Abstract
Metabolic networks are webs of integrated reactions organized to maximize growth and replication while minimizing the detrimental impact reactive metabolites can have on fitness. Enamines and imines, such as 2-aminoacrylate (2AA), are reactive metabolites produced as short-lived intermediates in a number of enzymatic processes. Left unchecked, the inherent reactivity of enamines and imines may perturb the metabolic network. Genetic and biochemical studies have outlined a role for the broadly conserved Rid (YjgF/YER057c/UK114) protein family, in particular RidA, in catalyzing the hydrolysis of enamines and imines to their ketone product. Herein, we discuss new findings regarding the biological significance of enamine and imine production and outline the importance of RidA in controlling accumulation of reactive metabolites.
Keywords: 2-aminoacrylate stress, enamine/imine metabolism, RidA, reactive metabolite
Metabolic networks must accommodate production of reactive metabolites
Metabolism consists of a network of biochemical reactions organized according to the chemical constraints of the cell and responsive to ever-changing environmental stimuli [1–3]. These networks are arranged to maximize the output of chemicals necessary for cell growth and survival, while minimizing the detrimental impact reactive metabolite accumulation can have on cell fitness [4–6],The biochemical pathways in central and secondary metabolism have been elucidated over the years by combinations of in vitro biochemistry and in vivo genetics. Importantly, a dichotomy exists where reactive metabolites act as obligatory catalytic intermediates, facilitating chemistry for synthesis of essential compounds. However, if left unchecked, their inherent reactivity can lead to detrimental disruption of the metabolic network. Examples of obligatory reactive intermediates include carbon monoxide in the Wood-Ljungdahl pathway of acetyl-CoA synthesis [7], acetaldehyde produced during ethanolamine catabolism [8], and nitric oxide generated by aerobic ammonia oxidation [9], among many others. Reactive metabolites can also contribute to metabolic robustness (see Glossary), leading to the emergence of non-native metabolic pathways. That is, the reactivity of certain metabolic intermediates or labile side-products, often acted upon by non-specific or promiscuous enzymes, can enable alternative methods of essential metabolite production [10, 11].
Enamines and imines are short-lived reactive metabolites produced as reaction intermediates by multiple enzymes central to amino acid metabolism [12–16]. Despite the prevalence of these reactive molecules in ubiquitous biochemical pathways, the potential for free enamine and imine species to persist in vivo was largely ignored prior to characterization of the RidA proteins as enamine deaminases [17–20]. This review summarizes the biochemical mechanisms for enamine and imine production, the in vivo consequences of enamine (specifically 2-aminoacrylate, or2AA) accumulation, and outlines the role of RidA in controlling enamine accumulation. We conclude with thoughts prompted by investigation of the RidA paradigm and propose challenges and opportunities that lie ahead in efforts to understand how reactive enamines and imines fit into emerging models of the cellular metabolic network.
Enamines and imines are reactive metabolites prevalent throughout metabolism
Enamines and imines contain unsaturated nitrogen often derived from the condensation of an aldehyde or ketone with a secondary or primary amine, respectively [21]. Peptide-bound enamines are produced biochemically via β-elimination of polypeptide residues, forming reactive peptide species that play an important role in pharmacology, enzyme engineering, and food science by facilitating formation of complex molecules [22–26]. Despite the practical use of peptide-bound enamines, the occurrence of free enamine (and imine) species in vivo and any physiological implications for the chemical make-up of the cell are poorly understood.
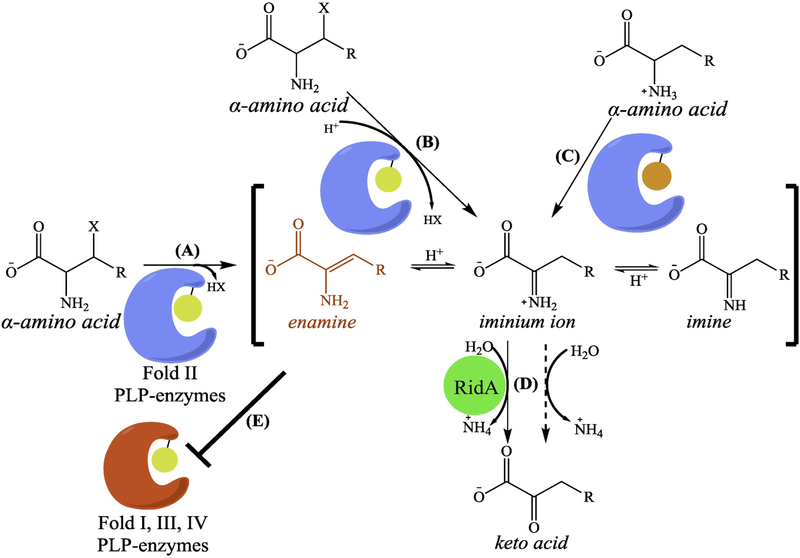
Biochemical studies show that many pyridoxal 5’-phosphate (PLP)-dependent enzymes generate enamine and/or imine intermediates from free α-amino acid substrates (Figure 1, Key Figure) [27]. These α-amino acid substrates contain electronegative side chains (e.g. L-serine, L-cysteine, 3-chloro-L-alanine, etc.), which are cleaved by dedicated (or promiscuous) α,β-eliminases to generate an enamine product. Some α,β-eliminases release an enamine directly into the reaction milieu [28, 29] (Figure 1A). This behavior is observed for a number of fold type II PLP-dependent α,β-eliminases (See Box 1 for a description of PLP enzyme fold type), such as biosynthetic serine/threonine dehydratase (IlvA, EC 4.3.1.19), cysteine desulfhydrase (EC 2.5.1.47), and diaminopropionate ammonia-lyase (EC 4.3.1.15) [15–17, 29, 30]. In some cases, the cofactor-bound enamine is protonated to an iminium ion prior to release from the enzyme (Figure 1B) [31, 32]. One example includes tryptophanase (TnaA; EC 4.1.99.1), a versatile fold type I PLP-dependent lyase responsible for tryptophan catabolism (via α,β-elimination) [33], cysteine detoxification (α,β-elimination) [34], and tryptophan synthesis (β-substitution) [35]. Iminium ions are also produced as catalytic intermediates by FAD-dependent dehydrogenases (Figure 1C). In most cases, the iminium ion intermediates are enzymatically hydrolyzed within the active site, but a number of FAD-dependent dehydrogenases release the iminium ion into the cytoplasm [19, 36]. The biological relevance of unbound enamine and imine species has generally been overlooked due to their short half-lives determined in vitro (less than 3 minutes) [37, 38]. However, recent studies have shown that accumulation of enamines, specifically 2AA, can alter the physiological state of an organism, most notably through covalent damage of PLP-dependent enzymes (Figure 1E) [39–43], To that end, many organisms encode RidA, which facilitates the catalysis of enamines and imines in vivo, as described below (Figure 1D).
Figure 1, Key Figure - Enzymatic production of free enamines and imines.
