Abstract
Hypoxia-inducible factor-2 (HIF-2) is a heterodimeric transcription factor formed through dimerization between an oxygen-sensitive subunit HIF-2α subunit and its obligate partner subunit ARNT. Enhanced HIF-2 activity drives some cancers, while reduced activity causes anemia in chronic kidney disease. Therefore, modulation of HIF-2 activity via direct-binding ligands could provide many new therapeutic benefits. Here, we explored HIF-2α chemical ligands using combined crystallographic, biophysical, and cell-based functional studies. We found chemically unrelated antagonists to employ the same mechanism of action. Their binding displaced residue M252 from inside the HIF-2α PAS-B pocket toward the ARNT subunit to weaken heterodimerization. We also identified first-in-class HIF-2α agonists and found they significantly displaced pocket residue Y281. Its dramatic side-chain movement increases heterodimerization stability and transcriptional activity. Our findings show that despite binding to the same HIF-2α PAS-B pocket, ligands can manifest as inhibitors versus activators by mobilizing different pocket residues to allosterically alter HIF-2α-ARNT heterodimerization.
INTRODUCTION
The basic helix-loop-helix-PER-ARNT-SIM (bHLH-PAS) family requires subunit dimerization between its members to form productive transcription factors. A common architectural feature of this family is their highly conserved DNA-binding domain, which must converge symmetrically through subunit dimerization to form a functional DNA-reading head1. Further unifying the family are their tandem PER-ARNT-SIM (PAS) domains (PAS-A and PAS-B). PAS domains in unrelated protein classes can function as molecular sensors, by binding to environmental and/or physiological ligands2. The PAS domains of mammalian bHLH-PAS members not only participate in heterodimer formation1, but also harbor unique pockets of various size3.
The hypoxia-inducible factors (HIFs) within the bHLH-PAS family function as sensors of low oxygen stress, and respond to hypoxia by coordinating genomic pathways in erythropoiesis, angiogenesis and cellular metabolism4,5. The HIF proteins function as obligate heterodimers consisting of one α subunit (any of HIF-1α, HIF-2α and HIF-3α)6,7, and a constitutively-expressed partner β subunit also known as ARNT (aryl hydrocarbon receptor nuclear translocator)4. The dimerization of HIF-α and ARNT results in an asymmetric quaternary architecture, creating a DNA-reading head for binding to hypoxia response elements8.
Molecular oxygen regulates the stability of HIF-α proteins through post-translational modifications. Under normoxia, prolyl hydroxylase domain (PHD) enzymes modify specific proline residues within HIF-1α and HIF-2α proteins9–11, leading to their subsequent proteasomal degradation. An asparagine residue in HIF-α is also targeted by factor inhibiting HIF (FIH) enzyme for hydroxylation under normoxia, further reducing its transcriptional activity12,13. Both of these oxygen-dependent mechanisms suppress HIF activities under normoxia, and are reversed under hypoxia to allow HIF-α protein accumulation and sustained activity.
Traditionally, transcription factors were considered difficult drug targets compared to enzymes, kinases, and G-protein coupled receptors. The nuclear receptor (NR) family has been a notable exception due to its conserved ligand-binding domains14. In the bHLH-PAS family, aryl hydrocarbon receptor (AHR) is known to bind diverse ligands15,16. A select few bHLH-PAS proteins were previously explored for ligand binding within their PAS-B domains, including HIF-2α17,18, HIF-1α19, ARNT20 and HIF-3α21. Based on systematic crystallographic and sequence comparisons, we previously suggested that their entire family harbors cavities for chemical ligands3. Accordingly, new studies are now needed to both identify specific ligands and to examine their functional consequences.
The HIF-α proteins serve as an excellent focal point in this regard, due to the recent characterizations of their multi-domain structures8. The PT2385 class of HIF-2α antagonists was recently developed and employed in animal clear cell renal cell carcinoma (ccRCC) tumorgraft models with promising results22–25. However, HIF-α small-molecule agonists have not been previously identified. Such ligands could prove desirable for anemia in the setting of chronic kidney disease (CKD). Currently, PHD enzyme inhibitors are undergoing clinical testing for anemia, as they upregulate HIF-2 activity26–28. Direct-binding HIF-2α agonists could provide advantages or complement the use of PHD inhibitors in CKD anemia, given that both approaches would enhance the expression of the target gene EPO29.
Here, we examined the ligand-binding capabilities of HIF-2α-ARNT using distinct chemical ligands, assessing their binding locations and functional manifestations. Relying on new co-crystal structures, hydrogen-deuterium exchange mass spectrometry (H/D-ex MS) data, time-resolved protein binding assays, and functional profiling of target gene expression, we identified allosteric mechanisms by which ligands can produce opposing activities, despite binding to a common binding site within the HIF-2α protein.
RESULTS
Biochemical and cellular activities of antagonist PT2385
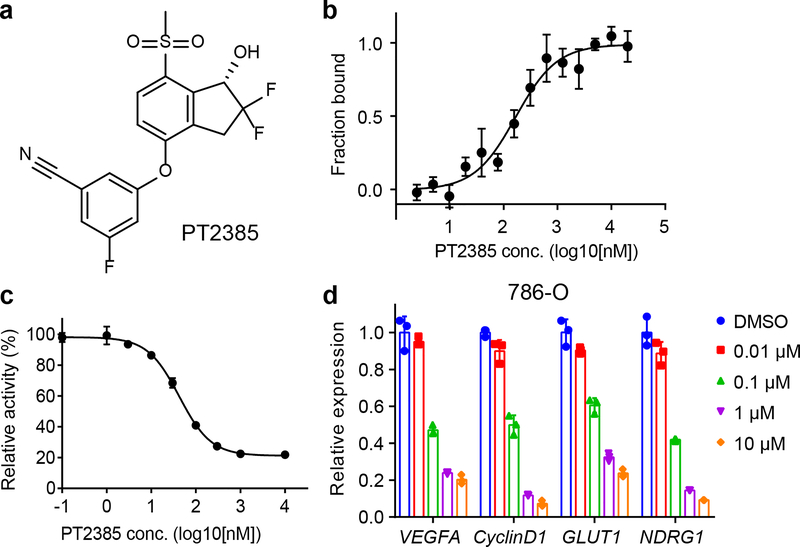
To assess the antagonistic properties of PT2385 (designated as compound 1 in this study) on HIF-2, we employed complementary biochemical and cell-based assays (Fig. 1). Using the recombinant PAS-B domain of HIF-2α, we found that PT2385 (Fig. 1a) binds with an equilibrium binding constant (Kd) of 167 nM, as measured by the MST (microscale thermophoresis) method30 (Fig. 1b). This affinity was slightly weaker than previously reported Kd of 50 nM measured by ITC (isothermal titration calorimetry)23, and the difference may be caused by variations in the protein constructs used, assay buffers, or methodologies (ITC vs. MST). Using a FP (fluorescence polarization) assay, we examined if PT2385 changes the binding affinity of HIF-2α-ARNT heterodimer for its hypoxia response element (HRE) (Supplementary Fig. 1a). In the presence of 0 μM, 0.1 μM, 1 μM and 10 μM PT2385, the DNA binding affinities were 65 nM, 96 nM, 118 nM and 339 nM, respectively, showing a dose-dependent inhibition. However, even at high and saturating concentrations of PT2385, DNA binding was not completely abolished.
Figure 1. Inhibition of HIF-2 by PT2385.
a, Chemical structure of PT2385. b, Binding of PT2385 to the HIF-2α PAS-B domain (Kd about 167 nM) as measured by MST. c, Dose-dependent inhibition of HRE luciferase reporter activity by PT2385. d, Expression of HIF-2 target genes in 786-O cells after treatment with PT2385 at various concentrations (0.01, 0.1, 1 or 10 μM). Error bars, mean ± s.d.; n=3 (distinct replicates for cell cultures).
