Significance
Telomeres protect the ends of linear chromosomes from illicit DNA processing events that threaten genome stability. Guanine-rich telomere DNA repeat sequences are protected by telomere-binding proteins, and are also prone to fold into structures called G quadruplexes (GQs). In highly proliferative cells, including the majority of human cancers, telomeres are maintained by the telomerase enzyme. Thus, telomerase and its telomere DNA substrates represent important targets for developing novel cancer drugs. The results of this study suggest GQ folding of newly synthesized DNA may occur within an actively extending telomerase enzyme. Our experiments highlight the importance of telomere DNA structure during the function of telomerase and its associated telomere-binding proteins.
Keywords: telomerase, telomere, G quadruplex, DNA structure, POT1–TPP1
Abstract
Telomerase reverse transcribes short guanine (G)-rich DNA repeat sequences from its internal RNA template to maintain telomere length. G-rich telomere DNA repeats readily fold into G-quadruplex (GQ) structures in vitro, and the presence of GQ-prone sequences throughout the genome introduces challenges to replication in vivo. Using a combination of ensemble and single-molecule telomerase assays, we discovered that GQ folding of the nascent DNA product during processive addition of multiple telomere repeats modulates the kinetics of telomerase catalysis and dissociation. Telomerase reactions performed with telomere DNA primers of varying sequence or using GQ-stabilizing K+ versus GQ-destabilizing Li+ salts yielded changes in DNA product profiles consistent with formation of GQ structures within the telomerase–DNA complex. Addition of the telomerase processivity factor POT1–TPP1 altered the DNA product profile, but was not sufficient to recover full activity in the presence of Li+ cations. This result suggests GQ folding synergizes with POT1–TPP1 to support telomerase function. Single-molecule Förster resonance energy transfer experiments reveal complex DNA structural dynamics during real-time catalysis in the presence of K+ but not Li+, supporting the notion of nascent product folding within the active telomerase complex. To explain the observed distributions of telomere products, we globally fit telomerase time-series data to a kinetic model that converges to a set of rate constants describing each successive telomere repeat addition cycle. Our results highlight the potential influence of the intrinsic folding properties of telomere DNA during telomerase catalysis, and provide a detailed characterization of GQ modulation of polymerase function.
Telomeres safeguard the ends of chromosomes from illicit DNA processing events that would otherwise threaten genome stability (1, 2). The foundation of telomere structure consists of short guanine (G)-rich DNA sequence repeats. The majority of mammalian telomeric DNA is double-stranded and can be up to several kilobases in length, whereas telomere ends are processed to terminate with a 3′ single-stranded G-rich overhang (∼50 to 500 nt in length) (3, 4). Repetitive G-rich DNA sequences are not unique to telomeres, and are found throughout the human genome (5). These G-rich repeats have the capacity to fold into G-quadruplex (GQ) structures composed of multiple Hoogsteen-bonded G-quartet motifs that stack together to yield stable DNA folds (6, 7). GQ folding has been implicated in a variety of biological processes. For example, replication of GQ-prone sequences is problematic, and requires contributions from specific DNA helicase enzymes to avoid replication-coupled DNA damage (8–10). Sequences with GQ-folding potential are enriched within promoter sequences of oncogenes, where they are thought to regulate gene expression (11). Finally, recent evidence suggests GQ folds can form in vivo in a spatially and temporally regulated manner (12–14). Thus, small molecules that bind and stabilize GQ folds hold promise as novel cancer drugs, motivating efforts to better understand how GQ structure can modulate enzyme function.
Telomerase is an RNA-dependent DNA polymerase that is uniquely adapted to synthesizing G-rich repetitive DNA sequences (15, 16). Telomerase activity combats gradual telomere shortening that occurs with each round of cellular division (17). While telomere shortening induces senescence or cell death in somatic tissues, highly proliferative cells such as stem cells rely upon telomerase activity to maintain telomeres to support continued rounds of cell division (15). Genetically inherited hypomorphic mutations in telomerase subunits cause human disorders characterized by deterioration of proliferative tissue types (18–21). In contrast, telomerase overexpression contributes to the immortal phenotype of ∼90% of human cancers, and is therefore an important target for development of novel cancer therapies (22).
Telomerase is a ribonucleoprotein (RNP) complex that includes the long noncoding telomerase RNA (TR) and the catalytic telomerase reverse transcriptase (TERT) protein subunit (23, 24). To initiate telomerase catalysis, the 3′ ssDNA telomeric tail base pairs with the TR template, forming a short RNA–DNA hybrid that is extended in the TERT active site (Fig. 1A). TERT utilizes a limited region of TR to direct synthesis of a defined telomere DNA repeat sequence (5′-GGTTAG-3′ in humans) with an associated rate constant, kpol (Fig. 1A). A unique property of telomerase is the ability to translocate on the DNA product (ktrans) to recycle the integral TR template during processive addition of multiple telomere repeats before dissociation from the DNA product (koff) (Fig. 1A) (25). This repeat-addition processivity (RAP) implicitly requires multiple points of contact between telomerase and its DNA substrate, a notion that is consistent with data from a variety of telomerase systems identifying “anchor-site” DNA interactions. Anchor-site interactions have been attributed to the TERT essential N-terminal (TEN) domain (26–31), a specific DNA-retention site near the TERT active site (32), as well as elements of the TR subunit itself (26–31, 33). Although the minimal telomerase RNP exhibits RAP, components of the telomere-associated shelterin complex can further enhance enzyme processivity. Specifically, the protection of telomeres 1 (POT1) protein binds to single-stranded telomeric DNA and together with its heterodimeric binding partner, telomere protection protein 1 (TPP1), is sufficient to enhance telomerase processivity in vitro (34). POT1–TPP1 is further required for telomerase recruitment to telomeres in vivo (35).
Fig. 1.
Human telomerase function. (A) Telomerase catalytic cycle. TERT and TR are shown simplified in gray and red, respectively. The telomere DNA is shown in blue and the telomerase anchor site is schematically represented by an anchor symbol. koff and kon represent the rate constants for dissociation from and annealing to the telomere, respectively. The rate constant for nucleotide addition during repeat synthesis is represented by kpol, and the translocation rate constant after the completion of each repeat is represented by ktrans. The rate constants governing nucleotide addition and translocation together define repeat-addition processivity. (B) Telomerase primer-extension assay with 50 nM 32P end-labeled (TTAGGG)3 primer. Nucleotide concentrations are indicated above the gel, and repeats added to the (TTAGGG)3 primer are indicated on the left. The R1/2 values are shown at the bottom of the gel. (C) Normalized gel band intensity (B) plotted as a function of repeat number. (C, Inset) Fraction left behind was calculated by dividing the sum of each RAP band and all bands below by the total intensity of a given lane. The plot of ln(1 − FLB) over repeat number was used to calculate R1/2 processivity values (B).
Model telomere DNA substrates harboring integer multiples of four consecutive telomere repeats are inefficient binding substrates for telomerase in vitro, while DNA primers with five, six, or seven consecutive repeats are efficiently bound and extended (36). Thus, while GQ structures can inhibit telomerase association (36, 37), the presence of a small single-stranded DNA overhang in the substrate appears sufficient to recover telomerase loading and function. While these previous findings illuminate DNA sequence determinants that mediate the initial binding of telomerase to its substrate, there remained an untested possibility that GQ structure may influence the behavior of an actively extending human telomerase–DNA complex, as was suggested by early studies of telomerase (38–40). Notably, the POT1–TPP1 heterodimer that decorates the G overhang of human telomeres resolves GQ structures in vitro through sequence-specific binding of the two POT1 oligonucleotide-binding (OB) fold domains (41–43). However, it remains unclear if POT1–TPP1 rapidly binds to newly synthesized DNA repeats to efficiently prevent the formation of GQs, or if POT1–TPP1 resolves GQs that form within the telomerase–telomere complex.
To study the relationship between DNA structure and human telomerase catalysis, we performed direct primer-extension assays using dNTP concentrations similar to those found in the cellular environment (44). Our experiments reveal a complex pattern of telomerase DNA product accumulation that indicates the efficiency of template recycling is dependent upon the number of synthesized repeats. Experiments using telomere DNA primers of varying sequence and salt conditions support the notion that a GQ can form within the telomerase–DNA complex. The addition of the POT1–TPP1 processivity factor alters the telomerase product profile but does not rescue full DNA-synthesis rates under GQ-destabilizing conditions. To estimate individual rate constants for successive repeat addition cycles, we performed global kinetic modeling of telomerase time-series data. Interestingly, our model converges to a unique solution of rate constants that provides a direct measure of processivity for each cycle of telomere repeat addition. Single-molecule Förster resonance energy transfer (smFRET) experiments reveal DNA structural dynamics during telomerase catalysis, supporting the notion that telomere DNA GQ folding modulates enzyme function. We present a working mechanistic model that provides a framework for understanding the delicate interplay of telomere DNA product folding and POT1–TPP1 during telomerase catalysis.
