Abstract
Three-dimensional (3D) multicellular aggregates, in comparison to two-dimensional monolayer culture, can provide tissue culture models that better recapitulate the abundant cell-cell and cell-matrix interactions found in vivo. In addition, aggregates are potentially useful building blocks for tissue engineering. However, control over the interior aggregate microenvironment is challenging due to inherent barriers for diffusion of biological mediators (e.g. growth factors) throughout the multicellular aggregates. Previous studies have shown that incorporation of biomaterials into multicellular aggregates can support cell survival and control differentiation of stem cell aggregates by delivering morphogens from within the 3D construct. In this study, we developed a highly efficient microparticle-based gene delivery approach to uniformly transfect human mesenchymal stromal cells (hMSC) within multicellular aggregates and cell sheets. We hypothesized that release of plasmid DNA (pDNA) lipoplexes from mineral-coated microparticles (MCMs) within 3D hMSC constructs would improve transfection in comparison to standard free pDNA lipoplex delivery in the media. Our approach increased transfection efficiency 5-fold over delivery of free pDNA lipoplexes in the media and resulted in homogenous distribution of transfected cells throughout the 3D constructs. Additionally, we found that MCMs improved hMSC transfection by specifically increasing macropinocytosis-mediated uptake of pDNA. Finally, we showed up to a three-fold increase of bone morphogenetic protein-2 (BMP-2) expression and enhanced calcium deposition within 3D hMSC constructs following MCM-mediated delivery of a BMP-2 encoding plasmid and culture in osteogenic medium. The technology described here provides a valuable tool for achieving efficient and homogenous transfection of 3D cell constructs and is therefore of particular value in tissue engineering and regenerative medicine applications.
Graphical Abstract

1. Introduction
Cell therapies have the potential to provide new treatments for a number of diseases and disorders by replacing damaged tissue with regenerated healthy tissue [1,2]. Many cell-based therapies use a scaffold to provide a structural template that supports tissue regeneration [3–5]. However, it is often difficult to match scaffold degradation rate with new tissue formation, and degradation byproducts can cause inflammation, cytotoxicity, or an immune response [6]. For example, degradation byproducts of poly(lactic-co-glycolic acid) (PLGA) scaffolds can lower the local pH to 4.0 [7], potentially triggering inflammation and edema. As a result, scaffold-based cell therapies can result in undesirable outcomes such as insufficient cell infiltration, limited vascular invasion, and impaired tissue remodeling. In contrast, scaffold-free tissue engineering strategies, in which cell aggregates are assembled without a scaffold material, can offer an attractive alternative approach for tissue regeneration. The high cell density within scaffold-free constructs enables abundant cell-cell and cell-extracellular matrix (ECM) interactions that may more closely mimic tissue morphogenesis. Indeed, previous studies have shown that culture of human mesenchymal stromal cells (hMSCs) as multicellular aggregates enhances in vitro differentiation, as shown by increased chondrogenic marker expression during directed chondrogenesis [8]. hMSC aggregates have also shown promise in tissue engineering, where they can be used as building blocks for de novo tissues [9]. However, it is challenging to deliver large molecules (e.g. growth factors) into these constructs due to an inherent transport barrier created by high cellular density, cell-mediated deposition of ECM, and cellular metabolic activity.
Previously, we and others have developed biomaterials-based approaches to overcome the diffusion barrier in cell aggregates, in which growth factor-laden microparticles were incorporated into the interior of 3D cell constructs [10,11] and promoted more homogenous differentiation [11–13]. However, this strategy was restricted to delivery of extracellular cues such as cytokines and growth factors, and could also suffer from the limited biological activity of recombinantly produced proteins. Alternatively, gene delivery to the interior of a cell aggregate could allow for sustained, local and bioactive production of a protein, as well as the ability to overexpress intracellular proteins such as transcription factors [14–18]. Non-viral gene delivery approaches have been specifically explored in cell-based therapies because of their desirable safety profiles and ease of synthesis relative to viral vectors [16,19]. Thus, non-viral gene delivery may represent an ideal strategy to direct cell function or influence cell response within cell aggregates [19,20]. However, non-viral gene delivery approaches typically suffer from low transfection efficiency in primary human cells, such as hMSCs, particularly in a 3D culture context. For example, while hMSC transfection with commercial reagents in standard two-dimensional (2D) cell culture on tissue culture polystyrene (TCPS) has resulted in greater than 25% transfection efficiency [21–25], there no reports of achieving similar transfection efficiencies in 3D using these commercial reagents. This is potentially due to the fact that the vectors frequently used for non-viral transfection (e.g. liposomes, polyplexes, nanoparticles) are designed for 2D culture platforms that do not account for three-dimensional (3D) gene delivery barriers. As an example of this, 3D cell aggregates and cell sheets introduce an inherent transport barrier that must be overcome to deliver the relatively large non-viral vector DNA complexes (~200–900 nm) to the interior [26,27]. Thus, non-viral gene delivery to the interior of scaffold-free human cell constructs is a significant challenge, and there remains a need for efficient and homogenous 3D transfection techniques.
Here, we describe and characterize a biomaterial-based, non-viral gene delivery approach that efficiently delivers genes throughout multicellular hMSC aggregates and cell sheets. Based on our previous reports[22,28–30], we hypothesized that incorporation of mineral-coated microparticles (MCM) loaded with plasmid DNA (pDNA) into cell aggregates or sheets would enable efficient homogeneous 3D gene delivery and superior efficiency than standard transfection via addition of free lipoplexes to the cell construct culture medium. Our results demonstrate that MCMs within 3D hMSC aggregates or sheets provided highly efficient and homogenous transfection. Characterizing the approach, we demonstrated that MCMs increased hMSC transfection efficiency by increasing macropinocytosis and corresponding uptake of pDNA. Optimization of this approach by varying the pDNA dosage and the amount of MCMs used led to a hMSC transfection efficiency up to 30%, which was 5-fold higher than our efficiency using standard non-viral transfection protocols on 2D substrates, and more than 20-fold higher than standard transfection protocols in 3D cell culture. Finally, to explore the utility of this approach for tissue engineering applications, we used these MCMs to deliver a plasmid encoding bone morphogenetic protein-2 (BMP-2) to enhance osteogenic differentiation of 3D hMSC constructs. Taken together, the biomaterial technology described here may have unique utility in achieving highly efficient and uniform transfection of scaffold-free 3D cell constructs.
2. Materials and Methods
2.1. MCMs preparation and characterization
Sintered hydroxyapatite microparticles (HA MPs, Plasma Biotal LTD) with particle size between 3–5 μm were used as the starting material for the MCMs and prepared as previously reported [22,30–32]. Briefly, the HA MPs were incubated in modified simulated body fluid (mSBF) at 37°C for 7 days with daily mSBF replacement. The mSBF was prepared by adding the following reagents into deionized water in the order shown: 141 mM NaCl, 4.0 mM KCl, 0.5 mM MgSO4, 1.0 mM MgCl2, 100 mM NaHCO3, 20.0 mM HEPES, 5.0 mM CaCl2, and 2.0 mM KH2PO4. The mSBF pH was then adjusted to 6.80. MCMs were harvested after seven days of incubation by rinsing with deionized water, filtering through a 40 μm strainer, and drying via lyophilization. Visual inspection and verification of mineral coating formation was done via a LEO 1530 scanning electron microscope (Gemini) at 3kv. The collected MCMs were sterilized by exposing under UV light for 20 min within a biosafety cabinet before incorporation into culture with hMSCs.