Using a 786-O cell line stably transfected with the HRE luciferase reporter, we measured an IC50 value of 42 nM for PT2385’s inhibition of transcriptional activity (Fig. 1c). A qPCR assay monitoring the expression of endogenous target genes of HIF-2 (e.g. VEGF, CyclinD1, GLUT1 and NDRG1) further revealed PT2385’s dose-dependent inhibitory activity in 786-O cells (Fig. 1d). Recent work demonstrated that PT2385 is selective for HIF-2α over HIF-1α, for binding and inhibition of transcriptional activity23. We previously showed that three residues differed between the PAS-B pockets of HIF-2α and HIF-1α, allowing chemical ligands to be highly selective for HIF-2α versus HIF-1α8. Therefore, it was not surprising that in Hep3B cells, PT2385 decreased the expression of HIF-2 specific genes (VEGF, EPO and NDRG1) but not the HIF-1 specific gene (PGK1) under hypoxia condition (Supplementary Fig. 1b).
Structural and allosteric effects of PT2385
To decipher how PT2385 physically binds to HIF-2α-ARNT to manifest its actions, we sought the ligand-bound complex structure. The co-crystallization of heterodimer and PT2385 was not successful, but the soaking method allowed us to obtain their complex structure at 3.0 Å resolution (Supplementary Table 1). The PT2385 binding location was unambiguously visualized within the PAS-B pocket of HIF-2α, based on “omit” electron density maps (Fig. 2a, b). PT2385 contacts multiple HIF-2α residues inside this pocket, and is stabilized through numerous hydrophobic interactions and a hydrogen bond formed between its hydroxyl group and the H293 side-chain nitrogen (Fig. 2c).
Figure 2. Allosteric mechanism of PT2385 revealed by the crystal structure and H/D-ex MS.
a, Binding position for PT2385 (circled in black) within the entire HIF-2α-ARNT crystal structure. b, Close-up look at the location of PT2385 inside the HIF-2α PAS-B domain, with green mesh showing the FO – FC omit map contoured at 2.7σ. c, Interactions of PT2385 (yellow) with surrounding residues in the pocket. d, The overall arrangement of ARNT and HIF-2α PAS-B domains is displayed on the left, with the HIF-2α PAS-B from PT2385-bound (magenta) or apo (orange) complexes superimposed. On the right, an enlarged and rotated view shows the different side-chain orientations of M252 in these two complexes. The side-chain movement of HIF-2α M252 caused by antagonist binding disrupts the dimerization of HIF-2α-ARNT complex at the PAS-B/PAS-B interface. e, H/D-ex MS results mapped on the crystal structure of HIF-2α-ARNT complex. The regions showing dynamic changes upon PT2385 binding are colored according to the maximum differences of deuteration levels (red for 40 to 50%, pink for 25 to 40%) as compared to the apo form complex (detailed in Supplementary Fig. 4), on top of the background colors of HIF-2α (gray) and ARNT (pale yellow). Enlarged and rotated views of dimer interfaces between the PAS-B domains, and among the PAS-A and bHLH domains are shown on the left and right, respectively.
To learn how PT2385 binding triggers the destabilization of HIF-2α-ARNT heterodimers, we compared this crystal structure to our previously reported apo HIF-2α-ARNT structure8 (Fig. 2d). We found a majority of HIF-2α residues within the pocket undergo only subtle conformational changes upon PT2385 binding, but dramatic changes occur in the positioning of H293 and M252 (Supplementary Fig. 2). The imidazole ring of H293 turns nearly 45° to form a hydrogen bond with PT2385. Ligand binding further causes a dramatic displacement of the M252 side-chain, forcing it outside the pocket (Fig. 2d). The M252 side-chain moves toward a heterodimeric junction between the PAS-B domain of HIF-2α and the PAS-B domain of ARNT. We previously showed that mutations introduced at this interface destabilize the HIF-2α-ARNT heterodimer8. Therefore, the repositioning of M252 to this junction could account for why PT2385 induces heterodimer destabilization23.
Effect of PT2385 on heterodimer stability in solution
Because our HIF-2α-ARNT-PT2385 structure was obtained by soaking instead of co-crystallization, we were concerned that our assessments of PT2385-induced changes could be limited. Therefore, we applied solution-based methods to further probe how PT2385 acts on the heterodimer. We used H/D-ex MS to initially characterize the dynamic features of HIF-2α-ARNT complex without any bound ligands (Supplementary Fig. 3a). The regions with high H/D exchange rates (indicating more flexible backbone structures), were largely located at the polypeptide termini and on loops connecting certain secondary structures within each domain (Supplementary Fig. 3b).
We then observed changes that PT2385 induced on HIF-2α-ARNT, by examining the differences in the deuterium exchange patterns of this complex in the presence versus the absence of PT2385 (Supplementary Fig. 4). PT2385 binding dramatically increased the exchange rates of several key regions located in both HIF-2α and ARNT, suggesting an allosteric effect in which protein regions distal to the ligand-binding site are impacted (Fig. 2d). By mapping these altered regions onto our crystal structure of HIF-2α-ARNT, we found changes are induced at domain-domain interfaces responsible for maintaining heterodimer stability (Fig. 2e).
Not surprisingly, the HI loop (located between Hβ and Iβ) of ARNT PAS-B domain interacting with HIF-2α PAS-B, which we had observed to be pushed away due to the PT2385-induced movement of M252 (Fig. 2d, right), displayed increased H/D exchange and enhanced dynamics upon PT2385 binding (Fig. 2e, left). The C-terminus of HIF-2α PAS-B domain, which contributes to the same PAS-B/PAS-B interface, also showed enhanced exchange, suggesting destabilization (Fig. 2e, left). Several α-helices mediating interactions between bHLH and PAS-A domains of both HIF-2α and ARNT, were also destabilized, as inferred from their higher exchange rates, suggesting even more distal effects by PT2385 (Fig. 2e, right).
These H/D-ex MS observations vividly illustrate how binding of PT2385 to the HIF-2α PAS-B domain impacts multiple domain interfaces simultaneously, all at a considerable distance to its physical binding pocket, extending as far away as the bHLH DNA reading head of the complex. Since the latter allosteric effects are less pronounced, they help explain why the DNA-binding affinity of the heterodimer are not completely eliminated (Supplementary Fig. 1a).
Effect of PT2385 on the affinity between HIF-2α and ARNT
We next sought to measure the binding affinity between HIF-2α and ARNT proteins in forming their heterodimer. As PT2385 dose-dependently reduced their interactions in cells (Fig. 3a), we assessed whether the complete or partial dissociation of the heterodimer would result with PT2385 addition in vitro. We developed a TR-FRET-based binding assay that could detect interactions between the two subunits (Supplementary Fig. 5a). Because the Kd values for ARNT and any of its heterodimerization partners were not previously determined, we first measured these equilibrium affinity constants of ARNT with a variety of known dimerization partners within the bHLH-PAS family, including HIF-2α, HIF-1α and NPAS3 (neuronal PAS domain protein 3).
Figure 3. PT2385 disrupts subunit-subunit interactions in HIF-2 heterodimers.
a, Co-IP results showing the destabilizing effect of PT2385 on the dimerization between overexpressed full-length HIF-2α and ARNT proteins. This experiment was independently repeated twice with similar results. b-c, Disrupting effects of PT2385 on the heterodimerization of HIF-2α-ARNT (b) and HIF-1α-ARNT (c), as measured by the TR-FRET-based assay. d, Interaction profile of ARNT and HIF-2α mutant M252A. e, Disrupting effect of PT2385 on the dimerization of ARNT and HIF-2α mutant M252A. f, Comparison of PT2385’s inhibitory effects at 10 μM on expression of HIF-2 target genes in HEK293T cells transfected with wild-type HIF-2α or M252A mutant. Error bars, mean ± s.d.; n=3 (distinct replicates for cell cultures). Gels for purified proteins and uncropped blots can be found in Supplementary Fig. 13.