Results
Telomerase Product Distribution Is Sensitive to dNTP Concentrations and Stoichiometry.
When measuring telomerase activity in vitro, it is common to employ direct primer-extension assays in the presence of [α32P]dGTP. This approach permits reactions to be performed with a large excess of unlabeled DNA substrate, benefits from very high sensitivity of product detection, and circumvents PCR-induced artifacts inherent to the telomere repeat amplification protocol assay. However, the use of [α32P]dGTP incorporation to detect product accumulation limits the amount of total dGTP that can be used in the assay, leading to the widely reported practice of using nonphysiological dNTP stoichiometry that has the potential to substantially alter the telomerase product distribution (40, 45). To circumvent this problem, we used 5′ radiolabeled DNA primers and cold dNTPs to monitor telomerase activity (Fig. 1B). For most experiments conducted in the present study, telomerase was reconstituted in vitro using a previously reported two-piece RNA strategy [human (h)TR 32 to 195 and hTR 239 to 328] in commercially available rabbit reticulocyte lysates (RRLs) (46, 47). The primary motivation for utilizing enzyme prepared in this manner is to facilitate direct comparison of biochemical data with results from single-molecule experiments, which require the use of the two-piece RNA system to permit site-specific modification of hTR (48) (see Methods below). Importantly, enzymes prepared in RRL with the two-piece RNA approach exhibit catalytic properties that are comparable to telomerase enzyme assembled with full-length hTR in HEK293T cells (SI Appendix, Fig. S1). We compared the results using 5′ end-labeled primers with standard assays performed with [α32P]dGTP using identical dNTP and primer concentrations (SI Appendix, Fig. S2). Although the reaction profiles are qualitatively distinct, we observed quantitatively similar product distributions for the two approaches when normalized for the amount of [α32P]dGTP incorporation. Importantly, the majority of the input DNA primers are not extended in our experiments, demonstrating our reaction conditions are sufficient to limit distributive telomerase activity (i.e., an individual primer being extended by multiple telomerase enzymes). Such processive telomerase activity is also evident when analyzing pulse–chase experiments in which longer DNA products continue to accumulate after addition of a 400-fold excess of cold DNA primer (SI Appendix, Fig. S3).
Having established that our end-labeled DNA primer assay is capable of accurately monitoring processive telomerase action, we next sought to analyze the influence of varying dNTP concentrations on the telomerase product distribution (Fig. 1B). Previous studies have defined telomerase processivity as the number of repeats corresponding to the point where the dissociated DNA represents 50% of the total population (i.e., median product length; R1/2) (34, 49) (SI Appendix). This number can be determined by fitting a linear regression to a plot of ln(1 − FLB) versus repeat number, where FLB is the fraction left behind (Fig. 1C, Inset and Methods). Titrating increasing amounts of dGTP in the presence of a large excess of dATP and dTTP yields a marked boost in RAP, as has been reported previously for both human and Tetrahymena telomerase (Fig. 1 B and C) (32, 40, 50–53). However, the use of a large excess of dATP and dTTP is not a good approximation for the physiological dNTP pool, which is generally closer to the ∼10 μM range (44). When assayed in the presence of equimolar dGTP, dATP, and dTTP, we observe the highest RAP of all conditions tested (Fig. 1B, lane 4 and Fig. 1C). Hence, we elected to perform all subsequent telomerase assays in our study under these optimized conditions of equimolar dNTPs.
G-Quadruplex Folding Varies the Pattern of Telomerase Product Accumulation.
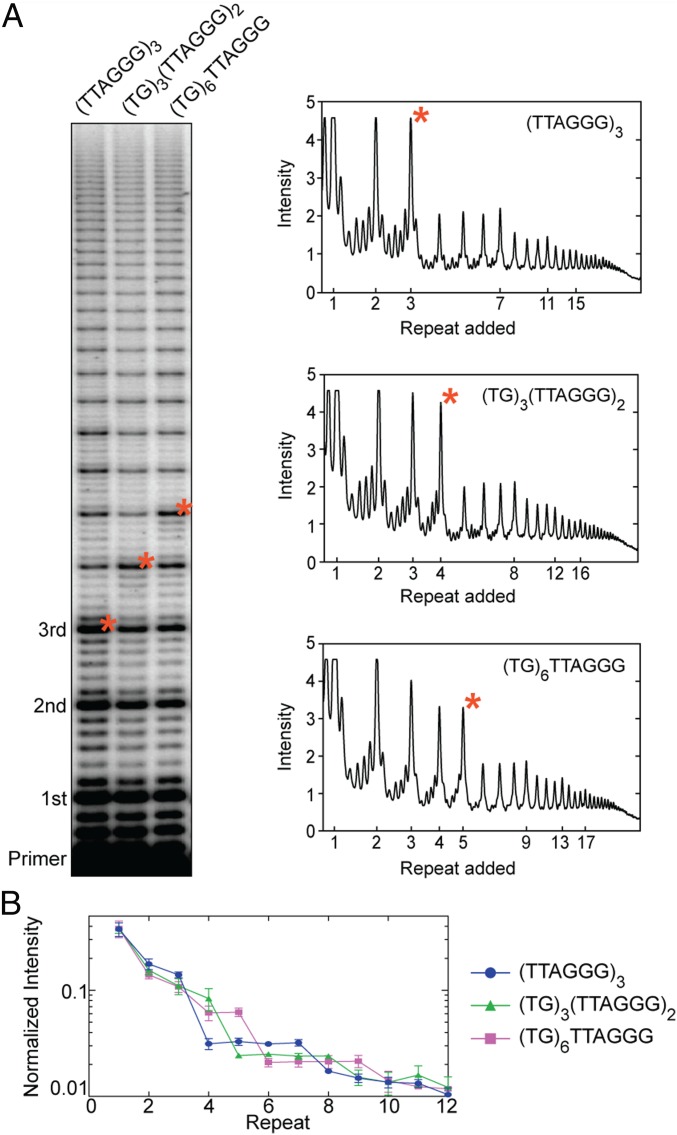
Established methods for approximating RAP using the R1/2 value described above assume an exponential decay in the distribution of accumulated telomerase product lengths with each telomere repeat added (34, 37, 49). In other words, the R1/2 value represents a weighted average processivity which is convenient for semiquantitative comparisons, but may mask underlying heterogeneity in the microscopic processivity associated with individual steps (SI Appendix). Indeed, we noted the appearance of plateaus in the product distribution when using equimolar concentrations of dNTPs in the end-labeled DNA primer-extension assay (Figs. 1C and 2). For example, when using a standard telomere DNA primer composed of the sequence (TTAGGG)3, we observed a sudden drop in product accumulation between the bands corresponding to the third and fourth telomere repeats added to the primer (Fig. 2A, lane 1, red asterisk). Further, the intensities of the subsequent four added repeats were approximately equal, until a second decrease in accumulation occurred between added repeats 7 and 8 (Fig. 2A). This pattern of four equally populated product lengths, followed by a decrease in accumulation, continued throughout the detectable range of telomere DNA products.
Fig. 2.
Telomerase product distribution profile varies with the number of consecutive TTAGGG DNA repeats. (A) Telomerase primer-extension assay with primers of varying TTAGGG composition. Primer variants are indicated at the top of the gel. Repeats added to the primer are indicated to the left. Lane profiles with raw intensity versus added repeat are shown for each primer variant (Right). Corresponding bands between the gel and lane profiles are indicated by red asterisks. (B) Gel band intensities were normalized to the total counts in each lane and are plotted as a fraction of the total counts versus repeat number. Data plotted represent the mean values from three independent experiments, and error bars are the SD.
Telomere DNA primers with at least four contiguous G-rich repeats can fold into a G quadruplex in vitro (54, 55), suggesting the observed RAP-associated “pattern of four” may be due to GQ folding of the DNA product within an actively extending telomerase complex. To test this hypothesis, we altered the 5′ end of the telomere DNA sequence so that it no longer harbored the requisite run of guanines needed to participate in GQ folding (Fig. 2A, lanes 2 and 3). Altering the primer in this way should change the product length where the pattern of four appears once the newly synthesized DNA folds into a GQ. Indeed, a modified DNA primer with a 5′ (TG)3 substitution supported telomerase RAP, but the plateaus in the product profile were delayed by one additional repeat, corresponding to the sequence needed to promote GQ formation in the product DNA (Fig. 2A, compare lanes 1 and 2). Similarly, when the first two repeats in the telomere DNA primer were substituted with a (TG)6 dinucleotide repeat sequence, the plateaus were delayed by two additional repeats (Fig. 2A, compare lanes 1 and 3). These results were highly reproducible across three independent experimental trials (Fig. 2B), and support the hypothesis that GQ folding within the nascent telomere DNA causes the telomerase product profile to deviate from a uniformly decreasing decay.
POT1–TPP1 Alters the Telomere DNA Product Distribution.
