SUMMARY
Zika virus (ZIKV) targets neural progenitor cells in the brain, attenuates cell proliferation, and leads to cell death. Here, we describe a role for the ZIKV protease NS2B-NS3 heterodimer in mediating neurotoxicity through cleavage of a host protein required for neurogenesis. Similar to ZIKV infection, NS2B-NS3 expression led to cytokinesis defects and cell death in a protease activity-dependent fashion. Among binding partners, NS2B-NS3 cleaved Septin-2, a cytoskeletal factor involved in cytokinesis. Cleavage of Septin-2 occurred at residue 306 and forced expression of a non-cleavable Septin-2 restored cytokinesis, suggesting a direct mechanism of ZIKV-induced neural toxicity.
In Brief
Mechanisms by which Zika virus leads to microcephaly are poorly understood. Here, Li et al. demonstrate the Zika protease, required for viral replication, associates with host proteins and cleaves Septin-2, a protein required for neural cell division.
Graphical Abstract

INTRODUCTION
Zika virus (ZIKV) represents a new threat to global health, due to the unexpected association with congenital neurodevelopmental birth defects observed in the fetuses of infected women during critical periods of pregnancy. During the peak of the epidemic in Brazil in 2016, the reported incidence of microcephaly increased 26-fold over baseline (Butler, 2016). ZIKV is capable of direct infection of neural progenitor cells in the fetal brain, leading to delayed mitosis, activation of p53, and apoptotic cell death (Ghouzzi et al., 2017; Li et al., 2016c; Ming et al., 2016; Zhang et al., 2016).
ZIKV contains a positive single-stranded 11-kb RNA genome encoding three structural (C, prM, and E) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5), providing functional diversity required for its life cycle. Individual ZIKV proteins tested to date, when introduced into human neural precursor cells (hNPCs) or fetal murine brain, can impact neurogenesis but are insufficient to mediate cell death (Liang et al., 2016; Yoon et al., 2017). Some ZIKV proteins can form heteromultimeric complexes, including prM and E, which assemble during viral packaging (Lorenz et al., 2002), and NS2B and NS3, which form a functional heterodimeric protease (Zuo et al., 2009). NS2B is a co-factor of NS3, which enhances its proteolytic activity and stabilizes its folding. NS3 is a multifunctional protein common to the flavivirus genus, with an N-terminal serine protease domain and a C-terminal nucleoside triphosphatase (NTPase) and RNA helicase domains (Wengler and Wengler, 1991).
Cytokinesis is the final stage of cell division whereby the two daughter cells physically separate, which requires contribution from the septin cytoskeleton. During cytokinesis, the contractile ring forms beneath the cell’s equatorial surface to form the cleavage furrow, and then ingression of the furrow results in the formation of an intercellular bridge called the midbody (D’Avino et al., 2015) and then finally abscission as the two daughter cells separate. Cytokinesis failure results in binucleated and multi-centrosome-containing cells, with resultant aneuploidy, genotoxic stress, and cell death (Hayashi and Karlseder, 2013). Septins are highly conserved guanosine triphosphate (GTP)-binding proteins that hetero-oligomerize (Mostowy and Cossart, 2012) and are crucial for midbody formation during cytokinesis (Shindo and Wallingford, 2014; Spiliotiset al., 2005) as well as membrane trafficking, spermatogenesis, and dendrite branching (Beites et al., 1999; Ihara et al., 2005; Xie et al., 2007). Of the 14 septin paralogs in humans, Septin 2, 6, and 7 bind in a heterotrimeric linear fashion (Weirich et al., 2008), which then binds other trimers to produce filaments and rings (Sirajuddin et al., 2007).
Congenital microcephaly refers to birth head circumference <2 SDs below mean, which can have genetic or environmental/ viral origins (Volpe et al., 2018), and ZIKV-related microcephaly shares many radiographic and pathological features with genetic forms of disease (Besnard et al., 2016; Chimelli et al., 2017; de Fatima Vasco Aragao et al., 2016). Two genes mutated in recessive microcephaly in particular, KIF14 (kinesin 14) and CIT(citron kinase), encode proteins that localize to the mitotic cleavage furrow. Loss of either demonstrates delayed mitosis due to failed abscission, supernumerary centrosomes, and multinucleated cells (LoTurco et al., 2003; Madaule et al., 1998; Moawia et al., 2017), resulting in chromosomal instability and p53-sensitive cell death (Bianchi et al., 2017). Although the ZIKV-related and KIF14/CIT-related microcephaly differ in that ZIKV cases show frequent brain calcification (de Fatima Vasco Aragao et al., 2016; Zare Mehrjardi et al., 2017), recent evidence suggests that ZIKV-infected cells strikingly share several of these mitotic defects with KIF14 or CIT deficiency (Carleton et al., 2006; Li et al., 2016b; Moawia et al., 2017; Onorati et al., 2016; Souza et al., 2016; Wolf et al., 2017).
Due to the similarities with these forms of microcephaly, and because ZIKV targets the radial glial neural stem cells of the brain (Garcez et al., 2016; Wu et al., 2016), we reasoned that ZIKV proteins might inadvertently impact the function of one or more proteins involved in neuronal cell division. Here, we show that the active NS2B3 ZIKV protease is capable of mediating cytotoxic effects of ZIKV, including delayed cytokinesis and failed mitotic abscission. We traced this to off-target direct cleavage of the host Septin 2 protein at residue 306, which leads to alterations of Septin cytoskeletal components. Introduction of a non-cleavable Septin 2 partially restores cytokinesis in cells expressing NS2B3. We thus isolate a ZIKV toxic component mediating cytokinesis defects. These results suggest a potential target for drugs to reduce the burden of disease.
RESULTS
ZIKV Protease NS2B3 Expression Results in Multipolar Spindles
To test for potential toxic effects of the ZIKV protease, we utilized just the N-terminal NS3 protease domain fused with the NS2B protein, termed NS2B3, derived from the American strain, which fully reconstitutes protease activity (Lei et al., 2016). We generated stable human 293T cell lines expressing NS2B3 and examined cell proliferation rates. We found that cells expressing NS2B3 exhibited a 33% reduction in EdU labeling as well as a 36% reduction in phosphor-histone H3 (pH3) immunoreactivity (Figures 1A–1D), suggesting reduced cell division. Correlated with this was evidence of profound cell death, evidenced by extensive cleaved caspase-3 positivity (Figures 1E and 1F). There was a dramatic 580% increase in the percent of cells with multipolar spindles and supernumerary centrosomes following NS2B3 expression (Figures 1G and 1H), similar to effects observed in ZIKV-infected cells (Figures S1A–S1C; Onorati et al., 2016; Souza et al., 2016; Wolf et al., 2017). None of these cellular phenotypes were observed in cells expressing proteolytically inactive NS2B3, containing a S135A (SA) mutation at the catalytic site (Zhou et al., 2006). Taken together, these results suggest that NS2B3 is sufficient to alter cell proliferation and survival, in a fashion similar to ZIKV infection, and may act to cleave one or more host proteins.
Figure 1. Active, but Not Protease-Dead, ZIKV NS2B3 Is Sufficientto Reduce Cell Proliferation, MediateApoptosis, and Result in Multipolar Spindles.
