Abstract
Meningiomas are the most common primary intracranial neoplasm. The current World Health Organization (WHO) classification categorizes meningiomas based on histopathological features, but emerging molecular data demonstrate the importance of genomic and epigenomic factors in the clinical behavior of these tumors. Treatment options for symptomatic meningiomas are limited to surgical resection where possible and adjuvant radiation therapy for tumors with concerning histopathological features or recurrent disease. At present, alternative adjuvant treatment options are not available in part due to limited historical biological analysis and clinical trial investigation on meningiomas. With advances in molecular and genomic techniques in the last decade, we have witnessed a surge of interest in understanding the genomic and epigenomic landscape of meningiomas. The field is now at the stage to adopt this molecular knowledge to refine meningioma classification and introduce molecular algorithms that can guide prediction and therapeutics for this tumor type. Animal models that recapitulate meningiomas faithfully are in critical need to test new therapeutics to facilitate rapid-cycle translation to clinical trials. Here we review the most up-to-date knowledge of molecular alterations that provide insight into meningioma behavior and are ready for application to clinical trial investigation, and highlight the landscape of available preclinical models in meningiomas.
Keywords: biomolecular, cell lines, epigenetic, genetic, sporadic meningioma, xenograft
Introduction
Meningiomas are tumors that originate from arachnoidal cap cells of the leptomeninges. With an incidence rate of 8.03 per 100 000 population, they account for over a third of primary central nervous system (CNS) tumors, making them the most common primary intracranial neoplasm.1 The incidence of meningiomas increases with age, with a sharp increase past 65 years, and with an aging population this tumor is becoming increasingly more prevalent in the neuro-oncology community. There is a need to raise awareness of the gaps in knowledge and needs for this patient population.
Meningiomas can occur throughout the craniospinal axis. Multiple anatomical classifications having been proposed based on the origin of dural attachment, with the most common locations being the convexity, olfactory groove, tuberculum sellae, parasagittal, parafalcine, sphenoid wing, petroclival, posterior fossa, or spinal.2 The 2016 World Health Organization (WHO) classification categorizes meningiomas into 3 grades and 15 histopathological subtypes.3 Classification of tumors as WHO grade I, II, or III is based on several histopathological features, including (i) mitotic activity (number of mitoses per 10 high-powered fields); (ii) brain invasion; and/or (iii) at least 3 other aggressive features such as sheet-like architecture, high nuclear-to-cytoplasm ratio, macronuclei, hypercellularity, and spontaneous necrosis. Specific histologies can render tumors as distinct grades regardless of other criteria. For example, tumors with predominantly clear-cell or chordoid histological morphology are by definition classified as grade II, while rhabdoid and papillary meningiomas are classified as grade III, although some grading controversies remain for these rare subtypes.3,4 Although the extent of mitotic activity observed is an essential grading criterion for this tumor, its detection is confounded by many factors, including sampling bias in a heterogeneous tumor, as well as technical factors and the experience of the pathologist.5 The degree of mitotic activity is an indicator of the proliferative potential of the tumor. Immunohistochemical staining of MIB-1/Ki-67 is often used to estimate the proliferative potential of the tumor. Moreover, the development of immunohistochemical staining for phospho-histone H3 at serine residue 10 has provided pathologists with an additional sensitive and more objective marker for mitotic figure detection; however, use of these markers is not formally included in the WHO grading system.6
The histopathological grade and the extent of resection of the tumor at surgery have been the most reliable clinical variables correlated with tumor recurrence. However, the accuracy of predicting recurrence remains a considerable challenge. The majority of meningiomas (80%) are classified as WHO grade I (“benign”) and have an indolent clinical course, with a 5–10% recurrence rate at 5 years.7 Conversely, there is a near universal recurrence pattern (80–100%) of WHO grade III anaplastic meningiomas with an overall extremely poor prognosis. WHO grade II atypical tumors (5-y recurrence rate of 50%) demonstrate the highest variability in recurrence, making them the most challenging tumor type to make the best evidence-based management decisions.4 It is commonly recognized that there exists a subset of clinically aggressive meningiomas that display early tumor recurrence, with grading often outside of the predicted WHO classification. These considerations highlight the need for more refined prognostication methods that facilitate individualized care.
In addition to tumor grade, the extent of resection (EOR) also correlates strongly with risk of tumor recurrence.8,9 The Simpson grade is the most widely adopted and accepted method to quantify the EOR in meningiomas. Simpson grades I–III are typically noted as gross total resection of tumor with varying degrees of dural and bony resection and resultant 9–29% estimated 10-year symptomatic recurrence rate. Simpson grades IV and V describe subtotal resections with an estimated 10-year symptomatic recurrence rate of 44–100%.2,8,9 Skull base meningiomas clearly illustrate the importance of EOR, as most are WHO grade I tumors but are associated with higher rates of recurrence, as these tumors frequently undergo subtotal resection due to their intimate relationship with critical neurovascular structures.
The current standard of care for most patients with symptomatic meningiomas, or those with considerable mass effect at the time of presentation and those that exhibit progressive growth on serial imaging, includes surgical resection of the lesion and involved dura where possible. Patients with WHO grade I tumors are traditionally managed in follow-up with surveillance imaging. For WHO grade II tumors, postoperative management remains highly controversial regardless of the extent of resection or presence of residual disease. The options are upfront adjuvant radiation therapy to prevent or delay recurrence or close surveillance imaging with delayed radiation at time of recurrence. To date, the verdict is not clear, as there have been no objective or randomized studies completed to demonstrate the superiority of either approach, with 2 international multi-institutional trials under way at present with the intent to address this critical clinical question in the management of WHO grade II meningiomas. For all grade III tumors, adjuvant radiation is strongly considered. With the limited efficacy and lack of any other chemotherapeutic or targeted therapies available, the majority of WHO grade III tumors continue to carry an extremely poor prognosis.10
Despite rigorous classification by histology, morphology, location, and EOR, there is still significant uncertainty in predicting tumor behavior and risk of recurrence for an individual patient, and thereby patients are typically monitored with long-term serial assessments. As biological data accumulate, molecular features will help refine prognostication beyond simple classifications toward more personalized approaches, and exploit targets for therapeutic advantage.
Faithful tumor models are critical to understanding the biological underpinnings of meningioma initiation and progression and in the assessment of the efficacy of potential targeted therapies. Therefore, in this article, we will review the recently established molecular landscape of meningiomas along with established preclinical models that may be used to better understand the biological underpinnings of different clinical behavior in meningiomas. Advances in our understanding of meningioma biology have ignited the development of several clinical trials utilizing novel targeted therapeutics, which will be discussed in detail in a separate review article within this supplement (“Advances in Multidisciplinary Therapy for Meningiomas”).
Genomic Alterations in Meningiomas
The 2016 revisions to the WHO CNS tumor classification adopted a novel integrated histologic-molecular approach to include molecular data into classifying some primary brain tumors, such as gliomas and medulloblastomas.3 However, the classification of meningiomas continues to be based entirely on the histopathological features, and this is in part due to the paucity of data in prior decades on the molecular landscape of meningiomas. It is only recently that several molecular alterations have been implicated in meningiomas.11–13 It is timely to digest and integrate the genomic findings that have the most impact on determining the clinical and biological behavior of meningiomas, with the view to incorporate these into the future pathological classification of these tumors.
Copy Number Alterations
A considerable body of data exists to establish the significance of copy number alterations (CNAs) in meningiomas. Somatic CNAs play a critical role in meningiomagenesis by dysregulating oncogene and tumor suppressor activity.14 Large sections of a chromosome can become duplicated or deleted, resulting in deviations from the normal diploid copy number of an allele. In many different tumor types, specific CNAs are associated with tumor aggressiveness.14–17
The first cytogenetic study on meningiomas, published in 1967, provided a breakthrough in our understanding of the genetic drivers of meningiomas, showing loss of a G-group chromosome (either chromosome 21 or 22) in all tumor samples under investigation and multiple chromosomal aberrations in half of the samples.18 Subsequent studies validated the increased incidence of monosomy 22 in meningiomas.19 These landmark studies led to the critical insight that loss of chromosome 22 is pivotal in meningiomagenesis for a large subset of the tumors. Loss of heterozygosity (LOH) studies further narrowed the region of interest to chromosome 22q, which was lost in 60–70% of sporadic meningiomas.20,21 The incidence of 22q LOH increases with WHO grade, with a 50% prevalence in WHO grade I tumors and 75–85% prevalence in WHO grades II and III tumors.21,22
Deletion of chromosome 1p is the second most common CNA identified and is mainly associated with higher WHO grade.21,23 Other recurrent chromosomal aberrations observed in meningiomas include loss of 4p, 6q, 7p, 9p, 10q, 11p, 14q, and 18q.21,24 These cytogenetic aberrations directed researchers to explore these chromosomal regions for potential meningioma driver genes that are discussed further below.