Various fold-type II PLP-dependent enzymes produce and release a reactive enamine intermediate following α,β-elimination of an amino acid precursor (A). The enamine is protonated and tautomerizes to form an iminium ion intermediate. A subset of PLP-dependent α,β-eliminases enzymatically facilitate iminium ion formation prior to its release from the active site (B). FAD- dependent enzymes can also produce an iminium ion directly (C). Equilibrium between enamine, imine, and iminium ion forms is largely dependent upon pH. In vitro, the iminium ion can react with free water, generating a stable keto acid product (D). RidA serves an important role in catalyzing the deamination of enamines and imines in vivo. Elimination of RidA leads to accumulation of free enamines, most notably 2-aminoacrylate (2AA) (R-group = H). Free 2AA can covalently modify and inactivate a number of PLP-dependent enzymes belonging to fold-type I, III, and IV (E). Enamine/imine generator and target enzymes are denoted using blue and red, respectively. Relevant coenzymes are denoted as bound circles, where PLP is colored yellow and FAD is orange.
Box 1. PLP-dependent enzymes are categorized according to fold type.
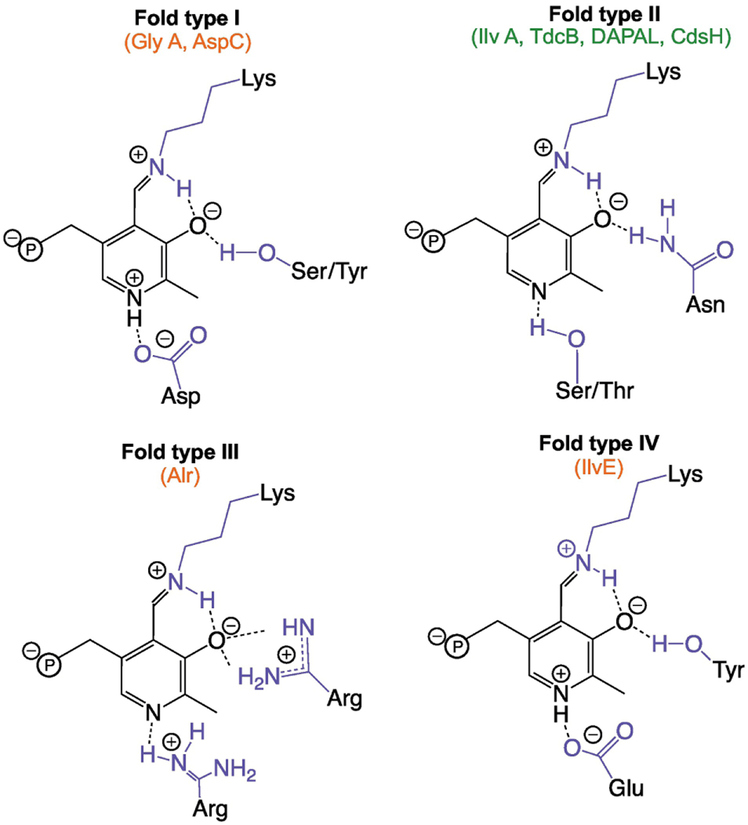
PLP-dependent enzymes are grouped according to structural similarities, which are generally indicative of catalytic mechanism and evolutionary relatedness [27]. Seven different fold types of PLP-dependent enzymes have been identified (See reviews by Schneider et al., and Percudani and Peracchi for a thorough coverage of PLP-dependent enzyme fold type) [68, 79]. Briefly, fold type I (aspartate aminotransferase family) is the most diverse and contains the largest number of aminotransferases, as well as a number of decarboxylases, lyases, and other enzymes. Fold type II (tryptophan synthase β family) contains alkyltransferases, ammonia lyases (α,β-eliminases), and a number of racemases, while fold type III (alanine racemase family) consists of all other PLP-dependent racemases and decarboxylases. The other class containing aminotransferases is fold type IV (D-amino acid aminotransferase family), which includes D-amino acid aminotransferases, L-branched chain aminotransferases, and 4-amino-4-deoxychorismate lyases. The three remaining fold types (V, glycogen phosphorylases; VI, D-lysine-5,6-aminomutases; and VII, L-lysine-2,3-aminomutases) each contain members belonging to one reaction type. The chemistry behind the breadth of reactions catalyzed by PLP-dependent enzymes, structural differences between fold-types, and their biological and industrial significance have been outlined in a number of reviews [27, 80–83]. Reaction specificity between PLP-dependent enzyme fold types is mediated through stereoelectronic control (cleaved bond must be aligned perpendicular to the PLP plane), electrophilic strength of the Schiff base, and catalytic side chain placement [83]. To the second point, the residues coordinating the pyridine nitrogen and the O3’ phenol oxygen from PLP appear to be largely responsible for controlling the electrophilic strength differences of the Schiff base between fold type I-IV enzymes, and their identities separate well with fold type and the respective reactions these enzymes catalyze (Figure I) [67, 84].
Characterization of RidA as a new enamine and imine deaminase
The Rid Superfamily and ability of RidA to hydrolyze enamine and/or imine species
The reactive intermediate deaminase (Rid) family of proteins (formally YjgF/YER057c/UK114) is a class of proteins identified by inter-subunit clefts made from seven well-conserved residues and found in all domains of life (Figure 2) [44, 45]. Recent phylogenetic analysis used synteny to split the Rid superfamily members into eight subfamilies: an archetypal RidA subfamily and seven other subfamilies (Rid1-Rid7) [36]. Rid1-Rid7 subfamily members are largely confined to bacteria and are often encoded in genomes also encoding RidA members [36]. Using the residue numbers from the Salmonella enterica ridA sequence, the most highly conserved residue across the Rid superfamily is glutamate 120 (E120). S30 and G31 are also common across all subfamilies and thus assumed to be significant in the function of Rid enzymes [36].
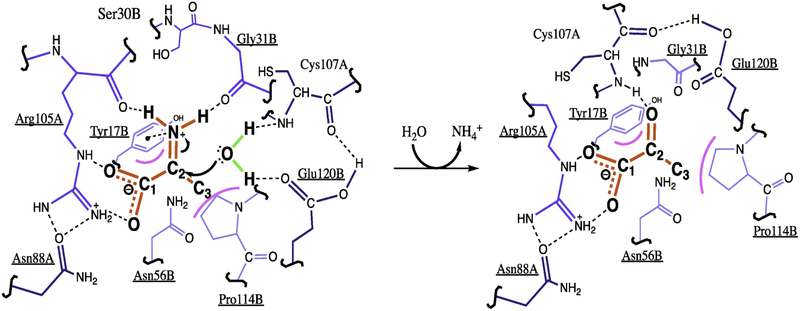
Figure 2 – Proposed reaction mechanism for RidA-mediated hydrolysis of 2-iminiopropionate (2IP).
A schematic for the proposed reaction mechanism of RidA using data extrapolated from the crystal structure of the RidA homolog, TdcF, with bound serine (2UYK) and 2-ketobutyrate (2UYN) [46]. The seven residues conserved among Rid family members are underlined. The substrate/product are shown in red, water is depicted in green, and RidA residues are shown in blue. The active site is at the interface of two subunits and residues from one subunit are labeled with an “A” while those from the other are labeled “B”. Darker blue colors indicate residues in the foreground and paler blue colors are used for those in the background. The carboxyl group of 2IP forms a salt bridge with the R105 side-chain, while the protonated imine is stabilized through hydrogen bonding with the backbone carbonyl groups from R105 and G31 (left). The imine is also stabilized through a cation-pi interaction with the phenyl group from Y17. The secondary amine from the C107 backbone and the carbonyl group from the E120 side chain activate water for its nucleophilic attack on C2 of the bound iminium ion. Following a rearrangement, pyruvate is produced in the active site, before it is released along with ammonium (right).