2.2. Plasmid DNA complex binding on MCMs
pMetLuc and pEGFP-N1 reporter vectors (Clontech) were amplified using competent TOF10F’ E. Coli (Life Technologies) and purified with a Giga plasmid purification kit (Qiagen) following the manufacturer’s protocol. pDNA lipoplexes were formed by mixing the pDNA with Lipofectamine 2000 (Lipo2000, Life Technologies) at N/P ratio of 2.0 in Opti-MEM® reduced serum medium (Life Technologies) according to the manufacturer’s protocol. 50, 100, or 200 μg of MCMs were added to 200 μL pDNA complex solution with various amount of pDNA lipoplexes for 2 h to allow the pDNA complexes to bind to the mineral surface. The pDNA complexes bound to MCMs were then centrifuged at 1200 RPM for 2 min and washed with 1.0 mL PBS once. The remaining pDNA complex in the supernatant was measured using a PicoGreen assay (Life Technologies) as previously described [22,32] and converted to the pDNA binding efficiency on MCMs.
2.3. hMSC culture
hMSCs (Lonza) were expanded at low density on T175 tissue culture polystyrene (TCPS) flasks to maintain their multipotency and used for experiments by passage 6. hMSCs were grown in growth medium comprising of α-Eagle Essential Media (α-MEM) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 U/mL penicillin/streptomycin at 37°C and 5% CO2. Media was exchanged every other day and hMSCs were passaged or harvested at 75% confluency.
2.4. hMSC aggregate formation and i) standard transfection with pDNA or ii) pDNA + MCMs
To evaluate the transfection efficiency of the i) standard and ii) pDNA+MCM approaches, 0.25 ×106 hMSCs were mixed with no MCMs, or with MCMs without pDNA at different ratios (50, 100 and 200 μg MCMs/Agg) incorporated into the aggregates similarly to previously described methods[33]. hMSCs were harvested by trypsinization and resuspended in growth medium with the varying MCM suspensions at 1.25×106 cells/mL (pDNA+MCMs), or with no MCMs (standard approach). 200 μL of these hMSC suspensions per well were added into V-shaped polypropylene microplates and centrifuged at 500g for 5 min. After 24 h of culture, the different amounts of pDNA lipoplexes were added to hMSC aggregates. For transfection of small aggregates during aggregation, we added 2 μg pEGFP lipoplexes to the 0.25×106 hMSC cell suspension and centrifuged the mixture into Aggrewell 800 (Stem Cell Technologies) microwell plates to yield aggregates with 6×103 rather than 250×103 cells per aggregate.
2.5. Incorporation of iii) pDNA/MCMs into hMSC aggregates
pDNA complexes bound to MCMs were resuspended in growth medium at 10, 20, 40 μg/mL, respectively. hMSCs were trypsinized and resuspended in the solutions above at a concentration of 1.25 × 106 cells/mL. pDNA/MCMs and hMSC were mixed at 1:1 volume ratio. The hMSC/MCM suspensions were then dispensed into sterile 96-well V-shaped polypropylene microplates at 200 μL/well after gently mixing. The plates were centrifuged at 500g for 5 min to pellet the hMSC/MCMs to form the MCMs-incorporated hMSC aggregates. hMSC aggregates were transfected for different time periods and their transfection efficiency with different plasmids were assessed by assays corresponding to each plasmid. pDNA/MCM transfection of small aggregates was performed as described in methods 2.4 using pDNA/MCMs in place of pDNA+MCMs.
2.6. Incorporation of pDNA lipoplex-loaded MCMs into hMSC sheets
hMSC suspensions (2×106 cells/mL) were uniformly mixed with pDNA lipoplex-bound MCMs (1.5 mg/sheet) with or without loaded pEGFP lipoplex or pBMP-2 lipoplex. For pEGFP lipoplexes, the pEGFP concentration were set at 0, 1, 5, 10 μg/sheet, respectively. For pBMP-2, the pBMP-2 concentrations were set at 0, 5, 10 μg/sheet, respectively. The mixture was then seeded onto the membrane of polycarbonate Transwell inserts (3 μm pore size, 12 mm diameter, Corning) and allowed to self-assemble as previously described[13]. hMSC sheets were transfected for different time periods and their transfection efficiency with different plasmids were assessed by assays corresponding to each plasmid.
2.7. Luminescence assay
Luciferase activity was measured using a Cell-Glow luciferase kit (Clontech). Briefly, 5 μL of luciferase substrate was added into 50 μL hMSC aggregates culture medium in a 96-well plate and incubated at room temperature for 5 min while protected from light. The luminescence was measured by a microplate reader (Fluoroskan Ascent FL). Cell metabolic activity in each aggregate after transfection was evaluated by the CellTiter-Blue assay kit (Promega) according to the manufacturer’s protocol and measured for fluorescence on a plate reader (Fluoroskan Ascent FL).
2.8. GFP expression analysis
For hMSC aggregates transfected with EGFP plasmid, aggregates were collected from the microplate after 48 hr of culture and fixed in 10% neutral buffered formalin for 30 min. The samples were then embedded in OCT embedding medium and cut into 5 μm-thick sections (Cryo Stat). The sections were mounted on to histology slides and DAPI counter staining was performed in certain experimental groups. Expression of EGFP in hMSC aggregates was determined using a Nikon Eclipse Ti inverted microscope equipped with a 10X PhL objective (Nikon), FITC fluorescence filter cube (Nikon), CoolSNAP HQ2 camera (Photometrics), and Elements analysis software (Nikon). For visualizing the MCMs embedded inside the hMSC aggregates, phase contrast images were captured with a 10× PhL objective.
2.9. Flow cytometry
For EGFP transfection efficiency determination, hMSCs were dissociated from the aggregates or cell sheets by incubating with PBS containing 0.25% Trypsin with 50 mM EDTA and while rotating for 15 min at room temperature. The dissociation mixture was then filtered through a 40 μm cell strainer to make the single cell suspension. Flow cytometry was performed with a FACSCalibur (BD) or Attune NxT (Thermo Fisher Scientific), with a minimum of 10,000 events per sample within the FSC/SSC gate for live cell populations. Untransfected hMSCs grown on TCPS were used as a negative control, and transfected hMSCs with pEGFP/Lipo2000 complex on TCPS were used as a positive control. Data analysis were performed via FlowJo software (Tree Star). pEGFP was labeled with CX-Rhodamine Label IT nucleic acid labeling kit (Mirus). hMSC aggregates were transfected with rhodamine-labeled pEGFP for 4 hr following the procedures described in section 2.4–2.5. The aggregates were dissociated using the aforementioned method, washed with 1% FBS in PBS, and analyzed for red fluorescence using the FACSCalibur (BD).
2.10. BMP-2 ELISA
hMSC aggregates were transfected with a plasmid encoding BMP-2 (pcDNA3.1/His/hBMP-2) similarly to the procedure described above for EGFP. After 72 h of transfection, the cell culture medium in each well was collected. The hMSC aggregates were then dissociated with 100 mM EDTA/PBS (pH=7.4) for 15 min with pipetting up and down every 5 min to ensure complete dissolution of the MCMs. The dissociated aggregates were centrifuged at 500g for 5 min and the supernatant was collected to measure the BMP-2 bound on the MCMs. The BMP-2 concentrations of the culture media and MCM fractions were determined via a Quantikine ELISA Kit (R&D Systems) following the manufacturer’s instructions.