Chromosomal aberrations increase in complexity with increasing tumor grade in meningiomas.25 In grade I tumors, the only recurring chromosomal change is the loss of 22q; however, higher-grade tumors demonstrate an increased accumulation of CNAs, with grades II and III tumors having a median of 3.0 and 9.5, respectively, chromosomal arm losses.21,24 The losses of 1p, 9p, and 14q are more frequently observed in higher-grade meningiomas that have higher rates of tumor recurrence.21,23,24,26–32 Interestingly, grade I meningiomas with a greater burden of genomic disruption overall, and specifically 1p deletion, have a higher likelihood of progression and recurrence, lending evidence that there are genomic factors not captured by histopathological grade that can inform on the clinical behavior of meningiomas.22,30,33 Additional evidence comes from findings of losses of chromosomes 1, 14, and 22 being more prevalent in recurrent and progressive tumors than de novo high-grade meningiomas.33,34 Although profiling the CNAs in meningiomas provides valuable information regarding propensity for an aggressive clinical course, whether these alterations remain independently prognostic after controlling for more newly discovered molecular alterations discussed in sections to follow remains unknown, and indicates the need for more studies that utilize integrative approaches employing multiple “omics” platforms to compare and weigh the prognostic importance of genomic alterations.32
Gene Mutation Signature
Leveraging of next-generation sequencing techniques has led to advances in establishing the mutational signature of meningiomas. Over the last two decades, a substantial body of evidence has focused on identifying mutations in genes driving meningiomagenesis and aggressive tumor behavior (Fig. 1). Efforts to further characterize the biological and clinical utility of these findings are ongoing, with several under investigation in clinical trials. Broadly the mutational landscape for meningiomas can be dichotomized as NF2 or non-NF2 based changes.
Fig. 1.
Summary of established epigenomic and genomic landscape of sporadic meningiomas. DNA methylation profiling distinguishes 2 distinct tumor subgroups with distinct risk profiles that refine risk of recurrence beyond standard-of-care histopathological grading and are associated with typical known mutations.
The NF2 gene was first implicated in meningiomas after it was found that the inactivation of NF2, a gene encoding the protein neurofibromin 2 (more commonly known as merlin or schwannomin), resulted in the genetic tumor predisposition syndrome of neurofibromatosis type 2.36,37 The hallmark of this genetic disease is the presence of bilateral vestibular schwannomas. However, roughly half of these patients also develop multiple meningiomas. Focal NF2 inactivating mutations are found in 30–50% of sporadic meningiomas and identified in meningiomas of all 3 histopathological grades.11–13,20
The NF2 gene contains 17 exons and is located on chromosome 22q12.2. NF2 produces a 69 kDa protein that functions as a membrane-cytoskeleton scaffolding protein involved in the regulation of multiple pathways, including the mammalian hippo, PI3K/mTORC1/Akt, and receptor-dependent mitogenic signaling pathways.38,39NF2 mutations frequently result in truncation of the protein, due to frameshift, nonsense, or splice-site mutations, producing a nonfunctional protein product.35 Consistent with Knudson’s 2-hit hypothesis, 97% of tumors with focal NF2 mutations have concomitant loss of chromosome 22q as the second hit.11–13,35 The loss of NF2 leads to dysregulation of these downstream pathways, resulting in cellular proliferation. NF2 mutant meningiomas indeed have a higher proliferation index and larger tumor size compared with other genotypic variants, corroborating this proliferative pathway dysregulation.36 Additionally, loss of NF2 gene product leads to overexpression of focal adhesion kinase (FAK), resulting in increased cellular migration and invasion.40,41 This association with overexpression of FAK may render these tumors sensitive to FAK inhibitors, which is currently under investigation in a clinical trial (NCT02523014).
It is clear that NF2 mutations and chromosome 22q loss play a significant role in the development of meningiomas. However, NF2 mutations are found in only half of all meningiomas.11–13,35 The development of next-generation sequencing technologies has helped usher in a renaissance in the field of meningioma biology, with the emergence of several additional genes that were recurrently mutated, with a pattern that is mostly mutually exclusive of NF2 mutation.
Approximately a fifth of meningiomas harbor mutations of TRAF7 (tumor necrosis factor [TNF] receptor associated factor 7).35TRAF7, located on chromosome 16p13, has been linked to modulation of the nuclear factor-kappaB transcription factor, activation of cellular stress pathways, ubiquitination of multiple cellular targets, and induction of apoptosis.37 Mutations in TRAF7 are mutually exclusive with NF2 mutations but often have a concomitant mutation in v-akt murine thymoma viral oncogene homolog 1 (AKT1) or Krüppel-like factor 4 (KLF4).12,13AKT1 (also known as protein kinase B) is a central cog in the oncogenic pathway controlled by phosphatidylinositol-3 kinase (PI3K), where it appears to suppress apoptosis.35AKT1 mutations occur in 10% of sporadic meningiomas, and all AKT1 mutations identified in meningioma (c.49G>A, AKT1E17K) lead to constitutive AKT1 activation independent of PI3K signaling and promote tumor growth.35,38 Similarly, most mutations in KLF4, a crucial regulator of cellular proliferation, have been activating (c.1225A>C, KLF4K409Q),35 resulting in tumor cell growth.39 Although AKT1 and KLF4 mutations can both concomitantly occur with TRAF7, they are mutually exclusive of each other.35,40
There are many significant clinical correlations in the mutational profile of meningiomas. For example, TRAF7, AKT1, and KLF4 mutations are almost exclusively observed in WHO grade I meningiomas.11–13,35 Moreover, secretory meningiomas invariably harbor concomitant TRAF7 and KLF4K409Q mutations,41 and similarly, meningiomas with AKT1E17K mutation tend to have meningothelial and transitional histopathological morphology.35,42 A strong correlation with tumor location has also been demonstrated, with TRAF7, AKT1, and KLF4 mutations more commonly located in anterior and middle fossa skull base meningiomas.11–13,35
The hedgehog (Hh) signaling pathway has also been implicated in the development of a subset of meningiomas, specifically via mutations in Smoothened (SMO; frizzled family receptors) and suppressor of fused homolog (SUFU) genes. SMO is a G-protein coupled-like receptor and part of the Hh signaling pathway. Mutations in this gene are found in 3–6% of meningiomas and are mutually exclusive with NF2, and other non-NF2 mutations.35,40SMO-mutant meningiomas are exclusively observed in WHO grade I meningiomas and occur in the anterior midline skull base.35,42 Most notably, the SMO mutation is one of the few targetable genetic alterations identified in meningiomas to date. A clinical trial with vismodegib, a competitive antagonist of the SMO receptor, is currently ongoing (NCT02523014). SUFU is also a member of the Hh family and mutated in less than 1% of sporadic meningiomas. However, familial cases of meningiomas are known to harbor germline mutations in SUFU.12 Details regarding clinical trials can be found in “Advances in Multidisciplinary Therapy for Meningiomas” within this supplement.
Another potential targetable mutation is phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA). This mutation is found in 4–7% of meningiomas.35,40,43PIK3CA is a known oncogene located on chromosome 3q26.3 that is mutated in approximately 15% of all human cancers.43PIK3CAH1047R and PIK3CAE545K, which are two of many mutations observed in PIK3CA, constitutively phosphorylate and activate AKT1.44PIK3CA mutations are mutually exclusive of NF2, SMO, and AKT1 and in very few cases co-occur with TRAF7 or KLF4 mutations.35,40
RNA polymerase II (POLR2A), the protein that mediates transcription of all protein-coding genes in eukaryotes, has been identified in approximately 6% of grade I meningiomas. The recurrent somatic hotspot mutations include G403K as well as in-frame deletion of residues Leu438 and His439.12 The functional role of POLR2A mutations in tumor development is not well understood. Similar to other non-NF2 meningiomas, POLR2A-mutant meningiomas are exclusively WHO grade I tumors that are most likely to be found in anterior skull base (tuberculum sellae) tumors and tend to harbor a meningothelial histopathological morphology.11–13,35
In addition to gene mutations that affect single factor dysregulation, disruptions of the machinery needed for gene processing are emerging as processes that are central to tumor progression. Converging lines of evidence suggest an essential role for the Switch/sucrose nonfermentable (SWI/SNF) chromatin-remodeling complex in meningioma formation and aggressiveness. Several families with multiple meningiomas and schwannomas harbor germline mutations in the SWI/SNF core complex unit SMARCB1.45,46 However, SMARCB1 mutations have also been observed in a small subset of sporadic grade I and grade II meningiomas, concurrently with NF2 mutations.11,47 Loss-of-function mutations in another core subunit of the SWI/SNF complex, SMARCE1, are a molecular signature of the clear-cell subtype of meningiomas.48–51
Overall, mutations in SWI/SNF complex genes are observed at higher frequency in anaplastic meningiomas compared with grades I and II meningiomas, and hence are associated with poor prognoses.52 Furthermore, high-grade meningiomas demonstrate upregulation of the polycomb repressive complex 2 (PRC2) and its catalytic domain, which function in balance with the SWI/SNF complex.47 Taken together, loss-of-function mutations in core members of the SWI/SNF complex and upregulation of its antagonist increasingly reveal the critical role of chromatin regulation in meningiomas.
