Abstract
Background
The low expression of miR93/25 (members of miR-106b~25 cluster) promoted the invasion and metastasis of colon cancer cells, which predicted poor survival. However, the role of miR-106b-5p, the member of miR-106b~25 cluster, in colorectal cancer (CRC) remains unclear.
Methods
Bioinformatics methods were used to predict the potential pairs of lncRNA-miRNA-mRNA. In situ hybridization and qPCR were used to evaluate the expression of MALAT1 and miR-106b-5p in the paraffin-embedded normal and CRC tissues. Kaplan–Meier analysis with the log-rank test was used for survival analyses. Immunohistochemistry staining was applied to investigate the expression of SLAIN2. Fluorescence recovery after photobleaching assay was applied to observe the microtubule (MT) mobility. In vitro and in vivo invasion and metastasis assays were used to explore the function of MALAT1/miR-106b-5p/SLAIN2 in the progression of CRC.
Findings
miR-106b-5p was identified as a suppressor in CRC. Functionally, ectopic or silencing the expression of miR-106b-5p inhibited or promoted the invasion and metastasis of CRC cells in vitro and in vivo. The long non-coding RNA MALAT1 regulated the miR-106b-5p expression and further mediated the mobility of SLAIN2-related MTs by functioning as a competing endogenous RNA in vitro and in vivo, which resulted in the progression of CRC. Clinically, low miR-106b-5p expression predicted poor survival of CRC patients, especially in combination with high MALAT1/ SLAIN2 expression.
Interpretation
miR-106b-5p served as a suppressor in combination with MALAT1/miR-106b-5p/SLAIN2, which might be a group of potential prognostic biomarkers in the prognosis of CRC.
Fund
This work was supported by National Program Project for Precision Medicine in National Research and Development Plan of China (2016YFC0905300), National Natural Science Foundation of China (81572930), National Key Research and Development Program of the Ministry of Science and Technology of China (2016YFC0905303, 2016YFC1303200), Beijing Science and Technology Program (D17110002617004), Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2018PT32012), CAMS Innovation Fund for Medical Sciences (CIFMS) (2016-I2M-1-001), Incentive Fund for Academic Leaders of Oncology Hospital, Chinese Academy of Medical Sciences (RC2016003), and Beijing Hope Run Special Fund from Cancer Foundation of China (LC2017A19). The project of Shanghai Jiaotong Univversity (YG2017QN30).
Keywords: miR-106ba-5p, MALAT1, Microtubules mobility, Colorectal cancer progression
Research in context.
Evidence before this study
CRC is the fourth leading cause of cancer-related deaths worldwide, which mainly resulted from tumor metastasis. The discovery of metastasis-associated markers and elucidating the mechanisms underlying the invasion and metastasis of CRC is urgently needed. Previous studies reported that the low expression of miR93/25 (members of miR-106b∼25 cluster) promoted the invasion and metastasis of colon cancer cells, which predicted poor survival. However, as the member of miR-106b~25 cluster, the role of miR-106b-5p in the metastasis of CRC is yet to be clarified.
Added value of this study
The in vivo and in vitro experiments identified miR-106b-5p as a suppressor, and MALAT1/miR-106b-5p/SLAIN2 axis played a significant role in promoting CRC.
Implications of all the available evidence
The MALAT1/miR-106b-5p/SLAIN2 axis consists of a group of potential prognostic and promising diagnostic biomarkers in the progression of CRC.
Alt-text: Unlabelled Box
1. Introduction
Colorectal cancer (CRC) is the fourth leading cause of cancer-related deaths worldwide [1], primarily originating from tumor metastasis [2], which is known to be involved in multi-genes and processes [3]. The aberrant regulation of oncogenes and suppressors play critical roles in the progressive cancers. Although several oncogenes or suppressors have been reported to guide the postoperative adjuvant chemotherapy, approximately 50% of CRC patients suffer from metastasis after the operation. Thus, identifying new prognostic markers and the mechanisms underlying the invasion and metastasis of CRC is an urgent requirement.
MicroRNAs (miRNAs, 18–24 nucleotides) are small non-coding RNAs that suppress the expression of targeted genes via binding to the 3-untranslated regions (3′UTRs) and regulate the progression of cancer [4]. Previously, we reported that as members of the miR-106b ∼ 25 cluster, miR25and miR93 were downregulated in colon cancer and the low expression of miR93/25 promoted the invasiveness and metastasis of colon cancer cells [5].Similarly, miR-106b-5p, also as the member of miR-106b ∼ 25 cluster, is overexpressed in various chemo/radioresistant cancers, including clear cell renal cell carcinoma (ccRCC) and CRC. Furthermore, miR-106b-5p overexpression induces cell radioresistance via the PTEN/PI3K/AKT pathways and p21 in colorectal cancer [6]. Moreover in ccRCC, characterized by exceptionally high resistance to radiation and chemotherapy, miR-106b-5p promotes renal cell carcinoma aggressiveness and stem-cell-like phenotype by activating Wnt/β-catenin signaling [7]. In addition, miR-106b-5p was found to be upregulated in ccRCC patients, and by inhibiting the function of the TRIM8 protein, the tumor suppressor activity of p53 is prevented, thereby enhancing the N-Myc pathway [8]. Thus, the miR-106b-5p might exert a dual role as a tumor suppressor or as an oncogene, thereby requiring further exploration of its roles in CRC.
Recently, accumulating evidence has shown that long non-coding RNAs (lncRNAs) are involved in cancer metastasis as competing endogenous RNAs (ceRNAs), followed by sponging and inhibiting the expression of miRNAs expression that activates the downstream targets [9]. Typically, the lncRNAs are longer than 200 nucleotides and lack the protein-coding functions. In addition, lncRNAs are known to be involved in a series of biological processes, such as chromatin modification, genomic imprinting, and the regulation of transcription and post-transcription [10]. In colon cancer, lncRNA HNF1A-AS1-mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression by functioning as a ceRNA [11]. Linc00152 functions as a ceRNA that confers oxaliplatin resistance with prognostic values in colon cancer [12]. In renal cancer, exposure-transmitted lncARSR promotes sunitinib resistance by acting as a ceRNA [13]. Therefore, the ce-lncRNAs that are involved in the progression of CRC are promising prognostic factors in the disease. Whether miR106b-5p is regulated by lncRNAs via ceRNA roles also need further exploration.
In the present study, we demonstrated that 1) miR-106b-5p has a lower expression in stage IV than in stage I-II in CRC tissues; 2) low miR-106b-5p indicated a short survival in CRC patients; 3) MALAT1 sponges miR-106b-5p to promote the invasion and metastasis of CRC via SLAIN2-enhanced mobility of the MTs; 4) MALAT1/miR-106b-5p-mediated SLAIN2 overexpression predicted the poor survival for CRC patients. The current data clarified the sponging role of MALAT1/miR-106b-5p in CRC metastasis, and thus, might provide novel prognostic factors for CRC metastasis.
2. Results
2.1. Low expression of miR-106b-5p is positively correlated to tumor progression and predicts poor survival in CRC patients
In order to understand the role of miR-106b-5p in CRC, we analyzed the expression of miR-106b-5p in CRC in The Cancer Genome Altas (TCGA) dataset. The level of miR-106b was significantly decreased in tumor tissues (n = 340) as compared to normal tissues (n = 11) (Fig. S1a). Subsequently, we assessed the level of miR-106b-5p in 95 pairs of CRC and normal samples obtained from Shanghai General Hospital and found that the expression was significantly lower in the tumor than that in the paired normal tissues (Fig. 1a). Furthermore, 52/95 patients were diagnosed as stage I-II, and the remaining 43 were diagnosed as stage III-IV according to the American Joint Committee on Cancer (AJCC). Based on the relapse, the 95 cases were also divided into two groups: relapse and non-relapse groups. Interestingly, qPCR showed that miR-106b-5p expression was much higher in both stage I-II (8.05 ± 2.06) and non-relapse group (8.02 ± 1.95) as compared to stage III-IV (6.81 ± 1.42) and relapse groups (6.57 ± 1.40) (Fig. 1b and c). Interestingly, the correlation between the clinicopathological parameters of the 95 CRC patients and miR-106b-5p expression revealed that the expression of miR-106b-5p was negatively associated with lymph node metastasis, distant metastasis, and tumor relapse (Table S1). These results indicated that low miR-106b-5p might be associated with CRC metastasis and relapse.
