STin2 10/10 genotype was less common among CD patients and controls. The same was true when we analyzed the corresponding female subgroups only (data not shown). However, even at very liberal correction for multiple comparisons of Pc = 0.008 (Pc = 0.05/N – number of genotypes analyzed), we did not find any significant difference in the distribution of individual STin2 VNTR genotypes between CD and controls (Table 3).
Table 3.
Logistic regression analysis of STIN2 VNTR genotype distribution in patients with Crohn’s disease and healthy controls*
| Polymorphism | No. (%) of participants | Logistic regression |
||||
|---|---|---|---|---|---|---|
| CD | controls | crude OR (95% CI) | P† | OR (95% CI)‡ | P† | |
| STin2 VNTR triallelic§ | 0.038II | 0.043II | ||||
| codominant model A | ||||||
| 12/12 | 76 (39.6) | 55 (35.0) | reference 1.0 | reference 1.0 | ||
| 12/10 | 89 (46.4) | 57 (36.3) | 1.13 (0.7-1.83) | 0.618 | 1.15 (0.71-1.86) | 0.570 |
| 10/10 | 26 (13.5) | 35 (22.3) | 0.54 (0.29-0.99) | 0.048 | 0.55 (0.30-1.02) | 0.056 |
| 12/9 | 1 (0.5) | 7 (4.5) | 0.10 (0.01-0.86) | 0.036 | 0.11 (0.013-0.89) | 0.040 |
| 10/9 | 0 (0.0) | 3 (1.9) | NA | 0.990 | NA | 0.100 |
| codominant model B¶ | 0.008** | 0.009** | ||||
| 12/12 | 76 (39.6) | 55 (35.0) | reference 1.0 | reference 1.0 | ||
| 12/10 | 89 (46.4) | 57 (36.3) | 1.13 (0.7-1.83) | 0.618 | 1.15 (0.71-1.86) | 0.600 |
| 10/10 | 26 (13.5) | 35 (22.3) | 0.54 (0.29-0.99) | 0.048 | 0.55 (0.3-1.02) | 0.060 |
| 9/others | 1 (0.5) | 10 (6.4) | 0.07 (0.01-0.58) | 0.014** | 0.073 (0.01-0.59) | 0.014** |
| STin2 VNTR biallelic | 0.041II | 0.045II | ||||
| codominant model | ||||||
| l/l | 76 (39.6) | 55 (35.0) | 2.02 (1.10-3.71) | 0.023 | 1.98 (1.08-3.64) | 0.028 |
| l/s | 90 (46.9) | 64 (40.8) | 2.06 (1.14-3.72) | 0.017 | 2.05 (1.13-3.71) | 0.018 |
| s/s | 26 (13.5) | 38 (24.2) | reference 1.0 | reference 1.0 | ||
| dominant | ||||||
| l/l | 76 (39.6) | 55 (35.0) | reference 1.0 | reference 1.0 | ||
| l/s-s/s | 116 (60.4) | 102 (65.0) | 0.82 (0.53-1.27) | 0.380 | 0.84 (0.54-1.30) | 0.400 |
| recessive | ||||||
| l/l-l/s | 166 (86.5) | 119 (75.8) | reference 1.0 | reference 1.0 | ||
| s/s | 26 (13.5) | 38 (24.2) | 0.49 (0.28-0.85) | 0.011** | 0.5 (0.29-0.86) | 0.013** |
*CD – Crohn’s disease; STin2 VNTR – variable number of tandem repeats (VNTR) found in intron 2 of serotonin reuptake transporter (SLC6A4) gene; OR – odds ratio; CI – confidence interval. l – Long (12) STin2 alleles; s – short (10, 9) STin2 alleles; 12, 10, 9: STin2 VNTR alleles.
†Bonferroni non-adjusted P values.
‡Logistic regression adjusted for age and sex.
§Overall STin2 VNTR genotype distribution in CD patients and healthy controls: χ2 = 15.857, df = 4, P = 0.003.
IIGlobal P value for logistic regression analysis.
¶Rare genotype variants grouped together.
**Significant P values (<Pc). Bonferroni correction for genotype distribution analysis Pc = 0.008 (0.05/6 – number of genotypes) and for logistic regression analysis Pc = 0.017 (0.05/3 – number of genetic model comparisons).
In logistic regression analysis, the co-dominant triallelic STin2 genotype model did not show a significant association of individual STin2 genotypes with CD (Table 3). However, when men and women were analyzed together under the recessive biallelic model (Table 3) or when women only were analyzed (data not shown), carriers of biallelic ss (s = STin2 10 or STin2 9), STin2 genotype form exhibited a significant negative association with CD compared with ll and ls carriers (female patients: Wald = 7.564, df = 1, P = 0.006, age-adjusted OR = 0.32, 95% CI = 0.14-0.72), which remained significant after sex and/or age adjustment (female patients: Wald = 7.564, df = 1, P = 0.006, OR adjusted by age = 0.32, 95% CI = 0.14-0.72). Nevertheless, only the variation of co-dominant STin2 model in which the rare genotypes (STin2 12/9 and 10/9) were grouped together showed significant global P values in crude and adjusted logistic regression analysis, and only when male and female patients were analyzed together (Table 3).