Inactivating germline and somatic mutations in the tumor suppressor breast cancer (BRCA)–associated protein 1 (BAP1) gene have been identified in a rare subset of aggressive meningiomas with rhabdoid morphology.53–55 Patients with germline BAP1 mutations are predisposed to multiple tumors, including melanoma, mesothelioma, and other cancers; identification of a germline BAP1 mutation, therefore, calls for increased vigilance in cancer surveillance in individuals who harbor this mutation. Mechanisms of BAP1 inactivation are diverse, including focal deletion, nonsense mutation, frameshift mutation, and intrachromosomal fusion. Additional loss of chromosome 3p, which harbors BAP1, results in full inactivation of this tumor suppressor. Importantly, BAP1-deficient mesothelioma tumor cells have been demonstrated to be sensitive to inhibitors of enhancer of zeste homolog 2, presenting an opportunity for potential pharmacologic inhibition in BAP1-mutant meningiomas.56
There is increasing awareness of the importance of studying the noncoding regions that affect gene expression. Similarly, in meningiomas, there has been recent recognition of the role of mutations in the telomerase reverse transcriptase (TERT) promoter. TERT maintains the length of the telomere ends by addition of a highly conserved sequence of TTAGGG at the end of each chromosome. Somatic cells normally repress TERT, leading to senescence. Mutations in the promoter region increase E-twenty-six transcription factor binding and upregulate TERT expression.57 The constitutive expression of TERT results in the immortalization of the cancer cell. TERT promoter mutations, occurring specifically in the hotspot regions C228T and C250T, are detected in 6.5–11% of meningiomas.58,59 In fact, TERT promoter mutations may be among the most common mutations in all human cancers, suggesting that presence of TERT promoter mutation is a potential biomarker for meningiomas with a higher likelihood of clinically aggressive behavior that exhibit higher rates of recurrence and shorter time to progression.57–59 TERT promoter mutations have been observed in both NF2-mutated and NF2-wildtype tumors; however, correlations with other genetic alterations have not been clearly established.58,59 Certainly, the presence of TERT promoter mutations should trigger clinicians to monitor the patient closely for transformation to a more aggressive tumor and potentially consider adjuvant intervention early in the course of the disease.
The identification of novel actionable molecular alterations that are drivers of tumor growth gives rise to the opportunity for developing targeted therapies toward genetic subsets of meningiomas. In keeping with this, several clinical trials—such as a phase II multicenter trial (NCT02523014) studying the effects of SMO receptor and FAK inhibitors on progression-free survival in meningiomas with SMO or NF2 mutations, respectively—are currently under way and will be discussed in more detail in our companion review article within this supplement, “Advances in Multidisciplinary Therapy for Meningiomas.”
The above highlights a key critical observation that while a high percentage of meningiomas have NF2 mutations, non-NF2 mutations identified each occur with a lower frequency than NF2 mutation. Interestingly, mutational signatures with driver mutations described above account for the vast majority (>90%) of meningiomas. While targeting each of these potentially druggable individual mutations offers invaluable treatment potential, looking to aberrations outside of the mutational landscape of meningiomas is essential, as few of the mutational alterations have been correlated with aggressive tumor behavior. Epigenetic alterations have gained considerable attention in the ability to classify and diagnose brain tumors with precision beyond standard-of-care clinical practice,60 and therefore epigenetic changes may also be the key to the next generation of diagnostics and therapeutics for meningiomas.
Epigenomic Alterations in Meningioma
DNA Methylation
DNA methylation is one of the best studied epigenetic regulators of gene transcription and plays a significant role in cancer biology. Methylation of DNA is catalyzed by the DNA methyltransferase (DNMT) family of enzymes, with DNMT3A and DNMT3B catalyzing de novo DNA methylation and DNMT1 mediating both de novo and maintenance methylation of DNA.61 Global DNA hypomethylation and focal DNA hypermethylation are associated with tumorigenesis.62 The most notable genes reported with differential DNA methylation in meningiomas are tissue inhibitor of metalloproteinase 3 (TIMP3), cyclin-dependent kinase inhibitor 2A (CDKN2A), and tumor protein 73 (TP73), which are hypermethylated in at least 10% of cases.63TIMP3 hypermethylation results in transcriptional downregulation and inhibits its tumor suppressor properties.64TIMP3 is frequently hypermethylated in higher-grade meningiomas, with a frequency of 40–60% in grade III tumors and associated with a shorter time to recurrence.63,65 In addition, TIMP3 is found on chromosome 22q12, and almost all cases with TIMP3 hypermethylation had a concurrent allelic loss of 22q.65 Seventy to eighty percent of high-grade tumors have TP73 promoter methylation, and this is not commonly seen in grade I tumors,66 suggesting the potential utility of TP73 promoter methylation as a biomarker for meningiomas of higher grade.
There has been an emerging recognition in the value of leveraging global methylation signatures of brain tumors beyond single gene methylation analysis.60 In this past year, 2 studies have provided evidence for the importance of global methylation profiles in molecular subclassification of meningiomas.67,68 Olar et al in 2017 first demonstrated that unsupervised clustering of DNA methylation data classified meningiomas into 2 distinct subgroups associated with recurrence-free survival. After adjusting for clinical factors, such as WHO grade and Simpson grade, a statistically significant association between DNA methylation subclasses and tumor recurrence was maintained.67 Similarly, Sahm et al in 2017 identified 2 major groups and 6 subgroups of meningiomas based on unsupervised clustering of DNA methylation data, with significantly different genomic makeup and clinical behaviors. Interestingly, most non-NF2 meningiomas clustered together into a single benign subgroup, further supporting the benign nature of these tumors and the difference from NF2-mutated tumors.68 These initial efforts suggest that epigenetic signatures may have strong clinical associations with tumor recurrence, to a more significant extent than can be correlated with mutational genetic analysis (Fig. 1), and that these may be applied clinically for individual patients. An additional manifestation of the importance of epigenetic changes in meningioma clinical behavior was recently shown, describing an increased risk of recurrence in tumors that show a loss of histone H3K27 trimethylation.69
Given the distinct clinical behavior of meningiomas based on global methylation signature, a validated methylome-based predictor for tumor recurrence would leverage knowledge gained from methylation analysis and translate it into clinical application to facilitate decision making regarding need for adjuvant therapy beyond standard of care. Additionally, conceivably, use of epigenetic modifiers to convert a poor prognosis meningioma to a more favorable methylation profile would be transformative in management options available for meningiomas.
Micro-RNA
Over the past decade, interest in the role of micro-RNA (miRNA) in carcinogenesis has been rapidly growing. MiRNAs are a subset of small (~22 nucleotides) noncoding RNAs that posttranscriptionally regulate the expression of target mRNA. The surge of miRNA research has been lagging in meningiomas, with only a few studies exploring the role of this epigenetic change in meningiomas. MiRNA-200a, which targets B-catenin, is downregulated approximately 25-fold in meningiomas and leads to abnormal WNT/B-catenin pathway signaling.70 MiRNA expression patterns can differentiate between tumors of different grades and those with different mutations (NF2 and non-NF2). They are also correlated with tumor growth.71 For example, miRNA-145 expression is markedly decreased in higher-grade tumors, and overexpression in cell lines has shown reduced proliferation and tumor growth.72 In addition to this, tumors that overexpress miRNA-109a and those that underexpress miRNA-29c-3p and miR-219-5p have been shown to have higher recurrence rates in meningiomas.73 Similar to miRNAs, long-noncoding RNAs (lncRNAs) are being explored as potential factors that can elucidate biological behavior of meningiomas. The ultimate clinical application of miRNAs and lncRNAs in understanding clinical behavior of meningiomas and possible exploitation as therapeutic prospects has yet to be established.
Transcriptomic Signatures in Meningioma
Given that signatures of epigenetic alterations, such as DNA methylation, have defined subgroups of meningiomas with important clinical implications, it is logical to investigate whether these signatures transcend to gene expression profiling to also recapitulate these important findings. Microarray gene expression analysis has identified potential biomarkers to distinguish benign meningiomas from those with higher risk of progression or recurrence. A 37-gene signature was identified to correlate well with overall survival,74 and recently an 18-gene signature was found to correlate with progression-free survival after validation in an independent cohort.75 These genes were found to be involved in pathways involving normal embryonic development, cell proliferation/invasion, and angiogenesis, among others. In addition to this, increased transglutaminase 2 (TGM2) expression has been described in higher-grade meningioma76 and the deleted in colorectal cancer (DCC) gene, which encodes a transmembrane receptor for netrin-1 and has also been suggested to be a marker for risk of meningioma progression.77 Most gene expression studies have been limited by their small sample size (10–50 tumors), with minimal overlap in the candidate genes identified between studies,64,74,76–80 and therefore further studies are needed to validate potential gene expression biomarkers.