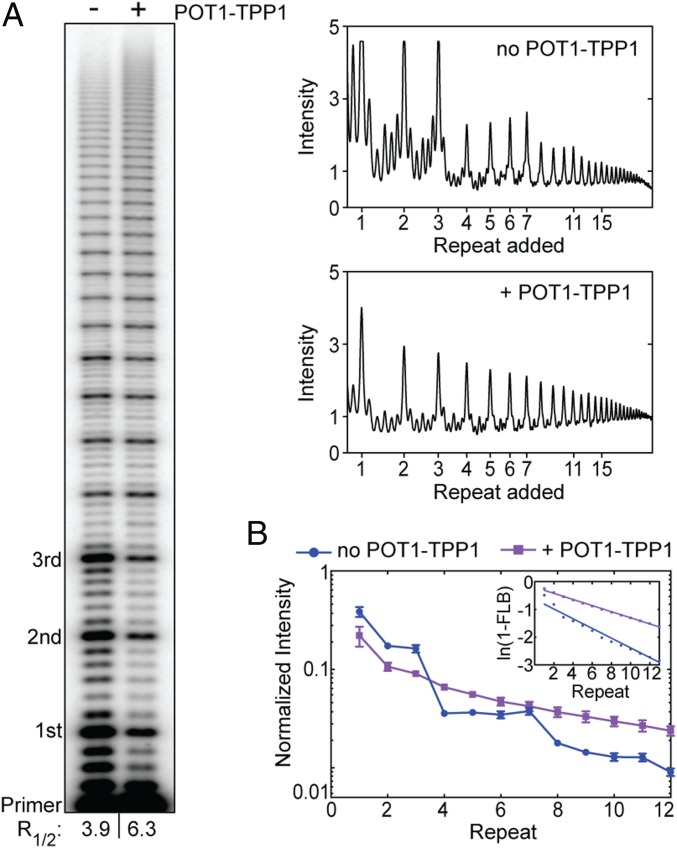
The POT1–TPP1 heterodimer binds to telomerase via the TEL-patch interaction between the TERT TEN domain and TPP1 (56). In addition, the POT1 subunit binds single-stranded telomeric DNA to provide additional anchor-site contacts that serve to promote telomerase RAP (34). POT1–TPP1 also binds and resolves telomere DNA GQ structures in vitro (42). To further evaluate GQ folding within the telomerase complex, we performed primer-extension assays in the absence and presence of POT1–TPP1 (Fig. 3). In addition to the expected enhancement of RAP, the presence of POT1–TPP1 abolished the four-repeat pattern in the distribution of telomerase products, which instead decay in a uniformly decreasing manner (Fig. 3). Notably, the effects of POT1–TPP1 on the DNA product distribution were the same for both the two-piece hTR reconstituted system and for endogenously reconstituted telomerase preparations from HEK293T cells (SI Appendix, Fig. S1). The effect of POT1–TPP1 on the telomere DNA product distribution is consistent with the ability of POT1–TPP1 to bind and remodel the nascent DNA product during active telomere extension. However, this result does not strictly differentiate between the possibilities of POT1–TPP1 preventing versus resolving a telomere GQ fold within the context of an actively extending telomerase complex.
Fig. 3.
POT1–TPP1 alters the telomerase product distribution profile. (A) Telomerase primer-extension assay in the absence and presence of POT1–TPP1. Numbers of added repeats are indicated to the left. The R1/2 values are shown at the bottom of the gel. Corresponding lane profiles with raw gel band intensities are plotted over added repeats (Right). (B) Gel band intensities in the absence (blue circles) or presence (purple squares) of POT1–TPP1 were normalized to the total counts in each lane and plotted as a fraction of the total counts versus repeat number. Data plotted represent the mean values from three independent experiments, and error bars are the SD. (B, Inset) The plot of ln(1 − FLB) over repeat number was used to calculate R1/2 processivity values shown in A.
Destabilization of G Quadruplexes by Li+ Cations Slows Telomere Repeat Synthesis.
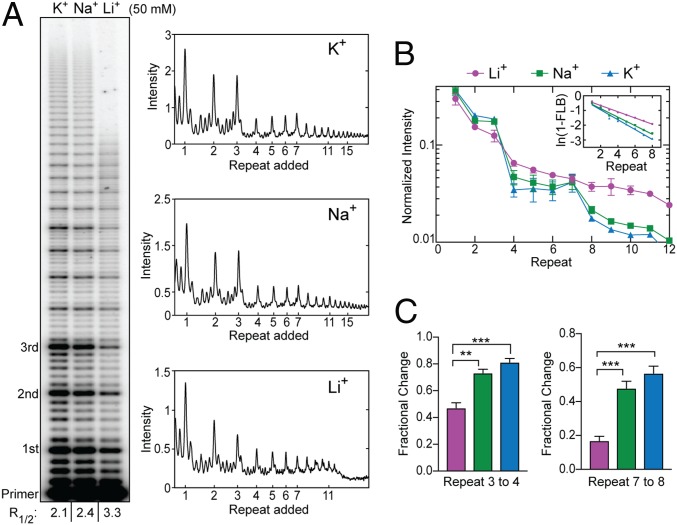
To ultimately address the question of whether GQ folding might impact telomerase function also in the presence of POT1–TPP1, we next set out to characterize the catalytic properties of telomerase under varying cation conditions. The H-bonding configuration of the G-quartet motifs within a GQ fold are differentially stabilized by coordination of specific monovalent cations, with a rank order of K+ > Na+ > Li+ in terms of degree of stabilization (57). We observed robust telomerase activity in direct primer-extension assays in all cation conditions tested (Fig. 4A). Interestingly, we did not observe the pattern-of-four RAP product distribution in the presence of Li+ (Fig. 4 A and B), the cation condition expected to least stabilize GQ folding during telomerase catalysis. This effect is most clearly evident when comparing the changes in band intensity between repeats 3 to 4 and 7 to 8 (Fig. 4C). Further, there was a clear reduction in total product accumulation in Li+ compared with Na+ and K+ (Fig. 4A). The lower total product accumulation in the presence of Li+ was a consequence of slower overall DNA-synthesis kinetics, as is evident from primer-extension time-course experiments (SI Appendix, Fig. S4). This observation is not completely unexpected, based on reported effects of Li+ on thermophilic DNA polymerases in the polymerase-chain reaction that describe slowed nucleotide incorporation (58). In our telomerase assays, such a general synthesis defect in Li+ should manifest as increased NAP (nucleotide-addition processivity) band intensities, since each nucleotide incorporation would make a larger contribution to the overall kinetics of each RAP cycle. Consistent with this notion, we observe an up to twofold increase in the ratio of NAP:RAP bands when comparing the product distributions in Li+ versus K+. However, in both cation conditions, the RAP products remain the most populated species across the gel (Fig. 4A), indicating that the catalytic substep(s) associated with product translocation and product repriming remains rate-limiting during RAP. These results demonstrate that GQ destabilization in the presence of Li+ negatively impacts the rate, and hence the product yield, of telomeric repeat synthesis.
Fig. 4.
Telomerase product distribution profile depends on monovalent cation identity. (A) Telomerase primer-extension assays in the presence of different monovalent cations. Repeats added to the primer are indicated to the left. The R1/2 values are shown at the bottom of the gel. Lane profiles with raw intensity versus repeat band for each lane are shown (Right). (B) Gel band intensities from experiments in KCl (blue triangles), NaCl (green squares), and LiCl (magenta circles) were normalized to the total counts in each lane and are plotted as a fraction of the total counts versus repeat number. Data plotted represent the mean values from three independent experiments, and error bars are the SD. (B, Inset) The plot of ln(1 − FLB) over repeat number was used to calculate the R1/2 processivity values shown in A. (C) Statistical analysis of fractional change in band intensities between repeats 3 to 4 (Left) and 7 to 8 (Right). Error bars represent the SD of experiments performed in triplicate (B). P values were calculated using a Welch’s t test; ***P < 0.001, **P < 0.01.