No significant difference was observed in 5-HTTLRP and rs25531 genotype distribution between CD group and controls (Table 4) or between sex subgroups and clinical CD subtypes (data not shown). The same was found when biallelic 5-HTTLRP and rs25531 genotype forms were analyzed under the codominant, dominant, or recessive genetic model. Nevertheless, we did observe more frequent, although non-significant, distribution of heterozygous (LS, LALG, L'S') and homozygous SS genotype forms in the CD group (Table 4).
Table 4.
Logistic regression analysis of 5-HTTLRP and rs25531 genotype distribution in patients with Crohn’s disease and healthy controls*
| Polymorphism | No. (%) of participants | Logistic regression |
||||
|---|---|---|---|---|---|---|
| CD | controls | crude OR (95% CI) | P† | OR (95% CI) ‡ | P† | |
| 5-HTTLRP biallelic§ | 0.180II | 0.200II | ||||
| codominant | ||||||
| L/L | 67 (34.9) | 70 (44.6) | reference 1.0 | reference 1.0 | ||
| L/S | 97 (50.5) | 68 (43.3) | 1.50 (0.94-2.35) | 0.090 | 1.48 (0.94-2.35) | 0.090 |
| S/S | 28 (14.6) | 19 (12.1) | 1.54 (0.79-3.02) | 0.210 | 1.52 (0.77-2.98) | 0.230 |
| dominant | ||||||
| L/L | 67 (34.9) | 70 (44.6) | reference 1.0 | reference 1.0 | ||
| L/S-S/S | 125 (65.1) | 87 (55.4) | 1.50 (0.97-2.31) | 0.070 | 1.49 (0.97-2.30) | 0.070 |
| recessive | ||||||
| L/L-L/S | 164 (85.4) | 138 (87.9) | reference 1.0 | reference 1.0 | ||
| S/S | 28 (14.6) | 19 (12.1) | 1.24 (0.66-2.32) | 0.500 | 1.22 (0.65-2.29) | 0.530 |
| 5-HTTLRP/rs25531 triallelic¶ | 0.670II | 0.670II | ||||
| codominant | ||||||
| LA/LA | 57 (29.7) | 57 (36.3) | reference 1.0 | reference 1.0 | ||
| LA/LG | 10 (5.2) | 11 (7.0) | 0.91 (0.36-2.31) | 0.840 | 0.91 (0.36-2.31) | 0.840 |
| LA/S | 88 (45.8) | 60 (38.2) | 1.47 (0.90-2.40) | 0.130 | 1.47 (0.90-2.41) | 0.130 |
| LG/S | 9 (4.7) | 8 (5.1) | 1.13 (0.41-3.12) | 0.820 | 1.07 (0.38-2.98) | 0.900 |
| LG/LG | 0 (0.0) | 2 (1.3) | NA | 0.990 | NA | 0.100 |
| S/S | 28 (14.6) | 19 (12.1) | 1.47 (0.74-2.93) | 0.270 | 1.46 (0.73-2.90) | 0.290 |
| 5-HTTLRP/rs25531 biallelic | 0.410II | 0.410II | ||||
| codominant | ||||||
| L'/L' | 57 (29.7) | 57 (36.3) | reference 1.0 | reference 1.0 | ||
| L'/S' | 98 (51.0 | 71 (45.2) | 1.38 (0.86-2.23) | 0.190 | 1.39 (0.86-2.24) | 0.180 |
| S'/S' | 37 (19.3) | 29 (18.5) | 1.28 (0.69-2.35) | 0.430 | 1.25 (0.68-2.31) | 0.470 |
| dominant | ||||||
| L'/L' | 57 (29.7) | 57 (36.3) | reference 1.0 | reference 1.0 | ||
| L'/S'- S'/S' | 135 (70.3) | 100 (63.7) | 1.35 (0.86-2.12) | 0.190 | 1.346 (0.86-2.11) | 0.200 |
| recessive | ||||||
| L'/L'- L'/S' | 155 (80.7) | 128 (81.5) | reference 1.0 | reference 1.0 | ||
| S'/S' | 37 (19.3) | 29 (18.5) | 1.05 (0.61-1.81) | 0.850 | 1.032 (0.60-1.77) | 0.910 |
*CD – Crohn’s disease; 5-HTTLPR – serotonin reuptake transporter (SLC6A4) length polymorphic region; rs25531 – single nucleotide (SNP) polymorphism found in the background of longer 5-HTTLPR allele variant; L’: LA; S’: LG or S (clustered biallelic 5-HTTLRP/rs25531 model based on transcriptional activity: L’/L’>L’/S’>S’/ S’) OR – odds ratio, CI – confidence interval.
†Bonferroni non-adjusted P values; Bonferroni correction for genotype distribution analysis Pc = 0.0083 (0.05/6 - number of genotypes) and for logistic regression analysis Pc = 0.0166 (0.05/3 - number of genetic model comparisons).