Proteomic Signatures in Meningiomas
Interest in broad protein profile screening in cancers has garnered significant momentum in the post-genome era. Proteomic-based approaches have tremendous clinical impact, as results may be validated via immunohistochemistry, allowing immediate facilitation for clinical use. Similar to transcriptomic analyses, proteomic research remains limited in meningiomas, with only 2 previous proteomic studies having established differential proteomic profiles between meningioma grades. Okamoto et al performed a comparative analysis of different grade meningiomas analyzing pure populations of tumor cells via gel electrophoresis combined with mass spectrometry and identified 15 proteins that were differentially expressed between benign and atypical tumors and 9 proteins that were differentially expressed between atypical and malignant tumors. Their analysis did not, however, consider a normal control, which would ideally be arachnoid cap cells. Furthermore, only one of the identified proteins was probed and validated by immunohistochemistry.81 Using more powerful high-throughput approaches (iTRAQ-based proteomics), Sharma et al identified over 2000 differentially expressed proteins in meningiomas associated with diverse signaling pathways such as integrin, Wnt, Ras, epidermal growth factor receptor, and Gardner-Rasheed feline sarcoma (FGR).82 Other proteomic studies have focused on grade I meningiomas and their differential expression to arachnoid tissue adjacent to tumor, identifying only 17 and 26 proteins to be downregulated and upregulated, respectively.83 Saydam et al performed proteomic analyses from a benign meningioma cell line and primary arachnoidal cells focusing on members of the minichromosome maintenance (MCM) family.84 Recognizing the importance of understanding the implication of genomic alterations in how it alters protein function, the field of proteomics has the potential to provide the next advance in knowledge of meningioma biology and translation to diagnostic, predictive, and therapeutic directions.
Molecular Alterations in Distinct Meningioma Populations
Pediatric Meningiomas
Meningiomas are exceedingly rare among children and adolescents (0–21 y), accounting for 0.4–4.1% of all pediatric brain tumors.85 One of the most intriguing differences from adult tumors is the frequent occurrence of aggressive histological subtypes.86–88 Rhabdoid or papillary meningiomas are observed in up to 13% of patients,86 and anaplastic tumors are diagnosed in about 6% of patients.87 Additionally, clear-cell meningiomas are found at a higher incidence in children compared with adults, perhaps as a consequence of germline SMARCE1 mutations, discussed in the above sections.86 Despite the greater proportion of higher-grade tumors, the long-term results for pediatric meningiomas are favorable, which raises questions regarding the relevance of aggressive histopathological features in meningiomas of this age group.89 An exploratory study using targeted Sanger sequencing did not identify the typical mutations found in adult meningiomas, such as mutations in SMO, AKT, KLF4, TRAF7 (exon 17), and TERT promoter.90 Of the 40 cases studied in this series, NF2 deletions were observed in 30 patients (75%). Future studies focusing on meningiomas should attempt to discover novel non-NF2 alterations, which may help explain both the predominance of specific histological subtypes as well as the lack of common non-NF2-associated mutations.
Radiation-Induced Meningiomas
Although cancer treatments have been advancing such that children are surviving their primary cancer in greater proportions, long-term complications of craniospinal radiation in the form of additional neoplasms in adult life are also increasing in proportion. Radiation-induced meningiomas (RIMs) are the most common brain neoplasm secondary to radiation and typically occur 10–30 years after radiotherapy.91,92 Children who have received only 1–2 Gy of radiation are at a 9.5-fold increased risk of developing a meningioma in their lifetime.93 These tumors exhibit concerning histopathological features, such as hypercellularity, pleomorphic nuclei, and increased mitotic index,91,94 and in line with this, display aggressive clinical phenotype with increased tumor recurrence rates.91,93,94 The mutational signature of RIMs is distinct from their sporadic counterparts.91,95 RIMs invariably have a more complex cytogenetic architecture, with higher rates of recurrent losses of chromosomes 1p and 22q in comparison to sporadic meningiomas.91 It is possible that the radiation therapy triggers double-stranded DNA breaks and subsequent error-prone repair, resulting in the increased CNAs observed in these tumors. Interestingly, the rates of NF2 mutations are significantly lower in RIMs in comparison to sporadic meningiomas (6% vs 30–50%).91,95,96 However, integrated multiplatform genomic interrogation of RIMs has uncovered promiscuous NF2 intronic genomic rearrangements in 40% of cases, exhibiting fusions with intra- and interchromosomal regions.95 Moreover, druggable targets found in sporadic meningiomas, such as AKT1 and SMO, were not observed in RIMs,95 highlighting that the biology of these tumors is distinct from sporadic counterparts. Additional investigations are needed to understand the risks for RIM formation as well as therapies that can be implemented early before tumor formation or progression.
Faithful Preclinical Models of Meningiomas
Tumor models are critical to facilitate investigation of mechanisms behind tumor initiation and progression as well as for testing and validation of new therapeutic strategies for clinical translation. Models are relevant if they recapitulate the disease as seen in humans from both a histological and a molecular perspective. To date, limited established preclinical models, both xenograft mouse models and genetically engineered models, have been developed for meningiomas and may be used to help translate discoveries for clinical utility.
Xenograft Models
The first reports of successful heterotopic engraftment of meningioma cells into athymic nude mice were in the 1970s.97,98 Since then, orthotopic xenograft models using either established cell lines, such as IOMM-Lee, or patient-derived xenograft (PDX) cells, have been used to fill large gaps in our understanding of meningioma biology, as well as to test drugs aimed at improving clinical outcomes.
Since detailed molecular analyses of patient meningioma specimens have only recently been completed, it is of little surprise that the genomics of PDX models have yet to be rigorously characterized to determine if they faithfully reproduce the features of the original tumors. Rath et al compared their meningioma PDX model with the parental grade II tumor and found that their PDX model mostly retained the original tumor’s chromosome profile, including 9p21.3 deletion and gains in chromosomes 3, 7, and 9, as well as intact NF2. Furthermore, the histologic and molecular features of the original tumor and in vitro passages were also successfully recapitulated in the subcutaneous flank xenografts.99
Established cell lines have been used to generate orthotopic xenograft models in the absence of patient-derived cells. The catalogue of available established cell lines in meningiomas, however, is limited. Perhaps the best characterized is the Ben-Men1 line, which is derived from a WHO grade I meningothelial meningioma with an NF2 mutation. IOMM-Lee and CH157-MN are both derived from higher-grade meningiomas, and IOMM-Lee was derived from a tumor that did not contain LOH of chromosome 22. Next-generation sequencing has demonstrated that cell lines retain their parental molecular features. For example, CH157 and Ben-Men1 harbor NF2 deletions like their parental tumors,100,101 whereas IOMM-Lee cells have intact NF2.102,103 CH157-MN and IOMM-Lee cells, both of which are harvested from aggressive primary tumors, also harbor TERT promoter mutations and harbor a copy number profile that is similar to malignant meningiomas. In addition to this, HBL-52 cell lines harbor the canonical TRAF7 mutation, potentially rendering them useful to studies targeting the TRAF7 mutation. However, cell lines carrying the other known non-NF2 mutations (SMO, AKT1, KLF4) have not been identified, and therefore, in vitro models that recapitulate these oncogenic alterations are in critical need.
Detailed genomic and epigenomic profiling of meningioma PDX and their matching original patient-derived tumors are critical next steps in advancing our understanding of meningioma biology and has a high likelihood of identifying novel mechanisms of aggressive tumor behavior that may be vulnerable to innovative treatment strategies.
Genetically Engineered Mouse Models
Although xenograft models have clear advantages, implantation of cells disrupts the tumor microenvironment, which may pose difficulties when evaluating treatment resistance. Genetically engineered models can accurately mimic the histology and clinical behavior of human tumors without disrupting the microenvironment and are gaining traction in meningiomas. Direct intrathecal injection of recombinant Cre adenovirus into NF2loxP/loxP mice has resulted in a model of mosaic NF2-driven benign meningiomas.104 Adenovirus Cre-mediated Cdkn2ab deletions in addition to the above NF2 biallelic inactivation have resulted in mice with a greater proportion of aggressive tumors with shorter overall survival and higher-grade tumors by histology.105 Although aggressive genetically engineered meningioma models can be developed, the alterations that are both necessary and sufficient to generate higher-grade, aggressive meningiomas remain unknown, and therefore the phenotype of the models developed can be variable. In addition to this, the rate of tumor induction is low in the established genetically engineered models, and a substantial period of time is needed until tumor burden is recognized. Lastly, due to alterations occurring in precursor cells, genetically engineered models do not recapitulate the phenotype of adult-onset meningiomas.
Preclinical Models: Junction of Genomics and Therapeutics
One of the earliest examples of using meningioma PDX to evaluate novel therapies in meningioma PDX was the use of the antiprogesterone RU-38486 against a primary human meningioma implanted into the subcutaneous flank of nu/nu Balb C mice that demonstrated inhibition of tumor growth in all mice.106 Similarly, because meningiomas often express estrogen and progesterone receptors, and high fatty acid synthase (FAS) had been associated with expression of such hormone-driven tumor cells, IOMM-Lee flank PDXs were treated with the FAS inhibitor cerulenin, which resulted in a 30% reduction in tumor mass.107 Recently it has been shown that merlin-deficient cells seem to be primarily dependent on the fatty acid malonyl-CoA for cellular metabolism, rendering grade I primary meningioma cells with NF2 deletion highly sensitive to the FAS inhibitor GSK2194069.108
Only a few preclinical studies using meningioma PDX models have generated data that can be linked to clinical trial concepts. For example, although several studies, using primary and IOMM-Lee meningioma flank xenografts, have suggested that hydroxyurea and calcium channel blockers may bear therapeutic potential,109,110 the combination of hydroxyurea and verapamil was ineffective in a phase I/II trial of treatment-resistant meningiomas (NCT00706810).111 Similarly, the cyclooxygenase-2 inhibitor celecoxib showed early promise against several subcutaneous flank PDX models, including IOMM-Lee, CH157-MN, and primary grade I meningioma.112 However, subsequent testing of intracranial PDX, developed from 4 separate primary meningiomas, showed no antitumor activity,113 and thus further clinical studies were not pursued. Given the strong association of epigenetic alterations with clinically relevant subtypes of meningiomas, the histone deacetylase inhibitor AR-42 had been tested in an intracranial Ben-Men1 PDX and was shown to inhibit pAkt, several cyclin-dependent kinases, and in vivo growth.102 This work has led to an ongoing proof-of-concept phase 0 clinical trial aimed at studying concentration, pAkt, and p16INKA after 3 weeks of AR-42 intake in patients with meningiomas (NCT02282917). Use of the mammalian target of rapamycin (mTOR) inhibitors sirolimus and everolimus has demonstrated reduced in vivo growth of IOMM-Lee cells implanted subcutaneously or intracranially.114 These findings contributed to the initiation of ongoing clinical trials aimed at evaluating the mTOR complex 1/2 inhibitor AZD2014 against recurrent sporadic grades II–III meningiomas (NCT03071874) and NF2-associated meningiomas (NCT02831257). Lastly, irinotecan, a topoisomerase inhibitor, was successfully used to arrest the subcutaneous growth of IOMM-Lee PDX.115 Four clinical trials have been completed but have not yet reported on topoisomerase inhibitor efficacy in refractory solid tumors, which included meningioma.
