Human cytomegalovirus is a leading cause of congenital disease. This reflects its capacity for systemic spread. A vaccine is needed, but the best viral targets are unclear. Attention has focused on the virion membrane fusion complex. It has 2 forms, so we need to know what each contributes to host colonization. One includes the virion glycoprotein O. We used murine cytomegalovirus, which has equivalent fusion complexes, to determine the importance of glycoprotein O after mucosal infection. We show that it drives local virus replication in epithelial cells. It was not required to infect myeloid cells, which establish systemic infection, but poor local replication reduced systemic spread as a secondary effect. Therefore, targeting glycoprotein O of human cytomegalovirus has the potential to reduce both local and systemic infections.
KEYWORDS: cytomegalovirus, dissemination, glycoproteins, host colonization, pathogenesis, tropism
ABSTRACT
Cytomegaloviruses (CMVs) establish systemic infections across diverse cell types. Glycoproteins that alter tropism can potentially guide their spread. Glycoprotein O (gO) is a nonessential fusion complex component of both human CMV (HCMV) and murine CMV (MCMV). We tested its contribution to MCMV spread from the respiratory tract. In vitro, MCMV lacking gO poorly infected fibroblasts and epithelial cells. Cell binding was intact, but penetration was delayed. In contrast, myeloid infection was preserved, and in the lungs, where myeloid and type 2 alveolar epithelial cells are the main viral targets, MCMV lacking gO showed a marked preference for myeloid infection. Its poor epithelial cell infection was associated with poor primary virus production and reduced virulence. Systemic spread, which proceeds via infected CD11c+ myeloid cells, was initially intact but then diminished, because less epithelial infection led ultimately to less myeloid infection. Thus, the tight linkage between peripheral and systemic MCMV infections gave gO-dependent infection a central role in host colonization.
IMPORTANCE Human cytomegalovirus is a leading cause of congenital disease. This reflects its capacity for systemic spread. A vaccine is needed, but the best viral targets are unclear. Attention has focused on the virion membrane fusion complex. It has 2 forms, so we need to know what each contributes to host colonization. One includes the virion glycoprotein O. We used murine cytomegalovirus, which has equivalent fusion complexes, to determine the importance of glycoprotein O after mucosal infection. We show that it drives local virus replication in epithelial cells. It was not required to infect myeloid cells, which establish systemic infection, but poor local replication reduced systemic spread as a secondary effect. Therefore, targeting glycoprotein O of human cytomegalovirus has the potential to reduce both local and systemic infections.
INTRODUCTION
Herpesviruses establish systemic infections that involve multiple cell types. Viral glycoproteins that alter tropism could help to guide how they spread. A common theme among beta- and gammaherpesviruses is the production of alternate membrane fusion complexes. Glycoprotein H (gH) and gB are essential for fusion. gL binds to gH and is often also essential, although not always (1). Most alternate complexes have additional, nonessential virion glycoproteins. They were first described for Epstein-Barr virus (EBV): its gp42 associates with gH/gL and promotes fusion with B cells rather than epithelial cells by binding to major histocompatibility complex (MHC) class II (2). The sequential production of gp42hi virions in cells lacking MHC class II and then of gp42lo virions in MHC class II-positive cells is hypothesized to switch EBV between B cells and epithelial cells during transmission (3). However, the restriction of EBV to humans makes this hypothesis hard to test. The related murid herpesvirus 4 (MuHV-4) makes alternate fusion complexes that incorporate gL or not (1). The gH/gL form promotes host entry by binding to olfactory epithelial heparan (4). However, both host entry and exit are epithelial infections (5, 6), and epithelial cells produce gLhi epithelium-tropic virions (7), so transmission does not involve tropism switching. gL-negative (gL−) fusion complexes may instead contribute to systemic spread, as myeloid cells produce gLlo virions and provide the viral gateway to B cells (7).
Among the betaherpesviruses, human herpesvirus 6 can alter its tropism by adding gO or gQ to gH/gL (8), while human cytomegalovirus (HCMV) can add either gO or a composite of UL128, UL130, and UL131 (9). UL128/UL130/UL131 promotes epithelial, endothelial, and myeloid cell tropisms, while its exclusion promotes fibroblast infection (10). Thus, there is again potential for tropism switching (11). A report that HCMV lacking gO (gO− HCMV) incorporates less gH/gL into virions and poorly infects endothelial and epithelial cell lines as well as fibroblasts (12) suggests a slightly different model, in which tropism for nonfibroblasts depends on UL128/UL130/UL131 displacing gO from just some complexes. As with EBV, the key unknown is the sequence of cell types normally infected during host colonization. HCMV presumably transmits via epithelial cells, and it persists in myeloid cells (13), but what fibroblast infection contributes to the viral life cycle is unclear. Normal and transformed epithelial cells may also differ in their infection requirements.
Murine CMV (MCMV) allows in vivo analysis. It adds to gH/gL to either gO or M131-129 (14). Like UL128, M131-129 has homology to chemokines and thus may both recruit myeloid cells and promote their infection directly. In vivo, M131− MCMV shows less monocyte recruitment and less salivary gland (SG) infection (15, 16), consistent with MCMV spreading via myeloid cell recirculation (17, 18). gO− MCMV propagates poorly in cultured fibroblasts (19). The failure of gO-positive (gO+) virus pseudotyping to rescue this defect was interpreted as impaired virion production. The same was proposed for HCMV lacking gO (20). However, a subsequent study identified instead poor gO− MCMV entry into NIH 3T3 fibroblasts and TCMK-1 epithelial cells (21). Entry into ANA-1 macrophages and MHEC5-T endothelial cells was preserved. Interestingly, gO− MCMV given intravenously (i.v.) poorly entered all cell types in the liver, including sinusoidal macrophages. gO+ pseudotyping restored this defect plus the virulence of intraperitoneal (i.p.) infection, suggesting a general entry defect of cell-free gO− virions but normal cell-to-cell spread. SG colonization showed no defect.
Inconsistencies between in vitro and in vivo conclusions emphasize the need to study physiologically relevant infections. i.p. and i.v. inoculations may not be optimal in this regard, as they bypass normal epithelial barriers. Hepatic infection appears to be a dead end for MCMV (22), so relating defects here to normal host colonization can be problematic. Natural postnatal infection must start at a mucosal surface. The surface targeted by HCMV is unknown. Oral entry is often assumed, but infected oral cells have not been identified, and infection by controlled oral inoculation has not been demonstrated. MCMV enters new hosts via the respiratory tract rather than orally (23). In the upper respiratory tract, it infects olfactory neurons, a tropism shared by MuHV-4 (4) and herpes simplex virus 1 (24); in the lower respiratory tract, it infects myeloid cells and type 2 alveolar epithelial cells (AEC2) (25). Cocaged mice transmit MCMV via the upper respiratory tract (23), so this is the likely natural entry route. Both upper and lower respiratory tract infections show myeloid cell-dependent spread via lymph nodes (LN), and the greater synchronicity of infection spread from the lungs makes it easier to track. We used lung infection by intranasal (i.n.) administration of gO− MCMV to understand what gO contributes to systemic spread after mucosal entry.
RESULTS
Generation of gO-deficient MCMV.
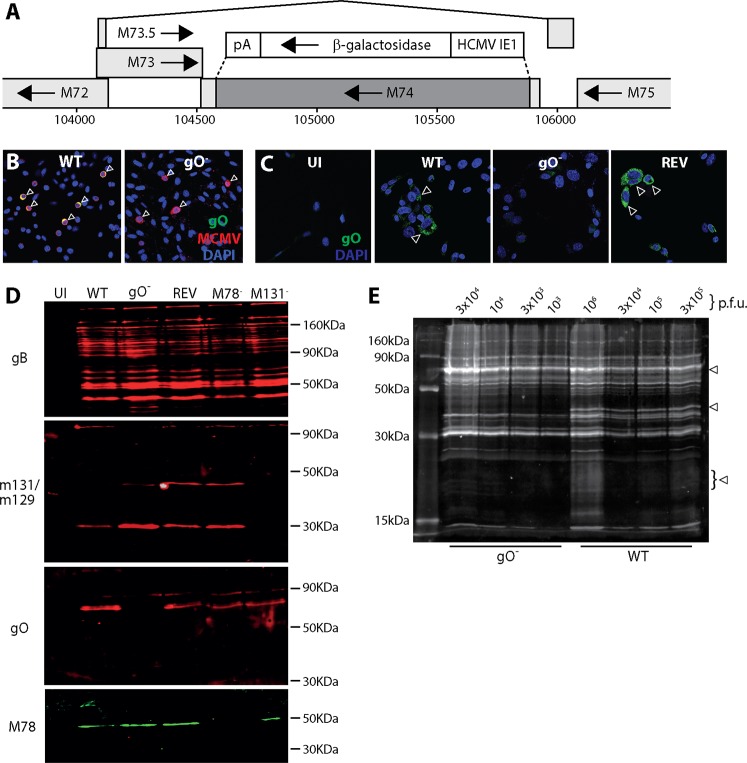
MCMV gO is encoded by M74 (genomic coordinates 104549 to 105865), which lies within an intron (positions 104214 to 105931) of M73.5 (26). To make gO− MCMV, we deleted most of M74 (genomic coordinates 104654 to 105816) and replaced it with a β-galactosidase (βgal) expression cassette (Fig. 1A). We made a revertant of this virus by recombining back in the unmutated genome segment. In an independently generated gO− mutant, luciferase expression was additionally linked to M78, an abundant early lytic gene (27). Correct mutagenesis was established by sequencing viral DNA across the mutation junctions. A lack of gO expression was confirmed by immunofluorescence (Fig. 1B and C) and by Western blotting of infected cell lysates (Fig. 1D) and purified virions (Fig. 1E).