Fig. S1.
The expression and prognosis analysis of miR-106b according to TCGA database. (a) The expression levels of miR-106b in the TCGA CRC dataset. (b) Kaplan-Meier curves for OS of CRC patients in the TCGA dataset (n = 328).
Fig. 1.
Low expression of miR-106b-5p is correlated to tumor progression and predicts poor survival in CRC patients. (a) The expression levels of miR-106b-5p in 95 pairs of CRC and normal tissues by qPCR analyses. (b) Relative miR-106b-5p expression levels in stage I-II and stage III-IV tumor tissues. (c) Relative miR-106b-5p expression levels in tumor relapse and non-relapse groups. (d–e) Kaplan–Meier analysis with a log-rank test for OS (d) and RFS (e) in 95 CRC patients according to miR-106b-5p expression. (f–g) Kaplan–Meier analysis with a log-rank test for OS in non-relapse groups (f) and relapse groups (g). Box and whiskers plot: min to max; **P < .01; ***P < .001, two-tailed Student t-test.
Next, Kaplan–Meier analysis with the log-rank test on the overall survival (OS) of the tumor, using the TCGA dataset, showed that low miR-106b-5p predicted a poor OS in CRC patients (Fig. S1b). Similarly, low miR-106b-5p also showed a poor OS and recurrence-free survival (RFS) in 95 CRC patients of Shanghai General Hospital dataset (Fig. 1d and e). Moreover, miR-106b-5p expression only affected the OS of the relapse group but not that of the non-relapse group (Fig. 1f and g). Taken together, these results indicated that low miR-106b-5p predicted the metastasis of CRC and poor prognosis.
2.2. Knockdown of miR-106b-5p promotes CRC invasion in vitro and in vivo
To explore the biological roles of miR-106b-5p in CRC progression, we analyzed the expression of miR-106b-5p in seven CRC cell lines (Caco2, HT-29, SW480, SW620, RKO, HCT-8, and LoVo) and one normal colon epithelial cell (NCM 460). As shown in Fig. S2a, the level of miR-106b-5p was lower in CRC cells than that in NCM 460 cells, and HCT-8 showed the lowest miR-106b-5p level, while SW480 presented a relatively high miR-106b-5p level; hence, these cells were selected for the following study (Fig. S2a). Subsequently, the mimics and inhibitors of miRNA-106b-5p were used to upregulate or knockdown miR-106b-5p in CRC cells, respectively (Fig. S2b and c, P < .01 for both, Student's t-test). Then, transwell assay was performed to measure the effects of miR-106b-5p on the CRC cells' migratory (without Matrigel) and invasiveness (with Matrigel) capacities. We found that the knockdown or overexpression of miR-106b-5p significantly increased or decreased the migratory and invasive capacities of SW480 or HCT-8 cells, respectively (Fig. 2a–d, both P < .05, Student's t-test). These data demonstrated that the low expression of miR-106b-5p promoted the migration and invasion of CRC cells in vitro.
Fig. S2.
miR-106b-5p is downregulated in CRC cell lines. (a) The expression of miR-106b-5p in one normal intestinal epithelial cell and 7 CRC cell lines by qPCR analyses. (b–c) Changes of miR-106b-5p expression in miR-106-5p-mimics treated HCT-8 cells (b) and in miR-106b-5p inhibitor treated SW480 cells (c). Error bars denote s.d. of triplicates. **P < .01. Student's t-test.
Fig. 2.
Low expression of miR-106b-5p promotes CRC migration and invasion in vitro and metastasis in vivo. (a–b) The effects of miR-106b-5p downregulation on migratory (a) and invasive (b) capabilities in SW480 cells. (c–d) The effects of miR-106b-5p overexpression on migratory (c) and invasive (d) abilities in HCT-8 cells. Original magnification × 200; scale bar 50 μm. (e) Representative image of ultrasound detection (left panel) and HE staining (right panel; original magnification × 400; scale bar 50 μm) of liver metastasis in nude mice three weeks after the injection of CRC cells via tail vein; tumor node (red and black arrows); normal liver cell (green arrow). (f) Nude mice was divided into four groups (n = 5) and injected with miR-106b-5p silenced-SW480, miR-106b-5p overexpressed-HCT-8 or their controls. The number of tumor colony in the liver of nude mice was counted with ultrasound detection. Error bars in Fig. f denote s.d. of each group (5 mice). In remaining cases, error bars denote s.d. of triplicates. *P < .05; **P < .01 ***P < .001, Student's t-test. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The liver metastasis of CRC is the most common metastasis in vivo. Thus, we performed an in vivo assay to investigate the effects of miR-106b-5p on tumor liver metastasis by injecting miR-106b-5p-silenced SW480 and miR-106b-5p -overexpressed HCT-8 cells as well as their controls into the tail veins of nude mice. Subsequently, ultrasonography was used to monitor the liver metastasis for three weeks and continually up to week 9 (Fig. 2e, left panel). Furthermore, hematoxylin-eosin (HE) staining was performed to count the number of tumor colonies in mice liver samples (Fig. 2e, right panel). The knockdown or overexpression of miR-106b-5p gave rise to a significant difference of colony number in the liver as compared to the controls in nude mice, respectively (Fig. 2f, both P < .05, Student's t-test). Taken together, these data further indicated that low miR-106b-5p expression also induced the invasion and metastasis of CRC cells in vivo.
2.3. MALAT1 functions as a ceRNA by sponging miR-106b-5p
Long non-coding RNAs are identified as a critical regulators in tumor genesis and metastasis. Accumulating evidence demonstrated that lncRNAs regulate miRNA and mRNA expression through competitively sponging miRNAs. To predict the potential lncRNAs that regulate miR-106b-5p, we selected the pairs of lncRNA-miRNA that were supported by at least three algorithms from StarBase and also filtered the pairs of lncRNA-miRNA from miRcode database. Next, we selected 16 lncRNAs from both public databases as the potential upstream regulators (Table S2). Among the 16 candidates, 4 lncRNAs (HOTAIR, MALAT1, XIST, and ZNRD1-AS1) have been widely reported to be cancer-related lncRNAs, which might be associated with miR-106b-5p. Furthermore, we analyzed these four lncRNAs in CRC tissues to investigate whether they could regulate miR-106b-5p. qPCR was used to analyze the level of these four lncRNAs in 20 pairs of normal and CRC tissues. We found that 3/4 lncRNAs have an elevated expression in the CRC samples as compared to the normal. The fold-change in the expression of four lncRNAs (HOTAIR, MALAT1, XIST, and ZNRD1-AS1) in CRC and normal tissues were 3.05, 3.46, 1.47, and 2.09, respectively (Fig. S3a). Strikingly, the level of MALAT1 in cases of stage III-IV was much higher than that of stage I-II, while HOTAIR did not show this difference (Fig. S3b). MALAT1 may be more closely related to CRC metastasis than HOTAIR. Then, we evaluated MALAT1 expression in eight cell lines (Fig. S3c). The further correlation analysis indicated that MALAT1 expression was negatively associated with miR-106b-5p expression (Fig. S3d). These results suggested that expression of MALAT1 was associated with miR-106b-5p level and MALAT1 may be a potential regulator of miR-106b-5p in CRC.
Fig. S3.