Despite disappointing results so far, some newer PDX-based studies offer hope for a new series of clinical trials, including those targeted against key molecular alterations in meningiomas. A screen of 11 chemotherapeutic agents commonly used against a variety of cancers, including mainstays such as temozolomide, platinum-based drugs, and methotrexate, has identified gemcitabine as being particularly effective against grade II M-16-N and grade III HKBMM meningioma cells in vitro and in vivo.116 A grade III anaplastic meningioma PDX model has also been shown to be sensitive to an oncolytic herpesvirus.117 Protein phosphatase 2A (PP2A) inhibitor LB-100 doubled the survival benefit of radiotherapy in mice with intracranial xenografts of human IOMM-Lee immortalized meningioma cells.118 As merlin inhibits p21-activated kinase 1 (Pak1), Pak inhibitors have also been shown to be able to suppress the growth of NF2-deficient meningioma KT21 and Ben-Men PDXs.119 Immune-based therapies are more difficult to test in PDX models, which are mostly based on immunocompromised mice. However, immunocompetent mice with humanized immune systems could be employed to study meningioma immunotherapies, as has been done in gliomas.120
Conclusions
Meningiomas are the most common primary brain tumor. However, they have not benefited from extensive research and investigative analysis. There have been outstanding recent large-scale genomic studies in meningiomas that have helped improve our understanding of the biology of meningiomas in addition to raising awareness of the need for further research on this tumor type. Translation of the emerging molecular, in particular genomic/epigenomic, knowledge into clinical management remains in its infancy and provides a tremendous opportunity to leverage and explore improved diagnostics of molecular pathology, predictive algorithms, and therapeutic strategies for meningiomas. Integrated histomolecular pathology reports that outline histopathological diagnosis and associated clinically important genetic and epigenetic changes within tumors will facilitate communication of crucial actionable findings between pathologists and clinicians and are a key step for the advancement of care of patients with meningiomas (Fig. 2). A comprehensive integrated multiplatform genomic and epigenomic characterization of clinically aggressive meningiomas will facilitate comprehensive and personalized care ranging from patient counseling to ultimately advancing treatment. Most of the available literature on meningioma genomics is centered around WHO grade I meningiomas, and those studies focusing on anaplastic or malignant meningioma have small cohort sizes. Harnessing collaborative science, the interdisciplinary team of clinicians and scientists within the International Consortium on Meningiomas (ICOM) will allow building of the critical large cohort sizes necessary for analysis as well as the merging of expertise, knowledge, and efforts to fill the gaps in knowledge needed to improve the management of meningiomas.
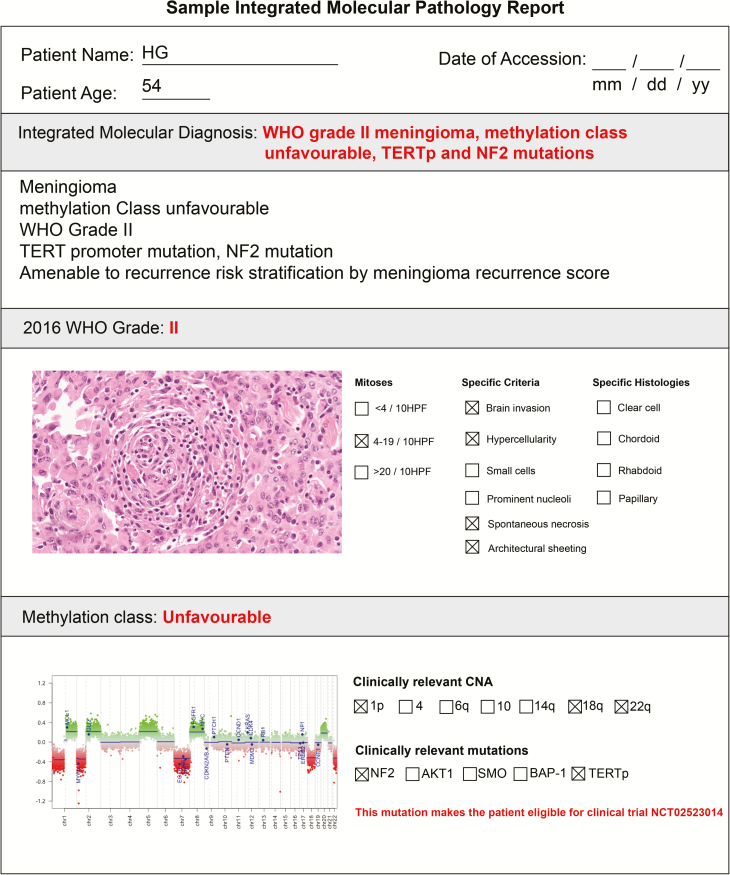
Fig. 2.
Sample integrated molecular-histopathological report. These reports can be used to facilitate communication of impactful histological (WHO grade, number of mitoses, specific histologies, and criteria) and key molecular findings (DNA methylation class, clinically relevant CNAs, and mutations).
Recommendations
To advance our understanding of the molecular biology of meningiomas, ICOM recommends:
• Ongoing collaboration and advocacy with the World Health Organization to integrate molecular alterations with the histopathological diagnosis of meningiomas (Fig. 2)
• Stronger emphasis on studying the epigenomic landscape of meningiomas to understand better critical alterations driving clinically aggressive behavior
• Incorporating genomic/epigenomic and key clinical factors into tools that may be used to establish predictors of tumor behavior and refine the precision in prognostication of meningiomas beyond general classifications
• Resource allocation and focused funding of interdisciplinary collaborative efforts focusing on uncovering the molecular alterations of higher-grade and aggressive meningiomas
• Building faithful and reproducible strong preclinical models that recapitulate the human disease from a histopathological, molecular, and clinical aspect, allowing for rapid translation of discoveries to the clinic
Funding
The collaborative effort of the International Consortium on Meningiomas is supported by The Brain Tumour Charity Quest for Cures: Collaborative Team Award and the Canadian Institute of Health Research.
Acknowledgments
We would like to acknowledge João-Paulo Almeida and Shirin Karimi for their assistance with the endoscopic and histopathological images found on the front cover of the journal.
Contributor Information
International Consortium on Meningiomas:
Kenneth Aldape, Karolyn Au, Jill Barnhartz-Sloan, Wenya Linda Bi, Priscilla K Brastianos, Nicholas Butowski, Carlos Carlotti, Michael D Cusimano, Francesco DiMeco, Katharine Drummond, Ian F Dunn, Evanthia Galanis, Caterina Giannini, Roland Goldbrunner, Brent Griffith, Rintaro Hashizume, C Oliver Hanemann, Christel Herold-Mende, Craig Horbinski, Raymond Y Huang, David James, Michael D Jenkinson, Christine Jungk, Timothy J Kaufman, Boris Krischek, Daniel Lachance, Christian Lafougère, Ian Lee, Jeff C Liu, Yasin Mamatjan, Alireza Mansouri, Christian Mawrin, Michael McDermott, David Munoz, Farshad Nassiri, Houtan Noushmehr, Ho-Keung Ng, Arie Perry, Farhad Pirouzmand, Laila M Poisson, Bianca Pollo, David Raleigh, Felix Sahm, Andrea Saladino, Thomas Santarius, Christian Schichor, David Schultz, Nils O Schmidt, Warren Selman, Andrew Sloan, Julian Spears, James Snyder, Suganth Suppiah, Ghazaleh Tabatabai, Marcos Tatagiba, Daniela Tirapelli, Joerg C Tonn, Derek Tsang, Michael A Vogelbaum, Andreas von Deimling, Patrick Y Wen, Tobias Walbert, Manfred Westphal, Adriana M Workewych, and Gelareh Zadeh
Conflict of interest statement. The authors have no personal, financial, or institutional interest in any of the drugs, materials, or devices described in this article.
