Abstract
One of the major challenges in evolutionary biology is the identification of the genetic basis of postzygotic reproductive isolation. Given its pivotal role in this process, here we explore the drivers that may account for the evolutionary dynamics of the PRDM9 gene between continental and island systems of chromosomal variation in house mice. Using a data set of nearly 400 wild-caught mice of Robertsonian systems, we identify the extent of PRDM9 diversity in natural house mouse populations, determine the phylogeography of PRDM9 at a local and global scale based on a new measure of pairwise genetic divergence, and analyze selective constraints. We find 57 newly described PRDM9 variants, this diversity being especially high on Madeira Island, a result that is contrary to the expectations of reduced variation for island populations. Our analysis suggest that the PRDM9 allelic variability observed in Madeira mice might be influenced by the presence of distinct chromosomal fusions resulting from a complex pattern of introgression or multiple colonization events onto the island. Importantly, we detect a significant reduction in the proportion of PRDM9 heterozygotes in Robertsonian mice, which showed a high degree of similarity in the amino acids responsible for protein–DNA binding. Our results suggest that despite the rapid evolution of PRDM9 and the variability detected in natural populations, functional constraints could facilitate the accumulation of allelic combinations that maintain recombination hotspot symmetry. We anticipate that our study will provide the basis for examining the role of different PRDM9 genetic backgrounds in reproductive isolation in natural populations.
Keywords: PRDM9, Mus musculus domesticus, Robertsonian fusion, postzygotic reproductive isolation, selection, recombination
Introduction
Understanding the genetic basis of speciation is a long-standing quest in biology. This entails the investigation of mechanisms responsible for postzygotic reproductive isolation, which can be attributable to genic (e.g., speciation genes) or chromosomal (e.g., inversions and fusions) factors (Coyne and Orr 1998; Orr and Turelli 2001; Faria and Navarro 2010; Farré et al. 2013; Capilla et al. 2014, 2016). Whether both types of mechanisms can influence speciation processes independently or in combination remains largely unexplored.
Genes that cause hybrid sterility have been described mostly in Drosophila and include the Odysseus-site Homeobox (OdsH) gene (Ting et al. 1998), the JYAlpha gene (Masly et al. 2006), Hmr (Barbash et al. 2003), nucleoporin Nup96 (Presgraves et al. 2003), and the Overdrive (Ovd) gene (Phadnis and Orr 2009). In mice, the PR domain zinc finger 9 (Prdm9) gene contributes to hybrid sterility in Musmusculus subspecies (Mus musculusdomesticus × Mus musculusmusculus) (Mihola et al. 2009). The Prdm9 gene is located within the proximal centromeric regions of mouse chromosome 17 and it is implicated in the genomic distribution of recombination hotspots. It codes for a meiotic-specific histone (H3) methyltransferase with a C-terminal tandem repeat zinc finger (ZnF) domain that adds H3K4me3 marks at nucleosomes close to double-strand breaks (DSBs) in early meiosis; it does so through its recognition of a species-specific and highly polymorphic repetitive DNA motif (Mihola et al. 2009; Baudat et al. 2010). The high amino acid variation that characterizes PRDM9 in mice and the repetitive nature of the DNA motif that it recognizes have suggested the existence of a positive molecular feedback implicated in the generation of recombination hotspots (Oliver et al. 2009; Jeffreys et al. 2013; Buard et al. 2014; Capilla et al. 2014; Kono et al. 2014; Schwartz et al. 2014; Baker et al. 2017). That is, the repair of DSBs in early meiosis can introduce mutations in preferred PRDM9 binding motifs, thus influencing the rapid evolution of novel binding domains (ZnFs) in the PRDM9 protein (Boulton et al. 1997; Coop and Myers 2007; Davies et al. 2016). Such evolutionary turnover has important implications for the establishment of reproductive postzygotic barriers since differences in recombination landscapes in hybrids can account for failure in chromosomal synapsis during meiosis (Qiao et al. 2012; Bhattacharyya et al. 2013; Smagulova et al. 2016). In this scenario, the presence of evolutionarily distinct heterozygous combinations of PRDM9 can result in asymmetric DSBs (i.e., differences in genomic distribution and number of DSBs) between homologous chromosomes, which depending on the interaction between different X-linked and autosomal loci (Balcova et al. 2016) can result in subfertile and even sterile phenotypes (Davies et al. 2016). Since interallelic PRDM9 incompatibilities can result in hybrid sterility due to failure in recognition of DNA-binding sites (Flachs et al. 2012), understanding PRDM9 variability in natural populations can reveal potential drivers behind the evolutionary dynamics of this gene.
With regards to the involvement of chromosomal rearrangements in evolution, house mice represent one of the most extraordinary models of chromosomal variation in mammals. The variation that exists in mice may promote speciation due to meiotic impairment and infertility associated with chromosomal heterozygosity (Capilla et al. 2014; Pavlova and Searle 2018). The standard karyotype of M. m. domesticus consists of 40 all-acrocentric chromosomes. However, numerous populations in Western Europe and North Africa show high variability in diploid numbers and karyotypes resulting from Robertsonian (Rb) fusions of nonhomologous acrocentric chromosomes and/or whole arm reciprocal translocations, giving rise to new metacentric chromosomes (Piálek et al. 2005). This includes mice from the northeast of the Iberian Peninsula, the so-called “Barcelona Rb system” (Adolph and Klein 1981; Gündüz, López-Fuster, et al. 2001; Medarde et al. 2012) and on the island of Madeira (Britton-Davidian et al. 2000), both Rb systems are characterized by distinct patterns of chromosomal variation. On the one hand, the Barcelona Rb system extends over an area of 5,000 km2 within the provinces of Barcelona, Tarragona, and Lleida (Spain) and includes individuals with diploid numbers (2n) ranging from 2n = 27 to 2n = 40 (Medarde et al. 2012, and references therein). This Rb system is characterized by the presence of seven different metacentric chromosomes (Rb[3.8], Rb[4.14], Rb[5.15], Rb[6.10], Rb[7.17], Rb[9.11], and Rb[12.13]), distributed in nongeographically coincident (staggered) clines leading to a progressive reduction in diploid numbers towards the center of the range, about 30-km west of the city of Barcelona (Gündüz, López-Fuster, et al. 2001; Medarde et al. 2012). The Barcelona Rb system is also noteworthy for the high levels of chromosomal polymorphism, which is consistent with the absence of a metacentric race (Medarde et al. 2012; Capilla et al. 2014; Sánchez-Guillén et al. 2015). On the other hand, mice from the Madeira archipelago (including the islands of Madeira and Porto Santo) show an extensive chromosomal radiation that includes mice with the standard 2n = 40 (i.e., the island of Porto Santo) and mice with a highly structured chromosomal race organization on the island of Madeira itself (Britton-Davidian et al. 2000). Madeira, an island with an extreme topography, has six well-established chromosomal races that have been described within a geographical range of only 742 km2. Diploid numbers vary from 2n = 22 to 2n = 28, with up to nine metacentric chromosomes accumulated within a maximum of 1,200 years (Förster et al. 2009). Importantly, most metacentric populations are geographically isolated and do not co-occur with others except for some cases of marginal overlap of the chromosomal races. The chromosomal differences between some of the Madeira races are so pronounced that hybrids are at very low frequency, presumably due to reduced F1 fitness associated with meiotic impairment (Britton-Davidian et al. 2000).
The contrasting nature of the chromosomal variation in the Madeira and Barcelona Rb systems provides a unique opportunity to investigate the mechanisms underlying PRDM9 variation. It permits the examination of PRDM9 evolutionary dynamics between a continental mouse population (with a distribution of metacentric chromosomes in a polymorphic state; i.e., the Rb Barcelona system) and those of an island’s chromosomal system (subdivided into entirely or partially reproductively isolated units with metacentric chromosomes in a homozygous state; i.e., the Madeira system). Here, using a phylogenetic approach on a data set of ∼400 wild-caught house mice from the Madeira archipelago (Madeira and Porto Santo Islands) and the Barcelona system (continental) (fig. 1 ) we 1) identify the extent of PRDM9 diversity in natural house mouse populations, 2) determine the phylogeography of PRDM9 at a local and global scale using a new method for the computation of genetic distances between complex repeats, and 3) analyze how selection might account for the PRDM9 natural diversity observed. In this context, our study provides the grounds for understanding the complexity of the mechanisms that can drive evolutionary dynamics of PRDM9 in natural populations.
Fig. 1.