‡Logistic regression adjusted for age and sex.
§Overall 5-HTTLRP genotype distribution between CD and healthy control group: χ2 = 3.410, df = 2, P = 0.182;
IIglobal P value of logistic regression analysis.
¶Overall triallelic 5-HTTLRP/rs25531 genotype distribution between CD and healthy control group: χ2 = 5.674, df = 5, P = 0.339.
Haplotype analysis
5-HTTLPR and rs25531 polymorphic region were in strong linkage disequilibrium, while LDs between them and STin2 VNTR polymorphic region were relatively weak (Figure 2). After Bonferroni correction, only S-S-STin2.12 haplotype showed a significantly higher frequency in CD group (Table 5). The higher frequency in CD group was also revealed for L-LG-STin2.10 haplotype, but it was not significant (Table 5).
Figure 2.
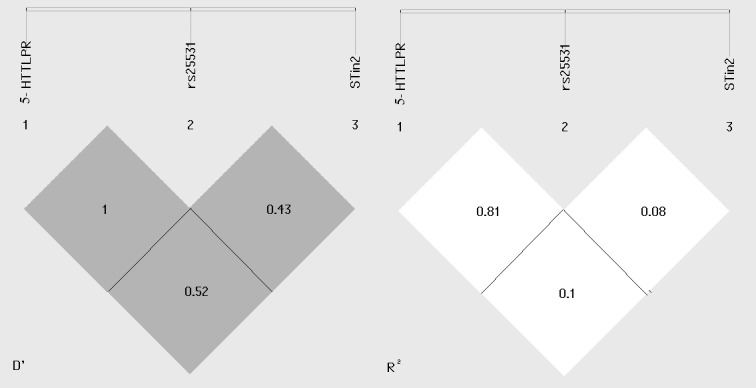
Linkage disequilibrium (LD) in SLC6A4 gene with D' and r2 values.
Table 5.
The frequency of SLC6A4 haplotypes in patients with Crohn’s disease and their healthy controls*
| SLC6A4 haplotypes | No. (%) of participants | χ2 | P† | OR (95% CI)† | |||
|---|---|---|---|---|---|---|---|
| 5-HTTLRP | rs25531 | STin2 VNTR | CD | controls | |||
| L | LA | STin2 10 | 110 (28.6) | 97 (30.8) | 0.417 | 0.518 | 0.89 (0.65-1.24) |
| S | - | STin2 12 | 131 (34.1) | 76 (24.2) | 8.133 | 0.004‡ | 1.62 (1.16-2.26) |
| L | LA | STin2 12 | 102 (26.5) | 88 (28.0) | 0.186 | 0.665 | 0.93(0.66-1.30) |
| L | LG | STin2 10 | 9 (2.3) | 6 (1.9) | 0.153 | 0.694 | 1.23 (0.43-3.50) |
| S | - | STin2 10 | 22 (5.7) | 27 (8.5) | 2.179 | 0.139 | 0.65 (0.36-1.16) |
| L | LG | STin2 12 | 9 (2.3) | 10 (3.1) | 0.461 | 0.496 | 0.73 (0.29-1.82) |
| L | LG | STin2 9 | 1 (0.2) | 7 (2.2) | 5.910 | 0.015 | 0.11 (0.01-0.94) |
| S | - | STin2 9 | 0 (0.0) | 3 (0.9) | 3.684 | 0.054 | NA |
* CD – Crohn’s disease; 5-HTTLPR – serotonin reuptake transporter (SERT/SLC6A4) length polymorphic region; STin2 VNTR – variable number of tandem repeats (VNTR) found in intron 2 of SLC6A4 gene; rs25531 – single nucleotide (SNP) polymorphism found in the background of longer 5-HTTLPR allele variant; LG – Long 5-HTTLPR allele exhibiting lower serotonin uptake and transcriptional activity equivalent to the short (S) allele of the 5-HTTLPR polymorphic region; LG – Long 5-HTTLPR allele exhibiting lower serotonin uptake and transcriptional activity equivalent to the short (S) allele of the 5-HTTLPR polymorphic region; Global result – Pearson χ2 = 18.29, Pearson P = 0.011; OR – odds ratio; CI – confidence interval.
†Bonferroni non-adjusted P values and confidence intervals; Bonferroni correction for haplotype analysis: Pc = 0.006 (0.05/8 – number of haplotype comparisons).
‡Significant P value (<Pc).
DISCUSSION
Our data revealed a significant difference in STin2 VNTR genotype and allele distribution between the overall CD group and healthy controls and between the female patient group and the corresponding control group. There was also a significant negative association of biallelic ss (STin2. 10/10 and STin2. 10/9 vs STin2. 12/12, 12/10 and 12/9 combined) genotype form and CD. We also found a significantly higher S-STin2.12 (5-HTTLPR/rs25531: S - STin2: STin2.12) haplotype distribution among CD patients. However, we did not find any association between 5-HTTLRP and rs25531 genotype or allele frequencies and CD or between any SLC6A4 polymorphic loci and clinical subtypes of CD. Nevertheless, our results indicate that STin2 VNTR polymorphism of SLC6A4 gene may contribute to CD pathogenesis.