Authorship statement. The generation of all manuscripts has been supported by the membership of the consortium, which at the time of supplement generation includes: Kenneth Aldape, Karolyn Au, Jill Barnhartz-Sloan, Wenya Linda Bi, Priscilla K. Brastianos, Nicholas Butowski, Carlos Carlotti, Michael D. Cusimano, Francesco DiMeco, Katharine Drummond, Ian F. Dunn, Evanthia Galanis, Caterina Giannini, Roland Goldbrunner, Brent Griffith, Rintaro Hashizume, C. Oliver Hanemann, Christel Herold-Mende, Craig Horbinski, Raymond Y. Huang, David James, Michael D. Jenkinson, Christine Jungk, Timothy J. Kaufman, Boris Krischek, Daniel Lachance, Christian Lafougère, Ian Lee, Jeff C. Liu, Yasin Mamatjan, Alireza Mansouri, Christian Mawrin, Michael McDermott, David Munoz, Farshad Nassiri, Houtan Noushmehr, Ho-Keung Ng, Arie Perry, Farhad Pirouzmand, Laila M Poisson, Bianca Pollo, David Raleigh, Felix Sahm, Andrea Saladino, Thomas Santarius, Christian Schichor, David Schultz, Nils O. Schmidt, Warren Selman, Andrew Sloan, Julian Spears, James Snyder, Suganth Suppiah, Ghazaleh Tabatabai, Marcos Tatagiba, Daniela Tirapelli, Joerg C. Tonn, Derek Tsang, Michael A. Vogelbaum, Andreas von Deimling, Patrick Y. Wen, Tobias Walbert, Manfred Westphal, Adriana M. Workewych, Gelareh Zadeh.
Sponsorship statement. This supplement was supported by an unrestricted grant from the MacFeeters Hamilton Neuro-Oncology Program at the Princess Margaret Cancer Center and by the Dr. Mary Hunter Brain Tumor Research Funds from the Toronto General & Western Hospital Foundation.
References
- 1. Ostrom QT, Gittleman H, Liao P, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro-Oncol. 2017;19(suppl_5):v1–v88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gallagher MJ, Jenkinson MD, Brodbelt AR, Mills SJ, Chavredakis E. WHO grade 1 meningioma recurrence: are location and Simpson grade still relevant?Clin Neurol Neurosurg. 2016;141:117–121. [DOI] [PubMed] [Google Scholar]
- 3. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 4. Vaubel RA, Chen SG, Raleigh DR, et al. Meningiomas with rhabdoid features lacking other histologic features of malignancy: a study of 44 cases and review of the literature. J Neuropathol Exp Neurol. 2016;75(1):44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jenkinson MD, Santarius T, Zadeh G, Aldape KD. Atypical meningioma—is it time to standardize surgical sampling techniques?Neuro Oncol. 2017;19(3):453–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Olar A, Wani KM, Sulman EP, et al. Mitotic index is an independent predictor of recurrence-free survival in meningioma. Brain Pathol. 2015;25(3):266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Adeberg S, Hartmann C, Welzel T, et al. Long-term outcome after radiotherapy in patients with atypical and malignant meningiomas—clinical results in 85 patients treated in a single institution leading to optimized guidelines for early radiation therapy. Int J Radiat Oncol Biol Phys. 2012;83(3):859–864. [DOI] [PubMed] [Google Scholar]
- 8. Simpson D. The recurrence of intracranial meningiomas after surgical treatment. J Neurol Neurosurg Psychiatry. 1957;20(1):22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gousias K, Schramm J, Simon M. The Simpson grading revisited: aggressive surgery and its place in modern meningioma management. J Neurosurg. 2016;125(3):551–560. [DOI] [PubMed] [Google Scholar]
- 10. Goldbrunner R, Minniti G, Preusser M, et al. EANO guidelines for the diagnosis and treatment of meningiomas. Lancet Oncol. 2016;17(9):e383–e391. [DOI] [PubMed] [Google Scholar]
- 11. Brastianos PK, Horowitz PM, Santagata S, et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet. 2013;45(3):285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clark VE, Harmancı AS, Bai H, et al. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet. 2016;48(10):1253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clark VE, Erson-Omay EZ, Serin A, et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339(6123):1077–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45(10):1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cheng I, Levin AM, Tai YC, et al. Copy number alterations in prostate tumors and disease aggressiveness. Genes Chromosomes Cancer. 2012;51(1):66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang Y, Martens JWM, Yu JX, et al. Copy number alterations that predict metastatic capability of human breast cancer. Cancer Res. 2009;69(9):3795–3801. [DOI] [PubMed] [Google Scholar]
- 18. Zang KD, Singer H. Chromosomal consitution of meningiomas. Nature. 1967;216(5110):84–85. [DOI] [PubMed] [Google Scholar]
- 19. Seizinger BR, de la Monte S, Atkins L, Gusella JF, Martuza RL. Molecular genetic approach to human meningioma: loss of genes on chromosome 22. Proc Natl Acad Sci U S A. 1987;84(15):5419–5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruttledge MH, Sarrazin J, Rangaratnam S, et al. Evidence for the complete inactivation of the NF2 gene in the majority of sporadic meningiomas. Nat Genet. 1994;6(2):180–184. [DOI] [PubMed] [Google Scholar]
- 21. Lee Y, Liu J, Patel S, et al. Genomic landscape of meningiomas. Brain Pathol. 2010;20(4):751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bi WL, Greenwald NF, Abedalthagafi M, et al. Erratum: genomic landscape of high-grade meningiomas. NPJ Genom Med. 2017;2:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cai DX, Banerjee R, Scheithauer BW, Lohse CM, Kleinschmidt-Demasters BK, Perry A. Chromosome 1p and 14q FISH analysis in clinicopathologic subsets of meningioma: diagnostic and prognostic implications. J Neuropathol Exp Neurol. 2001;60(6):628–636. [DOI] [PubMed] [Google Scholar]
- 24. Zang KD. Meningioma: a cytogenetic model of a complex benign human tumor, including data on 394 karyotyped cases. Cytogenet Cell Genet. 2001;93(3-4):207–220. [DOI] [PubMed] [Google Scholar]
- 25. Abedalthagafi MS, Merrill PH, Bi WL, et al. Angiomatous meningiomas have a distinct genetic profile with multiple chromosomal polysomies including polysomy of chromosome 5. Oncotarget. 2014;5(21):10596–10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. López-Ginés C, Cerdá-Nicolás M, Gil-Benso R, Barcia-Salorio JL, Llombart-Bosch A. Loss of 1p in recurrent meningiomas: a comparative study in successive recurrences by cytogenetics and fluorescence in situ hybridization. Cancer Genet Cytogenet. 2001;125(2):119–124. [DOI] [PubMed] [Google Scholar]
- 27. Ketter R, Henn W, Niedermayer I, et al. Predictive value of progression-associated chromosomal aberrations for the prognosis of meningiomas: a retrospective study of 198 cases. J Neurosurg. 2001;95(4):601–607. [DOI] [PubMed] [Google Scholar]
- 28. Leone PE, Bello MJ, de Campos JM, et al. NF2 gene mutations and allelic status of 1p, 14q and 22q in sporadic meningiomas. Oncogene. 1999;18(13):2231–2239. [DOI] [PubMed] [Google Scholar]
- 29. Bello MJ, de Campos JM, Kusak ME, et al. Allelic loss at 1p is associated with tumor progression of meningiomas. Genes Chromosomes Cancer. 1994;9(4):296–298. [DOI] [PubMed] [Google Scholar]
- 30. Al-Mefty O, Kadri PA, Pravdenkova S, Sawyer JR, Stangeby C, Husain M. Malignant progression in meningioma: documentation of a series and analysis of cytogenetic findings. J Neurosurg. 2004;101(2):210–218. [DOI] [PubMed] [Google Scholar]
- 31. Boström J, Meyer-Puttlitz B, Wolter M, et al. Alterations of the tumor suppressor genes CDKN2A (p16(INK4a)), p14(ARF), CDKN2B (p15(INK4b)), and CDKN2C (p18(INK4c)) in atypical and anaplastic meningiomas. Am J Pathol. 2001;159(2):661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aizer AA, Abedalthagafi M, Bi WL, et al. A prognostic cytogenetic scoring system to guide the adjuvant management of patients with atypical meningioma. Neuro Oncol. 2016;18(2):269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lamszus K, Kluwe L, Matschke J, Meissner H, Laas R, Westphal M. Allelic losses at 1p, 9q, 10q, 14q, and 22q in the progression of aggressive meningiomas and undifferentiated meningeal sarcomas. Cancer Genet Cytogenet. 1999;110(2):103–110. [DOI] [PubMed] [Google Scholar]
- 34. Krayenbühl N, Pravdenkova S, Al-Mefty O. De novo versus transformed atypical and anaplastic meningiomas: comparisons of clinical course, cytogenetics, cytokinetics, and outcome. Neurosurgery. 2007;61(3):495ü503. [DOI] [PubMed] [Google Scholar]
- 35. Yuzawa S, Nishihara H, Tanaka S. Genetic landscape of meningioma. Brain Tumor Pathol. 2016;33(4):237–247. [DOI] [PubMed] [Google Scholar]