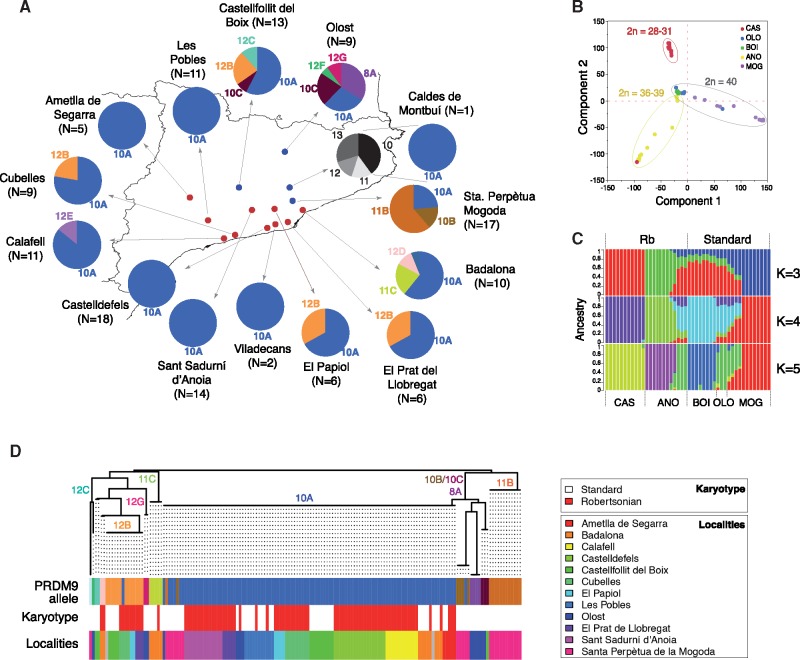
Geographical distribution and chromosomal characteristics of the Mus musculus domesticus populations analyzed. (A) Localities sampled in the Barcelona Rb system. Standard populations (blue dots) correspond to mice with 40 acrocentric chromosomes (standard karyotype) whereas Rb populations (red dots) had diploid numbers ranging from 2n = 39 to 2n = 28 (see supplementary table 1, Supplementary Material online, for further details of chromosomes involved in Rb fusions). (B) Localities sampled in the Madeira archipelago. Porto Santo Island is displayed in the inset; it contains standard mice with 40 acrocentric chromosomes (standard karyotype). Six metacentric races have been sampled on the Madeira Island: PEDC, PADC, PLDB, PPOD, PSAN, and PSVI (see supplementary tables 2 and 3, Supplementary Material online, for further details on the chromosomal composition of the chromosomal races). Following previous studies (Britton-Davidian et al. 2000), the chromosomal race PEDC is distributed on both on the northern (PEDC N.) and southern (PEDC S.) coasts of the Madeira Island.
Results
PRDM9 Diversity at a Local Scale
We successfully obtained polymerase chain reaction (PCR) products from the 395 wild-caught mice included in the study, detecting considerable variation in the number of ZnF repeats, depending on the region sampled (table 1 and supplementary tables 1–4 and figs. 1–4, Supplementary Material online). In the Barcelona Rb system, alleles with 10 ZnF repeats were the most frequently detected (73%), followed by alleles with 12 ZnF repeats (13.2%), 11 ZnF repeats (10%), 8 ZnF repeats (2%), and 13 ZnF repeats (1.8%). In the case of Madeira however, a higher diversity was detected when considering the number of ZnF repeats. PRDM9 alleles with 11 ZnF repeats were the most frequently detected (47%), followed by alleles with 10 ZnF repeats (25%), 13 ZnF repeats (17%), 12 ZnF repeats (6%), 15 ZnF repeats (2%), and 14 and 16 ZnF repeats (1% in each).
Table 1.
PRDM9 Variability Found in Wild Mus musculus domesticus including the Present Survey (Barcelona Rb system and Madeira archipelago) and Previous Studies in Eurasia (Buard et al. 2014; Kono et al. 2014).
| Barcelona | Madeira Archipelago |
Eurasiaa | |||
|---|---|---|---|---|---|
| Madeira | Porto Santo | ||||
| N | 185 | 199 | 11 | 76 | |
| PRDM9 alleles | 13 | 53 | 7 | 27 | |
| Min ZnF | 8 | 10 | 11 | 8 | |
| Max ZnF | 13 | 16 | 12 | 17 | |
Note.—N, number of wild mice included in the study; PRDM9 alleles, number of distinct PRDM9 alleles found based on the amino acid sequence; Min ZnF, minimum number of ZnF repeats; and Max ZnF, maximum number of ZnF repeats.
Data obtained from Kono et al. (2014) and Buard et al. (2014).
In order to investigate the source of contrasting variability patterns observed in the Madeira and Barcelona Rb systems, we sequenced the ZnF array in a total of 292 mice. This represented 74% of the sampled mice, a proportion that is consistent with previous studies on wild specimens (e.g., Kono et al. 2014). This resulted in the identification of 25 distinct ZnFs, four of which (#22, #26, #27, and #29) were newly identified for M. m. domesticus in this study (supplementary table 5, Supplementary Material online).
In the Barcelona Rb system, we obtained PRDM9 sequences from 132 mice (101 homozygous and 31 heterozygous) that represented 13 different PRDM9 alleles (figs. 2 and 3). These were classified according to both the sequence and number of ZnF repeats: one allele with eight ZnF repeats (8A), three alleles with 10 repeats (10A, 10B, and 10C), three alleles with 11 repeats (11B, 11C, and 11D), and six alleles with 12 repeats (12B, 12C, 12D, 12E, 12F, and 12G) (fig. 2). Eight of the 13 different PRDM9 alleles found in the Barcelona Rb system (8A, 10C, 11C, 11D, 12D, 12E, 12F, and 12G) were newly identified in this study. The diversity of PRDM9 alleles was widely variable among localities, particularly when comparing standard versus Rb populations, with the 10A allele the most commonly detected variant followed by 11B and 12B (fig. 3A). The highest variability was found in populations characterized by the standard karyotype (2n = 40; Castellfollit del Boix, Olost, and Santa Perpètua de Mogoda) where the majority of PRDM9 variants were represented. In addition, standard populations presented private alleles (8A, 10B, 11B, 10C, 12F, and 12G) that were undetected in Rb populations. In fact, only five alleles (10A, 11C, 12B, 12E, and 12D) were found in Rb mice. Among these, 10A was the most frequently distributed allele, with frequencies ranging from 23% to 100% in the localities Les Pobles, L’Ametlla de Segarra, Sant Sadurní d’Anoia, and Castelldefels (fig. 3A).
Fig. 3.
Allelic frequencies, population structure and phylogeny of the ZnFs found in the Barcelona Rb system. (A) Representation of the PRDM9 allelic frequencies in each locality (see table 1 and supplementary table 1, Supplementary Material online, for further details on the chromosomal composition of each locality). Legend—N, number of specimens sequenced per locality. (B) PCA in a subset of 50 mice from 5 localities of the Barcelona Rb system (CAS, Castelldefels; OLO, Olost; BOI, Castellfollit del Boix; ANO, Sant Sadurní d’Anoia; and MOG, Sta. Perpètua de Mogoda). (C) Plots showing the proportion of inferred ancestry for K = 3 to K = 5 in a subset of 50 mice from 5 localities of the Barcelona system (CAS, Castelldefels; OLO, Olost; BOI, Castellfollit del Boix; ANO, Sant Sadurní d’Anoia; and MOG, Sta. Perpètua de Mogoda). (D) Phylogenetic tree including all mice sequenced from the Barcelona Rb system (see text for further details). PRDM9 alleles are color coded following the pattern used in panel (A). For each individual, both the karyotype (white: standard; red: Rb fusions) and the locality (see inserted color legend) are indicated.
Fig. 2.
Natural PRDM9 allelic diversity found in the Barcelona Rb system and Madeira archipelago Mus musculus domesticus populations. (A) Representation of the mouse PRDM9Dom2 protein (reference genome, C57/BL6 strain). It consists of four domains: KRAB-like, SSXRD, PR/SET, and ZnF array. The underlined ZnFs represent repeats from position ZnF3 to ZnF6 that bind to the DNA in the mouse (Baker et al. 2015; Paigen and Petkov 2018). (B) Representation of the ZnF alignments for all PRDM9 alleles found in the present study. Each ZnF is color coded based on amino acid sequence affinity found at the most variable sites (−1, +3, +6) responsible for DNA binding. Additional information on ZnF amino acid sequences is provided in supplementary table 5, Supplementary Material online. Purple boxes encompass the more variable ZnF among individuals. Red asterisks indicate previously described PRDM9 alleles (Buard et al. 2014; Capilla et al. 2014; Kono et al. 2014). Geographical labels: (b) PRDM9 alleles found only in the Barcelona Rb system, (b/m) PRDM9 alleles found in both the Barcelona Rb system and the Madeira archipelago. PRDM9 alleles without geographical label correspond to alleles found only in the Madeira archipelago.
In the case of the Madeira archipelago, 160 mice were successfully sequenced (110 homozygous and 50 heterozygous), 152 of which were from the Madeira Island Rb system and 8 mice from Porto Santo Island (fig. 4). This represented 54 different alleles: six alleles with 10 ZnF repeats, 20 alleles with 11 ZnF repeats, 7 alleles with 12 ZnF repeats, 1 allele with 14 ZnF repeats, 6 alleles with 15 ZnF repeats, and 3 alleles with 16 ZnF repeats (figs. 2 and 4). Only five of them (10A, 11B, 11S, 11V, and 12C) were previously described in Eurasian populations and the Barcelona system (Buard et al. 2014; Capilla et al. 2014; Kono et al. 2014). The allele 10A corresponds to the alleles 48 and Ce3 described by Kono et al. (2014) and Buard et al. (2014) in M. m. domesticus and Mus musculuscastaneus, respectively. The allele 11B is homologous to the previously reported alleles 54, 55, and Db1 in M. m. domesticus (Buard et al. 2014; Kono et al. 2014), whereas 11S is the PRDM9 allele present on the t-haplotype (Kono et al. 2014). The 11V and 12C allele corresponded to allele 3 and 16, respectively (Buard et al. 2014).