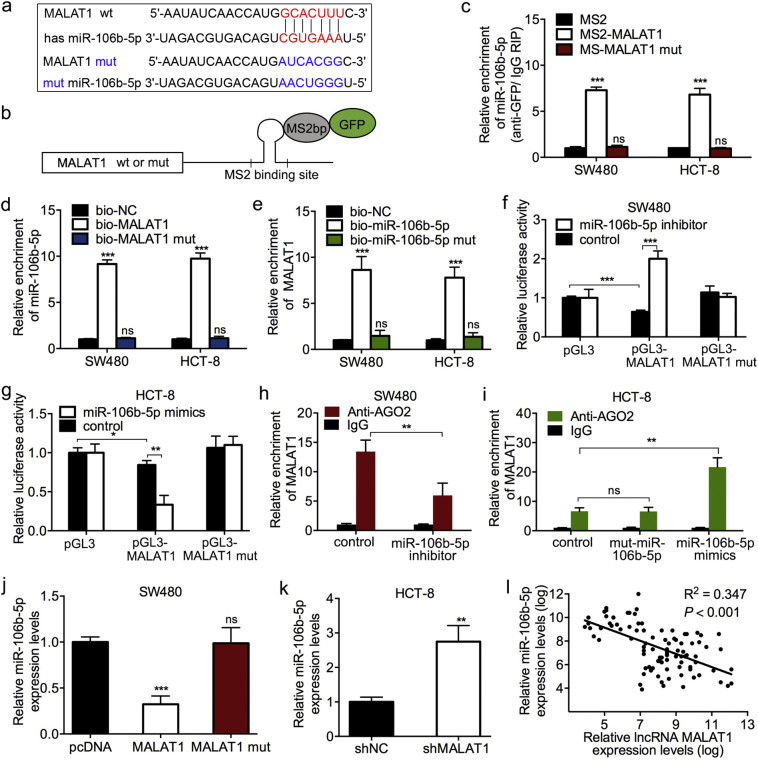
MALAT1 is the potential ceRNA of miR-106b-5p. (a) The expression of four miR-106b-5p associated lncRNAs predicted by bioinformatics in 20 pairs of CRC and normal tissues by qPCR analysis. (b) Relative HOTAIR and MALAT1 expression levels in stage I-II and stage III-IV tumor tissues. (c) MALAT1 expression levels in one normal intestinal epithelial cell and 7 CRC cell lines. (d) Correlation analysis (Pearson correlation) of MALAT1 and miR-106b-5p expression levels in 8 cell lines. (e–f) Changes of MALAT1 levels in MALAT1 plasmid transfected SW480 cells (e) and shMALAT1 transfected HCT-8 cells (f). (g) RNA fluorescence in situ hybridization (FISH) assay for MALAT1 and miR-106b-5p. Box and whiskers plot: min to max. Error bars denote s.d. of triplicates. *P < .05; ***P < .001. Student t-test.
In addition, SW480 and HCT8 cells were selected to establish the MALAT1-overexpression and knockdown cells (Fig. S3e–f). The bioinformatics analysis showed that MALAT1 contains a binding site of miR-106b-5p (Fig. 3a). We further mutated the binding sites of MALAT1 and miR-106b-5p, synthesizing the MALAT1 mut and miR-106b-5p mut plasmids. Also, the constructs containing MALAT1 (wild-type or mutant type) transcripts combined with the MS2 binding site were generated (Fig. 3b) and co-transfected into CRC cells with a construct containing MS2-binding protein (MS2bp, binds to the MS2 binding site) and GFP. An anti-GFP RNA immunoprecipitation (RIP) assay was performed, and the results showed that miR-106b-5p was only enriched in wild-type MALAT1, while the enrichment caused by mut MALAT1 was not significant as compared to the MS2 control (Fig. 3c). These results revealed that miR-106b-5p specifically binds to MALAT1. To further verify the correlation between miR-106b-5p and MALAT1, the RNA pull-down assay revealed that miR-106b-5p could be pulled down by biotin-labeled wild-type MALAT1. Also, MALAT1 could be pulled down by biotin-labeled wild-type miR-106b-5p, while the mutation in MALAT1 and miR-106b-5p resulted in the failure of pulldown (Fig. 3d and e). These data validated the binding roles between miR-106b-5p and MALAT1 in CRC cells.
Fig. 3.
MALAT1 functions as a competing endogenous RNA by sponging miR-106b-5p. (a) Predicted paired bases of miR-106b-5p in MALAT1. Mut type of MALAT1 and miR-106b-5p was generated by the mutation at the paired bases. (b) Schematic images of a construction containing MALAT1 wild or mutant type combined with MS2 binding sequence. (c) MS2-RIP followed by miR-106b-5p qPCR to measure miR-106b-5p endogenously associated with MALAT1. (d) SW480 and HCT-8 cell lysates were incubated with biotin-labeled MALAT1, qPCR analyses miR-106b-5p expression in the products of pulldown by biotin. (e) SW480 and HCT-8 cell lysates were incubated with biotin-labeled miR-106b-5p. qPCR analyses for MALAT1 expression in the products of pulldown. (f–g) Effects of miR-106b-5p knock down or overexpression on luciferase reporter activity with the wild-type and mutated MALAT1. (h–i) AGO2-RIP followed by qPCR to evaluate MALAT1 level after miR-106b-5p knockdown or overexpression. (j–k) Relative miR-106b-5p expression levels in SW480 cells transfected with MALAT1 or mut MALAT1 and HCT-8 cells transfected with shMALAT1. (l) The correlation analysis (Pearson's correlation) of MALAT1 and miR-106b-5p expression in 95 CRC tissues. Error bars in Fig. c–k denote s.d. of triplicates. **P < .01; ***P < .001; ns for no significance. Student's t-test.
Subsequently, a luciferase reporter construct containing MALAT1 (wild-type or miR-106b-5p binding site mutated type) was generated and co-transfected into SW480 cells or HCT-8 cells with miR-106b-5p inhibitor or miR-106b-5p mimics, respectively. Moreover, the luciferase assay showed that knockdown or overexpression of miR-106b-5p enhanced or inhibited the luciferase activity of wild-type MALAT1 but it does not affect the mutated type of MALAT1 (Fig. 3f and g). AGO2 is essential in miRNA-induced posttranscriptional repression or degradation of RNA to form an RNA-induced silencing complex (RISC) combined with the miRNA targets [14], which was confirmed with an anti-AGO2 RIP assay. Endogenous MALAT1 enrichment was decreased or increased after knockdown or overexpression of miR-106b-5p, which was not altered with the transfection of mut-miR-106b-5p (Fig. 3h and i). These results indicated that MALAT1 was recruited to AGO2-related miR-106b-5p-induced silencing complexes, thereby suggesting that MALAT1 interacts with miR-106b-5p.
To further elucidate the correlation between MALAT1 and miR-106b-5p, CRC cells were transfected with MALAT1 overexpression plasmids or shMALAT1 (Fig. S3e and f). The qPCR assay demonstrated that miR-106b-5p expression was reduced or upregulated in MALAT1-overexpressed SW480 or MALAT1-silenced HCT-8 cells but not changed in MALAT1-mut-transfected SW480 cells (Fig. 3j and k). Moreover, MALAT1 expression was negatively correlated with miR-106b-5p expression in 95 CRC tissues (Fig. 3l). RNA fluorescence in situ hybridization (FISH) assay was performed for their cellular localization. Even though MALAT1 is reported to be located in the nucleus, our results revealed MALAT1 and miR-106b-5p were highly enriched in the cytoplasm of CRC cells (Fig. S3g). Together, these data indicated that MALAT1 sponges miR-106b-5p and might function as ceRNA in CRC.
2.4. MALAT1 regulates the expression of SLAIN2 through competitive interaction with miR-106b-5p
To confirm the target gene of miR-106b-5p, we analyzed five miRNA datasets and predicted nine candidates that existed in all these five miRNA datasets (Fig. 4a). miR-106b-5p mimics were transfected into HCT-8 cells, followed with a qPCR analysis of the nine targets. As shown in Fig. S4a, SLAIN2 was the most significantly downregulated gene. Furthermore, we evaluated the expression of SLAIN2 mRNA in eight cell lines (Fig. S4b), and the correlation analysis showed that SLAIN2 was negatively associated with miR-106b-5p (Fig. S4c). Then, we established the luciferase reporter construct containing wild-type or mutant SLAIN 3′UTR (Fig. 4b). Also, we synthesized the mut-miR-106b-5p with the bases paired with mut-SLAIN2 3′UTR (Fig. 4b). The luciferase activity was significantly increased or decreased after treatment with miR-106b-5p inhibitor or mimics in a dose-dependent manner (Fig. 4c). Additionally, the luciferase activity of wild-type, but not the mutant type, SLAIN2 3′UTR could be enhanced or reduced by miR-106b-5p inhibitor or mimics (Fig. 4d and e). However, mut-miR-106b-5p could attenuate the luciferase activity of mutant SLAIN2 3′UTR (Fig. 4e). Furthermore, both mRNA and protein levels of SLAIN2 were upregulated or downregulated after cells treated with miR-106b-5p inhibitor or mimics (Fig. 4f–i). These results confirmed that SLAIN2 is a target of miR-106b-5p and the 3′UTR can specifically bind miR-106b-5p in CRC cells.
