Abstract
Bacterial RNA has emerged as an important activator of innate immune responses by stimulating Toll-like receptors TLR7 and TLR8 in humans. Guanosine 2′-O-methylation at position 18 (Gm18) in bacterial tRNA was shown to antagonize tRNA-induced TLR7/8 activation, suggesting a potential role of Gm18 as an immune escape mechanism. This modification also occurs in eukaryotic tRNA, yet a physiological immune function remained to be tested. We therefore set out to investigate the immune modulatory role of Gm18 in both prokaryotic and eukaryotic microorganisms, Escherichia coli and Saccharomyces cerevisiae, and in human cells. Using RiboMethSeq analysis we show that mutation of trmH in E. coli, trm3 in S. cereviase, and CRISPR/Cas9-induced knockout of TARBP1 in H. sapiens results in loss of Gm18 within tRNA. Lack of Gm18 across the kingdoms resulted in increased immunostimulation of peripheral blood mononuclear cells when activated by tRNA preparations. In E. coli, lack of 2′-O-methyltransferase trmH also enhanced immune stimulatory properties by whole cellular RNA. In contrast, lack of Gm18 in yeasts and human cells did not affect immunostimulation by whole RNA preparations. When using live E. coli bacteria, lack of trmH did not affect overall immune stimulation although we detected a defined TLR8/RNA-dependent gene expression signature upon E. coli infection. Together, these results demonstrate that Gm18 is a global immune inhibitory tRNA modification across the kingdoms and contributes to tRNA recognition by innate immune cells, but as an individual modification has insufficient potency to modulate recognition of the investigated microorganisms.
Keywords: immune stimulation, bacterial RNA, TLR, RNA modification, 2′-O-methylation
INTRODUCTION
To ensure a rapid immune response against a broad range of invading pathogens, cells of the host's innate immune system are equipped with a limited set of pattern recognition receptors (PRRs) that sense highly conserved microbial structures, so-called pathogen-associated molecular patterns (PAMPs). Bacterial RNA has been identified as a potent activator of innate immune responses and was shown to be sensed by Toll-like receptors (TLRs) localized in the endolysosome (Koski et al. 2004; Eberle et al. 2009; Eigenbrod et al. 2015). Within human peripheral blood mononuclear cells (PBMCs), monocytes recognize microbial RNA in a TLR8 dependent manner, triggering the release of proinflammatory mediators including TNF, IL-12p40, and IFN-β. In contrast, plasmacytoid dendritic cells (DCs) sense bacterial RNA via endosomal TLR7, culminating in the production of IFN-α (Diebold et al. 2004; Heil et al. 2004; Ablasser et al. 2009; Bergstrøm et al. 2015; Saitoh et al. 2017).
As both, bacterial and host RNA, are composed of the same building blocks, a reliable discrimination of self and non-self by the innate immune system is critical to ensure induction of an adequate antimicrobial defense in case of pathogen encounter while at the same time avoiding aberrant immune stimulation by host (“self”) RNA. Indeed, several auto-immune disorders including systemic lupus erythematosus have been associated with abnormal recognition of self nucleic acids (Barrat et al. 2005; Kono et al. 2009). Apart from compartmentalization—i.e., localization of nucleic acid sensing TLRs in the endolysosome to which self-RNA has limited access—and differences in the RNA sequence composition, another determinant for self/non-self discrimination has been attributed to RNA modification profiles. As such, both the extent and type of posttranscriptional RNA modifications differ between self and microbial RNA. In this regard, various studies have highlighted the importance of ribose 2′-O-methylation as a potent modification impeding immune stimulation by endosomal TLRs (Karikó et al. 2005; Robbins et al. 2007; Sioud et al. 2007; Gehrig et al. 2012; Jöckel et al. 2012). 2′-O-methylation not only impairs TLR7 and TLR8 activation but also acts as a TLR7/8 antagonist, thus inhibiting immune cell stimulation by otherwise immunogenic RNA species. Subsequent studies identified a DmDN (D = all but C; N = all) and a DmRC (D = all but C; R = G, A) motif as most efficient in antagonizing TLR7 and TLR8, respectively (Schmitt et al. 2017). Of note, 2′-O-methylation of cytosine consistently failed to impede TLR7/8 activation in previous studies (Robbins et al. 2007; Eberle et al. 2008; Kaiser et al. 2014).
Although the highest abundance of 2′-O-methylation has been described in eukaryotic rRNA, this modification is also found in certain bacterial tRNAs (Boccaletto et al. 2018). Specifically, tRNA 2′-O-methylation of guanosine at position 18 (Gm18), which is present in five tRNA species in Escherichia coli (E. coli), has been demonstrated to be necessary and sufficient to suppress immune stimulation via TLR7 by whole E. coli tRNA preparations. Total tRNA preparations derived from an E. coli mutant lacking Gm18 2′-O-methyltransferase activity (trmH knockout) elicited a stronger TLR7 response than the respective wild-type samples (Gehrig et al. 2012; Jöckel et al. 2012). These observations raised the hypothesis that Gm18, apart from its structural function, may also serve as an immune escape mechanism for E. coli or other microorganisms carrying this specific modification. Yet, this assumption is, thus far, exclusively based on experiments involving the transfection of purified tRNA preparations into immune cells. How E. coli tRNA 2′-O-methylation indeed shapes bacterial immunogenicity under physiological conditions, i.e., during infection with the whole, living microorganism, remains elusive. Moreover, Gm18 is present in the tRNA of lower and higher eukaryotes, including Saccharomyces cerevisiae (S. cerevisiae) as well as human cells. Its physiological significance for immune recognition via endosomal TLRs across the different kingdoms has not been studied so far. Given that eukaryotic RNA generally shows a much higher degree of posttranscriptional RNA modifications than prokaryotic RNA (Motorin and Helm 2011; Motorin 2015) Gm18 might play a more redundant role in eukaryotes as compared to bacteria but experimental evidence is lacking.
The aim of the present study was therefore to determine the role of Gm18 within tRNA in pro- and eukaryotic organisms, specifically E. coli, S. cerevisiae, and human cells, for modulating innate immune recognition. It should be tested whether Gm18 exerts an immune modulatory function across kingdoms and how this modification affects immune stimulation in general. We show that Gm18 deficiency augmented PBMC activation by purified total tRNA preparations from all investigated organisms. We also show for the first time an impact of Gm18 on the immune stimulatory properties of human tRNA. However, an impact of Gm18 in tRNA on immune activation by whole cellular RNA was only visible for E. coli but not for eukaryotes, presumably due to dominant differences in 2′-O-methylation in ribosomal RNA. In neither microbe did a knockout of the Gm18 methyltransferase impact the overall immune reaction, although we identified an RNA-TLR8 regulated gene signature in response to E. coli in general. Thus, Gm18 is a global immune modulatory tRNA modification but it does not seem to play a role as an immune escape mechanism during infection with the investigated microorganisms. The results suggest the existence of a threshold for Gm content in an invading pathogen's cellular RNA that governs TLR7/8 mediated immune responses.