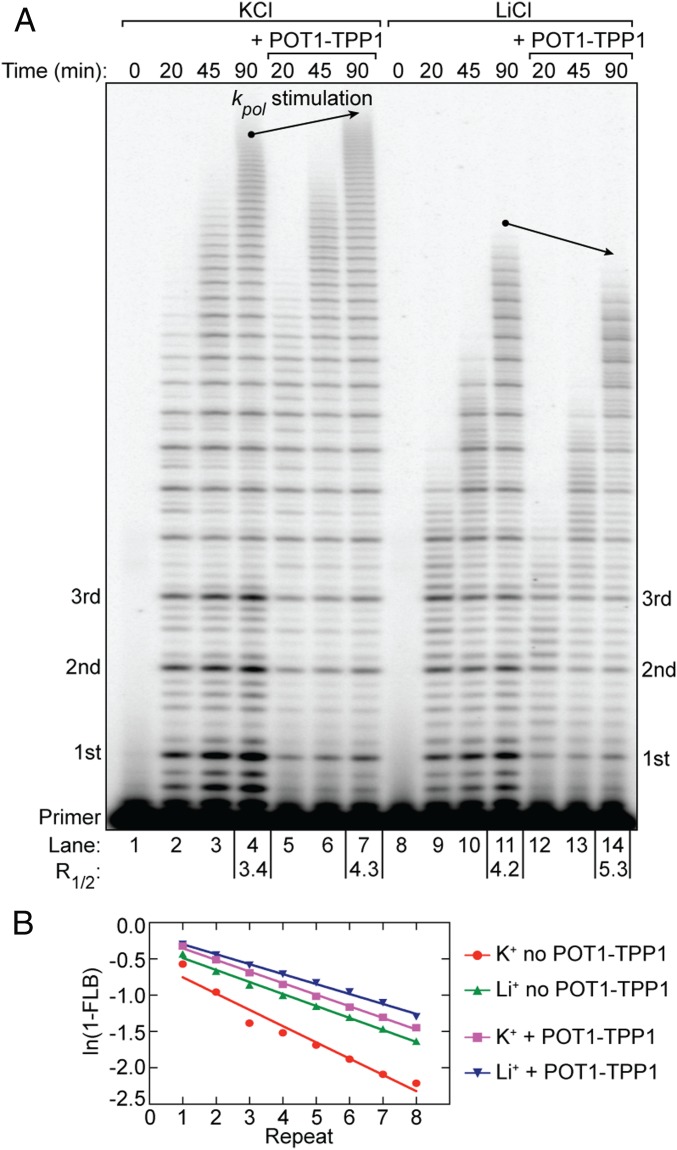
POT1–TPP1 Does Not Rescue DNA-Synthesis Rates in GQ-Destabilizing Conditions.
We next leveraged the ability to tune the degree of GQ stabilization in our telomerase assays to dissect the potential influence of nascent DNA product folding in the presence of POT1–TPP1. To this end, we performed time-course telomerase primer-extension assays using the established K+ and Li+ conditions in the absence or presence of POT1–TPP1 (Fig. 5). We reasoned if POT1–TPP1 acts by preventing GQ formation in nascent telomere DNA, then GQ destabilization in the presence of Li+ should not impact POT1–TPP1 enhancement of telomerase catalysis. Conversely, if GQ folding must occur in the presence of POT1–TPP1, then reduced DNA-synthesis rates under GQ-destabilizing conditions should not be rescued by POT1–TPP1 addition. As expected, primer-extension reactions in both K+ and Li+ in the presence of POT1–TPP1 exhibited stimulated processivity and lacked the pattern of four in product accumulation (Fig. 5). These results indicate that POT1–TPP1 can productively bind to the nascent product DNA and the telomerase enzyme in both cation conditions. Interestingly, POT1–TPP1 selectively failed to stimulate DNA-synthesis rates in the presence of Li+ (Fig. 5A, lanes 11 and 14 versus lanes 4 and 7). This result lends unanticipated support to the functional contribution of GQ folding for the POT1–TPP1–telomerase complex, and suggests that POT1–TPP1 may resolve, rather than prevent, formation of transient GQ structures within the actively extending enzyme complex (see Discussion for details).
Fig. 5.
POT1–TPP1 does not enhance telomerase catalysis rates in GQ-destabilizing conditions. (A) Telomerase primer-extension time-course assay in the absence and presence of POT1–TPP1 and under differential cation conditions. Time points of the reactions are indicated above the gel. POT1–TPP1–dependent differences in the maximum product length at 90-min reaction time (i.e., differences in synthesis rate) are indicated by arrows. R1/2 processivity values are given for reaction end points below the gel. (B) The plot of ln(1 − FLB) over repeat number was used to calculate R1/2 processivity values (A).
Changes in Reaction Rate Constants Govern the Modulation of Telomerase Microscopic Processivity.
To better understand the effects of GQ folding on the function of telomerase, we developed a detailed kinetic framework with which to model our experimental data (Fig. 6). We elected to perform our kinetic modeling on data generated in the absence of POT1–TPP1. This condition sensitizes our DNA primer-extension experiments to the influence of GQ folding on the observed product distribution, which is masked in the presence of POT1–TPP1 (Fig. 3). Telomerase processivity can be modeled as a series of consecutive reactions in which nucleotide addition is in competition with DNA dissociation at each step of the reaction (Fig. 6A). To simplify our telomerase kinetics analysis, we focus on the intense repeat addition bands, assuming the intervening nucleotide addition steps are relatively rapid and accompanied by little DNA dissociation (Fig. 6B) (see SI Appendix for justification). While the macroscopic processivity of telomerase can be conveniently described by the median product length (49), the experiments described in the present study provide clear evidence that telomerase products do not accumulate uniformly and display patterns dependent upon assay conditions, DNA sequence, and/or product length. We therefore developed a kinetic model that can be utilized to globally fit telomerase time-series data to extract individual rate constants for the forward progression and product dissociation at each RAP step that underlie the observed distributions of telomerase products (Fig. 6A). Using this scheme, we treat this multistep process as a first-order reaction with an effective forward rate constant (kf) for the transition between each repeat and a dissociation rate constant (kd) (Fig. 6A) (note that kf in this model reflects a combination of kpol and ktrans described in Fig. 1A). Using the kinetic scheme depicted in Fig. 6A, we can then define the microscopic processivity (p) at each step as p = kf/(kf + kd).
Fig. 6.
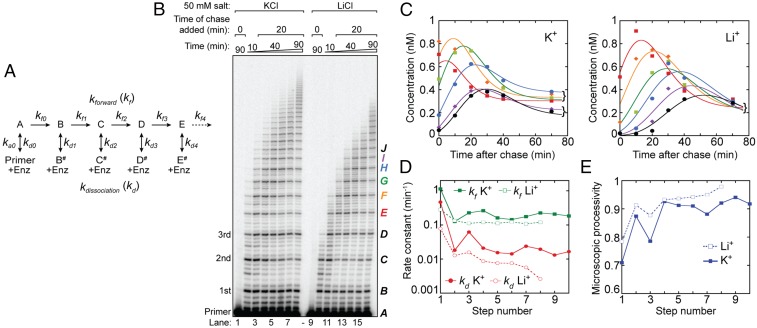
Human telomerase kinetics. (A) Kinetic mechanism for processive telomerase activity used to globally fit the primer-extension assay shown in B. The letters refer to the repeat band number (B, first added repeat; C, second repeat; etc.), and dissociated products are identified with the # symbol. Band intensities are proportional to the sum of products (e.g., B + B#). (B) Extending primer dissociation rate assay in the presence of 50 mM KCl or LiCl. Primer-extension assays were performed with 50 nM 32P end-labeled (TTAGGG)3 primer. Unlabeled chase (TTAGGG)3 primer (20 μM) was added to the reaction after 20 min of activity. A control reaction with 20 μM unlabeled chase primer added at the beginning of telomerase activity was included for both buffer conditions (lanes 1 and 9). Repeat number added to the primer is indicated on the left of the gel. Letters indicating band identity for kinetic modeling are indicated on the right of the gel. Bands E to J are colored according to the plot shown in C. (C) Representative global fits to bands E to J in KCl (Left) and LiCl (Right). The concentration of the products (see color code in B), based on band intensity relative to the initial 50 nM primer, was plotted against the time after the unlabeled chase. Note the clustering of bands E to H and I and J 70 min post chase in the presence of KCl, which corresponds to the four repeats of the first plateau and the first two repeats of the second plateau (cf. Fig. 2). This partitioning is not present in the presence of LiCl. See SI Appendix, Fig. S5 A and B for corresponding plots for bands B to D that precede the pattern-of-four bands. (D) Consecutive rate constant values for forward repeat addition (green squares; kf) and dissociation (red circles; kd) returned by DynaFit for data in the presence of KCl (solid symbols) and LiCl (open symbols). The step number refers to the rate constant subscripts shown in A. Note the overall reaction is slower in the presence of LiCl, and beyond band I (eighth step) the fitted rate constant values had a large error because the decay phase had barely started by 70 min. Therefore, these values were omitted. (E) Microscopic processivity [kf/(kf + kd)] at each step of the reaction calculated from the rate constants shown in D. Note the sawtooth structure in the presence of KCl (solid line) compared with the relative lack of structure in LiCl beyond the second step (dashed line). See also SI Appendix, Fig. S6C.