FIG 1.
Generation of gO-deficient MCMV. (A) Schematic map of M74 (genomic coordinates 104549 to 105865) showing the deleted region (coordinates 104564 to 105816) (dark shading) and the surrounding genes. M73 encodes gN. M75 encodes gH. The deleted region was replaced by an HCMV IE1-driven βgal expression cassette, as shown. A revertant virus was made by recombination back into the native M74 locus. (B) NIH 3T3 cells were infected (0.1 PFU/cell for 18 h) with wild-type (WT) or gO-deficient (gO−) viruses and then fixed and stained for gO with mAb Cl.M74.01 (mouse IgG1) and for MCMV virion antigens with a polyclonal rabbit serum. Nuclei were stained with DAPI. Arrows show example infected cells. WT-infected cells show both MCMV antigens and gO (colocalization appears yellow); gO− infected cells show only MCMV antigens (red). (C) NIH 3T3 cells were left uninfected (UI) or infected with WT, gO−, or revertant (REV) viruses (0.1 PFU/cell for 18 h) and then fixed and stained for gO. Nuclei were stained with DAPI. Arrows show example gO+ cells. No strong staining was seen in the UI and gO− samples. Some weak background staining probably reflects nonspecific mouse IgG binding by viral Fc receptors (46). (D) NIH 3T3 cells were infected or not (UI) with WT, gO−, REV, M78-deficient (M78−), or M131-deficient (M131−) MCMV (0.1 PFU/cell). M78− and M131− MCMV provided specificity controls. After 48 h, the cells were lysed and analyzed by SDS-PAGE for expression of gB (mAb Cl.M55.02; mouse IgG2a), M131-129 (mAb Cl.MCK-2.04; mouse IgG1), gO (mAb Cl.M74.01; mouse IgG1), and M78 (rabbit pAb). This confirmed a lack of gO in gO− MCMV. (E) Dilutions of WT and gO− virions recovered from infected NIH 3T3 cell supernatants were lysed, resolved by SDS-PAGE, and then immunoblotted with a rabbit pAb to MCMV virion antigens. The number of PFU loaded per lane is shown for each virus. Arrows show bands present in WT but not gO− samples. For the remaining bands, 105 PFU of WT MCMV was equivalent in protein content to 3 × 103 PFU gO− MCMV, implying a 30-fold excess of particles per PFU for gO− MCMV.
gO has a predicted molecular weight of 48 kDa after signal peptide cleavage and has consensus attachment sites for 6 N-linked and 12 O-linked glycans. A gO-specific monoclonal antibody (mAb) recognized a virion component with an apparent molecular weight of 70 kDa (Fig. 1D). A polyclonal rabbit serum raised against MCMV virions identified 3 bands present in gO+ but not gO− virions, at 70 kDa, 40 kDa, and 20 kDa, with the lower-molecular-weight bands being more obvious and the 20-kDa band being broad (Fig. 1E). gO has a consensus furin cleavage site (RRKR) that would generate 32-kDa and 16-kDa fragments, not counting glycans. Thus, gO may be cleaved by a furin-like enzyme, with the mAb recognizing mainly its full-length form and the polyclonal serum recognizing mainly its cleavage products. By both measures, the mutant virus lacked gO. gO− MCMV-infected cells showed some increase in M131-129 content, possibly because without gO, all gH/gL complexes were available for M131-129 binding.
gO− MCMV poorly infects fibroblasts.
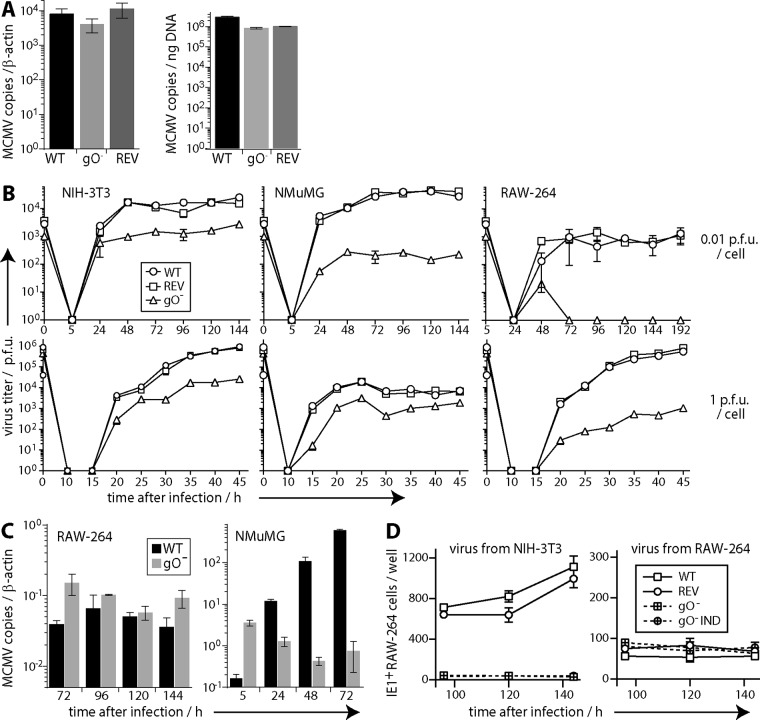
Our gO− MCMV grew poorly in NIH 3T3 fibroblasts, as noted for other gO mutants (20), with plaque titers 10- to 100-fold lower than those of gO+ wild-type (WT) or revertant (REV) viruses. These low titers possibly reflected defects in virion assembly, release, or plaque formation in murine embryonic fibroblasts (MEFs). gO− MCMV took longer to grow than gO+ MCMV, but stocks were harvested from supernatants of NIH 3T3 cells when they showed equivalent cytopathic effects. gO− stocks had at least 10-fold more viral protein per PFU than did gO+ stocks (Fig. 1E). We also compared gO+ and gO− stocks by quantitative PCR (QPCR) of viral DNA (Fig. 2A). They were equivalent in viral-to-cellular genome ratios and in viral DNA copies per nanogram of total DNA. However, equivalent titers of gO− MCMV consistently yielded 10- to 100-fold more total DNA. Therefore, gO− virus stocks contained 10- to 100-fold more viral genomes per PFU. These data argued that the low titers of gO− stocks were due to poor plaquing on MEFs rather than the stocks containing fewer virions due to defects in virion assembly or release.
FIG 2.
Replication of gO− MCMV in vitro. (A) DNA was extracted from WT, gO−, and REV virus stocks and compared for viral genome copies by QPCR. Viral copies were normalized by cellular copies of β-actin or by total DNA per reaction. Bars show means ± standard errors of the means (SEM) of data from triplicate samples. Across 3 independently produced stocks of each virus, there was no consistent difference in viral genome copies per cellular genome copy or per total amount of DNA. However, equivalent numbers of PFU yielded 10- to 100-fold more total DNA from gO− than from WT virus stocks. (B) NIH 3T3, NMuMG, or RAW-264 cells were infected with equivalent PFU of WT, REV, or gO− MCMV (0.01 or 1 PFU/cell for 2 h), washed with PBS, and cultured at 37°C. Triplicate samples at each time point were plaque assayed on MEFs. Each point shows the mean ± SEM. Time zero shows the redetermined titer of the input. For each cell type, the plaque yield of gO− MCMV was significantly reduced. (C) WT and gO− samples from the growth curves at 0.01 PFU/cell shown in panel B were assayed for viral genome loads by QPCR. Bars show means ± SEM of data from triplicate samples. Infected NMuMG cultures yielded significantly more gO− genomes at 5 h but significantly more WT genomes thereafter (P < 10−4). Infected RAW-264 cell cultures showed no consistent differences between WT and gO− genome loads. (D) Triplicate samples from growth curves at 0.01 PFU/cell on NIH 3T3 or RAW-264 cells were assayed for infectivity by incubation with 5 × 105 uninfected RAW-264 cells (18 h at 37°C) and then immunostaining for viral IE1 expression. gO+ MCMV (WT and REV) was compared with gO− MCMV, including an independently derived mutant (gO− IND). Each point shows the mean ± SEM of data from triplicate samples. Samples from NIH 3T3 cells showed significantly more gO+ than gO− infection (P < 10−6). Those from RAW-264 cells showed no significant difference.
gO− MCMV replicates poorly in fibroblasts and epithelial cells and normally in monocytes.