Fig. 4.
MALAT1 regulates the expression of SLAIN2 through competitive interaction with miR-106b-5p. (a) Potential targets of miR-106b-5p predicted with five miRNA target databases. (b) Schematic diagram of the luciferase reporter containing SLAIN2 3′UTR. Mutations were generated at the predicted miR-106b-5p-binding sites. (c) Roles of gradient concentration of miR-16b-5p inhibitor or mimics on the luciferase reporter activity with the wild-type SLAIN2 3′UTR. (d–e) Effects of miR-106b-5p overexpression or knock down on luciferase reporter activity with the wild-type and mutated SLAIN2 3′UTR. (f–g) SLAIN2 RNA expression in miR-106b-5p-overexpressed SW480 and silenced HCT-8 cells. (h–i) SLAIN2 protein levels in the above cells. (j) MALAT1 and SLAIN2 share the same miRNA binding site. (k–l) Luciferase activity of SLAIN2 wild-type or mutated 3′UTR in SW480 cells co-transfected with MALAT1, MALAT1 mut or MALAT1 + miR-106b-5p (k) and HCT-8 cells co-transfected with shNC, shMALAT1 or shMALAT1+ miR-106b-5p (l). (m–n) SLAIN2 RNA expression level in SW480 cells transfected with MALAT1, MALAT1 mut or MALAT1 + miR-106b-5p (m) and HCT-8 cells transfected with shNC, shMALAT1 or shMALAT1+ miR-106b-5p (n). (o) Western blot analysis of SLAIN2 expression in the above cells. (p) Representative IHC staining of SLAIN2 in 95 CRC tissues, original magnification × 200; scale bar 100 μm. (q–r) The correlation analyses (Pearson's correlation) of miR-106b-5p expression (q) or MALAT1 level (r) and SLAIN2 IHC scores in 95 CRC samples. Error bars denote s.d. of triplicates. *P < .05; **P < .01; ***P < .001; ns means no significance. Student's t-test.
Fig. S4.
SLAIN2 is the potential targets of miR-106b-5p. (a) qPCR analysis for the 9 predicted targets of miR-106b-5p in HCT-8 cells transfected with miR-106b-5p mimics. (b) Expression levels of miR-106b-5p in one normal intestinal epithelial cell and 7 CRC cell lines. (c) Correlation analysis (Pearson correlation) of miR-106b-5p and SLAIN2 mRNA expression levels in 8 cell lines.
Herein, we demonstrated that MALAT1 functioned as a ceRNA by sponging miR-106b-5p in the above assay. Interestingly, MALAT1 and SLAIN2 share the same miR-106b-5p binding site (Fig. 4j). We also found that miR-106b-5p regulated the SLAIN2 expression through binding to the 3′UTR. Consequently, we speculated whether MALAT1 regulated the SLAIN2 expression via sponging miR-106b-5p and promoted the progression of CRC. To verify this hypothesis, the luciferase assay was performed in CRC cells co-transfected with SLAIN2 3′UTR luciferase construct and MALAT1 or shMALAT1. The results showed that the overexpression of wild-type MALAT1, but not the mutant MALAT1, increased the luciferase activity of wild-type SLAIN2 3′UTR, and this promoted effect could be attenuated by miR-106b-5p mimics (Fig. 4k). Moreover, the luciferase activity of mut SLAIN2 3′UTR could not be regulated by MALAT1 (Fig. 4k). Conversely, the depletion of MALAT1 could decrease the luciferase activity of wild-type SLAIN2 3′UTR, but not mutant SLAIN2 3′UTR, and this decreasing effect could be reversed by miR-106b-5p inhibitors (Fig. 4l). To further understand the regulation of MALAT1 on the endogenous SLAIN2 in CRC cells, we conducted qPCR and Western blot assays and evaluated the roles of MALAT1 on the mRNA and protein levels of SLAIN2. Consecutively, the ectopic expression of wild-type MALAT1, but not the mutant MALAT1, could upregulate the mRNA level of SLAIN2, which could be attenuated by transfecting with miR-106b-5p mimics (Fig. 4m). In contrast, the knockdown of MALAT1 downregulated the SLAIN2 mRNA expression, and miR-106b-5p inhibitor partially rescued this downregulation effect (Fig. 4n). In addition, the protein levels of SLAIN2 similar to that of the mRNA (Fig. 4o). These data demonstrated that MALAT1 regulated the expression of SLAIN2 through competitive interaction with miR-106b-5p and inhibiting the miR-106b-5p activity in CRC cells.
Clinically, we measured the expression of SLAIN2 in 95 CRC tissues using immunohistochemistry (IHC) staining and found that the expression was positively correlated with the TNM stage (Fig. 4p and Table S4). A subsequent correlation analysis of SLAIN2 IHC scores and MALAT1 and miR-106b-5p expression indicated that SLAIN2 expression was negatively associated with miR-106b-5p expression, while positively correlated with MALAT1 expression (Fig. 4q and r, both P < .05, Pearson's correlation). Together, these results suggested that MALAT1 plays a critical role in the regulation of SLAIN2 expression by directly sponging miR-106b-5p in CRC.
2.5. MALAT1 sponges miR-106b-5p to promote the invasion and metastasis via SLAIN2-mediated the enhanced MTs mobility
Reportedly, SLAIN2 promotes mesenchymal cell invasion through the regulation of MTs growth [15]. MTs can modify cell morphology by regulating the polarity and adhesion of the cell, thereby resulting in pseudopod-based invasion [16]. The dynamics of MTs were associated with the migration and invasion of tumor cells [17]. Whether MALAT1 could mediate SLAIN2 involved in the enhancement of MTs mobility by sponging miR-106b-5p, followed by the invasion and metastasis of CRC is worth exploring. For further study, we firstly transfected Lv-SLAIN2 plasmid into SW480 cells to overexpress SLAIN2 (Fig. 5a) and performed transwell assays. Consequently, the overexpression of SLAIN2 increased the number of the migrated and invaded SW480 cells (Fig. 5b–c, both P < .001, Student's t-test). Conversely, the knockdown of SLAIN2 reduced the number of migrated and invasive HCT-8 cells. (Fig. 5d–f, both P < .01, Student's t-test). Moreover, in vivo assays verified that the ectopic expression or depletion of SLAIN2 increased or decreased the number of tumor colonies in the liver after injecting CRC cells into the tail vein of mice (Fig. 5g and h). Subsequently, the fluorescence recovery after photobleaching (FRAP) technique was employed to evaluate the speed of MT turnover to record the MT dynamics. GFP-α-tubulin and Lv-SLAIN or shSLAIN2 plasmids were co-transfected into CRC cells, resulting in stably expressed cell lines for the FRAP assay. The current data showed that the upregulation of SLAIN2 in SW480 cells promoted fluorescence recovery, including an increased mobile fraction and a decreased half-life after photobleaching, while the depletion of SLAIN2 in HCT-8 cells decreased the mobility and increased the half-life (Fig. 5i and j). Next, Western blot assay was used to measure the expression of the acetylated α-tubulin, a maker of MT stability. We found that acetylated α-tubulin was downregulated or upregulated after overexpression or knockdown of SLAIN2 in SW480 or HCT8 cells, respectively (Fig. 5k). Thus, these findings suggested that SLAIN2 facilitate the invasion and metastasis of CRC cells via regulation of the growth of MTs.