The apparent Kd values that we detected for ARNT binding to each of HIF-2α, HIF-1α and NPAS3, were 29.1 nM, 11.7 nM and 8.6 nM, respectively (Supplementary Fig. 5b-d). An important caveat to consider is the single nM-level affinity between the anti-His antibody and the His-tag protein subunit employed in this assay. Therefore, we may have been limited in detecting the actual Kd values if the true binding affinities are below the 10 nM level. Still, this assay provides an excellent means to monitor changes in affinity caused by ligand binding, or by point mutations on each protein.
We then used this assay to ask how PT2385 alters the dimerization affinities in each of HIF-2α-ARNT and HIF-1α-ARNT complexes (Fig. 3b,c). Ligand binding disrupted the dimerization between HIF-2α and ARNT with a Ki of 148 nM (Fig. 3b). However, the disruption of the subunits did not appear complete, as the TR-FRET value only changed 50%. This observation could be interpreted in two ways: either half of the complexes are fully dissociated and the remaining half are unaffected, or 100% of heterodimers undergo a partial separation of subunits. However, only the latter interpretation is consistent with our other solution-based studies. The H/D-ex MS data is inconsistent with the notion that the subunits actually dissociate completely into non-interacting monomers (i.e., we did not observe substantial increases in H/D patterns throughout the outside surfaces of both subunits). Moreover, we noted partial retention of DNA-binding activity even under saturating concentrations of PT2385, again inconsistent with full separation of the two subunits. Importantly, our TR-FRET studies detected no PT2385 effect on HIF-1α-ARNT subunit interactions (Fig. 3c), consistent with its HIF-2α selectivity.
Exploring PT2385’s allosteric mechanism by mutagenesis
As described above, our crystallographic studies suggested that the HIF-2α pocket residue, M252, mediates the allosteric effects of PT2385 binding to distal sites at the heterodimer junctions (Fig. 2d). To fully understand the importance of this residue, we generated a point mutation M252A in HIF-2α and conducted additional TR-FRET studies (Fig. 3d,e). We first detected how this mutation altered the ability of HIF-2α and ARNT to heterodimerize. The apparent Kd value for the mutant HIF-2α binding to ARNT was 1.4 nM (Fig. 3d), much lower than that of wild-type (WT) HIF-2α binding to ARNT (29.1 nM). We then examined PT2385’s effects on the mutant heterodimer. Compared to the WT HIF-2α (Fig. 3b), the M252A mutant was notably resistant to heterodimer disruption by PT2385 (Fig. 3e).
We employed cell-based functional studies to assess the differences in the function of WT and mutant heterodimers. Compared to the activity of WT HIF-2α transfected HEK293T cells, in which the expression of HIF-2 target genes (i.e. VEGFA, EPO and NDRG1) was substantially reduced by PT2385, the same cells transfected with HIF-2α mutant M252A showed no significant inhibitory effects by PT2385 (Fig. 3f). These functional studies with the M252A mutant, support our crystallographic inference that M252 is critical for mediating PT2385’s disruption of heterodimer.
In animal studies using the ccRCC tumorgraft model24, two point mutations (HIF-2α G323E and ARNT F446L) were previously reported to be acquired after prolonged treatment with PT2399 (PT2385 analog), causing drug resistance. PT2399 could not disassociate HIF-2α-ARNT in cells when either mutation was present. G323E locates inside the PAS-B pocket of HIF-2α, and the large side-chain of a glutamic acid replacement would occlude PT2399’s binding, accounting for drug resistance. However, ARNT F446L locates at a heterodimer interface, and its role in resistance to PT2399 was not obvious (Supplementary Fig. 5e).
To investigate the impact of these two resistance mutations on the association between HIF-2α and ARNT, we employed our TR-FRET-based assay. HIF-2α G323E mutation only modestly increased the affinity between two subunits (apparent Kd about 10.5 nM) (Supplementary Fig. 5f), but dramatically reduced PT2385’s ability to impact heterodimer stability (Supplementary Fig. 5g). We interpret these data as consistent with our structural finding that this mutation restricts PT2385’s access to its pocket.
Interestingly, the ARNT F446L mutation substantially enhanced the natural affinity of ARNT for HIF-2α (apparent Kd about 0.8 nM, compared to 29.1 nM in the WT) (Supplementary Fig. 5h), consistent with the positioning of this mutation at a dimer interface where it reinforces junctional stability. At the same time, PT2385 was able to still reduce the heterodimer stability in a dose-dependent manner with this mutation (Ki of 127 nM), consistent with unobstructed access of the drug to its pocket (Supplementary Fig. 5i). These data explain previously unknown aspects of in vivo resistance due to both mutations24.
Diverse antagonist chemotypes employ the same mechanism
While our data portray the mechanism of action used by PT2385 is allosteric and based on the partial destabilization of the heterodimer, it is unclear whether other small-molecule antagonists would also use this mechanism. We attempted to discover new HIF-2 antagonist chemotypes and to compare their mechanistic basis of action. Using purified HIF-2α-ARNT complex proteins, we created a high-throughput assay based on protein thermal shift changes31 for in vitro screening of chemical libraries. One hit that emerged from the LOPAC library was S(−)-p-bromotetramisole (designated as compound 2 or T1001, Fig. 4a), which increased the protein melting temperature of the complex (Tm) by approximately 1.5 °C (Supplementary Fig. 6). T1001 bound to the PAS-B domain of HIF-2α with a Kd of about 246 nM, as measured by MST (Fig. 4b). In studying its cellular effects on the expression of known HIF-2 target genes in 786-O cells (Fig. 4c), we found that compared to PT2385, T1001 was a weaker antagonist.
Figure 4. Newly identified antagonist T1001 points to a common inhibitory mechanism.
a, Chemical structure of T1001. b, Binding of T1001 to the HIF-2α PAS-B domain (Kd about 246 nM) measured by MST. c, Inhibition on the expression of HIF-2α target genes in 786-O cells by PT2385 (1 μM) and T1001 (10 μM). d, Binding site of T1001 within the HIF-2α PAS-B pocket, with green meshes showing the FO – FC omit map contoured at 3.0σ. e, Comparison of T1001’s inhibitory effects at 10 μM on expression of HIF-2 target genes in HEK293T cells transfected with wild-type HIF-2α or M252A mutant. f, Comparison of HIF-2α residues surrounding antagonists in the PAS-B structures. The ligands and residues are in magenta, pink and brown, for PT2385, 0X3 and T1001 respectively. g, A mechanistic diagram showing how antagonist binding leads to the movement of M252 to disrupt HIF-2α-ARNT heterodimers. The proximate positions of M252 side-chain in apo (orange) and antagonist-bound structures are compared and related to the disruption potency. Error bars, mean ± s.d.; n=3 (distinct replicates for cell cultures).
We obtained the co-crystal structure of HIF-2α-ARNT with T1001. The “omit” map in Fig. 4d shows that T1001 binds into the same PAS-B domain of HIF-2α as PT2385 (Supplementary Fig. 7). We asked if the HIF-2α residue M252 was also essential in mediating the inhibitory effects of T1001, and found that the expression of HIF-2 target genes (i.e. VEGFA and EPO) was suppressed by T1001 in the presence of WT HIF-2α, but not in the presence of the M252A mutant (Fig. 4e). These results confirmed that M252 residue is similarly important for T1001’s antagonistic activity.
Using H/D-ex MS studies, we found that T1001, like PT2385, produces a modest destabilizing effect at several junctions of the HIF-2α-ARNT heterodimer (Supplementary Fig. 8a). The locations of enhanced deuterium exchange rates on the crystal structure map to domain interfaces where PT2385 had also impacted (Fig. 2e and Supplementary Fig. 8b). Thus, our crystallographic, H/D-ex MS and mutation-based functional assays show a common allosteric mechanism employed by two chemically unrelated antagonists.