Altered function and down-regulation of SERT protein expression paralleled with abnormal 5-HT concentration, both locally in epithelial layer and in the circulation, has been documented in several human GI inflammatory conditions including CD, UC, and MC, as well as diverticulitis and active celiac disease (32-34). This was also documented in several animal models of intestinal inflammation (35-39). Pro-inflammatory mediators and growth factors released during IBD may down-regulate SLC6A4 transcription and decrease SERT protein expression and function (40-43).
Another possibility is that some individuals with IBD have a genetic predisposition leading to altered SERT expression with consequent changes in gastrointestinal 5-HT levels, which contributes indirectly to pro-inflammatory conditions in the affected intestinal mucosa.
However, despite the recognition that intestinal inflammatory diseases, such as CD, can have a strong genetic component, polymorphisms so far linked with SERT transporter have not been associated with CD.
Furthermore, in previous reports on GI diseases only STin2 VNTR was observed as an attractive candidate for a possible association of the SERT with IBS (10,44). Although IBD and IBS are usually viewed as dichotomous conditions, they exhibit similar alterations in serotonergic-signaling mechanisms (33,44-46). Due to the development of IBS symptoms in IBD patients in remission and the clinical overlap between IBD and IBS, some authors even argued that they may represent clinical manifestations of a pathophysiologic spectrum of the same disease (47,48). Several population studies showed that IBS patients have an increased risk of becoming IBD patients than patients without prior IBS history, and this effect may be greater for CD (49,50).
Reports regarding the link between STin2 polymorphism and IBS were controversial and inconclusive, with most of them indicating no association with IBS or its clinical subtypes (47). However, Wang et al (51) found that IBS patients had a greater frequency of STin2.12/10 and a lower frequency of STin2.12/12 genotype compared to controls, but identified no significant difference in STin2 polymorphism among different clinical subtypes of IBS.
Current research in functional implications of the STin2 polymorphism is also inconclusive. It is known that STin2 acts as a transcriptional regulator and has allele-dependent enhancer-like properties that may influence tissue-specific regulation of SLC6A4 gene (18,52). However, it seems that individual SLC6A4 polymorphisms have a weak influence compared with the combined effect of 5-HTTLPR and STin2 region (14,18,52,53). Therefore, their allelic combinations should be identified before concluding about functional and phenotypic associations. LD between these two loci, ranging from moderate (European) to very strong (Native Americans), was found in most of the studied populations (31). In addition, a partial linkage of STin2.12 allele with S allele of 5-HTTLPR (S12 haplotype combination) has been reported and had stronger enhancer-like properties on SLC6A4 transcription than L10 or S10 haplotype (53,54). We found a significantly higher distribution of S12 haplotype in CD group, showing its positive association with disease occurrence. However, since we did not measure the level of SERT expression or 5-HTT plasma or tissue levels, we were not able to indicate any functional consequences of this finding.
We did not confirm the association of 5-HTTLRP and rs25531 with CD occurrence, but we did observe the tendency toward higher frequencies of heterozygote 5-HTTLRP and rs25531 genotype forms in CD group. This effect of molecular heterosis was also described in some other studies of 5-HTTLPR (55-57).
To the best of our knowledge, there are no reports analyzing the relationship between the promoter polymorphic regions of the SLC6A4 gene with CD. Sikander et al (20) demonstrated a potential association between 5-HTTLPR polymorphism and UC and MC by finding a significantly lower frequency of SS vs non-SS (LL and LS combined) 5-HTTLPR genotypes in MC patients or UC patients in remission and significantly higher serum 5-HT levels (SS>LS>LL) in these two patient groups. This was especially the case in MC LS and SS genotype carriers compared with controls (20). In addition, UC patients with active disease and SS genotype also had increased 5-HT levels compared with control subjects expressing the same genotype (20). Similar to our results in CD patients, they did not observe any significant differences in 5-HTTLPR genotype and allele distributions between UC patients with active disease and healthy controls or between male and female patient subgroups (20). Likewise, Shiotani et al (34) also observed no significant association of 5-HTTLPR variants and UC. Our results were contrary to the findings of Sikander et al (20), as we found a higher frequency of SS genotype in CD group.
The observed discrepancies are possibly due to heterogeneous background of CD and other forms of IBD and/or ethnical or regional differences between the studied groups. They are also consistent with the previously mentioned hypothesis that 5-HTTLPR might represent only one of the contributing polymorphic factors responsible for the disease occurrence.
Regarding the IBS and 5-HTTLRP polymorphism, different associations were reported, but the results were contradictory and inconclusive even for the same population under the study. The most recent meta-analysis found no significant association between 5-HTTLPR and IBS in overall population, while in the IBS subtype- and ethnic subgroup-based analysis the LL genotype was demonstrated as a risk factor for constipation predominant IBS (58).