Fig. 5.
SLAIN2 promotes CRC invasion and metastasis via facilitates microtubules mobility. (a) The SLAIN2 expression evaluated by Western blot after transfected with Lv-SLAIN2 in SW480 cells. (b–c) The effects of SLAIN2 overexpression on migration and invasion.in SW480 cells. Original magnification × 200; scale bar 50 μm. (d) SLAIN2 expression levels were tested by western blot after transfected with three shRNAs in HCT-8 cells. (e–f) The roles of SLAIN2 knock-down on migratory and invasive capabilities in HCT-8 cells. Original magnification × 200; scale bar 50 μm. (g–h) The number of liver metastatic lesions in nude mice injected with the above SW480 (g) and HCT-8 (h) cells via tail vein. (i–j) FRAP assay analysis of mobility fraction and T1/2 (Time it took for half of the mobile fraction of microtubules to recover) after SLAIN2 overexpression and knock-down. (k) Western blot measured acetylated α-tubulin in SLAIN2 overexpressed-SW480 and SLAIN2-silenced HCT-8 cells. **P < .01; ***P < .001. Error bars in Fig. g–h denote s.d. of each group (5 mice). In remaining cases, error bars denote s.d. of triplicates. **P < .01 ***P < .001. P values were calculated with two-way ANOVA in Fig. i and Student's t-test in other panels.
The above results indicated that MALAT1 regulated SLAIN2 expression via sponging miR-106b-5p in CRC cells. Whether MALAT1 could sponge miR-106b-5p and promote the invasion and metastasis of CRC cells through SLAIN2-mediated enhancement of MTs mobility needs further investigation. The FRAP assay discovered that ectopic expression of wild-type MALAT1, but not the mutant form MALAT1, promote fluorescence recovery, including an increased mobile fraction and a decreased half-life after photobleaching in SW480 cells, while co-transfecting miR-106b-5p mimics into SW480 cells partially attenuate these effects (Fig. 6a). Conversely, the depletion of MALAT1 resulted in decreasing mobility and increasing half-life after photobleaching. However, co-transfecting the miR-106b-5p inhibitors into MALAT1-depleted HCT8 cells could partially abolish these effects (Fig. 6b). The overexpression of wild-type MALAT1, but not the mutant MALAT1, downregulated the acetylated α-tubulin while co-transfecting the miR-106b-5p mimics into MALAT1-overexpressed SW480 cells partially attenuated these effects (Fig. 6c). Conversely, the suppression of MALAT1 upregulated the acetylated α-tubulin, which could be partially rescued by co-transfecting the miR-106b-5p inhibitors (Fig. 6d). Taken together, these data indicated that MALAT1 could regulate the MTs' mobility in CRC cells via mediating the miR-106b-5p/SLIAN2 axis expression.
Fig. 6.
MALAT1 sponges miR-106b-5p to promote the invasion and metastasis via SLAIN2-mediated the enhanced MTs mobility. (a) FRAP assay analysis of mobility fraction and T1/2 (Time it took for half of the mobile fraction of microtubules to recover) in SW480 cells after transfected with MALAT1, MALAT1 mut or MALAT1 + miR-106b-5p. (b) Mobility fraction and T1/2 was measured with FRAP assay between the group treated with shNC, shMALAT1 and shMALAT1+ miR-106b-5p inhibitor in HCT-8 cells. (c–d) Western blot evaluated acetylated α-tubulin expression in MALAT1, MALAT1 mut or MALAT1 + miR-106b-5p transfected SW480 (c) and shNC, shMALAT1 and shMALAT1+ miR-106b-5p inhibitor treated HCT-8 cells (d). (e–f) Transwell assay analyzed the migratory and invasive abilities in MALAT1, MALAT1 mut or MALAT1 + miR-106b-5p transfected SW480 cells. (g–h) The changes of migratory and invasive abilities in shNC, shMALAT1 and shMALAT1+ miR-106b-5p inhibitor treated HCT-8 cells. (i–j) The number of liver metastatic lesions in nude mice injected with the above SW480 and HCT-8 cells via tail vein. Error bars in Fig. i–j denote s.d. of each group (5 mice). In remaining cases, error bars denote s.d. of triplicates. *P < .05; **P < .01; ***P < .001. P values were calculated with two-way ANOVA in left panel of a-b and Student's t-test in other panels.
Next, the transwell assay also showed that the overexpression of wild-type MALAT1, but not the mutant MALAT1, resulted in increased migration and invasive numbers of SW480 cells, which can be partially abrogated by co-transfecting the miR-106b-5p mimics in SW480 cells (Fig. 6e and f; Fig. S5a and b). However, the knockdown of MALAT1 reduced the number of migrated and invasive HCT-8 cells, which could be partially reversed by co-transfecting the miR-106b-5p inhibitors (Fig. 6g and h; Fig. S5c and d). These data indicated that MALAT1 could promote the CRC cells' migration and invasion by sponging miR-106b-5p in vitro.
Fig. S5.
MALAT1 promotes the migration and invasion by sponges miR-106b-5p in vitro. (a–b) Transwell assay for the migratory and invasive abilities in MALAT1, MALAT1 mut or MALAT1 + miR-106b-5p transfected SW480 cells. (c–d) The changes of migratory and invasive abilities in HCT-8 cells after treated with shNC, shMALAT1 and shMALAT1+ miR-106b-5p inhibitor. Original magnification ×200; scale bar 50 μm.
To further confirm the epigenetic roles of MALAT1on miR-106b-5p in vivo, the CRC cells were injected into tail vein of nude mice. Herein, we found that the overexpression of wild-type MALAT1 in SW480 cells, but not the mutant type, formed tumor colonies in the liver, while co-transfecting the miR-106b-5p mimics into SW480 partially reduced the colony number (Fig. 6l). Interestingly, the knockdown of wild-type MALAT1 in HCT-8 cells formed fewer tumor colonies in the liver, and these effects were reversed by co-transfecting the miR-106b-5p inhibitors (Fig. 6j). Thus, we revealed that MALAT1 promoted the CRC invasion and metastasis via sponging miR-106b-5p, and then regulated the SLAIN2-associated MT mobility.
2.6. High-level of MALAT1 is associated with CRC metastasis and predicts poor survival
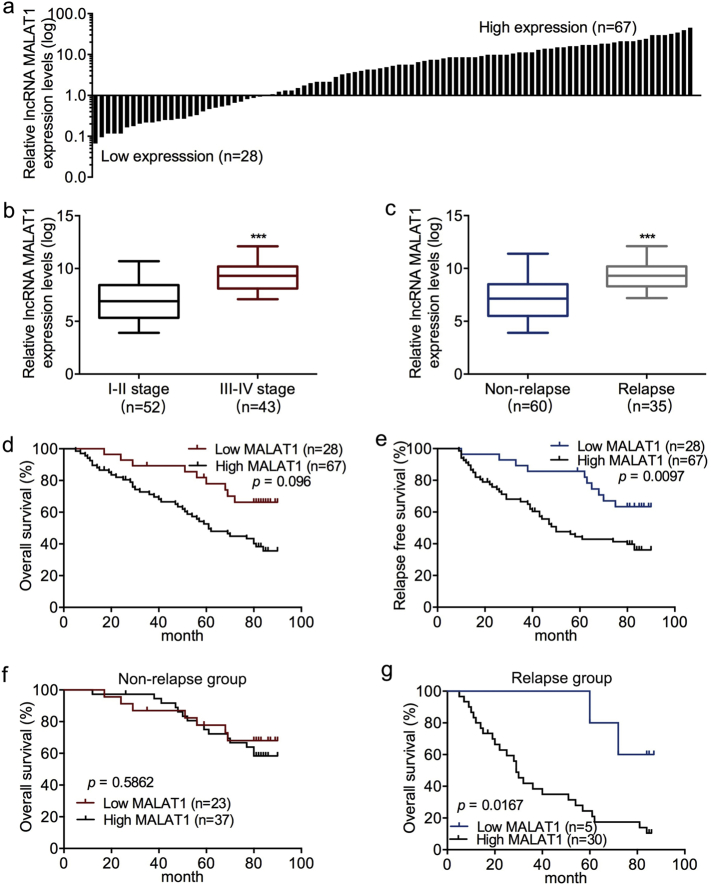
TCGA dataset was used to analyze MALAT1 expression in CRC. We found that the MALAT1 expression in CRC tissues was higher than that in the normal tissues, and levels of MALAT1 in stage IV was higher than that in stage I-II (Fig. S6a and b). Similarly, MALAT1 expression was upregulated in CRC samples as compared to paired normal controls from the dataset of Shanghai General Hospital (Fig. 7a). Importantly, MALAT1 was much higher in stage III-IV or relapse group patients as compared to stage I-II or non-relapse group, respectively (Fig. 7b and c). Subsequently, we analyzed the correlations between MALAT1 expression and clinicopathological parameters in the 95 CRC patients. The relative MALAT1 expression in CRC was described as high or low based on the comparison to the paired normal tissues. Also, high MALAT1 expression was positively associated with lymph node metastasis and distant metastasis (Table S3).