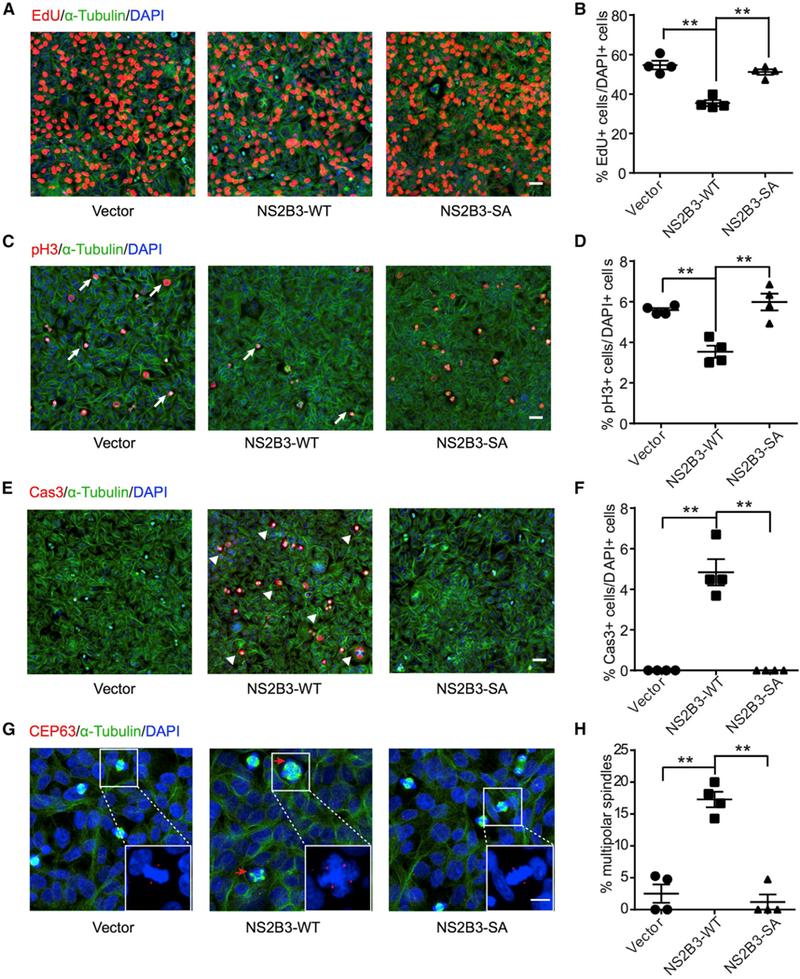
(A, C, and E) Cells stably expressing NS2B3 showed reduced incorporation of EdU (DNAsynthesis marker), reduced phospho-histone H3 (pH3) (mitotic marker), and increased caspase 3 (Cas3) (apoptotic marker) compared with vector and S135A (SA) enzymatic null from 293T cells. Scale bars represent 20 μm.
(G) NS2B3 expression leads to accumulation of cells with multipolar spindle morphology (arrows) and supernumerary centrosomes, as evident by centrosome marker CEP63. Right-bottom corner boxes present enlargement of cells with mitotic spindle. Scale bars represent 20 μm.
(B, D, F, and H) Distribution sample mean and SEM. N = 4 biological replicates. **p < 0.01.
ZIKV Proteaseome Includes Septin Family Members
To identify putative ZIKV protease targets, we performed co-affinity purification using recombinant glutathione S-transferase (GST)-tagged NS2B3 (Figure 2A) incubated with hNPC lysates, followed by mass spectrometry analysis. We utilized the S135A mutant, reasoning that this might prolong interactions with binding partners due to its inability to cleave substrates. The co-affinity purification identified a series of proteins interacting with NS2B3 (Figure 2B; Table S1; Student’s t test; p < 0.05), which we termed the ZIKV proteaseome (ZPO). Strikingly, these proteins clustered into seven specific interaction modules (Figure 2C): cytoskeleton; protein folding; proteasome; RNA binding and processing; chromosome maintenance; metabolism; and ribosome, suggesting one or more proteins within each module might be a protease target.
Figure 2. ZIKV Proteaseome Includes Septin Family Members.
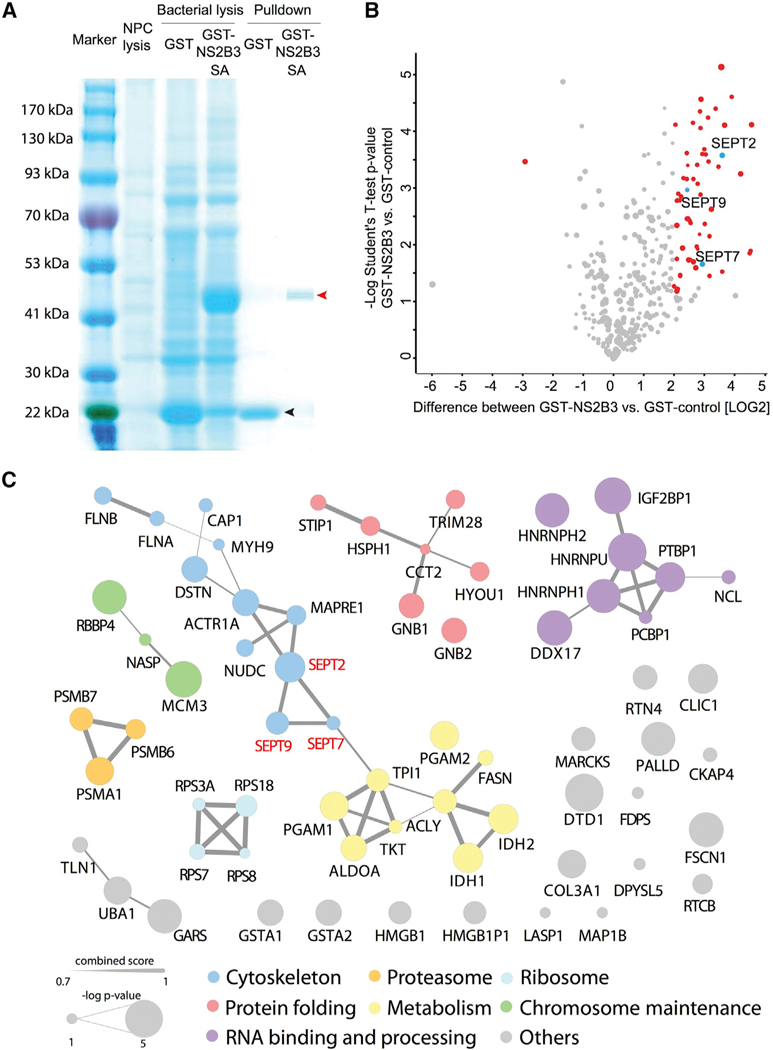
(A) Coomassie-stained gel comparing human neural precursor cell (hNPC) lysate, bacterial GST alone (black arrow), and GST-NS2B3 S135A (red arrow) prior to and after affinity purification.
(B) Label-free-quantitation-based volcano plot representing quantitative proteomic analysis of NS2B3 S135A pull-down. p values (—log10) are plotted as a function oflog2 fold changes (NS2B3-GST versus GST). Proteins >4-fold changed are marked in red (Student’s t test; p<0.05). See Table S1 for details. Septin 2, Septin 7, and Septin 9 are marked in blue. N = 3 biological replicates.
(C) Protein-protein interaction network analysis of ZIKV NS2B3-SA binding partners identified from MS/MS using String (https://string-db.org/; high confidence 0.700). Seven major networks with molecular functions were identified and labeled with different colors. Ball size represents the confidence level of each NS2B3-SA binging partner (—log p value). Line thickness represents the interaction strength (String combined score) between each protein. SEPT2, SEPT7, and SEPT9 are highlighted in red.
The cytoskeleton module within the ZPO had three hits in sub-units of the septin cytoskeleton, Septin-2 (SEPT2), Septin-7 (SEPT7), and Septin-9 (SEPT9). We confirmed the interaction between ZIKV NS2B3 and SEPT2/SEPT7 by western blot (Figure S1A). We found that native ZIKV NS3 interacts with SEPT2 directly, and the interaction was enhanced by the presence of NS2B (Figure S1B), excluding the possibility of an artificial interaction mediated by nonnative NS2B3 fusion protein. Septins form a key component of the contractile ring and midbody (Estey et al., 2010), which we confirmed in cultured cells (Figure S2C), and function with KIF14 and CIT in cytokinesis (El Amine et al., 2013; Kremeret al., 2005), encoded by two genes recently implicated in congenital microcephaly. The septin ring is critical for telophase abscission, during which the two daughter cells separate. This made septins attractive candidates in the pathogenesis of ZIKV-induced microcephaly.
