Abstract
Mycoplasmas are important model organisms for Systems and Synthetic Biology, and are pathogenic to a wide variety of species. Despite their relevance, many of the tools established for genome editing in other microorganisms are not available for Mycoplasmas. The Tn4001 transposon is the reference tool to work with these bacteria, but the transformation efficiencies (TEs) reported for the different species vary substantially. Here, we explore the mechanisms underlying these differences in four Mycoplasma species, Mycoplasma agalactiae, Mycoplasma feriruminatoris, Mycoplasma gallisepticum and Mycoplasma pneumoniae, selected for being representative members of each cluster of the Mycoplasma genus. We found that regulatory regions (RRs) driving the expression of the transposase and the antibiotic resistance marker have a major impact on the TEs. We then designed a synthetic RR termed SynMyco RR to control the expression of the key transposon vector elements. Using this synthetic RR, we were able to increase the TE for M. gallisepticum, M. feriruminatoris and M. agalactiae by 30-, 980- and 1036-fold, respectively. Finally, to illustrate the potential of this new transposon, we performed the first essentiality study in M. agalactiae, basing our study on more than 199,000 genome insertions.
Keywords: Mycoplasma, transposon, regulatory region, transformation efficiency, essentiality
1. Introduction
The Mollicutes class represents a taxonomic group of bacteria that has undergone an extreme genome downsizing process termed degenerative evolution.1 As a consequence of this evolutionary process, these bacteria are characterized by the lack of a cell wall, streamlined genomes and very limited biosynthetic pathways. All these features have turned Mollicutes, and particularly some species encompassed within the Mycoplasma genus, into appealing models to study basic principles of complex cellular processes, such as transcription and translation. For instance, Mycoplasma pneumoniae is an important model organism for Systems Biology as highlighted by the comprehensive knowledge acquired about its complete genome,2 transcriptome,3 proteome,4 DNA methylome5 and gene essentiality.6 In addition, the first whole-cell computational model developed was for Mycoplasma genitalium.7 At the same time, Mycoplasmas are also interesting organisms for the emerging field of Synthetic Biology and to study the minimal set of genes required to sustain life.6,8–10 Indeed, the genome of the first synthetic bacterium, JCVI Syn3.0, was inferred from the genome of Mycoplasma mycoides following a top-down approach in a design-build and test cycle.11 Aside from their relevance as model organisms, Mycoplasmas are also a serious concern for the medical and veterinary fields given their pathogenic effects on a wide variety of species. For instance, M. pneumoniae is a human pathogen that causes atypical pneumonia,7,12Mycoplasma gallisepticum causes pneumonia in poultry13 and Mycoplasma agalactiae causes contagious agalactia, a common disease in small ruminants with a tremendous economic impact.14
Identifying genes involved in pathogenicity is critical for generating new vaccines and therapies. Genes that are dispensable for in vitro growth (i.e. non-essential, NE) but essential (E) for the infection process could be target genes for vaccine design. Construction of essentiality maps is a fast way to decipher the essential or non-essential character of every single gene found in the genome of a given bacterium. Furthermore, these maps can also help in the identification of those genes that are not essential but whose disruption affects the fitness of the bacteria in a particular environment (i.e. fitness, F). The generation of these maps relies on the construction of saturating libraries of transposon mutants, followed by high-throughput insertion tracking by ultra sequencing (HITS) at different passages.6,15 Furthermore, construction of saturating libraries is also relevant for Haystack mutagenesis,16 a technique useful to establish gene–function relationships that is widely employed in Mycoplasmas, as a consequence of the paucity of other genome editing tools for these bacteria.17
Tn4001 is a gentamicin, tobramycin and kanamycin resistance-conferring transposon that was originally found in Staphylococcus aureus,18 but has been widely employed in Mycoplasmas.6,10,19 Few modifications have been made to this transposon to adapt it for use in Mycoplasmas aside from (i), placing the transposase coding gene outside the inverted repeats (in plasmids termed mini-transposons) to prevent re-excision from the transposon after the first transposition event,20 and (ii) replacing the original gentamicin resistance marker by tetracycline,18 chloramphenicol21 or puromycin22 resistance genes. Such a poor adaptation of the Tn4001 transposon to Mycoplasmas might partially explain the dramatic differences observed among transformation efficiencies (TEs) in species from this genus. For example, TE is consistently higher in those species belonging to the pneumoniae cluster (i.e. species closely related to M. pneumoniae)23 than in those species encompassed in the spiroplasma24 or hominis clusters.25
Here, we explored and identified the reasons underlying the lower transposon TE in a set of different Mycoplasma species that differ in their phylogenetic distance to M. pneumoniae. We hypothesized that poor recognition of certain gene regulatory regions (RRs) in the transposon vector might limit TE in some species. Therefore, we rationally engineered a vector variant that significantly increased the TE in all species tested except for M. pneumoniae, which was already transformed at high efficiencies with the unmodified vector. These species were selected to encompass the three different clusters described in the Mycoplasma phylogenetic tree, thereby opening up the door to more global studies of Mycoplasma species. Furthermore, a reporter assay allowed us to identify which of the RRs determining expression of the antibiotic resistance and transposase protein coding genes found in the native transposon vector were inefficiently recognized in each of the strains selected for this work. Finally, to show the potential of our transposon vector, we performed an essentiality study of M. agalactiae, obtaining an insertional coverage similar to the one reported for M. pneumoniae.6
2. Materials and methods
2.1. Bacteria strains and culture conditions
For wild-type M. pneumoniae, the strain M129 (ATTC 29342, subtype 1, broth passage no. 35) was used. Wild-type M. agalactiae 7784 was kindly provided by Christine Citti. Wild-type M. gallisepticum R high was kindly provided by Michael Szostak.25 Wild-type Mycoplasma feriruminatoris G5847 was kindly provided by Carole Lartigue.26 All strains were grown at 37°C in standard Hayflick medium supplemented with 100 µg/ml ampicillin and phenol red (0.005% w/v). M. agalactiae standard Hayflick culture was supplemented with sodium pyruvate (0.5%, pH 7.6, Sigma-Aldrich). M. feriruminatoris, M. agalactiae and M. gallisepticum were grown in suspension (180 rpm, 37°C). M. pneumoniae was grown without shaking at 37°C and with 5% CO2.
2.2. Plasmids
The plasmids were generated following the Gibson assembly method.27 The list of plasmids as well as a detailed description of the procedure followed to build all the constructs are described in Supplementary Table S1. For plasmid generation, DNA was isolated from NEB® 5-alpha High Efficiency (C2987P). The clones were isolated using LB agar + ampicillin (100 µg/ml) plates and confirmed by sequencing (GATC biotech). The list of all the primers used in this study for the generation of the plasmids can be found in Supplementary Table S2.
2.3. Transformation protocol
Transformation procedures were initially based on methods previously described but later slightly modified.24 For M. feriruminatoris, M. agalactiae and M. gallisepticum cultures, 10-ml log-phase cultures were harvested at 10,000 g for 10 min at 4°C. The medium was removed and 10 ml of fresh Hayflick was added. After 3 h, the culture was centrifuged at 10,000 g at 4°C and then washed three times with chilled electroporation buffer (EB; 272 mM sucrose, 8 mM HEPES, pH 7.4) before final resuspension in 300 µl chilled EB. After mixing 50 µl of cells with 1.5 µg of DNA and incubating for 20 min on ice, the mix was transferred into 0.1-cm electro cuvettes and electroporated in a BIO-RAD Gene Pulser Xcell apparatus. For M. pneumoniae (i.e. adherent strain) cells were grown in a 75-cm2 tissue flask containing 20 ml of fresh Hayflick and incubated at 37°C under 5% CO2 until late exponential phase. Cells were washed twice, resuspended in precooled EB, scraped off and passed through a 25-gauge (G25) syringe needle 10 times. Aliquots of 50 µl of cells in 0.1-cm cuvettes with 1.5 µg of the corresponding plasmid were kept on ice during 20 min. The electroporation settings were common for all strains: 1250 V/25 µF/100 Ω. Immediately after the pulse, 420 µl of fresh Hayflick was added to the cells. Subsequently, the cells were incubated at 37°C before seeding on agar plates. The incubation time was 30 min for M. feriruminatoris, 90 min for M. agalactiae and M. gallisepticum and 120 min for M. pneumoniae.