Fig. 4.
Allelic frequencies and phylogeny of the ZnFs found in the Madeira archipelago. (A) Representation of the PRDM9 allelic frequencies found in each metacentric race from the Madeira Island and Porto Santo Island (see supplementary tables 2 and 3, Supplementary Material online, for further details on chromosomal composition of metacentric races). Number of specimens sequenced per locality on Madeira Island: Achadas da Cruz (N = 7), Arco da Calheta (N = 11), Chão da Ribeira (N = 16), Estreito da Calheta (N = 3), Faial (N = 2), Fajã da Ovelha (N = 2), Levada Grande (N = 2), Lombada dos Cedros (N = 1), Lombada dos Marinheiros (N = 4), Lombo da Velha (N = 3), Lombo das Laranjeiras (N = 3), Lombo do Doutor (N = 3), Lugar da Raposeira (N = 2), Madalena do Mar (N = 2), Maloeira (N = 18), Moledos (N = 2), Ponta Delgada (N = 8), Ponta do Pargo (N = 18), Ponta do Sol (N = 3), Porto da Cruz (N = 1), Porto Moniz (N = 3), Prazeres (N = 2), Ribeira da Janela (N = 6), Ribeira da Laje (N = 3), Ribeira da Vaca (N = 3), Ribeira Funda (N = 3), Santa (N = 1), Santana (N = 4), São Vicente (N = 3), Seixal (N = 1), Sítio da Fajã (N = 3), Socorro (N = 7), Solar da Maloeira (N = 1), Solar dos Prazeres (N = 1), and Porto Santo Island (N = 8). (B) Phylogenetic tree including all mice sequenced from the Madeira archipelago (see text for further details). PRDM9 alleles are color coded following the pattern used in panel (A). For each individual, both the diploid number (2n) and the geographical distribution are indicated (see inserted color legend).
Among the 49 alleles newly identified in the Madeira archipelago, the most commonly represented on Madeira Island was 11K, followed by 10A, 10E, 11S, and 13C (fig. 4A). Moreover, some alleles were detected in all Rb races (supplementary table 2, Supplementary Material online) but absent in standard populations (40 all-acrocentric chromosomes) from Porto Santo (i.e., 10A and 13C). We found the greatest allele heterogeneity among the chromosomal races PADC (2n = 24–28) and PEDC (2n = 23–26), occurring in the western part of the island. In contrast, populations situated in eastern regions (PLDB, PPOD, PSAN, and PSVI) had a more homogeneous allele distribution (fig. 4A). Regarding standard populations (2n = 40) on the island of Porto Santo, the allele variability was lower than other populations from eastern Madeira. Among the alleles reported on Porto Santo, three of them (11X, 11V, and 11W) were private alleles that were undetected in Rb populations and two more (12C and 11F) were present in very low frequencies in the Madeira Island.
Genetic Diversity and Signal of Positive Selection on the ZnF Array Domain
Once the PRDM9 diversity was characterized at a local scale, we analyzed genetic diversity and the signal of positive selection on the ZnF array domain at a global scale, including previously described Eurasian populations (Buard et al. 2014; Kono et al. 2014). When comparing the overall genetic (nucleotide) variability of the ZnF array domain found in all three regions included in the study (Barcelona, Madeira, and Eurasia), we observed that the genetic diversity in Madeira was greater than in the Barcelona Rb system and comparable to the diversity across the whole of Eurasia (table 2). Moreover, the genetic differentiation between Madeira and the Barcelona Rb system (FST = 0.0295, P < 0.0001) or even between Madeira and Eurasia was low (FST = 0.0084, P < 0.0001).
Table 2.
Genetic Diversity Estimates of the ZnF Array Found in the Studied Areas (Madeira archipelago, Barcelona Rb system, and Eurasia).
| Madeira | Barcelona | Eurasia | |
|---|---|---|---|
| Average genetic distance | 5.406 | 2.307 | 6.123 |
| Average pairwise nucleotide diversity excluding hypervariable sites (−1, +3, +6) | 0.00456 | 0.002856 | 0.00416 |
| Average pairwise nucleotide diversity (nucleotide differences per base) | 0.02351 | 0.018626 | 0.02694 |
Understanding the evolutionary consequences of this ZnF array variability is important since polymorphisms in both number and sequence of the arrays might affect DNA-binding specificity resulting in changes in the recombination landscape of each individual (Davies et al. 2016). Thus, we analyzed whether selection played a role in shaping this variability by inferring dN/dS estimates averaged across all pairwise PRDM9 sequence alignments considering either hypervariable codons only (−1, +3, +6) or excluding them (see Material and Methods). We observed that the amino acids that recognize the specific DNA motif at positions −1, +3, and +6 are under strong positive selection, as shown by their extremely high dN/dS values (table 3). Position +6 showed less variability involving two of the residues described (Q and K) compared with positions −1 (A, E, Q, T, and V substitutions) and +3 (D, H, N, V, and S substitutions) (fig. 2). Interestingly, the rest of the PRDM9 sequence (the so-called nonhypervariable sites) was similarly subjected to positive selection in both the Madeira the Barcelona Rb systems, whereas it was under weak negative selection across Eurasia (table 3). Moreover, we detected differences in dN/dS values between areas of study, with the effects of selection being pronounced in the Madeira archipelago, intermediate in the Barcelona Rb system and weaker across Eurasia (table 3). To test whether these contrasting patterns in positive selection across regions was explained by the presence of Rb fusions, we calculated dN/dS values considering mice with and without Rb fusions in the Madeira and Barcelona Rb systems. Remarkably, individuals with Rb fusions showed higher dN/dS values than standard ones, both in Madeira and the Barcelona Rb system (table 3). This was consistent for both hypervariable sites and the rest of the ZnF array domain (table 3).
Table 3.
Average dN, dS, and dN/dS Values between Aligned ZnF Arrays in the Studied Areas (Madeira archipelago, Barcelona Rb system, and Eurasia).
| Madeira |
Barcelona |
Eurasia | All | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| All | St | Rb | St versus Rb | All | St | Rb | St versus Rb | |||
| dN(hs) | 0.181 (0.15–0.21) | 0.232 (0.19–0.28) | 0.175 (0.15–0.20) | 0.225 (0.19–0.26) | 0.089 (0.07–0.11) | 0.128 (0.10–0.15) | 0.0516 (0.03–0.07) | 0.0951 (0.07–0.12) | 0.218 (0.19–0.24) | 0.169 (0.15–0.19) |
| dS(hs) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0.00407 (0.001–0.008) | 0.00086 (0.0001–0.0019) |
| dN(nhs) | 0.0053 (0.004–0.007) | 0.00929 (0.006–0.013) | 0.00488 (0.003–0.006) | 0.00817 (0.005–0.011) | 0.00226 (0.001–0.004) | 0.00298 (0.002–0.005) | 0.00144 (0.0002–0.003) | 0.00237 (0.001–0.004) | 0.00362 (0.002–0.005) | 0.00413 (0.003–0.006) |
| dS(nhs) | 0.0006 (0.0003–0.001) | 0.00433 (0.001–0.008) | 0.00053 (0.0002–0.0009) | 0.00158 (0.0006–0.003) | 0.00028 (2.4–05–0.0007) | 0.00067 (9.1–05–0.001) | 0 (0–0) | 0.00032 (1.1–05–0.0008) | 0.00289 (0.002–0.004) | 0.00086 (0.0006–0.001) |
| dN(hs)/dS(hs) | ∞ (∞–∞) | ∞ (∞–∞) | ∞ (∞–∞) | ∞ (∞–∞) | ∞ (∞–∞) | ∞ (∞–∞) | ∞ (∞–∞) | ∞ (∞–∞) | 53.6 (24.8–236) | 197 (83–1110) |
| dN(hs)/dS(nhs) | 278 (159–566) | 53.7 (27.7–179) | 330 (189–708) | 142 (74.7–367) | 313 (113–3180) | 187 (78.5–1290) | ∞ (∞–∞) | 293 (100–8920) | 75.3 (51.5–115) | 196 (136–321) |
| dN(nhs)/dS(nhs) | 8.12 (3.99–17.9) | 2.15 (0.94–7.62) | 9.2 (4.65–20.7) | 5.16 (2.25–14.8) | 7.95 (2.45–83.7) | 4.34 (1.51–36.7) | ∞ (∞–∞) | 7.3 (2.2–182) | 1.25 (0.75–1.97) | 4.79 (2.82–8.13) |
Note.—Ninety-five percent confidence intervals for all estimates are indicated in parentheses and italics. hs, hypervariable sites (−1, +3, +6); nhs, nonhypervariable sites; Rb, individuals with Rb fusions; St, individuals with standard karyotype (i.e., absence of Rb fusions); and ∞, infinite values.
Differences in Heterozygous PRDM9 Allele Combinations
Even though the PRDM9 protein can use all its ZnFs to bind to the DNA in a sequence-specific manner, it has been suggested that only a subset of ZnFs within the array contributes to the binding of the specific DNA motif (Billings et al. 2013; Baker et al. 2015; Paigen and Petkov 2018). In the case of the house mouse, this hypothesis implies that the ZnF repeats located at positions ZnF3 to ZnF6 within the array would make primary contact with the DNA strand before recruiting the DSBs-induction machinery for the formation of recombination hotspots (review by Paigen and Petkov [2018]). Since the presence of PRDM9 heterozygous allelic combinations might result in subfertile phenotypes caused by asymmetric DSBs between homologous chromosomes, we sought to understand the functional constraints that might affect PRDM9 diversity in Barcelona and Madeira populations by analyzing the degree of conservation of positions ZnF3–ZnF6 within the array in PRDM9 heterozygous mice.