2.11. Endocytosis inhibition and analysis
hMSCs were cultured on TCPS in growth medium as described above. Cells were harvested by trypsinization and plated at 2000 cells per well in 96-well TCPS flat bottom plates. 36 hours after seeding, cells were treated with 0–12.8 μg MCMs per well and 100 μg/mL 10 kDA molecular weight Alex Fluor 594-conjudated dextran (Thermo Fisher Scientific) for 6 hours. Cells were then fixed in 10% neutral buffered formalin, washed 3X with PBS, and treated with 0.1 N HCl for 15 minutes to remove MCMs. Cells were then analyzed for Alexa Fluor594+ endosome area using a Nikon Eclipse Ti inverted microscope equipped with a 10X PhL objective (Nikon), TxRed fluorescence filter cube (Nikon), CoolSNAP HQ2 camera (Photometrics), and Elements analysis software (Nikon). Alternatively, cells were transfected with pEGFP or pEGFP/MCMs as described above and analyzed for expression of EGFP using a Nikon Eclipse Ti inverted microscope equipped with a 10X PhL objective (Nikon), FITC fluorescence filter cube (Nikon), CoolSNAP HQ2 camera (Photometrics), and Elements analysis software (Nikon). For endocytosis inhibition, cells were treated with 0.4 mM amiloride (Tocris Bioscience), 0.1 mM chlorpromazine (Alfa Aesar), or 5 mM methyl-β-cyclodextrin (Sigma Aldrich) in growth medium reduced to 0.5% serum for 1 hr prior to addition of MCMs and Alexa Fluor 594 dextran or transfection.
2.12. hMSC aggregate and sheet culture in osteogenic medium
hMSC aggregates and sheets were transfected with a plasmid encoding BMP-2 (pcDNA3.1/His/hBMP-2) as described above. After 72 h of transfection, the cell culture medium was removed from each well and replaced with osteogenic medium consisting of growth medium supplemented with 0.1 μM dexamethasone, 50 μg/mL ascorbic acid and 10mM β-glycerophosphate) for 5 weeks. The medium changes were performed every 3 days.
2.13. Histological analysis
Cultured aggregates were fixed in 4% neutral buffered formalin for 30 min and embedded in OCT media and kept frozen. The frozen embedded samples were sectioned into 5 μm and subjected to H&E staining and Alizarin red staining. Stained sections were imaged using a camera assisted microscope (Nikon Eclipse microscope, model E6000 with an Olympus camera, model DP79). Osteocalcin (OCN) was stained as a bone marker. Briefly, sections were blocked with 5% BSA in PBS. After blocking, primary antibody anti-OCN (Abcam, Ab52128) was applied to the sections. After washing with PBS, the staining signals were visualized with Donkey anti-Rabbit IgG H&L (DyLight® 488) (Abcam, Ab96891) and imaged using a Nikon Eclipse Ti inverted microscope equipped with a Nikon DS-Fi1 camera (Nikon).
2.14. Statistical analysis
Quantitative data are expressed as mean ± standard deviation. Statistical analysis was performed for all quantitative data using GraphPad Prism, with the statistical test used and significance reported in each figure.
3. Results
3.1. MCMs adsorb pDNA lipoplexes out of solution for incorporation into 3D constructs
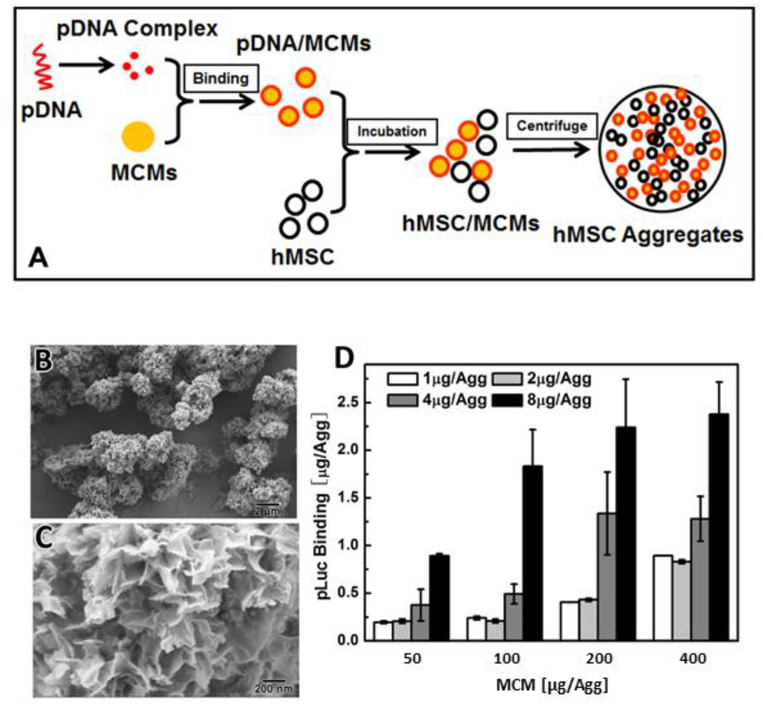
Mineral coated microparticles (MCMs) adsorbed pDNA complexed with Lipofectamine 2000 (Lipo2000) out of solution (Fig. 1A). The MCMs used here contained a layer of nanostructured mineral coating on the microparticle surface (Fig. 1B, C), generated by incubating 3–5 μm diameter hydroxyapatite microparticles in modified simulated body fluid (mSBF). This coating featured plate-like features, 200–400 nm in size (Fig. 1C). The resulting MCMs were capable of binding up to 2.38 μg of Metridia luciferase-encoding plasmid (pLuc) lipoplexes (Fig. 1-D), and the pLuc amount per aggregate could be varied between 0.19 to 2.38 μg pLuc by adjusting the initial amount of pLuc and MCMs in the binding solution. (Fig. 1-D)
Figure 1. Incorporation of pDNA-laden MCMs into hMSC aggregates:
(A) Schematic of the microparticle-based gene delivery approach: pDNA was condensed using Lipofectamine2000 and the resulting complex was adsorbed to the MCM surface. The pDNA-laden MCMs were mixed with hMSCs and centrifuged to transfect the aggregates. (B) Scanning electron micrograph (SEM) of MCMs at low magnification showing MCMs of 5–8 μm in diameter. (C) High magnification SEM micrograph of MCMs showing plate-like features around 200–400 nm in size. (D) MCM binding capacity of luciferase-encoding plasmid DNA lipoplexes.
3.2. MCM-mediated pDNA delivery improves transfection of hMSC aggregates
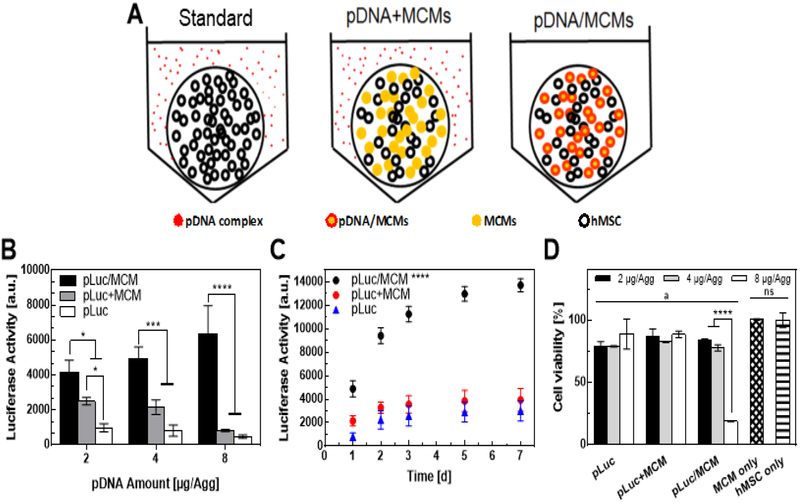
We compared three gene delivery approaches within hMSC aggregates: i) standard approach (pDNA): a standard transfection approach delivering free pDNA lipoplexes suspended in solution into the cell culture medium, ii) pDNA+MCMs: pDNA-free MCMs incorporated into aggregates, followed by a standard transfection approach delivering free pDNA lipoplexes suspended into the cell culture medium, iii) pDNA/MCMs: transfection via incorporation of pDNA-laden MCMs into aggregates as described in figure 1A (Fig. 2A). Aggregates contained 0.25×106 cells/aggregate, as this number of cells has been previously used in hMSC tissue engineering applications[11] and reliably produced aggregates of sufficient size such that the majority of cells were located in the aggregate interior. pLuc delivery via pDNA/MCMs (pLuc/MCMs) resulted in the greatest transgene expression among the approaches explored. Compared with the standard lipoplex delivery approach (pLuc), pLuc/MCM delivery increased transgene expression by up to 8-fold (Fig. 2B). Incorporation of pDNA-free MCMs followed by transfection of free lipoplexes (pLuc+MCMs) also increased transfection when compared with the standard approach, but not to the extent of the pLuc/MCM delivery within the aggregates. pLuc/MCM delivery also resulted in sustained gene expression for a longer timeframe (Fig. 2C). Specifically, luciferase activity in the pLuc and pLuc+MCMs groups plateaued 3 days post-transfection, while luciferase activity continued to increase in the pLuc/MCMs condition up to day 7 (Fig. 2C). Delivery of 8 μg/Agg pLuc/MCMs resulted the highest level of transgene expression, but also resulted in a > 80% reduction in aggregate viability relative to the no transfection control. In contrast, the 4 μg/Agg pLuc/MCMs condition resulted in 77% of the transgene expression level of the 8 μg/Agg but maintained up to 90% viability relative to the no transfection control (Fig. 2D).