2.4. Determination of transformation efficiency in Mycoplasma species
After incubating the transformations at 37°C during the above-mentioned time depending on the strain, 10-fold serial dilutions of the cultures were performed (from −1 to −8). The dilutions were made in a total volume of 100 µl and 10 µl of each dilution was plated onto Hayflick 0.8% agar plates. Transformations done with pMTnGm or pMTnGm-SynMyco vectors were counted on agar plates supplemented with 100 µg/ml gentamicin. Transformations done with pMTnCm vector were selected in agar plates supplemented with 20 µg/ml chloramphenicol. The mutant counts refer to mutants per transformation, where the final volume is 500 µl. TE is defined as the ratio of colony forming units (CFUs) counted on antibiotic supplemented plates (i.e. transformed cells carrying a transposon insertion) to non-supplemented plates (i.e. viable cells after transformation). To assess differences in TE between pMTnGm and pMTnCm, one-tailed paired t-tests were applied to three different experimental replicates for each strain employed in the study. As the mutants obtained for M. agalactiae, M. gallisepticum and M. feriruminatoris using pMTnCm transposon were below the limit of detection, for the statistical analysis, the number of mutants obtained was set to 49 CFU per batch, a value that corresponds to the maximum number of CFU under the limit of detection for each species. The P-values obtained for TE differences between pMTnGm and pMTnCm were 0.470, 0.186, 0.026 and 0.067 for M. feriruminatoris, M. agalactiae, M. gallisepticum and M. pneumoniae, respectively (for the raw data of these transformations as well as the statistical analysis, see Supplementary Table S3). The same statistical approach was applied to compare TE obtained with pMTnGm versus pMTnGm-SynMyco in three different experimental replicates for each species employed in the study (P-values of 1.49 × 10−4, 4.28 × 10−2, 4.97 × 10−2 and 0.325 for M. feriruminatoris, M. agalactiae, M. gallisepticum and M. pneumoniae, respectively). Data of TEs obtained with pMTnGm and pMTnGm-SynMyco as well as the statistical analysis are shown in Supplementary Table S4.
2.5. Phylogenetic analysis of selected Mycoplasma species
For the generation of the phylogenetic tree, 21 Mycoplasma species were selected. DNA sequences encoding the 16S rRNA of each species were aligned using multiple sequence alignment by ClustalW. For those species containing more than a single copy coding for the 16S rRNA, we selected one of the copies arbitrarily. The evolutionary history of these microorganisms was inferred using the neighbour-joining method.28 The optimal tree with the sum of branch length = 0.91 is shown. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (500 iterations) are shown next to the branches. The evolutionary distances were computed using the maximum composite likelihood method29,30 and are in the units of the number of base substitutions per site. Codon positions included were 1st + 2nd + 3rd + non-coding. All positions containing gaps and missing data were eliminated. There were a total of 1,464 positions in the final dataset. Evolutionary analyses were conducted in MEGA X.31 The 21 different DNA sequences coding for 16S rRNA and the accession number of the species to which they belong are listed in Supplementary Table S5, except for M. feriruminatoris whose assembled genome sequence was kindly provided by Dr Carole Lartigue.
2.6. Ribosome-binding site inclusion along different Mycoplasma species
As a reference we used the same set of 21 Mycoplasma species that were used to build up the phylogenetic tree. Specifically, for each species we extracted the 15 bases before the start codon of all their annotated genes. Within these sequences, we then checked for the presence of any of the subsequences reported to be RBS.32 This list included: GGA, GAG, AGG, AGGA, GGAG, GAGG, AGGAG, GGAGG, AGAAGG, AGCAGG, AGGAGG, AGTAGG, AGGCGG, AGGGGG and AGGTGG. Inclusion was represented as the percentage of genes in a bacterial species that included one of these RBS motifs (for the percentage of RBS inclusion of the species of the work, see Supplementary Table S6).
2.7. Strength evaluation of native regulatory regions driving the expression of the transposase and gentamicin resistance gene in pMTnGm, and comparison with SynMyco regulatory region in different Mycoplasma species
Three different reporter plasmids containing the mCherry coding sequence under the control of the three different RRs (i.e. pMTnGm-SynMyco+SynMyco RR-Venus+SynMyco RR-mCherry, pMTnGm-SynMyco+SynMyco RR-Venus+GmR-mCherry and pMTnGm-SynMyco+SynMyco RR-Venus+Tnp RR-mCherry) were generated. These plasmids contain genes coding for two fluorescent proteins (i.e. Venus and mCherry). These proteins have fused in its N-terminal part the mp-200 sequence, a 29 amino acid signal from M. pneumoniae MPN391a gene that has been previously fused to Venus and mCherry sequences to improve protein stability.33 Whereas the gene coding for the Venus fluorescent protein was included for normalization purposes in all constructs under the control of SynMyco RR, the gene coding for mCherry has been placed under the control of three different RRs depending on the construct: (i) the SynMyco RR, (ii) the 150 bp upstream region of the transposase coding gene (Tnp RR) and (iii) the 150 upstream region of the gentamicin resistance coding gene (Gm RR).
All constructs were transformed in M. agalactiae, M. gallisepticum, M. feriruminatoris and M. pneumoniae following the protocol described above. To generate a primary stock of the transformations, from each 500 µl batch 100 µl was inoculated in a flask containing 5 ml of Hayflick medium supplemented with 100 µg/ml gentamicin for M. pneumoniae, while for the other non-adherent strains the 100 µl was inoculated in 50 ml Falcon tubes (Fisher Scientific, 14-432-22) filled with 10 ml of Hayflick medium supplemented with 100 µg/ml gentamicin. When cultures were grown, the total biomass was resuspended in 1 ml of Hayflick medium and stored at −80°C.
To determine the fluorescence levels of each construct, cultures of the four different Mycoplasma strains carrying the three different reporter constructs plus a negative control for each strain not carrying any construct were used. The cultures were grown as described above using 10 µl of their respective primary stocks as inoculum (i.e. around 12 h for M. feriruminatoris, 48 h for M. gallisepticum and M. agalactiae and 72 h for M. pneumoniae) and cells were harvested and washed twice with chilled PBS buffer until final resuspension of the cultures in 500 µl of PBS. Five-fold serial dilutions of the final cell suspensions were done, and 100 µl of all dilutions were loaded in 96-Well Optical Btm Plt Polymerbase Black Lid plates (165305, Thermo Scientific).
The absorbance and fluorescence values were measured using Tecan I-control 1.9.17.0 Infinite 200. The settings were determined for optimal gain, 25 flashes and 20 µs of integration time. The fluorescence settings were ƛex = 514 nm and ƛem = 574 nm for Venus and ƛex = 550 nm and ƛem = 630 nm for mCherry. For each strain, the absorbance at ƛ = 600 nm was measured. For the fluorescence analysis we took the data of those wells (i.e. dilutions) in which OD600nm absorbance values were between 0.075–0.2; 0.2–0.35; 0.075–0.4 and 0.22–0.66 for M. feriruminatoris, M. agalactiae, M. pneumoniae and M. gallisepticum, respectively. These dilutions were selected so that fluorescent signals were clearly different from negative controls of each strain (i.e. not carrying fluorescent constructs) and proportional to the absorbance (i.e. not saturating signals). Fluorescence arbitrary units (AU) measured for Venus and mCherry were normalized to OD600nm for each condition (Venus AU/OD600 and mCherry AU/OD600) to obtain normalized fluorescence AU. For each of the four strains of the work, the mCherry and Venus fluorescence levels of WT cells (i.e. not carrying any fluorescent construct) were determined and subtracted from the fluorescence values obtained for each condition, to obtain subtracted AU. Finally, to compare the strength of all RRs analysed we calculated for each strain the ratio between normalized and subtracted mCherry UA/normalized and subtracted Venus UA. Statistical paired t-test analysis with one-tailed distribution was performed for each strain comparing the ratio of mCherry AU/Venus AU for (i) SynMyco-mCherry versus Gm RR-mCherry and (ii) SynMyco-mCherry versus Tnp RR-mCherry. For the data of the strength evaluation of SynMyco RR and the native RR driving the expression of the gentamicin and the transposase coding genes in the native pMTnGm as well as the statistical analysis, see Supplementary Table S7.