ZIKV NS2B3 Protease Mediates Cleavage of Septin-2 at Residue R306, Leading to Reduced Septin Levels
We next tested whether SEPT2 or SEPT7 was a substrate of the protease by transient transfection of ZIKV NS2B3 in 293T cells. Cells transfected with NS2B3 wild-type (WT), but not S135A, showed a down-shifted band of SEPT2 when probed with an anti-SEPT2 antibody (Figure 3A), suggesting cleavage by NS2B3. We compared SEPT2 levels between cells with and without NS2B3 expression in the same field of view by immunocytochemistry using an antibody against C-terminal SEPT2 and found NS2B3 WT, but not S135A, led to reduction of SEPT2 immunoreactivity (Figures S3A and S3B), suggesting loss of C-terminal SEPT2 from NS2B3-mediated cleavage. Because transient transfection would leave some cells untransfected, which could blunt the observed effects, we next studied SEPT2 levels in 293T cells stably expressing NS2B3. We observed a reduction in endogenous SEPT2 level, as well as presence of the cleaved SEPT2, which was notably more intense than following transient transfection (Figure S3C). SEPT7 did not show a cleavage product but exhibited a reduced level (Figure 3B), consistent with previous observations that stoichiometric levels are required for stability of the reciprocal septins (Kinoshita et al., 2002). SEPT9 level remained unchanged upon NS2B3 expression (Figure S3D).
Figure 3. ZIKV NS2B3 Protease Mediates Cleavage of Septin 2 at Residue R306.

(A and B) Cells transfected with ZIKV NS2B3 or NS2B3-S135A assessed by western blotting for SEPT2 and SEPT7. The lower-molecular-weight cleavage product was evident only with wild-type ZIKV NS2B3 and correlated with reduced SEPT7 levels, quantified below from N = 3.
(C and D) Cells infected with ZIKV showed the same SEPT2 cleavage band and reduced SEPT7 level, quantified below from N = 3 replicates.
(E) In vitro cleavage assay with recombinant NS2B3-WT or NS2B3-SA at low or high concentration (0.15 or 1.5 μM) with SEPT2 for1 h, analyzed by western with N- or C-terminal SEPT2 antibody. Cleaved SEPT2 is marked with red arrow, suggesting a C-terminal cleavage event.
(F) Schematic of SEPT2, GTPase, and coiled coil (CC) domains, with zoom in of residues 301–361 highlighting ZIKV NS2B3 polybasic motifs (tan).
(G) Extracted areas of SEPT2 peptides obtained by the three different digests (trypsin only or propionic anhydride labeling followed by trypsin or Asp-N digestion) applied to intact SEPT2 (light gray and gray outlines, respectively) or C-terminal SEPT2 fragment (C-term) from (E) (orange and red, respectively) were summed per residue and plotted as a function of SEPT2 sequence, normalized to signal of the most C-terminal residues. The signal of the N-terminal-labeled Asp-N generated peptide that defined the very N-terminal of the SEPT2 C-terminal fragment above.
(H) High-resolution/high-mass-accuracy tandem MS spectrum of the N-terminal and lysine propionylated (p)Asp-N peptide: pG307GRKpVENE(m/z = 500.75714 [1.5 ppm]; Mascot score of 37), consistent with R306 cleavage. B-, a-, and y-fragment ions are annotated.
(I) In vitro cleavage assay with recombinant NS2B3-WT and SEPT2-WT or-R306A, blotted with N- or C-terminal SEPT2 antibody. SEPT2-R306Afailed to cleave in the presence of active ZIKV NS2B3.
Next, we confirmed the presence of the down-shifted SEPT2 band and reduced SEPT7 in hNPCs infected with ZIKV comparing to cells with mock infection (Figures 3C and 3D), suggesting that the natural heterodimer produced by ZIKV infection is also capable of cleaving SEPT2. To exclude the possibility that SEPT2 was cleaved indirectly by a downstream protease, we repeated the experiment in vitro with recombinant proteins. ZIKV NS2B3 WT showed a dosage-dependent cleavage of human SEPT2, with molecular weight reduced by ~9 kDa from the C terminus, using anti-N-and C-terminal SEPT2 antibodies (Figure 3E). The sizes were consistent with the cleavage products observed following cellular transfection and suggest a single cleavage within the C-terminal domain of SEPT2 near the guanine nucleotide binding (G) domain, likely inactivating its function (de Almeida Marques et al., 2012; Meseroll et al., 2013; Sirajuddin et al., 2007).
Typical flavivirus proteases prefer substrates with basic residues (Arg or Lys) at the P1 and P2 sites and a short side chain (Gly, Ser, or Ala) at the P1′ site (Chambers et al., 1990; Yamshchikov and Compans, 1995). The C-terminal region of human SEPT2 revealed three such potential cleavage sites: R306; K310; and R341 (Figure 3F). To identify candidate cleavage sites, we performed liquid chromatography-tandem mass spectrometry (LC-MS/MS) on gel-extracted SEPT2 C-terminal cleavage product, together with a control using full-length SEPT2. We used ASP-N and trypsin digestion combined with propionyl labeling of primary amines to distinguish between ZIKV protease and ASP-N and trypsin protease cleavages (Yamshchikov and Compans, 1995; Figures 3G and S3E). The recovered peptide fragments from all three digest patterns suggested that C-terminal ZIKV protease cleavage occurred before residue 330, ruling out potential cleavage site R341. We also identified peptide K310pVENEDMNK (Figure S3F; where p represents propionate) from propionyl-trypsin digestion, which argued against K310 cleavage. We identified the N-terminal labeled peptide G307GRKVENE314 (Figure 3H) from propionyl-ASP-N digestion, which led to cleavage after glutamic acid (E), suggesting cleavage by NS2B3 at R306. We confirmed these results by introducing an alanine substitution at R306 of SEPT2 (SEPT2R306A), which was impervious to ZIKV NS2B3 in our in vitro protease assay (Figure 3I). To examine the impact of R306 cleavage on SEPT2 function, we transfected constructs expressing EGFP-fused full-length SEPT2 or with a C-terminal truncation at residue 306 (DC). EGFP-SEPT2 concentrated at the cleavage furrow during telophase as expected; however, EGFP-SEPT2ΔC was mislocalized throughout the cell cortex (Figure S3G). In addition, SEPT2 in cells engineered to stablyexpress NS2B3 failed to localize at the midbody during telophase, consistent with mislocalization as a result of NS2B3 cleavage (Figure S3H). We also found failed localization of SEPT2 and SEPT7 at the midbody in ZIKV-infected hNPCs (Figures S3I and S3J), suggesting disrupted septin complex upon ZIKV infection.
SEPT2 or SEPT7 Interference or ZIKV NS2B3 Expression Delays Cytokinesis, Partially Rescued by Non-cleavable SEPT2
Although septins are required for cytokinesis, their roles during neurogenesis are not known. We thus tested the roles of SEPT2 and SEPT7 during neurogenesis in cultured hNPCs (Figure S4A). We found that knockdown of SEPT2 or SEPT7 led to a 33% or 27% reduction in EdU-marked cells or 38% or 47% reduction in pH3-marked cells, respectively (Figures 4A, 4B, S4B, and S4C). The changes following SEPT2 or SEPT7 knockdown in hNPCs correlated with reduced survival and increased activated caspase-3 (Cas3), compared with control cells (Figure 4C), similar to what we found upon NS2B3 forced expression or published results following ZIKV infection. Considering the well-established role of septins in cytokinesis, we performed time-lapse recordings and observed ~6% of cells with SEPT2 knockdown completely failed abscission, and the remainder managed to complete cytokinesis but took 200% longer time than control. Knockdown of SEPT7 led to less severe cytokinesis delay, with a 123% increase in length of cytokinesis as comparing to control (Figures 4D and 4E). Similarly, a 160% increase in cytokinesis length was observed in 293T cells stably expressing NS2B3, but not S135A NS2B3 (Figures 4F and 4G). We also observed a lengthening of cytokinesis by 248% in hNPCs with NS2B3 expression (Figures S4D and S4E) and by 200% by ZIKV infection (Figures S4F–S4H). Given that SEPT2 R306A was impervious to ZIKV NS2B3 cleavage, we attempted to rescue the NS2B3-induced phenotype by co-expression of SEPT2-R306A. First, we confirmed that SEPT2 R306A produced a stable and properly localized protein in cells (Figure S4I). We found that cells expressing both ZIKV NS2B3 and SEPT2-R306A showed a partial rescue of cytokinesis delay (Figures 3H and 3I) and formation of multipolar spindle cells (Figures S4J and S4K) compared with ZIKV NS2B3 alone. In addition, co-expression of SEPT2-R306A restored SEPT7 level at the mid-body during cytokinesis in cells with NS2B3 expression (Figure S4L). The results suggest cleavage of SEPT2 by the NS2B3 protease results in failed cytokinesis and delayed cell division, events that are well established to induce genotoxic stress and cell death (Fuchs and Steller, 2011; Pucci et al., 2000).