Figure 2. Comparing transgene expression after plasmid DNA delivery into hMSC aggregates using different strategies:
A) Schematic of three pDNA delivery strategies: i) Standard: free pDNA lipoplexes added into the aggregate medium culture medium, ii) pDNA-free MCMs incorporated into hMSC aggregates, and then pDNA lipoplexes added into the aggregate culture medium, iii) pDNA lipoplexes adsorbed to the surface of MCMs, and then incorporated into hMSC aggregates. B) Luciferase activity of hMSC aggregate culture medium, 2 days after transfection using three different approaches. “*, ***, ****” represents p-value < 0.05, 0.001, and 0.0001 respectively for 2-way anova with Tukey post-hoc test. C) Luciferase activity of hMSC aggregate culture medium over time for 7 days after transfection using three different approaches. “****” represents p-value < 0.0001 for 2-way anova with Tukey post-hoc test. D) hMSC aggregates viability (cell-titer blue) after 2 days of transfection with the different approaches. “****” represents p-value < 0.0001 for 2-way anova with Tukey post-hoc test. “a” represents p-value < 0.05 for 2-way anova with Dunnet post-hoc test to hMSC only control. “ns” represents no significant difference.
3.3. MCMs improve spatial homogeneity of transfection of hMSC aggregates
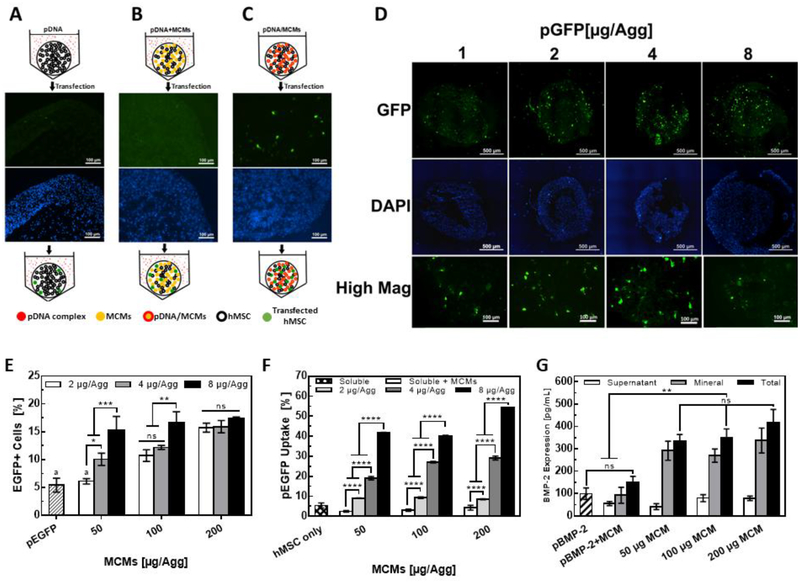
To evaluate the spatial distribution and homogeneity of transfected hMSCs within the aggregates, we delivered an EGFP-encoding plasmid (pEGFP) using the three approaches described previously. For both standard pDNA and pDNA+MCMs approaches, there were few EGFP-positive cells and, when present, they were located only at the aggregate periphery (Fig. 3A, B). In contrast, local delivery of pEGFP using MCMs from within aggregates (pDNA/MCMs) increased the number of EGFP-positive cells overall, and specifically increased the number of EGFP-positive cells in the aggregate interior (Fig. 3C). Delivering increasing amounts of pEGFP via the pDNA/MCM approach resulted in a dose-dependent increase in EGFP-positive cells in the aggregate interior (Fig. 3D). Specifically, the EGFP-positive cells within the aggregate interior increased as we increased the pDNA dose from 1–4 μg/Agg. At 8.0 μg/Agg, we observed a decrease in interior EGFP-positive cells (Fig 3D), which also coincided with the higher cytotoxicity in the aggregates previously observed for this condition (Fig 2D). Additionally, neither a reduction in aggregate size or transfection during aggregation with free pEGFP lipoplexes mixed with the cell suspension prior to centrifugation overcame this diffusion barrier to produce efficient and homogenous 3D transfection (Fig. S1). Interestingly, most EGFP-positive cells in pEGFP/MCM aggregates were in close proximity to MCMs (Fig. S2–3), indicating a local plasmid release from MCMs, similar to previously reported findings with these materials [22,32], was likely responsible for cell transfection within the aggregate.
Figure 3. MCM-mediated delivery increases transfection efficiency and homogeneity of hMSC aggregates.
hMSC aggregates (0.25×106 cell/Agg), transfected with (A) standard approach, (B) 100 μg pDNA-free MCMs and free pDNA lipoplexes suspended in the medium, and (C) the pDNA/MCM method, examined for distribution of EGFP-positive cells within epifluorescence micrographs of cryosectioned aggregates. (D) Examination of distribution of EGFP-positive cell in hMSC aggregates after transfected with increasing amounts of pEGFP with 100 μg MCMs for each aggregate. (E) Quantification of cellular uptake using flow cytometry and rhodamine-labeled pEGFP. Plasmid uptake increased in response to both the amount of plasmid and MCM delivered. “*, **, ***” represents p-value < 0.05, 0.01, and 0.001 respectively for 2-way anova with Tukey post-hoc test. “ns” represents no significant difference. (F) Optimization of pDNA/MCM method by varying pEGFP and MCMs amounts. hMSC transfection efficiencies increased with higher amounts of plasmid delivered for the 50 and 100 μg MCMs/Agg. All pDNA/MCM aggregate transfection efficiencies were higher than standard transfection in 2D except 2 μg pEGFP/50 μg MCM. “****” represents p-value < 0.0001 for 2-way anova with Tukey post-hoc test. (G) BMP-2 expression from hMSC aggregates transfected with pBMP-2/MCMs, measured via ELISA. “**” represents p-value < 0.01 for 2-way anova with Tukey post-hoc test. “ns” represents no significant difference.
Increases in the amount of plasmid delivered per aggregate also increased the percentage of EGFP-positive cells in conditions that included 50 and 100 μg MCMs per aggregate (Fig. 3E). Specifically, the transfection efficiency via the pDNA/MCM approach with 50 μg MCM per aggregate increased from 6.5% to 14.9% as the pEGFP dose increased from 2 to 8 μg/Agg (Figs. 3E, S4A). In comparison, the standard pEGFP delivery approach resulted in less than 1% GFP-positive cells (Fig. S4B). After optimization of the pEGFP and MCM doses (200 μg MCM, 8 μg pEGFP per aggregate), we achieved a 17.3% transfection efficiency in hMSC aggregates, which was over 3-fold higher than the 5.4% transfection efficiency achieved using Lipo2000 in 2D hMSC culture (Fig. 3E). In addition to the positive correlation between pEGFP dose and transfection efficiency, higher masses of MCMs also increased transfection efficiencies when delivering either 2 or 4 μg pEGFP/Agg (Fig. 3E), suggesting that the mineral coating itself may serve as a co-factor that facilitates transfection.