2.8. M. agalactiae pMTnGm-SynMyco transformation for essentiality study
M. agalactiae was transformed using the pMTnGm-SynMyco transposon following the protocol described above. Two hours after the transformation, ⅘ parts of the total 500 µl batch were inoculated into 10 ml Hayflick + 0.5% sodium pyruvate (Sigma-Aldrich P8574-5G) + 100 µg/ml gentamicin. After 24 h at 37°C, the culture was centrifuged for 10 min at 10,000 g. The supernatant was discarded and the cells were collected in 500 µl of Hayflick constituting passage 1 of the transformation. The procedure described above was repeated two more times until passage 3, using a volume of 8 µl of the immediately preceding passage, to grow all the passages (1/62.5 dilution). Taking into account the doubling time of M. agalactiae (4–5 h), and the passages performed the expected number of cell divisions in passage 3 is 20. From passage 3, 350 µl out of the total 500 µl were taken to extract genomic DNA using the MasterPure Complete DNA and RNA Purification Kit (MC85200, Lucigen) following the protocol described by the manufacturer.
2.9. Genome assembly and de novo annotation for M. agalactiae 7784
M. agalactiae was grown 48 h in 25 ml of Hayflick medium supplemented with sodium pyruvate at 37°C as described previously. After, the genomic DNA was isolated using the MasterPure Complete DNA and RNA Purification Kit (MC85200, Lucigen) following the protocol described by the manufacturer. The genomic DNA was sheared to 200–300 bp fragments using a Covaris S2 device. Then, paired-end Illumina libraries were created following previously described protocols34 and the size selected was 125 bp. The resulting libraries were quantified on an Agilent Bioanalyzer chip (Agilent Technologies). Double-stranded templates were amplified and sequenced on an Illumina GAII. Raw reads were analysed using the FastQC tool (website: http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc) for assessing the quality and the presence of adapters. Contigs were de novo assembled using ABySS35 and M. agalactiae 5632 genome as annotation reference resulting in 159 contigs with average length of 5,511 bp. In total, 503 genes present in M. agalactiae 5632 displayed homologs within contigs of M. agalactiae 7784. We increased this value up to 689 with a specific BlastN search where we restrictively selected hits with an alignment length greater than 95% and an e-value less than 1 × 10−5. This allowed us to provide a more accurate list of genes presented by the strain of interest to assign putative functions by homology as well as to detect gene duplications in the studied genome. We observed that 12 contigs were informative enough to capture a set of 689 genes. This set of contigs averaged 71,163 bp each in length and 853,960 bp in total. The latter value was considered to be the genome size for M. agalactiae 7784. The raw data of DNAseq, genome assembly and de novo annotation have been submitted as BioProject under accession PRJNA528179.
2.10. Sequencing of M. agalactiae 7784 transformed with pMTnGm-SynMyco
Genomic DNA sequencing was performed in the Genomics facility at Centre for Genomic Regulation in a HiSeq Sequencing v4 Chemistry controlled by Software HiSeq Control Software 2.2.58. Sequencing settings were fixed at 125 nucleotides in paired-end format. In the HiSeq sequencing technology from Illumina Genome Analyzer, the protocol starts with DNA fragmentation. Then, the fragmented DNA is amplified using oligos that add adapters allowing the subsequent binding of PCR products to the glass flow cell. Later, the sequencing is performed by synthesis cycles, in which a single complementary base for each deoxynucleotide (dNTP) is incorporated using a fluorescently labeled dNTP. Finally, lasers excite the fluorophores while a camera captures images of the flow cell.
2.11. Transposon mapping
For M. agalactiae 7784, paired-end sequencing raw reads were filtered to remove PCR duplicates using Fastuniq.36 Then, a specific inverted repeat (IR) associated to the transposon insertion process (TTTTACACAATTATACGGACTTTATC, length = 26) was trimmed by Trimmomatic37 and then mapped to the reference using Bowtie2 allowing 1 mismatch.38 Later, we selected paired reads mapped unambiguously with a minimum alignment quality of 30 using SAMtools.39 The last step relied on shell text processing tools (awk/grep/sed) to identify those pairs where one of the reads presented a shorter length than the original read length minus 26 (expected length if the IR was removed). Position reported represents the first mapped position in the chromosome contiguous to the IR.
2.12. Essentiality definition and training set generation for M. agalactiae and M. pneumoniae
The essentiality studies were developed using as reference the study previously done in M. pneumoniae.6 Specifically, we reanalysed the T4 dataset of M. pneumoniae.6 Transposition events are considered to behave as random events resembling a Poisson process over large portions of the chromosome with a uniform density. The analysis starts with two different linear insertion densities (r) for essential (rE) and non-essential genes (rNE). These values correspond to the total number of insertions mapped normalized by gene length for a training set of genes which, based on prior knowledge, we assume are either E or NE and can be used as reference to classify the rest of genes. For M. pneumoniae, we used as a training set the same group of genes that were defined in its essentiality study.6 In the case of M. agalactiae, a new E training set containing 32 genes was generated using the same criteria as for M. pneumoniae. For NE genes, we extracted 84 genes with no homolog in four closely related species to M. agalactiae: Mycoplasma hyosynoviae, Mycoplasma arthritidis, Mycoplasma pulmonis and Mycoplasma synoviae. Out of this group, we selected 17 genes that had been confirmed as NE in the essentiality study done in M. bovis PG45.10 The probability of a specific gene to be essential or not is evaluated predicting two values: PE and PNE using formula (1). In this formula, N represents the number of insertions mapped to a gene and L the length of that gene. L corresponds to the inner 90% part of each gene (removing the 5% in each terminus) since it has been observed that these regions allow a higher number of non-disruptive mutations as they would not be located in the core regions of the encoded protein. The value of r, inferred from the training sets, varies between essential (rE) and non-essential genes (rNE) and allows us to determine the probability of a particular gene to be essential or not with its specific N and L. If PE > 0.0 and PNE = 0.0, the gene is classified as E (essential), PE = 0.0 and PNE > 0.0 will correspond to NE (non-essential), and if both probabilities are non-zero we assume that the gene is F (fitness).
| (1) |
After Clusters of Orthologous Groups (COGs) were associated to the M. agalactiae genome using eggNOG-mapper40 utilizing the DIAMOND mapping mode, taxonomic scope set on Firmicutes and prioritizing quality over coverage. A subsequent step of manual curation of COG categories was performed. For the set of 689 genes found for M. agalactiae 7784, the genes included in the training list, the COG category assignment, their essentiality assigned class and the statistical analysis, see Supplementary Table S8.
3. Results and discussion
3.1. Transformation efficiencies in Mycoplasmas show dramatic differences depending on the strain
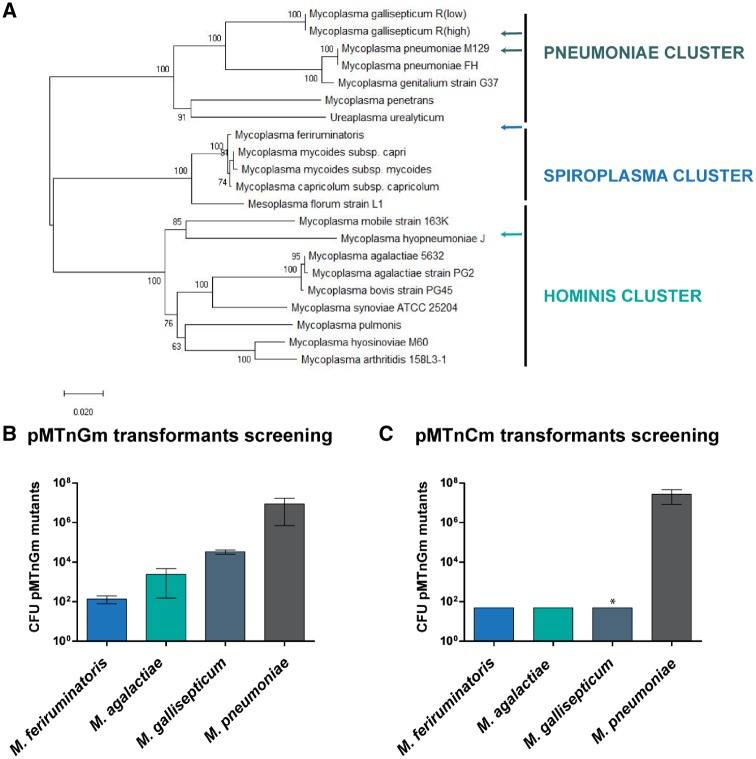
As a starting point of the project, we aimed to quantify the transposon TE in representative members of each cluster of the Mycoplasma genus (Fig. 1A). To this end, cultures of M. feriruminatoris (spiroplasma cluster), M. agalactiae (hominis cluster), M. pneumoniae and M. gallisepticum (pneumoniae cluster) were transformed with the pMTnGm vector,23 a mini-transposon plasmid derived from the original Tn4001 that confers resistance to gentamicin and is broadly employed in the Mycoplasma field. As expected from previous reports, M. pneumoniae showed the highest TE, with approximately one transformant for every 103 cells. M. pneumoniae was followed by M. gallisepticum and M. agalactiae, with three transformants for every 106 cells. Far behind these TEs was that of M. feriruminatoris, for which we found one transformant for every 109 cells (Fig. 1B).