With respect to rs25531 polymorphism, we found only two studies examining its association with IBS, and only one reported a positive association with disease occurrence with three times higher odds ratio for the LG allele distribution in IBS patients compared to controls (59,60).
There are several limitations to our study. The sample size was too small for a comprehensive genotype analysis, so the results cannot be generalized and should be interpreted cautiously. Second, we did not determine the 5-HT levels and were unable to evaluate the functional consequences of individual genotype or haplotype variants on SERT expression. Third, association studies of unrelated individuals, such as ours, warrant cautious interpretation as unknown sources of population stratification may affect the results.
In conclusion, we demonstrated significant differences in STin2 genotype and allele distribution between CD patients and healthy controls, and a negative association of biallelic ss STin2 genotype variant and a positive association of S12 haplotype with CD disease. Further large-scale studies in this and other populations aimed to confirm the obtained results and decipher the exact functional role of STin2 VNTR polymorphic region in intestinal inflammatory diseases are warranted.
Acknowledgments
Oxford Centre for Evidence-based Medicine level of evidence: 3b/4a.
Funding This work has been funded by Croatian Science Foundation; project name: Inflammatory Bowel Diseases (Crohn’s disease and ulcerative colitis); project number 108-1081874-1917.
Ethical approval The study protocol was approved by the Ethics Committee of the University Hospital Center Zagreb (Approval number 04/31-JG; contract number 108-1081874-1917, start date January 1, 2007).
Declaration of authorship All authors except BD conceived and designed the study; all authors acquired the data, analyzed and interpreted the data, drafted the manuscript; critically revised the manuscript for important intellectual content, gave approval of the version to be submitted; and agree to be accountable for all aspects of the work.
Competing interests All authors have completed the Unified Competing Interest form at www.icmje.org/coi_disclosure.pdf (available on request from the corresponding author) and declare: no support from any organization for the submitted work; no financial relationships with any organizations that might have an interest in the submitted work in the previous 3 years; no other relationships or activities that could appear to have influenced the submitted work.
References
- 1.Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12:205–17. doi: 10.1038/nrgastro.2015.34. [DOI] [PubMed] [Google Scholar]
- 2.Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J Clin Invest. 2007;117:514–21. doi: 10.1172/JCI30587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burisch J. Crohn’s disease and ulcerative colitis. Occurrence, course and prognosis during the first year of disease in a European population-based inception cohort. Dan Med J. 2014;61:B4778. [PubMed] [Google Scholar]
- 4.Xia B, Crusius J, Meuwissen S, Pena A. Inflammatory bowel disease: definition, epidemiology, etiologic aspects, and immunogenetic studies. World J Gastroenterol. 1998;4:446–58. doi: 10.3748/wjg.v4.i5.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraneveld AD, Rijnierse A, Nijkamp FP, Garssen J. Neuro-immune interactions in inflammatory bowel disease and irritable bowel syndrome: future therapeutic targets. Eur J Pharmacol. 2008;585:361–74. doi: 10.1016/j.ejphar.2008.02.095. [DOI] [PubMed] [Google Scholar]
- 6.El-Salhy M, Solomon T, Hausken T, Gilja OH, Hatlebakk JG. Gastrointestinal neuroendocrine peptides/amines in inflammatory bowel disease. World J Gastroenterol. 2017;23:5068–85. doi: 10.3748/wjg.v23.i28.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gershon MD. Serotonin is a sword and a shield of the bowel: serotonin plays offense and defense. Trans Am Clin Climatol Assoc. 2012;123:268–80. [PMC free article] [PubMed] [Google Scholar]
- 8.Terry N, Margolis KG. Serotonergic mechanisms regulating the GI tract: Experimental Evidence and Therapeutic Relevance. Handb Exp Pharmacol. 2017;239:319–42. doi: 10.1007/164_2016_103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shajib MS, Khan WI. The role of serotonin and its receptors in activation of immune responses and inflammation. Acta Physiol (Oxf) 2015;213:561–74. doi: 10.1111/apha.12430. [DOI] [PubMed] [Google Scholar]
