Abstract
Palmitate (PA) exposure induces stress conditions featuring ROS accumulation and upregulation of p62 expression, resulting in autophagic flux blockage and cell apoptosis. Sulfuretin (Sul) is a natural product isolated from Rhus verniciflua Stokes; the cytoprotective effect of Sul on human hepatic L02 cells and mouse primary hepatocytes under PA-induced stress conditions was investigated in this study. Sul induced mitophagy by activation of p-TBK1 and LC3 and produced a concomitant decline in p62 expression. Autophagosome formation and mitophagy were assessed by the sensitive dual fluorescence reporter mCherry-EGFP-LC3B, and mitochondrial fragmentation was analyzed using MitoTracker Deep Red FM. A preliminary structure–activity relationship (SAR) for Sul was also investigated, and the phenolic hydroxyl group was found to be pivotal for maintaining the cytoprotective bioactivity of Sul. Furthermore, experiments using flow cytometry and western blots revealed that Sul reversed the cytotoxic effect stimulated by the autophagy inhibitors 3-methyladenine (3-MA) and chloroquine (CQ), and its cytoprotective effect was almost eliminated when the autophagy-related 5 (Atg5) gene was knocked down. These studies suggest that, in addition to its antioxidative effects, Sul stimulates mitophagy and restores impaired autophagic flux, thus protecting hepatic cells from apoptosis, and that Sul has potential future medical applications for hepatoprotection.
Keywords: sulfuretin, palmitate, mitophagy, autophagic flux
Introduction
Mitochondria play a key role in ATP production and are crucial organelles for cell growth, apoptosis and death. Oxidative phosphorylation is the main, but not the only, function of mitochondria, and reactive oxygen species (ROS), including superoxide, hydrogen peroxide, and hydroxyl radicals, are formed in the process of mitochondrial respiratory activity [1]. Under physiological conditions, there is a balance between the formation of ROS and their elimination by antioxidant enzymes and other chemical substances [2, 3]. When the balance is disrupted by the excessive formation of ROS, oxidative stress (OS) occurs [4]. Palmitate (PA) triggers oxidative stress and mitochondrial oxidative damage and induces downstream apoptotic events by mitochondrial activation [5, 6]. Many diseases, such as aging, cancer, cardiovascular disease, diabetes, and neurodegenerative diseases, are associated with mitochondrial dysfunction [7–12]; in brief, mitochondrial function is closely related to health and disease development. Mitochondrial quality control (MQC) is one of the critical processes maintaining mitochondrial health, and autophagy is an indispensable cellular process that greatly contributes to MQC [13, 14].
Macroautophagy (often referred to as autophagy), a process of “self-eating”, is a response to starvation and stress [15] and removes aged and damaged organelles and proteins under normal conditions [16]. The autophagy process can be summarized by four major stages: induction, nucleation, expansion/completion, and termination/retrieval [17]. Cells generate double-membrane vesicles that elongate into what is defined as an autophagosome, which engulfs macromolecules or organelles and delivers them to the lysosome for degradation [18]. In virtually all cells, autophagy is a fundamental process that sustains basal functions, including protein and organelle turnover. Autophagy is a rapid response to sublethal stress so that cells can make changes in their metabolism and protect themselves from potential damage. Under stress, cells eliminate defective proteins and macromolecules, damaged organelles, and toxic aggregates through the autophagy process for survival [19, 20]. The blockage of autophagic flux can lead to cell apoptosis [21].
Selective degradation of mitochondria by autophagy, defined as mitophagy, is one of the crucial aspects of MQC and is important for the optimal maintenance of mitochondrial function [22, 23]. Mitophagy can be regulated by mitochondrial dynamics [24], mitochondrial membrane potential (MMP) [25], accumulation of full-length PINK1 to recruit PARKIN to mitochondria [26, 27], and phosphorylation of TBK1 [26]; mitophagy can be marked by the formation of mitophagosomes, decreased mitochondrial mass, and mitochondrial fragmentation [27]. Many studies have revealed that mitochondrial fission/fusion machinery participates in the regulation of mitophagy and have demonstrated that depolarized mitochondria are a substrate for mitophagy [28, 29], such that antimycin A and CCCP initiate mitophagosome formation in HCT-116 cells [24]. When threatened or dysfunctional, such as during OS, damaged and excess mitochondria are selectively eliminated through autophagy or mitophagy to maintain mitochondrial quality and quantity for survival [30].
Typically, to estimate overall autophagic flux, one must detect the autophagy marker LC3-II, and one should observe the formation of autophagosomes [31]. The increased synthesis or lipidation of LC3, corresponding to the increased formation of autophagosomes, is used as a standard way to evaluate autophagic activity [32]. Autophagy is a highly dynamic, multistep process; therefore, the number of autophagosomes is determined by both their formation and clearance [33]. To accurately assess autophagic activity, one should evaluate the digestion and degradation of contents and the breakdown products released [31, 32]. The accumulation of p62 is considered a marker of autophagic flux blockage [31, 33]. If autophagosomes have formed and LC3 is detected, but p62 has accumulated, this finding indicates that autophagosomes were formed but could not be degraded, which means that autophagic flux was blocked [32, 34]. As a result, the blockage of autophagic flux would lead to cell apoptosis [21].
Recent studies revealed that prolonged exposure of β cells to free fatty acids (FFAs), such as PA, leads to an increased number of autophagosomes due to blocked autophagic flux, which is clearly shown by increased LC3-II levels as well as p62 accumulation [35, 36]. When autophagy turnover was damaged in β cells, this effect subsequently led to apoptotic cell death. Under oxidative stress conditions, inhibition of autophagy promotes apoptosis of hepatocytes; in other words, the impaired autophagy function promotes oxidant-induced liver injury [37]. In addition, autophagy can regulate β-oxidation of fatty acids and may relieve lipotoxicity induced by free fatty acids such as PA in hepatic cells [38, 39]. Autophagy participates in liver metabolism through multiple pathways and plays diverse roles in the survival and function of hepatic cells [40]. Therefore, the restoration and promotion of autophagic flux may protect the cell from apoptosis under PA-induced OS conditions.
Sul is one of the natural products isolated from Rhus verniciflua and is known to have various biological activities, including anti-inflammatory [41] antimutagenic [42], anticancer [43], antioxidative stress [44], antiplatelet [45], and anti-rheumatoid arthritis effects [46]. In previous studies, most of the reports focused on the anti-inflammatory activity of Sul.
Here, we sought to validate the hepatoprotective effect of Sul on human hepatic L02 cells, and we found evidence that Sul promotes mitophagy, decreases p62 levels, and eliminates ROS to protect L02 cells from apoptosis. In addition, we made an effort to prove that Sul could be a potential mitophagy stimulator and restore autophagic flux to protect against cell apoptosis.
Materials and methods
Chemistry of Sul and its derivatives
Compounds S1, S2, S3, S4, and S5 were synthesized according to previously reported procedures [47, 48]. Sul was purchased from BioBioPha (China; #120–05–8). Starting materials, reagents and solvents were purchased from commercial suppliers and used without further purification. All nonaqueous reactions were performed under an inert atmosphere (argon) with rigid exclusion of moisture from reagents, and all reaction flasks were oven-dried. TLC was carried out on precoated TLC plates with silica gel HSGF 254. Spots were visualized under UV at 254 nm. 1H and 13C NMR spectra were measured on a Brucker AVANCE III 500 spectrometer using deuterated chloroform (CDCl3), deuterated methanol (CD3OD-d4), and deuterated dimethyl sulfoxide (DMSO-d6) as the solvents. HRMS were measured on a Micromass Ultra Q-Tof Ultima TM spectrometer.