RESULTS
Yeast tRNA fractions show inverse correlation of Gm content with type I interferon secretion by human peripheral blood mononuclear cells
First we intended to test whether 2′-O-methylation might affect the immune stimulatory potency of S. cerevisiae tRNA as had been described before for E. coli. As a starting point we investigated fractions of tRNA isoacceptors that had been obtained in counter current distribution experiments (Heitzler et al. 1992), a technique which separates tRNA according to their relative lipophilicity. Fractions were analyzed for their relative Gm, Am, Um, and Cm content by LC-MS/MS and compared to their IFN-α–inducing properties upon stimulation of human PBMCs. Due to highly cell-type specific receptor and cytokine expression profiles, IFN-α secretion within PBMCs is commonly used as a read-out for TLR7-dependent plasmacytoid DC activation. Gm-rich tRNAs were predominantly eluted in fractions 1–6 and 13–15, while relative Am levels showed a peak in fractions 13–15 (but were absent in the first ten fractions). Um was found prominently in fractions 3–6 and Cm in fractions 1–8 and 18–21 (Fig. 1A). Introduction of Gm, Am, and Um into synthetic RNA previously had been described to affect immune recognition whereas Cm consistently failed to impede TLR7/8 activation (Robbins et al. 2007; Eberle et al. 2008; Schmitt et al. 2017). When the individual tRNA fractions were tested for induction of type I IFN from PBMCs, they varied strongly in their IFN-α–inducing capacity (Fig. 1B), suggesting a negative correlation between 2′-O-methylation and IFN induction also in eukaryotic RNA. In a first approximation a correlation matrix showed a significant negative correlation between Gm content and IFN-α secretion (Fig. 1C), whereas Am, Cm, and Um content showed no relation. A closer examination confirmed a general negative impact of Gm on IFN secretion (Fig. 1D). We do not exclude that other modifications might contribute to immune modulation, e.g., Am and Um may affect the lowered IFN-α levels observed in fractions 13–15 and fractions 3–6, respectively.
FIGURE 1.
IFN-α production by PBMCs upon stimulation with fractionated tRNAs from S. cerevisiae inversely correlates with the Gm content. (A) Relative levels of 2′-O-methylated nucleotides quantified by LC-MS/MS in S. cerevisiae fractions that were generated as described in the Materials and Methods section. Values were normalized to highest relative amount of each modified nucleotide. (B) Human PBMCs (400,000/well in a total volume of 200 µL) were stimulated overnight in duplicate wells with fractionated and DOTAP-encapsulated tRNAs (c = 1 µg/mL). IFN-α release was measured in cell-free supernatants by ELISA. Bars represent pooled data from three individual donors +SEM. (C) A correlation matrix of IFN-α secretion and relative amount of Gm, Cm, Am, Um was calculated for Pearson's r including 95% confidence interval. (***) P < 0.001 (D) Individual correlations of IFN-α and Xm, normalized to the highest respective value are shown. For Gm, goodness of fit was calculated using an appropriated nonlinear dose response model (GraphPad Prism).
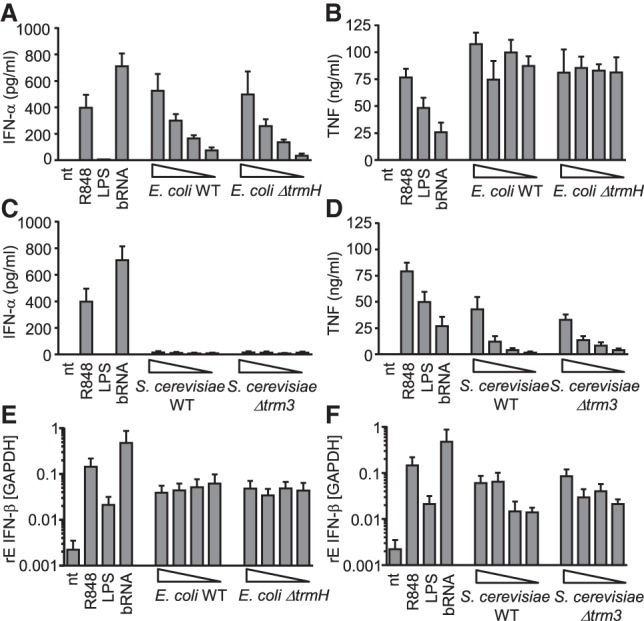
To confirm these starting observations of a negative regulatory function of Gm in S. cerevisiae we used a targeted, independent experimental approach: An isogenic mutant strain deficient in trm3, the trmH homolog in yeast, was analyzed. Total tRNA and whole cellular RNA preparations were purified from WT and mutant S. cerevisiae. We used unbiased RiboMethSeq analysis (Galvanin et al. 2019) to study the impact of loss of trm3 on the yeast tRNA modification pattern: Thereby we confirm the presence of Gm18 in eight tRNA isoacceptors corresponding to five tRNA species in WT samples (Boccaletto et al. 2018). trm3 deficient cells showed a lack of Gm18 in total tRNA extracts (Fig. 2A). We also used a complementary LC-MS/MS approach that demonstrated a residual Gm content in these preparations; this can be explained by the presence of Gm at position 34 in tRNAPhe (Fig. 2B; Boccaletto et al. 2018). In line with the results obtained with tRNA fractions, specific lack of Gm18 in trm3 mutant S. cerevisiae significantly enhanced TLR7 stimulatory properties of total tRNA preparations, although IFN-α secretion was generally lower as compared to stimulation with bacterial RNA, the latter serving as positive control (Fig. 2C). Thus, a physiological role of Gm18 modified tRNA in eukaryotes could be demonstrated. However, whole cellular RNA from both WT and Δtrm3 S. cerevisiae failed to induce any TLR7 activation (Fig. 2C). We also tested ΔtrmH E. coli lacking the corresponding Gm tRNA methyltransferase. Gm18 deficiency was also confirmed by RiboMethSeq and LC-MS/MS analysis. For ΔtrmH E. coli, not only enhanced IFN-α release by total tRNA, as published previously (Gehrig et al. 2012; Jöckel et al. 2012), but also by whole bacterial RNA preparations (Fig. 2D–F) was observed. Thus, in both microorganisms the presence of a few Gm18 modified tRNA isoacceptors was sufficient to manipulate recognition of whole tRNA preparations confirming a dominant-inhibitory effect of this modification. Yet, whole RNA preparations showed differing influence by loss of Gm18 in tRNA.
FIGURE 2.
Gm18 deficiency differentially regulates immune stimulation by isolated RNA species derived from S. cerevisiae and E. coli. (A,D) RiboMethSeq and (B,E) quantitative LC/MS-MS analysis of isolated S. cerevisiae (A,B) and E. coli (D,E) total tRNA preparations derived from WT strains and 2′-O-methyltransferase deficient mutants (Δtrm3, ΔtrmH). (A,D) Heatmaps display normalized mean methylation scores. tRNA isoacceptors and modified nucleotide positions are indicated on the right. (B) Percentage of Xm modified nucleotides normalized to total guanosine content. (C,F) Human PBMCs were transfected overnight with DOTAP-encapsulated total tRNAs and whole cellular RNA preparations (c = 0.5 µg/mL) from WT and 2′-O-methyltransferase deficient S. cerevisiae (Δtrm3) (C) or E. coli (ΔtrmH) (F). Stimulation with bacterial RNA derived from S. aureus served as positive control. IFN-α release was quantified in cell-free supernatants by ELISA. Each data point represents the mean value of one donor stimulated in duplicate wells. Error bars indicate mean ± SD of three to five individual donors. (*) P < 0.05; (***) P < 0.001.
Gm18 deficiency does not alter cytokine production in PBMCs upon infection with living E. coli and S. cerevisiae
In order to investigate whether Gm18 not only affects immune stimulation by purified RNA species but also by infection with the whole organism which is composed of a variety of PAMPs activating potentially a large set of PRRs, PBMCs were infected with E. coli and S. cerevisiae WT and Gm18 deficient mutant strains at various multiplicities of infection (MOIs) (Fig 3A–D). Although total cellular RNA preparations isolated from E. coli ΔtrmH–induced higher levels of IFN-α than their WT counterparts (Fig. 2F), no differences in IFN-α or TNF release were observed upon infection with the whole, living organism (Fig. 3A,B). Similar results were obtained for infection with WT and Δtrm3 S. cerevisiae (Fig. 3C,D). Of note, infection with S. cerevisiae entirely failed to induce IFN-α, as already observed for stimulation with total cellular RNA (Figs. 2C, 3C). While upon transfection of contaminant-free RNA into PBMCs TNF is commonly used as read-out for TLR8-dependent monocyte activation, this cytokine can also be induced by a variety of other PAMPs during infection with whole pathogens, for example via TLR4 by E. coli LPS or by yeast cell wall mannans (Beutler et al. 2001; Tada et al. 2002; Ferwerda et al. 2008). To better appreciate potential RNA-dependent effects, analysis was therefore expanded to the production of IFN-β, a mediator that is more prominently induced by RNA than by microbial cell wall components in both a TLR7- and TLR8-dependent manner. Yet, neither for E. coli nor for S. cerevisiae, lack of Gm18 affected IFN-β induction (Fig. 3E,F). Thus, the data do not provide evidence for a role of Gm18 as an immune escape mechanism under physiological conditions of infection within the context of the here analyzed living whole microorganisms.