Growth curves in NIH 3T3 fibroblasts, NMuMG epithelial cells, and RAW-264 monocytes all showed significant, gO-dependent reductions in virus titers (Fig. 2B). However, while infected RAW-264 cell cultures showed no consistent differences in viral genome loads, infected NMuMG cultures showed marked differences (Fig. 2C): the gO− cultures initially had more viral genomes, consistent with gO− stocks containing more genomes per PFU (viral inputs were normalized by PFU), but the representation of gO− genomes then declined, while that of gO+ genomes increased. As a further measure of virus propagation, we compared the infectivities of growth curve samples by single-cycle infection of RAW-264 cells. To do this, we added samples from NIH 3T3 or RAW-264 cell infections to uninfected RAW-264 cells and 18 h later stained them for MCMV IE1 expression with mAb CROMA-101 (Fig. 2D). The gO− samples from NIH 3T3 cells showed much less infectivity than the gO+ samples, but those from RAW-264 cells showed no defect. Therefore, gO− MCMV replicated normally in RAW-264 monocytes and poorly in NIH 3T3 fibroblasts, NMuMG epithelial cells, and MEFs.
gO− MCMV poorly enters fibroblasts.
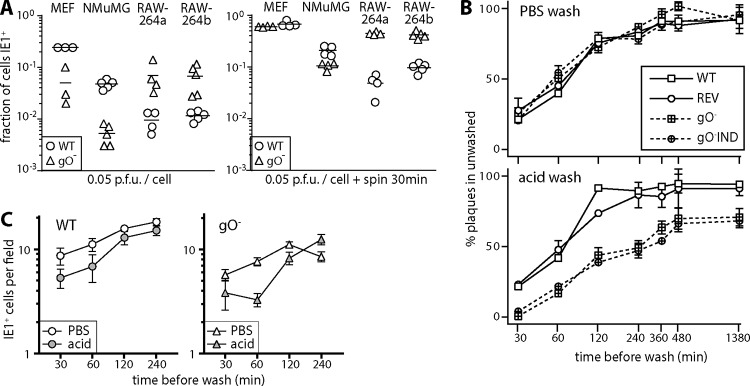
The capacity of gO− MCMV to release large amounts of viral protein and DNA from infected NIH 3T3 cells (Fig. 1E and 2A) was consistent with its low plaque titers being due to poor entry into fibroblasts rather than poor virion production or release. To test entry independently of new virion production, we compared gO+ (wild-type) and gO− MCMV stocks (grown in NIH 3T3 cells) for single-cycle infections of MEFs, NMuMG cells, and RAW-264 cells (Fig. 3A). As for Fig. 2D, we assayed infection by staining the exposed cells for viral IE1 expression. After 5 h, MEFs and NMuMG cells showed more infection by gO+ MCMV, while RAW-264 cells showed more infection by gO− MCMV. Similar results were obtained at 24 h. gO− MCMV did not necessarily infect RAW-264 cells better than gO+ MCMV did, as the inputs were normalized by PFU and thus contained more gO− viral DNA and protein. However, it was clear that the excess viral DNA and protein of gO− stocks did not represent noninfectious debris. Rather they represented particles that were poorly infectious for MEFs yet infectious for RAW-264 cells. Therefore, gO− MCMV showed defective entry specifically in NMuMG cells and MEFs.
FIG 3.
gO− MCMV poorly penetrates nonmyeloid cells. (A) MEFs, NMuMG cells, and RAW-264 cells (RAW-264a and RAW-264b are both RAW-264 cells but were obtained from separate sources and thus were many passages separate) were exposed to WT or gO− virions, with or without centrifugation (600 × g [spin 30 min]). Five hours later, the cells were fixed and stained for viral IE1 expression. Symbols show data for replicate infection wells, and bars show means. Without centrifugation, WT MCMV infected significantly more MEFs and NMuMG cells, and gO− MCMV infected significantly more RAW-264 cells (P < 0.01). With centrifugation, only NMuMG cells showed more WT infection (P < 0.05), but the excess gO− infection of RAW-264 cells was maintained (P < 0.01). (B) MEFs were adhered to 24-well plates and then incubated (37°C) with 200 PFU gO+ MCMV (WT and REV) or gO− MCMV (2 independent mutants). At the times indicated, the cells were washed with PBS or pH 3 citrate buffer (acid) and then incubated for 4 days to allow plaque formation. The number of plaques per well is shown relative to control unwashed samples for each virus. Points show means ± SEM of data from triplicate wells. At each time point, acid washes had a significantly greater effect on gO− titers than on gO+ titers. PBS washes showed no differential effect. Equivalent results were obtained in 2 repeat experiments. (C) In an experiment similar to the one in panel B, 500 PFU WT or gO− MCMV was added to 5 × 105 RAW-264 cells, and the cells were washed in PBS or pH 3 citrate buffer (acid) at the times indicated. After overnight incubation, the cells were fixed and stained for viral IE1 expression. Each point shows the mean ± SEM for triplicate wells. In the experiment shown, the level of gO− infection was somewhat lower than that of WT infection, as the input was reduced to give approximately equal infections, but gO− infection showed no greater susceptibility to the acid wash.
Cocentrifugation of cells and virus (600 × g for 30 min) increased all infections but reduced the gO-dependent defect in MEF and NMuMG cell infections while maintaining the difference in RAW-264 cell infection (Fig. 3A). As gO associates with gH/gL, it might be expected to promote viral membrane fusion (cell penetration). However, a possible alternative was that it promoted virion binding to nonmyeloid cells. To distinguish differences in binding and penetration, we added gO+ and gO− virions to MEFs and compared how quickly they acquired resistance to a phosphate-buffered saline (PBS) wash (which removes unbound virions) and to an acid wash (which inactivates all extracellular virions), reading out plaque formation after 4 days (Fig. 3B). gO+ virions rapidly acquired resistance to both PBS and acid washes. gO− virions showed a similar acquisition of resistance to the PBS wash but took significantly longer to resist the acid wash. Therefore, gO− MCMV was defective primarily in MEF penetration. We also compared how quickly gO+ and gO− virions acquired resistance to PBS and acid washes when added to RAW-264 cells (Fig. 3C), assaying infection by immunofluorescent staining for viral IE1 expression. Here gO+ and gO− virions showed no obvious difference. Therefore, gO significantly enhanced MCMV penetration of nonmyeloid cells.
gO− MCMV amplifies poorly in epithelial sites.
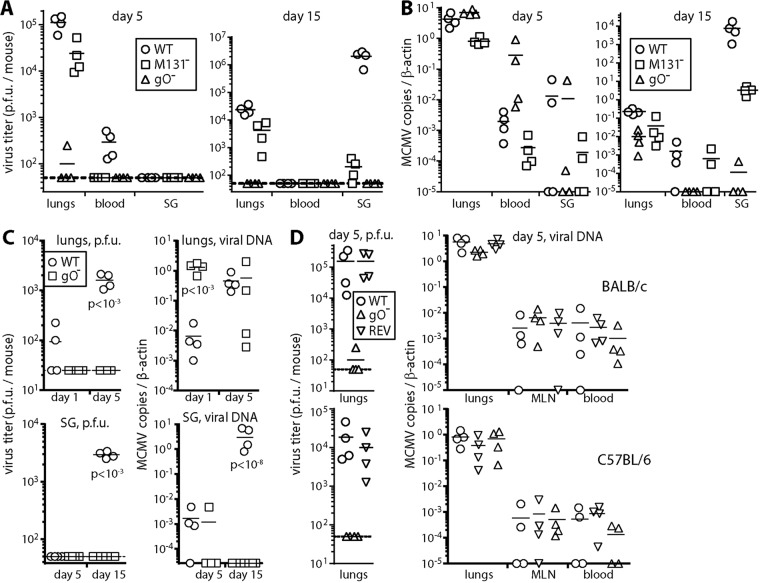
i.p. and i.v. MCMV inoculations establish cell-free viremias and reach the liver, presumably through cell-free virion uptake by macrophages lining its fenestrated capillaries. In contrast, respiratory infection spreads via CD11c+ cells with little cell-free viremia, and the liver is not a prominent target (18). Instead, CD11c+ cells deliver MCMV to diverse tissues by extravasation. In the SG, infected CD11c+ cells transfer MCMV to acinar cells. To establish the role of gO in this spread, we gave gO+ and gO− MCMV i.n. to BALB/c mice and then 5 and 15 days later assayed infections of the lungs, blood, and SG (Fig. 4). Comparison was made with MCMV lacking its myeloid cell-oriented M131-129 fusion complex component.