Figure 1.
Screening of transposon TEs across the mycoplasmal landscape. (A) Phylogenetic tree of 21 selected Mycoplasma species in which three main clusters (pneumoniae, spiroplasma and hominis) can be identified using the maximum composite likelihood method. The tree is drawn to scale with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree.29 (B) Bar plot showing the average of gentamicin resistant CFUs (in logarithmic scale) obtained for each of the indicated strains when using the pMTnGm vector (n = 3). (C) Bar plot showing the average of chloramphenicol resistant CFUs (in logarithmic scale) obtained for each of the indicated strains when using the pMTnCm vector. For the statistical analysis, for those species in which no mutants were detected the number of CFU was set to 49, the maximum value below the limit of detection. One-tailed t-test P-values are indicated with one asterisk (*) when P < 0.05 for TE obtained with pMTnGm vector compared to the TE obtained with pMTnCm vector.
Since the expression of the gentamicin resistance gene has been found to exert a detrimental effect on growth in M. genitalium even in the absence of gentamicin itself,23 we wanted to determine whether this effect was exacerbated in M. gallisepticum, M. agalactiae and M. feriruminatoris. This could be responsible for the low TE observed in these species with the pMTnGm vector. To this end, we repeated the same set of transformations, but this time with a modified version of pMTnGm vector termed pMTnCm. In this vector, the native gentamicin resistance gene as well as its RR were replaced by a chloramphenicol resistance under the control of the p438 RR, a minimal 22-bp sequence that controls the expression of a putative restriction enzyme in M. genitalium.
For the sake of clarity, from this point on, we will refer to the sequence immediately upstream of the translational start codon of each gene as the RR, comprising the promoter region involved in transcription as well as the 5′ UTR involved in translation.
We found that the number of M. pneumoniae transformants obtained with the pMTnCm vector was more than three times higher than the number obtained with the pMTnGm vector, thus falling in line with the previous reports of toxicity associated with expression of the gentamicin resistance gene.23 On the other hand, the TE was dramatically lower in all the other species tested, with the number of total transformants falling under our detection limit of 50 CFU per batch for M. gallisepticum, M. feriruminatoris and M. agalactiae (Fig. 1C). The most plausible explanation for these results is that only M. pneumoniae is able to efficiently recognize both the RR found upstream of the gentamicin resistance gene and the M. genitalium-derived p438 sequence driving the expression of the chloramphenicol resistance gene. In contrast, it seems that M. gallisepticum, M. feriruminatoris and M. agalactiae can recognize the RR controlling the gentamicin resistance gene but not the one controlling the chloramphenicol resistance gene.
These results suggest that TE might be directly related to the ability of the transcriptional/translational machinery of each strain to efficiently recognize the RRs controlling not only the antibiotic resistance gene, but also the transposase coding gene.
3.2. Design of an efficient regulatory region for a broad range of Mycoplasma species
Several aspects might influence the TE of a transposon vector and can be classified into two different categories. In the first category, we can include those factors that affect the TE of any vector, such as degradation of the plasmid by bacterial restriction machinery, or the optimality of the transformation protocol itself. Leaving these aside, there is a second group of factors that are specific for transposon vectors. Thus, once the vector has entered the cell, proper transcription/translation of the transposase coding gene is required for the correct insertion of the antibiotic resistance gene into the chromosome. Second, the insertion mutant would only be able to grow if the protein levels of the antibiotic resistance gene reach a certain threshold that is sufficient to promote growth on selective medium. Therefore, in an attempt to maximize the protein levels of both the transposase coding gene and the antibiotic resistance gene in all Mycoplasma species, we decided to design a RR termed SynMyco Regulatory Region (SynMyco RR) that would allow efficient transcription and translation in different mycoplasma species.
As a reference for the design, we chose the RR of MPN665, a gene whose transcript is in the top 5% of most transcribed genes in the transcriptome of M. pneumoniae.41 Also, its protein levels (2,646 copies per cell) are among the highest in the proteome of M. pneumoniae.4 MPN665 encodes for the elongation factor thermo-unstable (EF-Tu) protein. As a main component of the ribosome, the protein product of this gene has an essential role in translation and is universally conserved in the prokaryotic world.42,43 Thus, with the aim of identifying important elements, we aligned the RRs of MPN665 orthologues found in a representative set of Mycoplasma species (Fig. 2A). For the design of the SynMyco RR, we mainly kept those bases found in the MPN665 RR that are not changed to another base in other species. However, given that native RRs did not evolve to be the most productive transcriptional drivers, we also favoured those bases that were found to be optimal as transcription/translation determinants in a recent screen of synthetic sequences.44 This is exemplified by including an extended Pribnow box motif (TGN-Pribnow) within the SynMyco RR, a feature that is not found in any of the native EF-Tu RRs analysed but promotes higher transcription rates. Based on the screen mentioned above, we also changed the Pribnow box sequence from TAAAAT to the canonical TATAAT motif. Lastly, to ensure the functionality of the SynMyco RR in a broad range of Mycoplasma species, we paid special attention to other features found in the RRs such as the -35 box or the Shine-Dalgarno region (also known as, Ribosome Binding Site, RBS). In particular, the -35 box seems to have lost its prevalent role as a transcriptional driver during Mycoplasma evolution, as indicated by the absence of any consensus sequence among the most productive RRs found in the screen of synthetic sequences.44 In fact, only a degenerated -35 box with the sequence TTGANN can be found in the RRs of just 20% of the genes of M. pneumoniae.45 However, as the most prevalent sequence found at the -35 area of the EF-Tu RRs analysed was TGC, we included this in the SynMyco RR. On the other hand, the Shine-Dalgarno region, which is responsible for ribosome docking onto the mRNA, is present in only 26% of M. pneumoniae genes. In contrast, in other species such as M. agalactiae, it is found in as much as 73% of the coding sequences (Fig. 2B). For this reason, we also included a Shine-Dalgarno region in the design of the SynMyco RR, using the sequence that is most frequently found in the EF-Tu RRs analysed (i.e. 5′-AAAGGA-3′). The complete sequence of the SynMyco RR with its main sequence determinants indicated is shown in Fig. 2C.
Figure 2.
Analysis of RRs found in Mycoplasma species. (A) Sequence alignment of the EF-Tu RRs of 10 selected Mycoplasma species. Four main domains are highlighted in boxes: the -35 box, Pribnow box, the RBS sequence and the Translation Starting Codon. In addition, the experimentally determined transcriptional start site (TSS) is also shown for the M. pneumoniae RR. (B) Bar plot representing the fraction of RBS-positive genes normalized by the total number of genes per genome in 21 different Mycoplasma species. (C) Sequence of the SynMyco RR. The boxes highlight the same domains shown in panel A, plus the extended Pribnow domain, and the expected TSS inferred from the one experimentally determined in M. pneumoniae.