FIGURE 3.
Lack of Gm18 does not affect innate immune sensing of E. coli and S. cerevisiae upon infection of PBMCs. (A–D) Human PBMCs were infected in duplicate wells with E. coli (A,B) or S. cerevisiae (C,D) WT strain or methyltransferase deficient mutant at different MOIs (wedges indicate MOI 1, 0.3, 0.1, and 0.03), and supernatants were collected after overnight incubation. Stimulation with LPS (TLR4 ligand), R848 (TLR7/8 ligand), and bacterial RNA (bRNA, c = 0.5 µg/mL) derived from S. aureus served as controls. Levels of IFN-α (A,C) and TNF (B,D) were measured in cell-free supernatants by ELISA. (E,F) PBMCs were infected as in A–D and relative expression of IFN-β mRNA was assessed by qPCR 5 h postinfection with the indicated microorganisms. All results represent mean data (+SEM) of three independent experiments with different donors. Statistical analysis did not show any significant difference between infection with the WT strain and the respective Gm18 deficient mutant.
Gene expression profiling reveals TLR8-dependent pathways in E. coli sensing
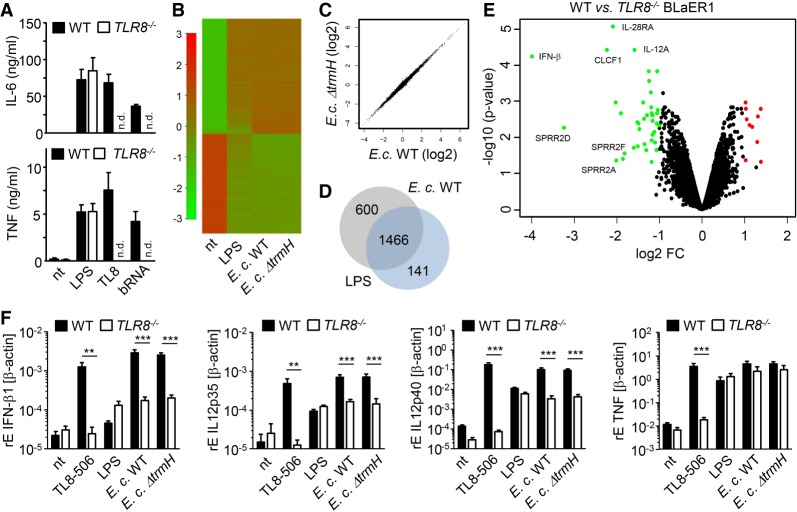
Next, we aimed to more broadly explore a potential impact of RNA sensing in general and of Gm18 in particular on E. coli mediated immune activation, as ΔtrmH E. coli gave differing results for whole RNA versus whole bacterial cell stimulation (Figs. 2, 3). To this end, microarray-based gene expression profiling was performed in trans-differentiated BLaER1 cells that recognize bacterial and archaeal RNA in a TLR8-dependent manner (Fig. 4A; Vierbuchen et al. 2017) and are used to model primary human monocyte responses (Rapino et al. 2013; Gaidt et al. 2018). In a first experiment, WT BLaER1 cells were infected with E. coli WT and trmH deficient mutant for 5 h to specifically detect Gm18 regulated pathways. This time point was evaluated as optimal for differential cytokine induction in preliminary time-course experiments (data not shown). Gene expression profiling revealed a total of 2066 and 2070 significantly regulated genes (defined as log2 fold-change ≥ +1 or ≤ −1 and P < 0.05) upon infection with WT or ΔtrmH E. coli, respectively, when compared to the untreated control (Fig. 4B). In both groups, the top regulated genes predominantly contained a variety of well-characterized inflammatory mediators involved in innate immunity, including IL-6, TNF and IFN-β (Supplemental Table 1A,B). However, analysis demonstrated that none of these genes were significantly different between WT and ΔtrmH E. coli (Fig. 4C), indicating that Gm18 does not influence overall immune stimulation by E. coli. Yet, it is well known that E. coli possesses a very bioactive LPS triggering TLR4 and indeed the gene signature induced by E. coli showed a large overlap with LPS, which was included as positive control (Fig. 4D).
FIGURE 4.
E. coli induces an RNA/TLR8-dependent gene signature which is not affected by Gm18. (A) BLaER1 cells were stimulated overnight with LPS, TLR8-specific ligand TL8-506 (TL8), and bacterial RNA from S. aureus. Levels of IL-6 and TNF were measured in cell-free supernatants by ELISA. (B–D) BLaER1 cell were stimulated with whole, live E. coli WT, E. coli ΔtrmH (MOI 10), or LPS for 5 h, and gene expression profiling was performed using an Illumina HT12 microarray. (B) Heatmap depicts 2304 significantly differentially regulated genes (defined as log2 fold-change >1 or < −1 and P < 0.05; mean of three independent experiments) for each condition as compared to the nontreated control. For each condition, data of three independent biological replicates were pooled. (C) Scatter plot displaying log2 intensity values of genes regulated by E. coli WT versus E. coli ΔtrmH relative to the unstimulated control. (D) Venn diagram of E. coli WT and LPS regulated genes. (E) Volcano plot of 482 regulated genes of BLaER1 WT compared to BLaER1 TLR8−/− upon stimulation with E. coli WT. Significantly differentially regulated genes (defined as log2 fold-change >1 or < −1 and P < 0.05) are highlighted in green and red. All microarray data were analyzed using Chipster software. (F) WT and TLR8−/− BLaER1 cells were stimulated for 5 h with TLR8-specific ligand TL8-506, LPS, E. coli WT, and E. coli ΔtrmH (MOI 10), and relative cytokine expression was determined by qPCR. Values represent mean data (+SEM) of three independent experiments. (***) P < 0.001.
We next asked the question to what extent an RNA-TLR8 axis contributed to E. coli–induced cell activation in general. To this end, a gene expression analysis was performed in WT and TLR8−/− BLaER1 cells upon infection with WT E. coli. Interestingly, 50 genes were significantly differentially regulated (40 down and 10 up) in TLR8−/− as compared to WT cells, including IFN-β, IL-12p35 and three members of the small proline-rich protein family (SPRRs) whose function in innate immunity is as yet insufficiently defined (Fig. 4E; Supplemental Table 1C). These results thus clearly point toward an RNA-TLR8 dependent gene signature in E. coli sensing even in the presence of a strong LPS response. Microarray data were further validated by qPCR, confirming a strong reduction of IFN-β and partial impairment of IL-12p35 and IL-12p40 expression in TLR8−/− cells upon E. coli exposure, while induction of TNF and IL-6 were not altered (Fig. 4F and data not shown). Together, these data demonstrate that despite the existence of TLR8-dependent pathways in E. coli recognition, Gm18 does not affect innate immune activation by this pathogen and thus does not serve as an immune escape mechanism under the here tested physiological conditions.