FIG 4.
gO− MCMV replication in vivo. (A) BALB/c mice given WT, gO−, or M131− MCMV i.n. (105 PFU) were plaque assayed for infectious virus 5 and 15 days later. gO− MCMV yielded negligible infectivity from all sites. M131− MCMV was readily detected in lungs but yielded significantly less infectivity than the WT in blood at day 5 and in SG at day 15 (P < 0.01). Points show data for individual mice, and bars show means. (B) Genome loads in organs of the mice from panel A were measured by QPCR. gO− genome loads were significantly lower than those of the WT at day 15 (P < 0.01) but not at day 5. (C) In a separate experiment, infectivity and viral genomes of BALB/c mice given WT or gO− MCMV i.n. (105 PFU) were measured in lungs after 1 or 5 days and in SG after 5 or 15 days. Again, gO− MCMV was undetectable by plaque assays. Lungs showed significantly more gO− viral genomes than WT viral genomes at day 1 but not at day 5. SG showed significantly more WT viral genomes at day 15 but not at day 5. (D) BALB/c and C57BL/6 mice given WT, gO−, or REV viruses i.n. (105 PFU) were assayed for infectivity and viral genomes 5 days later. gO− MCMV yielded negligible infectivity. However, genome loads were similar for all viruses in lungs, MLN, and blood.
At day 5, plaque assays (Fig. 4A) showed significantly lower gO− titers than gO+ titers in lungs and blood (P < 0.01). Viral genomes (Fig. 4B) were abundant in the same sites, so there was still virus entry and initial spread. However, at day 15, plaque assays did not detect gO− MCMV, and its genome loads in the lungs, blood, and SG were all significantly below those of the WT (P < 0.01). Therefore, longer-term dissemination was impaired. M131− MCMV was readily detected in the lungs, but its titers were reduced day 5 in blood (P < 0.05) and at day 15 in SG (P < 10−4).
Virus inputs were normalized by PFU, and at day 1, more gO− than gO+ genomes were detected in infected lungs, consistent with gO− virus stocks having more genomes per PFU (Fig. 4C). Parallel assays of day 5 infection showed that gO+ genome loads in the lungs increased after inoculation, whereas gO− loads declined. Therefore, gO− MCMV entering the lungs replicated poorly. Again, gO− genomes showed some initial seeding to the SG at day 5, but SG colonization was severely reduced at day 15. The capacity of i.n. gO− MCMV to disseminate acutely was confirmed in a repeat experiment in BALB/c and C57BL/6 mice (Fig. 4D), where day 5 gO− viral genome loads in mediastinal LN (MLN) and blood were equivalent to those of WT and REV viruses. Therefore, the impaired gO− MCMV spread at day 15 (Fig. 4B and C) seemed likely to reflect its poor amplification in the lungs at day 5 (Fig. 4C) rather than defective CD11c+ cell infection.
gO− MCMV poorly infects lung epithelial cells.
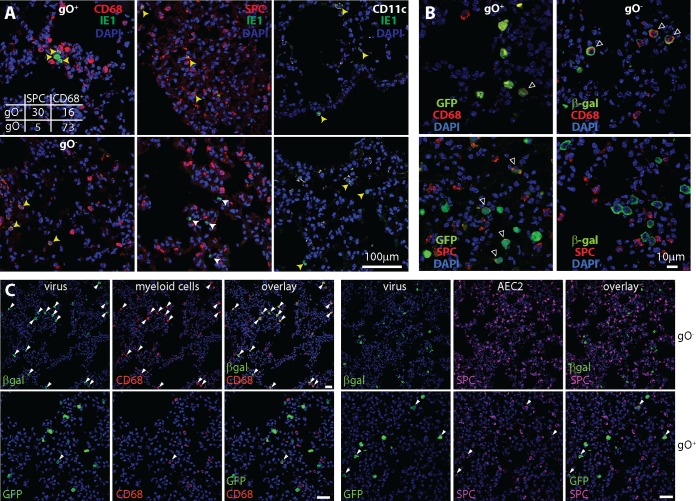
We explored gO− lung infection further by immunostaining tissue sections (Fig. 5). MCMV given i.n. directly infects alveolar myeloid cells (CD68+ CD11c+) and AEC2, which express the surfactant protein C precursor (SPC) (25). At day 5, gO+ MCMV was evident in both epithelial and myeloid cells, with most IE1+ cells being SPC+ AEC2 (Fig. 5A). In contrast, gO− MCMV infected very few AEC2: >90% of IE1+ cells were CD68+. At day 1, detecting infection by viral reporter gene expression, the difference was even more marked (Fig. 5B and C), with <25% WT infection and >95% gO− infection in CD68+ cells. Therefore, poor gO− MCMV amplification in the lungs was associated with a marked reduction in AEC2 infection. More CD68+ cell infection by gO− than by gO+ MCMV did not mean intrinsically enhanced infection, as poor entry into AEC2 would make more gO− virions available for myeloid infection. Both gO+ and gO− MCMV-infected cells showed punctate CD11c staining, consistent with MCMV infection redistributing this marker (18).
FIG 5.
gO− MCMV poorly infects lung epithelial cells. (A) BALB/c mice given gO+ MCMV (wild type) or gO− MCMV i.n. (105 PFU) were analyzed 5 days later by staining lung sections for viral IE1 expression and the following cellular markers: CD68 (alveolar macrophages and some dendritic cells), SPC (AEC2), and CD11c (dendritic cells and some alveolar macrophages). gO+ MCMV infected CD68+, SPC+, and CD11c+ cells; gO− MCMV infected CD68+ and CD11c+ cells but very few SPC+ cells. Representative images are shown. Yellow arrows show example IE1+ marker-positive cells. White arrows show example IE1+ marker-negative cells. The top left panel shows counts for IE1+ marker-positive cells across 3 representative sections for each of 3 mice per group. There was a significant bias of gO− MCMV infection toward CD68+ cells (P < 10−6 by Fisher’s exact test). The number of CD11c+ cells infected was not significantly different between gO+ and gO− MCMV. (B) BALB/c mice given gO+ or gO− MCMV i.n. (105 PFU) were analyzed 1 day later by costaining lung sections for viral reporter genes (GFP for WT and βgal for gO− MCMV) and cell markers as described above for panel A. Arrows show example dual-positive cells. (C) Lower-magnification images of mice infected as described above for panel B show the bias of gO+ MCMV toward and the strong bias of gO− MCMV against AEC2 infection across more infected cells. More than 95% of the cells infected by gO− MCMV were CD68+ (<5% SPC+), while >75% of the cells infected by gO+ MCMV were SPC+ (<25% CD68+), counting >200 cells for each virus (P < 10−6 by Fisher’s exact test). Arrows show examples of colocalization. Bars, 50 µm.
gO− MCMV spread via CD11c+ cells is functional, but reduced in amount.
CD11c+ myeloid cells infected by MCMV in the lungs migrate through the MLN to the blood (18). When blood leukocytes of mice at day 5 of infection were separated into CD11c+ and CD11c− fractions, gO− MCMV showed a distribution similar to that of gO+ MCMV with most viral genomes being in the CD11c+ fraction (Fig. 6A). Therefore, MCMV did not require gO to engage its normal pathway of spread. Infected CD11c+ cells migrate from the blood to the SG. In adult mice, they are the main infected cell type in SG for at least 2 weeks (18). At day 15, only gO+ MCMV showed CD11c+ infected cells on SG sections (Fig. 6B and C). Therefore, despite initially reaching CD11c+ cells, gO− systemic spread was impaired. This result was consistent with poor AEC2 infection in the lungs limiting local amplification and thus keeping infected CD11c+ cell numbers low.