Cell culture and plasmid transfection
The L02 cell line, a human hepatic cell line, was purchased from ATCC and maintained in RPMI 1640 (Corning; #1868882) with 10% FBS (Gibco) at 37 °C, 5% CO2 and 21% O2 in a regular incubator. L02 cells before passage 30 were used. The pBABE-puro mCherry-EGFP-LC3B plasmid was purchased from Addgene (Cambridge, MA, USA; Addgene ID: 22418). For imaging experiments, L02 cells were plated on coverslips in 12-well plates and incubated overnight to adhere. Cells were then transfected with the desired plasmid according to the manufacturer’s instructions and incubated for 48 h. In detail, L02 cells were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA; #11668019) containing the basic vector or mCherry-EGFP-LC3B in Opti-MEM (Invitrogen; #31985070). After 6 h of incubation, the medium was changed to RPMI 1640 with 10% FBS (Gibco; #10099141), cells were incubated for 2 days, and compounds were added into the medium 1 h before the laser confocal microscopy assay was performed.
Small interfering RNA (siRNA) transfection
L02 cells were plated in 6-well plates and incubated overnight to adhere. Cells were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA; #11668019) containing the scramble siRNA sequences or targeting Atg5 siRNA sequences in Opti-MEM (Invitrogen; #31985070). After 6 h of incubation, the medium was changed to RPMI 1640 with 10% FBS (Gibco; #10099141), and the cells were incubated for 2 days. Compounds were added to the medium 24 h before the flow cytometric (FCM) analysis and western blot were performed. The small interfering RNA sequences are supplied in supplementary information. All the experimental supplies used in this assay are RNase free.
Treatment of cells with palmitate and cell viability analysis
Cells were cultured in 12-well, 24-well, or 96-well plates with approximately 2 × 105 cells/mL in culture medium with 10% FBS. Palmitate (Sigma-Aldrich, St. Louis, MO, USA; final concentration, 0.4 mM; #P9767) was added to the wells to induce OS and cell apoptosis as indicated in each experiment. Cell viability was measured using an MTS Cell Proliferation Colorimetric Assay Kit (Promega, Madison, WI; #3581) according to the manufacturer’s instructions.
Mitochondrial membrane potential assay
Mitochondrial membrane potentials were measured using the fluorochrome tetramethylrhodamine ethyl ester perchlorate (TMRE; excitation wavelength: 540 nm; emission wavelength: 595 nm; Sigma-Aldrich, St. Louis, MO, USA; #87917). Cells were plated in 96-well plates and cultured with phenol red-free medium. TMRE was added at 40 min, and the cells were pretreated with Sul for 1 h. The fluorescence intensity of TMRE was measured with a microplate fluorescence reader.
Measurement of intracellular ROS production
The production of intracellular ROS was measured using the general oxidative stress indicator CM-H2DCFDA (Invitrogen, Carlsbad, CA, USA; #C6827) according to the manufacturer’s instructions. Briefly, cells were plated in 96-well plates, and after 24 h, they were cotreated with palmitate and Sul. The cells were washed twice with PBS and supplied with phenol red-free medium containing 10 μM CM-H2DCFDA dye, and then, the cells were incubated for 30 min at 37 °C in the dark. Cells were then washed with phenol red-free medium three times, and the DCF fluorescence intensity was measured using a microplate fluorescence reader (excitation wavelength: 488 nm; emission wavelength: 530 nm).
Antibodies and immunoblotting
Western immunoblotting was performed as described previously. In brief, cells were lysed, sonicated and boiled at 100 °C for 10 min in sample buffer (50 mM Tris-HCl, 2% w/v SDS, 10% glycerol, 1% β-mercaptoethanol, 0.01% bromophenyl blue pH 6.8). Cell lysates were separated by SDS-PAGE and transferred to nitrocellulose (NC) filter membranes. The membranes were first incubated with blocking buffer (TBS with 0.1% Tween 20 and 5% non-fat milk) for 1 h at room temperature and then incubated overnight at 4 °C in buffer containing primary antibodies. The membranes were washed three times and then incubated with secondary antibodies for 1 h. After three washes, immunostaining was visualized using an electrochemiluminescence and ChemiDoc imaging system (Bio-Rad). Anti-PARP (#9542), anti-cleaved caspase 3 (#9664), anti-cleaved caspase 9 (#9509) and anti-p-TBK1 (#5483) antibodies were purchased from Cell Signaling Technologies (Danvers, MA, USA). Anti-LC3B antibody (#L7543) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Anti-p62 antibody (#sc-25575) was purchased from Santa Cruz Biotechnology. Anti-β-actin (#AM1021B) antibody was purchased from ABGENT (San Diego, CA, USA).
FCM analyses
For cell flow cytometric (FCM) analysis, cells were trypsinized, and the single-cell suspensions were then stained with annexin V and propidium iodide (KeyGEN BioTECH; #KGA108) in binding buffer for 15 min at room temperature. Cells were then analyzed with a Guava Flow Cytometer (Millipore, St. Charles, MO, USA), and the data were collected with FlowJo software.
Fluorescence staining
For mitochondrial morphology analysis, cells were incubated with MitoTracker Deep Red FM (Invitrogen, Carlsbad, CA, USA; #M22426) for 20 min at room temperature. After washing with PBS, cells were fixed with 4% PFA (Sigma-Aldrich, St. Louis, MO, USA; #158127) for 10 min at room temperature. For autophagosome puncta formation analysis, nuclei were stained with Hoechst (Sigma-Aldrich, St. Louis, MO, USA; final concentration, 10 μg/mL; #14530) for 10 min. Cells were analyzed by laser confocal microscopy (Olympus, Tokyo, Japan).
Isolation of primary hepatocytes
Primary hepatocyte isolation was performed as described previously [49]. In detail, the male C57BL/6J mice at 6–8 weeks of age were anesthetized with 10% chloral hydrate, and 50 mL perfusion buffer was perfused through the portal vein at 37 °C. The perfusion buffer consisted of Krebs Ringer buffer, EGTA (0.19 mg/mL, Sangon Biotech, Shanghai; #A100732–0005) and Glucose (0.9 mg/mL). Then, 50 mL of collagenase-I (0.48 mg/mL, Worthington, Lakewood; #LS004196) dissolved in collagenase buffer was infused into the liver through the portal vein. The collagenase buffer consisted of Krebs Ringer buffer, HEPES (0.19 mg/mL, Sangon Biotech, Shanghai; #A600264–0250) and CaCl2 (2.5 mol/L) at 37 °C. The liver was aseptically removed to a sterile 6-cm cell culture dish containing 5 mL of cold perfusion buffer without collagenase. The excised liver was cut, and hepatocytes were dispersed by aspirating with a large-bore pipette, filtered through a 70-μm cell strainer (Thermo Fisher Scientific) into a 50-mL centrifuge tube and spun at 700 r/min for 5 min at 4 °C. The cells were then resuspended in HepatoZYME-SFM (Gibco, Grand Island, N.Y.; #17705021) medium containing 2 mM L-glutamine, 20 units/mL penicillin and 20 μg/mL streptomycin. Then, the cells were plated at 5 × 105 cells/mL in a 6-well culture plate that was precoated with gelatin.