To specifically explore why the pDNA/MCM method increased transfection efficiency in hMSC aggregates, we used rhodamine-labeled pEGFP to measure differences in cellular pEGFP uptake between the different transfection approaches. hMSCs in aggregates loaded with MCMs carrying increasing amounts of labeled plasmid increased their red fluorescence intensity 2 hours post-transfection (Fig. 3F). Specifically, delivery of 2, 4, 8 μg pEGFP/Agg resulted in 9.0%, 19.1%, and 41.6% of hMSC plasmid uptake (50 μg MCM/Agg). In comparison, hMSCs cultured in 2D resulted in 2.2% hMSC plasmid uptake. Using our previously described conditions for optimized transfection efficiency (200 μg MCM, 8 μg pEGFP per aggregate), we achieved 54.2% plasmid uptake. Also, increasing the MCM amount per hMSC aggregate, while keeping the plasmid amount fixed, increased the labeled pEGFP uptake (Fig. 3F). Interestingly, pDNA+MCMs increased plasmid uptake from 2.4% to 4.2%, further supporting the hypothesis that MCMs alone may serve as a co-factor that encourages plasmid uptake and subsequent transfection.
Lastly, we assessed the production of a secreted growth factor from hMSC aggregates transfected with a BMP-2 encoding plasmid (pBMP-2) using our MCM method. The total production of BMP-2 during the first 72 hr post-transfection with pDNA/MCMs was 3-fold higher than standard transfection of hMSC grown in 2D on TCPS (Fig. 3G). The pDNA+MCM condition also increased BMP-2 production relative to standard transfection, again illustrating the role of MCMs alone in improving transfection (Fig. 3G). Interestingly, up to 86% (50 μg MCM per aggregate) of the secreted BMP-2 protein was sequestered within the MCMs (Fig. 3G).
3.4. MCMs improve hMSC transfection through induction of macropinocytosis
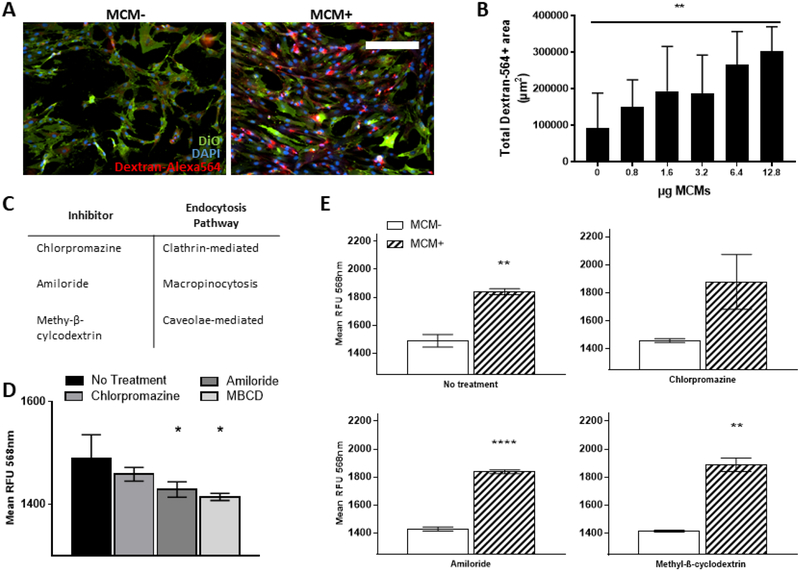
Non-viral transfection with pDNA is known to occur through various endocytosis pathways [34]. We cultured hMSCs in 2D with MCMs and Alexa Fluor-564 labeled 10 kDa molecular weight dextran to assess the effect of MCMs on endocytosis (Fig. 4A–E). MCMs increased the number of endosomes produced by hMSCs (Fig. 4A, B). Specifically, there was a dose dependent increase in the mean area of Alexa564-positive endosomes in hMSC culture with increases in MCM concentration (Fig. 4B). The highest concentration of MCMs resulted in a 330% increase in Alexa564-positive endosome area relative to hMSCs without MCMs (Fig. 4B). In addition, the mean fluorescence intensity of Alexa564-positive endosomes increased in a dose-dependent manner (Fig. S5), indicating that MCMs increase both the number of endosomes and the amount of material taken up into the endosome.
Figure 4. MCMs increase endocytosis in hMSCs and improve non-viral transfection through induction of macropinocytosis.
(A) Representative epifluorescence micrographs of hMSCs cultured in 2D with MCMs and Alexa Fluor 564-labeled 10k MW dextran. scale bar = 250μm (B) Quantification of increase in endocytosis in response to increasing MCM concentration. Endocytosis measured as total red fluorescence area. “**” represents p-value < 0.01 1-way anova with linear trend post-hoc test for dose-dependent response. (C) Table of small molecule inhibitors and specific endocytosis pathway inhibited. (D) Quantification of endocytosis in presence to small molecule inhibitors without MCMs. Endocytosis measured as mean red fluorescence intensity. “*” represents p-value < 0.05 1-way anova with Dunnet post-hoc test compared to no treatment (E) Quantification of endocytosis in presence to small molecule inhibitors with and without MCMs. Endocytosis measured as mean red fluorescence intensity. “**, ****” represents p-value < 0.01, 0.001 t-test.
To assess which endosomal pathway MCMs influence in hMSCs, we used small molecule inhibitors known to inhibit clathrin-dependent endocytosis (chlorpromazine), caveolae-mediated endocytosis (methyl-β-cyclodextrin), and macropinocytosis (amiloride) (Fig. 4C). In growth medium without MCMs, amiloride and methyl-β-cyclodextrin significantly reduced the number of Alexa564-positive endosomes (Fig. 4D), indicating that hMSCs use both macropinocytosis and caveolae-mediated endocytosis pathways under normal culture conditions. In the presence of MCMs, amiloride and methyl-β-cyclodextrin had no effect on endosome production (Fig. S6A), suggesting that MCMs upregulate both macropinocytosis and caveolae-mediated endocytosis. In comparing the effects of endocytosis inhibition during transfection, we observed that MCMs mitigated losses in transfection efficiency in hMSCs treated with either chlorpromazine or amiloride, but not methyl-β-cyclodextrin (Fig. S6B).
3.5. MCM-mediated delivery of BMP-2 plasmid enhances calcium deposition during osteogenic differentiation of 3D hMSC cultures
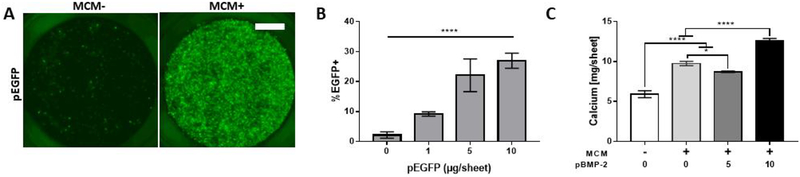
We next used the optimized pDNA/MCM method (200 μg MCM and 8 μg pBMP-2 per 250k cells) to deliver pBMP-2 during osteogenic differentiation of hMSC aggregates as well as larger cell sheets, in which we achieved up to 30% transfection efficiency (Fig. 5A, B). Cell aggregates (Fig. S7) in the pBMP-2-groups showed increased and more uniform ossification relative to the no transfection control after 5 weeks of culture, and increased calcium production in pBMP-2 transfected cell sheets (Fig. 5C). Cell aggregates showed abundant ECM deposition in all groups examined during the 5 weeks culture (Fig. S7), and Alizarin Red S staining showed increased mineralized tissue formed in aggregates transfected with 2 and 4 μg/Agg of pBMP-2/MCMs relative to no transfection (Fig. S7). In addition, increased osteocalcin (OCN) staining correlated with increased pBMP-2 delivered for 2 and 4 μg of total pDNA dosage. While aggregates transfected with 8 μg/Agg of pBMP-2 showed higher OCN staining, this was without a corresponding increase in Alizarin Red S staining (Fig. S7). For the larger cell sheets, pBMP-2/MCMs increased calcium production by 152% and 214% for the 5 and 10 μg pBMP-2 conditions, respectively, after 5 weeks in osteogenic medium when compared to the no transfection/no MCM control (Fig. 5C). Interestingly, MCMs alone also increased calcium production by the cell sheets relative to the MCM-free control after 5 weeks in osteogenic medium (Fig. 5C).