DNA dissociation is an effectively irreversible process when a large excess of unreacted primer remains, which outcompetes the rebinding of any product DNA. To ensure our experiments complied with this assumption, we analyzed telomerase kinetics following a chase with a 400-fold excess of unlabeled primer DNA, which serves to block reassociation of the labeled DNA primer following telomerase dissociation (Fig. 6B). Telomerase time-course assays were performed in the presence of either K+ or Li+ activity buffer conditions (Fig. 6B). Activity was initiated at time 0 in the presence of end-labeled telomere DNA primer and dNTPs, followed by addition of excess chase primer at 20 min. The presence of 400-fold excess cold primer before enzyme addition was sufficient to eliminate any observable extension of the 50 nM end-labeled DNA primer used in our assays (Fig. 6B, lanes 1 and 9). Time points were collected at regular intervals out to 90 min and the concentration of each repeat species (B + B#, C + C#, etc., in Fig. 6A) was determined at each time point from the band intensity, knowing that the intensity of the initial primer was 50 nM. Individual rate constants were estimated by fitting the concentration time courses globally, using DynaFit (59) (Fig. 6C and SI Appendix, Fig. S5 A and B). Global fitting converged to a set of rate constants that could be extracted with reasonable precision (see SI Appendix, Methods for details of kinetic modeling) (Fig. 6D). Comparison of the data obtained in the presence of K+ and Li+ revealed that the rate constants kf and kd decrease with increasing repeat number and result in slightly lower microscopic processivity values for short DNA products, as has been noted previously (49). However, in the presence of K+, the rate constants and microscopic processivity values show a sawtooth modulation that gives rise to the pattern-of-four clustering of products noted earlier (Fig. 6E, closed symbols). Although the effect is relatively small, it is robust as determined by Monte Carlo analysis and is reproducible between experiments and telomerase preparations (SI Appendix, Figs. S5 and S6). Interestingly, the rate constants for repeat addition (kf) and dissociation (kd) were greater in the presence of K+ than in Li+, but the resultant microscopic processivity was lower (Fig. 6 D and E). Microscopic processivity values were checked using the method of Peng et al. (60), and showed that those derived from kinetic analysis were self-consistent (SI Appendix, Fig. S6C).
Taken together, these results suggest that in the presence of K+, the ability of the DNA product to form a GQ fold (which first arises at step 3) promotes translocation (increased kf) but at the increased risk of DNA product dissociation (increased kd) (see Discussion for details). By implication, the absence of GQ folds in Li+ appears to slow product accumulation (decreased kf) (Figs. 4 and 5 and SI Appendix, Fig. S4) while decreasing dissociation rate constants (kd), the functional equivalent of which can be considered a more stable binding of the product DNA through the anchor site(s). The kinetic analysis and corresponding primer-extension assays hence raise questions as to the relationship between product DNA structure and anchor-site stability.
GQ Stabilization Alters DNA Product Handling and Structural Dynamics.
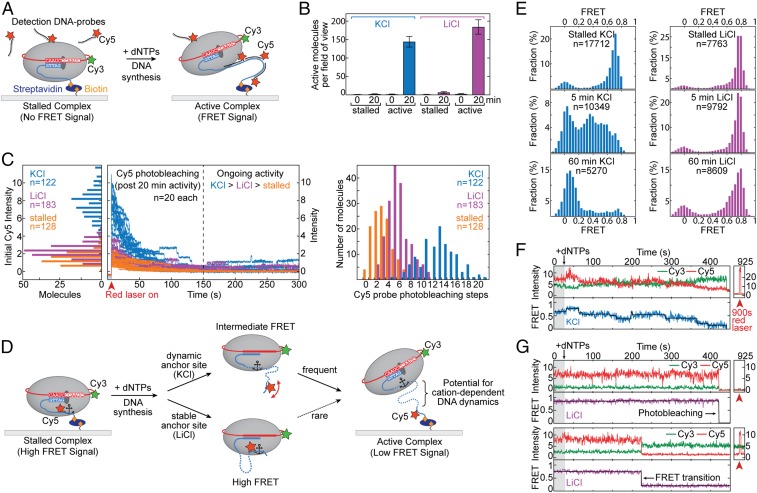
The results of our ensemble telomerase assays suggest that folding of the nascent DNA product can influence telomerase catalysis. To interrogate DNA conformation within an active RNP complex, we employed a single-molecule Förster resonance energy transfer assay that directly monitors DNA structure and dynamics within individual telomerase enzymes (48, 61). To simplify interpretation of our single-molecule experiments in the present study, we focus again on the telomerase catalytic core in the absence of POT1–TPP1. To ensure our smFRET assay supports telomerase activity in both K+ and Li+ buffers, we utilized a previously reported method for in situ detection of extended DNA products at the single-molecule level (62). Telomerase RNP complexes harboring a Cy3-modified telomerase RNA subunit were reconstituted in RRLs, bound to a biotinylated DNA primer, and surface-immobilized onto a streptavidin-coated quartz slide (Fig. 7A). Importantly, telomerase modified with Cy3 at hTR-U42 has been shown previously to retain wild-type catalytic function (48). The telomerase–DNA complexes were incubated in either K+ or Li+ activity buffer as well as with Cy5-labeled detection oligonucleotides, each with a sequence that is complementary to 2.5 repeats of the telomere product (SI Appendix, Table S1). In this way, telomere primers that are being actively extended by telomerase emerge from the enzyme and are detected as a FRET signal between the Cy3-labeled enzyme and the Cy5-labeled DNA probe (Fig. 7A). The appearance of the FRET signal was strictly dependent upon addition of activity buffer containing dNTPs and was time-dependent (Fig. 7B). After 20 min of incubation, comparable fractions of active telomerase molecules that produce a positive FRET signal were detected in both K+ and Li+ activity buffers (Fig. 7B), demonstrating that telomerase is catalytically active in either cation condition.
Fig. 7.
Single-molecule studies of telomerase in the presence of KCl and LiCl. (A) Schematic of the human telomerase smFRET activity assay. Purified telomerase is immobilized to a pegylated and biotinylated quartz slide through binding to a biotinylated telomere primer (blue). TERT is depicted as a gray oval, and hTR is shown in red. A Cy3 dye (green star) is conjugated to hTR. The binding of Cy5-labeled detection oligonucleotide probes (in black with red star) to newly synthesized telomere DNA is illustrated (Right). (B) Analysis of smFRET activity assays in KCl (blue) and LiCl (magenta) cation conditions. Negative controls were performed in the presence of Cy5 detection probes but in the absence of dNTPs (stalled). Time points and number of FRET-positive molecules per field of view imaged are indicated. Error bars are the SD across all fields imaged (n > 10). (C, Left) Histogram analysis of the initial Cy5 intensity distribution under direct laser excitation of active telomerase–DNA complexes (A, Right) in KCl (blue) and LiCl (magenta) cation conditions. Background signal was assessed in the absence of dNTPs (stalled; orange). (C, Center) Real-time traces showing time-dependent photobleaching of Cy5 dyes under direct laser excitation. Twenty representative traces are shown for simplification. Color coding is as indicated. (C, Right) Histogram analysis of the distribution of Cy5 photobleaching steps counted from each individual real-time trace. Conditions and number of traces are as indicated. (D) Schematic of human telomerase smFRET experiment designed to probe DNA structural dynamics. The telomere primer (blue) is conjugated to a Cy5 dye (red star). Telomere repeat synthesis impacts the FRET behavior depending upon anchor-site stability and identity of monovalent cations as illustrated. (E) Histogram analysis of smFRET assays (D) in KCl (blue) and LiCl (magenta). FRET distributions are shown for stalled telomerase complexes (Top) as well as at 5- and 60-min time points after addition of dNTPs (Middle and Bottom, respectively). (F and G) Representative real-time smFRET traces of individual telomerase complexes (D) in either KCl (F) or LiCl (G). Cy3 donor intensities are shown in green, and Cy5 acceptor intensities are in red. The corresponding FRET value (blue or magenta) was fit with steps (black) using an automated stepfinding algorithm in MATLAB (74). Direct laser excitation of the Cy5 dye in each trace is shown separately at a 900-s time point.
Next, we quantitatively monitored the total amount of product synthesized for individual telomerase–DNA complexes by incubating surface-immobilized telomerase enzymes in K+ and Li+ activity buffers for 20 min before the addition of Cy5 detection probe (Fig. 7 A and C). Using direct Cy5 laser excitation permitted a measure of the total fluorescence intensity value, which correlates with the number of Cy5 probes bound to the DNA product (Fig. 7C, Left, initial intensity). Moreover, prolonged exposure to direct laser excitation induced photobleaching of every DNA-bound Cy5 probe in a time-dependent manner (Fig. 7C, Center). We quantified the number of photobleaching steps for each single-molecule trace to yield a relative measure of DNA product length in either K+ or Li+ activity buffer (Fig. 7C, Right). As expected from our ensemble assays, histogram representations of the “initial Cy5 intensity” upon direct excitation, as well as the distribution of photobleaching steps, reflected a decreased DNA-synthesis rate in Li+ compared with K+ conditions (Fig. 7C). Interestingly, we occasionally observed upward steps in Cy5 intensity in either cation condition (K+ > Li+), consistent with real-time detection of Cy5 DNA probe binding to newly synthesized DNA repeats (Fig. 7C, Center). An equivalent but “stalled” (no dNTPs) telomerase complex served as background control. Note that nonzero amounts of initial intensity and Cy5 photobleaching steps were measured in this condition (Fig. 7C, orange); however, this was not due to Cy5 probe binding to the DNA primer, as confirmed earlier by the lack of a respective FRET signal (Fig. 7 A and B).