3.3. A transposon vector carrying the SynMyco regulatory region dramatically increases transformation efficiency
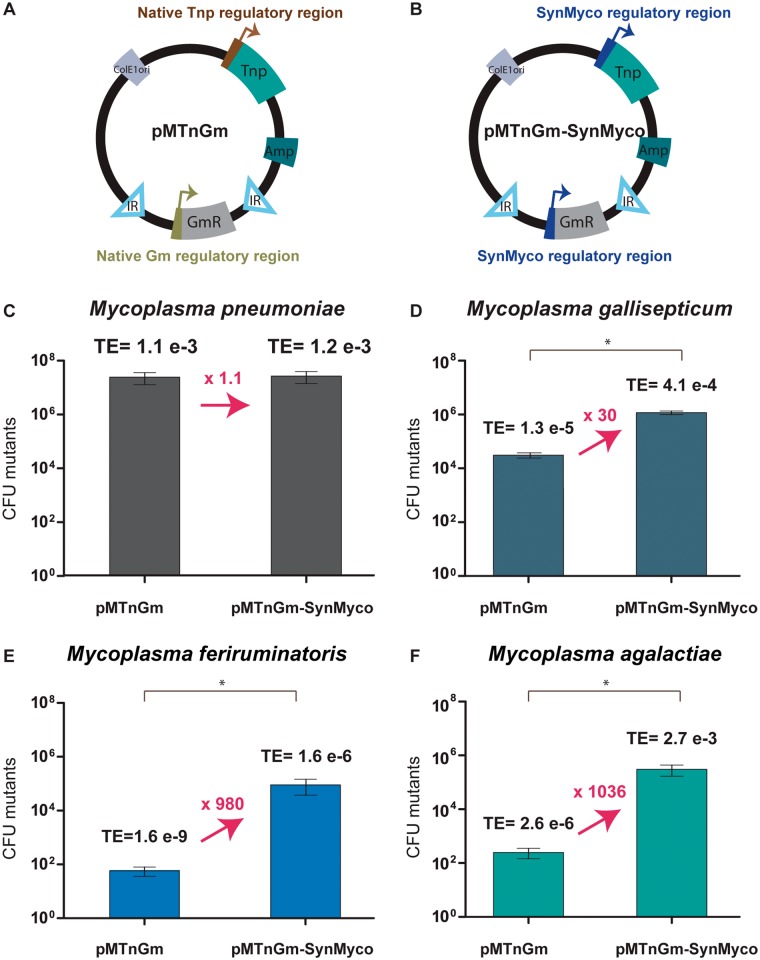
Taking the widely employed pMTnGm vector as reference (Fig. 3A), we constructed a new transposon vector termed pMTnGm-SynMyco in which both the transposase and the gentamicin resistance coding genes were placed under the control of the SynMyco RR (Fig. 3B). Subsequently, we used this vector to determine whether TE is higher with pMTnGm-SynMyco than with pMTnGm. To this end, cultures of M. pneumoniae, M. agalactiae, M. gallisepticum, and M. feriruminatoris were transformed in parallel with both vectors.
Figure 3.
Comparison between the transposon TE obtained with pMTnGm and pMTnGm-SynMyco in four different Mycoplasma species. (A) Scheme of the key modules of the pMTnGm transposon. (B) Scheme of the key modules of the pMTnGm-SynMyco transposon. For both (A) and (B) the abbreviations that appears in the figure are: Tnp for transposase coding gene, Amp for ampicillin resistance coding gene, IR for inverted repeats, ColE1 ori for ColE1 origin of replication and GmR for gentamicin resistance coding gene. (C) Bar plot representing the average CFUs of M. pneumoniae resistant to gentamicin (in logarithmic scale) obtained for three independent transformation replicates carried out with either pMTnGm (left side of each panel) or pMTnGm-SynMyco (right side of each panel). For each group of bars, the average of TE (CFU resistant to gentamicin / total CFU viable after transformation) is displayed on top. The fold change in TE is indicated in arrows connecting both sides of each panel. One-tailed t-test p-values are indicated with one asterisk (*) when P < 0.05 for TE obtained with pMTnGm-SynMyco vector compared to the TE obtained with pMTnGm vector. Similar bar plots are shown in (D) for M. gallisepticum, (E) for M. feriruminatoris and (F) for M. agalactiae.
We did not find significant differences in TE for M. pneumoniae when transformed with either of the two vectors (Fig 3C), which is not surprising taking into consideration that M. pneumoniae already showed a high TE with the pMTnGm vector. Therefore, these data suggest that the expression levels obtained with the RRs of the pMTnGm vector were already enough to saturate the system in this strain.
On the other hand, the TEs obtained with the pMTnGm-SynMyco vector were significantly higher (P < 0.05) in all other species when compared with the TEs obtained with the pMTnGm vector. In particular, for M. gallisepticum, we observed a 30-fold increase in TE, obtaining more than 106 mutants when transformed with pMTnGm-SynMyco (Fig. 3D).
For M. feriruminatoris, the strain showing the worst TE with pMTnGm (less than 100 total transformed cells per replicate), we observed a 980-fold increase in TE when transformed with pMTnGm-SynMyco, resulting in more than 90,000 individual clones carrying a transposon insertion (Fig. 3E).
Lastly, for M. agalactiae, we found that whereas with the pMTnGm vector we usually observed slightly more than 200 transformed cells per replicate, we obtained more than 3 × 105 insertion mutants when transformed with pMTnGm-SynMyco, a value that represents an increase in TE of more than 1036 times (Fig. 3F).
Thus, when transforming these Mycoplasmas strains with the pMTnGm vector, we can classify them into three different groups according to their TEs. The first group is the high efficiency group and contains M. pneumoniae, which showed a TE of 10−3. This is followed by the intermediate TE group (i.e. 10−5–10−6) composed of M. agalactiae and M. gallisepticum, and finally by the low TE group (i.e. 10−9) with M. feriruminatoris as the representative. In contrast, when transforming with the pMTnGm-SynMyco vector, we are only able to distinguish a high TE group (composed of M. pneumoniae, M. agalactiae and M. gallisepticum) and an intermediate TE group, (M. feriruminatoris alone).
It should be noted that although TE is consistently increased in M. gallisepticum, M. feriruminatoris and M. agalactiae when transformed with pMTnGm-SynMyco vector, the actual numbers of total mutants obtained might be further increased carefully by optimizing the transformation protocol for each species, something that we have not addressed in this study. In addition, while we have generated only a gentamicin version of the SynMyco transposon, we hypothesized that the other resistance markers available for Mycoplasmas (i.e. chloramphenicol, puromycin and tetracycline)18 and already implemented in native Tn4001 transposon-derived vectors, might in the future be included in pMTnGm-SynMyco vector in substitution of the gentamicin resistance gene. This would allow the generation of a set of four different transposon vectors highly efficient in a broad range of Mycoplasmas. Furthermore, as was recently shown for M. genitalium, resistance genes can also be flanked by lox sites in vectors carrying the SynMyco RR thereby allowing the recycling of antibiotic markers when employing protocols that require the iterative use of transposon insertion mutagenesis.46
3.4. Strength comparison of SynMyco regulatory region and those in the native pMTnGm vector
In all species tested, the number of transformed cells was higher when transformed with pMTnGm-SynMyco vector than with unmodified pMTnGm vector. These results suggested that the protein yields of the transposase and/or the protein conferring resistance to gentamicin were major determinants of the TE. In order to find out which of the two gene products, or both, were responsible for the increase in TE, we developed a reporter assay. This reporter assay allows quantification of the relative strength of SynMyco RR compared with the RRs driving the expression of the transposase (i.e. Tnp RR) and the gentamicin resistance gene (i.e. Gm RR) in the native pMTnGm vector. To this end, we created three different vectors derived from the pMTnGm-SynMyco in which the genes coding for the mCherry and Venus fluorescent proteins were introduced for quantification and normalization purposes, respectively. The region controlling the expression levels of the mCherry protein varies between the three different vectors, being either SynMyco RR, Tnp RR or Gm RR, whereas the RR controlling the expression of Venus reporter is constant in all the constructs, being always SynMyco RR (Fig. 4A). The three different constructs were transformed in all of the species studied in this work. Subsequently, the ratio between mCherry fluorescence and Venus fluorescence allowed us to determine the relative strength of each RR in M. pneumoniae (Fig. 4B), M. gallisepticum (Fig. 4C), M. feriruminatoris (Fig. 4D) and M. agalactiae (Fig. 4E).
Figure 4.