Figure 5. MCMs improve transfection and calcium production during osteogenic differentiation of 10 mm hMSC sheets.
(A) Epifluorescence micrographs of 10 mm hMSC sheets (2×106 cells per sheet) transfected and formed in 12-well transwells. Scale bar = 2.5 mm (B) Quantification of pEGFP hMSC sheet transfection via flow cytometry using the optimized pDNA/MCM condition. “****” represents p-value < 0.0001 1-way anova with linear trend post-hoc test for dose-dependent response. (C) hMSC sheet calcium production, measured Arsenazo III colorimetric detection after EDTA chelation of cell sheets transfected and cultured in osteogenic medium for 5 weeks. “*, ****” represents p-value < 0.05, and 0.0001 respectively for 2-way anova with Tukey post-hoc test.
4. Discussion
Scaffold-free cell constructs (e.g. cell aggregates and cell sheets) are an attractive strategy for tissue engineering due to their abundant cell-cell interactions, which mimic aspects of tissue development and regeneration. Within these constructs, 3D delivery of proteins is critical to direct cell differentiation and tissue morphogenesis. However, high cellular density and ECM production in 3D cell constructs creates an inherent diffusion limitation that limits protein delivery [35,36]. Previously, we have used biomaterials to successfully deliver growth factors within hMSC aggregates, but limitations inherent to recombinant proteins motivated us to explore a strategy to instead deliver genes within 3D constructs.
In conventional non-viral gene delivery approaches, pDNA complexes are directly taken up across the cell membrane. In transfection of 3D tissues, these complexes, which have diameters of 200–900 nm, must pass through multiple cell layers to reach the aggregate interior where the majority of cells reside [27,37]. This creates a significant challenge for efficient transfection of cell aggregates. Indeed, in our current study, transfection of 3D cell aggregates using standard pDNA delivery resulted in only a few successfully transfected cells, which only appeared at the periphery of the aggregates (Figs. 2, 3A). Additionally, we observed that neither reducing the aggregate size or attempting to transfect the cells with free pDNA lipoplexes during aggregation was an effective strategy to transfect 3D constructs (Fig. S1). In contrast, aggregates transfected with pDNA-laden MCMs showed up to an 8-fold increase in transgene expression (Fig. 2B), with homogenous transfection throughout the aggregate interior (Fig. 3C). Thus, pDNA complex delivery within the aggregate eliminated the need for pDNA complexes to be transported to the interior cells through diffusion (Figs. S2–3), which led to substantially improved 3D transfection.
After optimizing the pDNA/MCM approach to maximize transfection efficiency (Fig. 3E), we explored how MCMs promoted increased transfection in these hMSC constructs. First, MCMs served as gene carrier by binding pDNA complexes to the nanometer-scale mineral coating (Fig. 1), and then releasing pDNA via dissolution of the mineral coating. We have described this general mechanism of binding and release of pDNA-lipoplexes with calcium dissolution previously [22,32], and observed that pDNA release and resulting transfection was a function of calcium content of the mineral coating and culture medium, pH, and amount of pDNA originally bound. With respect to pBMP-2 specifically, we have previously shown that this coating applied to PLGA scaffolds bound pBMP-2 lipoplexes with greater than 80% percent binding efficiency and was capable of releasing lipoplexes to transfect hMSCs seeded in the coated construct [30]. Although we have previously observed and characterized sustained release kinetics of pDNA from these coatings over a three-week time period [32], we have not observed sustained transfection. Importantly, our previous observations and findings suggest that only the initially released lipoplexes are capable of transfecting cells, and therefore the plasmid uptake results described here should not be extrapolated or imply that transfection within the aggregates continues to occur beyond the two-hour time period we examined. Releasing pDNA from within cell aggregates increased cellular pDNA lipoplex uptake by 25-fold when compared to the standard pDNA delivery approach (Fig. 3F). We also observed that increasing the amounts of MCMs, either with or without bound pDNA, improved transfection efficiencies (Figs. 2B, 3E–G). This observation indicated that MCMs alone influence transfection, which is consistent with our previous study showing that calcium ions released from mineral coatings can serve as a co-factor for transfection of cells in 2D cell culture [22]. Here, the dissolution of the mineral coating likely led to simultaneously high local concentrations of both pDNA complexes and MCMs in 3D, further potentiating transfection within the cell constructs (Figs. S2–3).
Cellular uptake of pDNA lipoplexes is the first step in non-viral transfection and is known to occur through different endocytosis pathways in different cell types [34]. We transfected hMSCs while inhibiting specific pathways of endocytosis using small molecules to determine how MCMs mechanistically improve transfection of hMSCs. We established that hMSCs utilize both macropinocytosis and caveolae-mediated endocytosis to sample their soluble microenvironment under normal culture conditions, and that MCMs both increased the number of endosomes produced by these pathways (Fig. 4A–D) under normal conditions and in the presence of their respective inhibitors (Fig. 4E). In comparing the transfection efficiency with and without MCMs in the presence of inhibitors, we found that MCMs significantly mitigated losses in transfection efficiencies for chlorpromazine- and amiloride-treated hMSCs, but not for methyl-β-cyclodextrin-treated hMSCs (Fig. S6B). Taken together, the increase in endocytosis in amiloride- and methyl-β-cyclodextrin-treated hMSCs cultured MCMs (Fig. 4E) and mitigation of transfection losses in amiloride-treated cells (Fig. S6B) indicates that MCMs specifically upregulate macropinocytosis in order to improve hMSC transfection. This finding adds further mechanistic insight into our previous studies exploring mineral-based materials for gene delivery [22,28,29] by identifying endocytosis pathways which can be targeted by this material to improve non-viral gene delivery, and contributes to a growing body of literature describing how material surface properties such as charge [38] and nanotopography[39] can influence how hMSCs sample their microenvironment.
BMP-2 is a potent osteogenic factor known to enhance hMSC differentiation toward an osteogenic fate [40]. Indeed, previous studies have reported that delivery of BMP-2 plasmid can help promote osteogenic differentiation of hMSCs and increase development of bone-like tissue [41–43]. Here, we delivered a BMP-2 encoding plasmid to explore the functional utility of MCM-based gene delivery to 3D scaffold-free cell constructs. We hypothesized that the improved 3D transfection observed in the pDNA/MCM method would result in increased secretion of BMP-2 from within hMSC constructs, which would in turn enhance osteogenic differentiation of hMSC aggregates and cell sheets. pBMP-2 delivery allowed for robust and homogeneous calcium deposition within large hMSC constructs, including 10 mm-diameter cell sheets, indicating enhanced 3D osteogenic differentiation (Fig. 5C, D). Interestingly, the majority of the BMP-2 produced in the 3D hMSC constructs after transfection was sequestered within the mineral coating (Fig. 3B). In considering previous demonstrations that mineral coatings can be used for controlled release of recombinant BMP-2 [3,31], it is possible that sequestering of overexpressed BMP-2 here turned the MCMs into in situ depots, providing sustained release of BMP-2 from within the aggregates, further potentiating osteogenic differentiation (Figs. 5C–D, S7). This sequestering mechanism and the full potential of this approach in bone tissue engineering warrant additional inquiry in future studies.