Gm18 deficiency in human cells enhances immune cell activation by total tRNA
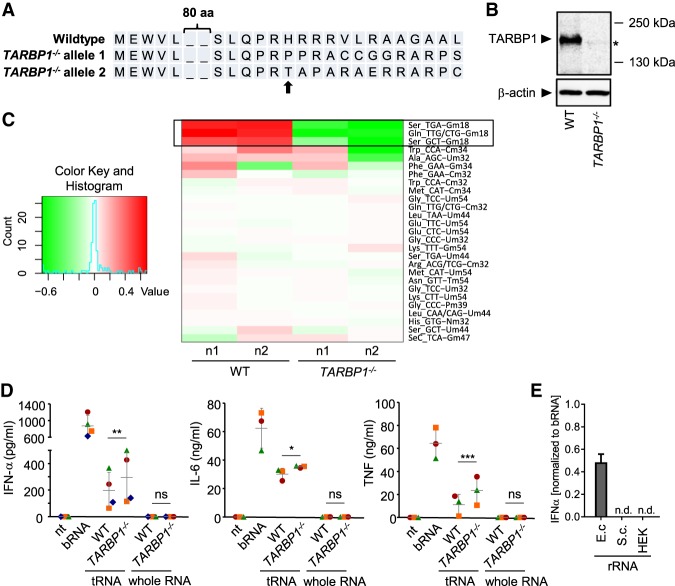
In a next step, we further wanted to analyze the influence of Gm18 on immune stimulation in the human system (higher eukaryote). Therefore, TARPB1, the proposed human homolog of trmH, was targeted by CRISPR/Cas9 in HEK293 cells, a cell line that is easy to transfect and to genetically manipulate. Successful knockout of TARBP1 was confirmed by sequencing, demonstrating biallelic frame-shift–inducing insertions resulting in altered amino acid sequences (Fig. 5A), as well as on the protein level (Fig. 5B). Lack of Gm18 in tRNA derived from TARBP1-deficient cells was then analyzed by RiboMethSeq analysis (Fig. 5C). Indeed we can show that TARBP1 in human cells has the proposed 2′-O-methyltransferase activity. However, surprisingly, Gm18 was detected in only three isoacceptors corresponding to two tRNA species (Ser, Gln) in HEK WT cells but not in further tRNAs. Yet, despite the relatively low abundance of Gm18 in human tRNA, preparations derived from TARBP1−/− cells still caused a slight but statistically significant increase in immune stimulation by total tRNA preparations (Fig. 5D). In contrast, whole cellular RNA was in general nonstimulatory and no differences were observed between RNA from WT and TARBP1−/− cells. This pattern was similar to the results obtained with S. cerevisae (Fig. 2C) but different to E. coli (Fig. 2F). We speculated that within whole cellular RNA the high content of ribosomal RNA might explain the differences. Indeed, when testing rRNA preparations we observed a clear discrimination of prokaryotic rRNA, which was stimulatory, from eukaryotic rRNA, that was inactive (Fig. 5E), indicating a dominant contribution of immune silencing modifications present in rRNA.
FIGURE 5.
Gm18 deficiency in TARBP1−/− HEK cells enhances immune stimulation by isolated tRNAs but not by whole cellular RNA. (A) HEK cells in which TARBP1 was targeted by CRSIPR/Cas9 were analyzed by Sanger sequencing, confirming biallelic frame-shift–inducing indels resulting in an altered amino acid sequence. Arrow indicates cleavage site. (B) Western blot analysis of WT and TARBP1−/− HEK cells. (*) Indicates unspecific band. β-actin was used as loading control. (C) RiboMethSeq analysis of total tRNA preparations from WT and TARBP1−/− HEK cells. Heatmap indicates 2′-O-methylation sites. Analysis was performed from two independent biological samples (n1 and n2). (D) Human PBMCs were stimulated overnight with 0.5 µg/mL DOTAP-encapsulated RNA samples. IFN-α, IL-6, and TNF release were measured in cell-free supernatants by ELISA. Each data point represents the mean value of one donor stimulated in duplicate wells. Error bars indicate mean ± SD of three to four individual donors. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (E) rRNA fractions were prepared from E. coli, S. cerevisae, and HEK WT cells and used for stimulation of PBMCs. RNA was used at 0.5 µg/mL and secreted IFN-α was normalized to stimulation by 1 µg/mL whole bacterial RNA (n = 3, mean + SD; n.d., not detectable).
DISCUSSION
RNA modifications have been recognized as critical modulators of RNA-mediated innate immune responses and a number of studies have highlighted the TLR7/8 antagonistic properties of ribose 2′-O-methylation in this context (Robbins et al. 2007; Hamm et al. 2010; Jung et al. 2015; Schmitt et al. 2017). Yet, it was unknown whether Gm in its natural context plays a comparable immune-modifying role across different kingdoms. We now show that deletion of Gm18 by knockout of the modifying enzymes trmH, trm3, and TARBP1 in prokaryotes, lower and higher eukaryotes results in an increased immune stimulation by tRNA. Using the unbiased RiboMethSeq approach to study frequencies of Gm, this immune modulatory effect could be observed although only a few isoacceptors had a Gm modification, thus demonstrating a potent antagonistic activity of naturally occurring Gm18.
Apart from its role in microorganisms, we also investigated the significance of Gm18 in the human system. Surprisingly, RiboMethSeq analysis identified Gm18 in only three isoacceptors corresponding to two tRNA species (Ser and Gln). This finding was rather unexpected as human tRNA is generally believed to show the highest degree of posttranslational RNA modifications. Yet, despite its low abundance, Gm18 deficiency still enhanced cytokine release by total tRNA, pointing toward a potential role of this modification for immune modulation. Mitochondrial tRNA, which accounts for ∼13 ± 5% of total tRNA in our study, may at least partially contribute to the overall immune stimulation observed by tRNA preparations (Karikó et al. 2005). Despite an effect of TARBP1 that could only be observed for tRNA but not whole RNA, links to a physiological contribution of Gm18 to self/foreign discrimination have been published: Rare single-nucleotide variants were found in TARBP1 in exome sequencing studies associated with autoimmune psoriasis (Tang et al. 2014) and TARBP1 is differentially regulated between healthy patients and those with autoimmune SLE or ANCA vasculitis (Lyons et al. 2010). Moreover, HIV has been recently shown to use TARBP1 to recruit 2′-O-methyltransferase activity to its RNA to hide the RNA from immune recognition (Ringeard et al. 2019).
Human and S. cerevisae rRNA and total cellular RNA preparations lacked immune stimulatory properties entirely, suggesting the presence of relevant immune silencing modifications within rRNA. Of note, rRNA from S. cerevisiae and especially from Homo sapiens has been reported to be heavily 2′-O-methylated on both guanosine and adenosine residues by the 2′-O-methyltransferases NOP1 in yeast and its homolog fibrillarin in humans (Jansen et al. 1991; Tollervey et al. 1991; Marchand et al. 2016; Erales et al. 2017). Studying fibrillarin and rRNA 2′-O-methylation would be interesting but is challenging as NOP1 was described to be crucial for cell viability in yeast (Tollervey et al. 1991). Interestingly, antifibrillarin antibodies are detected in the serum of patients suffering from autoimmune diseases (Van Eenennaam et al. 2002; Barrat et al. 2005; Hagberg and Rönnblom 2015).
The fact that 2′-O-methylation is also naturally occurring in a subset of tRNA isoacceptors at position 18 (Gm18) in E. coli gave rise to the hypothesis that this modification, beyond its structural role, may have acquired a secondary function as an immune escape mechanism in organisms bearing this modification (David 2012; Gehrig et al. 2012; Jöckel et al. 2012). Yet, this idea was based on the transfection of purified tRNAs into PBMCs, a situation not adequately reflecting the physiological conditions of infection with the live microorganism. In the present study, we demonstrate that Gm18 deficiency in E. coli indeed enhances immune stimulation by isolated total tRNA and even whole cellular RNA preparations, yet does not interfere with innate immune sensing of live E. coli by both PBMCs and BLaER1 cells. Similar results were obtained for S. cerevisiae. These results therefore argue against a role of Gm18 as a relevant immune escape mechanism under physiological conditions at least in the microorganisms tested. As tRNA constitutes only 10%–20% of total E. coli RNA with only five tRNA species being Gm18 methylated, it is conceivable that the ratio of Gm18 methylated to unmethylated RNA might not be sufficient to suppress immune activation during infection when rRNA is also present. Due to the dominant action of LPS when studying E. coli (Fig. 4D), a role of Gm18 might be more prominent in other bacteria. Another influencing factor might be accessibility of different RNA species.