Comparison of the SynMyco RR efficiency versus the native regulatory regions of pMTnGm transposon. (A) Scheme of the key modules of (i) pMTnGm-SynMyco+SynMyco RR-Venus+SynMycoRR-mCherry transposon, (ii) pMTnGm-SynMyco+SynMycoRR-Venus+Gm RR-mCherry transposon and (iii) pMTnGm-SynMyco+SynMyco-Venus+TnpRR-mCherry transposon. The abbreviations that appear in the figure are: Tnp for transposase coding gene, Amp for ampicillin resistance coding gene, IR for inverted repeats, ColE1 ori for ColE1 origin of replication, GmR for gentamicin resistance coding gene, Venus for Venus protein coding gene and mCherry as mCherry protein coding gene. (B) Bar plot representing the ratio obtained in M. pneumoniae for mCherry/Venus fluorescence using transposons represented in panel A. While Venus reporter is always under control of SynMyco RR, the mCherry gene is controlled by SynMyco RR in the left bar, Gm RR in the central bar and Tnp RR in the right bar. On top of each bar is represented the average ratio of mCherry/Venus fluorescence obtained for each of the constructs in three replicates. One-tailed t-test p-values are indicated with one asterisk (*) when P < 0.05 for the ratio of mCherry/Venus under the control of SynMyco RR versus either Gm RR or Tnp RR. Similar bar plots are shown in (C) for M. gallisepticum, (D) for M. feriruminatoris and (E) for M. agalactiae.
In all species, the ratio between mCherry and Venus fluorescence is close to 1 (i.e. 0.9 for M. pneumoniae, 1.1 for M. gallisepticum, 1.1 for M. feriruminatoris and 1.3 for M. agalactiae) when both reporters were under control of the SynMyco RR. Interestingly, in M. gallisepticum and M. feriruminatoris, the ratio is also close to 1 when mCherry is under control of Gm RR (i.e. 1 for M. gallisepticum and 1.1 for M. feriruminatoris) but drastically drops when the expression of mCherry is driven by Tnp RR (i.e. 0.2 for M. gallisepticum and 0.03 for M. feriruminatoris). Altogether, these data suggest that in these two species, when transforming with pMTnGm or pMTnGm-SynMyco vectors, the difference in TE would be due to the expression level of the transposase gene and not the antibiotic resistance gene.
Poor performance of Tnp RR was also observed in M. pneumoniae and M. agalactiae as indicated by the low fluorescence ratio observed when mCherry is under control of Tnp RR (i.e. 0.1 for M. pneumoniae and 0.1 for M. agalactiae). Moreover, in contrast to M. gallisepticum and M. feriruminatoris where Gm RR and SynMyco RR provide similar protein yields, in M. pneumoniae and M. agalactiae the fluorescence ratio is also reduced when the expression of mCherry is driven by Gm RR (i.e. 0.1 for M. pneumoniae and 0.2 for M. agalactiae). Thus, in M. pneumoniae and M. agalactiae, SynMyco RR outperforms not only Tnp RR but also GmRR in terms of protein yields.
It has not escaped our notice that the reporter assay shows the same expression profile for M. agalactiae and M. pneumoniae, whereas TE upon using pMTnGm-SynMyco is only drastically increased in M. agalactiae (i.e. 1036-fold change) but not in M. pneumoniae (i.e. 1.1-fold change). We hypothesized that this observation might be related with the different doubling times of the species. Slow dividing species, such as M. pneumoniae, would have more time to deliver the transposon cargo into the chromosome before cell duplication starts to dilute the plasmid within the growing population. In contrast, in species dividing faster, such as M. agalactiae, a quick transposition into the chromosome mediated by high expression levels of the transposase coding gene would represent an advantage to avoid the dilution effect associated with cell division and thus would lead to an increased TE. An alternative or complementary hypothesis is that M. pneumoniae might be somehow more permissive than the other species for the presence of extrachromosomal elements inside the cell. In this scenario, a fast transposition into the chromosome mediated by high expression levels of the transposase coding gene would represent a greater advantage for the other mycoplasma strains than for M. pneumoniae.
In summary, our reporter assay shows that SynMyco RR is efficiently recognized in all the species tested, and suggests that the factor limiting the TE with native pMTnGm vector in most Mycoplasma species is the expression of the transposase coding gene.
3.5. Unblocking the global study of essential and dispensable genes in M. agalactiae
To the best of our knowledge, studies regarding essentiality in Mycoplasmas have only been published so far for M. genitalium,8M. pneumoniae,6Mycoplasma pulmonis,47Mycoplasma bovis10 or Mesoplasma florum.48 However, while for M. pneumoniae almost 350,000 unique transposon insertion mutants have been tracked, the other studies analysed a substantially lower number of clones. For instance, the M. bovis study only obtained 319 mutants, representing one insertion every 3,145 bp of the total genome,10 and the study in M. genitalium analysed around 3,300 mutants, showing an insertional coverage of one disruption over every 175 bp.8 Obviously, the accuracy of the assignment of genes to each one of the three categories of essentiality [i.e. essential (E), non-essential (NE) and fitness (F)] is directly related to the insertional coverage of the transposon mutagenesis. Moreover, the existence in bacteria of small open reading frame-encoded polypeptides (i.e. short open reading frame-encoded proteins, also known as SEPs) as short as 11 amino acids in length is becoming widely accepted.49 Thus, high transposon TE is necessary to study the essentiality of these SEPs.6,50
To illustrate the potential that our pMTnGm-SynMyco vector could have in the study of a broad range of Mycoplasma species, we decided to perform an essentiality study for M. agalactiae 7784, a strain for which no essentiality map is currently available. First, we sequenced, assembled and annotated for the first time the genome of this strain. Then, after transforming the strain with pMTnGm-SynMyco vector we were able to map 199,723 transposon insertions to the M. agalactiae genome, with an estimated genome size of 853,960. This represents ∼23.3 insertions over every 100 bp, which is around half of the coverage observed in M. pneumoniae6 (354,447 transposon insertions mapped, representing ∼43 insertions over every 100 bp) but still substantially higher than the ones reported in other essentiality studies (Fig. 5A and B).
Figure 5.
Essentiality study in M. agalactiae using the pMTnGm-SynMyco transposon and a comparison with previous studies in M. pneumoniae. (A) Genome disruption profile for M. pneumoniae. The y-axis represents the logarithmic average of total reads covering a window of 1,000 bp (x-axis). (B) Genome disruption profile for M. agalactiae representing the same information as in the previous panel. (C) Insertion density by gene distribution in M. pneumoniae and M. agalactiae as indicated. The x-axis represents the percentage of bp in a gene that is disrupted and the y-axis the frequency of densities in the distribution. To better compare M. pneumoniae and M. agalactiae transposon insertion distributions, we standardized both distributions using min-max scaling. (D) Box-plot representing the statistical comparison of specific subsets of genes expected to be essential (E) and non-essential (NE) in M. pneumoniae and M. agalactiae as indicated. The asterisk represents P-value < 0.05 (3.62 e−41 and 1.20 e−20) when comparing density of insertions of E and NE coding genes in M. pneumoniae and M. agalactiae respectively.
When considering the frequency of insertions per gene, we observed a bimodal distribution separating essential from non-essential genes in both strains (Fig. 5C). Next, we explored the insertional profile at the gene level using sets of known E and NE genes as inferred from the reference essentiality study in M. pneumoniae.6 Whereas for M. pneumoniae the training sets for E and NE genes comprise 27 and 28 genes, respectively, for M. agalactiae we generated dedicated training sets based on bibliography containing 32 E genes and 17 NE genes. When comparing the average density of insertions between E and NE genes in M. pneumoniae and M. agalactiae, we observed that the two groups were significantly different within each species (two-tailed t-test with equal variances, P-values equal to 1.42e−47 and 1.05e−20, respectively; Fig. 5D). Taken altogether, we were able to assign one category for 689 genes found in the M. agalactiae 7784 genome: E (43.98% of genes), F (25.25% of genes) or NE (30.77% of genes) (see Supplementary Table S8), in line with the percentage of E (49.28%), F (13.40%) and NE (37.32%) genes obtained for M. pneumoniae.6
When M. agalactiae essential genes were classified according to the COGs functional categories annotation system, we found that the categories that were significantly enriched in essential genes were: (i) protein coding genes involved in translation, ribosomal structure and biogenesis, (ii) functional RNAs, (iii) protein coding genes without assigned COG category and (iv) protein coding genes with unknown function (see Supplementary Table S8).