Fig. S6.
The expression and prognosis analysis of MALAT1 according to TCGA database. (a–b) Comparisons of the expression of MALAT1 in the TCGA CRC dataset (a, normal VS tumor; b, I-II stage VS IV stage). (c–d) Kaplan-Meier curves for OS (c, n = 370) and RFS (d, n = 340) of CRC patients in the TCGA dataset.
Fig. 7.
High-level of MALAT1 is associated with CRC metastasis and predicts poor survival. (a) The expression of MALAT1 in 95 pairs of CRC and normal tissues by qPCR analyses. Data was calculated with 2−ΔΔCt method. (b) Relative MALAT1 expression levels in stage I-II and stage III-IV tumor tissues. (c) Relative MALAT1 expression levels in tumor relapse and non-relapse groups. (d–e) Kaplan–Meier analysis with a log-rank test for OS (d) and RFS (e) in 95 CRC patients according to MALAT1 expression. (f–g) Kaplan–Meier analysis with a log-rank test for OS in non-relapse (f) and relapse groups (g). Box and whiskers plot: min to max; ***P < .001, two-tailed Student t-test.
Kaplan–Meier analysis with a log-rank test on tumor OS and RFS using TCGA dataset and our dataset showed that high MALAT1 resulted in poor OS and RFS in CRC patients (Fig. S6c and d; Fig. 7d and e). Moreover, high MALAT1 expression predicted poor OS in the relapse group but not in the non-relapse group (Fig. 7f and g). Thus, high MALAT1 is associated with CRC metastasis and predicted poor survival.
3. Discussion
Previously, the low expression of miR25 and miR93, the members of miR-106b~25 cluster family, has been demonstrated to promote the invasion and metastasis of colon cancer cells [5]. In this study, we verified that the level of another member of the cluster, miR-106b-5p was downregulated in CRC tissues, which in turn, predicted poor prognosis for CRC patients. The downregulation of miR-106b-5p promoted the CRC cell invasion and metastasis in vitro and in vivo, which indicated the significant roles of miR-106b-5p in the development of CRC. Moreover, the present study also demonstrated that lncRNA MALAT1 sponges miR-106b-5p, followed by regulating the SLAIN2-associated MT mobility. The high risk of tumor recurrence and metastasis suggested that CRC patients with low miR-106b-5p expression and high MALAT1/SLAIN2 levels should receive an enhanced treatment post-surgery.
Even though increasing mechanisms concerning genetic and epigenetic alterations in CRC have been widely revealed, the survival rate of CRC patients is still poor [18]. Tumor recurrence and metastasis are ascribed for the poor survival of CRC patients after radical operation. Thus, identifying novel biomarkers that can efficiently predict the risks of tumor recurrence and metastasis might provide a better approach for an improved prognosis of CRC. Herein, we showed that miR-106b-5p acted as a repressor in CRC and high miR-106b-5p expression predicted an improved survival for CRC patients. These findings are consistent with the previous reports wherein miR-106b-5p was shown to function as a suppressor in papillary thyroid cancer and bladder cancer [19,20]. Reportedly, the miR-106b/25 cluster (consisting of miR-106b, miR-93, and miR-25) is localized on chromosome 7 (7q22.1) within intron 13 of the mini-chromosome maintenance gene, MCM7 that is transcriptionally activated by N-Myc. The MCM7 protein possesses the vital function of “licensing” the DNA synthesis during the transition from G1 to S phase [21,22]. Interestingly, miR-106b-5p promotes the aggressiveness and stem-cell-like phenotype of ccRCC cells by activating the Wnt/β-catenin signaling pathway [7]. Also, the upregulated level of miR-106b-5p prevents the p53 tumor suppressor activity, thereby enhancing the N-Myc pathway [8]. These dual roles as a tumor suppressor or oncogene indicated that miR-106b-5p might function in various cancers through different physiological behaviors. The investigation of ce-lncRNA provided insights into the mechanisms of dysregulation of miRNAs in CRC carcinogenesis and development to discover novel treatment approaches. Therefore, we hypothesized that endogenous lncRNAs regulated the miR-106b-5p expression in CRC.
The current bioinformatics analyses and luciferase reporter assays identified that MALAT1, a lncRNA, might be a potential ceRNA by sponging miR-106b-5p in CRC. These findings suggested that MALAT1 might promote the invasion and metastasis by regulating miR-106b-5p, which is in agreement with the previous findings [[23], [24], [25], [26]]. In hepatocellular carcinoma, MALAT1 is upregulated and acts as a proto-oncogene via the activation of the Wnt pathway and induction of the oncogenic splicing factor SRSF [27]. The upregulation of the lncRNA MALAT1 also contributes to the proliferation and metastasis in esophageal squamous cell carcinoma [28]. In gastric cancer, MALAT1 could drive the development of cancer and promote peritoneal metastasis [23,29]. However, these findings were challenged by the study by Han et al., which showed that MALAT1 was functional as a tumor suppressor by attenuating the ERK/MAPK-mediated growth and MMP2-mediated invasiveness in glioma [30]. Thus, the different roles of MALAT1 were exerted on different tumors. The specific lncRNA was valuable in predicting the prognosis of CRC by investigating the correlations and between MALAT1 and miR-106b-5p.
High invasiveness and metastatic abilities are regarded as the main reasons for poor prognosis in malignant tumors [31]. In the present study, the overexpression of MALAT1 also promoted the invasion and metastasis of CRC cells in vitro and in vivo by sponging miR-106b-5p. Clinically, MALAT1 expression was negatively correlated with miR-106b-5p in CRC tissues. These findings were in agreement with those reported previously that MALAT1 enhanced the migration and invasiveness of CRC cells [25]. These results were similar to another study, wherein lncRNA NEAT1_2 regulated the expression of ATAD2 by sponging miR-106b-5p in papillary thyroid cancer and promoted the progression of bladder cancer [32]. Consequently, the current study verified that miR-106b-5p was sponged by MALAT1, which in turn, promoted the invasion and metastasis of CRC cells in vitro and in vivo.
Furthermore, onco-miRNAs functioned by downregulating the expression of the tumor suppressors in cancers [33]. The subsequent bioinformatics analyses and luciferase reporter assays demonstrated that SLAIN2 was the potential downstream target in CRC. It has been reported as an essential gene for mesenchymal cell invasion both in vitro and in the mouse tumor model via vesicle transport regulation and focal adhesion formation [15]. As a key component of MT plus end interaction networks, SLAIN2 affects the MT activity and promotes the MT-dependent membrane deformation, which is essential for mesenchymal cell invasion [15,17]. The current functional studies demonstrated that MALAT1 upregulated the SLAIN2 expression by sponging miR-106b-5p and also enhanced the SLAIN2-associated MT activity. These findings revealed the mechanism of dysregulation and the promoting roles of miR-106b-5p on CRC. Based on the lncRNA prediction programs analysis, we only selected MALAT1 and identified SLAIN2 as the functional up- and downstream regulator of miR-106b-5p in CRC progression; however, whether other promising regulators of miR-106b-5p in CRC progression occur necessitate further investigation.
