Abstract
DNA mismatches are highly polymorphic and dynamic in nature, albeit poorly characterized structurally. We utilized the antitumour antibiotic CoII(Chro)2 (Chro = chromomycin A3) to stabilize the palindromic duplex d(TTGGCGAA) DNA with two G:G mismatches, allowing X-ray crystallography-based monitoring of mismatch polymorphism. For the first time, the unusual geometry of several G:G mismatches including syn–syn, water mediated anti–syn and syn–syn-like conformations can be simultaneously observed in the crystal structure. The G:G mismatch sites of the d(TTGGCGAA) duplex can also act as a hotspot for the formation of alternative DNA structures with a GC/GA-5′ intercalation site for binding by the GC-selective intercalator actinomycin D (ActiD). Direct intercalation of two ActiD molecules to G:G mismatch sites causes DNA rearrangements, resulting in backbone distortion to form right-handed Z-DNA structures with a single-step sharp kink. Our study provides insights on intercalators-mismatch DNA interactions and a rationale for mismatch interrogation and detection via DNA intercalation.
INTRODUCTION
DNA mismatches are potentially mutagenic and thus must be repaired by the mismatch repair system to maintain the integrity of the genetic information within the cell (1). Failure to correct for mismatched base pairs eventually leads to abnormal functioning of the cell and constitutes a major mechanism behind an increasing number of genetic defects and cancers (2–4). The presence of mismatches does not normally dramatically modify overall DNA helix geometry; however, it may exert a significant impact on local base step geometry and accordingly alters the structural topology of DNA (5,6). Structural analyses of the mismatched base pairs in various sequence contexts show that mismatches are highly polymorphic in nature. For example, many of the mismatched base pairs can exist as wobble pairs, protonated bases, bifurcated hydrogen bonds and various pairing conformations involving anti–syn and anti–anti isomerization (7–9).
The small molecules that recognize these mismatched DNA duplexes can induce various degrees of structural deformations, with many having pharmaceutical and/or diagnostic potential (10,11). The presence of unstable mismatches in DNA structures may cause base flipping into extrahelical positions, which itself is an important phenomenon observed upon small molecule ligand binding to DNA duplexes (12–14). DNA bending has also been recognized as an important consequence of the action of those small molecules that intercalate into DNA duplexes (15–17). Several studies have shown that large-scale deformations and structural rearrangements occur in DNA duplexes in order to achieve more energetically favourable and stable structures (18–20). Notably, many of the structural features observed for small molecule–DNA complexes have also been reported in DNA–protein complexes, suggesting that in some cases these may share similar interaction mechanisms (21,22).
In the current study, we utilize a self-complementary ‘GC rich’ DNA sequence, d(TTGGCGAA)2 containing two guanine-guanine (G:G) mismatches, in order to understand the polymorphism that exists in the mismatches and its structural consequences for DNA duplexes. A previous study reported that G:G mismatches can adopt either a symmetric or an asymmetric structure with two guanosines in anti and syn conformations, and the mismatch being held by two hydrogen bonds between the N1 imino protons and carbonyl oxygen atoms of guanine (23). Chromomycin A3 (Chro), is a tetrahydroanthracene-based glycoside antibiotic belonging to the aureolic acid family. It contains di- and trisaccharide components linked to a tetrahydroanthracen-2-yl chromophore via O-glycosidic bonds at position 2 and 6, respectively (24,25). It has been shown that the dimeric Chro complex with CoII ion, CoII(Chro)2, can interact with those GC sites having flanking G:G mismatches (26,27). Here, we use CoII(Chro)2 as a mild stabilizer in order to stabilize and enable crystallization of this G:G mismatch-containing duplex. The structure shows, for the first time, that different arrangements of G:G mismatches can be tolerated in a single duplex, along with the more common symmetric anti:syn type of arrangement around the glycosidic angle conformations. On the other hand, the anti-cancer GC-selective DNA intercalator, actinomycin D (ActiD), is known to interfere with the replication and transcription of DNA in tumour cells by intercalating its phenoxazone ring at a GpC step in genomic DNA (28–32). We find that the two G:G mismatches provide an excellent binding environment for the ActiD. We have solved the crystal structures of the ActiD–d(TTGGCGAA)2 complex, which show large-scale structural rearrangements in the DNA duplex. Notably, these lead to the formation of a right-handed ‘zigzag’ (Z) DNA-type of backbone structure containing a G(anti):A(syn) mispair instead of the standard G:C base pair at the intercalation site. This causes a sharp kinking of the DNA helix that is more marked than that induced by sequence-specific DNA-binding drugs. These structures altogether display extraordinary features that can be exploited as unique mismatch DNA recognition characteristics revealed through DNA intercalation.