While multiple crystal structures of RidA homologs proved useful in identifying the putative active site of this class of proteins [46, 47], a number of genetic observations ultimately led to the identification of the biochemical function and physiological role for RidA [48–51]. Mentioned previously, S. enterica IlvA is an α,β-eliminase with activity on serine (KM = 43 mM) or threonine (KM = 6 mM) as substrates [52]. In vitro, IlvA alone produces α-ketobutyrate from threonine, or pyruvate from serine, showing that solvent water is sufficient for hydrolysis of the enamine to the corresponding keto acid [17]. The addition of purified RidA to the IlvA reactions increased the rate of ketoacid formation, showing that RidA catalyzes the hydrolysis of enamines derived from threonine (2-aminocrotonate; 2AC) and serine (2AA) [17]. These and other data culminated in the designation of RidA as an enamine/imine deaminase [49, 50]. Purified RidA orthologs from organisms across the tree of life displayed conserved enamine hydrolysis-enhancing activity in vitro, confirming the designation of this protein subfamily as enamine deaminases [17].
RidA has a broad substrate range, acting not only on 2AA and 2AC, but also on the imines of glutamine, histidine, leucine, methionine, or phenylalanine generated by FAD-dependent oxidases [19, 36]. Currently, a physiological role for RidA in the deamination of 2AC and 2AA has been identified in S. enterica and other organisms [39, 43, 53, 54], but the significance of its activity on other substrates remains to be determined.
Proposed mechanism of RidA-dependent enamine and imine catalysis
Efforts to define the catalytic residues involved in deaminase activity of RidA by site-directed mutagenesis identified a conserved arginine (R105) as the sole residue essential for function in vivo; variants altered at this site had greatly decreased activity in vitro [17]. RidA variants E120A and Y17F also had decreased deaminase activity in vitro, but retained the ability to complement ridA mutant phenotypes [17]. These data, the conserved residue observations above, and the crystal structure of the RidA homolog TdcF [46], suggested a mechanism for how RidA catalyzes the hydrolysis of iminium ion substrates. A representation of this mechanism using 2-iminiopropionate (2IP) as an example substrate is shown in Figure 2. In the mechanistic model, R105 forms a bidentate salt bridge with the carboxylic acid of the substrate. A substitution of this residue is expected to disrupt the critical interaction with the substrate carboxylic acid, consistent with the loss of activity of the RidAR105A variant [17]. R105 is absolutely conserved in RidA and Rid1-Rid3 subfamily members, predicting that members of these protein subfamilies act on amino acid-derived substrates [36]. Furthermore, Rid4–7 subfamilies lack R105 and no amino acid-derived enamine/imine deaminase activity has been detected for these subfamilies [19]. Based on the crystal structure from TdcF, the backbone carbonyl groups of R105 and G31 appear to stabilize the iminium ion (e.g. 2IP) formed from the substrate, while the backbone of C107 and the side chain of Glu-120 coordinate the water involved in hydrolysis of 2IP [46]. Although the TdcF crystal structure suggests the hydroxyl group of Y17 is too far away to hydrogen bond with the substrate, this residue may directly interact with substrate/product or play an important role in proper folding of the active site. The TdcF crystal structure also shows a number of solvent water molecules localized near Y17, making it reasonable that the Tyr hydroxyl group coordinates a water molecule near the imine nitrogen, where it acts as a proton donor for formation of ammonia [46]. Importantly, this modelproposes a mechanism for iminium ion hydrolysis. Consistent with RidA enzymes acting upon iminium ion substrates, when iminium ions are generated directly via FAD-dependent oxidases (Figure 1C), inclusion of RidA increases the rate of hydrolysis in vitro [19, 20]. However, it remains unclear whether RidA decreases enamine accumulation by binding the enamine and facilitating both iminium ion formation and subsequent hydrolysis, or whether RidA decreases enamine levels simply through increased consumption of its iminium ion tautomer [17].
Chemistry of free enamine species: from in vitro discovery to biological relevance
Inactivation of PLP-dependent enzymes by 2-aminoacrylate
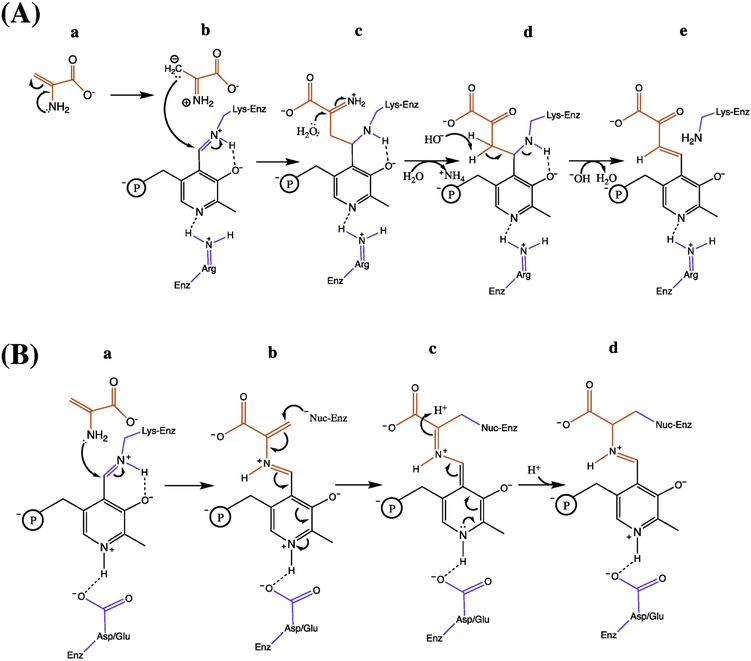
Many non-eliminase PLP-dependent enzymes are promiscuous and capable of catalyzing adventitious α,β-elimination reactions using amino acids substrates with strong electronegative leaving groups (e.g.−Cl, −SO3−) bound to their β-carbon [27]. In vitro studies showed that the 2AA generated during these promiscuous elimination reactions can immediately and irreversibly damage the source PLP-dependent enzymes before 2AA escapes the active site [55–57]. The inactivation could be caused by 1) release of 2AA from PLP and subsequent nucleophilic attack of the electrophilic enzyme-bound PLP Schiff base by the β-carbon of 2AA [56] (Figure 3A), or 2) attack of the 2AA/PLP adduct by active site nucleophilic residues [57] (Figure 3B). Both scenarios produce an irreversibly inactivated enzyme [58]. Critically, these in vitro systems contained a single enzyme that both generated, and was inactivated, by 2AA. The precedence for 2AA inactivation of PLP-dependent enzymes, coupled with biochemical characterization of RidA and genetic observations in ridA mutant backgrounds, led to a model of the biologically relevant function of RidA [17, 39]. An extrapolation of the in vitro results suggested that in vivo, PLP-dependent α,β-eliminases generated 2AA that could diffuse from the generator enzyme(s) and covalently modify susceptible PLP-dependent target enzymes [17]. The critical assumption was that, in the absence of RidA, 2AA would persist long enough in the cellular milieu to interact with and inactivate target enzymes. Given this assumption, several phenotypes of ridA mutant strains were predicted to be the consequence of 2AA-dependent inactivation of PLP-enzymes [39, 41, 42].
Figure 3 – Free 2-aminoacrylate inhibits PLP-dependent enzymes by two mechanisms.