Figure 4. SEPT2 or SEPT7 Interference or ZIKV NS2B3 Expression Delays Cytokinesis, Partially Rescued by Non-cleavable SEPT2.

(A–C) Knockdown of SEPT2 orSEPT7 showed reduced incorporation of EdU, reduced pH3, and increased cleaved Cas3 compared with scrambled short hairpin RNA (shRNA) from hNPCs. Bar represents 50 μm. From N = 4.
(D and E) Time-lapse frames showed knockdown of SEPT2 or SEPT7 lengthened cytokinesis. For all images, arrows track to a single cell undergoing mitosis into two cells. Note the timescale is expanded in the frames to capture completion of telophase. Scale bars represent 15 μm. n = 15–20 cells in each group.
(F and G) Transfection of NS2B3, but not NS2B3-SA, showed similar lengthening of cytokinesis. Bar represents 15 μm. n = 15–20 cells in each group.
(H and I) Co-transfection with non-cleavable SEPT2-R306A partially rescued lengthened cytokinesis induced by NS2B3 transfection. Bar represents 15 μm. n = 12–18 cells in each group.
*p < 0.05; **p < 0.01.
DISCUSSION
Here, we describe a potential mechanism by which a single ZIKV component, the protease, can induce toxicity in hNPCs. ZIKV protease displays specific catalytic efficiency many times higher than for the West Nile protease, attributed to sequence divergence or the ability of ZIKV protease to dimerize, which may potentiate this mechanism in neural cells (Lei et al., 2016). We found that expression of NS2B3 was sufficient to mediate cellular toxicity, an effect that depended upon protease activity. ZIKV protease expression causes delayed cell division, increased cellular apoptosis, and multipolar spindles, effects attributable to a defect in cytokinesis. We found that ZIKV NS2B3 bound to and cleaved host SEPT2 and led to disruption of SEPT2 and SEPT7 containing septin complex at the midbody during cytokinesis.
Previous work identified other host substrates of flavivirus proteases, which could impact both viral replication and host response: FAM134B is a reticulophagy receptor that is cleaved by flaviviral proteases at a polybasic residue hinge domain (Lennemann and Coyne, 2017). FAM134B cleavage inhibits reticulophagy, leading to enhanced viral replication presumably by mobilizing endoplasmic reticulum (ER) membrane for efficient viral budding. Further, both ZIKV and the closely related Dengue virus proteases can cleave human, but not mouse, Sting protein (Aguirre et al., 2012; Ding et al., 2018; Yu et al., 2012), a positive regulator of the interferon response, and provides a potential explanation for why humans, but not mice, are susceptible to flaviviral infection.
Whether protease from other flaviviruses target septins remains to be tested. No other flavivirus have been yet reported to cause microcephaly; thus, it is also possible that the special tropism toward hNPCs, or the ability of ZIKV to penetrate the placental and fetal brain, may contribute to microcephaly mechanisms (Cugola et al., 2016; Martines et al., 2016; Miner et al., 2016; Tabata et al., 2016). We compared the sequence of ZIKV NS2 and NS3 protease domains between different ZIKV isolates. ZIKV was first isolated from Uganda in 1947, followed by small outbreaks in Southeast Asia and the Pacific Islands prior to the recent epidemic. We found that the Asian and American strains share the same sequence; however, the African strain differs by one amino acid in NS2B and three amino acids in the NS3 protease domain within the protease-relevant regions (Figure S4M). We thus cloned and transfected these American and African strain-specific NS2B3 isoforms in human cells and found similar cleavage efficiency of SEPT2 (Figure S4N). This argues that the propensity of the ZIKV protease toward SEPT2 is not limited to strains linked to microcephaly.
Multiple studies have shown that ZIKV infection in human NPCs leads to growth impairment, Cas3 activation, and apoptosis, but mechanisms of toxicity are still under investigation. Robust transcriptional changes have been reported in NPCs following ZIKV infection, predominantly upregulation in apoptotic and immune-response pathways and downregulation of cell-cycle pathways (Dang et al., 2016; Li et al., 2016a; McGrath et al., 2017). Interestingly, transcriptional changes were noted in genes previously found mutated in primary microcephaly (Li et al., 2016a). One way this occurs is through a direct effect of the ZIKV transcript on the ability of the neural RNA-binding protein Musashi-1 to bind to endogenous targets, increasing viral load and reducing expression of factors implicated in neural stem cell function (Chavali et al., 2017). ZIKV proteins NS4A and NS4B can directly suppress neurogenesis through inhibition of Akt-mTOR signaling (Liang et al., 2016). ZIKV NS2A expressed in mouse embryonic brain leads to reduced proliferation through an effect on cellular adherens junctions (Yoon et al., 2017). ZIKV infection leads to mitochondrial sequestration of phospho-TBK1 and centrosomal depletion (Onorati et al., 2016). Finally, not all ZIKV pathology is mediated by infection of NPCs, as there is a robust vascular and inflammatory response in the brain, leading to leaky blood brain barrier and macrophage infiltration (Shao et al., 2016). These factors likely combine to produce neurotoxic effects, limiting NPC health, ultimately resulting in microcephaly.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Joseph G. Gleeson (jogleeson@ucsd.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
HEK293T and HeLa cells were purchased from ATCC and grown at 37°C in DMEM medium supplemented with 10% fetal bovine serum and Penicillin-Streptomycin. Aedes albopictus C6/36 cells (ATCC CRL-1660) were maintained in RPMI-1640 medium with 2% FBS at 28°C. Neural progenitor cells derived from human H9 ES cells were cultured at 37°C in DMEM/F12 media, supplemented with 0.5X N2, 0.5X B27, 20 ng/μl bFGF.
Generation of stable 293T cell lines
Lentivirus using pCDH-CMV-MCS-EF1-copGFP-NS2B3 was generated with WT or S135AZIKV NS2B3, then transduced into 293T cells, FACS-sorted for GFP-positive cells 4d after infection, propagated and verified for GFP-positivity, then used for cellular and biochemistry assays.
Human neural progenitor cells culture
Neural progenitor cells (NPCs) were obtained from embryoid bodies (EBs) originating from human H9 ES cells formed by mechanical dissociation of cell clusters and plated in suspension in differentiation medium (DMEM F12,1X N2,1μM Dorsomorphin (Tocris), 2 μM A8301 (Tocris) and rotated at 95 rpm for 7d as described (Li et al., 2016b), then EBs plated onto Matrigel (BD Biosciences) coated dishes in NBF medium (DMEM F12, 0.5X N2, 0.5X B27, 20 ng/μl bFGF). Rosettes were collected after 5–7d, dissociated with Accutase (Millipore), and resultant NPCs plated onto poly-ornithine/laminin (Sigma) dishes with NPC medium, and used within 1wk.