In the case of the Madeira system, we detected a clear deficit of heterozygotes with respect to Hardy–Weinberg equilibrium within Rb races, with PRDM9 heterozygotes reduced to about half of the expected (supplementary table 6, Supplementary Material online). These deviations from Hardy–Weinberg equilibrium were not found in standard mice in Porto Santo, suggesting a relationship between the presence of Rb fusions and the selection against PRDM9 heterozygotes on Madeira. A contrasting pattern was found in the Barcelona Rb system where both standard and Rb populations show a reduction in the number of heterozygote individuals for PRDM9 (supplementary table 6, Supplementary Material online).
As a way to explain this reduction in heterozygosity, we observed that not all heterozygous PRDM9 combinations were equally represented among populations (fig. 5 and supplementary table 7 and fig. 4, Supplementary Material online). The most common PRDM9 allele combinations in the Barcelona Rb system were 10A/12B, 10B/11B, and 10A/11C, whereas in the Madeira archipelago, most frequent alleles were 11K/13C, 10A/11K, and 10A/13C. Although in the Madeira archipelago no heterozygous combinations were shared between standard and Rb mice, only one allele combination (10A/12B) was present in both standard and Rb mice in the Barcelona Rb system (fig. 5A). In both Rb systems (Barcelona and Madeira), differences in population allelic frequency were significant between standard and Rb mice (Pearson test, P ≤ 0.05). When analyzing the conservation in amino acid sequence of the ZnF repeats located at positions ZnF3–ZnF6 along the ZnF array in the different heterozygous combinations, we detected higher sequence conservation in Rb mice when compared with standard mice in both the Madeira and Barcelona Rb systems (fig. 5). In the case of the Barcelona Rb system, an average conservation of 94.4% was found in Rb mice versus 75% in standard mice, this difference being statistically significant (Wilcoxon test, P = 0.0075). These values were lower in the Madeiran archipelago, where the average sequence conservation of repeats from ZnF3–ZnF6 in Rb mice was 70.5% versus 44.4% in mice with no Rb fusions, though the differences in conservation were statistically significant (Wilcoxon test, P = 0.0252) (fig. 5 and supplementary fig. 4, Supplementary Material online).
Fig. 5.
Conservation in amino acid sequence of the ZnF repeats located in positions 3–6 (ZnF3–ZnF6) along the ZnF array in PRDM9 heterozygous combinations found in the study. (A) Bubble chart representing population frequency (size of the bubble) and the percentage of amino acid (Aa) sequence conservation of the highly variable positions (−1, +3, and +6) of the initial repeats along the array (from ZnF3 to ZnF6) (purple intensity). Data are shown for each heterozygous combination found in Standard and Rb mice in the Barcelona Rb system and the Madeira archipelago (see supplementary table 7, Supplementary Material online, for further details). (B, C) Examples of PRDM9 alleles in heterozygous combination and the predicted distribution of recombination hotspots. Yellow starts represent the location of recombination hotspots across homologous chromosomes (red and blue lines). (B) The ZnF repeats located in positions 3–6 (ZnF3–ZnF6) in the 10A/12E combination present 100% of Aa sequence conservation. This can result in symmetric distribution of recombination hotspot in homologous chromosomes. (C) The ZnF repeats located in positions 3–6 (ZnF3–ZnF6) in the 10B/11B combination present 33.3% of Aa sequence conservation. This can result in asymmetric distribution of recombination hotspot in homologous chromosomes.
Phylogeography of PRDM9
In order to investigate whether the observed variability of PRDM9 was reflective of population structure, we studied the phylogeography of the different alleles detected in the Rb systems individually (Barcelona and Madeira) and in relation to previously described alleles in Eurasian populations (Buard et al. 2014; Capilla et al. 2014; Kono et al. 2014). Although previous allozyme and microsatellite analysis (Britton-Davidian et al. 2007; Förster et al. 2013) provided information regarding the origin of the chromosomal radiation of Madeiran mice, the population structure of the Barcelona system is not entirely known at this stage. Thus, we first determined genetic diversity and population differentiation in this system based on the single nucleotide polymorphism (SNP) genotyping of a subset of 50 mice using the Mega Mouse Universal Genotyping Array. We applied a maximum likelihood method to infer the genetic ancestry of each individual, where the individuals are assumed to have originated from K ancestral clusters. The results of the principal component analysis (PCA) and the plots for K = 3 to K = 5 are shown in figure 3B and C. We observed clustering for the two Rb populations (Castelldefels and Sant Sadurní d’Anoia) when compared with the standard populations (Olost, Castellfollit del Boix, and Santa Perpètua de Mogoda). In fact, the PCA revealed that Olost and Castellfollit del Boix populations, both located at the northern area of the Rb system distribution, clustered together. Compared with these, both Rb populations (Castelldefels and Sant Sadurní d’Anoia) showed progressive genetic differentiation, being more pronounced in Castelldefels (fig. 3B), a population with the lowest diploid number (2n = 28–31) and with the presence of a high number of heterozygous Rb fusions (supplementary table 1, Supplementary Material online).
Likewise, estimation of population structure revealed heterogeneity of genetic clusters according to populations (fig. 3C). The genetic structure based on the ADMIXTURE analysis (Alexander et al. 2009) revealed that mice from Castelldefels (one of the populations with the highest number of Rb fusions detected) showed high levels of ancestry (0.99%), forming a cohesive cluster (fig. 3C). An exception to this pattern was a single individual (specimen 955), which shared ancestry with mice from Sant Sadurní d’Anoia. This individual was characterized by the highest diploid number in Castelldefels (2n = 32) and by the absence of Rb fusion 3.18, characteristic of the remaining mice from this population (supplementary table 1, Supplementary Material online). Within the Sant Sadurní d’Anoia population (2n ranging from 35 to 39), mice were distributed into two different groups, with three individuals sharing ancestry with the standard populations Olost and Castellfollit, both populations highly homogenous according to the PCA. The Santa Perpètua Mogoda population was very unusual, despite its standard condition (i.e., the absence of Rb fusions), it was highly differentiated from Olost and Castellfollit del Boix in both the PCA and the estimates of individual ancestry.
We then constructed phylogenetic trees for both the Madeira and Barcelona Rb systems by the Neighbor-Joining approach using a new measure of pairwise genetic divergence (see Material and Methods). The key feature of our approach is that it considers point mutations, insertions, and deletions of whole repeat units, as well as duplications of single repeat units as a consequence of nonhomologous recombination, slippage, or related biological processes. This is of relevance since single-unit duplications and indels have different weights in their contribution to the genetic distance. This approach allowed us to effectively estimate genetic distances among the polymorphic repeats of PRDM9 detected in the Madeira and the Barcelona Rb systems. The distribution of haplotypes obtained showed greater diversification on the island of Madeira than the Barcelona system (figs. 3D and 4B). Although the phylogenetic analysis grouped PRDM9 alleles into three major phylogroups in the Barcelona system (those representing the alleles 10A, 11B, and 12B; fig. 3D), the allelic diversification was much higher in the Madeira Island with alleles 10A, 11B, 11S, 11K, and 13C representing the most common haplotypes (fig. 4B). Differences between both Rb systems were also exemplified by higher genetic diversity estimates of the ZnF array on Madeira (5.406) than in the Barcelona Rb system (2.307), the former being comparable to the diversity across the whole Eurasia (6.123) (table 2). When plotting the chromosomal configuration in the phylogenetic trees we detected that in both Rb systems (Madeira and Barcelona), standard mice showed a tendency to carry private alleles. This was exemplified by alleles 8A, 10B, 10C, 11B, 12C, 12G, and 12F in Olost, St. Perpètua de Mogoda, and Castellfollit del Boix (fig. 3D), and 11W and 11X in Porto Santo (fig. 4A).
Taking advantage of previous surveys of wild captured mice in Europe and Asia, we further compared the allele variation found in the Madeira and Barcelona Rb systems with those described in Eurasia (fig. 6). When the geographical distribution was plotted onto the global phylogenetic tree, Madeira haplotypes showed mixed origins. The phylogenetic reconstruction of all three sampled regions (and informed by evidence that the genetic differentiation between Madeira and the Barcelona Rb system or even Eurasia is small, table 2) suggests that Madeira was colonized by a highly different population, most likely as a result of multiple colonization/introgression events from multiple parts of Eurasia.
Fig. 6.
Phylogeographic depiction of PRDM9 at a global scale. Phylogenetic reconstruction including mice sequenced from the Madeira archipelago, Barcelona Rb system and Eurasia (data extracted from Kono et al. [2014] and Buard et al. [2014]). For each individual, both the diploid number (2n) and the geographical distribution (Madeira, Barcelona, and Eurasia) are indicated (see inserted legends). The diploid number is not known for Eurasian samples so they are represented in gray.