MATERIALS AND METHODS
Drugs and oligonucleotides
The self-complementary synthetic DNA oligonucleotide sequence d(TTGGCGAA) used for crystallization was commercially synthesised by MDBio, Inc. with purification performed by polyacrylamide gel electrophoresis. The drugs chromomycin A3 (Chro) and actinomycin D (ActiD) were purchased from Sigma Chemical Co. All the experimental procedures were performed using de-ionized water from a Milli-Q system. Absorbance measurements were carried out in a quartz cuvette using a JASCO UV/VIS spectrophotometer and the concentrations of oligonucleotides were determined according to Beer's law (A = ϵ.b.c, where A is the optical density at 260 nm, ϵ is the extinction coefficient, b is the cell path length (1 cm), and c is the molar DNA concentration). The concentration of Chro and ActiD solutions were determined from the optical density (ϵ405 nm = 8,800 M−1 cm−1 for Chro and ϵ224 nm = 35 280 M−1 cm−1 for ActiD). Oligomer extinction coefficients (ϵ) were calculated according to tabulated values of monomer and dimer extinction coefficients with reasonable assumptions (33).
Crystallization of CoII(Chro)2–d(TTGGCGAA)2 and ActiD–d(TTGGCGAA)2 complexes
The CoII(Chro)2–d(TTGGCGAA)2 and ActiD–d(TTGGCGAA)2 complexes were crystallized using the sitting drop vapour diffusion method. The crystals of CoII(Chro)2–d(TTGGCGAA)2 were obtained from a solution of 0.5 mM single-stranded DNA, 1 mM Chro, 2 mM cobalt(II) chloride, 5 mM sodium cacodylate (pH 7.3), 7 mM magnesium chloride, 12 mM spermine, 4% PEG400 and 4% 1-propanol, equilibrated against 500 μl of 30% PEG400 at 4°C. Because Chro is yellow, crystals with yellowish coloured and square-shaped morphology, implying formation of the CoII(Chro)2–d(TTGGCGAA)2 complex, were harvested after 4 days. Crystals of the ActiD–d(TTGGCGAA)2 complex were obtained from solutions containing 0.5 mM single-stranded DNA, 0.5 mM ActiD, 10 mM 2-(N-morpholino) ethane sulfonic acid (MES) buffer (pH 6.5), 1.5 M sodium malonate, 100 mM lithium chloride and 10 mM MnCl2 5 mM CaCl2. The solutions were equilibrated at 4°C with 500 μl of 1.5 M sodium malonate using the vapour diffusion method. Long hexagonal-prism-shaped, yellow-coloured crystals were obtained within 5–7 days implying the formation of the ActiD–DNA complex.
X-ray data collection, phasing and structure refinement of CoII(Chro)2–d(TTGGCGAA)2 and ActiD–d(TTGGCGAA)2 complexes
X-ray diffraction data of the CoII(Chro)2–d(TTGGCGAA)2 and ActiD–d(TTGGCGAA)2 complexes were collected on synchrotron radiation facilities. Diffraction data integration and data reduction were processed using the HKL-2000 program package (34). The diffraction data for the CoII(Chro)2–d(TTGGCGAA)2 crystal in space group P2221 with unit-cell parameters a = 47.82, b = 48.09, c = 96.72 Å, was collected at 110 K on a Bruker DIP-6040 detector at beamline BL44XU of the SPring-8. The phases for the CoII(Chro)2–d(TTGGCGAA)2 complex structure was determined by molecular replacement with Phaser MR in PHENIX suite (v1.8.4–1496) using the partial structure of the [Mg2+-(chromomycin A3)2]–d(TTGGCCAA)2 complex (PDB ID: 1VAQ) as a template (35). The diffraction data in space group I4122 with unit-cell parameters a = b = 59.35, and c = 93.66 Å, for the ActiD–d(TTGGCGAA)2 complex crystal was collected using the Rayonix MX300HS CCD Area Detector at the TPS 05A beamline of the National Synchrotron Radiation Research Center. Single-wavelength anomalous diffraction (SAD) data were collected at a resolution of 2.12 Å from single peak wavelength using manganese (MnII) as the anomalous scattering atom. The diffraction data were indexed, integrated, and scaled using the HKL-2000 software package, followed by MnII substructure localization using SHELX C/D/E (36). The resulting well-defined SAD electron density maps were used to build initial models using the molecular graphics programs MIFit (version 2010.10) and WinCoot (version 0.8.4) (37). The built modelled structure of the ActiD–d(TTGGCGAA)2 complex was used as a template to determine the phases of the ActiD–d(TTGGCGAA)2 complex at the higher resolution of 1.52 Å. Structure refinements were performed using the PHENIX (v1.8.4–1496) package; the crystallographic and refinement statistics of these complexes are listed in Supplementary Table S1. All atoms in the molecules were modelled in and are well defined, as indicated by the quality of the electron density maps (Supplementary Figure S1a and b). The DNA nucleotide geometry parameters reported by Parkinson et al. were used (38) in the refinement. The final 2Fo-Fc electron density maps were created using CCP4i and PyMOL (version 2.2.3) was used to draw the graphical representations of the refined structures (39). DNA helical parameters were analysed using Web-3DNA (40) and the CURVES+ program (41).