Cell oxygen consumption rate analysis
Cells were cultured in Seahorse 96-well plates (Seahorse Bioscience, USA; #W21715) with approximately 1 × 104 cells in each well. Cells were pretreated with multidose Sul for 1 h, and OCR was measured using a Seahorse XFe96 analyzer according to the manufacturer’s instructions. Oligomycin, FCCP, and antimycin A & Rotenone were added during the assay.
Quantitative real-time PCR (RT-qPCR)
Total mRNA was isolated using Trizol (Thermo Fisher Scientific-Invitrogen; #15596018) according to the manufacturer’s instructions. Then, the PrimeScript RT reagent kit (Takara Bio; #RR036A) was used to reverse 1 μg of RNA to cDNA according to the manufacturer’s instructions. Real-time PCR was performed using AceQ Qpcr SYBR Green Master Mix (Vazyme; #Q131–02) and a StratageneMx 3000 P thermal cycler. Primer sequences are supplied in supplementary information.
Statistical analyses
Values are reported as the means ± SEM. P-values were calculated by Student’s t-test, and P < 0.05 was considered statistically significant. All graphs were plotted with GraphPad Prism software.
Results
Sul decreases ROS levels in human hepatic L02 cells and provides cytoprotection against palmitate treatment
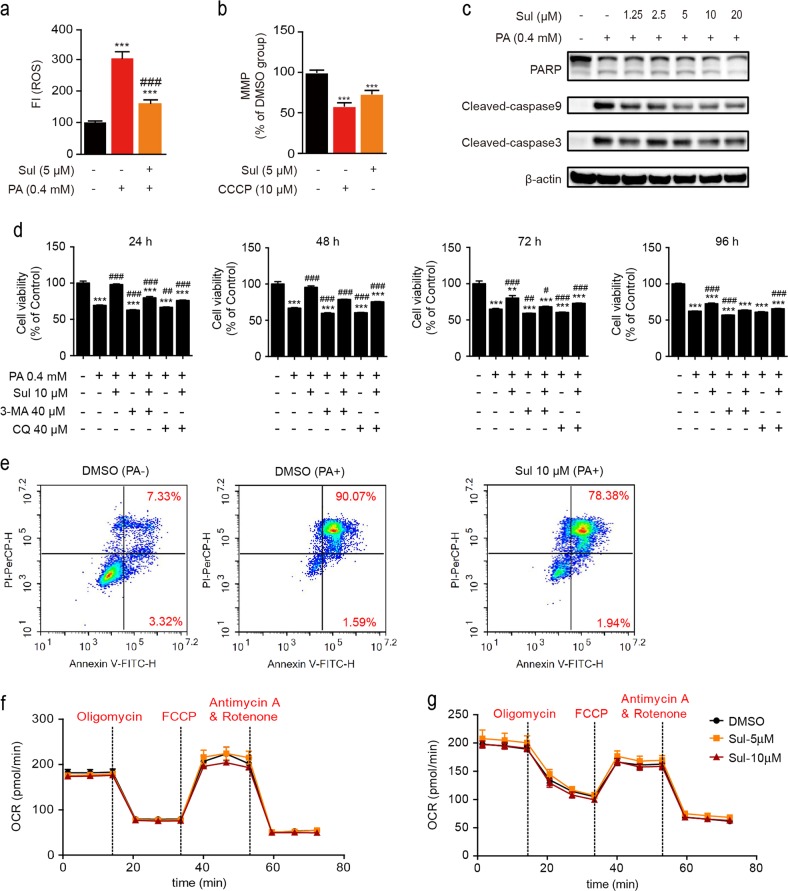
Free fatty acids such as PA induce lipotoxicity in hepatic cells [38, 39], and ROS accumulation plays an important role in cytotoxicity and cell death. Sul was reported to have cytoprotective effects against tert-butyl hydroperoxide-induced hepatotoxicity [50], so the antioxidant effect of Sul was initially investigated under PA-induced oxidative stress conditions in our study. The intracellular ROS level was determined by the fluorescence intensity of the fluorescent probe CM-H2DCFDA. After exposure to 0.4 mmol/L PA for 24 h, ROS levels were 3-fold higher than the ROS levels in the control cells (Fig. 1a), and cotreatment with Sul dramatically reduced the ROS levels (P < 0.001). Because the majority of ROS are produced by mitochondria, we then investigated the effect of Sul on mitochondrial function. TMRE was used to detect the mitochondrial membrane potential (mt∆ψ, MMP) of L02 cells in response to Sul. After treatment with 5 μM Sul, L02 cells showed an ~25% decrease in mt∆ψ compared to cells treated with DMSO (P < 0.001), and the chemical uncoupler CCCP showed an ~45% reduction (Fig. 1b). Western blot assays revealed that PA-induced cell death was apoptotic, as indicated by upregulated levels of cleaved PARP, cleaved caspase 3, and cleaved caspase 9 protein, and the above cell apoptosis could be prevented by Sul treatment in a dose-dependent manner (Fig. 1c).
Fig. 1.
Sul decreases ROS levels in human hepatic L02 cells and provides cytoprotection against palmitate treatment. a ROS level is measured in L02 cells induced with palmitate and treated with Sul for 24 h. b Mitochondrial membrane potential (MMP) detection while CCCP as a positive control. c Western blot analysis of apoptosis marker proteins including cleaved-PARP, cleaved-caspase3 and cleaved caspase9 while in response to PA and Sul treatments in L02 cells. d Time-course cell viability analysis after PA treatment with or without Sul or autophagy inhibitors CQ and 3-MA. e FCM analysis of apoptotic rate induced by PA and with or without Sul for 24 h in L02 cells. Representative plots from three independent experiments are shown. f Oxygen consumption experiment performed in L02 cells with Sul treatment. g Oxygen consumption experiment performed in hepatocytes with Sul treatment. Student’s t-test. All datas were shown as means ± SD of three independent replications. Asterisks indicate P-values (**P < 0.01, ***P < 0.001) of control versus treated groups, Croisillons indicate P-values (#P < 0.05, ##P < 0.01, ###P < 0.001) of PA and compounds co-treated groups versus PA treated DMSO group. PA, palmitate; Sul, sulfuretin; CQ, chloroquine; 3-MA, 3-Methyladenine; CCCP, carbonyl cyanide m-chlorphenyl-hydrazone. FCCP, Carbonyl cyanide-p-trifluoromethoxyphenylhydrazone
The dramatic decrease in MMP triggers mitophagy, which is one of the important ways to MQC which influence the cell viability. Therefore, we applied the autophagy inhibitors 3-MA and CQ to evaluate whether there was a correlation between Sul-induced MMP decrease and cell viability. Then, the time-course cytotoxic effect of PA was evaluated by cell viability assays, which showed that PA treatment caused cell death obviously, and Sul protected the cell viability to 48 h. In addition, Sul reversed cell viability damaged by autophagy inhibitors (Fig. 1d). FCM analysis revealed that PA induced a remarkable cell apoptosis rate after 24 h with 91.66% apoptotic cells, while cells cotreated with Sul reduced the apoptotic rate to 80.32% (Fig. 1e). To ensure the effect of mitochondrial electron transport chain (ETC) function we investigated the oxygen consumption rate by Sul treatment. The results show that the respiratory rate of ETC neither L02 cells (Fig. 1f) nor hepatocytes (Fig. 1g) was affected by multidose Sul treatments for 1 h.
These data suggest that Sul may interfere with the function of mitochondria through promoting mitophagy rather than respiratory function to exert its cytoprotective activity.