Discussion
Distinctive Phylogeographic Patterns of ZnF Variability and the Evolution of PRDM9 in the House Mouse across Eurasia
Our study includes a wide survey of nearly 400 wild-caught mice representing two highly distinct house mouse Rb systems: the Madeira archipelago (Madeira and Porto Santo Islands) and the Barcelona system (continental). We identified 49 newly described alleles on the Madeira archipelago, and 8 new alleles in the continental (Barcelona) Rb system, revealing that intraspecific PRDM9 diversity in M. m. domesticus is far greater than previously reported. This adds substantially to previously described allelic diversity in natural populations of Mus musculus subspecies (28 distinct PRDM9 alleles in M. m. domesticus, 34 in Mus musculusmusculus, and 37 in M. m. castaneus) (Buard et al. 2014; Capilla et al. 2014; Kono et al. 2014).
Most importantly, we detected contrasting evolutionary patterns in the continental versus island chromosomal races of the house mouse. This was reflected by unprecedented high levels of PRDM9 diversity in Madeiran house mice, an insular Rb system characterized by distinct metacentric races, when compared with the Barcelona Rb system (where Rb fusions are not yet fixed within populations and are present in polymorphic state). Our analysis of genetic diversity and positive selection, together with the phylogeographic reconstruction of PRDM9, suggests that the variability observed could be the result of, at least, two possible (not mutually exclusive) scenarios in the Madeira system: 1) the current populations of Rb mice could reflect a complex pattern of introgression or multiple colonization events into the island and 2) meiotic impairment on hybridization of different populations of Rb mice allowed PRDM9 alleles to diverge among metacentric populations.
Initial studies based on mtDNA lineages (Bonhomme et al. 2011) indicated a recent common ancestry of all extant house mice populations, as well as a complex history owing to founder events, genetic drift and secondary admixture. This was suggestive of two phases of mouse colonization westward across the Mediterranean basin; an initial event during the early progression of Neolithic human expansion (starting 12,000 years ago) followed by a second, more recent (some hundred years ago), related to maritime trade. This progression can explain the presence of Mediterranean alleles in the Barcelona system and subsequent in situ diversification. Madeira, on the other hand, has a more recent history of mouse colonization originated first from Danish Vikings in the 9th century followed by a second incursion by 15th century Portuguese settlers (Gündüz, Auffray, et al. 2001; Britton-Davidian et al. 2007; Förster et al. 2009, 2013). Our analyses of genetic diversity reveal a complex colonization pattern in Madeira, much more so than in the Barcelona Rb system and leading to a PRDM9 diversity comparable to the diversity across the whole of Eurasia. Additionally, the phylogenetic reconstruction of PRDM9 in Madeira indicated that the same group of alleles appeared in different clusters independently. Altogether, our results suggest multiple colonization/introgression events into Madeira from Eurasia (including the Mediterranean basin).
Under this scenario of multiple colonization events and together with the extreme topography of Madeira (i.e., chromosomal races separated by mountain barriers), the presence of Rb fusions might have resulted in meiotic impairment in hybrids that subsequently contributed to the high rate of diversification of PRDM9. Our analysis of genetic diversity and positive selection supports this view. Since chromosomal fusions can act as postzygotic barriers and can restrict gene flow among chromosomal races (Franchini et al. 2010; Giménez et al. 2013; Capilla et al. 2014), this can result in high genetic differentiation between populations, thus facilitating the appearance of new PRDM9 variants. In fact, in the case of Madeira, each chromosomal race is geographically localized and homogeneous with respect to the constitution of Rb fusions, and chromosomal hybrids are rare (Britton-Davidian et al. 2000; Nunes et al. 2005). On the other hand, Madeira is extremely variable regarding its genetic structure, as can be appreciated both from the PRDM9 phylogenetic tree and from the high genetic differentiation between chromosomal races. Moreover, the relative deficit of heterozygous for PRDM9 (despite the strong genetic mixing) points to strong selection for karyotype compatibility. In this context, differences in positive selection can be explained by the presence of Rb fusions since dN/dS values were higher in Rb mice than standard mice, both in Madeira and the Barcelona Rb systems. This was true both for hypervariable codons and for the rest of PRDM9 sequence.
An association between Rb fusions and PRDM9 diversity was also observed in the Barcelona Rb system, although the effects were mild, probably related to a differential demographic history. The Barcelona system represented approximately 10% of the novel PRDM9 alleles found in our study, with the highest diversity observed in standard mice located in the northern-eastern distribution of the sampled populations. The allelic distribution together with the pattern of genetic diversity detected by the PCA and ancestry analyses was suggestive of a larger connectivity among populations in the Barcelona Rb system than in the case of Madeira. This was exemplified by the 10A allele, which was present in all populations especially in Rb mice, representing the greatest allele homogeneity (nearly 95% of mice from Rb populations carried 10A). Considering the factors that might be responsible for the widespread distribution of this allele in the Barcelona system, it is possible that the PRDM9 allele frequency pattern observed in this system could result from a population bottleneck that facilitated the formation and subsequent expansion of Rb fusions. According to the PCA and ancestry analysis, standard mouse populations within the Barcelona system, such as Olost and Castellfollit del Boix, may represent the origin of the genetic diversity detected out of which Rb populations such as Gavà and Sant Sadurní d’Anoia subsequently appeared as differentiated populations. Consequently, Rb mice may have diverged into different, genetically distinct populations resulting in a homogeneous distribution of PRDM9 alleles with a predominance of the 10A allele. This is consistent with suggestions by Gündüz, López-Fuster, et al. (2001) for the origin of the Barcelona system by primary intergradation. This Rb system may therefore represent an example of a radiation that could eventually lead to the formation of a metacentric race without geographic isolation (Sans-Fuentes et al. 2010).
Functional Implications of Maintaining Conserved DNA-Binding Repeats
Despite the ZnF variability observed in both Rb systems, we detected significant deviations from Hardy–Weinberg equilibrium (i.e., a deficit of heterozygotes) where the PRDM9 heterozygous combinations most frequently encountered maintained amino acid sequence similarity from position ZnF3 to ZnF6 in the ZnF array. These results suggest that observed PRDM9 variability (either as a result of phylogeographic dynamics and/or population isolation triggered by meiotic impairment of hybrids), can be subjected to functional constraints that facilitate the accumulation of allelic combinations that maintain recombination hotspot symmetry.
Under this scenario, it is important to take into account the particular features of the PRDM9 protein (Davies et al. 2016; Tiemann-Boege et al. 2017; Paigen and Petkov 2018), which include 1) its DNA-binding properties, 2) the rapid coevolution of the ZnF domain and the DNA motif that it recognizes, 3) asymmetric DSBs due to preferential binding of PRDM9 to new target sequences, and 4) hybrid sterility triggered by asymmetric DSBs and interallelic PRDM9 combinations. Previous work on house mouse inbred strains found that male sterility is a complex process that can be influenced by the heterozygous allelic combination of PRDM9 (Dzur-Gejdosova et al. 2012; Gregorová et al. 2018) and their interaction with different X-linked and autosomal loci (Balcova et al. 2016). Likewise, the presence of highly divergent PRDM9 alleles in a heterozygous state can be detrimental due to elevated levels of asymmetric DSBs between homologous chromosomes (Davies et al. 2016). This is especially important since allelic combinations of PRDM9 with conserved DNA-binding repeats were maintained in metacentric populations, suggestive of the presence of functional constraints.
In the light of our findings, the rapid formation of new PRDM9 alleles in the presence of Rb fusions, can act synergistically with the formation of new binding motifs distributed across the genome in different metacentric populations. This could result in functional constraints that might facilitate the accumulation of PRDM9 heterozygous combinations that maintain a high degree of similarity in the amino acids responsible for protein–DNA binding, thus resulting in recombination hotspot symmetry in homologous chromosomes. Mounting evidence in humans and mice has shown that variation in both the sequence and number of ZnF repeats strongly influence the distribution of meiotic DSBs (Berg et al. 2010; Grey et al. 2011; Brick et al. 2012; Baker et al. 2015). Small variations in the amino acid sequence of ZnF repeats can modulate PRDM9 binding affinities to specific DNA motifs (Berg et al. 2010). In fact, differences in a single ZnF repeat can affect the specificity of over 70% of the meiotic DSBs, leading to the redistribution of recombination sites in a single generation (Smagulova et al. 2016). The rapid evolution of ZnF sequences would therefore lead to rapid changes in the distribution of recombination sites across the genome by creating new DNA motifs that would be recognized with stronger affinity by the new PRDM9 allelic variants. This will, in turn, compensate the loss of the primary DNA motifs by gene conversion during the repairing of crossovers during the early stages of meiosis (Smagulova et al. 2016).
Although little is known at this stage about the dynamics and the mechanistic constraints that affect recombination landscapes at a finer scale in metacentric wild populations, our results nonetheless suggest that Rb fusions might play a role in bringing new selective pressures on PRDM9. These outcomes provide impetus to examine the effect of genetic backgrounds on PRDM9 and the development of reproductive isolation between natural populations.