2AA is highlighted in red and enzyme active site residues are shown in blue. The phosphate group of PLP is shown with ‘℗’. (A) A PLP/pyruvate adduct is formed following addition of 2AA via a C-C linkage. (a) Tautomeric rearrangement of 2AA generates a carbanion intermediate that (b) acts as a nucleophile to attack the 4’C of PLP. (c) Hydrolysis of the imine releases ammonium and creates a PLP/pyruvate adduct covalently bound to the active site lysine residue. (d) The adduct is susceptible to base-catalyzed formation of (e) a free PLP/pyruvate adduct. (B) 2AA/PLP covalently modifies a nucleophilic active site residue. (a) 2AA forms an external aldimine with PLP. (b) The alkene from 2AA participates in the conjugated π-bond system with PLP, strengthening the electrophilic nature of Cβ and allowing nucleophilic attack by an active site residue. (c) This forms a quinonoid intermediate, which can be rearranged and protonated to produce (d) a 2AA/PLP external aldimine covalently bound to a nucleophilic active site residue.
IlvA is the primary source of 2-aminoacrylate stress in S. enterica
The ridA mutant phenotypes were considered first with the purpose of identifying the proposed enzyme generator(s) of 2AA. S. enterica ridA mutants display a number of phenotypes, as shown in Table 1. It was particularly telling that the strain was sensitive to exogenous serine, and this sensitivity was reversed by the addition of isoleucine [48]. Isoleucine controls the activity of the serine/threonine dehydratase IlvA by feedback inhibition [59]. When the wild-type ilvA allele was replaced with an allele (ilvA219) encoding the feedback-insensitive IlvAL447F variant, isoleucine failed to reverse the serine sensitivity, indicating the effect of isoleucine was through allosteric regulation of IlvA [49, 50]. These and other data led to the key conclusion that multiple phenotypes of a ridA mutant in S. enterica were due to the IlvA-dependent generation of 2AA from serine [17, 39]. Although IlvA-dependent dehydration of L-serine is the predominant source of 2AA in S. enterica under standard growth conditions, other PLP-dependent α,β-eliminases are capable of generating substantial levels of free 2AA, when their substrates are supplied exogenously [15, 16] (Table 1).
TABLE 1.
Characterized generators and targets of 2-aminoacrylate stress in Salmonella enterica
| A | |||
|---|---|---|---|
| Endogenous 2AA generators | Gene Product |
Stressa | Reference |
| Anabolic Ser/Thr dehydratase [EC: 4.3.1.19] |
IlvA | Serine | [49] |
| Catabolic Ser/Thr dehydratase [EC: 4.3.1.19] |
TdcBb | ||
| Cysteine desulfhydratase [EC: 2.5.1.47] |
CdsH | Cysteine | [15] |
| Diaminopropionate NH4 lyase [EC: 4.3.1.15] |
DAPAL | Diaminopropionate | [16] |
| B | |||
| Endogenous 2AA targets | Gene Product |
Phenotypec | Reference |
| Serine hydroxymethyl transferase [EC: 2.1.2.1] |
GlyA | Gly limitation in minimal medium |
[85] |
| BCAA Aminotransferase [EC: 2.6.1.42] |
IlvE | Ile requirement on pyruvate carbon source |
[49] |
| Alanine racemase [EC: 5.1.1.1] |
Alr | Ala is a poor nitrogen source |
[42] |
| Alanine racemase [EC: 5.1.1.1] |
DadX | Ala is a poor nitrogen source |
[42] |
In the absence of RidA, if the listed metabolite is added to minimal glucose growth medium, the relevant generator enzyme uses it to produce 2AA and prevent cell growth.
TdcB is only produced anaerobically in response to threonine and does not contribute to 2AA stress under standard aerobic growth conditions.
2AA-dependent damage of the relevant enzyme is responsible for the listed growth phenotype in aS. enterica ridA mutant. Abbreviations: Gly:glycine, Ile:isoleucine, Ala:alanine
Free 2-aminoacrylate inactivates multiple cellular PLP-dependent enzymes
While a serine/threonine dehydratase is the dominant 2AA generator in S. enterica and in most other organisms tested thus far [43, 54, 60], multiple enzymes are potentially damaged by 2AA. For instance, branched chain amino acid aminotransferase (IlvE) activity is decreased by 30–50% in ridA mutants; this decreased activity was confirmed to be due to covalent modification of the IlvE active site by 2AA [39]. Importantly, the generation of 2AA (by IlvA), inactivation of IlvE, and preventative quenching of 2AA by RidA were all reconstituted in vitro [39]. Growth phenotypes showed that multiple metabolic pathways were compromised in a ridA strain experiencing 2AA stress (Table 1). Thus far, assays have shown that serine hydroxymethyltransferase (GlyA; EC 2.1.2.1), aspartate aminotransferase (AspC; EC 2.6.1.1), and alanine racemases (Alr/DadX; EC 5.1.1.1), in addition to branched-chain amino acid aminotransferase (IlvE; EC 2.6.1.42), have lower specific activity in ridA mutant strains than in wild type [39, 41, 42, 53]. The mechanism(s) of inactivation has not been rigorously defined for target PLP-enzymes other than IlvE. Significantly, in a ridA mutant, the activity of multiple target proteins is compromised but not eliminated, resulting in the perturbation of the metabolic network that can result in multiple pleiotropic phenotypes. For some of these phenotypes, (i.e., motility defect) the presumed target enzyme has not been identified [53, 60].
The work summarized above established the RidA paradigm in S. enterica, which in its simplest form states that 2AA produced by PLP-dependent α,β-eliminases can be deaminated by RidA. In the absence of RidA, the 2AA can 1) persist in vivo, and 2) diffuse into and inactivate distinct target PLP-dependent enzymes. This definition provided a starting point to explore the causes and implications of 2AA stress in a variety of organisms. Beyond S. enterica, enamine-related stress extends into other species, and RidA also plays an important part in controlling enamine stress for these organisms [19, 36, 43, 54, 60,61].
The conundrum of PLP enzymes being both generators and targets for 2AA.
Enzymes differ in their susceptibility to 2-aminoacrylate damage.
A dichotomy exists between PLP-dependent enzymes that generate 2AA, and those damaged by free 2AA. Presumably, 2AA generators are largely recalcitrant to 2AA-dependent damage, though these enzymes do not appear entirely immune to mechanism-based inactivation by 2AA [13, 62]. This form of inactivation involves the enzyme-catalyzed generation of 2AA from a 2AA precursor (β-substituted alanine species); 2AA is then displaced and nucleophilically attacks the enzyme used to generate it, rendering it permanently inactive. Some PLP-dependent enzymes can become sensitized to mechanism-based 2AA inactivation through site-directed mutagenesis [63]. This is exemplified by tryptophan synthase (EC 4.2.1.20), which catalyzes the last two reactions in the biosynthesis of tryptophan [64]. The PLP-dependent β-subunit of tryptophan synthase catalyzes both β-elimination and β-substitution reactions, but variant β-subunits bias reactions towards β-elimination relative to wild-type tryptophan synthase [65]. These same variants are susceptible to inactivation by 2AA produced from L-serine or 3-chloro-L-alanine, whereas the wild-type enzyme was undamaged by serine and less-damaged than the variant enzyme by 3-chloro-L-alanine [66]. These data showed that changing a single active site residue profoundly influenced susceptibility to 2AA inactivation, suggesting a fine line exists between substrate-based inactivation and successful elimination reactions. This work provides insights about how enzymes routinely exposed to 2AA could be protected from damage. The fact that enzymes proficient at α,β-elimination and 2AA generation are still susceptible tomechanism-based inactivation [13, 62], albeit less so than promiscuous eliminases [62], highlights the reactivity and potential toxicity of 2AA.