The results are compatible with a model of a certain threshold for Gm content in an organism's cellular RNA to modulate TLR7/8 mediated immune responses: In S. cerevisae and human cells, the overall Gm content is high, especially due to its abundance in rRNA, and exceeds a certain threshold, thus resulting in lack of immune recognition. Deletion of Gm only in tRNA does not increase sensing of the whole organism, as the overall Gm content is still sufficiently high (rRNA). In contrast in E. coli, Gm levels are generally low due to the lack of rRNA 2′-O-methylation, resulting in immune stimulation by all RNA species. However, when only tRNA is analyzed, a situation of Gm enrichment is achieved which now reveals the antagonistic function of this modification.
E. coli is known to be recognized in a predominantly TLR4-dependent manner (Takeuchi et al. 1999; Roger et al. 2009), in line with a large overlap of the E. coli– and LPS-induced gene expression profiles observed in our study. Yet, microarray-based analysis of WT and TLR8−/− BLaER1 cells clearly identified an additional TLR8- and thus RNA-dependent gene signature triggered by E. coli infection, including regulation of classical proinflammatory mediators like IFN-β and IL-12 as well as proteins with less well-defined functions in innate immunity (e.g., small proline-rich proteins, SPRRs). Interestingly, an RNA-dependent induction of specific cytokines by E. coli was also described by the groups of L. Sander and J. Blander who raised the concept of bacterial RNA as so-called “vita-PAMP”: Due to the rapid degradation of RNA after bacterial cell death, RNA was proposed to signify microbial viability and thus a persistent infectious threat to the host, culminating in more robust innate and adaptive immune responses (Sander et al. 2011; Barbet et al. 2018; Ugolini et al. 2018). Indeed, the here observed genes that are strongly dependent on TLR8 (e.g., IL-12 and IFN-β) have also be described by the Blander and Sander groups. While initial investigations emphasized the relevance of E. coli RNA for the induction of IFN-β and activation of the NLRP3 inflammasome with subsequent production of bioactive IL-1β (Sander et al. 2011), recent studies highlighted the contribution of microbial viability sensing via TLR8 for T follicular helper cell differentiation and antibody responses in a manner dependent on IL-12 and on an IFN-β/IL-1β axis (Barbet et al. 2018; Ugolini et al. 2018). It might therefore be interesting to analyze T helper cell differentiation instead of global gene expression of monocytes by stimulation with trmH mutants as those might affect only a small, yet important part of the gene signature.
In summary, the findings of this study demonstrate that Gm18 is an immune inhibitory RNA modification across organisms of different kingdoms, yet it does not serve as an immune evasion mechanism in the studied organisms. The results also identify an impact of Gm18 on immune stimulatory properties within human tRNA; in conjunction with published data on differential TARBP1 regulation and SNPs in autoimmune diseases a potential contribution in tolerance to self-RNA can be envisioned.
MATERIALS AND METHODS
Reagents
RPMI 1640 containing stable glutamine was purchased from Anprotec; FCS was obtained from Gibco; Ficoll (1.078 g/mL), DPBS w/o Ca2+ and Mg2+ from Pan Biotech; TRIzol from Thermo Fisher Scientific; R848 and TL8-506 from InvivoGen; DOTAP (N-[1-(2, 3-dioleoyloxy)propyl]-N, N, N-trimethylammonium methylsulfate) from Roth; peptone (Sigma-Aldrich), yeast extract (AppliChem) Nanosept MF, bio-inert membrane, purple, 0.45 µm were from Pall Laboratory; and ammonium acetate was from J.B. Baker.
Preparation of RNA samples from E. coli, S. cerevisiae, and HEK cells
WT (parental strain: BW25113) and trmH deficient (JW3626-1) Escherichia coli (provided from E. coli Genetic Stock Center CGSC, Yale University) were grown in Luria-Bertani (LB) medium (Merck) and harvested within the mid-log phase growth, pelleted, and the cell wall was digested by lysozyme (c = 40 mg/mL, 20 min at 37°C). WT (parental strain: Y00000) and trm3 deficient (Y03809) S. cerevisiae (purchased from Euroscarf, Johann Wolfgang Goethe-University, Frankfurt, Germany) were grown in YPD-Medium (20 g peptone; 10 g yeast extract; ad 975 mL dH2O; pH 6.5), harvested in the mid-log phase growth, pelleted and the cell wall was digested by Lyticase (100 U, 20 min at 37°C). HEK cells were harvested using Trypsin-solution and pelleted at 1300 rpm for 15 min. Total RNA of all species was isolated using TRIzol reagent according to the manufacturer's protocol. The obtained RNA was further separated by agarose gel electrophoresis, and RNA bands corresponding to tRNA and/or rRNA were excised. Gel slices were transferred to a 2 mL reaction tube and mashed, and 500 µL ammonium acetate 0.5 M was added. Samples were stored for 1 h at −80°C and shaken overnight at room temperature at 500 rpm to extract RNA. The next day, samples were transferred to a Nanosept filter and flow through was mixed with 1250 µL ethanol to precipitate RNA. RNA preparations were stored for 1–2 h at −80°C and centrifuged for 60 min at 10,000g, 4°C. Pellets were washed in 70% ethanol and RNA was dissolved in nuclease-free water. Purity of the RNA preparations was validated by determining the 260/230 nm and 260/280 nm extinction ratio by NanoDrop (Thermo Scientific). Only RNA preparations with ratios >1.8 were used for stimulation experiments.
Fractionation of S. cerevisiae tRNA
The fractions of tRNA yeast used in experiments shown in Figure 1 were generated several decades ago using fractions counter current distribution, an old separation technique that separated tRNA according to their relative lipophilicity. Despite long-term storage, analysis of the fractions by high-resolution capillary electrophoresis (QIAxcel, Qiagen) did not indicate significant degradation. According to a mapping of the tRNA distribution (Heitzler et al. 1992), the relevant fractions 1–6 contain Gm-containing tRNAs accepting the aminoacids Phe (Gm34), Ser, Tyr, Trp, and Leu (all Gm18) and fractions 13–15 contain His (Gm18, Am4) and Val (no Gm). Fractions 4–5 contained Ser (Um44) (Boccaletto et al. 2018). All analyses on the 2′-O-methylation status of the preexisting tRNA fractions using LC-MS and RiboMethSeq analysis, as described below, were performed during the course of the present study.
LC-MS measurements of Gm, Am, Um, and Cm levels
tRNA samples were digested into nucleotides using 0.6 U nuclease P1 from P. citrinum (Sigma-Aldrich), 0.2 U snake venom phosphodiesterase from C. adamanteus (Worthington), 2 U FastAP (Thermo Scientific), 10 sU benzonase (Sigma-Aldrich), 200 ng Pentostatin (Sigma-Aldrich), and 500 ng Tetrahydrouridine (Merck-Millipore) in 25 mM ammonium acetate (pH 7.5; Sigma-Aldrich) overnight at 37°C. The nucleosides were then spiked with internal standard (13C stable isotope-labeled nucleosides from S. cerevisiae, SIL-IS) and subjected to analysis. Technical triplicates with 100 ng digested RNA and 50 ng internal standard were analyzed via LC-MS (Agilent 1260 series and Agilent 6460 Triple Quadrupole mass spectrometer equipped with an electrospray ion source [ESI]). The solvents consisted of 5 mM ammonium acetate buffer (pH 5.3; solvent A) and LC-MS grade acetonitrile (solvent B; Honeywell). The elution started with 100% solvent A with a flow rate of 0.35 mL/min, followed by a linear gradient to 8% solvent B at 10 min and 40% solvent B after 20 min. Initial conditions were regenerated with 100% solvent A for 10 min. The column used was a Synergi Fusion (4 µM particle size, 80 Å pore size, 250 × 2.0 mm; Phenomenex). The UV signal at 254 nm was recorded via a diode array detector (DAD) to monitor the main nucleosides. ESI parameters were as follows: gas temperature 350°C, gas flow 8 L/min, nebulizer pressure 50 psi, sheath gas temperature 350°C, sheath gas flow 12 L/min, capillary voltage 3000 V. The MS was operated in the positive ion mode using Agilent MassHunter software in the dynamic MRM (multiple reaction monitoring) mode. For quantification, a combination of external and internal calibration was applied as described previously (Kellner et al. 2014).