It is also noteworthy that MCMs alone improved calcium production in cell sheets, suggesting that this microparticle based non-viral gene delivery strategy might have unique potential for enhancing cell differentiation in bone engineering applications. Future studies could use this approach to deliver other therapeutic genes – such as other growth factors or transcription factors - into 3D constructs to engineer scaffold-free tissues for a range of clinical needs. For example, vascular endothelial growth factor (VEGF) is a potent angiogenesis promoter [44,45], and delivery of a VEGF-encoding plasmid could be used to promote angiogenesis in engineered 3D tissues. The generic binding mechanism of pDNA lipoplexes to the mineral coating [46] makes it rather simple to modify our approach to overexpress any protein of interest. It is also possible to deliver multiple genes together to achieve synergistic effects from more than one factor. As a result, this microparticle-based non-viral gene delivery strategy may have broad applicability in tissue engineering and biofabrication using 3D cell constructs.
5. Conclusions
In the current study, we describe and characterize a microparticle based non-viral gene delivery approach to efficiently and homogenously transfect 3D hMSC constructs. We found that the transport barrier inherent to 3D constructs rendered non-viral based cell transfection of hMSC aggregates virtually unachievable, and that we could circumvent this obstacle by incorporating pDNA-laden MCMs into the aggregate interior. Compared with the standard gene delivery approach, release of pDNA from inside of the aggregates increased transfection efficiency and uniformity of transfected cell distribution. Examining which endocytosis pathways interact with MCMs suggested that the observed improvement in transfection was due to increases in macropinocytosis triggered by the presence of the MCMs. As a demonstration of the utility of this approach, MCM-delivery of a BMP-2 plasmid led to production of bioactive BMP-2, which then promoted increased uniform calcium deposition in 3D hMSC constructs cultured in osteogenic medium. Taken together, this material represents a versatile tool that offers better control over 3D differentiation of human cells, and more broadly allows for presentation of biological cues from within any 3D constructs that are relevant for tissue engineering, but present significant diffusion barriers that otherwise can be difficult to overcome.
Supplementary Material
Statement of Significance:
This original research describes a materials-based approach, whereby use of mineral-coated microparticles improves the efficiency of non-viral gene delivery in three three-dimensional human mesenchymal stromal cell constructs. Specifically, it demonstrates the use of mineral-coated microparticles to enable highly efficient transfection of human mesenchymal stromal cells in large, 3D culture formats. The manuscript also identifies specific endocytosis pathways that interact with the mineral coating to afford the improved transfection efficiency, as well as demonstrates the utility of this approach toward improving differentiation of large cell constructs. We feel that this manuscript will impact the current understanding and near-term development of materials for non-viral gene delivery in broad tissue engineering and biofabrication applications, and therefore be of interest to a diverse biomaterials audience.
6. Acknowledgement
The authors would like to thank the financial support provided by the AO Foundation, the National Institutes of Health (R01AR059916), the National Science Foundation (Collaborative Research Proposal DMR 1106165/1105591), and a generous philanthropic gift from The Shannon Family in support of the Musculoskeletal Regeneration Partnership to the laboratory of W. Murphy, as well as funding provided to A. Khalil by the National Institute of General Medical Sciences’ Biotechnology Training Program (Grant: NIH 5 T32 GM008349) and National Science Foundation (Grant: DGE-1256259). In addition, the authors would like to thank the financial support provided by the National Institutes of Health (R01AR063194) to P. Dang and the laboratory of E. Alsberg.
The authors would like to acknowledge use of facilities and instrumentation supported by the National Science Foundation through the University of Wisconsin-Madison Materials Research Science and Engineering Center (Grant: DMR-1121288, 0079983, and 0520057), University of Wisconsin-Madison Nanoscale Science and Engineering Center (Grant: DMR-0832760 and 0425880), and University of Wisconsin Carbone Cancer Center Support Grant P30 CA014520.
Footnotes
Disclosures and competing financial interests
W. Murphy is a founder and stockholder for Stem Pharm, Inc. and Tissue Regeneration Systems, Inc.
8. References
- [1].Mooney DJ, Vandenburgh H, Cell Delivery Mechanisms for Tissue Repair, Cell Stem Cell. 2 (2008) 205–213. [DOI] [PubMed] [Google Scholar]
- [2].Brindley DA, Davie NL, Sahlman WA, Bonfiglio GA, Culme-Seymour EJ, Reeve BC, Mason C, Promising Growth and Investment in the Cell Therapy Industry during the First Quarter of 2012, Cell Stem Cell. 10 (2012) 492–496. [DOI] [PubMed] [Google Scholar]
- [3].Suárez-González D, Barnhart K, Migneco F, Flanagan C, Hollister SJ, Murphy WL, Controllable mineral coatings on PCL scaffolds as carriers for growth factor release., Biomaterials. 33 (2012) 713–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Li W-J, Laurencin CT, Caterson EJ, Tuan RS, Ko FK, Electrospun nanofibrous structure: a novel scaffold for tissue engineering., J. Biomed. Mater. Res 60 (2002) 613–21. [DOI] [PubMed] [Google Scholar]
- [5].Parekh SH, Chatterjee K, Lin-Gibson S, Moore NM, Cicerone MT, Young MF, Simon CG, Modulus-driven differentiation of marrow stromal cells in 3D scaffolds that is independent of myosin-based cytoskeletal tension., Biomaterials. 32 (2011) 2256–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Murphy WL, McDevitt TC, Engler AJ, Materials as stem cell regulators, Nat. Mater 13 (2014) 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ding AG, Schwendeman SP, Acidic microclimate pH Distribution in PLGA microspheres monitored by confocal laser scanning microscopy, Pharm. Res 25 (2008) 2041–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Johnstone B, Hering TM, Caplan AI, Goldberg VM, Yoo JU, In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells, Exp. Cell Res. 238 (1998) 265–272. [DOI] [PubMed] [Google Scholar]
- [9].Mironov V, Boland T, Trusk T, Forgacs G, Markwald RR, Organ printing: Computer-aided jet-based 3D tissue engineering, Trends Biotechnol. 21 (2003) 157–161. [DOI] [PubMed] [Google Scholar]
- [10].Carpenedo RL, Bratt-Leal AM, a Marklein R, a Seaman S, Bowen NJ, McDonald JF, McDevitt TC, Homogeneous and organized differentiation within embryoid bodies induced by microsphere-mediated delivery of small molecules., Biomaterials. 30 (2009) 2507–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Solorio LD, Fu AS, Hernández-Irizarry R, Alsberg E, Chondrogenic differentiation of human mesenchymal stem cell aggregates via controlled release of TGF-beta1 from incorporated polymer microspheres., J. Biomed. Mater. Res. A 92 (2010) 1139–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bratt-Leal AM, Nguyen AH, Hammersmith K. a., Singh A, McDevitt TC, A microparticle approach to morphogen delivery within pluripotent stem cell aggregates, Biomaterials. (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Solorio LD, Vieregge EL, Dhami CD, Dang PN, Alsberg E, Engineered cartilage via self-assembled hMSC sheets with incorporated biodegradable gelatin microspheres releasing transforming growth factor-β1, J. Control. Release 158 (2012) 224–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Evans CH, Gene therapy for bone healing, Expert Rev. Mol. Med 12 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shea LD, Smiley E, Bonadio J, Mooney DJ, DNA delivery from polymer matrices for tissue engineering, Nat Biotechnol. 17 (1999) 551–554. [DOI] [PubMed] [Google Scholar]