Chemical modification and SAR of Sul
To further understand the structure and activity relationship, we investigated the preliminary SAR of Sul. Because MMP is a comprehensive indicator of mitochondrial function, an MMP assay of L02 cells was used to evaluate the activity of modified Sul compounds (Table 1). We mainly focused on the multiple phenolic hydroxyl structures of Sul to verify their effects on the MMP model. The results showed that when all of the phenolic hydroxyl groups were protected by methyl groups, compound S3 completely lost its activity. Protection of both 3′- and 4′-hydroxyl groups could not be tolerated. However, compound S1, which had only the 6-hydroxyl group protected, maintained a certain degree of potency in the MMP model, although the potency was decreased compared to the potency of Sul. Removing one of the hydroxyl groups on ring B was allowed, but compound S4 and compound S5 also showed no higher potency than Sul. Compound S2, which expanded ring C, had only weak activity. These results indicated that the hydroxyl group on ring B is important and greatly contributes to the activity, while the hydroxyl group on ring A has little influence. Based on the SAR results, we chose Sul as a tool because Sul had the highest potency and had been demonstrated to have cytoprotective effects [50, 51].
Table 1.
The compounds synthesized and their activity in MMP model
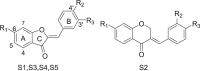
| |||||
|---|---|---|---|---|---|
| Compound | R1 | R2 | R3 | Concentration (μM) | MMP (% DMSO) |
| Sulfuretin | OH | OH | OH | 5 | 65.63 ± 0.31*** |
| S1 | OMe | OH | OH | 5 | 85.69 ± 2.56*** |
| S2 | OH | OH | OH | 5 | 91.91 ± 3.71* |
| S3 | OMe | OMe | OMe | 5 | 111.17 ± 3.20** |
| S4 | OH | H | OH | 5 | 76.52 ± 5.59*** |
| S5 | OH | OH | H | 5 | 79.76 ± 9.09** |
| CCCP | 10 | 22.64 ± 0.31*** | |||
Carbonyl cyanide m-chlorphenyl-hydrazone (CCCP) is a positive control. Asterisks indicate p-values (*P < 0.05, **P < 0.01, ***P < 0.001) of DMSO group versus treated groups, Student’s t-test. All datas were shown as means ± SD of three independent replications. Sul, sulfuretin; CCCP; S1-S5, Sul derivatives
Sul promotes mitophagy in human hepatic L02 cells
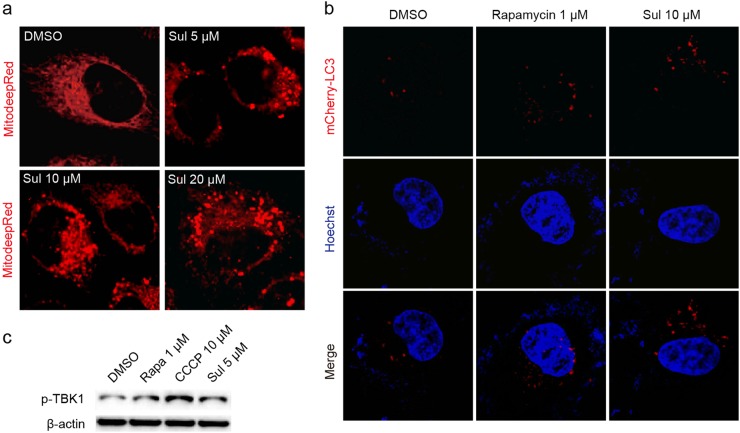
ROS are mainly produced in mitochondria, and mitophagy is important for cell quality control processes [1, 22]. The decrease in MMP is the prerequisite for mitophagy, so we then tested whether Sul influences mitophagy in L02 cells, and laser confocal microscopy was used to observe the mitochondrial fragmentation that occurred with mitophagy [25]. By staining with MitoTracker Deep Red FM, a commercially available mitochondrial indicator, the DMSO group showed classic mitochondrial structures (Fig. 2a). However, when treated with 5, 10, or 20 μM of Sul, the mitochondrial network structures were fragmented, as determined by laser confocal microscopy (Fig. 2a). This finding suggests that Sul may activate the mitophagy process to form autophagosomes. We also examined the effect of Sul on mitochondrial morphology (fragmentation) at different time points, and the same phenomenon was observed (Figure S1). To further confirm whether Sul can induce mitophagy, the pBABE-puro mCherry-EGFP-LC3B plasmid was used to investigate LC3-II puncta formation. The formation of mCherry fluorescent spots (red puncta) represents the formation of autophagosomes. The mTOR inhibitor rapamycin promoted autophagy, and the formation of autophagosomes was facilitated (Fig. 2b). As expected, Sul significantly induced autophagy and was much more effective than rapamycin (Fig. 2b).
Fig. 2.
Sul promotes mitophagy in human hepatic L02 cells. a Fluorescent staining of mitochondria by MitoTracker Deep Red for 20 min with or without multi-dose Sul treatment. b Autophagosome observation by laser confocal microscopy after pBABE-puro mCherry-EGFP-LC3B plasmid transfected into L02 cell and treated with Sul. Nuclei are stained with Hoechst. c Phosphorylation state of TBK1 detected by western blot assay while rapamycin and CCCP as positive control. Representative plots from three independent experiments are shown. Sul, sulfuretin; CCCP, carbonyl cyanide m-chlorphenyl-hydrazone. See also Figure S1
In addition, the phosphorylation of TBK1, which is considered one of the criteria of mitophagy, was evaluated [26]. CCCP is recognized as a positive control compound that can trigger mitophagy by lowering the MMP [25]. The results showed that Sul, as well as CCCP, could significantly promote the phosphorylation of TBK1 compared to the phosphorylation observed in the DMSO group (Fig. 2c). This result was consistent with the mitochondrial fragmentation phenomenon observed in the fluorescence images showing that Sul promoted mitophagy.