Surface plasmon resonance (SPR) analysis
The affinity between ActiD and the DNA duplexes was measured using a BIAcore 3000 A surface plasmon resonance (SPR) instrument (Pharmacia, Uppsala, Sweden) with a SensorChip SA5 (Pharmacia) by monitoring the change in the refractive index of the sensor chip surface. In general, these changes are assumed to be proportional to the mass of the molecules bound to the chip and are recorded in resonance units (RU). The 5′-biotin labelled hairpin DNA duplexes used in the SPR experiments were purified by polyacrylamide gel electrophoresis. To control the amount of DNA that bound to the streptavidin SA chip surface, the biotinylated oligomer was manually immobilized on the chip surface. ActiD was prepared in a solution of 30 mM Tris–HCl buffer (pH 7.3) and 100 mM NaCl. Different concentrations of drugs were passed over the chip surface for 90 s at a flow rate of 30 μl min−1 to reach equilibrium. One of the flow cells was used as a blank control. Then, the blank buffer solution was passed over the chip to initiate the dissociation reaction, and this flow was continued for an additional 600 s to complete the reaction. Next, the surface was recovered by washing with 10 ml of 2 mM HCl. Sensorgrams for the interactions between the hairpin DNA duplexes and the drug were analysed using the BIA evaluation software, version 3. The SPR-binding constants of ActiD bound to the G:G mismatched DNA were calculated using a bivalent ligand model (42), which describes the two sequential binding events for ActiD binding to DNA duplex as described below:
 |
This model usually takes cooperative effects into account. The fit was considered acceptable when chi-square values were <3.
Fluorescence measurements
Fluorescence spectra were used to monitor the interactions between d(TTGGCGAA)2 duplex and 7-amino-actinomycin D (7-AActiD). 7-AActiD is a structural analogue of ActiD which shows fluorescence properties. Under similar conditions, it is proposed that the intercalation ability of 7-AActiD remains similar to that of native ActiD (43). The binding of 7-AActiD to DNA causes enhanced fluorescence emission exhibiting a maximum near 650 nm and a fluorescence intensity enhancement near 520 nm in the excitation spectrum with 650-nm emission monitoring (43). The total concentration of DNA and 7-AActiD was set at 2 μM and incubated at 25°C for 2 h. All spectra were recorded at 25°C in 50 mM Tris–HCl, pH 7.3 and 100 mM NaCl. Fluorescence spectra were measured at 25°C with a JASCO model FP-4500 spectrofluorophotometer (44). Experiments were performed using a 3 × 3 mm quartz cell cuvette with a 1 cm path length. The emission wavelength was set at 650 nm, and the excitation wavelengths were 420–600 nm with the scanning speed of 50 nm mn−1.