However, for the purpose of this work it is more relevant to compare the essentiality map of M. agalactiae at high density of insertions with one obtained with low density to illustrate the importance of high coverage. Unfortunately, there is no essentiality study done in M. agalactiae, but there is a low coverage analysis in its closely related specie M. bovis PG4510 (319 transposon insertional mutants individually sequenced, versus more than 199,000 transposon insertions analysed by deep sequencing in our study). Sequencing of individual insertion clones is useful to classify the genes within the F-NE categories (i.e. genes disrupted by a transposon insertion) but cannot distinguish between both. Examples of this limitation are the genes coding for a tRNA modification GTPase and for the deoxyribonuclease IV. In the M. bovis essentiality study, both genes (i.e. MBOVPG45_0060 and MBOVPG45_0301) were classified as NE given the isolation of individual clones carrying insertions within their coding region. However, our essentiality study based on the analysis of at least 600 times more mutants than the one of M. bovis and mapped by ultra sequencing classified these two genes (i.e. MAGA7784_RS00280 and MAGA7784_RS02715) as F in M. agalactiae. This implies that although these genes are non-essential, their disruption cause a growth impairment of these particular clones at least for M. agalactiae, something that cannot be directly measured by genomic sequencing of transposon insertion in a limited number of clones.
Moreover, aside from its lack of accuracy in distinguishing between NE and F genes, the main limitation of small transposon libraries is related with E genes. Specifically, the low coverage of these libraries makes it difficult to ascertain whether a gene is free of insertions because of its essential character or as a result of the randomness of the integration and/or low sampling of the mutants. Indeed, only two genes were suggested by the authors as highly probable essential genes in the M. bovis study, MBOVPG45_0337 and MBOVPG45_0710, coding for an ATP-binding protein and SGNH/GDSL hydrolase, respectively. This assumption was based on the lack of clones carrying a transposon insertion within the coding regions of these genes in spite of their large size (i.e. 3420 bp for MBOVPG45_0337 and 8013 bp for MBOVPG45_0710). Nonetheless, it should be noted that the insertional coverage of the mutant library of this study, with one insertion every 3145 bp of the genome on average, is in the same range of these genes in terms of size. In our M. agalactiae essentiality study, the orthologue of the SGNH/GDSL hydrolase coding gene (i.e. MAGA7784_RS03410) was found E, confirming the hypothesis of the M. bovis report. In contrast, the gene coding for the ATP-binding protein (i.e. MAGA7784_RS02615) was determined as NE as indicated by the presence of 891 transposon insertions within its sequence. This suggests that the absence of clones carrying an insertion within MBOVPG45_0337 gene is most likely a consequence of the low coverage of the insertional library, rather than the gene being truly essential in M. bovis. Furthermore, the limitation of low coverage libraries to assign a truly E character for a given gene is even more evident with small coding sequences. For instance, in M. bovis there were no clones found to carry a transposon insertion within the coding sequences of genes MBOVPG45_0043 (582 bp) or MBOVPG45_0596 (963 bp), coding for sigma-70 RNA polymerase factor and Holliday junction branch migration DNA helicase ruvB, respectively. This might indicate that these genes are E. However, homologues of these genes are fully covered with transposon insertions not only in M. agalactiae (MAGA7784_RS00215 and MAGA7784_RS01205), but also in M. pneumoniae (MPN626 and MPN536), suggesting the lack of insertions in the M. bovis study is more likely related with the small size of the genes rather than with their essential character. All of these examples illustrate the importance of high coverage transposon insertion libraries for the appropriate category assignment of a given gene, something that can be only achieved with efficient transposon vectors such as pMTnGm-SynMyco.
In conclusion, we demonstrate that the RRs driving the expression of the transposase and the resistance genes have a tremendous impact on the TE achieved after vector transformation. Although problems derived from poor recognition of vector RRs can be avoided with transformation procedures based on purified transposases,51 it should be noted that not all transposases are commercially available and their cost might limit use in many research groups. Indeed, screening libraries of RRs for the key elements of transposon vectors has already been shown to be a useful strategy to produce a moderate increase (i.e. around one order of magnitude) in the TE of different bacteria.52 However, an approach such as the one followed in our study involving rational design of RRs rather than the screen of a limited number of variants might be more effective in increasing TE. In fact, we have seen that the pMTnGm-SynMyco vector significantly increases the TE in three phylogenetically distant Mycoplasma species. For this reason, we hypothesize that this vector might improve the reported TE in all species belonging to the Mycoplasma genus. Even though we have shown that the SynMyco RR is efficiently recognized in different species, it cannot be excluded that other designs of RRs could also lead to an increase in TE. As exemplified in this work with M. agalactiae, the higher TE obtained using this vector may unblock the development of new essentiality maps for other Mycoplasma species, and thereby promote global knowledge of these interesting microorganisms. In addition, the higher insertional coverage obtained with the pMTnGm-SynMyco vector should facilitate the isolation of clones of interest from libraries of insertional mutants, an advancement which may boost gene-function assignments in Mycoplasma species that have been poorly studied so far.
Supplementary Material
Acknowledgements
We thank Dr Sarah A. Head for critical manuscript revision. We also thank Tony Ferrar for language editing (http://www.theeditorsite.com). We acknowledge the staff of the CRG Genomics Unit and the CRG Bioinformatics Unit, especially Jochen Hecht and Luca Cozzuto for their assistance and fruitful discussions.
Funding
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement 634942 (MycoSynVac) and was also financed by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme, under grant agreement 670216 (MYCOCHASSIS). We also acknowledge support of the Spanish Ministry of Economy, Industry and Competitiveness (MEIC) to the EMBL partnership, the Centro de Excelencia Severo Ochoa and the CERCA Programme/Generalitat de Catalunya.
Accession numbers
The raw data of DNAseq, genome assembly and de novo annotation was submitted as BioProject under accession PRJNA528179. The transposon sequencing in Mycoplasma agalactiae dataset can be found in ArrayExpress with accession number: E-MTAB-7425.
Conflict of interest
None declared.