- [16].Storrie H, Mooney DJ, Sustained delivery of plasmid DNA from polymeric scaffolds for tissue engineering., Adv. Drug Deliv. Rev 58 (2006) 500–14. [DOI] [PubMed] [Google Scholar]
- [17].Yang F, Cho SW, Son SM, Bogatyrev SR, Singh D, Green JJ, Mei Y, Park S, Bhang SH, Kim BS, Langer R, Anderson DG, Genetic engineering of human stem cells for enhanced angiogenesis using biodegradable polymeric nanoparticles, Proc Natl Acad Sci U S A. 107 (2010) 3317–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zandstra PW, Bauwens C, Yin T, Liu Q, Schiller H, Zweigerdt R, Pasumarthi KBS, Field LJ, Scalable Production of Embryonic Stem Cell-Derived Cardiomyocytes, Tissue Eng. 9 (2003) 767–778. [DOI] [PubMed] [Google Scholar]
- [19].Akinc A, Lynn DM, Anderson DG, Langer R, Parallel synthesis and biophysical characterization of a degradable polymer library for gene delivery., J. Am. Chem. Soc 125 (2003) 5316–23. [DOI] [PubMed] [Google Scholar]
- [20].Bonadio J, Smiley E, Patil P, Goldstein S, Localized, direct plasmid gene delivery in vivo: prolonged therapy results in reproducible tissue regeneration, Nat. Med 5 (1999) 753–759. [DOI] [PubMed] [Google Scholar]
- [21].Yang F, Green JJ, Dinio T, Keung L, Cho S-W, Park H, Langer R, Anderson DG, Gene delivery to human adult and embryonic cell-derived stem cells using biodegradable nanoparticulate polymeric vectors., Gene Ther. 16 (2009) 533–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Choi S, Yu X, Jongpaiboonki L, Hollister SJ, Murphy WL, Inorganic coatings for optimized non-viral transfection of stem cells, Sci. Rep 3 (2013) 1567–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].de Carvalho TG, Pellenz FM, Laureano A, da Rocha Silla LM, Giugliani R, Baldo G, Matte U, A simple protocol for transfecting human mesenchymal stem cells, Biotechnol. Lett 40 (2018) 617–622. [DOI] [PubMed] [Google Scholar]
- [24].Gonzalez-Fernandez T, Sathy BN, Hobbs C, Cunniffe GM, McCarthy HO, Dunne NJ, Nicolosi V, O’Brien FJ, Kelly DJ, Mesenchymal stem cell fate following non-viral gene transfection strongly depends on the choice of delivery vector, Acta Biomater. 55 (2017) 226–238. [DOI] [PubMed] [Google Scholar]
- [25].Gheisari Y, Soleimani M, Azadmanesh K, Zeinali S, Multipotent mesenchymal stromal cells: Optimization and comparison of five cationic polymer-based gene delivery methods, Cytotherapy. 10 (2008) 815–823. [DOI] [PubMed] [Google Scholar]
- [26].Luo D, Saltzman WM, Synthetic DNA delivery systems, Nat Biotech. 18 (2000) 33–37. [DOI] [PubMed] [Google Scholar]
- [27].Ma B, Zhang S, Jiang H, Zhao B, Lv H, Lipoplex morphologies and their influences on transfection efficiency in gene delivery, J. Control. Release 123 (2007) 184–194. [DOI] [PubMed] [Google Scholar]
- [28].Khalil AS, Yu X, Xie AW, Fontana G, Umhoefer JM, Johnson HJ, Hookway TA, McDevitt TC, Murphy WL, Functionalization of microparticles with mineral coatings enhances non-viral transfection of primary human cells, Sci. Rep 7 (2017) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].McMillan A, Nguyen MK, Gonzalez-Fernandez T, Ge P, Yu X, Murphy WL, Kelly DJ, Alsberg E, Dual non-viral gene delivery from microparticles within 3D high-density stem cell constructs for enhanced bone tissue engineering, Biomaterials. 161 (2018) 240–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yu X, Murphy WL, 3-D Scaffold Platform for Optimized Non-viral Transfection of Multipotent Stem Cells., J. Mater. Chem. B. Mater. Biol. Med 2 (2014) 8186–8193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yu X, Khalil A, Dang PN, Alsberg E, Murphy WL, Multilayered Inorganic Microparticles for Tunable Dual Growth Factor Delivery., Adv. Funct. Mater 24 (2014) 3082–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Choi S, Murphy WL, Sustained plasmid DNA release from dissolving mineral coatings., Acta Biomater. 6 (2010) 3426–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Dang PN, Dwivedi N, Yu X, Phillips L, Bowerman C, Murphy WL, Alsberg E, Guiding Chondrogenesis and Osteogenesis with Mineral-Coated Hydroxyapatite and BMP-2 Incorporated within High-Density hMSC Aggregates for Bone Regeneration, ACS Biomater. Sci. Eng 2 (2016) 30–42. [DOI] [PubMed] [Google Scholar]
- [34].Khalil IA, Kogure K, Akita H, Harashima H, Uptake Pathways and Subsequent Intracellular Trafficking in Nonviral Gene Delivery, Pharmacol. Rev 58 (2006) 32–45. [DOI] [PubMed] [Google Scholar]
- [35].Van Winkle AP, Gates ID, Kallos MS, Mass transfer limitations in embryoid bodies during human embryonic stem cell differentiation., Cells. Tissues. Organs 196 (2012) 34–47. [DOI] [PubMed] [Google Scholar]
- [36].Sachlos E, Auguste DT, Embryoid body morphology influences diffusive transport of inductive biochemicals: a strategy for stem cell differentiation., Biomaterials. 29 (2008) 4471–80. [DOI] [PubMed] [Google Scholar]
- [37].He CX, Tabata Y, Gao JQ, Non-viral gene delivery carrier and its three-dimensional transfection system, Int. J. Pharm 386 (2010) 232–242. [DOI] [PubMed] [Google Scholar]
- [38].Chung TH, Wu SH, Yao M, Lu CW, Lin YS, Hung Y, Mou CY, Chen YC, Huang DM, The effect of surface charge on the uptake and biological function of mesoporous silica nanoparticles in 3T3-L1 cells and human mesenchymal stem cells, Biomaterials. 28 (2007) 2959–2966. [DOI] [PubMed] [Google Scholar]
- [39].Teo BKK, Goh S, Kustandi TS, Wei W, Yee H, Yim EKF, Biomaterials The effect of micro and nanotopography on endocytosis in drug and gene delivery systems, Biomaterials. 32 (2011) 9866–9875. [DOI] [PubMed] [Google Scholar]
- [40].De Biase P, Capanna R, Clinical applications of BMPs, Injury. 36 Suppl 3 (2005) S43–6. [DOI] [PubMed] [Google Scholar]
- [41].Nie H, Ho M-L, Wang C-K, Wang C-H, Fu Y-C, BMP-2 plasmid loaded PLGA/HAp composite scaffolds for treatment of bone defects in nude mice, Biomaterials. 30 (2009) 892–901. [DOI] [PubMed] [Google Scholar]
- [42].Wegman F, Bijenhof A, Schuijff L, Oner FC, a Dhert WJ, Alblas J, Osteogenic differentiation as a result of BMP-2 plasmid DNA based gene therapy in vitro and in vivo., Eur. Cell. Mater 21 (2011) 230–42. [DOI] [PubMed] [Google Scholar]
- [43].Rose LC, Kucharski C, Uludağ H, Protein expression following non-viral delivery of plasmid DNA coding for basic FGF and BMP-2 in a rat ectopic model, Biomaterials. 33 (2012) 3363–3374. [DOI] [PubMed] [Google Scholar]
- [44].Ferrara N, Gerber H-P, LeCouter J, The biology of VEGF and its receptors., Nat. Med 9 (2003) 669–76. [DOI] [PubMed] [Google Scholar]
- [45].Carmeliet P, Jain RK, Angiogenesis in cancer and other diseases., Nature. 407 (2000) 249–57. [DOI] [PubMed] [Google Scholar]
- [46].Ginebra M-P, Traykova T, Planell JA, Calcium phosphate cements: Competitive drug carriers for the musculoskeletal system?, Biomaterials. 27 (2006) 2171–2177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.