FIG 6.
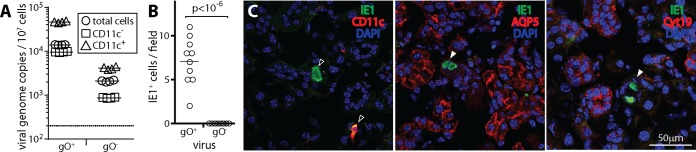
Poor SG infection by gO− MCMV. (A) BALB/c mice were given gO+ MCMV (wild type) or gO− MCMV i.n. (105 PFU). Five days later, blood leukocytes were separated by antibody capture into CD11c+ and CD11c− fractions and then assayed for infection by QPCR of viral DNA. Total indicates unfractionated leukocytes. In this experiment, gO− viral loads in the blood were lower than gO+ viral loads but were still readily detectable. Symbols show data for individual mice, bars show means, and the dotted line shows the limit of detection. For both gO+ and gO− MCMV, CD11c+ cell enrichment significantly increased viral genome representation, while CD11c+ cell depletion significantly reduced it (P < 0.01). (B) SG sections at day 15 after i.n. infection show infected (IE1+) cells for gO+ but not gO− MCMV. Counts were obtained from 3 sections for each of 3 mice per group. (C) Costaining for IE1 and cell type-specific markers shows that the infected cells in SG of gO+ MCMV-infected mice were predominantly CD11c+ (dendritic cells) (>90% CD11c+, counting >50 cells) rather than being positive for Aquaporin 5 (AQP5), a marker of SG acinar cells, or for cytokeratin-19 (Cyt19), a marker of SG duct cells, consistent with previously reported data (18). Open arrows show example marker-positive IE1+ cells, and filled arrows show example IE1+ marker-negative cells.
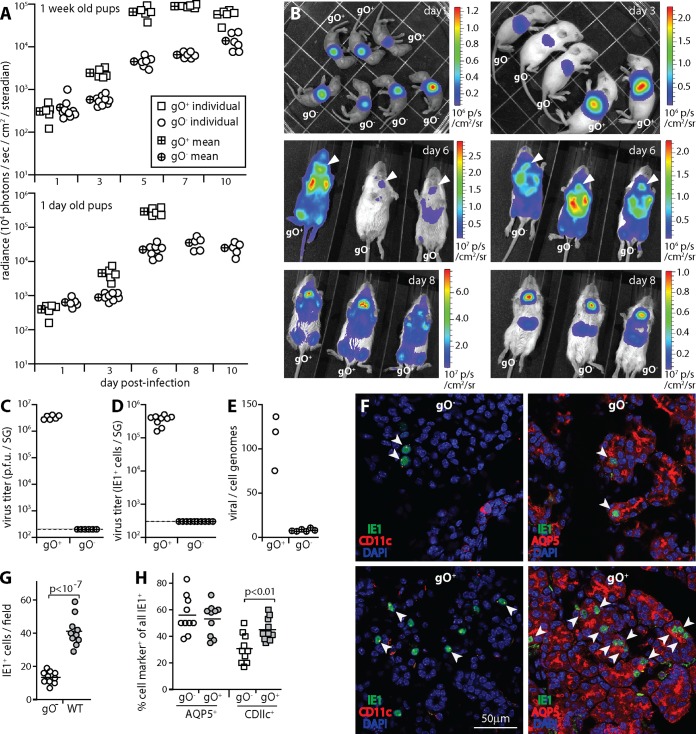
To explore systemic spread further, we gave gO+ and gO− MCMV i.n. to pups, in which it replicates to higher titers (Fig. 7). We tracked luciferase-expressing gO+ and gO− MCMV in 6- to 8-day-old pups by live imaging (Fig. 7A and B). At infection day 1, gO+ and gO− signals were equivalent. From day 3 onward, gO+ signals were significantly higher. However, gO− MCMV still colonized SG. Pups infected when they were 1 day old had stronger signals with a similar distribution. The 1-day-old pups infected by gO+ MCMV grew poorly and so were euthanized at day 6. Those infected by gO− MCMV, and 6- to 8-day-old pups infected by either virus, showed no obvious ill effects.
FIG 7.
The spread of gO− MCMV in pups is quantitatively impaired. (A) BALB/c pups were given luciferase-positive gO+ or gO− MCMV i.n. (104 PFU). Infection was tracked by luciferin injection and live imaging of light emission for the whole mouse. In 6- to 8-day-old pups, gO+ signals were significantly higher than gO− signals (P < 10−4) from infection day 3 onward. At infection day 1, there was no difference. Pups infected when they were 1 day old showed a similar pattern, but those infected with gO+ MCMV failed to thrive and thus were sacrificed at infection day 6. gO− infection had no noticeable ill effects. (B) Example images of pups infected when they were 6 to 8 days old show that gO+ and gO− luciferase signals had similar distributions, but from infection day 3 onward, gO+ signals were greater. Note the different sensitivity scales. Arrows at infection day 6 show SG signals. (C) SG taken 10 days after i.n. infection of 6- to 8-day-old pups with gO+ or gO− MCMV were plaque assayed for infectious virus. Symbols show data for individual mice. The dashed line shows the limit of assay sensitivity. (D) SG taken 10 days after i.n. infection of 6- to 8-day-old pups were assayed for infection by overnight incubation of homogenate dilutions with 5 × 105 RAW-264 cells and then staining for viral IE1 expression and counting positive cells. Symbols show data for individual mice. The dashed line shows the limit of assay sensitivity. (E) SG taken 10 days after i.n. infection of 6- to 8-day-old pups were assayed for viral genomes by QPCR. Viral DNA copy numbers were normalized by the cellular DNA copy number for each sample. Symbols show data for individual mice. (F) SG sections from 6- to 8-day-old pups taken 10 days after i.n. infection with gO+ or gO− MCMV were stained for viral IE1 antigen plus markers of dendritic cells (CD11c) or SG acinar cells (AQP5). Arrows show example dual-positive cells. (G) Total cell counts for randomly selected fields of view from 3 sections for each of 3 mice per group, as illustrated in panel F, show significantly more infected cells for gO+ than for gO− MCMV. Symbols show data for individual sections, and bars show means. (H) The cell type of infection in SG, as illustrated in panel F, was determined by counting IE1+ CD11c+ and IE1+ AQP5+ cells across sections from 3 mice per group (>300 cells for gO+ MCMV and >100 cells for gO− MCMV). Symbols show data for individual sections, with the number of marker-positive IE1+ cells expressed as a percentage of all IE1+ cells on the section. Bars show means. gO+ MCMV showed a higher proportion of CD11c+ infected cells than did gO− MCMV and no difference in AQP5+ infected cells, so gO was not required for MCMV transfer to acinar cells.
gO+ MCMV reached high plaque titers in pup SG. gO− MCMV was undetectable by a plaque assay (Fig. 7C). Assaying infectivity by incubating SG homogenates with RAW-264 cells and then staining for viral IE1 expression also showed a large gO-dependent defect (Fig. 7D), and while gO− genomes were detected in SG, gO+ genomes were significantly more abundant (Fig. 7E). Therefore, by all measures, gO− SG infection was significantly impaired. Day 10 SG sections (Fig. 7F) consistently showed more gO+ than gO− MCMV-infected cells. However, both gO+ and gO− MCMV reached acinar cells, with similar ratios of infected CD11c+ cells to acinar cells, so there did not appear to be a gO-dependent defect in CD11c+ cell-to-acinar cell virus transfer. As in adult mice, the reduced SG colonization by gO− MCMV seemed most likely to reflect its reduced upstream amplification in the lungs. The marked defect in infectious virus recovery from SG suggested that there might also be a defect in gO− virion production by acinar cells.
DISCUSSION
Systemic spread by HCMV underlies its capacity to transmit and cause disease, so it is an important vaccine target. Most antiviral vaccines rely on neutralizing antibodies. As beta- and gammaherpesviruses infect different cell types in different ways, neutralization can be cell type dependent. Predicting its impact requires an understanding of how individual viral glycoproteins contribute to host colonization. This cannot necessarily be extrapolated from in vitro infections. For example, gp350 is a prominent neutralization target for B cell infection by EBV in vitro, but a gp350 vaccine did not reduce B cell infection in vivo (28). Similarly, vaccination with HCMV gB, another prominent in vitro neutralization target, only modestly reduced infection in vivo (29), and what protection there was probably did not work through neutralization (30). HCMV vaccines based on the pentameric form of gH (gH/gL/UL128/UL130/UL131) are now being developed, again based on neutralization in vitro (31). As herpesviruses transmit from immune hosts, key neutralization targets might be expected to show antigenic diversity. UL128/UL130/UL131 is less diverse than gO (32). Therefore, it is important to understand how gO and UL128/UL130/UL131 contribute to host colonization. Analyzing the homologous glycoproteins of MCMV can provide clues. Here gO-dependent defects in MCMV lung and SG infections showed the importance of this glycoprotein for host colonization after mucosal entry.