Authentication of cell lines used
We used HEK293T cells (sex typed as female) and H9 ESC (sex typed as female) that were obtained directly from ATCC, and thus they were not further authenticated. All cell lines under active culture were screened monthly for the presence of mycoplasma, and all tests were negative throughout the course of data acquisition.
METHOD DETAILS
DNA construction
The ORF of ZIKV NS2B3 WT/S135A (Shiryaev et al., 2017) was cloned to pCDH-CMV-MCS-EF1-copGFP using Gibson cloning method. SEPT2 and SEPT7 ORFs were amplified human NPC cDNA and cloned into pcDNA3.1(+) and pGEX-6P. Mutations were introduced with site-directed mutagenesis and all cloning was performed with Gibson isothermal method, then sequence-verified. Knockdown of human SEPT2 and SEPT7 was performed with Sigma MISSION® shRNA reagents. Flag-ZIKV NS3 expression construct is a gift from Hongjun Song’s lab.
Immunofluorescence
Cultured cells were permeabilized with 1% Triton X-100 in PBS (PBT), blocked with Donkey serum (Millipore) and immunostained with antibodies against flavivirus envelope protein (MAB10216, 4G2 clone 1, EMD Millipore 1:500), pH3-Ser10 (06–570, Millipore, 1:500), α-tubulin (sc-53030, Santa Cruz, 1:500), CEP63 (06–1292, Millipore, 1:250), active-Caspase-3 (AB3623, Millipore, 1:250), GFP (MA1–952, Thermo-Fisher, 1:500), SOX2 (MA1–014, Thermo-Fisher, 1:500), Nestin (NES, Aves, 1:500). Alexa Fluor® secondary antibodies (Invitrogen, 1:500) were used for detection. EdU labeling and staining were performed with the Click-iT EdU Kit (Thermo-Fisher). Cultured cells were labeled with 10μM EdU for 2 hours before fixation. EdU was detected using Alexa Fluor-488 or555-azide.
Generation of NPC cells lines with shRNA
NPC lines with SEPT2 or SEPT7 knockdown were generated via lentiviral transduction encoding shRNA against human SEPT2 or SEPT7, followed by puromycin selection (1 mg/μl) for one week. We generated two different shRNAs for each gene. Knockdown was confirmed with western blotting before using the cells for cellular and biochemistry assays.
GST-pulldown assay and proteomics
pGEX-6P-NS2B3-S135A was introduced into E. coli strain BL21(DE3), induced with 100 μM IPTG for 4h at 30oC, then purified using Pierce GST reagents, incubated with human NPC lysis to capture interacting partners, washed and eluted under manufacturer’s protocols. Eluted samples (n = 3) were SDS-PAGE-purified and excised protein bands were reduced (10mM DTT) and alkylated (30mM iodoacetamide) and digested using a mix of endopeptidase Lys-C (Wako) and trypsin (Promega). Peptides were extracted followed by StageTips (Rappsilber et al., 2007) based desalting. Each sample was analyzed using a 70 min gradient delivered at 200nL/min, increasing from 1% Bto 40% B in 70 min. Extracted peptides were analyzed by LC-MS/MS (Dionex 3000 coupled toQ-Exactive Plus, Thermo Scientific). Data were analyzed using MaxQuant v. 1.6.0.13. Co-affinity purification was analyzed by label free quantitation (LFQ). Peptide and protein FDR was 2% and 1%, respectively. Match between runs was used. Matched proteins were filtered for potential contaminations and reverse database matches. Log2 transformed LFQ values were used and it was required that any protein was measured in at least 2-of-3 replicates for at least one condition. Significant changing proteins were called using a permutation based False Discovery Rate (FDR) t test (p > 0.05) and requiring a fold difference of 4. Proteins fulfilling the above criteria were analyzed by STRING (https://string-db.org). For LC-MS/MS, peptides were separated on a packed-in-emitter C18 column (12 cm / 75 μm, 3 μm beads, Nikkyo Technologies, Japan) at 200 or 300 nl/min. Buffer A was 0.1% formic acid and Buffer B was 80% acetonitrile in 0.1% formic acid. The mass spectrometers were operated in data dependent (DDA) mode using high resolution/high mass accuracy for both MS and MS/MS, 60,000 resolution and 17,500 resolution respectively. Normalized collision energy was set at 31 μV with AGC target and maximum injection time was 2e5, and 54ms, respectively.
Mass Spectrometry Analysis of Septin-2 C-terminal fragment
Proteins were separated by SDS-PAGE. After aligning of anti-SEPT2 western blot, gel bands corresponding to full length SEPT2 and C-terminal SEPT2 fragment were excised and digested using trypsin then propionic anhydride labeling followed by digestion by either trypsin or Asp-N (Promega). Extracted peptides were analyzed by LC-MS/MS (EasyLC 1200 coupled to Fusion Lumos, Thermo Scientific), separated by reversed phase with a gradient increasing from 1% B to 40% B in 40 min. MS data were queried against Uniprots Human database (March 2016) using no enzymatic (Asp-N experiment) or semi-tryptic constraints using Proteome Discoverer ver1.4 (Thermo Fisher)/Mascot 2.4 (Matrix Science). Oxidation of methionine was allowed. For samples treated with propionic anhydride, propionylation of N-termini and lysine were allowed as variable modifications. Matched peptides were filtered using < 5% FDR (Percolator 2) and mass accuracy < 5 ppm in addition to manual validation of tandem spectra. Extracted areas of Septin-2 peptides obtained by the three different digestion strategies for intact SEPT2 and C-terminal SEPT2 were summed per residue and plotted as a function of SEPT2 using PepEx 3, after normalization to the signal of the most C-terminal residues.
In vitro protease assay
Recombinant human SEPT2 was purchased from Abcam (ab99296). Recombinant WT and S135A ZIKV NS2B3, WT and R306A human SEPT2 were purified using Pierce GST Protein Interaction Pull-down kit or Dynabeads His-Tag Isolation and Pulldown kit. Purified enzyme and substrate, after dialysis, were incubated in protease assay buffer (Tris-HCl pH8.0, 20% Glycerol) at 37oC for 1h, then analyzed by western blotting with anti-SEPT2 antibodies. Benzoyl-norleucine-lysine-lysine-arginine 7-amido-4-methylcou-marin (Bz-Nle-Lys-Lys-Arg-AMC; Genscript) was used as substrate to measure protease activity and the efficacy of different NS3 inhibitors. The fluorescence signal from released AMC was monitored at 460 nm with excitation at 360 nm.
Co-immunoprecipitation
For co-immunoprecipitation, two days after transfection, 293T cells were lysed in cell lysis buffer (50mM Tris-HCl, pH8.0, 170mM NaCl, 0.5% NP-40, protease inhibitor (Roche). Cells were continued lysed with rotation on a nutator at 4°C for 30 minutes. Anti-FLAG® M2 Magnetic Beads (M8823, Sigma) were incubated with cell lysate on a nutator at 4°C overnight. The beads were washed with lysis buffer for 3 times and finally boiled in Laemmli Sample Buffer (Bio-Rad) with 0.5% 2-Mercaptoethanol at 95°C for 10 minutes.
Western blotting
Western blotting was performed with the following antibodies: anti-SEPT2 (Proteintech, 60075, 1:2000), anti-SEPT2 (N-terminal) (Santa Cruz, 20408, 1:500), anti-SEPT2 (C-terminal) (Abcam, ab185998, 1:1000), anti-SEPT7 (Abcam, ab158073, 1:1000), anti-SEPT9 (ThermoScientific, PA5–13200, 1:1000) anti-GAPDH (EMD Millipore, MAB374, 1:2000), anti-NS2B (GeneTex, GTX124246, 1:500). Quantification of each band was performed using ImageJ software.
Time-lapse analysis
Time-lapse recording was performed using LCV110 “VivaView” Incubator epifluorescent microscope (Olympus) with 2 min-interval DIC or GFP fluorescence images taken consecutively for 24h. Images were analyzed using ImageJ software. Cytokinesis length was defined as the time period between the formation of the mitotic spindle until full separation of the two daughter cells.