PLP-enzyme active site residues may determine susceptibility to 2-aminoacrylate damage
PLP-dependent enzymes are generally grouped into one of seven fold types according to structural similarities (Box 1) [67, 68]. Pairing 2AA interaction profiles with the relevant PLP-dependent enzymes revealed a correlation between fold type and 2AA production/inactivation; these data included four enzymes inactivated by 2AA and four that generated 2AA (Table 1). Generators, or enzymes most immune to 2AA damage, belonged to fold type II [15–17]. Conversely, enzymes damaged via the covalent addition of 2AA/PLP mechanism (Figure 3B) were fold type I and IV enzymes [39, 42], and those damaged via the PLP/pyruvate adduct mechanism (Figure 3A) were fold type III racemases [39, 41, 42]. Reactions catalyzed by fold type I and IV PLP-dependent enzymes involve a quinonoid intermediate [69]. Enzymes belonging to these fold types satisfy this requirement through placement of a negatively charged amino acid adjacent to the pyridine N from PLP, which promotes the nitrogen’s protonation and allows subsequent quinonoid formation [69]. Alternatively, L-alanine racemization (fold type III) and α,β-elimination (fold type II) do not require formation of a quinonoid intermediate [69]. Formation of a quinonoid intermediate is an essential step in the mechanism of damage via 2AA/PLP (Figure 3B), and may explain why the enzymes believed to be damaged in this way belong to fold type I and IV. In contrast, the PLP/pyruvate adduct damage mechanism (Figure 3A) requires the 4’C of PLP to act as an electrophile for successful attack by the 2AA-derived carbanion [58]. Therefore, the finding that fold type III racemases resist formation of a quinonoid intermediate may help explain why these enzymes are damaged via the PLP/pyruvate adduct mechanism and not the 2AA/PLP mechanism [70]. These data support a model where fold type II enzymes are more recalcitrant to 2AA damage via the 2AA/PLP mechanism due to their decreased propensity for forming a quinonoid species. In contrast, differences in the relative electrophilic strength of the C4’ may account for fold type II enzymes being more immune than fold type III to the PLP/pyruvate adduct mechanism of 2AA-dependent damage. Further work using variant enzymes will be valuable in better defining features of the active site that determine the susceptibility of an enzyme to 2AA damage.
Understanding the global scope of enamine/imine reactivity
The global metabolic effects of 2-aminoacrylate are largely unexplored
A number of PLP-enzymes targeted by 2AA have been identified, primarily by work in S. enterica [39, 41, 42, 61]. However, in the context of metabolic network configurations, the large number of potential targets (>40 PLP-dependent enzymes are encoded in S. enterica [67, 71]) complicates efforts to define the global physiological consequences of 2AA stress. Moreover, the detectable physiological impact (e.g., phenotype) of 2AA stress varies by organism, reflecting how organism-specific metabolic network architecture dictates 2AA damage outcome [54, 60, 61]. As a first step toward understanding the global impact of 2AA stress in S. enterica, one study used differential gene expression in a ridA mutant as a downstream indicator of the metabolic perturbation caused by 2AA stress [53]. The resulting data identified a previously unknown 2AA-dependent motility defect, but failed to reflect dramatic changes in the expected metabolic pathways linked to 2AA production or damage. These initial global efforts haveemphasized the need for innovative technical approaches to better capture the complex metabolic environment in a living cell and identify the potentially subtle consequences of 2AA stress.
RidA-independent mechanisms for quenching 2-aminoacrylate
Studies of RidA highlight the importance of reactive metabolite control mechanisms in vivo despite the spontaneous lability of some reactive metabolites in vitro. Multi-copy suppressor analyses revealed alternative, RidA-independent mechanisms of reactive enamine control. For example, when overexpressed, cystathionine β-lyase (MetC; EC 4.4.1.8) and aspartate/glutamate racemase (YgeA, MMP0739; E.C 5.1.1.13) quench 2AA in S, enterica mutants lacking RidA [30, 72]. This work suggests that these enzymes do not catalyze enamine hydrolysis but instead produce another reactive metabolite that reacts with and quenches 2AA. In the case of MetC, the data support a model where MetC acts upon a non-cystathionine sulfur-containing substrate to generate a free thiol group. The free thiol is then thought to generate a stable adduct with 2AA [30] Similar thiol-mediated reactions with enzyme-bound enamines, such as during lanthionine formation, are well established [24, 73, 74]. Alternatively, YgeA and MMP0739 contain a threonine in place of an active site cysteine residue found in well-characterized PLP-independent racemases [72]. The absence of this cysteine residue is proposed to allow formation of a carbanion intermediate that participates in a nucleophilic attack of 2AA, quenching it through derivatization [72]. This model is reminiscent of alkyl radical quenching of enzyme-bound dehydroalanine used for post-translational enzyme engineering [22, 23]. The discovery of these two alternative systems for quenching 2AA underscores a number of points. First, RidA may represent one of several strategies that have evolved to limit accumulation of reactive enamines/imines in vivo. In fact, a handful of non-RidA enzymes have been reported to quenchimines including aldehyde oxidase (AOX; EC 1.2.3.1) [75], pyrroline 5-carboxylate reductase (ProC; EC 1.5.1.2) [76], and glutamate dehydrogenase (GdhA; EC 1.4.1.4) [77]. Second, while 2AA has a demonstrated role in damaging PLP-dependent enzymes, this molecule may also perturb the metabolic network through direct interaction with other free reactive metabolites.
Concluding Remarks
The study of RidA and its role in catalyzing hydrolysis of enamines and imines has spurred a new interest in the prevalence and impact of these and other reactive species in the metabolic network. New questions have emerged concerning the strategies used by cells to control endogenous levels of enamines and imines as well as the metabolic fate and fitness impact if their accumulation is left unchecked (see Outstanding Questions). The prevalence and consequence of enamine and/or imine species in organisms remain largely unknown, and is an exciting area for future studies. The focus of this review was limited to the deleterious consequences of uncontrolled enamine accumulation. However, less-explored physiological benefits of enamine accumulation have been described [17, 78], suggesting there may be additional instances where enamines positively influence metabolic robustness. Future work probing the generation and consequences of enamines and imines in wild-type organisms, characterizing other Rid family proteins, and Rid-independent mechanisms of quenching enamines and imines will provide insights about the role of reactive metabolites and their control in the maintenance of a robust metabolic network.
Outstanding Questions.
What is the catalytic mechanism used by RidA to deaminate enamines and imines?
Do RidA/Rid family proteins act on substrates other than amino acid-derived enamines and imines?
Many organisms contain multiple Rid homologs; do these homologs serve as specialized deaminases within specific metabolic pathways, and in the case or Rid4-Rid7 enzymes, what is the catalytic function they have?
What structural features in PLP-dependent enzyme targets of 2-amnoacrylate (2AA) dictate the mechanism of damage and what factors minimize 2AA damage of the PLP-dependent enzymes that generate it?
Enzyme-bound enamines can interact with nucleophilic metabolites; does 2AA perturb the metabolic network through direct interactions with free metabolites?