RiboMethSeq analysis
Total tRNA RiboMethseq analysis was performed essentially as described previously (Marchand et al. 2017). Briefly, about 100 ng of total tRNA either from E. coli, S. cerevisiae, or human HEK cells were randomly fragmented in bicarbonate buffer for different times (6–10 min for E. coli, 8–12 min for S. cerevisiae, and 14 min for human HEK, at 95°C). The reaction was stopped by ethanol precipitation as previously described in Marchand et al. (2016). RNA fragments were then subjected to end-repair and converted to a library using NEBNext Small RNA Library kit (ref#E7330S, New England Biolabs) following the manufacturer's instructions. Library sequencing was performed using Illumina HiSeq 1000 sequencer in a single read mode for 50 nt. Analysis of human tRNA 2′-O-methylation was performed by trimming of raw sequencing reads using Trimmomatic v0.32, followed by end-to-end alignment by Bowtie 2.0 to nonredundant collection of human intronless tRNA genes. Reference sequence was created by collapsing of identical tRNA sequences and deduction of the consensus sequences from closely related species. The resulting reference allowed unambiguous alignment of >80% of tRNA reads. Further data analysis was done by previously described bioinformatic pipeline (Galvanin et al. 2019). Briefly, RiboMethSeq scores were calculated from the cleavage profiles derived from cumulated 5′- and 3′-end counts. Candidate 2′-O-methylated sites in tRNA species were identified by values of Score A >0.5 and Score Mean >0.90, followed by visual inspection of the coverage profiles to exclude irregularly cleaved regions. MethScore (Score C2) was used to quantify the 2′-O-methylation level. Differential heatmaps for figures were constructed by normalization of MethScore levels for every given position to the average value observed in the series of samples. Positive (shown in red) values indicate higher methylation compared to the average (overmethylation), while negative (shown in green) positions have MethScore below the average (undermethylation).
Generation of TARBP-1 deficient HEK cells using CRISPR–Cas9
TARBP1-deficient HEK cells were created via a S. pyogenes derived CRISPR/Cas9 system using a NGG PAM sequence. gRNAs were designed by using a Blue Heron webpage (www.blueheronbio.com) with a GC content between 60% and 80% and blasted to control for off target effects. gRNAs were custom synthesized by Eurofins Genomics and cloned into a GFP-encoding vector backbone using the restriction enzyme BbsI (Chen et al. 2013). Plasmids were kindly provided by Professor Dr. Dirk Grimm, BioQuant, University of Heidelberg, Heidelberg, Germany. The TARBP1−/− clone used in the current study was generated using the following gRNA: fw: 5′-CACCGCGGCGCCAGGCGCGCGCCAC-3′; rev: 5′-AAACGTGGCGCGCGCCTGGCGCCGC-3′. Nucleotides highlighted in gray indicate the overhang required for cloning into the BbsI digested vector. Resulting plasmids were transfected into HEK293 cells using Lipofectamine 2000, and GFP-positive cells were sorted into 96-well plates 24 h posttransfection. After 1–2 wk, single clones were analyzed for successful targeting of TARBP1 by both Sanger sequencing (primer sequences for PCR and sequencing: fw 5′- ACTCCTGGCCTTACGTCTAAATC-3′, rev 5′- ACGGCTGCTAGCACTTCCAC-3′) and western blot analysis (anti-TARBP1 antibody [EPR8727] Abcam). Sequences were analyzed using CRISP-ID software (Dehairs et al. 2016).
Isolation and stimulation of human peripheral blood mononuclear cells (PBMCs)
Human PBMCs were isolated from heparinized blood of healthy volunteers upon informed consent and approval by the local ethical committee (admission number: S-716/2017) by standard Ficoll-Hypaque density gradient centrifugation (Ficoll 1.078 g/mL). PBMCs were resuspended in RPMI 1640 supplemented with 2% heat-inactivated human serum. For transfection experiments, all RNAs were encapsulated with DOTAP at a ratio of 3 µL DOTAP per 1 µg of RNA in serum-free medium according to the manufacturer's protocol. Unless otherwise indicated, cells were stimulated with RNA preparations at a final concentration of 500 ng/mL, and transfection was performed at a density of 2 × 105 PBMCs per well in a 96-well flat bottom plate at a final volume of 100 µL. Cells were incubated overnight in a humidified 5% CO2 atmosphere at 37°C. For cytokine measurement, levels of TNF, IL-6 (BD), and IFN-α (eBioscience) were determined in cell-free supernatants by ELISA.
Trans-differentiation and stimulation of BLaER1 cells
WT and TLR8−/− B-cell like BLaER1 cells, the latter generated as previously described (Vierbuchen et al. 2017), were kindly provided by Holger Heine, Research Center Borstel, Germany. BlaER1 cells were trans-differentiated into monocyte/macrophage-like cells by seeding 1 million cells/well into a six-well in 3 mL/well RPMI 1640 medium supplemented with 10% FCS, 1% Pen/Strep, 10 ng/mL IL-3, 10 ng/mL M-CSF, and 150 nM β-Estradiol. Plates were incubated at 37°C and 5% CO2. At days 2 and 5 of trans-differentiation, cells were collected by pipetting up and down, centrifuged at 1300 rpm for 10 min, and fresh medium with the aforementioned supplements was added. After 1 wk, BLaER1 cells were sorted using the AutoMACS magnetic cell sorter system and the human CD19 MicroBeads kit according to manufacturer's instructions (Miltenyi Biotech) to deplete residual CD19+ BLaER1 cells. CD19 negative, trans-differentiated monocyte/macrophage like BlaER1 cells were resuspended in RPMI 1640 supplemented with 10% FCS. For experiments involving measurement of IL-6 and TNF in cell-free supernatants by ELISA (BD), 1 × 105 cells were transferred into a 96-well flat bottom plate and stimulated overnight in duplicate wells with LPS (c = 100 ng/mL), TL8-506 (c = 100 ng/mL), and bacterial RNA derived from S. aureus complexed with DOTAP at a ratio of 3 µL DOTAP per 1 µg RNA (c = 500 ng/mL). S. aureus RNA was isolated as described previously (Schmitt et al. 2017). For microarray analysis and for assessment of cytokine induction by quantitative real-time PCR, 2 × 105 cells were seeded in a 48-well plate in a final volume of 250 µL. To avoid preactivation of BLaER1 cells due to MACS sorting, cells were rested overnight at 37°C, 5% CO2 prior to stimulation with LPS, TL8-506, or infection with live WT and ΔtrmH E. coli (MOI 10) for 5 h in duplicate wells. At 1.5 h postinfection, penicillin/streptomycin was added to kill extracellular bacteria.