RESULTS
Structure of the d(TTGGCGAA)2 DNA duplex with G:G mismatches stabilized by CoII(Chro)2
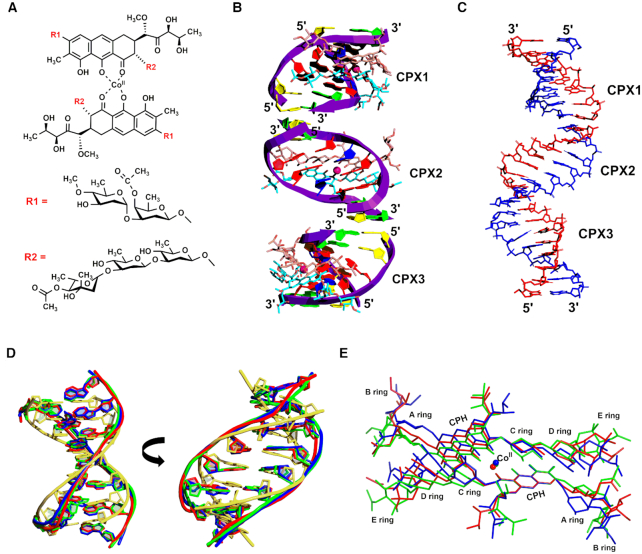
In order to understand the structural basis of G:G mismatch flexibility, we solved the crystal structure of d(TTGGCGAA)2 containing two G:G mismatches flanked by GC base pairs at 2.50 Å resolution. As the DNA duplex containing the mismatch was highly unstable, CoII-(Chro)2 was used to stabilize and crystallize the overall complex structure (Figure 1A). The oligonucleotide self assembles into an antiparallel duplex comprising three continuous independent duplexes in one asymmetric unit (Figure 1b); these duplexes were labelled as CPX1, CPX2 and CPX3, respectively (Figure 1C). The DNA duplexes were further stabilized by coordination of the N7 of guanines G4 and G12 with the CoII ions along with water-mediated interactions with the guanine O6 and N1 atoms in the three duplexes (Supplementary Figure S2). The details of CoII-(Chro)2 binding to the DNA were found to be similar to those reported previously for drug-DNA interactions, which showed some differences in the three complexes (26). These differences in drug–DNA interactions might be due to the presence of a G:G mismatch and its conformational diversity and heterogeneity in the crystal structures. The details of CoII-(Chro)2 binding to the d(TTGGCGAA)2 duplex are described in the Supplementary Note and in Supplementary Figure S3 and Table S2.
Figure 1.
Overall structure of the CoII-(Chro)2–d(TTGGCGAA)2 complex. (A) Chemical structure of the CoII-(Chro)2 dimer complex. (B) Refined structure of the CoII-(Chro)2–d(TTGGCGAA)2 complex viewed from the major groove. The asymmetric unit contains three independent complexes (CPX1, CPX2, and CPX3). The DNA duplex is represented in the purple cartoon, cobalt(II) ions are shown as pink spheres, and (Chro)2 are shown in brown and sky-blue sticks. Thymines are coloured in yellow, guanines in red, cytosines in blue, and adenines in green. (C) The d(TTGGCGAA)2 duplex backbone is represented as red and blue sticks in all three complexes (CPX1, CPX2, and CPX3). (D) Superimposition of the overall structure between CPX1 (green), CPX2 (red), and CPX3 (blue) of d(TTGGCGAA)2 duplexes with modelled B-form DNA d(TTGGCGAA/TTCGCCAA) (yellow). (E) Superimposition of the [CoII(Chro)2] ligand from three independent complexes: CPX1 (green), CPX2 (red) and CPX3 (blue).
The self-assembled asymmetric unit of three duplexes is held together by van der Waals contacts between the stacking bases to form a pseudo-continuous double helix organized as a right-handed plectonemic supercoil. Even though the three duplexes have different packing environments, they are virtually superimposable, with a root-mean-square deviation of 0.67 Å (Figure 1D). The central 4 bp segment of each duplex adopting a B-like character comprises two G:G mismatches separated by two C:G base pairs. The roll angles between the central GpC base pairs are negative, which results in curvature of the DNA structures toward the major groove (Supplementary Figure S4). As a consequence widening of the minor groove was observed, which is a result of CoII(Chro)2 binding. The average minor-groove width for each complex is 14.2–14.6 Å at the central GpC base pairs whereas the minor groove slightly widens at G:G mismatch sites up to 17.2 Å in each complex, which differs substantially from the average width of B-DNA (45) (6.0 Å) (Figure 1D). Nevertheless, although the three DNA duplexes show overall structural similarity, conformational flexibility was observed for the CoII(Chro)2 dimers in all three complexes (Figure 1E) (Supplementary Figure S5). The two chromophores in each of these complexes are at a dihedral angle of c.a. 85°–104°, which is substantially different from that in the previously reported crystal structures of MetalII–(Chro)2 complexes (26) (Figure 1E), indicating the degree of conformational change required to accommodate the G:G mismatch-containing DNA duplexes.