Do imines or iminium ions formed from PLP-, or FAD-dependent enzymes damage enzymes or react directly with other metabolites in vivo?
Figure I.
Generalized scheme showing coordination of pyridine N and 3’O from pyridoxal 5’-phosphate (black) for the four main fold types of PLP-dependent enzymes. Since fold types V-VII contain members belonging to only one reaction type, these were excluded from the scheme. Enzyme active site residues are shown in blue and the phosphate group of PLP is shown with ‘℗’. Beneath the fold type label, generators of free 2AA are highlighted in green and enzymes damaged by free 2AA are shown in red. Note, the identified residues are provided using the listed enzymes as templates; for example, fold type III decarboxylases coordinate the pyridine N with a glutamate residue replacing the listed arginine residue. A comprehensive list of PLP coordinating residues is provided by Singh et al. [86].
Highlights.
Multiple PLP-dependent α,β-eliminases use α-amino acid substrates to generate and release enamine intermediates, specifically 2-aminoacrylate (2AA).
In the absence of RidA, 2AA accumulates and damages PLP-dependent enzymes through covalent modification.
Rid proteins are conserved in all domains of life and split into an archetypal RidA subfamily and seven other subfamilies (Rid1-Rid7). Rid4-Rid7 proteins are missing an active site arginine essential for the enamine/imine deaminase activity seen in the other subfamilies, suggesting additional uncharacterized roles for Rid enzymes.
Non-RidA enzymes, such as S. enterica cystathionine β-lyase, MetC, and the putative aspartate/glutamate racemase from S. enterica (YgeA) or Methanococcus maripaludis (MMP0739) quench 2AA in vivo, likely by producing a reactive intermediate that interacts with free 2AA.
Acknowledgements
This work was supported by the competitive grants program at the NIH (GM095837) to DMD.
Conflict of Interest
The authors wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Glossary
- α,β-eliminases
Also known as β-lyases; these enzymes abstract a proton bound to the α-carbon of amino acid substrates, leading to cleavage of a bond formed with the β-carbon, generating an enamine intermediate.
- Aminotransferases
Also known as transaminases; this group of PLP-dependent fold type I and IV enzymes exchange an amino group on one molecule with a keto group from a different molecule.
- β-elimination
A reaction where two atoms or molecular groups are cleaved from adjacent carbon atoms, resulting in formation of a new pi bond between the adjacent atoms.
- Dehydrogenases
A group enzymes that oxidize a substrate by reducing an electron acceptor, which is often in the form of NAD+/NADP+or a flavin coenzyme.
- Enamines
Unsaturated compounds containing a functional group made from an alkene covalently bound to an amine.
- Exogenous
Originating outside an organism.
- Feedback inhibition
The end product of a metabolic reaction inhibits an enzyme involved in the generation of that product by binding a site independent from the active site.
- Imines
Also known as Schiff bases, imines are unsaturated compounds containing a functional group made from a double bond between carbon and an unprotonated nitrogen.
- Iminium ion
A protonated imine, or an unsaturated compound containing a functional group made from a double bond between carbon and a protonated nitrogen.
- Mechanism-based inactivation
Enzyme inactivation that involves the catalytically-induced bioactivation of a compound to a reactive molecule, which then covalently modifies an active site residue and/or a prosthetic group of the enzyme, rendering it inactive.
- Metabolic robustness
A qualitative assessment of alternative/additional metabolic mechanisms in place enabling an organism to withstand genetic/environmental perturbations.
- Pleiotropic
An effect where a single gene impacts multiple ostensibly distinct pathways, often resulting in multiple phenotypic outcomes.
- Promiscuous enzymes
Enzymes harboring activities in addition to their dedicated activity/activities, where a dedicated activity is defined by its ability to provide an observable physiological benefit to the organism. A promiscuous enzyme is defined within a specific environmental and genetic context.
- Pyridoxal 5’-phosphate
The active form of vitamin B6, this coenzyme is used in a large number of diverse enzymatic reactions.
- Racemases
Also known as isomerase enzymes, these enzymes invert the stereochemistry around the asymmetric carbon of the substrate.
- Synteny
The physical co-localization of genetic loci on a chromosome.
- Quinonoid intermediate
A mesoionic zwitterion intermediate relating in structure to a quinone, and where the negative charge is delocalized due to the heterocyclic electron-accepting nature of the pyridine ring from PLP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Keller MA et al. (2015) The widespread role of non-enzymatic reactions in cellular metabolism. Curr. Opin. Biotechnol 34, 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Lorenzo V et al. (2015) Chemical reactivity drives spatiotemporal organisation of bacterial metabolism. FEMS Microbiol. Rev 39 (1), 96–119. [DOI] [PubMed] [Google Scholar]
- 3.Albert R et al. (2000) Error and attack tolerance of complex networks. Nature 406 (6794), 378–382. [DOI] [PubMed] [Google Scholar]
- 4.de Crecy-Lagard V et al. (2018) Newly-discovered enzymes that function in metabolite damage-control. Curr. Opin. Chem. Biol 47, 101–108. [DOI] [PubMed] [Google Scholar]
- 5.Imlay JA (2013) The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol 11 (7), 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semchyshyn HM (2014) Reactive carbonyl species in vivo: generation and dual biological effects. Sci. World J 2014, 417842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menon S and Ragsdale SW (1996) Evidence that carbon monoxide is an obligatory intermediate in anaerobic acetyl-CoA synthesis. Biochemistry 35 (37), 12119–12125. [DOI] [PubMed] [Google Scholar]
- 8.Brinsmade SR et al. (2005) Minimal functions and physiological conditions required for growth of Salmonella enterica on ethanolamine in the absence of the metabolosome. J. Bacteriol 187 (23), 8039–8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caranto JD and Lancaster KM (2017) Nitric oxide is an obligate bacterial nitrification intermediate produced by hydroxylamine oxidoreductase. Proc. Natl. Acad. Sci. U. S. A 114 (31), 8217–8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J and Copley SD (2012) Inhibitory cross-talk upon introduction of a new metabolic pathway into an existing metabolic network. Proc. Natl. Acad. Sci. U. S. A 109 (42), E2856–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bazurto JV et al. (2016) An unexpected route to an essential cofactor:Escherichia coli relies on threonine for thiamine biosynthesis. MBio 7 (1), e01840–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chargaff E and Sprinson DB (1943) Studies on the mechanism of deamination of serine and threonine in biological systems. J. Biol. Chem 151, 273–280. [Google Scholar]
- 13.Phillips AT and Wood WA (1965) The mechanism of action of 5’-adenylic acid-activated threonine dehydratase. J. Biol. Chem 240 (12), 4703–4709. [PubMed] [Google Scholar]
- 14.Clausen T et al. (1996) Crystal structure of the pyridoxal-5’-phosphate dependent cystathionine beta-lyase from Escherichia coli at 1.83 A. J. Mol. Biol 262 (2), 202–224. [DOI] [PubMed] [Google Scholar]