In conclusion, the current study revealed the significant roles of MALAT1-mediated SLIAN2-associated MT mobility of CRC cells via sponging miR-106b-5p on the invasion and metastasis of CRC. This study not only provided new insight into the mechanism of low expression of miR-106b-5p in CRC but also indicated that the combination of MALAT1/miR-106b-5p might provide a precise prognostic marker for CRC patients.
4. Materials and methods
4.1. Patient samples
A total of 95 CRC patients without preoperative anti-cancer treatment were enrolled in this study. All the cases were diagnosed pathologically and treated in Shanghai General Hospital during 2010. The patients with advanced-stage disease received standard postoperative 5-fluorouracil-based chemotherapy. The present study was approved by the Ethics Committee of Shanghai General Hospital and National Cancer Center/National Clinical Research Center for Cancer/ Cancer Hospital, Chinese Academy of Medical Sciences, and Peking Union Medical College.
4.2. Cell culture, overexpression, and RNA interference
Seven CRC cell lines (Caco2, HT-29, SW480, SW620, RKO, HCT-8, and LoVo) and one normal colon epithelial cell (NCM 460) were purchased from the Cell Bank of Chinese Academy of Sciences and cultured in DMEM medium with 10% fetal bovine serum (FBS, Gibco) in an incubator at 37 °C with 5% CO2. All the cell lines were authenticated through short tandem repeat analysis and used within 6 months. The last authentication was carried out in February 2018.
The mimics and inhibitor of miR-106b-5p used to overexpress or knockdown the expression of miR-106b-5p were obtained from RiBoBio (Guangzhou, China). The MALAT1, MALAT1-mut, shMALAT1 plasmids were synthesized by GenPharma (Shanghai, China). The plasmids and oligonucleotides were transfected into the cells using Lipofectamine 3000 (Invitrogen). The vectors with overexpression or knockdown of SLAIN2 were constructed and packaged as virus by GenPharma. SW480 cells stably expressing MALAT1, MALAT1-mut, or Lv-SLAIN2, and HCT-8 cells stably expressing shMALAT1 or shSLAIN2 were selected under puromycin antibiotic.
4.3. Cell migration and invasion assay
Transwell chambers (with inserts of 8-mm pore size) uncoated or coated with Matrigel were utilized for migration or invasion assay, respectively. Briefly, 1 × 105 cells were seeded in the upper chamber with FBS-free culture medium while the culture medium with 20% FBS was added in the bottom chamber. After 24 h incubation, the cells were fixed and stained with crystal violet.
4.4. In vivo experiment
105 CRC cells/mL in a 200 μL volume were injected into the tail veins of nude mice. Three weeks after injection, ultrasonography was used to monitor liver metastasis continuously for 8 weeks. Subsequently, all mice were sacrificed at 8 weeks, and the livers were retrieved for HE staining. The tumor colonies formed in the livers were observed and counted under a microscope. The experimental procedures were approved by the Animal Experiment Ethics Committee of the Cancer Hospital, Chinese Academy of Medical Sciences.
4.5. RNA extraction and quantitative PCR
The total RNA was extracted using TRizol reagent (Invitrogen, CA, USA) and reverse transcribed using the PrimeScript RT reagent kit (TaKaRa, Dalian, China). Quantitative PCR was performed using SYBR Premix Ex Taq II (TaKaRa), and U6 or GAPDH was used as an internal control.
4.6. Protein extraction and western blot
Whole cell lysates were extracted using RIPA, separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to PVDF membranes. Subsequently, the membrane was probed with an acetylated α-tubulin antibody (1:1000), followed by incubation with secondary antibody and detected by chemiluminescence system.
4.7. HE staining, IHC, and FISH assays
For HE staining, paraffin-embedded tissues were dewaxed, rehydrated, HE stained, and dehydrated.
For IHC staining, after dewaxing and hydration, antigen retrieval was performed in EDTA. The tissues were stained with diaminobenzidine after sequential incubation with SLAIN2 antibody and secondary antibody as reported previously [34]. Then, the tissues were counterstained with hematoxylin and hydrated. The MALAT1 and miR-106B-5P oligodeoxynucleotide probes were obtained from RiboBio (Guangzhou, China). Moreover, FISH was performed as our reported previously [11].
4.8. RNA immunoprecipitation (RIP) assay
Cells were co-transfected with pMS2bp-GFP and MS2, MS2-MALAT1, or MS2-MALAT1 mut. After 48 h, RIP assay was performed using Magna RIP™ Kit (Millipore) according to the manufacturer's instructions. The cell lysates were incubated with antibodies against GFP and IgG. After reverse transcription, the miR-106b-5p level was analyzed by qPCR.
For anti-AGO2 RIP, cells were transfected with miR-106b-5p mimics or miR-106b-5p inhibitor or controls for 48 h. Then, RIP assay was performed using AGO2 antibody. qPCR was performed to estimate the level of MALAT1 in RIP products.
4.9. RNA pull-down assay
Biotin-labeled miR-106b-5p and MALAT1 were synthesized by Genepharm and transfected into cells for 48 h. Then, the cells lysates were incubated with M-280 streptavidin magnetic beads (Sigma). TRIzol was used to purify the bound RNAs, and qPCR was employed to measure the level of MALAT1 or miR-106b-5p.
4.10. Luciferase reporter assay
The 3′-UTR of SLAIN2- or MALAT1-containing miR-106b-5p putative binding sites were amplified and cloned into a pGL3 vector. The mutation at the miR-106b-5p binding site was performed using a QuikChange Site-Directed Mutagenesis Kit (Stratagene). The cells were co-transfected with the wild-type or mutant luciferase vectors and miR-106b-5p, MALAT1, or the controls for 48 h. A dual-luciferase reporter gene assay system was utilized (Promega, Madison, WI, USA), and the relative luciferase activity was normalized to that of Renilla luciferase activity.
4.11. FRAP
Confocal laser-scanning microscopy was used for photobleaching for 10 s in cells stably expressing the GFP-labeled α-tubulin plasmid. At least 15 regions/sample were selected for bleaching and observed for 2 min to acquire the images every 2 s. The fraction of MTs that recovered within 2 min (mobility fraction), as well as the time required for half of the mobile fraction of MTs to recover (t1/2), was recorded. The recovered fluorescence intensities were normalized to that of the background and unbleached regions within the observed cells.
4.12. Bioinformatics analysis
The TCGA colorectal cancer RNAseq (Illumina HiSeq) dataset was used to compare the expression of miR-106b-5p or MALAT1 in normal and CRC tissues. Kaplan–Meier survival curves were drawn using the KMplot program (http://kmplot.com/analysis/).
4.13. Statistical analysis
SPSS 21.0 statistical software (SPSS Inc., Chicago, IL, USA) was applied for data analysis in this study. All in vitro assays were conducted for three biological and technical replicates. The data are represented as mean ± standard deviation (SD). P < .05 indicates statistical significance. Student's t-test or one-way ANOVA was used to compare the continuous variables in two or multiple groups, respectively. Fisher's exact test or chi-square test was applied to calculate the differences in the categorical variables. Correlations analyses were performed using Pearson's or Spearman's rank correlation test. RFS and OS were analyzed by Kaplan–Meier analysis with log-rank tests.
The following are the supplementary data related to this article.
The correlation between the clinicopathological characteristics and miR-106b-5p expression in 95 CRC patients.
Predicted potential lncRNAs associated with miR-106b-5p.
MALAT1 expression with relation to metastasis in 95 CRC patients.
The correlation between SLAIN2 expression level and TNM stage and differentiation of 95 CRC tissues.
Acknowledgments
Acknowledgements
The authors wish to thank all the patients enrolled in this study. This work was supported by National Program Project for Precision Medicine in National Research and Development Plan of China (2016YFC0905300), National Natural Science Foundation of China (81572930), National Key Research and Development Program of the Ministry of Science and Technology of China (2016YFC0905303, 2016YFC1303200), Beijing Science and Technology Program (D17110002617004), Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences(2018PT32012), CAMS Innovation Fund for Medical Sciences (CIFMS) (2016-I2M-1-001), Incentive Fund for Academic Leaders of Oncology Hospital, Chinese Academy of Medical Sciences (RC2016003),and Beijing Hope Run Special Fund from Cancer Foundation of China (LC2017A19).