Materials and Methods
Sampling and DNA Extraction
Our data set comprised a total of 395 wild-caught house mice, 185 mice from the Barcelona Rb system (Medarde et al. 2012), 199 mice from Madeira Island, and 11 from Porto Santo Island (table 1, fig. 1, and supplementary tables 1 and 2, Supplementary Material online). Mice were karyotyped previously (Medarde et al. 2012; Chmátal et al. 2014, supplementary fig. 1, Supplementary Material online). Animals were housed and treated in strict accordance with ethical guidelines approved by the Universitat Autònoma de Barcelona (Spain) and University of Lisbon (Portugal). Genomic DNA was extracted from tissue biopsies preserved in absolute ethanol or fresh tissue using proteinase K digestion (Sambrook et al. 1989).
The Barcelona Rb system included mice from 15 localities selected for their geographical distribution and distinctive chromosomal configurations (fig. 1 and supplementary table 1, Supplementary Material online). This covered the full extent of the chromosomal polymorphism previously described for the Barcelona Rb system (Medarde et al. 2012). This included localities with high frequencies of almost all metacentric chromosomes (2n = 28–39), localities in the vicinity of Rb areas but without chromosomal fusions (2n = 40), and localities geographically located between these population groups containing intermediate diploid numbers (2n = 39–40) (supplementary table 1, Supplementary Material online). Four of the localities considered in the study had mice with standard karyotypes (2n = 40, Castellfollit del Boix, Santa Perpètua de Mogoda, Olost, and Caldes de Montbuí) and defined here as standard populations (supplementary table 1, Supplementary Material online). The remaining localities (N = 11) included mice with Rb fusions, defined here as Rb populations (2n = 28–39, Badalona Les Pobles, L’Ametlla de Segarra, El Papiol, Calafell, Sant Sadurní d’Anoia, Cubelles, La Granada, El Prat de Llobregat, Viladecans, and Castelldefels). Mice from Rb populations were characterized by having between one to seven Rb fusions involving 14 different chromosomes (Rb[3.8], Rb[4.14], Rb[5.15], Rb[6.10], Rb[7.17], Rb[9.11], and Rb[12.13]), either in heterozygous or homozygous states (supplementary table 1, Supplementary Material online). The Rb fusions were present as chromosomal polymorphisms in the Barcelona Rb system, with varying frequencies (supplementary table 1, Supplementary Material online).
The Madeira system was represented by mice from 34 localities that included all six chromosomal races occurring on the island of Madeira (as originally defined by Britton-Davidian et al. [2000]) (supplementary table 2, Supplementary Material online): PSAN (2n = 22), PADC (2n = 24–28), PEDC (2n = 23–26), PLDB (2n = 24), PPOD (2n = 27–28), and PSVI (2n = 26–27). A total of 20 different metacentrics in homozygous state are described in Madeira, 9 of which have not been detected elsewhere (Rb[2.19], Rb[4.5], Rb[4.16], Rb[5.18], Rb[9.18], Rb[11.12], Rb[11.19], Rb[13.17], and Rb[15.18]), highlighting the uniqueness of this system (see supplementary table 3, Supplementary Material online, for details on the chromosomal composition of chromosomal races). Mice with the standard karyotype (2n = 40) from the neighboring island of Porto Santo (four localities) were also included in our investigation (fig. 1 and supplementary table 2, Supplementary Material online).
Prdm9 Sequencing
The entire ZnF array is encoded by the last exon of the Prdm9 gene that extends from the second ZnF repeat towards the C-terminal domain. We amplified this region in all mice by PCR using the primers ZFA-L_F (forward) and ZFA-L_R (reverse) described by Kono et al. (2014) (supplementary table 4, Supplementary Material online) using the ExTaq (TaKaRa) following the protocol in Capilla et al. (2014). Briefly, the PCR conditions were 95 °C (3 min), 30 cycles of 95 °C (30 s), 64 °C (30 s), and followed by 72 °C (90 s). The ZnF array exhibits not only length polymorphisms (variation of the number of ZnF repeats) but also amino acid variation (alleles with the same number of ZnF repeats but different amino acid composition). As a consequence, it is necessary in each case to identify the length of the allele and subsequently the amino acidic composition of the sequence. To this end, PCR products were separated on 1% agarose gels. This allowed both the ZnF array length to be discerned (homozygous and heterozygous) and optimal DNA recovery from bands for subsequent sequencing (supplementary fig. 2, Supplementary Material online).
To determine the sequences of ZnF repeats, PCR products of the homozygous mice were subjected to bidirectional Sanger sequencing with the same primers used for amplification: primers ZFA-L_F and ZFA-L_R. Only sequences reproducible in at least three independent amplification reactions were included in the data analyses. Bands of heterozygous mice were purified from the gels using the Nucleospin Gel and PCR Clean-up kit (Macherey-Nagel) and subsequently sequenced. Sequences from heterozygous mice were analyzed using the Bioedit 7.2.5 package. Two different software packages were used to distinguish allelic variants and to define haplotypes of heterozygous mice: Phase (http://stephenslab.uchicago.edu/software.html#phase) and Champuru v1.0 (http://www.mnhn.fr/jfflot/champuru/). This allowed inferences on whether a sample was homozygous or heterozygous for its amino acid composition (Capilla et al. 2014). All allele sequences were translated using ExPasy: SIB bioinformatics resource portal (Artimo et al. 2012).
Additionally, we included 118 published PRDM9 sequences of wild-derived M. m. domesticus and M. m. castaneus specimens drawn from various locations in Eurasia (Buard et al. 2014; Capilla et al. 2014; Kono et al. 2014) for the phylogeographic analysis of PRDM9 at a global scale. Overall, the phylogenetic comparative study included three M. m. domesticus sampling regions: the Barcelona Rb system, the Madeira archipelago (including both Madeira and Porto Santo Islands) and Eurasia (previously published data; Buard et al. 2014; Kono et al. 2014) (table 1).
PRDM9 Allele Classification
Following the nomenclature used in previous studies (Buard et al. 2014; Capilla et al. 2014; Kono et al. 2014), PRDM9 alleles were classified using the number of ZnF repeats and the extent of amino acid variation in the highly variable positions −1, +3, and +6 of each ZnF repeat (table 1 and supplementary table 5, Supplementary Material online). Each ZnF repeat was identified by a number (from #3 to #34) based on its amino acid sequence (supplementary table 5, Supplementary Material online). Subsequently, the combination of different of ZnF repeats was classified as distinct PRDM9 alleles (fig. 2). Moreover, for each PRDM9 allele found in heterozygous state, the amino acid conservation of the ZnF repeats located in positions ZnF3–ZnF6 of the array was calculated based on the hypervariable sites (−1, +3, and +6; Kono et al. 2014) (supplementary table 6, Supplementary Material online).
SNP Genotyping
SNP genotyping data included 50 mice from 5 localities of the Barcelona Rb system: 26 mice from three standard populations (Castellfollit del Boix, N = 8; Santa Perpètua de Mogoda, N = 13; and Olost, N = 5) and 24 mice from two Rb populations (Sant Sadurní d’Anoia, N = 12 and Castelldefels, N = 12). Genomic DNA was extracted using a standard protocol with proteinase K (Sambrook et al. 1989). Subsequently, mice were genotyped using the Mega Mouse Universal Genotyping Array (Morgan et al. 2015), which consist of 77,808 evenly distributed SNP markers built on the Illumina Infinium platform. SNPs were filtered to remove markers with missing values >5% threshold (i.e., markers that do not fit Hardy–Weinberg expectations, P < 10−5) using PLINK version 1.9 (Purcell et al. 2007). The proportion of missing data and heterozygosity per locus and per sample were also calculated to evaluate possible bias. This resulted in a final data set of 63,344 informative SNPs distributed across all chromosomes, with the exception of the Y. Using this data set, the ADMIXTURE software (Alexander et al. 2009) was used to estimate individual ancestry and admixture proportions assuming K populations based on a maximum likelihood method. Analyses were run only for SNPs with a greater than 95% genotype call. The numbers of clusters (K) evaluated here ranged from 1 to 10. To further evaluate the final K value, an Evanno’s ΔK was applied (Evanno et al. 2005). Subsequently, three different K values (K = 3, 4, 5) showed the lowest likelihood values (supplementary fig. 3, Supplementary Material online). ADMIXTURE analyses were plotted using an R framework. In addition, PCA was implemented using a module of PLINK 1.9.
Phylogenetic Analysis of PRDM9 Alleles
Phylogenetic trees for the nucleotide sequences of PRDM9 alleles were built via a Neighbor-Joining approach informed by a new measure of pairwise genetic divergence between ZnF repeats. Briefly, divergence between pairs of ZnF repeats was computed by first masking codons corresponding to the hypervariable amino acid positions (−1, +3, +6). All repeat units share a high sequence similarity and have the same length, it was therefore straightforward to align these units and compute the pairwise genetic (Hamming) distances μ(υ,υ′) between any two units υ and υ′. Then, the evolution of the ZnF repeats was modeled as a Markov process involving nucleotide mutation, insertion and deletions of a single repeat unit as well as single-unit duplication/slippage. Under this framework, we defined the genetic distance between repeats as an edit distance, that is, as the minimum cost to change a repeat to another through a series of elementary operations. The edit distance is minimized by one or possibly several alignments between repeats r = (r1; r2; r3; …) and r′ = (; ; ; …). The cost was defined as the weighted sum of all contributing elements: 1) point mutations, small indels, and other within-unit processes (cost μ[rj, ] for an alignment between the jth and the jth units of the two repeats, weight wm); 2) insertions and deletions of whole units (cost 1, weight wi); and 3) single-unit duplication/slippage (cost 1, weight ws; plus an extra cost μ[rj, rj−1] or μ[, ] for the point mutations and small indels between neighboring duplicated units, which was weighted by wm). An R implementation of the algorithms described here is provided at https://github.com/lucaferretti/RepeatDistance.