References
- 1. Woese C.R., Maniloff J., Zablen L.B.. 1980, Phylogenetic analysis of the mycoplasmas, Proc. Natl. Acad. Sci. USA, 77, 494–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Himmelreich R., Hilbert H., Plagens H., Pirkl E., Li B.C., Herrmann R.. 1996, Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae, Nucleic Acids Res., 24, 4420–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guell M., van Noort V., Yus E., et al. 2009, Transcriptome complexity in a genome-reduced bacterium, Science, 326, 1268–71. [DOI] [PubMed] [Google Scholar]
- 4. Maier T., Schmidt A., Guell M., et al. 2011, Quantification of mRNA and protein and integration with protein turnover in a bacterium, Mol. Syst. Biol., 7, 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lluch-Senar M., Luong K., Lloréns-Rico V., et al. 2013, Comprehensive methylome characterization of Mycoplasma genitalium and Mycoplasma pneumoniae at single-base resolution, PLoS Genet., 9, e1003191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lluch-Senar M., Delgado J., Chen W.-H., et al. 2015, Defining a minimal cell: essentiality of small ORFs and ncRNAs in a genome-reduced bacterium, Mol. Syst. Biol., 11, 780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Karr J.R., Sanghvi J.C., Macklin D.N., et al. 2012, A whole-cell computational model predicts phenotype from genotype, Cell, 150, 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Glass J.I., Assad-Garcia N., Alperovich N., et al. 2006, Essential genes of a minimal bacterium, Proc. Natl. Acad. Sci. USA, 103, 425–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. French C.T., Lao P., Loraine A.E., Matthews B.T., Yu H., Dybvig K.. 2008, Large-scale transposon mutagenesis of Mycoplasma pulmonis, Mol. Microbiol., 69, 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sharma S., Markham P.F., Browning G.F.. 2014, Genes found essential in other mycoplasmas are dispensable in Mycoplasma bovis, PLoS One, 9, e97100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hutchison C.A. 3rd, Chuang R.-Y., Noskov V.N., et al. 2016, Design and synthesis of a minimal bacterial genome, Science, 351, aad6253. [DOI] [PubMed] [Google Scholar]
- 12. Waites K.B., Talkington D.F.. 2004, Mycoplasma pneumoniae and its role as a human pathogen, Clin. Microbiol. Rev., 17, 697–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Levisohn S., Kleven S.H.. 2000, Avian mycoplasmosis (Mycoplasma gallisepticum), Rev. Sci. Technol., 19, 425–42. [PubMed] [Google Scholar]
- 14. Kumar A., Rahal A., Chakraborty S., Verma A.K., Dhama K.. 2014, Mycoplasma agalactiae, an etiological agent of contagious agalactia in small ruminants: a review, Vet. Med. Int., 2014, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Jesus M.A., Gerrick E.R., Xu W., et al. 2017, Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis, MBio, 8, e02133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Halbedel S., Busse J., Schmidl S.R., Stülke J.. 2006, Regulatory protein phosphorylation in Mycoplasma pneumoniae. A PP2C-type phosphatase serves to dephosphorylate HPr (Ser-P), J. Biol. Chem., 281, 26253–9. [DOI] [PubMed] [Google Scholar]
- 17. Halbedel S., Stülke J.. 2007, Tools for the genetic analysis of Mycoplasma, Int. J. Med. Microbiol., 297, 37–44. [DOI] [PubMed] [Google Scholar]
- 18. Lyon B.R., May J.W., Skurray R.A.. 1984, Tn4001: a gentamicin and kanamycin resistance transposon in Staphylococcus aureus, Mol. Gen. Genet., 193, 554–6. [DOI] [PubMed] [Google Scholar]
- 19. Dybvig K., French C.T., Voelker L.L.. 2000, Construction and use of derivatives of transposon Tn4001 that function in Mycoplasma pulmonis and Mycoplasma arthritidis, J. Bacteriol., 182, 4343–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pour-El I., Adams C., Minion F.C.. 2002, Construction of mini-Tn4001tet and its use in Mycoplasma gallisepticum, Plasmid., 47, 129–37. [DOI] [PubMed] [Google Scholar]
- 21. Hahn T.W., Mothershed E.A., Waldo R.H. 3rd, Krause D.C.. 1999, Construction and analysis of a modified Tn4001 conferring chloramphenicol resistance in Mycoplasma pneumoniae, Plasmid, 41, 120–4. [DOI] [PubMed] [Google Scholar]
- 22. Algire M.A., Lartigue C., Thomas D.W., Assad-Garcia N., Glass J.I., Merryman C.. 2009, New selectable marker for manipulating the simple genomes of Mycoplasma species, Antimicrob. Agents Chemother., 53, 4429–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pich O.Q., Burgos R., Planell R., Querol E., Piñol J.. 2006, Comparative analysis of antibiotic resistance gene markers in Mycoplasma genitalium: application to studies of the minimal gene complement, Microbiology, 152, 519–27. [DOI] [PubMed] [Google Scholar]
- 24. Chopra-Dewasthaly R., Zimmermann M., Rosengarten R., Citti C.. 2005, First steps towards the genetic manipulation of Mycoplasma agalactiae and Mycoplasma bovis using the transposon Tn4001mod, Int. J. Med. Microbiol., 294, 447–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beaman K.D., Pollack J.D.. 1983, Synthesis of adenylate nucleotides by Mollicutes (mycoplasmas), J. Gen. Microbiol., 129, 3103–10. [DOI] [PubMed] [Google Scholar]
- 26. Jores J., Fischer A., Sirand-Pugnet P., et al. 2013, Mycoplasma feriruminatoris sp. nov., a fast growing Mycoplasma species isolated from wild Caprinae, Syst. Appl. Microbiol., 36, 533–8. [DOI] [PubMed] [Google Scholar]
- 27. Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A. 3rd, Smith H.O.. 2009, Enzymatic assembly of DNA molecules up to several hundred kilobases, Nat. Methods, 6, 343–5. [DOI] [PubMed] [Google Scholar]
- 28. Saitou, N. and Nei, M. 1987, The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol., 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 29. Felsenstein J. 1985, Confidence limits on phylogenies: an approach using the bootstrap, Evolution, 39, 783. [DOI] [PubMed] [Google Scholar]
- 30. Tamura K., Nei M., Kumar S.. 2004, Prospects for inferring very large phylogenies by using the neighbor-joining method, Proc. Natl. Acad. Sci. USA., 101, 11030–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kumar S., Stecher G., Li M., Knyaz C., Tamura K.. 2018, MEGA X: molecular evolutionary genetics analysis across computing platforms, Mol. Biol. Evol., 35, 1547–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Omotajo D., Tate T., Cho H., Choudhary M.. 2015, Distribution and diversity of ribosome binding sites in prokaryotic genomes, BMC Genomics, 16, 604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zimmerman C.-U., U. Zimmerman C., Herrmann R.. 2005, Synthesis of a small, cysteine-rich, 29 amino acids long peptide in Mycoplasma pneumoniae, FEMS Microbiol. Lett., 253, 315–21. [DOI] [PubMed] [Google Scholar]
- 34. Bentley D.R., Balasubramanian S., Swerdlow H.P., et al. 2008, Accurate whole human genome sequencing using reversible terminator chemistry, Nature, 456, 53–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jackman S.D., Vandervalk B.P., Mohamadi H., et al. 2017, ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter, Genome Res., 27, 768–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xu H., Luo X., Qian J., et al. 2012, FastUniq: a fast de novo duplicates removal tool for paired short reads, PLoS One, 7, e52249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bolger A.M., Lohse M., Usadel B.. 2014, Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics, 30, 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Langmead B., Salzberg S.L.. 2012, Fast gapped-read alignment with Bowtie 2, Nat. Methods, 9, 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li H., Handsaker B., Wysoker A., et al. 2009, The sequence alignment/map format and SAM tools, Bioinformatics, 25, 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huerta-Cepas J., Forslund K., Coelho L.P., et al. 2017, Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper, Mol. Biol. Evol., 34, 2115–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Junier I., Unal E.B., Yus E., Lloréns-Rico V., Serrano L.. 2018, Insights into the mechanisms of basal coordination of transcription using a genome-reduced bacterium, Cell Syst., 7, 227–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schrader J.M., Uhlenbeck O.C.. 2011, Is the sequence-specific binding of aminoacyl-tRNAs by EF-Tu universal among bacteria? Nucleic Acids Res., 39, 9746–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cammarano P., Tiboni O., Sanangelantoni A.M.. 1989, Phylogenetic conservation of antigenic determinants in archaebacterial elongation factors (Tu proteins), Can. J. Microbiol., 35, 2–10. [DOI] [PubMed] [Google Scholar]
- 44. Yus E., Yang J.-S., Sogues A., Serrano L.. 2017, A reporter system coupled with high-throughput sequencing unveils key bacterial transcription and translation determinants, Nat. Commun., 8, 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lloréns-Rico V., Lluch-Senar M., Serrano L.. 2015, Distinguishing between productive and abortive promoters using a random forest classifier in Mycoplasma pneumoniae, Nucleic Acids Res., 43, 3442–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mariscal A.M., González-González L., Querol E., Piñol J.. 2016, All-in-one construct for genome engineering using Cre-lox technology, DNA Res., 23, 263–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dybvig K., Lao P., Jordan D.S., Simmons W.L.. 2010, Fewer essential genes in mycoplasmas than previous studies suggest, FEMS Microbiol. Lett., 311, 51–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Baby V., Lachance J.-C., Gagnon J., et al. 2018, Inferring the minimal genome of Mesoplasma florum by comparative genomics and transposon mutagenesis, mSystems, 3, e00198–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li H., Xiao L., Zhang L., et al. 2018, FSPP: a Tool for genome-wide prediction of smORF-encoded peptides and their functions, Front. Genet., 9, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Miravet-Verde S., Ferrar T., Espadas-García G., et al. 2019, Unraveling the hidden universe of small proteins in bacterial genomes, Mol. Syst. Biol., 15, e8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Goryshin I.Y., Reznikoff W.S.. 1998, Tn5 in vitro transposition, J. Biol. Chem., 273, 7367–74. [DOI] [PubMed] [Google Scholar]
- 52. Liu H., Price M.N., Waters R.J., et al. 2018, Magic pools: parallel assessment of transposon delivery vectors in bacteria, mSystems, 3, e00143–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.