In addition to these two antagonists, we previously described the binding mode of antagonist 0X38,32. We made further comparisons among HIF-2α-ARNT crystal structures bound to each of PT2385, T1001 and 0X3. These ligands rely on a similar constellation of amino-acid residues for their binding, but their abilities to physically displace M252 from the pocket are notably different (Fig. 4f). PT2385 causes the most significant displacement for M252 side-chain, forcing it completely outside of the PAS-B pocket. 0X3 has a moderate impact, and T1001 produces the least displacement. The extents of movement by M252 from inside the pocket and toward the dimer interface correlate with the relative potencies of these antagonists (Fig. 4g). This key information should guide the future development of new antagonists.
Identification of HIF-2 agonists
We sought first-in-class HIF-2α binding ligands that could increase the transcriptional activity of HIF-2. Although agonists were not previously identified, the possibility that PAS pockets would allow upregulation of HIF-2 activity was plausible. This is because several cancer-related missense mutations in the pockets of PAS-A or PAS-B domains of HIF-2α protein8,33 increased transcriptional activity (Supplementary Fig. 9a,b). Therefore, we reasoned that the pockets were wired to enhance transcriptional activity through their interior contact residues. Interestingly, these cancer mutations did not result in detectible changes in stabilities of HIF-2α and ARNT heterodimers, based on our Co-IP studies (Supplementary Fig. 9c).
Our previous protein thermal shift screens had employed small chemical libraries, leading us not to find agonists. Therefore, we took a more efficient approach based on affinity selection-mass spectrometry (AS-MS) screening34 employing a larger chemical library of 32,000 compounds. M1001 (designated as compound 3, Fig. 5a) emerged as the only hit with agonistic activity. M1001 binding increased the Tm of HIF-2α-ARNT complex by 0.8 °C in the thermal shift assay (Supplementary Fig. 6), and it was subsequently found to bind to the HIF-2α PAS-B domain by MST (Fig. 5b). 786-O cells treated with M1001 showed modestly increased expression of HIF-2 target genes, producing the opposite response of PT2385 (Fig. 5c). These data confirmed that M1001 directly binds to HIF-2α and has the properties of a weak agonist.
Figure 5. The allosteric mechanism of agonist M1001.
a, Chemical structure of M1001. b, Binding of M1001 to the HIF-2α PAS-B domain (Kd about 667 nM) measured by MST. c, Opposite effects on the expression of HIF-2 target genes in 786-O cells by PT2385 (1 μM) and M1001 (10 μM). d, Binding position for M1001 (circled in black) within the entire HIF-2α-ARNT crystal structure. The visible loop region between ARNT PAS-A and PAS-B domains (A/B loop) is also indicated. e, Close-up look at the location of M1001 inside the HIF-2α PAS-B domain, with green meshes showing the FO – FC omit map contoured at 2.7σ. f, Interactions of M1001 (magenta) with surrounding residues in the pocket. g, The overall arrangement of ARNT and HIF-2α PAS-B domains is displayed in the middle, with the HIF-2α PAS-B domains from M1001-bound (cyan) or apo (orange) complexes superimposed; while enlarged or rotated views on both sides show the conformational changes due to M1001 binding. The side-chain movement of HIF-2α Y281 not only promotes an additional hydrogen bond between HIF-2α Y281 and ARNT Y456 (right), but also altered the confirmation of HIF-2α E287 and enabled its interaction with ARNT N350 on the PAS-A/B loop (left). h, H/D-ex MS results mapped on the structure of HIF-2α-ARNT complex. The regions showing dynamic changes upon M1001 binding are colored according to the maximum differences of deuteration levels (dark blue for −50 to −40%, pale purple for −40 to −25%, and pink for 25 to 40%) as compared to the apo form complex (detailed in Supplementary Fig. 11), on top of the background colors of HIF-2α (gray) and ARNT (pale yellow). Enlarged and rotated views of dimer interfaces between the two PAS-B domains, and between HIF-2α PAS-A and PAS-B domains are shown on the left and right, respectively. Error bars, mean ± s.d.; n=3 (distinct replicates for cell cultures).
Structure-function studies to reveal agonistic mechanism
We solved the complex structure of the heterodimer bound to M1001, observing the ligand inside the PAS-B domain of HIF-2α (Fig. 5d), as seen in “omit” maps (Fig. 5e). Like the antagonists, M1001 binding was stabilized through hydrophobic interactions and a single hydrogen bond with the backbone oxygen of residue A277 (Fig. 5f). Most of the residues surrounding M1001 adopt similar conformations as in the apo HIF-2α-ARNT complex, except for Y281 that undergoes a striking displacement (Supplementary Fig. 10a). Upon M1001 binding, the side-chain ring of Y281 flips out from inside the HIF-2α pocket and moves towards the ARNT subunit, creating a new hydrogen bond between HIF-2α and ARNT through the latter’s Y456 residue (Fig. 5g, right). This new contact appears to confer added stability to the heterodimer.
In all of our previous structural studies with the unliganded or antagonist-bound forms of the HIF-2α-ARNT complexes, we had found a loop connecting ARNT PAS-A and PAS-B domains (A/B loop) to be disordered (lacking electron density). However, in the M1001-bound complex, we detected clear electron density for the first time, consistent with a ligand-stabilized ARNT A/B loop (Supplementary Fig. 10b,c). We could assign nearly all the residues of this loop. Aside from M1001 stabilizing this segment, the Fα helix of M1001-bound HIF-2α PAS-B domain also moved towards the A/B loop (Fig. 5g left), further enhancing the subunit interface between HIF-2α and ARNT A/B loop (now with newly formed contact between E287 from HIF-2α Fα helix and N350 from the A/B loop of ARNT). These observations indicate that M1001’s binding to HIF-2α allosterically enhances the stabilities of protein segments, and enhances the interfacial junctions connections of HIF-2α and ARNT.
To assess if the solution state of the complex was also more stable in the presence of M1001, we employed the H/D-ex MS method to examine the altered dynamics of HIF-2α-ARNT. In clear opposition to antagonists PT2385 and T1001, M1001 produced a more stabilizing effect on various segments of the HIF-2α-ARNT complex (Supplementary Fig. 11). Closely examining the individual regions undergoing reduced H/D exchange, two critical locations are worth noting (Fig. 5h). One region is the PAS-B/PAS-B dimer interface, where the HI loop of ARNT PAS-B and the C-terminus of HIF-2α PAS-B are stabilized by M1001 binding (Fig. 5h, left), in opposition to the actions of antagonists (Fig. 2e and Supplementary Fig. 8b, both left). The second region is the PAS-A/PAS-B interface within the HIF-2α subunit, where several secondary structural elements become stabilized, including the second half of Fα, Gβ and Hβ in PAS-B domain, and the first half of Gβ in PAS-A domain (Fig. 5h, right). These solution-based data confirm that M1001 enhances the stabilities of subunit interactions at multiple locations, in contrast to antagonists.
Since agonism by M1001 was modest, we relied on a structure-activity relationship (SAR) approach to search through chemically related M1001 analogs commercially available, and to identify molecules with increased potency and/or efficacy. One analog, M1002 (designated as compound 4, Fig. 6a), increased the Tm value of HIF-2α-ARNT complex by approximately 1.8 °C in the thermal shift assay (Supplementary Fig. 6), substantially more than the 0.8 °C seen with M1001. In 786-O cells, M1002 enhanced the expression of HIF-2 target genes with greater efficacy than M1001 (Fig. 6b and Fig. 5c).