ZIKV generation and infection
Stocks and strains were generated according to published methods: for in vitro experiments we used MR-766 African strain (Tang et al., 2016). ZIKV stocks were generated from virus seeds (passage 2) to infect Aedes albopictus C6/36 cells (ATCC CRL-1660), maintained in RPMI-1640 medium with 2% FBS (PAA) at 28°C for 6d, then harvested by freeze-thaw, centrifuged at 6,000 g for 15 min to remove cell debris, dispersed into single-use aliquots, and stored at −80°C. Infectious viral titer was determined by plaque assay as described (Yuan et al., 2017). hNPC were infected for 2 hours at 5.33E+06 IFU/ml and further cultured in NPC media for 3 days before analysis.
Strategy for randomization and/or stratification
No data randomization was used for this study. In Figures 4D–4H, all cells in the field of view that completed mitosis during the timelapse acquisition were included in the measurement of averages.
Blinding of datasets
In Figures 1 and 4A–4C, cells were counted by a blinded observer. In Figure 2 sample identification for mass spectrometry were blinded to the Mass Spectrometry Core prior to analysis.
Inclusion and exclusion criteria of data
We did not set prior inclusion or exclusion criteria for data.
QUANTIFICATION AND STATISTICAL ANALYSIS
Unless otherwise specified, data are presented as mean ± standard error of the mean (SEM). Significance between two groups was assessed by the Student’s two-tailed t test. Datasets consisting of more than 2 groups were analyzed by analysis of variance (ANOVA). In animal experiments, n represents number of animals utilized in each treatment group. In cytokinesis time-lapse experiment, n represents number of cells captured and analyzed in each group. Immunofluorescence analysis, quantitative PCRs, western blots and biochemical assays were performed in triplicate or four replicates. A p value that was less than 0.05 was considered statistically significant for all datasets. Bonferroni correction was used for all multiple comparisons. All statistical analysis was performed using GraphPad Prism software. No statistical methods were used to predetermine sample size.
DATA AND SOFTWARE AVAILABILITY
Protein-protein interaction network analysis of ZIKV NS2B3-SA binding partners identified from MS/MS using String (https://string-db.org/cgi/input.pl).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Flavivirus envelope protein, 4G2 clone 1 | EMD Millipore | MAB10216; RRID:AB_827205 |
| Phopho-H3-Ser10 | EMD Millipore | 06–570; RRID:AB_310177 |
| α-tubulin | Santa Cruz | sc-53030; RRID:AB_2272440 |
| CEP63 | EMD Millipore | 06–1292; RRID:AB_10918481 |
| active-Caspase-3 | EMD Millipore | AB3623; RRID:AB_303959 |
| SEPT2 | Proteintech | 60075; RRID:AB_2187016 |
| SEPT2 (N-terminal) | Santa Cruz | 20408; RRID:AB_2187133 |
| SEPT2 (C-terminal) | Abcam | ab185998 |
| SEPT7 | Abcam | ab158073 |
| SEPT9 | Thermo-Fisher | PA5–13200; RRID:AB_10987482 |
| GAPDH | EMD Millipore | MAB374; RRID:AB_2107445 |
| NS2B | GeneTex | GTX124246; RRID:AB_11170698 |
| NeuN | Abcam | ab104224; RRID:AB_10711040 |
| Bacterial and Virus Strains | ||
| Zika virus Strain FSS13025 | WRCEVA | FSS13025 |
| Zika virus Strain MR-766 | WRCEVA | MR-766 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| human SEPT2 | Abcam | ab99296 |
| Dorsomorphin | Tocris | 3093 |
| A8301 | Tocris | 2939 |
| bFGF | Thermo-Fisher | PHG0261 |
| Matrigel | BD Biosciences | 356234 |
| Accutase | EMD Millipore | SCR005 |
| TRIzol | Thermo-Fisher | 15596026 |
| laminin | Sigma | L2020 |
| puromycin | Sigma | P8833 |
| Critical Commercial Assays | ||
| Gibson Assembly® Master Mix | New England Biolabs | E2611S |
| Q5® Site-Directed Mutagenesis Kit | New England Biolabs | E0554S |
| Pierce GST Protein Interaction Pull-Down Kit | Thermo-Fisher | 21516 |
| RNeasy Mini Kit | QIAGEN | 74104 |
| SuperScript III First-Strand Synthesis System | Thermo-Fisher | 18080051 |
| iTaq Universal SYBR® Green Supermix | Bio-rad | 1725120 |
| Experimental Models: Cell Lines | ||
| HEK293T | ATCC | CRL-3216; RRID:CVCL_0063 |
| HeLa | ATCC | CCL-2; RRID:CVCL_0030 |
| C6/36 | ATCC | CRL-1660; RRID:CVCL_Z230 |
| H9 | WiCell | WA09; RRID:CVCL_9773 |
| Recombinant DNA | ||
| pCDH-ZIKV-NS2B3 | This paper | N/A |
| pCDH-ZIKV-NS2B3-S135A | This paper | N/A |
| pGEX-6P-ZIKV-NS2B3 | (Shiryaev et al., 2017) | N/A |
| pGEX-6P-ZIKV-NS2B3-S135A | (Shiryaev et al., 2017) | N/A |
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
| pSG5-FLAG-ZIKV-NS3 | (Yoon et al., 2017) | N/A |
| pcDNA3.1(+)-SEPT2 | This paper | N/A |
| pcDNA3.1(+)-SEPT2-R306A | This paper | N/A |
| MISSION® shRNA-SEPT2 | Sigma | 004404 and 010891 |
| MISSION® shRNA-SEPT7 | Sigma | 001788 and 009859 |
| pEGFP-SEPT2 | (DeMay et al., 2011) | N/A |
| pEGFP-SEPT2-R306A | This paper | N/A |
| pGEX-6P-SEPT2 | This paper | N/A |
| pGEX-6P-SEPT2-R306A | This paper | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.net/Fiji |
| GraphPad Prism | GraphPad | https://www.graphpad.com |
| String | String | https://string-db.org/cgi/input.pl |
Highlights.
Zika protease NS2B3 is a heterodimer that cleaves host proteins in hNPCs
NS2B3 expression causes protease-dependent cytokinesis defects and apoptosis
NS2B3 associates with the septin cytoskeleton and cleaves Septin-2
Non-cleavable Septin-2 partially restores cytokinesis in NS2B3-expressing cells
ACKNOWLEDGMENTS
We thankAlex Strongin from Sanford-Burnham-Prebys Discovery Institute for the ZIKV NS2B3 clone; Guo-li Ming and Hongjun Song from University of Pennsylvania for NS2B and NS3 clones; Amy Gladfelter from UNC for Septin clones; the Rockefeller University Bio-imaging Resource Center; Kevin Cannon at The University of North Carolina at Chapel Hill; Oswald Quehenberger, Aaron Armando, and MildaSimonaitis atthe UCSD Lipidomics Core; and Alysson Muotri, AlexeyTerskikh, Charles Rice, Margaret MacDonald, and Edward Holmes for discussions. H.L. is supported by a Druckenmiller Fellowship from the New York Stem Cell Foundation. The work was supported by NIH grant R01 NS106387 to J.G.G., A.V.T., and S.S. J.G.G. is an Investigatorwith the Howard Hughes Medical Institute. The Sohn Conference Foundation and the Leona M. and Harry B. Helmsley Charitable Trust are acknowledged for mass spectrometry instrumentation.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and one table and can be found with this article online at https://doi.org/10.1016Zj.neuron.2019.01.010.