MCMV host colonization has 2 main components: replication in peripheral sites and transfer between peripheral sites. The latter manifests as a myeloid cell-associated viremia (18). HCMV-infected blood monocytes are hypothesized to derive from proliferating, latently infected stem cells (33). However, stem cell transplantation rarely transfers HCMV infection (34), and rapid reductions in HCMV blood loads by inhibitors of lytic infection (35) argue against a large contribution from latency-associated proliferation. MCMV does not colonize bone marrow stem cells (36). It peripherally infects mature dendritic cells, which then recirculate (18). Most peripheral replication occurs in nonmyeloid cells (25). The lack of sustained systemic spread of gO− MCMV, despite its initiation, supported the idea that peripheral replication in epithelial cells and fibroblasts plays a key role in seeding dendritic cell infection. A similar scheme for HCMV would explain why it has a fibroblast-tropic form. gO+ HCMV targets cells that express platelet-derived growth factor receptor α (37, 38). Therefore, the expression of this receptor may identify cells important for amplifying HCMV loads in peripheral tissues.
MuHV-4 spreads in vivo by sequential switching between B cell and myeloid cell infections (24). However, gp42hi EBV can still infect epithelial cells (39), and myeloid cells pass MuHV-4 to non-B cells as well as B cells (40), broadening viral tropism rather than switching it. M131-129 or UL128/UL129/UL131 may analogously divert just some stromal CMV infection into myeloid cells for systemic redistribution. MCMV host colonization is compromised less by disrupting the M131-129 myeloid tropism determinant than by disrupting M33 (41), which impairs infected myeloid cell recirculation (18). Thus, we envisage that CMV tropism operates more as a preference than as exclusive switching. If so, vaccination against pentameric HCMV gH/gL may only partially reduce host colonization.
gO deficiency did not completely prevent epithelial cell infection by MCMV. It had a large effect on AEC2 infection by cell-free virions but seemed not to reduce dendritic cell-to-SG acinar cell virus transfer, as their infection ratio was unchanged. This could reflect different gO dependencies for infecting different epithelial cell types or that cell-to-cell virus spread, with likely high multiplicity and enhancement by intercellular contacts, is inherently more difficult to block than infection by cell-free virions. The limited impact of gO deficiency on MCMV spread in the liver (21) could be similarly explained by infection multiplicity rather than involving a qualitatively different form of fusion.
For gO− MCMV in pup SG, the greater defect in infectious virus recovery than in viral genome loads suggested a particular importance for gO in the infectivity of virions derived from acinar cells, that is, those destined for transmission. However, differences in infection of different acinar cell subsets are also possible. In general, the details of MCMV spread to and from particular cell types in tissues remain ill defined. Local, basolateral epithelial spread is likely, as observed for other herpesviruses (5, 24, 42), but remains unproven. Whether there might be qualitative differences in membrane fusion between cell-free virions and those passing directly between cells also needs further study. Our key findings were that gO is important directly for local MCMV amplification after mucosal entry and indirectly for systemic spread. These results validated the idea that host colonization is driven by local lytic infection and redistribution (18) rather than by latency-associated cell proliferation. As local lytic infection also drives disease, gO merits consideration as a component of HCMV vaccines.
MATERIALS AND METHODS
Mice.
BALB/c and C57BL/6 mice were given MCMV i.n. under isoflurane anesthesia, either as adults (6 to 12 weeks old) (105 PFU in 30 µl) or as pups (104 PFU in 5 µl). For live imaging, luciferase was expressed from the MCMV genome in tandem with M78 by cotranslation and autocatalytic release (27). Infected mice were given 1 mg d-luciferin i.p. and then scanned for light emission with a charge-coupled-device camera under isoflurane anesthesia (IVIS-2000; Xenogen). All experiments were approved by the University of Queensland Animal Ethics Committee in accordance with Australian National Health and Medical Research Council guidelines (projects 301/13 and 479/15) (47). Experimental groups were compared by Student’s 2-tailed unpaired t test unless otherwise stated. Differences were considered significant at >95% confidence.
Cells and viruses.
NIH 3T3 fibroblasts (ATCC CRL-1658), RAW-264 monocytes (ATCC TIB-71), NMuMG epithelial cells (ATCC CRL-1636), and MEFs from day 13 to 14 CD1 mouse embryos (43) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 2 mM glutamine, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 10% fetal calf serum (complete medium). All viruses were grown on NIH 3T3 cells. Infected cultures were harvested when >50% of cells showed cytopathic effects. They were cleared of cell debris by low-speed centrifugation (500 × g for 10 min), and virus was then concentrated by ultracentrifugation (35,000 × g for 2 h) and stored in aliquots at −80°C. To disrupt MCMV M74, which encodes gO (genomic coordinates 105865 to 104549), a βgal expression cassette (15) was cloned between genomic flanks to replace coordinates 104564 to 105816. The plasmid was cotransfected with MCMV genomic DNA into NIH 3T3 cells using Lipofectamine (Life Technologies), and βgal+ plaques were picked to homogeneity. Correct mutagenesis was confirmed by PCR and DNA sequencing of viral DNA across the mutation boundaries. A revertant was made by cotransfecting gO− MCMV DNA with an unmutated genomic segment flanking M74 and then picking βgal− plaques. An independent mutant (gO− IND) was made by recombining the same M74 disruption into an MCMV derivative in which luciferase is expressed downstream of M78 (27). M131− (15), M78− (43), and green fluorescent protein-positive (GFP+) (44) MCMVs were described previously. All viruses were derived from MCMV strain K181.
Virus assays.
Infectious virus was plaque assayed on MEFs. Subconfluent cell monolayers were cocentrifuged with virus or tissue homogenate dilutions (600 × g for 30 min) and then overlaid with complete medium plus 0.3% carboxymethylcellulose. After culture at 37°C for 4 days, the cells were fixed in 1% formaldehyde and stained with 0.1% toluidine blue for plaque counting. To assay infectivity independently of replication in MEFs, virus dilutions were overlaid onto RAW-264 cells and cultured for 18 h at 37°C. The cells were then fixed and stained for viral IE1 expression with mAb CROMA-101 (45) plus Alexa 488-goat polyclonal antibody (pAb) to mouse IgG. IE1+ cells were counted under UV illumination. To enumerate MCMV genome copies in extracted DNA (Wizard genomic DNA purification; Promega), genomic coordinates 111218 to 111461 were amplified by QPCR (LightCycler 480 SYBR green; Roche Diagnostics) and converted to genome copies by comparison with plasmid standards amplified in parallel. Cellular DNA loads were quantified for each sample by amplifying a β-actin gene fragment. Viral DNA loads were then normalized by cellular DNA loads.
Purification of CD11c+ cells from blood.
Mice were bled by cardiac puncture into PBS–10 mM EDTA. Mononuclear cells were recovered by centrifugation on Ficoll-Paque (GE Healthcare) and incubated with anti-mouse CD11c microbeads (MACS; Miltenyi Biotec) for 1 h at 4°C. CD11c− and CD11c+ cells were then separated on a MACS LS column according to the manufacturer’s instructions.
Immunostaining.
Organs were fixed in 1% formaldehyde–10 mM sodium periodate–75 mM l-lysine (18 h at 4°C), equilibrated in 30% sucrose (18 h at 4°C), and then frozen in OCT. Sections (6 μm) were air dried (1 h at 23°C), washed three times in PBS, and blocked with 0.1% Triton X-100 plus 5% horse serum. To detect viral antigens, sections were incubated (18 h at 4°C) with mouse IgG1 mAb CROMA-101 (45), a polyclonal rabbit serum raised by three subcutaneous inoculations with MCMV K181 (44), or a chicken pAb to virus-expressed βgal (Abcam). Virus-expressed GFP was visualized directly. To detect cellular antigens, sections were incubated (18 h at 4°C) with antibodies to SPC (goat pAb; Santa Cruz Biotechnology), cytokeratin-19 (Cyt19) (goat pAb N13; Santa Cruz Biotechnology), CD68 (rat mAb FA-11; Abcam), CD11c (hamster mAb HL-3; BD Biosciences), or Aquaporin V (AQP5) (rabbit pAb; Alamone Labs). After incubation with primary antibodies, sections were washed three times in PBS; incubated (1 h at 23°C) with combinations of Alexa 488-, Alexa 568-, or Alexa 647-conjugated donkey pAb to goat, mouse, rabbit, or rat IgG (Abcam); washed three times in PBS; stained with DAPI (4′,6-diamidino-2-phenylindole); and mounted in ProLong Gold (Life Technologies). Images were acquired with a Zeiss LSM510 microscope and analyzed with Zen software or ImageJ.