Sul reverses PA-induced or autophagy inhibitor-induced autophagic flux blockage to protect cells from apoptosis
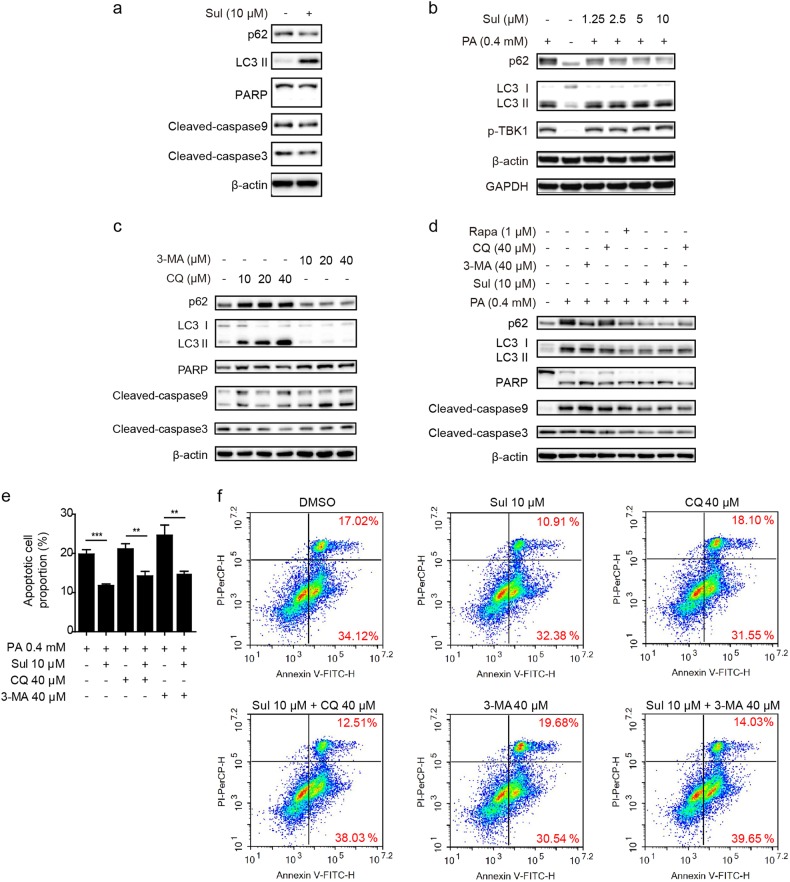
Based on our data showing that Sul protected cells from apoptosis and promoted mitophagy, we hypothesized that mitophagy activation may be a novel mechanism through which Sul exerts its hepatoprotective function. To confirm this hypothesis, the effects of Sul on mitophagy, with or without PA-induced OS conditions were assessed. The results showed that when cells were treated with Sul for 24 h, the autophagy flux evidenced by upregulated LC3-II levels and reduced p62 levels for autophagy processing, accompanying the downregulated apoptosis signals, cleaved caspase 9 and cleaved caspase 3, were clearly observed (Fig. 3a). In addition, PA caused not only LC3-II production and TBK1 phosphorylation but also p62 protein accumulation, indicating that autophagic flux and mitophagy were blocked (Fig. 3b). In addition, Sul treatment restored p62 protein accumulation in a dose-dependent manner (Fig. 3b). The autophagy inhibitors CQ and 3-MA have been widely used to verify the inhibition of autophagy. CQ prevents the fusion of the autophagosome and lysosome, and the degradation of LC3 and p62 proteins would be blocked, resulting in the observed accumulation of these two proteins; 3-MA inhibits the initiation of autophagy, so neither LC3 nor p62 was increased (Fig. 3c). Regarding cleaved caspases and cleaved PARP as apoptotic signals, inhibition of autophagy by CQ and 3-MA both led to increased levels of L02 apoptosis (Fig. 3c).
Fig. 3.
Sul reverses PA or autophagy inhibitors induced autophagic flux blockage to protect cells from apoptosis. a Western blot analysis of autophagy and apoptosis marker proteins in response to Sul treatment in L02 cells. b Western blot analysis of autophagy and mitophagy marker proteins in response to various treatments in L02 cells. c Western blot analysis of autophagy and apoptosis marker proteins in response to multi-dose of autophagy flux inhibitors (CQ and 3-MA) in L02 cells. d Western blot analysis of autophagy and apoptosis marker proteins in response to various treatments in L02 cells. e Statistical data of the late stage apoptotic cell proportion under conditions performed in f. f Representative FCM analysis of apoptotic rate with PA induced in response to various treatments in L02 cells for 12 h. Student’s t-test. Datas were shown as means ± SD of three independent replications. Asterisks indicate P-values (**P < 0.01, ***P < 0.001). PA, palmitate; Sul, sulfuretin; CQ, chloroquine; 3-MA, 3-Methyladenine. See also Figure S2 and Figure S4
Based on the results of Fig. 3a–c, we next used CQ, 3-MA and rapamycin to detect the effects of the mitophagy modulators under PA-induced L02 cell apoptotic conditions. As shown in Fig. 3d, the autophagy-inducer rapamycin reduced the cleaved form of PARP and the caspases, and the autophagy inhibitor further aggravated cell apoptosis. Cotreatment with Sul partially restored the autophagic flux marker p62 levels and reduced the cleaved PARP and caspase levels produced by PA.
To better illustrate the cell protective effect of Sul and confirm its relationship to autophagy, we performed FCM analyses. According to the statistical data, Sul displayed good cytoprotective efficacy (Fig. 3e). In detail, cells were incubated with PA (0.4 mM) for 12 h, and the proportion of nonviable apoptotic (late stage of apoptosis) cells increased dramatically to 19.84% (Fig. 3e, f). Treatment with Sul alleviated cell apoptosis, and there was an approximately 8% decline (Fig. 3e, f). As expected, based on the PA-induced lipotoxicity, the autophagy inhibitors CQ or 3-MA further increased the proportion of nonviable apoptotic cells (21.16% and 24.67%, respectively). Sul coincubation with CQ or 3-MA recovered the apoptotic rate to 14.27% and 14.81%, respectively (Figs. 3e, f).
Subsequently, cells were incubated with 0.4 mmol/L PA for 18 h, and the apoptotic rate of cells was remarkably increased compared to the rate at 12 h of treatment (Figure S2); similar to the results shown in Fig. 3, Sul showed protective activity against the PA-induced apoptotic rate from 80.05% to 71.75%. Treatment with the autophagy inhibitor 3-MA for 18 h aggravates cell apoptosis, and Sul reduced the apoptotic cell proportion from 89.55% to 72.57% (Figure S2).
Taken together, these results revealed that Sul may exert hepatoprotective activity by promoting autophagy, specifically mitophagy.
Sul eliminates ROS induced by PA and reverses autophagy inhibitors induced cell apoptosis in primary hepatocytes
To further validate the cytoprotective effect of Sul on hepatocytes, we isolated primary mouse hepatocytes for further study. ROS levels were seriously increased after PA treatment, and the autophagy inhibitors could not further increase ROS, and Sul eliminated ROS in various conditions remarkable in hepatocytes (Fig. 4a). The time-course cell viability assays showed that Sul exhibited a well cytoprotective effect on PA treatment and that autophagy inhibitors caused cell death (Fig. 4b). According to FCM analysis, based on the PA-induced lipotoxicity, the autophagy inhibitors CQ or 3-MA sharply increased the proportion of nonviable apoptotic cells (Fig. 4c). Sul coincubation with CQ or 3-MA reversed the apoptotic cell rate (Fig. 4c). The statistical data of FCM analyses are shown in Fig. 4d. The promoting effect of Sul on autophagy or mitophagy is positively correlated with its cytoprotective effect, which is represented by the intensity of the cleaved form of PARP and the caspases (Fig. 4e). In summary, Sul promoted autophagic flux to reduce cell apoptosis induced by PA and autophagy inhibitors.
Fig. 4.
Sul eliminates ROS induced by PA and reverse autophagy inhibitors induced cell apoptosis in primary hepatocytes. a Time-course ROS detection in response to various treatments in hepatocyte. b Time-course cell viability measurement in response to various treatments in hepatocyte. c Representative FCM analysis of apoptotic rate with PA induced in response to various treatments in hepatocyte for 24 h. Representative plots from three independent experiments are shown. d Statistical data of the whole apoptotic cell proportion under conditions what mentioned in c. e Western blot analysis of autophagy, mitophagy and apoptosis marker proteins in response to various treatments in hepatocytes. Student’s t-test. All datas were shown as means ± SD of three independent replications. Asterisks indicate P-values (*P < 0.05, **P < 0.01, ***P < 0.001) of control versus treated groups, Croisillons indicate P-values (#P < 0.05, ##P < 0.01, ###P < 0.001) of PA and compounds co-treated groups versus PA treated DMSO group. PA, palmitate; Sul, sulfuretin; CQ, chloroquine; 3-MA, 3-Methyladenine. See also Figure S3 and Figure S4
In addition, after PA treatment, the morphology of primary hepatocytes was seriously impaired, and Sul remarkably improved the phenotype of hepatocytes (Figure S3). The autophagy inhibitors aggravated cell morphology damage, and Sul mitigated this phenomenon (Figure S3).