Figure 6. The agonistic effects of M1002.
a, Chemical structure of M1002. b, Clear agonistic effects on the expression of HIF-2 target genes in 786-O cells by M1002 (10 μM) as compared with the antagonist PT2385 (1 μM). c, TR-FRET-based protein binding assays testing the effects of M1002 on the associations between ARNT and its partners HIF-2α, HIF-1α and NPAS3, respectively. d, Comparison of M1002’s agonistic effects at 10 μM on HIF-2 target genes in HEK293T cells transfected with wild-type HIF-2α or Y281A mutant. Error bars, mean ± s.d.; n=3 (distinct replicates for cell cultures). e, Co-IP results showing the effects of HIF-2α ligands (PT2385, T1001, M1001 and M1002 at various concentrations) on the association between full-length HIF-2α and VHL proteins. This experiment was independently repeated twice with similar results.
To see how the M1001/M1002 agonist class could impact the physical dimerization between isolated HIF-2α and ARNT proteins in solution, we used the TR-FRET-based assay. As shown in Fig. 6c, M1002 enhanced the physical association of HIF-2α and ARNT in a dose-dependent fashion. To assess its selectivity for HIF-2α, we studied its effects on both HIF-1α-ARNT and NPAS3-ARNT complexes. M1002 could increase the interactions between HIF-1α and ARNT, but only at higher compound concentrations. But M1002 had no discernible effect on the association of NPAS3 with ARNT (Fig. 6c).
To address the mechanistic question of whether the agonistic effect of M1002 was dependent on HIF-2α residue Y281 (Fig. 5g), we made a point mutation Y281A and compared activities with WT when overexpressing each protein in HEK293T cells. M1002 treatment increased the expression of HIF-2 target genes (i.e. VEGA, EPO and NDRG1) with the WT protein, but not with the Y281A mutant (Fig. 6d), pointing to Y281 as a critical element in its allosteric action.
As the von Hippel-Lindau (VHL) tumor suppressor protein binds to HIF-α proteins to regulate their stability, we further tested if HIF-2α ligands might change the extent of interaction between VHL and HIF-2α. We co-transfected HEK293T cells with plasmids encoding both full-length proteins, and treated cells with different ligands. As shown by the co-IP results in Fig. 6e, in contrast to the antagonists (PT2385 and T1001), two agonists (M1001 and M1002) could reduce the physical association between HIF-2α and VHL. The reduced VHL binding to HIF-2α in the presence of agonists could be due to the intrinsically weakened interactions between their proteins caused by the conformational changes induced in HIF-2α, or due to secondary effects the compounds may have in reducing proline hydroxylation levels on HIF-2α.
Potential synergy of HIF-2α agonists and PHD inhibitors
The activation of HIF pathways may be clinically desirable in renal anemia and ischemia26,35. Several small-molecule compounds are in clinical trials to treat anemia in CKD patients, notably roxadustat (FG-4592)36, daprodustat (GSK1278863)37, vadadustat (AKB-6548)38 and molidustat (BAY 85–3934)39. All of these are PHD inhibitors which boost HIF-2α protein levels, leading to increased HIF-2 activity and the production of endogenous EPO29.
Since PHD inhibitors activate the HIF-2 pathway indirectly, in contrast to the agonists we described here that bind directly to HIF-2α, we asked whether the combination of these two types of approaches could synergistically stimulate EPO expression. We selected two known PHD inhibitors (molidustat and roxadustat), and examined their effects on the expression of HIF target genes in Hep3B cells co-treated with M1002 (Supplementary Fig. 12). The PHD inhibitors increased the expression of HIF-2 target genes (EPO and NDRG1) about 3–5 fold, but had relatively small effects on HIF-1 selective target gene (PGK1). M1002 alone at low concentration produced little enhancement on gene expression. However, co-treatment of this same concentration of M1002 together with PHD inhibitors elevated the expression of EPO and NDRG1 but not that of PGK1, suggesting synergy between HIF-2α agonists and PHD inhibitors.
DISCUSSION
We investigated the activities, binding sites, and allosteric mechanisms of known and newly discovered HIF-2 modulators, and found all to influence the stability of HIF-2α-ARNT complex allosterically. By comparing the crystal structures of HIF-2α-ARNT in complex with various antagonists (including PT2385, T1001 and 0X3), we found that ligand-induced movement of HIF-2α pocket residue M252’s side-chain was critical in destabilizing the heterodimeric complex formed between HIF-2α and ARNT. We further found that the relative potency of the antagonists correlated with the degree to which they could physically displace the M252 side-chain from inside of pocket and point it towards the dimer interface.
While PT2385 is a potent inhibitor, the resistance mutations that arise due to its extended exposure in vivo could limit its long-term effectiveness and ultimately necessitate the use of new classes of antagonists for prolonged therapy24. In addition to the PAS-B domain binding compounds emphasized here and in prior studies32,40–42, new antagonists that bind to other pockets of HIF-2α-ARNT complex (e.g. PAS-A domain) could prove equally promising8. In this regard, our thorough structural description of this ligand pocket, together with residue M252’s allosteric role should facilitate the discovery of many novel and potent antagonists. 0X3, PT2385 and T1001 are chemically unrelated to each other, yet all bind effectively with an overlapping footprint to the same pocket. This observation shows the PAS-B domain pocket of HIF-2α to be highly accommodative for a wide-variety of distinct chemical ligands and ideal for further ligand discovery.
In terms of agonists, we showed for the first time that small-molecules capable of activating HIF-2α protein could be discovered from high-throughput screens. These agonists could provide an alternative approach to treating renal anemia. Some PHD inhibitors are currently in Phase III clinical trials, validating the HIF-2/PHD axis as an effective target for increasing EPO protein levels and ameliorating anemia26–28. However, a possible disadvantage of PHD inhibitors may be that their other targets, unrelated to the HIF-2α protein, could create undesired side-effects during anemia therapy. Direct-binding HIF-2α agonists should be explored given their likelihood to produce fewer side-effects. The agonist class we discovered here binds to the same HIF-2α PAS-B pocket as the antagonists. However, we observed agonists trigger the movement of an altogether different residue from inside the pocket, namely Y281. This displaced residue stabilizes the overall conformations and enhances subunit dimerization, in contrast to role of M252 in mediating the effects of antagonists.
Our studies here also show for the first time that HIF-2 can be functionally modulated in a bidirectional manner (agonists versus antagonists) through chemical ligands, even when these ligands bind to precisely the same PAS-B pocket of HIF-2α. The gain-of-function cancer related mutations that mapped directly to HIF-2α pockets foreshadowed the possibility of upregulating functional activity using agonists that contact these pockets. In the future, similar sets of studies with other mammalian bHLH-PAS family members are needed to assess if they too are amenable to activation and inhibition via direct-binding chemical ligands.
ONLINE METHODS
Plasmid construction and site-directed mutagenesis.
Recombinant expression plasmids containing the N-terminal bHLH-PAS-A-PAS-B regions of mouse HIF-2α and ARNT proteins, pSJ2-HIF-2α (3–361) and pMKH-ARNT (82–464), as well as the full-length plasmids pCMV-Tag4-HIF-2α (Flag-tagged) and pCMV-Tag1-ARNT (Myc-tagged) used in co-immunoprecipitation, were described previously8. To express the single PAS-B domain of HIF-2α, its DNA segment coding for residues 241–361 was cloned into the pSJ2 vector. For the plasmid construction of GFP-tagged ARNT used in the TR-FRET assay, we cloned ARNT DNA segment coding for residues 82–470, and amplified the DNA of Aequorea GFP from a pAdTrack-CMV plasmid (gifted by Dr. Denny Liu at SBP). Two DNA segments were then further digested and ligated using a BamHI cutting site designed at the C-terminus of ARNT and N-terminus of GFP, before inserting together into the pMKH vector to produce a chimeric ARNT-GFP protein with no purification tags. Site-directed mutagenesis was performed on corresponding wild-type HIF-2α or ARNT plasmids and confirmed by DNA sequencing.