To directly monitor DNA structural dynamics, we next performed a variation of our smFRET assay in the absence of detection probe, that instead pairs Cy3-labeled telomerase together with site-specifically Cy5-labeled telomere primer. This assay further interrogates anchor-site stability that may underlie the altered microprocessivity values extracted from our kinetic modeling (Fig. 7D). Data collected on surface-immobilized stalled telomerase–DNA complexes in the absence of dNTPs yielded a predominant FRET distribution centered at ∼0.75, together with a minor zero-FRET population that represents telomerase–DNA complexes lacking a functional acceptor dye (Fig. 7E, Top). Next, the telomerase complexes were activated for DNA synthesis by introducing dNTPs in telomerase activity buffer, resulting in an increase in populations of lower FRET values over time (Fig. 7E, Middle and Bottom). These FRET changes upon activation of DNA synthesis are consistent with the Cy5 label on the telomere DNA moving farther away from telomerase (Fig. 7E). On the timescale of minutes to hours, both K+ and Li+ buffers support multiple rounds of DNA repeat synthesis sufficient to accumulate a FRET∼0 state, in which the dyes are separated beyond their FRET range (Fig. 7F, Bottom). We note that every data point in the accumulating FRET∼0 state is derived from primer-bound telomerase (i.e., requires the presence of a Cy3 dye) and is not due to telomerase dissociation. Interestingly, despite robust telomerase activity in Li+ buffer (Fig. 7 B and C), we observed an unanticipated stable high-FRET population over the course of a 1-h experiment in this cation condition (Fig. 7E). Furthermore, in contrast to K+ cation conditions, the accumulation of the FRET∼0 population occurred in apparent absence of any detectable intermediate-FRET states at the time resolution of our measurements (Fig. 7E).
Consistent with these findings, analysis of real-time single-molecule FRET trajectories collected on actively extending telomerase complexes revealed substantially different FRET behaviors in each cation condition. In K+ buffer, dynamic transitions between discrete FRET states were observed (Fig. 7F and SI Appendix, Fig. S7). The FRET dynamics generally progressed to lower values but included transient excursions to higher-FRET states, indicative of complex DNA conformational dynamics. In contrast, in Li+ buffer, we did not observe a substantial drop in FRET in the majority of traces before photobleaching of the Cy5 dye occurred, which can be unambiguously confirmed using direct Cy5 laser excitation at the end of data acquisition (Fig. 7G, Top and SI Appendix, Fig. S7). According to the FRET population analysis in a 1-h time window (Fig. 7E), the bona fide FRET transition to an ∼0 value in Li+ buffer (i.e., not due to Cy5 photobleaching) should be a rare event on the timescale of the experiment. Nevertheless, we did observe occasional FRET traces that captured abrupt transitions from the high- to low-FRET states, despite the presence of an active Cy5 dye (Fig. 7G, Bottom). The prolonged high-FRET state observed in Li+ buffer conditions suggests the presence of a stable anchor-site contact during DNA synthesis that, once disrupted, results in a rapid and large displacement of product DNA away from the telomerase enzyme (Fig. 7D, Bottom pathway). In contrast, GQ-stabilizing conditions give rise to complex FRET dynamics, implying that nascent DNA folding may facilitate efficient extrusion of the telomere DNA away from the telomerase enzyme.
Discussion
The foundation of telomere structure consists of short G-rich repeat sequences, GGTTAG in humans, that have the propensity to fold into G-quadruplex structures in vitro and in vivo (6, 7). Previous studies have suggested that multiple copies of the telomerase processivity factor and shelterin component POT1–TPP1 decorate single-stranded telomeric DNA and thereby hinder the formation of GQ structures (41). POT1–TPP1 further facilitates the recruitment of telomerase to the telomere through interactions between the TPP1 TEL patch and the TERT TEN domain (56, 63). However, the interplay between newly synthesized G-rich repeats and the telomerase–POT1–TPP1 complex remains poorly understood.
Here, we present evidence that GQ formation can occur within an actively extending telomerase complex in vitro and that formation of such GQs affects the kinetic properties of telomerase. Interestingly, the addition of POT1–TPP1 stimulates telomerase processivity, but does not rescue decreased telomere synthesis rates caused by GQ-destabilizing conditions. This finding suggests that GQ folding may also occur in the context of the POT1–TPP1–telomerase complex and modulates telomerase activity. We describe a detailed kinetic framework for telomerase catalysis and use this model to globally fit telomerase time-series data. Our kinetic modeling reveals small but significant GQ-dependent changes in the rate constants describing the telomere synthesis reaction and permits us to calculate microscopic processivity values for each cycle of telomere DNA repeat synthesis (see SI Appendix, Methods for details). Moreover, single-molecule FRET experiments reveal that GQ folding impacts the dynamic handling of newly synthesized DNA by the telomerase complex. We note that DNA structures other than GQs, such as two strictly consecutive G hairpins, could also in principle give rise to the pattern-of-four DNA product distributions and kinetic effects described in this study. However, to our knowledge, such structures would not be expected to display the observed cation-dependent effects, which are pronounced in the coordination of G-quartet planes. In the presence of physiologically high K+ concentrations, two closely spaced G hairpins can, moreover, be expected to adopt a GQ structure when confined in the telomerase complex. Nevertheless, while we favor a GQ-dependent model to explain our data, future experiments are warranted to directly test for the presence of GQ structures within the telomerase complex. Collectively, our study reveals the delicate interplay between telomere DNA structure and the actively extending human telomerase complex.
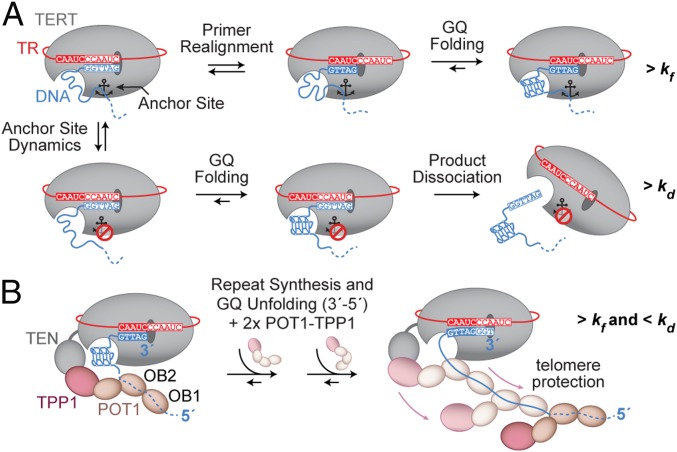
We present a working model for the mechanism of GQ-dependent effects on telomerase repeat-addition processivity, as well as the interplay between GQ structures and the POT1–TPP1 processivity factor (Fig. 8). The complex rearrangements necessary for template recycling during multiple rounds of telomere repeat synthesis require multiple anchor sites between telomerase and its DNA substrate to prevent product dissociation (Fig. 1A). In the schematic model depicted in Fig. 8A, two principal pathways are shown that depend on the register of the 3′ end of the telomere as well as on the state of anchor-site interactions during the formation of a GQ. GQ folding may bias the positioning of the 3′ end of the primer to favor realignment for a subsequent round of repeat synthesis, provided anchor-site contacts remain intact (Fig. 8A, Top pathway). This outcome is mechanistically similar to DNA hairpin-induced translocation models proposed for diverse telomerase systems (64, 65). Alternately, GQ folding within the DNA product may compete with anchor-site contacts to promote dissociation (Fig. 8A, Bottom pathway), as was suggested in previous telomerase studies that investigated the effect of putative GQ folding during active telomere elongation (38–40).
Fig. 8.
Model of GQ folding within the actively extending telomerase complex. (A) At the completion of a telomere repeat, telomerase (with TERT depicted in gray and hTR in red) is annealed to the telomere DNA (blue). The schematically drawn anchor site (anchor symbol) is either engaged with the telomere DNA (Top pathway) or disengaged (Bottom pathway). Sequential repeat synthesis and primer realignment extrude telomere DNA from the active site to eventually allow for GQ formation. In the case where anchor-site contacts are maintained at this stage (Top, Middle and Right cartoons), the formation of a GQ may bias the enzyme complex toward another round of telomere repeat addition. If the anchor-site contacts are instead broken when GQ formation occurs (Bottom, Left and Middle cartoons), primer realignment results in product dissociation at this stage (Bottom, Right cartoon). (B) Extension of the mechanistic GQ model (A) illustrating the possibility and functional contribution of GQ formation in the presence of the GQ resolvase POT1–TPP1. A telomere-bound POT1–TPP1 unit engages the hTERT TEN domain via TPP1. Upon GQ formation, further POT1–TPP1 units are recruited to resolve the GQ in a 3′-to-5′ direction, resulting in fully protected telomere DNA. Concurrent telomere synthesis generates additional G-rich repeats for the process to continue in a four-repeat periodicity.