- 15.Ernst DC et al. (2014) Endogenous synthesis of 2-aminoacrylate contributes to cysteine sensitivity in Salmonella enterica. J. Bacteriol 196 (18), 3335–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ernst DC et al. (2016) L-2,3-diaminopropionate generates diverse metabolic stresses in Salmonella enterica. Mol. Microbiol 101 (2), 210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambrecht JA et al. (2012) Conserved YjgF protein family deaminates reactive enamine/imine intermediates of pyridoxal 5’-phosphate (PLP)-dependent enzyme reactions. J. Biol. Chem 287 (5), 3454–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niehaus TD et al. (2014) Arabidopsis and maize RidA proteins preempt reactive enamine/imine damage to branched-chain amino acid biosynthesis in plastids. Plant Cell 26 (7), 3010–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodge-Hanson KM and Downs DM (2017) Members of the Rid protein family have broad imine deaminase activity and can accelerate the Pseudomonas aeruginosa D-arginine dehydrogenase (DauA) reaction in vitro. PLoS One 12 (9), e0185544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Degani G et al. (2018) Imine deaminase activity and conformational stability of UK114, the mammalian member of the Rid protein family active in amino acid metabolism. Int. J.Mol. Sci 19 (4) 945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams JP (2000) Imines, enamines and oximes.J. Chem. Soc. Perkin Trans 1 (2), 125–139. [Google Scholar]
- 22.Wright TH et al. (2016) Posttranslational mutagenesis: A chemical strategy for exploring protein side-chain diversity. Science 354 (6312), aag1465. [DOI] [PubMed] [Google Scholar]
- 23.Yang A et al. (2016) A chemical biology route to site-specific authentic protein modifications. Science 354 (6312), 623–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Field D et al. (2015) Bioengineering lantibiotics for therapeutic success. Front Microbiol. 6,1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seebeck FP and Szostak JW (2006) Ribosomal synthesis of dehydroalanine-containing peptides. J. Am. Chem. Soc 128 (22), 7150–7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorham SD et al. (1992) Effect of chemical modifications on the susceptibility of collagen to proteolysis. II. Dehydrothermal crosslinking. Int. J. Biol. Macromol 14 (3), 129–138. [DOI] [PubMed] [Google Scholar]
- 27.Eliot AC and Kirsch JF (2004) Pyridoxal phosphate enzymes:mechanistic, structural, and evolutionary considerations. Annu. Rev. Biochem 73, 383–415. [DOI] [PubMed] [Google Scholar]
- 28.Flavin M and Slaughter C (1969) Enzymic reactions of enamines with N-ethylmaleimide.J. Biol. Chem 244 (6), 1434–1444. [PubMed] [Google Scholar]
- 29.Datta P and Bhadra R (1978) Biodegradative threonine dehydratase. Reduction of ferricyanide by an intermediate of the enzyme-catalyzed reaction. Eur. J. Biochem 91 (2), 527–532. [DOI] [PubMed] [Google Scholar]
- 30.Ernst DC et al. (2018) Increased activity of cystathionine beta-lyase suppresses 2-aminoacrylate stress in Salmonella enterica. J. Bacteriol 200 (9), e00040–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hillebrand GG et al. (1979) Formation of an intermediate and its rate of conversion to pyruvate during the tryptophanase-catalyzed degradation of S-o-nitrophenyl-L-cysteine.Biochemistry 18 (9), 1751–1755. [DOI] [PubMed] [Google Scholar]
- 32.Vederas JC et al. (1978) Stereochemistry and mechanism of reactions catalyzed by tryptophanase from Escherichia coli. J. Biol. Chem 253 (15), 5350–5354. [PubMed] [Google Scholar]
- 33.Newton WA and Snell EE (1964) Catalytic properties of tryptophanase, a multifunctional pyridoxal phosphate enzyme. Proc. Natl. Acad. Sci. U. S. A 51, 382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Awano N et al. (2005) Identification and functional analysis of Escherichia coli cysteine desulfhydrases. Appl. Environ. Microbiol 71 (7), 4149–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan RK et al. (1972) Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II Properties of a high transducing lysate. Virology 50, 883–898. [DOI] [PubMed] [Google Scholar]
- 36.Niehaus TD et al. (2015) Genomic and experimental evidence for multiple metabolic functions in the RidA/YjgF/YER057c/UK114 (Rid) protein family. BMC Genomics 16, 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flavin M and Slaughter C (1964) An intermediate trapped by maleimides in a pyridoxal-phosphate potentiated enzymatic elimination reaction. Biochemistry 3, 885–93. [DOI] [PubMed] [Google Scholar]
- 38.Hillebrand GGD, James L; Suelter, Clarence H (1979) Formation of an intermediate and its rate of conversion to pyruvate during the tryptophanase-catalyzed degradation of S-o-Nitrophenyl-L-cysteine. Biochemistry 18 (9), 1751–1755. [DOI] [PubMed] [Google Scholar]
- 39.Lambrecht JA et al. (2013) RidA proteins prevent metabolic damage inflicted by PLP-dependent dehydratases in all domains of life. MBio 4 (1), e00033–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lambrecht JA and Downs DM (2013) Anthranilate phosphoribosyl transferase (TrpD) generates phosphoribosylamine for thiamine synthesis from enamines and phosphoribosyl pyrophosphate. ACS Chem. Biol 8 (1), 242–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flynn JM et al. (2013) Decreased coenzyme A levels in ridA mutant strains of Salmonella enterica result from inactivated serine hydroxymethyltransferase. Mol. Microbiol 89 (4), 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flynn JM and Downs DM (2013) In the absence of RidA, endogenous 2-aminoacrylate inactivates alanine racemases by modifying the pyridoxal 5’-phosphate cofactor. J Bacteriol. 195 (16), 3603–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niehaus TD et al. (2014) Arabidopsis and maize RidA proteins preempt reactive enamine/imine damage to branched-chain amino acid biosynthesis in plastids. Plant Cell 26 (7), 3010–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leitner-Dagan Y et al. (2006) CHRD, a plant member of the evolutionarily conserved YjgF family, influences photosynthesis and chromoplastogenesis. Planta 225 (1), 89–102. [DOI] [PubMed] [Google Scholar]
- 45.Parsons L et al. (2003) Solution structure and functional ligand screening of HI0719, a highly conserved protein from bacteria to humans in the YjgF/YER057c/UK114 family.Biochemistry 42 (1), 80–89. [DOI] [PubMed] [Google Scholar]