- 10.Makker J, Chilimuri S, Bella JN. Genetic epidemiology of irritable bowel syndrome. World J Gastroenterol. 2015;21:11353–61. doi: 10.3748/wjg.v21.i40.11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldner D, Margolis KG. Association of serotonin transporter promoter polymorphism (5HTTLPR) with microscopic colitis and ulcerative colitis: time to be asSERTive? Dig Dis Sci. 2015;60:819–21. doi: 10.1007/s10620-015-3598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, et al. Association of anxiety related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274:1527–31. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- 13.Heils A, Teufel A, Petri S, Stöber G, Riederer P, Bengel D, et al. Allelic variation of human serotonin transporter gene expression. J Neurochem. 1996;66:2621–4. doi: 10.1046/j.1471-4159.1996.66062621.x. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura M, Ueno S, Sano A, Tanabe H. The human serotonin transporter linked polymorphism (5HTTLPR) shows ten novel allelic variants. Mol Psychiatry. 2000;5:32–8. doi: 10.1038/sj.mp.4000698. [DOI] [PubMed] [Google Scholar]
- 15.Lesch KP, Wolozin BL, Murphy DL, Reiderer P. Primary structure of human platelet serotonin (5HT) uptake site-identity with the brain 5-HT transporter. J Neurochem. 1993;60:2319–22. doi: 10.1111/j.1471-4159.1993.tb03522.x. [DOI] [PubMed] [Google Scholar]
- 16.De Luca V, Tharmalingam S, King N, Strauss J, Bulgin N, Kennedy JL. Association study of a novel functional polymorphism of the serotonin transporter gene in bipolar disorder and suicidal behaviour. Psychopharmacology (Berl) 2005;182:128–31. doi: 10.1007/s00213-005-0046-z. [DOI] [PubMed] [Google Scholar]
- 17.Wendland JR, Martin BJ, Kruse MR, Lesch KP, Murphy DL. Simultaneous genotyping of four functional loci of human SLC6A4, with a reappraisal of 5-HTTLPR and rs25531. Mol Psychiatry. 2006;11:224–6. doi: 10.1038/sj.mp.4001789. [DOI] [PubMed] [Google Scholar]
- 18.Hranilovic D, Stefulj J, Furac I, Kubat M, Balija M, Jernej B. Serotonin transporter gene promoter (5-HTTLPR) and intron 2(VNTR) polymorphisms in Croatian suicide victims. Biol Psychiatry. 2003;54:884–9. doi: 10.1016/S0006-3223(03)00179-3. [DOI] [PubMed] [Google Scholar]
- 19.Battersby S, Ogilvie AD, Smith CA, Blackwood DH, Muir WJ, Quinn JP, et al. Structure of a variable number tandem repeat of the serotonin transporter gene and association with affective disorder. Psychiatr Genet. 1996;6:177–81. doi: 10.1097/00041444-199624000-00001. [DOI] [PubMed] [Google Scholar]
- 20.Sikander A, Sinha SK, Prasad KK, Rana SV. Association of serotonin transporter promoter polymorphism (5-HTTLPR) with microscopic colitis and ulcerative colitis. Dig Dis Sci. 2015;60:887–94. doi: 10.1007/s10620-014-3482-y. [DOI] [PubMed] [Google Scholar]
- 21.Feuerstein JD, Cheifetz AS. Crohn disease: epidemiology, diagnosis, and management. Mayo Clin Proc. 2017;92:1088–103. doi: 10.1016/j.mayocp.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 22.Markeljevic J, Sarac H, Bozina N, Henigsberg N, Simic M, Cicin Sain L. Serotonin transporter gene polymorphisms: Relation with platelet serotonin level in patients with primary Sjogren’s syndrome. J Neuroimmunol. 2015;282:104–9. doi: 10.1016/j.jneuroim.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Rausch JL, Johnson ME, Fei YJ, Li JQ, Shendarkar N, Hobby HM, et al. Initial conditions of serotonin transporter kinetics and genotype: influence on SSRI treatment trial outcome. Biol Psychiatry. 2002;51:723–32. doi: 10.1016/S0006-3223(01)01283-5. [DOI] [PubMed] [Google Scholar]
- 24.Bozina N, Medved V, Kuzman MR, Sain I, Sertic J. Association study of olanzapine-induced weight gain and therapeutic response with SERT gene polymorphisms in female schizophrenic patients. J Psychopharmacol. 2007;21:728–34. doi: 10.1177/0269881106072750. [DOI] [PubMed] [Google Scholar]
- 25.Hu XZ, Lipsky RH, Zhu G, Akhtar LA, Taubman J, Greenberg BD, et al. Serotonin transporter promoter gain-of-function genotypes are linked to obsessive-compulsive disorder. Am J Hum Genet. 2006;78:815–26. doi: 10.1086/503850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito K, Yoshida K, Sato K, Takahashi H, Kamata M, Higuchi H, et al. A variable number of tandem repeats in the serotonin transporter gene does not affect the antidepressant response to fluvoxamine. Psychiatry Res. 2002;11:235–9. doi: 10.1016/S0165-1781(02)00141-5. [DOI] [PubMed] [Google Scholar]
- 27.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–8. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 28.Faul F, Erdfelder E, Lang AG, Buchner AG. *Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39:175–91. doi: 10.3758/BF03193146. [DOI] [PubMed] [Google Scholar]