Protein expression and purification.
To obtain HIF-2α-ARNT complex proteins, the pSJ2-HIF-2α plasmid was co-transformed along with pMKH-ARNT into BL21-CodonPlus (DE3)-RIL competent cells (Agilent Technologies). Following 0.1 mM IPTG induced protein expression overnight at 16 °C, cell pellets were lysed by sonication, and supernatants were applied onto pre-packed His∙Bind resin (Novagen). The bound proteins were further purified using SP Sepharose (GE Healthcare), and the eluted fractions were then loaded on a HiLoad 16/60 Superdex 200pg gel filtration column (GE Healthcare) equilibrated in 20 mM Tris (pH 8.0) and 400 mM NaCl. DTT was added to the pooled protein peak fractions at 10 mM. The heterodimeric proteins of HIF-2α and ARNT-GFP were prepared similarly as described above, except that the pMKH-ARNT-GFP plasmid was used in the place of pMKH-ARNT. The ARNT-GFP protein was co-expressed and purified in complex with HIF-1α and NPAS3 (plasmids made in our previous studies3,8), respectively. The single PAS-B domain of HIF-2α was produced by transformation of pSJ2-HIF-2α (241–361) into BL21-CodonPlus (DE3)-RIL, followed by overnight expression, and purification using His-tag affinity chromatography and gel filtration chromatography.
MST (microscale thermophoresis) binding assay.
The binding affinities of HIF-2α ligands were measured using MST method. Purified HIF-2α protein (PAS-B domain) was labeled with the Monolith NT™ Protein Labeling Kit RED. Compounds were diluted in a range of concentrations and mixed with labeled protein at room temperature in the assay buffer containing 20 mM Tris (pH 7.5), 50 mM NaCl, 5 mM β-mercaptoethanol, 0.05% Tween-20 and 1% DMSO. Mixed samples were loaded into Monolith TM standard-treated capillaries, and the thermophoresis was carried out on a Monolith NT.115 instrument (NanoTemper Technologies GmbH). Binding was measured with 20% LED power and “medium” MST power, with an optimized time setting (5s Fluo, Before; 20s MST On; 5s Fluo, After). Kd values were obtained by fitting the MST data in the GraphPad Prism 7 software.
FP (fluorescence polarization) DNA binding assay.
The preparation of fluoresceinated double-strand HRE DNA (6-FAM labeled F-strand: 5’-GGCTGCGTACGTGCGGGTCGT-3’; unlabeled R-strand: 5’-ACGACCCGCACGTACGCAGCC-3’) was described previously8. HIF-2α-ARNT protein complex was diluted in a 2-fold series in binding buffer (20 mM Tris pH 8.0, 20 mM NaCl), and each sample was incubated with 0.1 μM, 1 μM or 10 μM PT2385 (MedChemExpress HY-12867, 99.48% purity) for 1 hour (with the same amount of 0.1% DMSO as a control), before binding assays started by the addition of 2 nM DNA. The FP signals were recorded using black 96-well plates on a Synergy H1 microplate reader (BioTek Instruments), and Kd values were calculated by fitting curves in GraphPad Prism 7.
Luciferase reporter assay.
786-O cells with a stable-transfected HRE-luc reporter were kindly provided by Dr. Ian Pass at SBP. These cells were seeded in RPMI1640 medium with 10% FBS in 96-well plates at 5 × 104 per well, and one day later treated with PT2385 at different concentrations (final DMSO concentration kept at 0.1%). After 24-hour incubation, cells were lysed and analyzed using Steady-Glo Luciferase Assay System (Promega). To test the transcriptional activities of HIF-2α harboring the cancer-related mutations, HEK293T cells (ATCC CRL-3216) were seeded in 24-well plates in DMEM containing 10% FBS, and one day later transfected with 200 ng of pCMV-Tag4-HIF-2α (WT, mutants or empty plasmid), 1 ng of pRL (control Renilla luciferase) and 100 ng of HRE-luc reporter using 0.6 μL jetPRIME regent for each well. Medium was refreshed after overnight transfection, and luciferase activity was measured another 24 hours later using the Dual-Glo Luciferase Assay System (Promega). Final data were normalized by the relative ratio of firefly and Renilla luciferase activity.
Real-time PCR.
786-O cells (ATCC CRL-1932) were cultured in RPMI1640 medium with 10% FBS in 6-well plates at 37°C in 5% CO2. For the treatment, antagonists or agonists were added at various concentrations with the equal amount of 0.1% DMSO as a control. After 24 hours, cells were collected, and RNA was isolated with EZNA Total RNA Kit (Omega Bio-tek), followed by cDNA synthesis using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). HEK293T cells were seeded in 6-well plates and transfected with pCMV-Tag4-HIF-2α plasmids (WT, mutants M252A or Y281A). Medium was refreshed after overnight transfection, and cells were treated with 10 μM compounds (PT2385, T1001 or M1002) in 0.1% DMSO for 24 h before RNA isolation.
Hep3B cells (ATCC HB-8064) were cultured in DMEM medium with 10% FBS in 6-well plates. To test the inhibitory effects of PT2385, cells were first placed overnight in normoxia (21% O2) and hypoxia (1% O2) conditions, respectively. Then they were treated with 0.1% DMSO or 10 μM PT2385 for 8 h before RNA isolation. For the co-treatment of PHD inhibitors and HIF-2α agonist, Hep3B cells were incubated with 10 μM of molidustat (Cayman Chemical 15297) or roxadustat (Cayman Chemical 15294) for 8 h before 5 μM of M1002 (Molport 001–006-026, >95% purity) was added into the medium (final DMSO concentration all kept at 0.1%). After another 24 hours, cells were collected for RNA isolation and cDNA synthesis.
Real-time PCR was performed on the StepOnePlus system using Fast SYBR Green Master Mix (Thermo Fisher Scientific). The targeted gene expressions were normalized to the expression of Beta Actin (ACTB) in the same sample. PCR primers were synthesized by Integrated DNA Technologies as follows: ACTB (F: GCACAGAGCCTCGCCTT, R: GTTGTCGACGACGAGCG); VEGFA (F: TACCTCCACCATGCCAAGTG, R: ATGATTCTGCCCTCCTCCTTC); NDRG1 (F: CTCCTGCAAGAGTTTGATGTCC, R: TCATGCCGATGTCATGGTAGG); GLUT1 (F: TCTGGCATCAACGCTGTCTTC, R: CGATACCGGAGCCAATGGT); CyclinD1 (F: TGGAGCCCGTGAAAAAGAGC, R: TCTCCTTCATCTTAGAGGCCAC); EPO (F: GGAGGCCGAGAATATCACGAC, R: CCCTGCCAGACTTCTACGG); and PGK1 (F: TTAAAGGGAAGCGGGTCGTTA, R: TCCATTGTCCAAGCAGAATTTGA).
Co-IP (immunoprecipitation).
Similar to our previous work8, HEK293T cells were seeded in 10 cm dishes and cultured in DMEM containing 10% FBS at 37°C with 5% CO2. One day later, cells were transfected with 2 μg pCMV-Tag4-HIF-2α (wild-type or mutants) and 6 μg pCMV-Tag1-ARNT (or pCMV-Tag1-VHL) plasmids by 16 μL jetPRIME regent (Polyplus-transfection). After overnight incubation, medium was refreshed with 50 μM 2,2’-dipyridine and various concentrations of PT2385 (or other ligands) in 0.15% DMSO. Another 24 hours later, cells were harvested and sonicated in 700 μL lysis buffer (1X TBS with 1 mM EDTA, 1% TRITON X-100 and 1X protease inhibitor cocktail). About 40 μg of supernatant was saved as input for western blot using Monoclonal ANTI-FLAG M2 antibody (Sigma F1804) and anti-Myc rabbit mAb (Cell Signaling #2278). Immunoprecipitation was performed with 1 mg of supernatant and 40 μL of ANTI-FLAG M2 affinity gel suspension (Sigma A2220), followed by western blot using the anti-Myc antibody.