Any contact with the growing product DNA must be dynamic in the course of activity, and might involve further elements of telomerase or associated factors, such as POT1–TPP1 that binds to telomeric DNA and enhances RAP (34, 41). Indeed, recent studies using reconstituted human shelterin complexes showed that a TRF2–TIN2–TPP1 protein bridge stimulates processivity through anchoring of telomerase to DNA substrates that possessed both duplex and ssDNA telomere DNA sequence (66). However, the presence of the POT1 protein within this shelterin core complex was necessary to achieve maximal stimulation of telomerase processivity. Our data indicate that GQ formation within actively extending telomerase might be required for correct POT1–TPP1 function, in particular the stimulation of telomeric DNA-synthesis rates during processive enzyme action. We propose a speculative model, wherein GQ formation within the telomerase complex, followed by POT1–TPP1 binding and GQ unfolding, serves as a mechanism to ensure the concerted decoration and protection of nascent single-stranded telomeric DNA (Fig. 8B). This model is consistent with previous studies reporting a preference of POT1–TPP1 binding to telomeric DNA in a 3′-to-5′ direction (34, 67). This directionality was reported also for the unfolding of GQs by POT1, whereby its individual OB-fold domains engage the GQ in a stepwise manner (Fig. 8B, OB2 before OB1) (42, 43). Interestingly, the binding of multiple POT1–TPP1 units to telomeric DNA might occur in a pairwise and cooperative fashion, whereby each pair of POT1–TPP1 dimers spans four consecutive G-rich repeats of the unfolded telomeric DNA (34, 67) (Fig. 8B). These lines of evidence are inconsistent with the binding of POT1–TPP1 to each telomeric repeat as it emerges from the telomerase enzyme (i.e., 5′-to-3′ directionality), but rather describe a “lagging” type of protein decoration, in which multiple telomeric repeats are produced before POT1–TPP1 binding in a reverse direction (Fig. 8B).
The recently reported cryo-EM structures of the human and Tetrahymena telomerase enzymes provide a platform for investigating the potential for GQ folding within the actively extending telomerase complex (33, 68). Interestingly, when analyzing the EM density of the human telomerase complex, we noticed a structural pocket immediately proximal to the path of the nascent DNA emerging from the active site (SI Appendix, Fig. S8A). This pocket is flanked by two evolutionarily conserved and potentially dynamic elements that are essential for telomerase processivity: the TEN domain and the hTR P6.1 stem loop (45, 69). The volume of this pocket is compatible with the dimensions of a GQ fold, and raises the possibility that the nascent DNA product has ample space to assume a GQ structure within the confines of the telomerase RNP complex before POT1–TPP1 binding to the DNA. In the Tetrahymena structure, the C-terminal domain of the processivity factor, Teb1, is positioned as a lid to the DNA exit pocket, forming an enclosed cavity that is also sufficient to accommodate a single GQ fold (SI Appendix, Fig. S8B). We propose the hypothesis that the Teb1-related shelterin component POT1 may occupy a similar position in the human telomerase complex. Thus, the anticipated binding of POT1 to the DNA as it is threaded out of this DNA exit channel is not mutually exclusive with the possibility of GQ folding within this protected cavity (33, 55, 68).
Our proposed model, while speculative, provides a functional explanation for the evolutionary conservation of G-rich telomere DNA by providing a possible mechanistic link between GQ folding in telomere maintenance and telomerase function. Future experiments should continue to investigate the influence of the shelterin proteins POT1–TPP1 on the folding properties of the nascent telomere DNA product, as well as to evaluate the in vivo significance of telomerase-confined GQ folding and its suitability for GQ-targeted anticancer therapies.
Methods
Preparation of RNAs.
Telomerase RNA fragments [hTR CR4/5 (hTR 239 to 328) and hTR template/pseudoknot (hTR t/PK; 32 to 195)] were prepared using standard in vitro transcription protocols. For smFRET experiments, preparation of dye-labeled RNA fragments was performed as described previously (70) (see SI Appendix, Methods for details).
Telomerase Expression and Purification.
Human telomerase was reconstituted with purified RNA fragments in rabbit reticulocyte lysate using the Promega TnT Quick Coupled Transcription/Translation System (47). FLAG-TERT was expressed off the pNFLAG-hTERT plasmid (71) (details of RRL reconstitution are described in SI Appendix, Methods). For telomerase expression in HEK293T cells, lysate was provided by Samantha Sanford and Patricia Opresko, University of Pittsburgh, Pittsburgh, PA, prepared as described previously (72). Telomerase enzyme was purified via the N-terminal FLAG tag on hTERT using ANTI-FLAG M2 beads (Sigma-Aldrich), and eluted with 3×FLAG peptide (48) (see SI Appendix, Methods for details).
32P End Labeling of DNA Primers.
Fifty picomoles of the indicated DNA primer was labeled with [γ32P]ATP using T4 polynucleotide kinase (NEB). For generating experimental data for kinetic modeling, end-labeled primers were PAGE-purified and DNA concentration was precisely determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific).
Primer-Extension Assays.
Telomerase activity assays were performed in 1× TB [50 mM Tris⋅HCl, pH 8.3, 50 mM KCl (NaCl or LiCl when indicated), 1 mM MgCl2, 2 mM DTT] as described previously (48) (see SI Appendix, Methods for details). Where indicated, POT1–TPP1 was added to the reaction mixture at a final concentration of 500 nM. The “fraction left behind” for a given lane was calculated by summing each RAP band and all RAP bands below it divided by the total RAP band intensity counts for that lane. The natural logarithm of (1 − FLB) was then plotted against repeat number and fitted by linear regression (see main text and SI Appendix, Methods for details and caveats). The slope value of the linear fit was used to determine processivity R1/2 values from −ln(2)/slope (49).
POT1–TPP1 Expression and Purification.
Procedures for insect cell expression were according to the Bac-to-Bac Baculovirus Expression System (Thermo Fisher Scientific) using the pFEV-POT1 and pFastBac-TPP1N (TPP1 residues 89 to 334) plasmids (see SI Appendix, Methods for details). POT1–TPP1N was purified via a GST tag on POT1 using Glutathione Sepharose 4B resin (GE Healthcare) and eluted with glutathione, and the affinity tag was removed by TEV-protease cleavage.
Kinetic Analysis.
For kinetic analysis, the primer-extension assay described above was modified by chasing with 20 μM cold (TTAGGG)3 DNA primer. The chase primer was added after 20 min of initiating the reaction, to prevent radiolabeled DNA primer and radiolabeled DNA product rebinding within the subsequent 70-min time course. The intensities of the RAP bands on the gel electrophoretogram were converted to absolute concentrations based on the intensity of the initial radiolabeled primer band (i.e., 50 nM). Multiple exposures of the gel to the phosphor screen were analyzed to ensure intense and weak bands remained within the linear range of the phosphorimager. These data were analyzed globally according to the sequential model (Fig. 6A) using DynaFit (59, 73). Further details are given in SI Appendix, Methods.
Single-Molecule Experiments.
Unless indicated otherwise, all single-molecule experiments were performed as previously described (48). Detailed experimental methods describing microscope slide preparation, telomerase enzyme immobilization, in situ telomerase activity assays, and data acquisition/analysis can be found in SI Appendix, Methods.
Supplementary Material
Acknowledgments
We thank Dr. Harry Noller, Dr. Alan Zahler, Dr. Patricia Opresko, and Samantha Sanford for critical reading of the manuscript. We also thank Dr. Tim Sterne-Weiler for his help reviewing the statistical analyses presented in the manuscript. This work was supported by National Institutes of Health Grants F99CA212439 (to L.I.J.) and R01GM095850 (to M.D.S.). J.H. was supported by a Swiss National Science Foundation Early Postdoc Mobility Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814777116/-/DCSupplemental.