- 29.Bozina N, Mihaljević-Peles A, Sagud M, Jakovljević M, Sertić J. Serotonin transporter polymorphism in Croatian patients with major depressive disorder. Psychiatr Danub. 2006;18:83–9. [PubMed] [Google Scholar]
- 30.Noskova T, Pivac N, Nedic G, Kazantseva A, Gaysina D, Faskhutdinova G, et al. Ethnic differences in the serotonin transporter polymorphism (5-HTTLPR) in several European populations. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32:1735–9. doi: 10.1016/j.pnpbp.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 31.Gelernter J, Cubells JF, Kidd JR, Pakstis AJ, Kidd KK. Population studies of polymorphisms of the serotonin transporter protein gene. Am J Med Genet. 1999;88:61–6. doi: 10.1002/(SICI)1096-8628(19990205)88:1<61::AID-AJMG11>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 32.Tada Y, Ishihara S, Kawashima K, Fukuba N, Sonoyama H, Kusunoki R, et al. Downregulation of serotonin reuptake transporter gene expression in healing colonic mucosa in presence of remaining low-grade inflammation in ulcerative colitis. J Gastroenterol Hepatol. 2016;31:1443–52. doi: 10.1111/jgh.13268. [DOI] [PubMed] [Google Scholar]
- 33.Coates MD, Mahoney CR, Linden DR, Sampson JE, Chen J, Blaszyk H, et al. Molecular defects in mucosal serotonin content and decreased serotonin reuptake transporter in ulcerative colitis and irritable bowel syndrome. Gastroenterology. 2004;126:1657–64. doi: 10.1053/j.gastro.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 34.Shiotani A, Kusunoki H, Kimura Y, Ishii M, Imamura H, Tarumi K, et al. S100A expression and interleukin-10 polymorphisms are associated with ulcerative colitis and diarrhea predominant irritable bowel syndrome. Dig Dis Sci. 2013;58:2314–23. doi: 10.1007/s10620-013-2677-y. [DOI] [PubMed] [Google Scholar]
- 35.Linden DR, Foley KF, McQuoid C, Simpson J, Sharkey KA, Mawe GM. Serotonin transporter function and expression are reduced in mice with TNBS-induced colitis. Neurogastroenterol Motil. 2005;17:565–74. doi: 10.1111/j.1365-2982.2005.00673.x. [DOI] [PubMed] [Google Scholar]
- 36.Linden DR, Chen JX, Gershon MD, Sharkey KA, Mawe GM. Serotonin availability is increased in mucosa of guinea pigs with TNBS-induced colitis. Am J Physiol Gastrointest Liver Physiol. 2003;285:G207–16. doi: 10.1152/ajpgi.00488.2002. [DOI] [PubMed] [Google Scholar]
- 37.Bertrand PP, Barajas-Espinosa A, Neshat S, Bertrand RL, Lomax AE. Analysis of real-time serotonin (5-HT) availability during experimental colitis in mouse. Am J Physiol Gastrointest Liver Physiol. 2010;298:G446–55. doi: 10.1152/ajpgi.00318.2009. [DOI] [PubMed] [Google Scholar]
- 38.Oshima S, Fujimura M, Fukimiya M. Changes in number of serotonin-containing cells and serotonin levels in the intestinal mucosa of rats with colitis induced by dextran sodium sulfate. Histochem Cell Biol. 1999;112:257–63. doi: 10.1007/s004180050445. [DOI] [PubMed] [Google Scholar]
- 39.Matsumoto K. Immunohistochemical analysis of altered serotonin signaling and transient receptor potential vanilloid 1 channels in experimental colitis mice. Yakugaku Zasshi. 2014;134:1165–70. doi: 10.1248/yakushi.14-00189. [DOI] [PubMed] [Google Scholar]
- 40.Barbaro MR, Di Sabatino A, Cremon C, Giuffrida P, Fiorentino M, Altimari A, et al. Interferon-γ is increased in the gut of patients with irritable bowel syndrome and modulates serotonin metabolism. Am J Physiol Gastrointest Liver Physiol. 2016;310:G439–47. doi: 10.1152/ajpgi.00368.2015. [DOI] [PubMed] [Google Scholar]
- 41.Nazir S, Kumar A, Chatterjee I, Anbazhagan AN, Gujral T, Priyamvada S, et al. Mechanisms of intestinal serotonin transporter (SERT) upregulation by TGF-β1 induced non-Smad pathways. PLoS One. 2015;10:e0120447. doi: 10.1371/journal.pone.0120447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gill RK, Anbazhagan AN, Esmaili A, Kumar A, Nazir S, Malakooti J, et al. Epidermal growth factor upregulates serotonin transporter in human intestinal epithelial cells via transcriptional mechanisms. Am J Physiol Gastrointest Liver Physiol. 2011;300:G627–36. doi: 10.1152/ajpgi.00563.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foley KF, Pantano C, Ciolino A, Mawe GM. IFN-gamma and TNF-alpha decrease serotonin transporter function and expression in Caco2 cells. Am J Physiol Gastrointest Liver Physiol. 2007;292:G779–84. doi: 10.1152/ajpgi.00470.2006. [DOI] [PubMed] [Google Scholar]