Crystallization and X-ray data collection.
HIF-2α-ARNT protein complexes were crystallized by mixing equal volume of protein and the reservoir containing tacsimate (pH 7.0) and PEG3350 using sitting-drop vapor diffusion method at 16 °C as before8. For PT2385 and M1001 (MolPort 001-008-374, >95% purity), crystals of HIF-2α-ARNT in complex with ligands were obtained by adding compounds (100 μM) to the drops containing apo protein crystals and soaking overnight before crystal harvest. For T1001 (Santa Cruz Biotechnology sc-201431, ≥98% purity), co-crystallization of HIF-2α-ARNT with ligands (100 μM) was successful. 30% PEG400 was added into the reservoir solution to cryoprotect the crystals before flash frozen. Diffraction data were collected at 100 K at the Argonne National Laboratory SBC-CAT 19ID beamline, and processed using the HKL3000 program43.
Structure determination and refinement.
The structures of HIF-2α-ARNT in complex with ligands were solved by molecular replacement with the program Phaser44, using the apo HIF-2α-ARNT structure (PDB code 4ZP4) as the search model. Further manual model building was facilitated using Coot45, combined with the structure refinement using phenix.refine46. The diffraction data and final statistics are summarized in Supplementary Table 1. The Ramachandran statistics, as calculated by Molprobity47, are 92%/0, 93%/0.2% and 93%/0.2% (favored/outliers) for complex structures of HIF-2α-ARNT-PT2385, HIF-2α-ARNT-T1001 and HIF-2α-ARNT-M1001, respectively. All the structural figures were prepared using PyMol (The PyMol Molecular Graphics System, Version 1.3, Schrödinger, LLC).
H/D-ex MS (hydrogen-deuterium exchange mass spectrometry) assay.
Hydrogen-deuterium exchange experiments were performed using the in-house deuterium exchange system48,49. In this system, the enzymatic digestion, peptide separation and MS analysis are all fully automated. To start the H/D-ex reaction, 3 μl pre-chilled protein stock solution at 19.5 μM (free HIF-2 protein complex, HIF-2 protein complex combined with agonists or antagonists) was diluted into 9 μl D2O buffer (8.3 mM Tris, 250 mM NaCl, in D2O, pDREAD 7.2), and incubated at 0 °C for 30, 100, 300, 1000, 3000 and 10000 s, respectively. At indicated times, 18 μl cold acidic buffer (1.6 M GuHCl, 100 mM Glycine, 16.6% (v/v) Glycerol, pH 2.4) was added to quench the exchange reaction. To maximize sequence coverage of HIF2α-ARNT complex, various concentrations of GuHCl (0.08, 0.8, 1.6 and 3.2 M) were used to optimize quench condition, and the best coverage map was obtained using 1.6 M GuHCl quench buffer. Quenched samples were then frozen on dry ice and were passed through an immobilized pepsin column (16 μl bed volume) on ice in H2O containing 0.05% (v/v) trifluoroacetic acid at 20 μl min−1 for pepsin digestion. The pepsin column was made by conjugating pepsin onto POROS 20 AL self-pack aldehyde-activated resin at 40 mg/ml via couple chemistry and packing at 5 ml min−1 into one 1×20 mm guard column. The proteolytic products were processed on a C18 trap column for desalting (Optimizetechnologies, Magic C18 AQ, 0.2×2 mm) and on a Michrom Magic C18 (3 μm, 0.2×50 mm, 200 Å) with a 30-min linear acetonitrile gradient (6.4–38.4%) for separation. The eluted samples were subjected to an OrbiTrap Elite mass spectrometer (Thermo Fisher Scientific), which was optimized for H/D-ex analysis as previously reported50. The identification of peptides was performed using Proteome Discoverer (Thermo Fisher Scientific), and their deuteration levels were determined by HDEXaminer (Sierra Analytics Inc). In addition, H/D-ex analysis was also conducted on both non-deuterated and fully deuterated samples to correct the back-exchange51. The Peptide coverage map for HIF-2α-ARNT is shown in Supplementary Fig. 14.
TR-FRET (time-resolved fluorescence energy transfer)-based in vitro binding assay.
This method was developed based on the LanthaScreen technology (Thermo Fisher Scientific). Purified protein complexes of GFP-tagged ARNT and His-tagged HIF-2α, HIF-1α or NPAS3 were dispensed into 1536-well plates with serial dilution, in the assay buffer containing 20 mM Tris (pH 7.4), 100 mM NaCl, 1mM DTT and 0.005% tween-20. After addition of the LanthaScreen Elite Terbium-labeled anti-His antibody (Thermo Fisher Scientific PV5863) into each well at 5 nM, the plate was centrifuged at 800 rpm for 1 min and kept in dark for 1 h. Then the protein interactions were monitored via the energy transfer signal with an EnVision multilabel plate reader (PerkinElmer). The TR-FRET value was determined as a ratio of the signal measured at 520 nm (GFP) to the signal measured at 492 nm (terbium). The apparent Kd values of each protein complex were calculated by plotting the ratios (520 nm/492 nm) against the protein concentrations in GraphPad Prism 7. To test effects of antagonists or agonists on heterodimerization, similar protocols were used except that the protein concentration was kept at 50 nM, while the compounds were added in a wide range of serial dilution with 1% DMSO.
Thermal shift binding assays.
The initial screening based on thermal shift method was carried out in 384-well plates with a LightCycler 480 instrument (Roche). The concentrations for protein and compounds used in the screening were 2 μM and 20 μM (with 0.1% DMSO), respectively. To get quantitative thermal shift (ΔTm) values, we ran this assay in 96-well format using the Protein Thermal Shift (PTS) Dye Kit on a StepOnePlus qPCR machine (Thermo Fisher Scientific), according to the manufacturer’s instructions. For each well, the concentration of protein complex was about 1.5 μM, and all the antagonists or agonists were tested at 10 μM in the assay buffer containing 20 mM Tris (pH 8.0), 400 mM NaCl and 0.1% DMSO. Final data were analyzed using the PTS software v1.2 to calculate the Derivative curve determined Tm values and to compare ΔTm values of different ligands.
AS-MS (affinity selection-mass spectrometry) screening.
We employed a 32,000-chemical library consisting of 80 distinct pools of 400 compound mixtures, to rapidly identify the preferential binding of candidate molecules to our protein targets. Diluted HIF-2α-ARNT protein was dispensed into wells with a final protein concentration of 10 μM and a final compound concentration of 2 μM each in about 1.5% DMSO. The plate was kept in a shaker at room temperature for 1 h with gentle mixing, to allow fully interactions between protein and compounds. After spinning the plate at 1500 g for 1 min to precipitate the insoluble components, supernatant from each well was subjected to a size exclusion PolyHYDROXYETHYL A column (PolyLC Inc.) to separate the protein-compound complexes and unbound components. Isolated protein-compound complexes were then passed into a reversed phase Accucore C18 LC column (Thermo Fisher Scientific) in a heat chamber. Heat-released compounds were further separated and analyzed by TOF-MS system (Agilent Technologies). The key parameters of above two screens (PTS and AS-MS) are summarized in Supplementary Table 2.
Data availability.
The three sets of structural data from HIF-2α-ARNT crystals in complex with PT2385, T1001 and M1001 are available in wwPDB under accession codes 6E3S, 6E3T and 6E3U, respectively. Other data shown in the article are available from the correspond authors upon reasonable request.