- 36. Yuzawa S, Nishihara H, Yamaguchi S, et al. Clinical impact of targeted amplicon sequencing for meningioma as a practical clinical-sequencing system. Mod Pathol. 2016;29(7):708–716. [DOI] [PubMed] [Google Scholar]
- 37. Zotti T, Vito P, Stilo R. The seventh ring: exploring TRAF7 functions. J Cell Physiol. 2012;227(3):1280–1284. [DOI] [PubMed] [Google Scholar]
- 38. Bleeker FE, Felicioni L, Buttitta F, et al. AKT1(E17K) in human solid tumours. Oncogene. 2008;27(42):5648–5650. [DOI] [PubMed] [Google Scholar]
- 39. Tetreault MP, Yang Y, Katz JP. Krüppel-like factors in cancer. Nat Rev Cancer. 2013;13(10):701–713. [DOI] [PubMed] [Google Scholar]
- 40. Abedalthagafi M, Bi WL, Aizer AA, et al. Oncogenic PI3K mutations are as common as AKT1 and SMO mutations in meningioma. Neuro-Oncol. 2016;18(5):649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reuss DE, Piro RM, Jones DT, et al. Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations. Acta Neuropathol. 2013;125(3):351–358. [DOI] [PubMed] [Google Scholar]
- 42. Sahm F, Bissel J, Koelsche C, et al. AKT1E17K mutations cluster with meningothelial and transitional meningiomas and can be detected by SFRP1 immunohistochemistry. Acta Neuropathol. 2013;126(5):757–762. [DOI] [PubMed] [Google Scholar]
- 43. Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br J Cancer. 2006;94(4):455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci U S A. 2005;102(3):802–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Christiaans I, Kenter SB, Brink HC, et al. Germline SMARCB1 mutation and somatic NF2 mutations in familial multiple meningiomas. J Med Genet. 2011;48(2):93–97. [DOI] [PubMed] [Google Scholar]
- 46. Bacci C, Sestini R, Provenzano A, et al. Schwannomatosis associated with multiple meningiomas due to a familial SMARCB1 mutation. Neurogenetics. 2010;11(1):73–80. [DOI] [PubMed] [Google Scholar]
- 47. Harmancı AS, Youngblood MW, Clark VE, et al. Integrated genomic analyses of de novo pathways underlying atypical meningiomas. Nat Commun. 2017;8:14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smith MJ, O’Sullivan J, Bhaskar SS, et al. Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nat Genet. 2013;45(3):295–298. [DOI] [PubMed] [Google Scholar]
- 49. Smith MJ, Wallace AJ, Bennett C, et al. Germline SMARCE1 mutations predispose to both spinal and cranial clear cell meningiomas. J Pathol. 2014;234(4):436–440. [DOI] [PubMed] [Google Scholar]
- 50. Raffalli-Ebezant H, Rutherford SA, Stivaros S, et al. Pediatric intracranial clear cell meningioma associated with a germline mutation of SMARCE1: a novel case. Childs Nerv Syst. 2015;31(3):441–447. [DOI] [PubMed] [Google Scholar]
- 51. Tauziede-Espariat A, Parfait B, Besnard A, et al. Loss of SMARCE1 expression is a specific diagnostic marker of clear cell meningioma: a comprehensive immunophenotypical and molecular analysis. Brain Pathol. 2018;28(4):466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Collord G, Tarpey P, Kurbatova N, et al. An integrated genomic analysis of anaplastic meningioma identifies prognostic molecular signatures. Sci Rep. 2018;8(1):13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wadt K, Choi J, Chung JY, et al. A cryptic BAP1 splice mutation in a family with uveal and cutaneous melanoma, and paraganglioma. Pigment Cell Melanoma Res. 2012;25(6):815–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shankar GM, Santagata S. BAP1 mutations in high-grade meningioma: implications for patient care. Neuro Oncol. 2017;19(11):1447–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Abdel-Rahman MH, Pilarski R, Cebulla CM, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet. 2011;48(12):856–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cheung M, Testa JR. BAP1, a tumor suppressor gene driving malignant mesothelioma. Transl Lung Cancer Res. 2017;6(3):270–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Koelsche C, Sahm F, Capper D, et al. Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta Neuropathol. 2013;126(6):907–915. [DOI] [PubMed] [Google Scholar]
- 58. Sahm F, Schrimpf D, Olar A, et al. TERT promoter mutations and risk of recurrence in meningioma. J Natl Cancer Inst. 2016;108(5):djv377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Goutagny S, Nault JC, Mallet M, Henin D, Rossi JZ, Kalamarides M. High incidence of activating TERT promoter mutations in meningiomas undergoing malignant progression. Brain Pathol. 2014;24(2):184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22(22):4632–4642. [DOI] [PubMed] [Google Scholar]
- 62. Gao F, Shi L, Russin J, et al. DNA methylation in the malignant transformation of meningiomas. PLoS One. 2013;8(1):e54114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bello MJ, Amiñoso C, Lopez-Marin I, et al. DNA methylation of multiple promoter-associated CpG islands in meningiomas: relationship with the allelic status at 1p and 22q. Acta Neuropathol. 2004;108(5):413–421. [DOI] [PubMed] [Google Scholar]
- 64. He S, Pham MH, Pease M, et al. A review of epigenetic and gene expression alterations associated with intracranial meningiomas. Neurosurg Focus. 2013;35(6):E5. [DOI] [PubMed] [Google Scholar]
- 65. Barski D, Wolter M, Reifenberger G, Riemenschneider MJ. Hypermethylation and transcriptional downregulation of the TIMP3 gene is associated with allelic loss on 22q12.3 and malignancy in meningiomas. Brain Pathol. 2010;20(3):623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nakane Y, Natsume A, Wakabayashi T, et al. Malignant transformation-related genes in meningiomas: allelic loss on 1p36 and methylation status of p73 and RASSF1A. J Neurosurg. 2007;107(2):398–404. [DOI] [PubMed] [Google Scholar]
- 67. Olar A, Wani KM, Wilson CD, et al. Global epigenetic profiling identifies methylation subgroups associated with recurrence-free survival in meningioma. Acta Neuropathol. 2017;133(3):431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sahm F, Schrimpf D, Stichel D, et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. doi: 10.1016/S1470-2045(17)30155-9 [DOI] [PubMed] [Google Scholar]
- 69. Katz LM, Hielscher T, Liechty B, et al. Loss of histone H3K27me3 identifies a subset of meningiomas with increased risk of recurrence. Acta Neuropathol. 2018;135(6):955–963. [DOI] [PubMed] [Google Scholar]
- 70. Saydam O, Shen Y, Würdinger T, et al. Downregulated microRNA-200a in meningiomas promotes tumor growth by reducing E-cadherin and activating the Wnt/beta-catenin signaling pathway. Mol Cell Biol. 2009;29(21):5923–5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Harmancı AS, Youngblood MW, Clark VE, et al. Integrated genomic analyses of de novo pathways underlying atypical meningiomas. Nat Commun. 2017;8:14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kliese N, Gobrecht P, Pachow D, et al. miRNA-145 is downregulated in atypical and anaplastic meningiomas and negatively regulates motility and proliferation of meningioma cells. Oncogene. 2013;32(39):4712–4720. [DOI] [PubMed] [Google Scholar]
- 73. Zhi F, Zhou G, Wang S, et al. A microRNA expression signature predicts meningioma recurrence. Int J Cancer. 2013;132(1):128–136. [DOI] [PubMed] [Google Scholar]
- 74. Chen F, Xiang CX, Zhou Y, et al. Gene expression profile for predicting survival of patients with meningioma. Int J Oncol. 2015;46(2):791–797. [DOI] [PubMed] [Google Scholar]
- 75. Olar A, Goodman LD, Wani KM, et al. A gene expression signature predicts recurrence-free survival in meningioma. Oncotarget. 2018;9(22):16087–16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Huang YC, Tsai YH, Lee JD, et al. Hemodynamic factors may play a critical role in neurological deterioration occurring within 72 hrs after lacunar stroke. PLoS One. 2014;9(10):e108395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Schulten HJ, Hussein D, Al-Adwani F, et al. Microarray expression data identify DCC as a candidate gene for early meningioma progression. PLoS One. 2016;11(4):e0153681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Serna E, Morales JM, Mata M, et al. Gene expression profiles of metabolic aggressiveness and tumor recurrence in benign meningioma. PLoS One. 2013;8(6):e67291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tabernero MD, Espinosa AB, Maillo A, et al. Patient gender is associated with distinct patterns of chromosomal abnormalities and sex chromosome linked gene-expression profiles in meningiomas. Oncologist. 2007;12(10):1225–1236. [DOI] [PubMed] [Google Scholar]