- 46.Burman JD et al. (2007) The crystal structure of Escherichia coli TdcF, a member of the highly conserved YjgF/YER057c/UK114 family. BMC Struct. Biol 7,30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Volz K (1999) A test case for structure-based functional assignment: the 1.2 A crystal structure of theyjgFgene product from Escherichia coli. Prot. Sci 8 (11), 2428–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Enos-Berlage JL et al. (1998) Complex metabolic phenotypes caused by a mutation in yjgF, encoding a member of the highly conserved YER057c/YjgF family of proteins. J. Bacteriol 180 (24), 6519–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmitz G and Downs DM (2004) Reduced transaminase B (IlvE) activity caused by the lack of yjgF is dependent on the status of threonine deaminase (IlvA) in Salmonella enterica Serovar Typhimurium. J. Bacteriol 186 (3), 803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Christopherson MR et al. (2008) YjgF is required for isoleucine biosynthesis when Salmonella enterica is grown on pyruvate medium. J. Bacteriol 190 (8), 3057–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christopherson MR et al. (2012) Suppressor analyses identify threonine as a modulator of ridA mutant phenotypes in Salmonella enterica. PLoS One 7 (8), e43082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borchert AJ and Downs DM (2018) Analyses of variants of the Ser/Thr dehydratase IlvA provide insight into 2-aminoacrylate metabolism in Salmonella enterica. J. Biol. Chem 293 (50), 19240–19249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borchert AJ and Downs DM (2017) Endogenously generated 2-aminoacrylate inhibits motility in Salmonella enterica. Sci Rep 7 (1), 12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ernst DC and Downs DM (2018) Mmf1p couples amino acid metabolism to mitochondrial DNA maintenance in Saccharomyces cerevisiae. MBio 9 (1), e00084–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walsh C (1982) Suicide substrates: mechanism-based enzyme inactivators.Tetrahedron 38, 871–909. [DOI] [PubMed] [Google Scholar]
- 56.Likos JJ et al. (1982) A novel reaction of the coenzyme of glutamate decarboxylase with L-serine O-sulfate. Biochemistry 21 (18), 4377–4386. [DOI] [PubMed] [Google Scholar]
- 57.Ueno H et al. (1982) Chemistry of the inactivation of cytosolic aspartate aminotransferase by serine O-sulfate. Biochemistry 21 (18), 4387–4393. [DOI] [PubMed] [Google Scholar]
- 58.Esaki N and Walsh CT (1986) Biosynthetic alanine racemase of Salmonella typhimurium: purification and characterization of the enzyme encoded by the alr gene. Biochemistry 25 (11), 3261–3267. [DOI] [PubMed] [Google Scholar]
- 59.Eisenstein E (1995) Allosteric regulation of biosynthetic threonine deaminase from Escherichia coli: effects of isoleucine and valine on active-site ligand binding and catalysis. Arch. Biochem. Biophys 316 (1), 311–8. [DOI] [PubMed] [Google Scholar]
- 60.Irons J et al. (2018) PA5339, a RidA homolog, is required for full growth in Pseudomonas aeruginosa. J. Bacteriol 200 (22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Borchert AJ and Downs DM (2017) The response to 2-aminoacrylate differs in Escherichia coli and Salmonella enterica, despite shared metabolic components. J. Bacteriol 199 (14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arfin SM and Koziell DA (1971) Inhibition of growth of Salmonella typhimurium and of threonine deaminase and transaminase B by beta-chloroalanine. J. Bacteriol 105 (2), 519–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ahmed SA et al. (1991) Mechanism of mutual activation of the tryptophan synthase-alpha and beta-subunits - analysis of the reaction specificity and substrate-induced inactivation of active-site and tunnel mutants of the beta-subunit. J. Biol. Chem 266 (32), 21548–21557. [PubMed] [Google Scholar]
- 64.Schleicher E et al. (1976) Letter: Stereochemistry and mechanism of reactions catalyzed by tryptophanase and tryptophan synthetase. J. Am. Chem. Soc 98 (4), 1043–1044. [DOI] [PubMed] [Google Scholar]
- 65.Jhee KH et al. (1998) Mutation of an active site residue of tryptophan synthase (beta-serine 377) alters cofactor chemistry. J. Biol. Chem 273 (19), 11417–11422. [DOI] [PubMed] [Google Scholar]
- 66.Jhee KH et al. (1998) Tryptophan synthase mutations that alter cofactor chemistry lead to mechanism-based inactivation. Biochemistry 37 (41), 14591–14604. [DOI] [PubMed] [Google Scholar]
- 67.Percudani R and Peracchi A (2003) A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 4 (9), 850–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schneider G et al. (2000) The manifold of vitamin B-6 dependent enzymes. Struct. Fold. Des 8 (1), R1–R6. [DOI] [PubMed] [Google Scholar]
- 69.Griswold WR and Toney MD (2011) Role of the pyridine nitrogen in pyridoxal 5′-phosphate catalysis: activity of three classes of PLP enzymes reconstituted with deazapyridoxal 5’-phosphate. J. Am. Chem. Soc 133 (37), 14823–14830. [DOI] [PubMed] [Google Scholar]
- 70.Watanabe A et al. (2002) Reaction mechanism of alanine racemase from Bacillus stearothermophilus: x-ray crystallographic studies of the enzyme bound with N-(5′-phosphopyridoxyl)alanine. J. Biol. Chem 277 (21), 19166–19172. [DOI] [PubMed] [Google Scholar]
- 71.Karp PD et al. (2017) The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hodge-Hanson KM et al. (2018) Expression of PLP-independent racemases can reduce 2-aminoacrylate stress in Salmonella enterica. J. Bacteriol 200 (9), e00751–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cotter PD et al. (2005) Bacterial lantibiotics: strategies to improve therapeutic potential. Curr. Protein Pept. Sci 6 (1), 61–75. [DOI] [PubMed] [Google Scholar]
- 74.Cavera VL et al. (2015) Bacteriocins and their position in the next wave of conventional antibiotics. Int J Antimicrob Agents 46 (5), 494–501. [DOI] [PubMed] [Google Scholar]
- 75.Brandange S and Lindblom L (1979) The enzyme “aldehyde oxidase” is an iminium oxidase. Reaction with nicotine delta 1’(5’) iminium ion.Biochem. Biophys. Res Commun 91 (3), 991–996. [DOI] [PubMed] [Google Scholar]
- 76.Isenberg S and Newman EB (1974) Studies on L-serine deaminase in Escherichia coli K-12. J. Bacteriol 118 (1), 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fisher HF et al. (1982) Glutamate dehydrogenase catalyzes the reduction of a Schiff base (delta 1-pyrroline-2-carboxylic acid) by NADPH. J. Biol. Chem 257 (22), 13208–13210. [PubMed] [Google Scholar]
- 78.Bazurto JV et al. (2017) Untargeted metabolomics confirms and extends the understanding of the impact of aminoimidazole carboxamide ribotide (AICAR) in the metabolic network of Salmonella enterica. Microb. Cell 5 (2), 74–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Percudani R and Peracchi A (2009) The B6 database: a tool for the description and classification of vitamin B6-dependent enzymatic activities and of the corresponding protein families. BMC Bioinformatics 10, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Merigliano C et al. (2018) The relationship between vitamin B6, diabetes and cancer Front. Genet. 9, 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Phillips RS (2015) Chemistry and diversity of pyridoxal-5’-phosphate dependent enzymes. Biochim. Biophys. Acta 1854 (9), 1167–1174. [DOI] [PubMed] [Google Scholar]
- 82.Steffen-Munsberg F et al. (2015) Bioinformatic analysis of a PLP-dependent enzyme superfamily suitable for biocatalytic applications. Biotechnol. Adv 33 (5), 566–604. [DOI] [PubMed] [Google Scholar]
- 83.Toney MD (2011) Controlling reaction specificity in pyridoxal phosphate enzymes BBA-Proteins Proteom. 1814 (1), 1407–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Casasnovas R et al. (2012) C-H activation in pyridoxal-5’-phosphate Schiff bases: the role of the imine nitrogen. A combined experimental and computational study. J. Phys Chem. B 116 (35), 10665–10675. [DOI] [PubMed] [Google Scholar]
- 85.Ernst DC and Downs DM (2016) 2-aminoacrylate stress induces a context-dependent glycine requirement in ridA strains of Salmonella enterica. J. Bacteriol 198 (3), 536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Singh R et al. (2013) Chemogenomics of pyridoxal 5 ‘-phosphate dependent enzymes. J Enzym. Inhib. Med. Ch 28 (1), 183–194. [DOI] [PubMed] [Google Scholar]