Supplementary Material
Acknowledgements
We thank members of the Structural Biology Center at Argonne National Laboratory for their help with data collection at the 19-ID beamline, and Garib N. Murshudov at University of Cambridge for the help with ligand building at the 2016 CCP4/APS summer school. This work was supported by Welcome Trust, and by grants from the National Institutes of Health (R01GM117013, R01DK118297) and U.S. ARMY Medical Research (W81XWH-16-1-0322) to F.R., as well as grants from Shandong University (Qilu Young Scholar 86963072), National Natural Science Foundation of China (31700114), Natural Science Foundation of Jiangsu Province (BK20170399), and the 111 Project (B16030) to D.W.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Wu D & Rastinejad F Structural characterization of mammalian bHLH-PAS transcription factors. Curr Opin Struct Biol 43, 1–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moglich A, Ayers RA & Moffat K Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17, 1282–94 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu D, Su X, Potluri N, Kim Y & Rastinejad F NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors. Elife 5, e18790 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McIntosh BE, Hogenesch JB & Bradfield CA Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu Rev Physiol 72, 625–45 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Schito L & Semenza GL Hypoxia-Inducible Factors: Master Regulators of Cancer Progression. Trends Cancer 2, 758–770 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Keith B, Johnson RS & Simon MC HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer 12, 9–22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ravenna L, Salvatori L & Russo MA HIF3alpha: the little we know. FEBS J 283, 993–1003 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Wu D, Potluri N, Lu J, Kim Y & Rastinejad F Structural integration in hypoxia-inducible factors. Nature 524, 303–8 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Ivan M et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–8 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Jaakkola P et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–72 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Yu F, White SB, Zhao Q & Lee FS HIF-1alpha binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc Natl Acad Sci U S A 98, 9630–5 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lando D, Peet DJ, Whelan DA, Gorman JJ & Whitelaw ML Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science 295, 858–61 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Lando D et al. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev 16, 1466–71 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang P, Chandra V & Rastinejad F Structural overview of the nuclear receptor superfamily: insights into physiology and therapeutics. Annu Rev Physiol 72, 247–72 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denison MS & Nagy SR Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol 43, 309–34 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Denison MS, Soshilov AA, He G, DeGroot DE & Zhao B Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol Sci 124, 1–22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheuermann TH et al. Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor. Proc Natl Acad Sci U S A 106, 450–5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Key J, Scheuermann TH, Anderson PC, Daggett V & Gardner KH Principles of ligand binding within a completely buried cavity in HIF2alpha PAS-B. J Am Chem Soc 131, 17647–54 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardoso R et al. Identification of Cys255 in HIF-1alpha as a novel site for development of covalent inhibitors of HIF-1alpha/ARNT PasB domain protein-protein interaction. Protein Sci 21, 1885–96 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Y et al. Regulating the ARNT/TACC3 axis: multiple approaches to manipulating protein/protein interactions with small molecules. ACS Chem Biol 8, 626–35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fala AM et al. Unsaturated fatty acids as high-affinity ligands of the C-terminal Per-ARNT-Sim domain from the Hypoxia-inducible factor 3alpha. Sci Rep 5, 12698 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hewitson KS & Schofield CJ The HIF pathway as a therapeutic target. Drug Discov Today 9, 704–11 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Wallace EM et al. A Small-Molecule Antagonist of HIF2alpha Is Efficacious in Preclinical Models of Renal Cell Carcinoma. Cancer Res 76, 5491–500 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Chen W et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature 539, 112–117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho H et al. On-target efficacy of a HIF-2alpha antagonist in preclinical kidney cancer models. Nature 539, 107–111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maxwell PH & Eckardt KU HIF prolyl hydroxylase inhibitors for the treatment of renal anaemia and beyond. Nat Rev Nephrol 12, 157–68 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Yousaf F & Spinowitz B Hypoxia-Inducible Factor Stabilizers: a New Avenue for Reducing BP While Helping Hemoglobin? Curr Hypertens Rep 18, 23 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Gupta N & Wish JB Hypoxia-Inducible Factor Prolyl Hydroxylase Inhibitors: A Potential New Treatment for Anemia in Patients With CKD. Am J Kidney Dis 69, 815–826 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Yeh TL et al. Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials. Chem Sci 8, 7651–7668 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seidel SA et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 59, 301–15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciulli A & Abell C Fragment-based approaches to enzyme inhibition. Curr Opin Biotechnol 18, 489–96 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheuermann TH et al. Allosteric inhibition of hypoxia inducible factor-2 with small molecules. Nat Chem Biol 9, 271–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forbes SA et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 43, D805–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Annis DA, Nickbarg E, Yang X, Ziebell MR & Whitehurst CE Affinity selection-mass spectrometry screening techniques for small molecule drug discovery. Curr Opin Chem Biol 11, 518–26 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Bonomini M, Del Vecchio L, Sirolli V & Locatelli F New Treatment Approaches for the Anemia of CKD. Am J Kidney Dis 67, 133–42 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Besarab A et al. Roxadustat (FG-4592): Correction of Anemia in Incident Dialysis Patients. J Am Soc Nephrol 27, 1225–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brigandi RA et al. A Novel Hypoxia-Inducible Factor-Prolyl Hydroxylase Inhibitor (GSK1278863) for Anemia in CKD: A 28-Day, Phase 2A Randomized Trial. Am J Kidney Dis 67, 861–71 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Pergola PE, Spinowitz BS, Hartman CS, Maroni BJ & Haase VH Vadadustat, a novel oral HIF stabilizer, provides effective anemia treatment in nondialysis-dependent chronic kidney disease. Kidney Int 90, 1115–1122 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Beck H et al. Discovery of Molidustat (BAY 85–3934): A Small-Molecule Oral HIF-Prolyl Hydroxylase (HIF-PH) Inhibitor for the Treatment of Renal Anemia. ChemMedChem 13, 988–1003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogers JL et al. Development of inhibitors of the PAS-B domain of the HIF-2alpha transcription factor. J Med Chem 56, 1739–47 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheuermann TH et al. Isoform-Selective and Stereoselective Inhibition of Hypoxia Inducible Factor-2. J Med Chem 58, 5930–41 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Wehn PM et al. Design and Activity of Specific Hypoxia-Inducible Factor-2alpha (HIF-2alpha) Inhibitors for the Treatment of Clear Cell Renal Cell Carcinoma: Discovery of Clinical Candidate ( S)-3-((2,2-Difluoro-1-hydroxy-7-(methylsulfonyl)-2,3-dihydro-1 H-inden-4-yl)oxy)-5-fluorobenzonitrile (PT2385). J Med Chem (2018). [DOI] [PubMed] [Google Scholar]
- 43.Minor W, Cymborowski M, Otwinowski Z & Chruszcz M HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62, 859–66 (2006). [DOI] [PubMed] [Google Scholar]
- 44.McCoy AJ et al. Phaser crystallographic software. J Appl Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marsh JJ et al. Structural insights into fibrinogen dynamics using amide hydrogen/deuterium exchange mass spectrometry. Biochemistry 52, 5491–502 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woods VL Jr. & Hamuro Y High resolution, high-throughput amide deuterium exchange-mass spectrometry (DXMS) determination of protein binding site structure and dynamics: utility in pharmaceutical design. J Cell Biochem Suppl Suppl 37, 89–98 (2001). [DOI] [PubMed] [Google Scholar]
- 50.Walters BT, Ricciuti A, Mayne L & Englander SW Minimizing back exchange in the hydrogen exchange-mass spectrometry experiment. J Am Soc Mass Spectrom 23, 2132–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S et al. Mechanism of intracellular cAMP sensor Epac2 activation: cAMP-induced conformational changes identified by amide hydrogen/deuterium exchange mass spectrometry (DXMS). J Biol Chem 286, 17889–97 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The three sets of structural data from HIF-2α-ARNT crystals in complex with PT2385, T1001 and M1001 are available in wwPDB under accession codes 6E3S, 6E3T and 6E3U, respectively. Other data shown in the article are available from the correspond authors upon reasonable request.