Sul protects cell apoptosis via mitophagy, which is blunted by autophagy-related 5 gene deficiency
Because Sul exerted cytoprotective activity both in human hepatic L02 cells and primary mouse hepatocytes, the results suggested that mitophagy induction may be the underlying mechanism. To confirm whether the reduction of p62 was due to accelerated protein degradation or transcription inhibition, we detected the p62 mRNA level under PA-induced conditions with or without autophagy inhibitors in L02 cells and hepatocytes, respectively (Figure S4). As the results show, PA dramatically increased p62 mRNA levels in both L02 (Figure S4a) and primary hepatocytes (Figure S4b), and Sul treatment did not reduce the transcription level of p62 in L02 cells but a slight decrease in hepatocytes. There is a tendency for autophagy inhibitors to up-regulate the p62 mRNA level, and there is a light decrease after Sul co-incubation. Our data suggested that Sul may accelerate the degradation of p62 by promoting autophagy rather than inhibiting p62 transcription under PA-induced conditions.
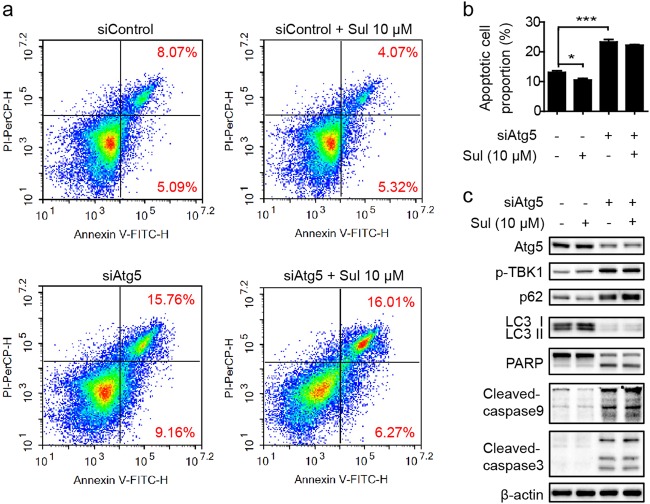
To further validate that the cytoprotective effects of Sul were mainly due to autophagic flux promotion, we used small interfering RNA (siRNA), which targeted and knock down human ATG5 so that the autophagy process would be interrupted. While ATG5 was knocked down, a similar phenomenon with autophagy inhibitor treatment has been observed. Lack of but not total deficiency of ATG5 leads to severe cell apoptosis, and Sul slightly affected the apoptotic process (Fig. 5a, b). In addition, siAtg5 triggered autophagy blockage, and the cytoprotective effect of Sul was barely detectable (Fig. 5c). These data demonstrated that the function of Sul is dependent on the promotion of autophagic flux.
Fig. 5.
Sul protects cell apoptosis via mitophagy would be blunted by autophagy related gene 5 deficiency. a Representative FCM analysis of apoptotic rate after Atg5 knock-down in response to Sul treatment in L02 cells for 24 h. Representative plots from three independent experiments are shown. b Statistical data of the whole apoptotic cell proportion under conditions what mentioned in a. c Western blot analysis of autophagy, mitophagy and apoptosis marker proteins in response to Atg5 knock-down and Sul treatment in L02 cells. Student’s t-test. Datas were shown as means ± SD of three independent replications. Asterisks indicate P-values (*P < 0.05, ***P < 0.001). Sul, sulfuretin; siAtg5, sequences targeted for human ATG5 small interfering RNA (siRNA)
Taken together, these results show that Sul promoted autophagy, specifically mitophagy, to protect cells from apoptosis under PA-induced oxidative stress or autophagy inhibitor-induced autophagic flux blockage. These results suggest that Sul may have potential hepatoprotective applications for diverse mechanisms of hepatotoxicity.
Discussion
It is well known that ROS are mainly produced by mitochondria [1]. ROS maintain balance under physiological conditions, and disruptions in this balance may lead to oxidative stress, resulting in mitochondrial or cell dysfunction and damage to lipids, proteins and DNA [2–4, 6]. Palmitate (PA) triggers oxidative stress and mitochondrial oxidative damage and induces downstream apoptotic events through mitochondrial activation [5, 6]. Here, we investigated the natural product Sul and validated its hepatoprotective effect in the human hepatic cell line L02 and primary hepatocytes. In our study, we found that Sul significantly reduced ROS accumulation and the apoptotic signals induced by PA. These results suggested that the hepatoprotective effect of Sul may partially come from its antioxidant activity by reducing intracellular ROS levels. Previous research has also verified that the protective effect of Sul against t-BHP-induced oxidative damage in human liver-derived HepG2 cells and the cytoprotective effects of Sul against accidental cell death against hydrogen peroxide (H2O2) are due to ROS scavenging [51].
As the mitochondria are the main producing organelles of ROS, we then evaluated the effect of Sul on mitochondrial functions and surprisingly found that Sul could lower MMP. In other words, Sul could likely regulate mitochondrial functions because MMP depolarization is considered as a trigger of mitophagy according to our knowledge [22, 24]. In this case, we subsequently focused on mitochondria and found that Sul promotes the occurrence of mitophagy, activates mitophagy processes to form autophagosomes, significantly promotes the phosphorylation of TBK1, upregulates LC3-II levels and reduces p62 protein levels (Figs. 2–4). Because Sul could not downregulate the p62 mRNA level induced by PA (Figure S4), the p62 mRNA level plus the p62 protein level collectively represents the autophagy flux. The p62 protein level is highly dynamic processed, which includes formation and degradation [52]. When the autophagy flux was blocked, it caused blockage of p62 protein degradation, which leads to p62 accumulation, and the mRNA level may be maintained at a normal level or compensatory elevated [52]. This means that Sul may accelerate the degradation of p62 by promoting autophagy rather than inhibiting p62 transcription under PA-induced conditions.
Regarding the cytoprotective activity of Sul on impaired hepatocytes, we speculated that the decrease in MMP may stimulate the mitophagy, which process will recycle the damaged mitochondria and improve the quality control of healthy mitochondria and that this process will potentially contribute to the protective activity of PA induced in hepatocytes. Taken together, our study and results suggest that Sul protects hepatic cell injury by regulating autophagic flux, especially mitophagy.
Autophagy is essential for survival and many physiological processes. It occurs as a fundamental process in cells to sustain basal functions, including protein and organelle turnover. Disruption or blockade of autophagy may participate in the pathogenesis of many diseases. It has been well demonstrated that autophagy protects against diverse neurodegenerative diseases [19]. Autophagy participates in liver metabolism through multiple pathways and plays diverse roles in the survival and function of hepatic cells [40]. Under oxidative stress conditions, impaired autophagy function would promote oxidant-induced apoptosis of hepatocytes and lead to liver injury [37]. Autophagy can regulate β-oxidation of fatty acids and may reduce lipotoxicity induced by free fatty acids such as PA in hepatic cells [38, 39]. Rapamycin is an inhibitor of mTOR that activates autophagy and is widely used in the treatment of diseases. Many studies have shown that rapamycin produces its effects through the activation of autophagy; rapamycin is used against peritendinous fibrosis [53], to facilitate fracture healing [54], to reduce PA-induced ER stress in adipocytes [55], to protect against OVA-induced asthma in mice [56], to prevent cerebral stroke in rats [57], to protect the liver from ischemia and reperfusion injury [58], and for other purposes.