References
- 1.de Lange T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 2.de Lange T. How shelterin solves the telomere end-protection problem. Cold Spring Harb Symp Quant Biol. 2010;75:167–177. doi: 10.1101/sqb.2010.75.017. [DOI] [PubMed] [Google Scholar]
- 3.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 4.McElligott R, Wellinger RJ. The terminal DNA structure of mammalian chromosomes. EMBO J. 1997;16:3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maizels N, Gray LT. The G4 genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: The structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saharia A, et al. Flap endonuclease 1 contributes to telomere stability. Curr Biol. 2008;18:496–500. doi: 10.1016/j.cub.2008.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.León-Ortiz AM, Svendsen J, Boulton SJ. Metabolism of DNA secondary structures at the eukaryotic replication fork. DNA Repair (Amst) 2014;19:152–162. doi: 10.1016/j.dnarep.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 10.London TB, et al. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J Biol Chem. 2008;283:36132–36139. doi: 10.1074/jbc.M808152200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat Rev Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaffitzel C, et al. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lam EYN, Beraldi D, Tannahill D, Balasubramanian S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat Commun. 2013;4:1796. doi: 10.1038/ncomms2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez R, et al. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol. 2012;8:301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blackburn EH, et al. Recognition and elongation of telomeres by telomerase. Genome. 1989;31:553–560. doi: 10.1139/g89-104. [DOI] [PubMed] [Google Scholar]
- 16.Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 17.Allsopp RC, et al. Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alder JK, et al. Ancestral mutation in telomerase causes defects in repeat addition processivity and manifests as familial pulmonary fibrosis. PLoS Genet. 2011;7:e1001352. doi: 10.1371/journal.pgen.1001352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parry EM, Alder JK, Qi X, Chen JJ, Armanios M. Syndrome complex of bone marrow failure and pulmonary fibrosis predicts germline defects in telomerase. Blood. 2011;117:5607–5611. doi: 10.1182/blood-2010-11-322149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gramatges MM, Qi X, Sasa GS, Chen JJ, Bertuch AA. A homozygous telomerase T-motif variant resulting in markedly reduced repeat addition processivity in siblings with Hoyeraal Hreidarsson syndrome. Blood. 2013;121:3586–3593. doi: 10.1182/blood-2012-08-447755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaug AJ, Crary SM, Fioravanti MJ, Campbell K, Cech TR. Many disease-associated variants of hTERT retain high telomerase enzymatic activity. Nucleic Acids Res. 2013;41:8969–8978. doi: 10.1093/nar/gkt653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim NW, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 23.Feng J, et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura TM, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 25.Greider CW. Telomerase is processive. Mol Cell Biol. 1991;11:4572–4580. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaug AJ, Podell ER, Cech TR. Mutation in TERT separates processivity from anchor-site function. Nat Struct Mol Biol. 2008;15:870–872. doi: 10.1038/nsmb.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- 28.Lai CK, Mitchell JR, Collins K. RNA binding domain of telomerase reverse transcriptase. Mol Cell Biol. 2001;21:990–1000. doi: 10.1128/MCB.21.4.990-1000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robart AR, Collins K. Human telomerase domain interactions capture DNA for TEN domain-dependent processive elongation. Mol Cell. 2011;42:308–318. doi: 10.1016/j.molcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lue NF. A physical and functional constituent of telomerase anchor site. J Biol Chem. 2005;280:26586–26591. doi: 10.1074/jbc.M503028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lue NF, Li Z. Modeling and structure function analysis of the putative anchor site of yeast telomerase. Nucleic Acids Res. 2007;35:5213–5222. doi: 10.1093/nar/gkm531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu RA, Tam J, Collins K. DNA-binding determinants and cellular thresholds for human telomerase repeat addition processivity. EMBO J. 2017;36:1908–1927. doi: 10.15252/embj.201796887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang J, et al. Structure of telomerase with telomeric DNA. Cell. 2018;173:1179–1190.e13. doi: 10.1016/j.cell.2018.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang F, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt JC, Zaug AJ, Kufer R, Cech TR. Dynamics of human telomerase recruitment depend on template-telomere base pairing. Mol Biol Cell. 2018:mbc.E17-11-0637. doi: 10.1091/mbc.E17-11-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang H, et al. Telomeric overhang length determines structural dynamics and accessibility to telomerase and ALT-associated proteins. Structure. 2014;22:842–853. doi: 10.1016/j.str.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lei M, Zaug AJ, Podell ER, Cech TR. Switching human telomerase on and off with hPOT1 protein in vitro. J Biol Chem. 2005;280:20449–20456. doi: 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- 38.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 39.Min B, Collins K. Multiple mechanisms for elongation processivity within the reconstituted Tetrahymena telomerase holoenzyme. J Biol Chem. 2010;285:16434–16443. doi: 10.1074/jbc.M110.119172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun D, Lopez-Guajardo CC, Quada J, Hurley LH, Von Hoff DD. Regulation of catalytic activity and processivity of human telomerase. Biochemistry. 1999;38:4037–4044. doi: 10.1021/bi982249n. [DOI] [PubMed] [Google Scholar]
- 41.Taylor DJ, Podell ER, Taatjes DJ, Cech TR. Multiple POT1-TPP1 proteins coat and compact long telomeric single-stranded DNA. J Mol Biol. 2011;410:10–17. doi: 10.1016/j.jmb.2011.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang H, Buncher N, Opresko PL, Myong S. POT1-TPP1 regulates telomeric overhang structural dynamics. Structure. 2012;20:1872–1880. doi: 10.1016/j.str.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 44.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 45.Chen JL, Greider CW. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J. 2003;22:304–314. doi: 10.1093/emboj/cdg024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen JL, Greider CW. Template boundary definition in mammalian telomerase. Genes Dev. 2003;17:2747–2752. doi: 10.1101/gad.1140303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weinrich SL, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 48.Parks JW, Stone MD. Coordinated DNA dynamics during the human telomerase catalytic cycle. Nat Commun. 2014;5:4146. doi: 10.1038/ncomms5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Latrick CM, Cech TR. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 2010;29:924–933. doi: 10.1038/emboj.2009.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hammond PW, Cech TR. dGTP-dependent processivity and possible template switching of Euplotes telomerase. Nucleic Acids Res. 1997;25:3698–3704. doi: 10.1093/nar/25.18.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maine IP, Chen SF, Windle B. Effect of dGTP concentration on human and CHO telomerase. Biochemistry. 1999;38:15325–15332. doi: 10.1021/bi991596+. [DOI] [PubMed] [Google Scholar]
- 52.Hardy CD, Schultz CS, Collins K. Requirements for the dGTP-dependent repeat addition processivity of recombinant Tetrahymena telomerase. J Biol Chem. 2001;276:4863–4871. doi: 10.1074/jbc.M005158200. [DOI] [PubMed] [Google Scholar]
- 53.Chen Y, Podlevsky JD, Logeswaran D, Chen JJL. A single nucleotide incorporation step limits human telomerase repeat addition activity. EMBO J. 2018;37:e97953. doi: 10.15252/embj.201797953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moye AL, et al. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat Commun. 2015;6:7643. doi: 10.1038/ncomms8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci USA. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nandakumar J, et al. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature. 2012;492:285–289. doi: 10.1038/nature11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 58.Klenow H, Henningsen I. Effect of monovalent cations on the activity of the DNA polymerase of Escherichia coli B. Eur J Biochem. 1969;9:133–141. doi: 10.1111/j.1432-1033.1969.tb00586.x. [DOI] [PubMed] [Google Scholar]
- 59.Kuzmic P. DynaFit—A software package for enzymology. Methods Enzymol. 2009;467:247–280. doi: 10.1016/S0076-6879(09)67010-5. [DOI] [PubMed] [Google Scholar]
- 60.Peng Y, Mian IS, Lue NF. Analysis of telomerase processivity: Mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol Cell. 2001;7:1201–1211. doi: 10.1016/s1097-2765(01)00268-4. [DOI] [PubMed] [Google Scholar]
- 61.Wu JY, Stone MD, Zhuang X. A single-molecule assay for telomerase structure-function analysis. Nucleic Acids Res. 2010;38:e16. doi: 10.1093/nar/gkp1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hwang H, Opresko P, Myong S. Single-molecule real-time detection of telomerase extension activity. Sci Rep. 2014;4:6391. doi: 10.1038/srep06391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xin H, et al. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 64.Yang W, Lee YS. A DNA-hairpin model for repeat-addition processivity in telomere synthesis. Nat Struct Mol Biol. 2015;22:844–847. doi: 10.1038/nsmb.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jarstfer MB, Cech TR. Effects of nucleotide analogues on Euplotes aediculatus telomerase processivity: Evidence for product-assisted translocation. Biochemistry. 2002;41:151–161. doi: 10.1021/bi011649z. [DOI] [PubMed] [Google Scholar]
- 66.Lim CJ, Zaug AJ, Kim HJ, Cech TR. Reconstitution of human shelterin complexes reveals unexpected stoichiometry and dual pathways to enhance telomerase processivity. Nat Commun. 2017;8:1075. doi: 10.1038/s41467-017-01313-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Corriveau M, Mullins MR, Baus D, Harris ME, Taylor DJ. Coordinated interactions of multiple POT1-TPP1 proteins with telomere DNA. J Biol Chem. 2013;288:16361–16370. doi: 10.1074/jbc.M113.471896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen THD, et al. Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature. 2018;557:190–195. doi: 10.1038/s41586-018-0062-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O’Connor CM, Lai CK, Collins K. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J Biol Chem. 2005;280:17533–17539. doi: 10.1074/jbc.M501211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Akiyama BM, Stone MD. Assembly of complex RNAs by splinted ligation. Methods Enzymol. 2009;469:27–46. doi: 10.1016/S0076-6879(09)69002-9. [DOI] [PubMed] [Google Scholar]
- 71.Drosopoulos WC, Direnzo R, Prasad VR. Human telomerase RNA template sequence is a determinant of telomere repeat extension rate. J Biol Chem. 2005;280:32801–32810. doi: 10.1074/jbc.M506319200. [DOI] [PubMed] [Google Scholar]
- 72.Fouquerel E, et al. Oxidative guanine base damage regulates human telomerase activity. Nat Struct Mol Biol. 2016;23:1092–1100. doi: 10.1038/nsmb.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 74.Kerssemakers JW, et al. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.