- 44.Jin DC, Cao HL, Xu MQ, Wang SN, Wang YM, Yan F, et al. Regulation of the serotonin transporter in the pathogenesis of irritable bowel syndrome. World J Gastroenterol. 2016;22:8137–48. doi: 10.3748/wjg.v22.i36.8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moses PL, Coates MD, Mahoney CR, Sampson JE, Linden D, Chen JJ, et al. Key elements of serotonin signaling are altered in IBD and IBS: support for a molecular basis of the irritable bowel syndrome. Am J Gastroenterol. 2003;98(suppl 1):S262–3. doi: 10.1111/j.1572-0241.2003.08529.x. [DOI] [Google Scholar]
- 46.Galligan JJ. 5-hydroxytryptamine, ulcerative colitis, and irritable bowel syndrome: molecular connections. Gastroenterology. 2004;126:1897–9. doi: 10.1053/j.gastro.2004.04.028. [DOI] [PubMed] [Google Scholar]
- 47.Spiller R, Major G. IBS and IBD - separate entities or on a spectrum? Nat Rev Gastroenterol Hepatol. 2016;13:613–21. doi: 10.1038/nrgastro.2016.141. [DOI] [PubMed] [Google Scholar]
- 48.Bercik P, Verdu EF, Collins SM. Is irritable bowel syndrome a low-grade inflammatory bowel disease? Gastroenterol Clin North Am. 2005;34:235–45. doi: 10.1016/j.gtc.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 49.Garcia Rodriguez LA, Ruigomez A, Wallamander MA, Johansson S, Olbe L. Detection of colorectal tumors and inflammatory bowel disease during follow up of patients with initial diagnosis of irritable bowel syndrome. Scand J Gastroenterol. 2000;35:306–11. doi: 10.1080/003655200750024191. [DOI] [PubMed] [Google Scholar]
- 50.Porter CK, Cash BD, Pimentel M, Akinseye A, Riddle MS. Risk of inflammatory bowel disease following a diagnosis of irritable bowel syndrome. BMC Gastroenterol. 2012;12:55. doi: 10.1186/1471-230X-12-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang BM, Wang YM, Zhang WM, Zhang QY, Liu WT, Jiang K, et al. Serotonin transporter gene polymorphism in irritable bowel syndrome. Zhonghua Nei Ke Za Zhi. 2004;43:439–41. [PubMed] [Google Scholar]
- 52.Fiskerstrand CE, Lovejoy EA, Quinn JP. An intronic polymorphic domain often associated with susceptibility to affective disorders has allele dependent differential enhancer activity in embryonic stem cells. FEBS Lett. 1999;458:171–4. doi: 10.1016/S0014-5793(99)01150-3. [DOI] [PubMed] [Google Scholar]
- 53.Hranilovic D, Stefulj J, Schwab S, Borrmann-Hassenbach M, Albus M, Jernej B, et al. Serotonin transporter promotor and intron 2 polymorphisms: relationship between allelic variants and gene expression. Biol Psychiatry. 2004;55:1090–4. doi: 10.1016/j.biopsych.2004.01.029. [DOI] [PubMed] [Google Scholar]
- 54.Ali FR, Vasiliou SA, Haddley K, Paredes UM, Roberts JC, Miyajima F, et al. Combinatorial interaction between two human serotonin transporter gene variable number tandem repeats and their regulation by CTCF. J Neurochem. 2010;112:296–306. doi: 10.1111/j.1471-4159.2009.06453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeo A, Boyd P, Lumsden S, Saunders T, Handley A, Stubbins M, et al. Association between a functional polymorphism in the serotonin transporter gene and diarrhoea predominant irritable bowel syndrome in women. Gut. 2004;53:1452–8. doi: 10.1136/gut.2003.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sonuga-Barke EJ, Kumsta R, Schlotz W, Lasky-Su J, Marco R, Miranda A, et al. A functional variant of the serotonin transporter gene (SLC6A4) moderates impulsive choice in attention-deficit/hyperactivity disorder boys and siblings. Biol Psychiatry. 2011;70:230–6. doi: 10.1016/j.biopsych.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steffens DC, Taylor WD, McQuoid DR, Krishnan KR. Short/long heterozygotes at 5HTTLPR and white matter lesions in geriatric depression. Int J Geriatr Psychiatry. 2008;23:244–8. doi: 10.1002/gps.1869. [DOI] [PubMed] [Google Scholar]
- 58.Zhang ZF, Duan ZJ, Wang LX, Yang D, Zhao G, Zhang L. The serotonin transporter gene polymorphism (5-HTTLPR) and irritable bowel syndrome: a meta-analysis of 25 studies. BMC Gastroenterol. 2014;14:23. doi: 10.1186/1471-230X-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kohen R, Jarrett ME, Cain KC, Jun SE, Navaja GP, Symonds S, et al. The serotonin transporter polymorphism rs25531 is associated with irritable bowel syndrome. Dig Dis Sci. 2009;54:2663–70. doi: 10.1007/s10620-008-0666-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farjadian S, Fakhraei B, Moeini M, Nasiri M, Fattahi MR. Serotonin transporter gene polymorphisms in Southwestern Iranian patients with irritable bowel syndrome. Arab J Gastroenterol. 2013;14:59–62. doi: 10.1016/j.ajg.2013.03.001. [DOI] [PubMed] [Google Scholar]