In the present work, we found that the polyphenol natural product Sul not only scavenges PA-induced ROS but also activates mitophagy in L02 cells and primary hepatocytes through a mechanism of reversing the blockage of autophagic flux and reducing apoptosis events induced by PA. This was further confirmed by the demonstration that apoptosis induced by the autophagy inhibitors CQ and 3-MA was reversed by Sul. When knockdown of ATG5 interrupted the autophagy process, the cytoprotective effect of Sul was almost eliminated (Fig. 5). These results further demonstrated that the cytoprotective effect of Sul was mainly due to autophagic flux promotion. Our study thus identified Sul as attenuating ROS accumulation and regulating autophagic flux to protect cells from apoptosis. The unique hepatoprotective mechanisms, including ROS homeostasis and restoration of autophagic flux, could be a new strategy for hepatic cell protection.
Electronic supplementary material
Acknowledgements
This work was supported by a grant from the Shanghai Commission of Science and Technology (16JC1405000) and K.C. Wong Education Foundation.
Author contributions
YTL and YFL conducted the biological experiments, analyzed the results and wrote the paper; YFX conducted the chemical experiments, analyzed the results and wrote the paper; JYL, FJN, and JL conceived the idea for the project, analyzed the results and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Jia Li, Email: jli@simm.ac.cn.
Fa-jun Nan, Email: fjnan@simm.ac.cn.
Jing-ya Li, Email: jyli@simm.ac.cn.
Electronic supplementary material
The online version of this article (10.1038/s41401-018-0193-5) contains supplementary material, which is available to authorized users.
References
- 1.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rocha M, Esplugues JV, Hernandez-Mijares A, Victor VM. Mitochondrial-targeted antioxidants and oxidative stress: a proteomic prospective study. Curr Pharm Des. 2009;15:3052–62. doi: 10.2174/138161209789058138. [DOI] [PubMed] [Google Scholar]
- 3.Gutteridge JMC, Mitchell J. Redox imbalance in the critically Ill. Brit Med Bull. 1999;55:49–75. doi: 10.1258/0007142991902295. [DOI] [PubMed] [Google Scholar]
- 4.Gutteridge JMC. Lipid-peroxidation and antioxidants as biomarkers of tissue-damage. Clin Chem. 1995;41:1819–28. [PubMed] [Google Scholar]
- 5.Wei Y, Wang D, Topczewski F, Pagliassotti MJ. Saturated fatty acids induce endoplasmic reticulum stress and apoptosis independently of ceramide in liver cells. Am J Physiol Endocrinol Metab. 2006;291:E275–81. doi: 10.1152/ajpendo.00644.2005. [DOI] [PubMed] [Google Scholar]
- 6.Egnatchik RA, Leamy AK, Noguchi Y, Shiota M, Young JD. Palmitate-induced activation of mitochondrial metabolism promotes oxidative stress and apoptosis in H4iiec3 rat hepatocytes. Metabolism. 2014;63:283–95. doi: 10.1016/j.metabol.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernyhough P, Roy Chowdhury SK, Schmidt RE. Mitochondrial stress and the pathogenesis of diabetic neuropathy. Expert Rev Endocrinol Metab. 2010;5:39–49. doi: 10.1586/eem.09.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira IL, Resende R, Ferreiro E, Rego AC, Pereira CF. Multiple defects in energy metabolism in Alzheimer’s disease. Curr Drug Targets. 2010;11:1193–206. doi: 10.2174/1389450111007011193. [DOI] [PubMed] [Google Scholar]
- 9.Kones R. Parkinson’s disease: mitochondrial molecular pathology, inflammation, statins, and therapeutic neuroprotective nutrition. Nutr Clin Pract. 2010;25:371–89. doi: 10.1177/0884533610373932. [DOI] [PubMed] [Google Scholar]
- 10.Rocha M, Apostolova N, Hernandez-Mijares A, Herance R, Victor VM. Oxidative stress and endothelial dysfunction in cardiovascular disease: mitochondria-targeted therapeutics. Curr Med Chem. 2010;17:3827–41. doi: 10.2174/092986710793205444. [DOI] [PubMed] [Google Scholar]
- 11.Rosenstock TR, Duarte AI, Rego AC. Mitochondrial-associated metabolic changes and neurodegeneration in Huntington’s disease—from clinical features to the bench. Curr Drug Targets. 2010;11:1218–36. doi: 10.2174/1389450111007011218. [DOI] [PubMed] [Google Scholar]
- 12.Wende AR, Young ME, Chatham J, Zhang JH, Rajasekaran NS, Darley-Usmar VM. Redox biology and the interface between bioenergetics, autophagy and circadian control of metabolism. Free Radic Bio Med. 2016;100:94–107. doi: 10.1016/j.freeradbiomed.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang JH. Teaching the basics of autophagy and mitophagy to redox biologists-mechanisms and experimental approaches. Redox Biol. 2015;4:242–59. doi: 10.1016/j.redox.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Redmann M, Benavides GA, Wani WY, Berryhill TF, Ouyang X, Johnson MS, et al. Methods for assessing mitochondrial quality control mechanisms and cellular consequences in cell culture. Redox Biol. 2018;17:59–69. doi: 10.1016/j.redox.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J, Guan KL. Regulation of the autophagy initiating kinase Ulk1 by nutrients roles of Mtorc1 and Ampk. Cell Cycle. 2011;10:1337–8. doi: 10.4161/cc.10.9.15291. [DOI] [PubMed] [Google Scholar]
- 16.Redmann M, Darley-Usmar V, Zhang JH. The role of autophagy, mitophagy and lysosomal functions in modulating bioenergetics and survival in the context of redox and proteotoxic damage: implications for neurodegenerative diseases. Aging Dis. 2016;7:150–62. doi: 10.14336/AD.2015.0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee MS. Role of islet beta cell autophagy in the pathogenesis of diabetes. Trends Endocrinol Metab. 2014;25:620–7. doi: 10.1016/j.tem.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 18.He CC, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Yang D, Wang W, Piao S, Zhou J, Saiyin W, et al. Inhibition of autophagy by 3-Ma enhances Il-24-induced apoptosis in human oral squamous cell carcinoma cells. J Exp Clin Cancer Res. 2015;34:97. [DOI] [PMC free article] [PubMed]
- 22.Redmann M, Dodson M, Boyer-Guittaut M, Darley-Usmar V, Zhang JH. Mitophagy mechanisms and role in human diseases. Int J Biochem Cell B. 2014;53:127–33. doi: 10.1016/j.biocel.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 24.Shen QF, Yamano K, Head BP, Kawajiri S, Cheung JTM, Wang CX, et al. Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Mol Biol Cell. 2014;25:145–59. doi: 10.1091/mbc.e13-09-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–46. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. The Pink1-parkin mitochondrial ubiquitylation pathway drives a program of Optn/Ndp52 recruitment and Tbk1 activation to promote mitophagy. Mol Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pickrell AM, Youle RJ. The roles of Pink1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85:257–73. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–25. doi: 10.1016/S1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 29.Twig G, Shirihai OS. The interplay between mitochondrial dynamics and mitophagy. Antioxid Redox Signal. 