A video abstract is available at https://doi.org/10.1016/j.neuron.2019.01.010#mmc4.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, et al. (2012). DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 8, e1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beites CL, Xie H, Bowser R, andTrimble WS (1999). The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat. Neurosci. 2, 434–439. [DOI] [PubMed] [Google Scholar]
- Besnard M, Eyrolle-Guignot D, Guillemette-Artur P, Lastère S, Bost-Bezeaud F, Marcelis L, Abadie V, Garel C, Moutard ML, Jouannic JM, et al. (2016). Congenital cerebral malformations and dysfunction in fetuses and newborns following the 2013 to 2014 Zika virus epidemic in French Polynesia. Euro Surveill. 21, 13. [DOI] [PubMed] [Google Scholar]
- Bianchi FT, Tocco C, Pallavicini G, Liu Y, Vernì F, Merigliano C, Bonaccorsi S, El-Assawy N, Priano L, Gai M, et al. (2017). Citron kinase deficiency leads to chromosomal instability and TP53-sensitive microcephaly. Cell Rep. 18, 1674–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler D (2016). Zika virus: Brazil’s surge in small-headed babies questioned by report. Nature 530, 13–14. [DOI] [PubMed] [Google Scholar]
- Carleton M, Mao M, Biery M, Warrener P, Kim S, Buser C, Marshall CG, Fernandes C, Annis J, and Linsley PS (2006). RNA interference-mediated silencing of mitotic kinesin KIF14 disrupts cell cycle progression and induces cytokinesis failure. Mol. Cell. Biol. 26, 3853–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers TJ, Hahn CS, Galler R, and Rice CM (1990). Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44, 649–688. [DOI] [PubMed] [Google Scholar]
- Chavali PL, Stojic L, Meredith LW, Joseph N, Nahorski MS, Sanford TJ, Sweeney TR, Krishna BA, Hosmillo M, Firth AE, et al. (2017). Neurodevelopmental protein Musashi-1 interacts with the Zika genome and promotes viral replication. Science 357, 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimelli L, Melo ASO, Avvad-Portari E, Wiley CA, Camacho AHS, Lopes VS, Machado HN, Andrade CV, Dock DCA, Moreira ME, et al. (2017). The spectrum of neuropathological changes associated with congenital Zika virus infection. Acta Neuropathol. 133, 983–999. [DOI] [PubMed] [Google Scholar]
- Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JL, Guimaraes KP, Benazzato C, Almeida N, Pignatari GC, Romero S, et al. (2016). The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534, 267–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino PP, Giansanti MG, and Petronczki M (2015). Cytokinesis in animal cells. Cold Spring Harb. Perspect. Biol. 7, a015834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang J, Tiwari SK, Lichinchi G, Qin Y, Patil VS, Eroshkin AM, and Rana TM (2016). Zikavirusdepletes neural progenitors in human cerebral or-ganoidsthrough activation ofthe innate immune receptorTLR3. Cell Stem Cell 19, 258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida Marques I, Valadares NF, Garcia W, Damalio JC, Macedo JN, de Araújo AP, Botello CA, Andreu JM, and Garratt RC (2012). Septin C-terminal domain interactions: implications for filament stability and assembly. Cell Biochem. Biophys. 62, 317–328. [DOI] [PubMed] [Google Scholar]
- de Fatima Vasco Aragao M, van der Linden V, Brainer-Lima AM, Coeli RR, Rocha MA, Sobral da Silva P, Durce Costa Gomes de Carvalho M, van der Linden A, Cesario de Holanda A, and Valenca MM (2016). Clinical features and neuroimaging (CT and MRI) findings in presumed Zika virus related congenital infection and microcephaly: retrospective case series study. BMJ 353, i1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMay BS, Bai X, Howard L, Occhipinti P, Meseroll RA, Spiliotis ET, Oldenbourg R, and Gladfelter AS (2011). Septin filaments exhibit a dynamic, paired organization that is conserved from yeast to mammals. J. Cell Biol. 193, 1065–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Gaska JM, Douam F, Wei L, Kim D, Balev M, Heller B, and Ploss A (2018). Species-specific disruption of STING-dependent antiviral cellular defenses by the Zika virus NS2B3 protease. Proc. Natl. Acad. Sci. USA 115, E6310–E6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Amine N, Kechad A, Jananji S, and Hickson GR (2013). Opposing actions of septins and Sticky on Anillin promote the transition from contractile to midbody ring. J. Cell Biol. 203, 487–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estey MP, Di Ciano-Oliveira C, Froese CD, Bejide MT, and Trimble WS (2010). Distinct roles of septins in cytokinesis: SEPT9 mediates midbody abscission. J. Cell Biol. 191, 741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Y, and Steller H (2011). Programmed cell death in animal development and disease. Cell 147, 742–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, and Rehen SK (2016). Zika virus impairs growth in human neurospheres and brain organoids. Science 352, 816–818. [DOI] [PubMed] [Google Scholar]
- Ghouzzi VE, Bianchi FT, Molineris I, Mounce BC, Berto GE, Rak M, Lebon S, Aubry L, Tocco C, Gai M, et al. (2017). ZIKA virus elicits P53 activation and genotoxic stress in human neural progenitors similar to mutations involved in severe forms of genetic microcephaly and p53. Cell Death Dis. 8, e2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi MT, and Karlseder J (2013). DNA damage associated with mitosis and cytokinesis failure. Oncogene 32, 4593–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara M, Kinoshita A, Yamada S, Tanaka H, Tanigaki A, Kitano A, Goto M, Okubo K, Nishiyama H, Ogawa O, et al. (2005). Corticalorganization by the septin cytoskeleton is essential for structural and mechanical integrity of mammalian spermatozoa. Dev. Cell 8, 343–352. [DOI] [PubMed] [Google Scholar]
- Kinoshita M, Field CM, Coughlin ML, Straight AF, and Mitchison TJ (2002). Self- and actin-templated assembly of Mammalian septins. Dev. Cell 3, 791–802. [DOI] [PubMed] [Google Scholar]
- Kremer BE, Haystead T, and Macara IG (2005). Mammalian septins regulate microtubule stability through interaction with the microtubule-binding protein MAP4. Mol. Biol. Cell 16, 4648–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J, Hansen G, Nitsche C, Klein CD, Zhang L, and Hilgenfeld R (2016). Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor. Science 353, 503–505. [DOI] [PubMed] [Google Scholar]
- Lennemann NJ, and Coyne CB (2017). Dengue and Zika viruses subvert reticulophagy by NS2B3-mediated cleavage of FAM134B. Autophagy 13, 322–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Xu D, Ye Q, Hong S, Jiang Y, Liu X, Zhang N, Shi L, Qin CF, and Xu Z (2016a). Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell 19, 120–126. [DOI] [PubMed] [Google Scholar]
- Li H, Bielas SL, Zaki MS, Ismail S, Farfara D, Um K, Rosti RO, Scott EC, Tu S, Chi NC, et al. (2016b). Biallelic mutations in citron kinase link mitotic cytokinesis to human primary microcephaly. Am. J. Hum. Genet. 99, 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Saucedo-Cuevas L, Shresta S, and Gleeson JG (2016c). The neurobiology of Zika virus. Neuron 92, 949–958. [DOI] [PubMed] [Google Scholar]
- Liang Q, Luo Z, Zeng J, Chen W, Foo SS, Lee SA, Ge J, Wang S, Goldman SA, Zlokovic BV, et al. (2016). Zika virus NS4A and NS4B proteins deregulate Akt-mTOR signaling in human fetal neural stem cells to inhibit neurogenesis and induce autophagy. Cell Stem Cell 19, 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz IC, Allison SL, Heinz FX, and Helenius A (2002). Folding and dimerization of tick-borne encephalitis virus envelope proteins prM and E in the endoplasmic reticulum. J. Virol. 76, 5480–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoTurco JJ, Sarkisian MR, Cosker L, and Bai J (2003). Citron kinase is a regulator of mitosis and neurogenic cytokinesis in the neocortical ventricular zone. Cereb. Cortex 13, 588–591. [DOI] [PubMed] [Google Scholar]