Declaration of interests
The authors declare no conflict of interest.
Author contributions
Meng Zhuang, Senlin Zhao, Zheng Jiang, Dongwang Yan, Xishan Wang conceived and designed the research study; Meng Zhuang, Senlin Zhao, Song Wang, Peng Sun, Jichuan Quan analyzed the data; Meng Zhuang, Senlin Zhao, Zheng Jiang, Dongwang Yan, Xishan Wang wrote and revised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Dongwang Yan, Email: yandw70@163.com.
Xishan Wang, Email: wxshan1208@126.com.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer Statistics. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Brouwer N.P.M., Bos A.C.R.K., Lemmens V.E.P.P. An overview of 25 years of incidence, treatment and outcome of colorectal cancer patients. Int J Cancer. 2018 Aug 10 doi: 10.1002/ijc.31785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Adams B.D., Parsons C., Walker L., Zhang W.C., Slack F.J. Targeting noncoding RNAs in disease. J Clin Invest. 2017;127(3):761–771. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Q., Zou C., Zou C. MicroRNA-25 functions as a potential tumor suppressor in colon cancer by targeting Smad7. Cancer Lett. 2013;335(1):168–174. doi: 10.1016/j.canlet.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 6.Zheng L., Zhang Y., Liu Y. MiR-106b induces cell radioresistance via the PTEN/PI3K/AKT pathways and p21 in colorectal cancer. J Transl Med. 2015 Aug 4;13 doi: 10.1186/s12967-015-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jun Lu, Wei Jin-Huan, Feng Zi-Hao. miR-106b-5p promotes renal cell carcinoma aggressiveness and stem-cell-like phenotype by activating Wnt/β-catenin signalling. Oncotarget. 2017;8(13):21461–21471. doi: 10.18632/oncotarget.15591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mastropasqua F., Marzano F., Valletti A. TRIM8 restores p53 tumour suppressor function by blunting N-MYC activity in chemo-resistant tumours. Mol Cancer. 2017;16(1):67. doi: 10.1186/s12943-017-0634-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulitsky I., Shkumatava A., Jan C.H., Sive H., Bartel D.P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147(7):1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang C., Qiu S., Sun F. Long non-coding RNA HNF1A-AS1 mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer Lett. 2017;410:50–62. doi: 10.1016/j.canlet.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 12.Chen P., Fang X., Xia B., Zhao Y., Li Q., Wu X. Long noncoding RNA LINC00152 promotes cell proliferation through competitively binding endogenous miR-125b with MCL-1 by regulating mitochondrial apoptosis pathways in ovarian cancer. Cancer Med. 2018;7(9):4530–4541. doi: 10.1002/cam4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu L., Ding J., Chen C. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29(5):653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Bose M., Barman B., Goswami A., Bhattacharyya S.N. Spatiotemporal uncoupling of microRNA-mediated translational repression and target RNA degradation controls MicroRNP recycling in mammalian cells. Mol Cell Biol. 2017;37(4) doi: 10.1128/MCB.00464-16. pii: e00464-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouchet B.P., Noordstra I., van Amersfoort M. Mesenchymal Cell Invasion Requires Cooperative Regulation of Persistent Microtubule Growth by SLAIN2 and CLASP1. Dev Cell. 2016;39(6):708–723. doi: 10.1016/j.devcel.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Etienne-Manneville S. Microtubules in cell migration. Annu Rev Cell Dev Biol. 2013;29:471–499. doi: 10.1146/annurev-cellbio-101011-155711. [DOI] [PubMed] [Google Scholar]
- 17.Friedl P., Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147(5):992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 18.Dienstmann R., Salazar R., Tabernero J. Personalizing colon cancer adjuvant therapy: selecting optimal treatments for individual patients. J Clin Oncol. 2015;33(16):1787–1796. doi: 10.1200/JCO.2014.60.0213. [DOI] [PubMed] [Google Scholar]
- 19.Lee E., Collazo-Lorduy A., Castillo-Martin M. Identification of microR-106b as a prognostic biomarker of p53-like bladder cancers by ActMiR. Oncogene. 2018 doi: 10.1038/s41388-018-0367-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian C., Yan W., Li T. Differential expression of MiR-106b-5p and MiR-200c-3p in newly diagnosed versus chronic primary immune thrombocytopenia patients based on systematic analysis. Cell Physiol Biochem. 2018;45(1):301–318. doi: 10.1159/000486811. [DOI] [PubMed] [Google Scholar]
- 21.Koppen A., Ait-Aissa R., Koster J. Direct regulation of the minichromosome maintenance complex by MYCN in neuroblastoma. Eur J Cancer. 2007;43(16):2413–2422. doi: 10.1016/j.ejca.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 22.Shohet J.M., Hicks M.J., Plon S.E. Minichromosome maintenance protein MCM7 is a direct target of the MYCN transcription factor in neuroblastoma. Cancer Res. 2002;62(4):1123–1128. [PubMed] [Google Scholar]
- 23.YiRen H., YingCong Y., Sunwu Y. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16(1):174. doi: 10.1186/s12943-017-0743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang S.M., Hu W.W. Long non-coding RNA MALAT1 promotes oral squamous cell carcinoma development via microRNA -125b/STAT3 axis. J Cell Physiol. 2018;233(4):3384–3396. doi: 10.1002/jcp.26185. [DOI] [PubMed] [Google Scholar]
- 25.Yang M.H., Hu Z.Y., Xu C. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015;1852(1):166–174. doi: 10.1016/j.bbadis.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S.S., Harford J.B., Moghe M., Rait A., Pirollo K.F., Chang E.H. Targeted nanocomplex carrying siRNA against MALAT1 sensitizes glioblastoma to temozolomide. Nucleic Acids Res. 2018;46(3):1424–1440. doi: 10.1093/nar/gkx1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malakar P., Shilo A., Mogilevsky A. Long noncoding RNA MALAT1 promotes hepatocellular carcinoma development by SRSF1 upregulation and mTOR activation. Cancer Res. 2017;77(5):1155–1167. doi: 10.1158/0008-5472.CAN-16-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu L., Wu Y., Tan D. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L., Geng Y., Feng R. The human RNA surveillance factor UPF1 modulates gastric cancer progression by targeting long non-coding RNA MALAT1. Cell Physiol Biochem. 2017;42(6):2194–2206. doi: 10.1159/000479994. [DOI] [PubMed] [Google Scholar]
- 30.Han Y., Wu Z., Wu T. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis. 2016;7:e2123. doi: 10.1038/cddis.2015.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson K., Lewalle A., Fritzsche M., Thorogate R., Duke T., Charras G. Mechanisms of leading edge protrusion in interstitial migration. Nat Commun. 2013;4:2896. doi: 10.1038/ncomms3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun W., Lan X., Zhang H. NEAT1_2 functions as a competing endogenous RNA to regulate ATAD2 expression by sponging microRNA-106b-5p in papillary thyroid cancer. Cell Death Dis. 2018;9(3):380. doi: 10.1038/s41419-018-0418-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bu P., Wang L., Chen K.Y. miR-1269 promotes metastasis and forms a positive feedback loop with TGF-beta. Nat Commun. 2015;6:6879. doi: 10.1038/ncomms7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao S., Jiang T., Tang H. Ubiquitin D is an independent prognostic marker for survival in stage IIB-IIC colon cancer patients treated with 5-fluoruracil-based adjuvant chemotherapy. J Gastroenterol Hepatol. 2015;30(4):680–688. doi: 10.1111/jgh.12784. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The correlation between the clinicopathological characteristics and miR-106b-5p expression in 95 CRC patients.
Predicted potential lncRNAs associated with miR-106b-5p.
MALAT1 expression with relation to metastasis in 95 CRC patients.
The correlation between SLAIN2 expression level and TNM stage and differentiation of 95 CRC tissues.