2011;14:1939–51. doi: 10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scherz-Shouval R, Elazar Z. Ros, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007;17:422–7. doi: 10.1016/j.tcb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Kliosnky D. Guidelines for the use and interpretation of assays for monitoring aAutophagy (3rd Edition) (Vol 12, Pg 1, 2015) Autophagy. 2016;12:443. doi: 10.1080/15548627.2016.1147886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, Chang JT, Guo B, Hansen M, Jia K, Kovacs AL, et al. Guidelines for monitoring. Autophagy Caenorhabditis Elegans Autophagy. 2015;11:9–27. doi: 10.1080/15548627.2014.1003478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang XJ, Chen S, Huang KX, Le WD. Why should autophagic flux be assessed? Acta Pharmacol Sin. 2013;34:595–9. doi: 10.1038/aps.2012.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jangamreddy JR, Panigrahi S, Los MJ. Monitoring of autophagy is complicated—Salinomycin as an example. Biochim Biophys Acta. 2015;1853:604–10. doi: 10.1016/j.bbamcr.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 35.Las G, Serada SB, Wikstrom JD, Twig G, Shirihai OS. Fatty acids suppress autophagic turnover in beta-cells. J Biol Chem. 2011;286:42534–44. doi: 10.1074/jbc.M111.242412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mir SUR, George NM, Zahoor L, Harms R, Guinn Z, Sarvetnick NE. Inhibition of autophagic turnover in beta-cells by fatty acids and glucose leads to apoptotic cell death. J Biol Chem. 2015;290:6071–85. doi: 10.1074/jbc.M114.605345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Singh R, Xiang Y, Czaja MJ. Macroautophagy and chaperone-mediated autophagy are required for hepatocyte resistance to oxidant stress. Hepatology. 2010;52:266–77. doi: 10.1002/hep.23645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sinha RA, You SH, Zhou J, Siddique MM, Bay BH, Zhu X, et al. Thyroid hormone stimulates hepatic lipid catabolism via activation of autophagy. J Clin Invest. 2012;122:2428–38. doi: 10.1172/JCI60580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egnatchik RA, Leamy AK, Jacobson DA, Shiota M, Young JD. Er calcium release promotes mitochondrial dysfunction and hepatic cell lipotoxicity in response to palmitate overload. Mol Metab. 2014;3:544–53. doi: 10.1016/j.molmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madrigal-Matute J, Cuervo AM. Regulation of liver metabolism by autophagy. Gastroenterology. 2016;150:328–39. doi: 10.1053/j.gastro.2015.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee DS, Jeong GS, Li B, Park H, Kim YC. Anti-Inflammatory effects of sulfuretin from Rhus Verniciflua stokes via the induction of heme oxygenase-1 expression in murine macrophages. Int Immunopharmacol. 2010;10:850–8. doi: 10.1016/j.intimp.2010.04.019. [DOI] [PubMed] [Google Scholar]
- 42.Park KY, Jung GO, Lee KT, Choi J, Choi MY, Kim GT, et al. Antimutagenic activity of flavonoids from the heartwood of Rhus Verniciflua. J Ethnopharmacol. 2004;90:73–9. doi: 10.1016/j.jep.2003.09.043. [DOI] [PubMed] [Google Scholar]
- 43.Kim JM, Noh EM, Kwon KB, Kim JS, You YO, Hwang JK, et al. Suppression of Tpa-induced tumor cell invasion by sulfuretin via inhibition of Nf-Kappab-dependent Mmp-9 expression. Oncol Rep. 2013;29:1231–7. doi: 10.3892/or.2012.2218. [DOI] [PubMed] [Google Scholar]
- 44.Lee JC, Lim KT, Jang YS. Identification of Rhus Verniciflua stokes compounds that exhibit free radical scavenging and anti-apoptotic properties. Biochim Biophys Acta. 2002;1570:181–91. doi: 10.1016/S0304-4165(02)00196-4. [DOI] [PubMed] [Google Scholar]
- 45.Jeon WK, Lee JH, Kim HK, Lee AY, Lee SO, Kim YS, et al. Anti-platelet effects of bioactive compounds isolated from the bark of Rhus Verniciflua stokes. J Ethnopharmacol. 2006;106:62–9. doi: 10.1016/j.jep.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 46.Lee YR, Hwang JK, Koh HW, Jang KY, Lee JH, Park JW, et al. Sulfuretin, a major flavonoid isolated from Rhus Verniciflua, ameliorates experimental arthritis in mice. Life Sci. 2012;90:799–807. doi: 10.1016/j.lfs.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 47.Shin SY, Shin MC, Shin JS, Lee KT, Lee YS. Synthesis of aurones and their inhibitory effects on nitric oxide and Pge(2) productions in Lps-induced raw 264.7 cells. Bioorg Med Chem Lett. 2011;21:4520–3. doi: 10.1016/j.bmcl.2011.05.117. [DOI] [PubMed] [Google Scholar]
- 48.Siddalah V, Rao CV, Venkateswarlu S, Krishnaraju AV, Subbaraju GV. Synthesis, stereochemical assignments, and biological activities of homoisoflavonoids. Bioorgan Med Chem. 2006;14:2545–51. doi: 10.1016/j.bmc.2005.11.031. [DOI] [PubMed] [Google Scholar]
- 49.Wang Q, Jiang L, Wang J, Li SF, Yu Y, You J, et al. Abrogation of hepatic Atp-citrate lyase protects against fatty liver and ameliorates hyperglycemia in leptin receptor-deficient mice. Hepatology. 2009;49:1166–75. doi: 10.1002/hep.22774. [DOI] [PubMed] [Google Scholar]
- 50.Lee DS, Kim KS, Ko W, Li B, Jeong GS, Jang JH, et al. The cytoprotective effect of sulfuretin against tert-butyl hydroperoxide-induced hepatotoxicity through Nrf2/Are and Jnk/Erk Mapk-mediated heme oxygenase-1 expression. Int J Mol Sci. 2014;15:8863–77. doi: 10.3390/ijms15058863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee DS, Kim KS, Ko W, Keo S, Jeong GS, Oh H, et al. Cytoprotective effects of sulfuretin from Rhus Verniciflua through regulating of heme oxygenase-1 in human dental pulp cells. Nat Prod Sci. 2013;19:54–60. [Google Scholar]
- 52.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Arozena AA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd Edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng W, Qian Y, Chen S, Ruan H, Fan C. Rapamycin protects against peritendinous fibrosis through activation of autophagy. Front Pharmacol. 2018;9:402. [DOI] [PMC free article] [PubMed]
- 54.Yin ZY, Yin J, Huo YF, Yu J, Sheng LX, Dong YF. Rapamycin facilitates fracture healing through inducing cell autophagy and suppressing cell apoptosis in bone tissues. Eur Rev Med Pharm. 2017;21:4989–98. [PubMed] [Google Scholar]
- 55.Yin Jiajing, Gu Liping, Wang Yufan, Fan Nengguang, Ma Yuhang, Peng Yongde. Rapamycin Improves Palmitate-Induced ER Stress/NFκB Pathways Associated with Stimulating Autophagy in Adipocytes. Mediators of Inflammation. 2015;2015:1–12. doi: 10.1155/2015/272313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang D, Mou Y, Ye L, Jin M, Bai C. Rapamycin protects ova induced asthma in mice through regulating autophagy. Eur Respir J. 2015;46:PA598. [Google Scholar]
- 57.Wu ML, Zhang HD, Kai JJ, Zhu F, Dong JY, Xu ZW, et al. Rapamycin prevents cerebral stroke by modulating apoptosis and autophagy in penumbra in rats. Ann Clin Transl Neur. 2018;5:138–46. doi: 10.1002/acn3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu JJ, Lu TF, Yue S, Shen XD, Gao F, Busuttil RW, et al. Rapamycin protection of livers from ischemia and reperfusion injury is dependent on both autophagy induction and mammalian target of rapamycin complex 2-Akt activation. Transplantation. 2015;99:48–55. doi: 10.1097/TP.0000000000000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.