Microarray analysis and quantitative real-time PCR
For microarray analysis and quantitative real-time PCR, total RNA was extracted 5 h poststimulation or infection using the RNeasy Mini Kit including an on-column DNase treatment according to the manufacturer's instructions (Qiagen). For microarray-based gene expression profiling, biotin-labeled cRNA of three independent experiments was hybridized to a whole-genome HumanHT-12 array from Illumina according to the manufacturer's instructions, and Microarray scanning was carried out using an iScan array scanner. As test for significance, a Bayes test was used on the bead expression values of the groups of interest. The average expression value was calculated as mean of the measured expressions of beads together with the standard deviation of the beads. Genes with a P-value <0.05 were selected and log2 fold expression changes of differentially regulated genes were calculated. All analyses were performed using Chipster Software. Microarray data have been uploaded to GEO (accession number GSE125797). For quantitative real-time PCR, total RNA was reverse transcribed using 10 µL isolated RNA in a total reaction volume of 20 µL with a High Capacity cDNA RT Kit (Applied Biosystems) according to the manufacturer's instructions. For quantitative real-time PCR analysis, SYBRGreen PCR Master Mix Fast (Applied Biosystems) was used. noRT and nontemplate controls as well as melt curve analysis served to specify control. Relative expression of target gene mRNA (for Primers see Table 1) was calculated by ΔCT compared to the indicated housekeeping gene.
TABLE 1.
List of primer sequences used for quantitative real-time PCR
Statistical analysis
Statistical significance between groups was determined by the two-tailed Student's t-test or two-way ANOVA with Bonferroni posttest. For experiments comparing immune stimulation by RNA species derived from WT cells as compared to the respective Gm18 deficient mutant, significance was calculated using the linear mixed-effects model with Tukey's rank test to account for donor variation. Differences were considered significant for (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. Analyses were performed using Graph Pad Prism 7 (Graph Pad Software) or using R version 3.3.0 with the package nlme.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Suzan Leccese and Selina Hassel for excellent technical support. We are grateful to Gérard Keith (IBMC Strasbourg, France) for yeast tRNA fractions. This work was supported by German Research Foundation (DFG) grants DA592/5 and DA592/7 (to A.D.) and He 3397/18-1 (to M.H.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.070243.118.
REFERENCES
- Ablasser A, Poeck H, Anz D, Berger M, Schlee M, Kim S, Bourquin C, Goutagny N, Jiang Z, Fitzgerald KA, et al. 2009. Selection of molecular structure and delivery of RNA oligonucleotides to activate TLR7 versus TLR8 and to induce high amounts of IL-12p70 in primary human monocytes. J Immunol 182: 6824–6833. 10.4049/jimmunol.0803001 [DOI] [PubMed] [Google Scholar]
- Barbet G, Sander LE, Geswell M, Leonardi I, Cerutti A, Iliev I, Blander JM. 2018. Sensing microbial viability through bacterial RNA augments T follicular helper cell and antibody responses. Immunity 48: 584–598 e585. 10.1016/j.immuni.2018.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrat FJ, Meeker T, Gregorio J, Chan JH, Uematsu S, Akira S, Chang B, Duramad O, Coffman RL. 2005. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J Exp Med 202: 1131–1139. 10.1084/jem.20050914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrøm B, Aune MH, Awuh JA, Kojen JF, Blix KJ, Ryan L, Flo TH, Mollnes TE, Espevik T, Stenvik J. 2015. TLR8 senses Staphylococcus aureus RNA in human primary monocytes and macrophages and induces IFN-β production via a TAK1-IKKβ-IRF5 signaling pathway. J Immunol 195: 1100–1111. 10.4049/jimmunol.1403176 [DOI] [PubMed] [Google Scholar]
- Beutler B, Du X, Poltorak A. 2001. Identification of Toll-like receptor 4 (Tlr4) as the sole conduit for LPS signal transduction: genetic and evolutionary studies. J Endotoxin Res 7: 277–280. 10.1177/09680519010070040901 [DOI] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, Piątkowski P, Bagiński B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A, et al. 2018. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46: D303–D307. 10.1093/nar/gkx1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al. 2013. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155: 1479–1491. 10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David R. 2012. Immune evasion: Gm18, a bacterial ‘invisibility cloak’. Nat Rev Microbiol 10: 238–239. 10.1038/nrmicro2767 [DOI] [PubMed] [Google Scholar]
- Dehairs J, Talebi A, Cherifi Y, Swinnen JV. 2016. CRISP-ID: decoding CRISPR mediated indels by Sanger sequencing. Sci Rep 6: 28973 10.1038/srep28973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. 2004. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303: 1529–1531. 10.1126/science.1093616 [DOI] [PubMed] [Google Scholar]
- Eberle F, Giessler K, Deck C, Heeg K, Peter M, Richert C, Dalpke AH. 2008. Modifications in small interfering RNA that separate immunostimulation from RNA interference. J Immunol 180: 3229–3237. 10.4049/jimmunol.180.5.3229 [DOI] [PubMed] [Google Scholar]
- Eberle F, Sirin M, Binder M, Dalpke AH. 2009. Bacterial RNA is recognized by different sets of immunoreceptors. Eur J Immunol 39: 2537–2547. 10.1002/eji.200838978 [DOI] [PubMed] [Google Scholar]
- Eigenbrod T, Pelka K, Latz E, Kreikemeyer B, Dalpke AH. 2015. TLR8 senses bacterial RNA in human monocytes and plays a nonredundant role for recognition of Streptococcus pyogenes. J Immunol 195: 1092–1099. 10.4049/jimmunol.1403173 [DOI] [PubMed] [Google Scholar]
- Erales J, Marchand V, Panthu B, Gillot S, Belin S, Ghayad SE, Garcia M, Laforêts F, Marcel V, Baudin-Baillieu A, et al. 2017. Evidence for rRNA 2′-O-methylation plasticity: control of intrinsic translational capabilities of human ribosomes. Proc Natl Acad Sci 114: 12934–12939. 10.1073/pnas.1707674114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferwerda G, Meyer-Wentrup F, Kullberg BJ, Netea MG, Adema GJ. 2008. Dectin-1 synergizes with TLR2 and TLR4 for cytokine production in human primary monocytes and macrophages. Cell Microbiol 10: 2058–2066. 10.1111/j.1462-5822.2008.01188.x [DOI] [PubMed] [Google Scholar]
- Gaidt MM, Rapino F, Graf T, Hornung V. 2018. Modeling primary human monocytes with the trans-differentiation cell line BLaER1. Methods Mol Biol 1714: 57–66. 10.1007/978-1-4939-7519-8_4 [DOI] [PubMed] [Google Scholar]
- Galvanin A, Ayadi L, Helm M, Motorin Y, Marchand V. 2019. Mapping and quantification of tRNA 2′-O-methylation by RiboMethSeq. Methods Mol Biol 1870: 273–295. 10.1007/978-1-4939-8808-2_21 [DOI] [PubMed] [Google Scholar]
- Gehrig S, Eberle ME, Botschen F, Rimbach K, Eberle F, Eigenbrod T, Kaiser S, Holmes WM, Erdmann VA, Sprinzl M, et al. 2012. Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J Exp Med 209: 225–233. 10.1084/jem.20111044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg N, Rönnblom L. 2015. Systemic lupus erythematosus—a disease with A dysregulated type I interferon system. Scand J Immunol 82: 199–207. 10.1111/sji.12330 [DOI] [PubMed] [Google Scholar]
- Hamm S, Latz E, Hangel D, Müller T, Yu P, Golenbock D, Sparwasser T, Wagner H, Bauer S. 2010. Alternating 2′-O-ribose methylation is a universal approach for generating non-stimulatory siRNA by acting as TLR7 antagonist. Immunobiology 215: 559–569. 10.1016/j.imbio.2009.09.003 [DOI] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. 2004. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303: 1526–1529. 10.1126/science.1093620 [DOI] [PubMed] [Google Scholar]