Subsequently, each pair of ZnF repeats was aligned using a modified Needleman–Wunsch algorithm corresponding to the model considered, with different costs for each type of mutations (wm for point mutations, wi for insertions/deletions, and ws for slippage). The genetic distance between two sequences was defined as the weighted sum of the cost of each mutation between the sequences. Our selection of costs was as follows: 1) Mismatch between units rju and : wmμ(rj, ) and 2) insertion of a whole unit rj in repeat r: min[wi, ws + wmμ(rj, rj−1), ws + wmμ(, )]. The best alignment according to this choice of costs corresponds precisely to the alignment that minimizes the sum of costs of mutations between sequences; hence the minimum cost obtained from the Needleman–Wunsch algorithm provides the distance between repeats defined above.
Weight parameters were chosen among the ones that maximized the agreement between mean genetic distances in the subset of 50 mice from the Barcelona system included in the SNP genotyping (computed from the 20 Mb around the PRDM9 nucleotide sequence) and the mean ZnF genetic distance for the same individuals. Agreement between the genetic distances was defined in terms of Pearson correlation of the pairwise distances of all individuals for which both genotype and PRDM9 sequence data were available. The correlation was computed numerically for a grid of weight values with a step of 0.25. In case of ties, the combination with the lowest ratios wi/wm and ws/wm was chosen. The final choice of weights maximizing the agreement between genetic distances is wm = 1 for nucleotide point mutations, wi = 3.5 for unit insertions/deletions, and ws = 1.75 for single-unit duplications. All pairs of sequences were pairwise aligned with our approach, using the genetic distance corresponding to these weights.
The PRDM9 phylogenetic trees were reconstructed from the molecular distance discussed above, using the Neighbor-Joining method implemented in the R library APE (Paradis et al. 2004). Trees were rooted at midpoint using the R package phangorn (Schliep 2011).
Genetic Diversity and Positive Selection
The PRDM9 alleles were tested for deviations from Hardy–Weinberg proportions in all populations with more than ten sequenced individuals. The significance of the deficit of heterozygous individuals for PRDM9 alleles with respect to Hardy–Weinberg predictions was assessed by an exact permutation test based on 1,000,000 permutations.
Pairwise nucleotide diversity was estimated as the mean number of nucleotide differences per base averaged across all pairs of aligned nucleotide sequences, ignoring gaps in the pairwise sequence alignment. FST values between multiple population/chromosomal races were computed using the formula described by Hudson et al. (1992). This computed the within-population diversity as the average of the within-population diversities of all population (in order to weight the contribution of each population equally) and permitted the removal of hypervariable codons from the sequence.
Selection was inferred from dN/dS estimates averaged across all pairwise sequence alignments as follows. First, we computed all the pairwise alignments using the Needleman–Wunsch algorithm described before. For each pair, we estimated dN as the number of nonsynonymous mutations per nonsynonymous site, considering either hypervariable codons only (−1, +3, +6) or excluding them, and ignoring gaps in the pairwise alignment. The fraction of nonsynonymous sites was estimated by randomization conditional on the nucleotide frequencies of the sequence. Then, dS was similarly estimated as the number of synonymous mutations per synonymous site (since there were often no synonymous mutations in hypervariable codons, we used synonymous mutations in the other codons as an alternative estimate of neutral rates). Finally, we averaged dN and dS across all sequence pairs and estimated dN/dS as the ratio of these averages. This corresponds to the approximate Maximum Likelihood estimate of dN/dS for Poisson-distributed mutations. Neutral evolution would result in an expected value of dN/dS around 1. The 95% confidence intervals were computed by bootstrapping (sampling 500 random sets of codons with replacement). We computed dN/dS both across all sequences and within/between specific subsets (such as sequences from individuals with standard karyotype or with Rb fusions). All analyses of genetic diversity were implemented via custom R scripts.
Data Accessibility
Data underlying this article are available on GeneBank (from MK848086 to MK848149).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
C.V. and L.C. were supported by FPI predoctoral fellowships from the Ministerio de Economía y Competitividad (MINECO) (BES-2011-047722 and BES-2015-072924, respectively). This study was partially supported by grants from MINECO to J.V. (CGL2010-15243) and to A.R.-H. (CGL2010-20170, CGL2014-54317-P, CGL2017-83802-P, and BFU2015-71786-REDT). L.F. is supported by funding from BBSRC grant BBS/E/I/00007039. S.I.G. (fellowship SFRH/BPD/88854/2012), J.B.S. and M.L.M. were supported by Fundação para a Ciência e a Tecnologia (FCT, PTDC/BIA-EVF/116884/2010). Financial support from “Alianza 4 Universidades” to R.A.S.-G. is also acknowledged. Thanks are due for the financial support of CESAM (UID/AMB/50017 - POCI-01-0145-FEDER-007638), FCT/MCTES through national funds (PIDDAC), and the cofunding by the FEDER, within the PT2020 Partnership Agreement and Compete 2020. The authors acknowledge F. Pardo-Manuel de Villena for his assistance in SNP genotyping. We also acknowledge the valuable help in field work and/or karyotyping and genotyping of Joaquim Tapisso, Ana Cerveira, Maria da Graça Ramalhinho, Janice Britton-Davidian, Pedro Martins da Silva, Rita Monarca, Nuria Medarde, Marta Pla-Bagaria, and Laura Palacios-Fernández.
Author Contributions
A.R.-H. conceived and devised the study. C.V., L.C., L.F., A.L., R.A.S.-G., S.I.G., M.L.M., J.B.S., and A.R.-H. contributed to the design of the methodological approaches. C.V., L.C., R.A.S.-G., B.F.-S., J.B.-R., and A.R.-H. contributed to PRDM9 sequencing experiments and allele classification. C.V., G.A.-L., L.F., and A.R.-H. contributed to SNP genotyping and phylogenetic analysis of PRDM9 alleles. L.F. and A.L. developed the approach for the computation of genetic distances between complex repeats. S.I.G., J.V., J.B.S., M.L.M., and A.R.-H. contributed to reagents and data collection. C.V., L.C., and A.R.-H. wrote the first draft of the manuscript with contributions of all authors. All authors read and approved the final version of the manuscript.
References
- Adolph S, Klein J.. 1981. Robertsonian variation in Mus musculus from Central Europe, Spain, and Scotland. J Hered. 723:219–221. [DOI] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 199:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artimo P, Jonnalagedda M, Arnold K, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, et al. 2012. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40:597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CL, Kajita S, Walker M, Saxl RL, Raghupathy N, Choi K, Petkov PM, Paigen K.. 2015. PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination. PLoS Genet. 111:e1004916.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker Z, Schumer M, Haba Y, Bashkirova L, Holland C, Rosenthal GG, Przeworski M.. 2017. Repeated losses of PRDM9-directed recombination despite the conservation of PRDM9 across vertebrates. eLife 6:e24133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcova M, Faltusova B, Gergelits V, Bhattacharyya T, Mihola O, Trachtulec Z, Knopf C, Fotopulosova V, Chvatalova I, Gregorova S, et al. 2016. Hybrid sterility locus on chromosome X controls meiotic recombination rate in mouse. PLoS Genet. 124:e1005906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash DA, Siino DF, Tarone AM, Roote J.. 2003. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc Natl Acad Sci U S A. 1009:5302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B.. 2010. Prdm9 is a major determinant of meiotic recombination hotspots in human and mice. Science 21:836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IL, Neumann R, Lam K-W, Sarbajna S, Odenthal-Hesse L, May CA, Jeffreys AJ.. 2010. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 4210:859–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya T, Gregorova S, Mihola O, Anger M, Sebestova J, Denny P.. 2013. Mechanistic basis of infertility of mouse intersubspecific hybrids. Proc Nat Acad Sci U S A. 110:468–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billings T, Parvanov ED, Baker CL, Walker M, Paigen K, Petkov PM.. 2013. DNA binding specificities of the long zinc-finger recombination protein PRDM9. Genome Biol. 144:R35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhomme F, Orth A, Cucchi T, Rajabi-Maham H, Catalan J, Boursot P, Auffray J-C, Britton-Davidian J.. 2011. Genetic differentiation of the house mouse around the Mediterranean basin: matrilineal footprints of early and late colonization. Proc Biol Sci. 2781708:1034–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton A, Myers RS, Redfield RJ.. 1997. The hotspot conversion paradox and the evolution of meiotic recombination. Proc Natl Acad Sci U S A. 9415:8058–8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV.. 2012. Genetic recombination is directed away from functional genomic elements in mice. Nature 4857400:642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton-Davidian J, Catalan J, Lopez J, Ganem G, Nunes AC, Ramalhinho MG, Auffray JC, Searle JB, Mathias ML.. 2007. Patterns of genic diversity and structure in a species undergoing rapid chromosomal radiation: an allozyme analysis of house mice from the Madeira archipelago. Heredity 994:432–442. [DOI] [PubMed] [Google Scholar]