Immunofluorescence.
Cells were fixed in 1% paraformaldehyde (PFA) (23°C for 30 min), washed three times in PBS, and permeabilized/blocked by incubation in 0.1% Triton-TX100 plus 5% goat serum. They were then stained for MCMV virion antigens with a rabbit pAb and for gO with mouse IgG1 mAb Cl.M74.01. Secondary detection was done with Alexa 488-goat pAb to mouse IgG and Alexa 594-goat pAb to rabbit IgG. Nuclei were stained with DAPI, samples were mounted with ProLong Gold, and fluorescence was visualized with a Zeiss LSM510 confocal microscope.
Western blotting.
Virus stocks, infected cells, or purified virions were lysed directly in Laemmli’s buffer with 2-mercaptoethanol, heated (50°C for 10 min), resolved by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad), and then immunoblotted with antibodies to MCMV virion antigens (rabbit pAb), M78 (rabbit pAb) (43), gB (Cl.M55.02; mouse mAb IgG2a), open reading frame 74 (ORF74) (Cl.M74.01; mouse mAb IgG1), or M131-129 (Cl.MCK-2.04; mouse mAb IgG1). All MCMV-specific mAbs were produced by the University of Rijeka Center for Proteomics. Secondary detection was done with far-red fluorescence anti-mouse IgG and anti-rabbit IgG antibodies from Li-Cor Biosciences. Signals were visualized on an Odyssey imager.
ACKNOWLEDGMENTS
This work was supported by the Australian National Health and Medical Research Council (grants 1079180, 1122070, and 1140169) and by Queensland Health. J.Y. was the recipient of a University of Queensland Ph.D. scholarship. B.A. was supported by the Deutsche Forschungsgemeinschaft (AD 131/4-1). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Gillet L, May JS, Colaco S, Stevenson PG. 2007. Glycoprotein L disruption reveals two functional forms of the murine gammaherpesvirus 68 glycoprotein H. J Virol 81:280–291. doi: 10.1128/JVI.01616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Kenyon WJ, Li Q, Müllberg J, Hutt-Fletcher LM. 1998. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J Virol 72:5552–5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hutt-Fletcher LM. 2016. The long and complicated relationship between Epstein-Barr virus and epithelial cells. J Virol 91:e01677-16. doi: 10.1128/JVI.01677-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillet L, May JS, Stevenson PG. 2009. In vivo importance of heparan sulfate-binding glycoproteins for murid herpesvirus-4 infection. J Gen Virol 90:602–613. doi: 10.1099/vir.0.005785-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milho R, Frederico B, Efstathiou S, Stevenson PG. 2012. A heparan-dependent herpesvirus targets the olfactory neuroepithelium for host entry. PLoS Pathog 8:e1002986. doi: 10.1371/journal.ppat.1002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.François S, Vidick S, Sarlet M, Desmecht D, Drion P, Stevenson PG, Vanderplasschen A, Gillet L. 2013. Illumination of murine gammaherpesvirus-68 cycle reveals a sexual transmission route from females to males in laboratory mice. PLoS Pathog 9:e1003292. doi: 10.1371/journal.ppat.1003292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frederico B, Milho R, May JS, Gillet L, Stevenson PG. 2012. Myeloid infection links epithelial and B cell tropisms of murid herpesvirus-4. PLoS Pathog 8:e1002935. doi: 10.1371/journal.ppat.1002935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mori Y, Akkapaiboon P, Yonemoto S, Koike M, Takemoto M, Sadaoka T, Sasamoto Y, Konishi S, Uchiyama Y, Yamanishi K. 2004. Discovery of a second form of tripartite complex containing gH-gL of human herpesvirus 6 and observations on CD46. J Virol 78:4609–4616. doi: 10.1128/JVI.78.9.4609-4616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerna G, Revello MG, Baldanti F, Percivalle E, Lilleri D. 2017. The pentameric complex of human cytomegalovirus: cell tropism, virus dissemination, immune response and vaccine development. J Gen Virol 98:2215–2234. doi: 10.1099/jgv.0.000882. [DOI] [PubMed] [Google Scholar]
- 10.Hahn G, Revello MG, Patrone M, Percivalle E, Campanini G, Sarasini A, Wagner M, Gallina A, Milanesi G, Koszinowski U, Baldanti F, Gerna G. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J Virol 78:10023–10033. doi: 10.1128/JVI.78.18.10023-10033.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scrivano L, Sinzger C, Nitschko H, Koszinowski UH, Adler B. 2011. HCMV spread and cell tropism are determined by distinct virus populations. PLoS Pathog 7:e1001256. doi: 10.1371/journal.ppat.1001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wille PT, Knoche AJ, Nelson JA, Jarvis MA, Johnson DC. 2010. A human cytomegalovirus gO-null mutant fails to incorporate gH/gL into the virion envelope and is unable to enter fibroblasts and epithelial and endothelial cells. J Virol 84:2585–2596. doi: 10.1128/JVI.02249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turtinen LW, Saltzman R, Jordan MC, Haase AT. 1987. Interactions of human cytomegalovirus with leukocytes in vivo: analysis by in situ hybridization. Microb Pathog 3:287–297. doi: 10.1016/0882-4010(87)90062-3. [DOI] [PubMed] [Google Scholar]
- 14.Wagner FM, Brizic I, Prager A, Trsan T, Arapovic M, Lemmermann NA, Podlech J, Reddehase MJ, Lemnitzer F, Bosse JB, Gimpfl M, Marcinowski L, MacDonald M, Adler H, Koszinowski UH, Adler B. 2013. The viral chemokine MCK-2 of murine cytomegalovirus promotes infection as part of a gH/gL/MCK-2 complex. PLoS Pathog 9:e1003493. doi: 10.1371/journal.ppat.1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleming P, Davis-Poynter N, Degli-Esposti M, Densley E, Papadimitriou J, Shellam G, Farrell H. 1999. The murine cytomegalovirus chemokine homolog, m131/129, is a determinant of viral pathogenicity. J Virol 73:6800–6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saederup N, Aguirre SA, Sparer TE, Bouley DM, Mocarski ES. 2001. Murine cytomegalovirus CC chemokine homolog MCK-2 (m131-129) is a determinant of dissemination that increases inflammation at initial sites of infection. J Virol 75:9966–9976. doi: 10.1128/JVI.75.20.9966-9976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collins TM, Quirk MR, Jordan MC. 1994. Biphasic viremia and viral gene expression in leukocytes during acute cytomegalovirus infection of mice. J Virol 68:6305–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrell HE, Bruce K, Lawler C, Oliveira M, Cardin R, Davis-Poynter N, Stevenson PG. 2017. Murine cytomegalovirus spreads by dendritic cell recirculation. mBio 8:e01264-17. doi: 10.1128/mBio.01264-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scrivano L, Esterlechner J, Mühlbach H, Ettischer N, Hagen C, Grünewald K, Mohr CA, Ruzsics Z, Koszinowski U, Adler B. 2010. The m74 gene product of murine cytomegalovirus (MCMV) is a functional homolog of human CMV gO and determines the entry pathway of MCMV. J Virol 84:4469–4480. doi: 10.1128/JVI.02441-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang XJ, Adler B, Sampaio KL, Digel M, Jahn G, Ettischer N, Stierhof YD, Scrivano L, Koszinowski U, Mach M, Sinzger C. 2008. UL74 of human cytomegalovirus contributes to virus release by promoting secondary envelopment of virions. J Virol 82:2802–2812. doi: 10.1128/JVI.01550-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemmermann NA, Krmpotic A, Podlech J, Brizic I, Prager A, Adler H, Karbach A, Wu Y, Jonjic S, Reddehase MJ, Adler B. 2015. Non-redundant and redundant roles of cytomegalovirus gH/gL complexes in host organ entry and intra-tissue spread. PLoS Pathog 11:e1004640. doi: 10.1371/journal.ppat.1004640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sacher T, Podlech J, Mohr CA, Jordan S, Ruzsics Z, Reddehase MJ, Koszinowski UH. 2008. The major virus-producing cell type during murine cytomegalovirus infection, the hepatocyte, is not the source of virus dissemination in the host. Cell Host Microbe 3:263–272. doi: 10.1016/j.chom.