- 80. Schmidt M, Mock A, Jungk C, et al. Transcriptomic analysis of aggressive meningiomas identifies PTTG1 and LEPR as prognostic biomarkers independent of WHO grade. Oncotarget. 2016;7(12):14551–14568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Okamoto H, Li J, Vortmeyer AO, et al. Comparative proteomic profiles of meningioma subtypes. Cancer Res. 2006;66(20):10199–10204. [DOI] [PubMed] [Google Scholar]
- 82. Sharma S, Ray S, Mukherjee S, Moiyadi A, Sridhar E, Srivastava S. Multipronged quantitative proteomic analyses indicate modulation of various signal transduction pathways in human meningiomas. Proteomics. 2015;15(2–3):394–407. [DOI] [PubMed] [Google Scholar]
- 83. Cui GQ, Jiao AH, Xiu CM, et al. Proteomic analysis of meningiomas. Acta Neurol Belg. 2014;114(3):187–194. [DOI] [PubMed] [Google Scholar]
- 84. Saydam O, Senol O, Schaaij-Visser TB, et al. Comparative protein profiling reveals minichromosome maintenance (MCM) proteins as novel potential tumor markers for meningiomas. J Proteome Res. 2010;9(1):485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tufan K, Dogulu F, Kurt G, Emmez H, Ceviker N, Baykaner MK. Intracranial meningiomas of childhood and adolescence. Pediatr Neurosurg. 2005;41(1):1–7. [DOI] [PubMed] [Google Scholar]
- 86. Perry A, Giannini C, Raghavan R, et al. Aggressive phenotypic and genotypic features in pediatric and NF2-associated meningiomas: a clinicopathologic study of 53 cases. J Neuropathol Exp Neurol. 2001;60(10):994–1003. [DOI] [PubMed] [Google Scholar]
- 87. Arivazhagan A, Devi BI, Kolluri SV, Abraham RG, Sampath S, Chandramouli BA. Pediatric intracranial meningiomas–do they differ from their counterparts in adults?Pediatr Neurosurg. 2008;44(1):43–48. [DOI] [PubMed] [Google Scholar]
- 88. Hui M, Uppin MS, Saradhi MV, Sahu BP, Purohit AK, Sundaram C. Pediatric meningiomas an aggressive subset: a clinicopathological and immunohistochemical study. J Postgrad Med. 2015;61(1):32–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Dudley RWR, Torok MR, Randall S, et al. Pediatric versus adult meningioma: comparison of epidemiology, treatments, and outcomes using the Surveillance, Epidemiology, and End Results database. J Neurooncol. 2018;137(3):621–629. [DOI] [PubMed] [Google Scholar]
- 90. Battu S, Kumar A, Pathak P, et al. Clinicopathological and molecular characteristics of pediatric meningiomas. Neuropathology. 2018;38(1):22–33. [DOI] [PubMed] [Google Scholar]
- 91. Al-Mefty O, Topsakal C, Pravdenkova S, Sawyer JR, Harrison MJ. Radiation-induced meningiomas: clinical, pathological, cytokinetic, and cytogenetic characteristics. J Neurosurg. 2004;100(6):1002–1013. [DOI] [PubMed] [Google Scholar]
- 92. Godlewski B, Drummond KJ, Kaye AH. Radiation-induced meningiomas after high-dose cranial irradiation. J Clin Neurosci. 2012;19(12):1627–1635. [DOI] [PubMed] [Google Scholar]
- 93. Lee JW, Wernicke AG. Risk and survival outcomes of radiation-induced CNS tumors. J Neurooncol. 2016;129(1):15–22. [DOI] [PubMed] [Google Scholar]
- 94. Yamanaka R, Hayano A, Kanayama T. Radiation-induced meningiomas: an exhaustive review of the literature. World Neurosurg. 2017;97:635–644.e8. [DOI] [PubMed] [Google Scholar]
- 95. Agnihotri S, Suppiah S, Tonge PD, et al. Therapeutic radiation for childhood cancer drives structural aberrations of NF2 in meningiomas. Nat Commun. 2017;8(1):186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lillehei KO, Donson AM, Kleinschmidt-DeMasters BK. Radiation-induced meningiomas: clinical, cytogenetic, and microarray features. Acta Neuropathol. 2008;116(3):289–301. [DOI] [PubMed] [Google Scholar]
- 97. Ueyama Y, Morita K, Ochiai C, Ohsawa N, Hata J, Tamaoki N. Xenotransplantation of a human meningioma and its lung metastasis in nude mice. Br J Cancer. 1978;37(4):644–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Rana MW, Pinkerton H, Thornton H, Nagy D. Heterotransplantation of human glioblastoma multiforme and meningioma to nude mice. Proc Soc Exp Biol Med. 1977;155(1):85–88. [DOI] [PubMed] [Google Scholar]
- 99. Rath P, Miller DC, Litofsky NS, et al. Isolation and characterization of a population of stem-like progenitor cells from an atypical meningioma. Exp Mol Pathol. 2011;90(2):179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Tanaka K, Sato C, Maeda Y, et al. Establishment of a human malignant meningioma cell line with amplified c-myc oncogene. Cancer. 1989;64(11):2243–2249. [DOI] [PubMed] [Google Scholar]
- 101. Püttmann S, Senner V, Braune S, et al. Establishment of a benign meningioma cell line by hTERT-mediated immortalization. Lab Invest. 2005;85(9):1163–1171. [DOI] [PubMed] [Google Scholar]
- 102. Burns SS, Akhmametyeva EM, Oblinger JL, et al. Histone deacetylase inhibitor AR-42 differentially affects cell-cycle transit in meningeal and meningioma cells, potently inhibiting NF2-deficient meningioma growth. Cancer Res. 2013;73(2):792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Mei Y, Bi WL, Greenwald NF, et al. Genomic profile of human meningioma cell lines. PloS One. 2017;12(5):e0178322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kalamarides M, Niwa-Kawakita M, Leblois H, et al. Nf2 gene inactivation in arachnoidal cells is rate-limiting for meningioma development in the mouse. Genes Dev. 2002;16(9):1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Peyre M, Stemmer-Rachamimov A, Clermont-Taranchon E, et al. Meningioma progression in mice triggered by Nf2 and Cdkn2ab inactivation. Oncogene. 2013;32(36):4264–4272. [DOI] [PubMed] [Google Scholar]
- 106. Olson JJ, Beck DW, Schlechte JA, Loh PM. Effect of the antiprogesterone RU-38486 on meningioma implanted into nude mice. J Neurosurg. 1987;66(4):584–7. [DOI] [PubMed] [Google Scholar]
- 107. Haase D, Schmidl S, Ewald C, et al. Fatty acid synthase as a novel target for meningioma therapy. Neuro Oncol. 2010;12(8):844–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Stepanova DS, Semenova G, Kuo YM, et al. An essential role for the tumor-suppressor merlin in regulating fatty acid synthesis. Cancer Res. 2017;77(18):5026–5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Jensen RL, Wurster RD. Calcium channel antagonists inhibit growth of subcutaneous xenograft meningiomas in nude mice. Surg Neurol. 2001;55(5):275–283. [DOI] [PubMed] [Google Scholar]
- 110. Ragel BT, Gillespie DL, Kushnir V, Polevaya N, Kelly D, Jensen RL. Calcium channel antagonists augment hydroxyurea- and ru486-induced inhibition of meningioma growth in vivo and in vitro. Neurosurgery. 2006;59(5):1109–20; discussion 1120. [DOI] [PubMed] [Google Scholar]
- 111. Karsy M, Hoang N, Barth T, et al. Combined hydroxyurea and verapamil in the clinical treatment of refractory meningioma: human and orthotopic xenograft studies. World Neurosurg. 2016;86:210–219. [DOI] [PubMed] [Google Scholar]
- 112. Ragel BT, Jensen RL, Gillespie DL, Prescott SM, Couldwell WT. Celecoxib inhibits meningioma tumor growth in a mouse xenograft model. Cancer. 2007;109(3):588–597. [DOI] [PubMed] [Google Scholar]
- 113. Friedrich S, Schwabe K, Grote M, Krauss JK, Nakamura M. Effect of systemic celecoxib on human meningioma after intracranial transplantation into nude mice. Acta Neurochir (Wien). 2013;155(1):173–182. [DOI] [PubMed] [Google Scholar]
- 114. Pachow D, Andrae N, Kliese N, et al. mTORC1 inhibitors suppress meningioma growth in mouse models. Clin Cancer Res. 2013;19(5):1180–1189. [DOI] [PubMed] [Google Scholar]
- 115. Gupta V, Su YS, Samuelson CG, et al. Irinotecan: a potential new chemotherapeutic agent for atypical or malignant meningiomas. J Neurosurg. 2007;106(3):455–462. [DOI] [PubMed] [Google Scholar]
- 116. Takeda H, Okada M, Kuramoto K, et al. Antitumor activity of gemcitabine against high-grade meningioma in vitro and in vivo. Oncotarget. 2017;8(53):90996–91008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Nigim F, Esaki S, Hood M, et al. A new patient-derived orthotopic malignant meningioma model treated with oncolytic herpes simplex virus. Neuro Oncol. 2016;18(9):1278–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Ho WS, Sizdahkhani S, Hao S, et al. LB-100, a novel protein phosphatase 2A (PP2A) inhibitor, sensitizes malignant meningioma cells to the therapeutic effects of radiation. Cancer Lett. 2018;415:217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Chow HY, Dong B, Duron SG, et al. Group I Paks as therapeutic targets in NF2-deficient meningioma. Oncotarget. 2015;6(4):1981–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Zhai L, Ladomersky E, Lauing KL, et al. Infiltrating T cells increase IDO1 expression in glioblastoma and contribute to decreased patient survival. Clin Cancer Res. 2017;23(21):6650–6660. [DOI] [PMC free article] [PubMed] [Google Scholar]