- Britton-Davidian J, Catalan J, Ramalhinho MG, Ganem G, Auffray J-C, Capela R, Biscoito M, Searle JB, Mathias ML.. 2000. Rapid chromosomal evolution in island mice. Nature 4036766:158.. [DOI] [PubMed] [Google Scholar]
- Buard J, Rivals E, Dunoyer de Segonzac D, Garres C, Caminade P, de Massy B, Boursot P.. 2014. Diversity of Prdm9 zinc finger array in wild mice unravels new facets of the evolutionary turnover of this coding minisatellite. PLoS One 91:e85021.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capilla L, Garcia Caldés M, Ruiz-Herrera A.. 2016. Mammalian meiotic recombination: a toolbox for genome evolution. Cytogenet Genome Res. 1501:1–16. [DOI] [PubMed] [Google Scholar]
- Capilla L, Medarde N, Alemany-Schmidt A, Oliver-Bonet M, Ventura J, Ruiz-Herrera A.. 2014. Genetic recombination variation in wild Robertsonian mice: on the role of chromosomal fusions and Prdm9 allelic background. Proc R Soc B 281:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmátal L, Gabriel SI, Mitsainas GP, Martínez-Vargas J, Ventura J, Searle JB, Schultz RM, Lampson MA.. 2014. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr Biol. 2419:2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Myers SR.. 2007. Live hot, die young: transmission distortion in recombination hotspots. PLoS Genet. 3:377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA.. 1998. The evolutionary genetics of speciation. Philos Trans R Soc Lond B Biol Sci. 3531366:287–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AB, Hatton E, Altemose N, Hussin JG, Pratto F, Zhang G, Hinch AG, Moralli D, Biggs D, Camerini-Otero RD, et al. 2016. Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature 5307589:171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzur-Gejdosova M, Simecek P, Gregorova S, Bhattacharyya T, Forejt J.. 2012. Dissecting the genetic architecture of F1 hybrid sterility in house mice. Evolution 6611:3321–3335. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J.. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 148:2611–2620. [DOI] [PubMed] [Google Scholar]
- Faria R, Navarro A.. 2010. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol Evol (Amst). 2511:660–669. [DOI] [PubMed] [Google Scholar]
- Farré M, Micheletti D, Ruiz-Herrera A.. 2013. Recombination rates and genomic shuffling in human and chimpanzee—a new twist in the chromosomal speciation theory. Mol Biol Evol. 304:853–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flachs P, Mihola O, Šimeček P, Gregorová S, Schimenti JC, Matsui Y, Baudat F, de Massy B, Piálek J, Forejt J, et al. 2012. Interallelic and intergenic incompatibilities of the Prdm9 (Hst1) gene in mouse hybrid sterility. PLoS Genet. 811:e1003044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster DW, Günduz İ, Nunes AC, Gabriel S, Ramalhinho MG, Mathias ML, Britton-Davidian J, Searle JB.. 2009. Molecular insights into the colonization and chromosomal diversification of Madeiran house mice. Mol Ecol. 1821:4477–4494. [DOI] [PubMed] [Google Scholar]
- Förster DW, Mathias ML, Britton-Davidian J, Searle JB.. 2013. Origin of the chromosomal radiation of Madeiran house mice: a microsatellite analysis of metacentric chromosomes. Heredity (Edinb). 1104:380–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini P, Colangelo P, Solano E, Capanna E, Verheyen E, Castiglia R.. 2010. Reduced gene flow at pericentromeric loci in a hybrid zone involving chromosomal races of the house mouse Mus musculus domesticus. Evolution 64:2020–2032. [DOI] [PubMed] [Google Scholar]
- Giménez MD, White TA, Hauffe HC, Panithanarak T, Searle JB.. 2013. Understanding the basis of diminished gene flow between hybridizing chromosome races of the house mouse. Evolution 675:1446–1462. [DOI] [PubMed] [Google Scholar]
- Gregorová S, Gergelits V, Chvatalova I, Bhattacharyya T, Valiskova B, Fotopulosova V, Jansa P, Wiatrowska D, Forejt J.. 2018. Modulation of Prdm9-controlled meiotic chromosome asynapsis overrides hybrid sterility in mice. eLife 7:e34282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey C, Barthès P, Chauveau-Le Friec G, Langa F, Baudat F, de Massy B.. 2011. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol. 910:e1001176.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gündüz İ, Auffray J-C, Britton-Davidian J, Catalan J, Ganem G, Ramalhinho MG, Mathias ML, Searle JB.. 2001. Molecular studies on the colonization of the Madeiran archipelago by house mice. Mol Ecol. 108:2023–2029. [DOI] [PubMed] [Google Scholar]
- Gündüz I, López-Fuster MJ, Ventura J, Searle JB.. 2001. Clinal analysis of a chromosomal hybrid zone in the house mouse. Genet Res. 771:41–51. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP.. 1992. Estimation of levels of gene flow from DNA sequence data. Genetics 1322:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys AJ, Cotton VE, Neumann R, Lam KG.. 2013. Recombination regulator PRDM9 influences the instability of its own coding sequence in humans. Proc Natl Acad Sci U S A. 1102:600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono H, Tamura M, Osada N, Suzuki H, Abe K, Moriwaki K, Ohta K, Shiroishi T.. 2014. Prdm9 polymorphism unveils mouse evolutionary tracks. DNA Res. 213:315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masly JP, Jones CD, Noor MAF, Locke J, Orr HA.. 2006. Gene transposition as a cause of hybrid sterility in Drosophila. Science 3135792:1448–1450. [DOI] [PubMed] [Google Scholar]
- Medarde N, López-Fuster MJ, Muñoz-Muñoz F, Ventura J.. 2012. Spatio-temporal variation in the structure of a chromosomal polymorphism zone in the house mouse. Heredity (Edinb). 1092:78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J.. 2009. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 3235912:373–375. [DOI] [PubMed] [Google Scholar]
- Morgan AP, Fu C-P, Kao C-Y, Welsh CE, Didion JP, Yadgary L, Hyacinth L, Ferris MT, Bell TA, Miller DR.. 2015. The mouse universal genotyping array: from substrains to subspecies. G3 6:263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes AC, Britton-Davidian J, Catalan J, Ramalhinho MG, Capela R, Mathias ML, Ganem G.. 2005. Influence of physical environmental characteristics and anthropogenic factors on the position and structure of a contact zone between two chromosomal races of the house mouse on the island of Madeira (North Atlantic, Portugal). J Biogeogr. 3212:2123–2134. [Google Scholar]
- Oliver PL, Goodstadt L, Bayes JJ, Birtle Z, Roach KC, Phadnis N, Beatson SA, Lunter G, Malik HS, Ponting CP.. 2009. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 512:e1000753.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA, Turelli M.. 2001. The evolution of postzygotic isolation: accumulating Dobzhansky–Muller incompatibilities. Evolution 556:1085–1094. [DOI] [PubMed] [Google Scholar]
- Paigen K, Petkov PM.. 2018. PRDM9 and its role in genetic recombination. Trends Genet. 344:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K.. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 202:289–290. [DOI] [PubMed] [Google Scholar]
- Pavlova SV, Searle JB.. 2018. Chromosomes and speciation in mammals In: Zachos FE, Asher RJ, editors. Mammalian evolution, diversity and systematics. Berlin (Germany: ): De Gruyter; p. 17–38. [Google Scholar]
- Phadnis N, Orr HA.. 2009. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 3235912:376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piálek J, Hauffe HC, Searle JB.. 2005. Chromosomal variation in the house mouse. Biol J Linn Soc. 843:535–563. [Google Scholar]
- Presgraves DC, Balagopalan L, Abmayr SM, Orr HA.. 2003. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 4236941:715–719. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 813:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Chen JK, Reynolds A, Höög C, Paddy M, Hunter N.. 2012. Interplay between synaptonemal complex, homologous recombination, and centromeres during mammalian meiosis. PLoS Genet. 86:e1002790.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T.. 1989. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sánchez-Guillén RA, Capilla L, Reig-Viader R, Martínez-Plana M, Pardo-Camacho C, Andrés-Nieto M, Ventura J, Ruiz-Herrera A.. 2015. On the origin of Robertsonian fusions in nature: evidence of telomere shortening in wild house mice. J Evol Biol. 281:241–249. [DOI] [PubMed] [Google Scholar]
- Sans-Fuentes MA, García-Valero J, Ventura J, López-Fuster MJ.. 2010. Spermatogenesis in house mouse in a Robertsonian polymorphism zone. Reproduction 1404:569–581. [DOI] [PubMed] [Google Scholar]
- Schliep KP. 2011. Phangorn: phylogenetic analysis in R. Bioinformatics 274:592–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JJ, Roach DJ, Thomas JH, Shendure J.. 2014. Primate evolution of the recombination regulator PRDM9. Nat Commun. 5:4370.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F, Brick K, Pu Y, Camerini-Otero RD, Petukhova GV.. 2016. The evolutionary turnover of recombination hot spots contributes to speciation in mice. Genes Dev. 303:266–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemann-Boege I, Schwarz T, Striedner Y, Heissl A.. 2017. The consequences of sequence erosion in the evolution of recombination hotspots. Philos Trans R Soc B 3721736:20160462.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CT, Tsaur SC, Wu ML, Wu CI.. 1998. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 2825393:1501–1504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data underlying this article are available on GeneBank (from MK848086 to MK848149).