- Heitzler J, Maréchal-Drouard L, Dirheimer G, Keith G. 1992. Use of a dot blot hybridization method for identification of pure tRNA species on different membranes. Biochim Biophys Acta 1129: 273–277. 10.1016/0167-4781(92)90503-R [DOI] [PubMed] [Google Scholar]
- Jansen RP, Hurt EC, Kern H, Lehtonen H, Carmo-Fonseca M, Lapeyre B, Tollervey D. 1991. Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. J Cell Biol 113: 715–729. 10.1083/jcb.113.4.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jöckel S, Nees G, Sommer R, Zhao Y, Cherkasov D, Hori H, Ehm G, Schnare M, Nain M, Kaufmann A, et al. 2012. The 2′-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition. J Exp Med 209: 235–241. 10.1084/jem.20111075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, von Thülen T, Laukemper V, Pigisch S, Hangel D, Wagner H, Kaufmann A, Bauer S. 2015. A single naturally occurring 2′-O-methylation converts a TLR7- and TLR8-activating RNA into a TLR8-specific ligand. PLoS One 10: e0120498 10.1371/journal.pone.0120498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser S, Rimbach K, Eigenbrod T, Dalpke AH, Helm M. 2014. A modified dinucleotide motif specifies tRNA recognition by TLR7. RNA 20: 1351–1355. 10.1261/rna.044024.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K, Buckstein M, Ni H, Weissman D. 2005. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23: 165–175. 10.1016/j.immuni.2005.06.008 [DOI] [PubMed] [Google Scholar]
- Kellner S, Ochel A, Thüring K, Spenkuch F, Neumann J, Sharma S, Entian KD, Schneider D, Helm M. 2014. Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. Nucleic Acids Res 42: e142 10.1093/nar/gku733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono DH, Haraldsson MK, Lawson BR, Pollard KM, Koh YT, Du X, Arnold CN, Baccala R, Silverman GJ, Beutler BA, et al. 2009. Endosomal TLR signaling is required for anti-nucleic acid and rheumatoid factor autoantibodies in lupus. Proc Natl Acad Sci 106: 12061–12066. 10.1073/pnas.0905441106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski GK, Karikó K, Xu S, Weissman D, Cohen PA, Czerniecki BJ. 2004. Cutting edge: innate immune system discriminates between RNA containing bacterial versus eukaryotic structural features that prime for high-level IL-12 secretion by dendritic cells. J Immunol 172: 3989–3993. 10.4049/jimmunol.172.7.3989 [DOI] [PubMed] [Google Scholar]
- Lyons PA, McKinney EF, Rayner TF, Hatton A, Woffendin HB, Koukoulaki M, Freeman TC, Jayne DR, Chaudhry AN, Smith KG. 2010. Novel expression signatures identified by transcriptional analysis of separated leucocyte subsets in systemic lupus erythematosus and vasculitis. Ann Rheum Dis 69: 1208–1213. 10.1136/ard.2009.108043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand V, Blanloeil-Oillo F, Helm M, Motorin Y. 2016. Illumina-based RiboMethSeq approach for mapping of 2′-O-Me residues in RNA. Nucleic Acids Res 44: e135 10.1093/nar/gkw547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand V, Pichot F, Thüring K, Ayadi L, Freund I, Dalpke A, Helm M, Motorin Y. 2017. Next-generation sequencing-based RiboMethSeq protocol for analysis of tRNA 2′-O-methylation. Biomolecules 7: E13 10.3390/biom7010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y. 2015. RNA modification. In eLS, pp. 1–18. Wiley Online Library. [Google Scholar]
- Motorin Y, Helm M. 2011. RNA nucleotide methylation. Wiley Interdiscip Rev RNA 2: 611–631. 10.1002/wrna.79 [DOI] [PubMed] [Google Scholar]
- Rapino F, Robles EF, Richter-Larrea JA, Kallin EM, Martinez-Climent JA, Graf T. 2013. C/EBPα induces highly efficient macrophage transdifferentiation of B lymphoma and leukemia cell lines and impairs their tumorigenicity. Cell Rep 3: 1153–1163. 10.1016/j.celrep.2013.03.003 [DOI] [PubMed] [Google Scholar]
- Ringeard M, Marchand V, Decroly E, Motorin Y, Bennasser Y. 2019. FTSJ3 is an RNA 2′-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature 565: 500–504. 10.1038/s41586-018-0841-4 [DOI] [PubMed] [Google Scholar]
- Robbins M, Judge A, Liang L, McClintock K, Yaworski E, MacLachlan I. 2007. 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol Ther 15: 1663–1669. 10.1038/sj.mt.6300240 [DOI] [PubMed] [Google Scholar]
- Roger T, Froidevaux C, Le Roy D, Reymond MK, Chanson AL, Mauri D, Burns K, Riederer BM, Akira S, Calandra T. 2009. Protection from lethal gram-negative bacterial sepsis by targeting Toll-like receptor 4. Proc Natl Acad Sci 106: 2348–2352. 10.1073/pnas.0808146106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh SI, Abe F, Kanno A, Tanimura N, Mori Saitoh Y, Fukui R, Shibata T, Sato K, Ichinohe T, Hayashi M, et al. 2017. TLR7 mediated viral recognition results in focal type I interferon secretion by dendritic cells. Nat Commun 8: 1592 10.1038/s41467-017-01687-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander LE, Davis MJ, Boekschoten MV, Amsen D, Dascher CC, Ryffel B, Swanson JA, Müller M, Blander JM. 2011. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature 474: 385–389. 10.1038/nature10072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt FCF, Freund I, Weigand MA, Helm M, Dalpke AH, Eigenbrod T. 2017. Identification of an optimized 2′-O-methylated trinucleotide RNA motif inhibiting Toll-like receptors 7 and 8. RNA 23: 1344–1351. 10.1261/rna.061952.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud M, Furset G, Cekaite L. 2007. Suppression of immunostimulatory siRNA-driven innate immune activation by 2′-modified RNAs. Biochem Biophys Res Commun 361: 122–126. 10.1016/j.bbrc.2007.06.177 [DOI] [PubMed] [Google Scholar]
- Tada H, Nemoto E, Shimauchi H, Watanabe T, Mikami T, Matsumoto T, Ohno N, Tamura H, Shibata K-i, Akashi S, et al. 2002. Saccharomyces cerevisiae- and Candida albicans-derived mannan induced production of tumor necrosis factor α by human monocytes in a CD14- and Toll-like receptor 4-dependent manner. Microbiol Immunol 46: 503–512. 10.1111/j.1348-0421.2002.tb02727.x [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. 1999. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11: 443–451. 10.1016/S1074-7613(00)80119-3 [DOI] [PubMed] [Google Scholar]
- Tang H, Jin X, Li Y, Jiang H, Tang X, Yang X, Cheng H, Qiu Y, Chen G, Mei J, et al. 2014. A large-scale screen for coding variants predisposing to psoriasis. Nat Genet 46: 45–50. 10.1038/ng.2827 [DOI] [PubMed] [Google Scholar]
- Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt EC. 1991. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J 10: 573–583. 10.1002/j.1460-2075.1991.tb07984.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugolini M, Gerhard J, Burkert S, Jensen KJ, Georg P, Ebner F, Volkers SM, Thada S, Dietert K, Bauer L, et al. 2018. Recognition of microbial viability via TLR8 drives TFH cell differentiation and vaccine responses. Nat Immunol 19: 386–396. 10.1038/s41590-018-0068-4 [DOI] [PubMed] [Google Scholar]
- Van Eenennaam H, Vogelzangs JHP, Bisschops L, Te Boome LCJ, Seelig HP, Renz M, De Rooij DJ, Brouwer R, Pluk H, Pruijn GJM, et al. 2002. Autoantibodies against small nucleolar ribonucleoprotein complexes and their clinical associations. Clin Exp Immunol 130: 532–540. 10.1046/j.1365-2249.2002.01991.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Bang C, Rosigkeit H, Schmitz RA, Heine H. 2017. The human-associated archaeon Methanosphaera stadtmanae is recognized through its RNA and induces TLR8-dependent NLRP3 inflammasome activation. Front Immunol 8: 1535 10.3389/fimmu.2017.01535 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.