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 23.Farrell HE, Lawler C, Tan CS, MacDonald K, Bruce K, Mach M, Davis-Poynter N, Stevenson PG. 2016. Murine cytomegalovirus exploits olfaction to enter new hosts. mBio 7:e00251-16. doi: 10.1128/mBio.00251-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shivkumar M, Milho R, May JS, Nicoll MP, Efstathiou S, Stevenson PG. 2013. Herpes simplex virus 1 targets the murine olfactory neuroepithelium for host entry. J Virol 87:10477–10488. doi: 10.1128/JVI.01748-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farrell HE, Lawler C, Oliveira MT, Davis-Poynter N, Stevenson PG. 2015. Alveolar macrophages are a prominent but nonessential target for murine cytomegalovirus infecting the lungs. J Virol 90:2756–2766. doi: 10.1128/JVI.02856-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scalzo AA, Dallas PB, Forbes CA, Mikosza AS, Fleming P, Lathbury LJ, Lyons PA, Laferté S, Craggs MM, Loh LC. 2004. The murine cytomegalovirus M73.5 gene, a member of a 3′ co-terminal alternatively spliced gene family, encodes the gp24 virion glycoprotein. Virology 329:234–250. doi: 10.1016/j.virol.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 27.Farrell H, Oliveira M, Macdonald K, Yunis J, Mach M, Bruce K, Stevenson P, Cardin R, Davis-Poynter N. 2016. Luciferase-tagged wild-type and tropism-deficient mouse cytomegaloviruses reveal early dynamics of host colonization following peripheral challenge. J Gen Virol 97:3379–3391. doi: 10.1099/jgv.0.000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sokal EM, Hoppenbrouwers K, Vandermeulen C, Moutschen M, Léonard P, Moreels A, Haumont M, Bollen A, Smets F, Denis M. 2007. Recombinant gp350 vaccine for infectious mononucleosis: a phase 2, randomized, double-blind, placebo-controlled trial to evaluate the safety, immunogenicity, and efficacy of an Epstein-Barr virus vaccine in healthy young adults. J Infect Dis 196:1749–1753. doi: 10.1086/523813. [DOI] [PubMed] [Google Scholar]
- 29.Pass RF. 2009. Development and evidence for efficacy of CMV glycoprotein B vaccine with MF59 adjuvant. J Clin Virol 46:S73–S76. doi: 10.1016/j.jcv.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson CS, Huffman T, Jenks JA, Cisneros de la Rosa E, Xie G, Vandergrift N, Pass RF, Pollara J, Permar SR. 2018. HCMV glycoprotein B subunit vaccine efficacy mediated by nonneutralizing antibody effector functions. Proc Natl Acad Sci U S A 115:6267–6272. doi: 10.1073/pnas.1800177115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wussow F, Chiuppesi F, Martinez J, Campo J, Johnson E, Flechsig C, Newell M, Tran E, Ortiz J, La Rosa C, Herrmann A, Longmate J, Chakraborty R, Barry PA, Diamond DJ. 2014. Human cytomegalovirus vaccine based on the envelope gH/gL pentamer complex. PLoS Pathog 10:e1004524. doi: 10.1371/journal.ppat.1004524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dolan A, Cunningham C, Hector RD, Hassan-Walker AF, Lee L, Addison C, Dargan DJ, McGeoch DJ, Gatherer D, Emery VC, Griffiths PD, Sinzger C, McSharry BP, Wilkinson GW, Davison AJ. 2004. Genetic content of wild-type human cytomegalovirus. J Gen Virol 85:1301–1312. doi: 10.1099/vir.0.79888-0. [DOI] [PubMed] [Google Scholar]
- 33.Sinclair J. 2008. Human cytomegalovirus: latency and reactivation in the myeloid lineage. J Clin Virol 41:180–185. doi: 10.1016/j.jcv.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 34.Griffiths PD, Cope AV, Hassan-Walker AF, Emery VC. 1999. Diagnostic approaches to cytomegalovirus infection in bone marrow and organ transplantation. Transpl Infect Dis 1:179–186. doi: 10.1034/j.1399-3062.1999.010306.x. [DOI] [PubMed] [Google Scholar]
- 35.Emery VC, Cope AV, Bowen EF, Gor D, Griffiths PD. 1999. The dynamics of human cytomegalovirus replication in vivo. J Exp Med 190:177–182. doi: 10.1084/jem.190.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seckert CK, Renzaho A, Reddehase MJ, Grzimek NK. 2008. Hematopoietic stem cell transplantation with latently infected donors does not transmit virus to immunocompromised recipients in the murine model of cytomegalovirus infection. Med Microbiol Immunol 197:251–259. doi: 10.1007/s00430-008-0094-1. [DOI] [PubMed] [Google Scholar]
- 37.Kabanova A, Marcandalli J, Zhou T, Bianchi S, Baxa U, Tsybovsky Y, Lilleri D, Silacci-Fregni C, Foglierini M, Fernandez-Rodriguez BM, Druz A, Zhang B, Geiger R, Pagani M, Sallusto F, Kwong PD, Corti D, Lanzavecchia A, Perez L. 2016. Platelet-derived growth factor-α receptor is the cellular receptor for human cytomegalovirus gHgLgO trimer. Nat Microbiol 1:16082. doi: 10.1038/nmicrobiol.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y, Prager A, Boos S, Resch M, Brizic I, Mach M, Wildner S, Scrivano L, Adler B. 2017. Human cytomegalovirus glycoprotein complex gH/gL/gO uses PDGFR-α as a key for entry. PLoS Pathog 13:e1006281. doi: 10.1371/journal.ppat.1006281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feederle R, Neuhierl B, Bannert H, Geletneky K, Shannon-Lowe C, Delecluse HJ. 2007. Epstein-Barr virus B95.8 produced in 293 cells shows marked tropism for differentiated primary epithelial cells and reveals interindividual variation in susceptibility to viral infection. Int J Cancer 121:588–594. doi: 10.1002/ijc.22727. [DOI] [PubMed] [Google Scholar]
- 40.Frederico B, Chao B, May JS, Belz GT, Stevenson PG. 2014. A murid gamma-herpesviruses exploits normal splenic immune communication routes for systemic spread. Cell Host Microbe 15:457–470. doi: 10.1016/j.chom.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 41.Davis-Poynter NJ, Lynch DM, Vally H, Shellam GR, Rawlinson WD, Barrell BG, Farrell HE. 1997. Identification and characterization of a G protein-coupled receptor homolog encoded by murine cytomegalovirus. J Virol 71:1521–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson DC, Webb M, Wisner TW, Brunetti C. 2001. Herpes simplex virus gE/gI sorts nascent virions to epithelial cell junctions, promoting virus spread. J Virol 75:821–833. doi: 10.1128/JVI.75.2.821-833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yunis J, Farrell HE, Bruce K, Lawler C, Sidenius S, Wyer O, Davis-Poynter N, Stevenson PG. 2018. Murine cytomegalovirus degrades MHC class II to colonize the salivary glands. PLoS Pathog 14:e1006905. doi: 10.1371/journal.ppat.1006905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farrell HE, Davis-Poynter N, Bruce K, Lawler C, Dolken L, Mach M, Stevenson PG. 2015. Lymph node macrophages restrict murine cytomegalovirus dissemination. J Virol 89:7147–7158. doi: 10.1128/JVI.00480-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trgovcich J, Stimac D, Polić B, Krmpotić A, Pernjak-Pugel E, Tomac J, Hasan M, Wraber B, Jonjić S. 2000. Immune responses and cytokine induction in the development of severe hepatitis during acute infections with murine cytomegalovirus. Arch Virol 145:2601–2618. doi: 10.1007/s007050070010. [DOI] [PubMed] [Google Scholar]
- 46.Thäle R, Lucin P, Schneider K, Eggers M, Koszinowski UH. 1994. Identification and expression of a murine cytomegalovirus early gene coding for an Fc receptor. J Virol 68:7757–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.National Health and Medical Research Council. 2013. Australian code for the care and use of animals for scientific purposes, 8th ed. National Health and Medical Research Council publication no. EA28. National Health and Medical Research Council, Canberra, Australia: https://nhmrc.gov.au/about-us/publications/australian-code-care-and-use-animals-scientific-purposes. [Google Scholar]