- Madaule P, Eda M, Watanabe N, Fujisawa K, Matsuoka T, Bito H, Ishizaki T, and Narumiya S (1998). Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature 394, 491–494. [DOI] [PubMed] [Google Scholar]
- Martines RB, Bhatnagar J, Keating MK, Silva-Flannery L, Muehlenbachs A, Gary J, Goldsmith C, Hale G, Ritter J, Rollin D, et al. (2016). Notes from the field: evidence of Zika virus infection in brain and placental tissues from two congenitally infected newborns and two fetal losses-Brazil, 2015. MMWR Morb. Mortal. Wkly. Rep. 65, 159–160. [DOI] [PubMed] [Google Scholar]
- McGrath EL, Rossi SL, Gao J, Widen SG, Grant AC, Dunn TJ, Azar SR, Roundy CM, Xiong Y, Prusak DJ, et al. (2017). Differential responses of human fetal brain neural stem cells to Zika virus infection. Stem Cell Reports 8, 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meseroll RA, Occhipinti P, and Gladfelter AS (2013). Septin phosphorylation and coiled-coil domains function in cell and septin ring morphology in the filamentous fungus Ashbya gossypii. Eukaryot. Cell 12, 182–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner JJ, Cao B, Govero J, Smith AM, Fernandez E, Cabrera OH, Garber C, Noll M, Klein RS, Noguchi KK, et al. (2016). Zika virus infection during pregnancy in mice causes placental damage and fetal demise. Cell 165, 1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming GL, Tang H, and Song H (2016). Advances in Zika virus research: Stem cell models, challenges, and opportunities. Cell Stem Cell 19, 690–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moawia A, Shaheen R, Rasool S, Waseem SS, Ewida N, Budde B, Kawalia A, Motameny S, Khan K, Fatima A, et al. (2017). Mutations of KIF14 cause primary microcephaly by impairing cytokinesis. Ann. Neurol. 82, 562–577. [DOI] [PubMed] [Google Scholar]
- Mostowy S, and Cossart P (2012). Septins: the fourth component of the cytoskeleton. Nat. Rev. Mol. Cell Biol. 13, 183–194. [DOI] [PubMed] [Google Scholar]
- Onorati M, Li Z, Liu F, Sousa AMM, Nakagawa N, Li M, Dell’Anno MT, Gulden FO, Pochareddy S, Tebbenkamp ATN, et al. (2016). Zika virus disrupts phospho-TBK1 localization and mitosis in human neuroepithelial stem cells and radial glia. Cell Rep. 16, 2576–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucci B, Kasten M, and Giordano A (2000). Cell cycle and apoptosis. Neoplasia 2, 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, and Ishihama Y (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906. [DOI] [PubMed] [Google Scholar]
- Shao Q, Herrlinger S, Yang SL, Lai F, Moore JM, Brindley MA, and Chen JF (2016). Zika virus infection disrupts neurovascular development and results in postnatal microcephaly with brain damage. Development 143, 4127–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo A, and Wallingford JB (2014). PCP and septins compartmentalize cortical actomyosin to direct collective cell movement. Science 343, 649–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiryaev SA, Farhy C, Pinto A, Huang CT, Simonetti N, Elong Ngono A, Dewing A, Shresta S, Pinkerton AB, Cieplak P, et al. (2017). Characterization of the Zika virus two-component NS2B-NS3 protease and structure-assisted identification of allosteric small-molecule antagonists. Antiviral Res. 143, 218–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M, Farkasovsky M, Hauer F, Kuhlmann D, Macara IG, Weyand M, Stark H, and Wittinghofer A (2007). Structural insight into filament formation by mammalian septins. Nature 449, 311–315. [DOI] [PubMed] [Google Scholar]
- Souza BS, Sampaio GL, Pereira CS, Campos GS, Sardi SI, Freitas LA, Figueira CP, Paredes BD, Nonaka CK, Azevedo CM, et al. (2016). Zika virus infection induces mitosis abnormalities and apoptotic cell death of human neural progenitor cells. Sci. Rep. 6, 39775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliotis ET, Kinoshita M, and Nelson WJ (2005). A mitotic septin scaffold required for Mammalian chromosome congression and segregation. Science 307, 1781–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata T, Petitt M, Puerta-Guardo H, Michlmayr D, Wang C, Fang-Hoover J, Harris E, and Pereira L (2016). Zika virus targets different primary human placental cells, suggesting two routes for vertical transmission. Cell Host Microbe 20, 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, Yao B, Shin J, Zhang F, Lee EM, et al. (2016). Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell 18, 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe J, Inder T, Darras B, de Vries LS, du Plessis A, Neil J, and Perlman J (2018). Volpe’s Neurology of the Newborn (Elsevier; ). [Google Scholar]
- Weirich CS, Erzberger JP, and Barral Y (2008). The septin family of GTPases: architecture and dynamics. Nat. Rev. Mol. Cell Biol. 9, 478–489. [DOI] [PubMed] [Google Scholar]
- Wengler G, and Wengler G (1991). The carboxy-terminal part of the NS 3 protein of the West Nileflavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology 184, 707–715. [DOI] [PubMed] [Google Scholar]
- Wolf B, Diop F, Ferraris P, Wichit S, Busso C, Missé D, and Gönczy P (2017). Zika virus causes supernumerary foci with centriolar proteins and impaired spindle positioning. Open Biol. 7, 160231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KY, Zuo GL, Li XF, Ye Q, Deng YQ, Huang XY, Cao WC, Qin CF, and Luo ZG (2016). Vertical transmission of Zika virus targeting the radial glial cells affects cortex development of offspring mice. Cell Res. 26, 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Vessey JP, Konecna A, Dahm R, Macchi P, and Kiebler MA (2007). The GTP-binding protein Septin 7 is critical for dendrite branching and dendritic-spine morphology. Curr. Biol. 17, 1746–1751. [DOI] [PubMed] [Google Scholar]
- Yamshchikov VF, and Compans RW (1995). Formation of the flavivirus envelope: role of the viral NS2B-NS3 protease. J. Virol. 69, 1995–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Song G, Qian X, Pan J, Xu D, Rho HS, Kim NS, Habela C, Zheng L, Jacob F, et al. (2017). Zika-virus-encoded NS2Adisruptsmamma-lian cortical neurogenesis by degrading adherensjunction proteins. Cell Stem Cell 21, 349–358.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CY, Chang TH, Liang JJ, Chiang RL, Lee YL, Liao CL, and Lin YL (2012). Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog. 8, e1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Huang XY, Liu ZY, Zhang F, Zhu XL, Yu JY, Ji X, Xu YP, Li G, Li C, et al. (2017). A single mutation in the prM protein of Zika virus contributes to fetal microcephaly. Science 358, 933–936. [DOI] [PubMed] [Google Scholar]
- Zare Mehrjardi M, Poretti A, Huisman TA, Werner H, Keshavarz E, and Araujo Junior E (2017). Neuroimaging findings of congenital Zika virus infection: a pictorial essay. Jpn. J. Radiol. 35, 89–94. [DOI] [PubMed] [Google Scholar]
- Zhang F, Hammack C, Ogden SC, Cheng Y, Lee EM, Wen Z, Qian X, Nguyen HN, Li Y, Yao B, et al. (2016). Molecular signatures associated with ZIKV exposure in human cortical neural progenitors. Nucleic Acids Res. 44, 8610–8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Singh NJ, and Kim KS (2006). Homology modeling and molecular dynamics study ofWest Nilevirus NS3 protease: a molecular basisforthe catalytic activity increased by the NS2B cofactor. Proteins 65, 692–701. [DOI] [PubMed] [Google Scholar]
- Zuo Z, Liew OW, Chen G, Chong PC, Lee SH, Chen K, Jiang H, Puah CM, and Zhu W (2009). Mechanism of NS2B-mediated activation of NS3pro in dengue virus: molecular dynamics simulations and bioassays. J. Virol. 83, 1060–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Protein-protein interaction network analysis of ZIKV NS2B3-SA binding partners identified from MS/MS using String (https://string-db.org/cgi/input.pl).