Asymmetric G:G mismatch base-pairing and its effects on DNA duplex geometry
The duplex contains six G:G mismatched pairs in three individual complexes (Figure 2A–C). The glycosidic torsion angles, χ, of the majority of the residues in the central GpGpCpG core occur in anti conformation ranges (χ ∼ +90° to +180°; −90° to −180°) with the exception of G3, G11 and G14, which adopt syn conformations in CPX1, and G11 and G14 in CPX2 and CPX3 adopting syn conformations as well (χ ∼ −90° to +90°) (Supplementary Table S3). The G6:G11 mismatched base pairs adopt anti–syn conformations in all three complexes with the N1 of G6 (anti), showing a bifurcated Hoogsteen type of hydrogen bonding with O6 and N7 of G11 (syn), which has been identified in a previous study (23). The G3:G14 mismatched base pairs, which have some H-bonded distances >3.2 Å compared to that of the G6:G11 base pairs, are therefore more flexible and adopt more diverse conformations including a syn–syn form in CPX1, a water-stabilized anti–syn type in CPX2 and an unusual syn–syn base pair type in CPX3. The syn–syn type in CPX1 shows a distinct interaction pattern with the van der Waals interactions between the C6 carbonyl groups of G3 and G14 bases (Figure 2A). In CPX2, the G3:G14 mismatched base pairs adopt an anti–syn conformation with a single hydrogen bond between N2 of G3 (anti) and O6 of G14 (syn) along with water-mediated hydrogen bonding between the N2 of G3 and N7 of the G14 in the mismatch (Figure 2B). The interactions between the water molecule and the mismatch create an unusual base-pair geometry by pushing the guanines toward the exterior of the helix to create a single hydrogen bond between the bases. Notably, in CPX3, the G3 nucleotide has a glycosidic torsion angle (χ) of 147° (Figure 2C). This unusual conformation for G3 is stabilized by a hydrogen bond between the N1 atom of G3 and O6 of G14 (syn) (3.2 Å). However, the two backbone dihedral parameters ϵ and ζ between G3:G14 residues in CPX3 undergo drastic transitions (−6° and 150°, respectively) compared with G3:G14 of CPX1 (166° and −84° respectively) and CPX2 (175° and −67°). This causes G3 to be flipped in the opposite orientation to produce ‘syn-like’ geometry for G3 with a higher degree of distortion whilst forming a base pair with G14 (syn). Furthermore, the syn conformations in the G:G mismatches prefer C1′- and C4′-exo puckers, with the exception of G14 (syn) in CPX2, which adopts a C3′-endo pucker. The anti-form guanine bases in the G:G mismatches exhibit a range of sugar puckers including C1′-exo, C4′-endo, C2′-endo and C3′-endo conformations.
Figure 2.
Geometry of G:G mismatches in CoII-(Chro)2–DNA complex. (A–C) Cartoon representations of all three independent DNA duplexes present in an asymmetric unit of the CoII-(Chro)2–DNA complex. The DNA backbone is represented as silver arrows, adenine in green, thymine in orange, cytosine in red and guanine in blue. The refined (2Fo-Fc) difference Fourier electron density map shows G:G mismatched pairs of the CoII-(Chro)2–d(TTGGCGAA)2 complex, contoured at 1.0 σ. Hydrogen bonds are represented by dotted lines, with numbers indicating the distance between the two contributing atoms in Angstroms (Å).
Increased base pair opening and stretch distance were observed at the G3:G14 and G6:G11 mismatched pairs. Notably, the syn–syn and syn–syn-like conformations at G3:G14 show larger opening angles than those of anti–syn, suggesting greater flexibility. Furthermore, the syn–syn and syn–syn-like types of mismatches in CPX1 and CPX3 show distinctive high shear distance and negative tilt angle compared to those of the anti–syn and are distinct from those observed in native B- or A-DNA (Supplementary Figure S6). The local structural heterogeneity at the mismatch sites may thus function as a hotspot to destabilize the duplex and enable the formation of alternative DNA structures.
DNA binding affinity analysis of ActiD to the G:G mismatched DNA duplex
Numerous DNA intercalators have been developed to recognize mismatched sites with high selectivity. They induce nucleotide flip-out in addition to more moderate structural perturbations such as kinking (11,46–47). We therefore examined the features of the ‘hotspot’ function of the G:G mismatch structural polymorphism to obtain the structural details following DNA intercalator recognition. Initially, the binding stoichiometry of ActiD was analysed by Job titration using fluorescence spectroscopy (48). As 7-AActiD is a derivative of ActiD, the results of 7-AActiD are likely to closely match those with ActiD. A Job-type titration plot showed a distinctive maximum at c.a. 0.6 (mole fraction of 7-AActiD), identifying the 2:1 stoichiometry of the complex in solution (Supplementary Figure S7). To characterize the binding affinity of ActiD to G:G mismatched DNA duplex, ActiD was allowed to interact with biotin-labelled hairpin DNA duplexes at various concentrations, and the maximum binding capacity (Rmax) (in RU) was measured by SPR. Based on the Job titration analysis and structural results, biotin-labelled hairpin DNA duplex, b-GGCG, provided two ActiD DNA-binding sites with the trinucleotide 5′-TGT-3′ as the loop region (Supplementary Figure S8a). The DNA hairpin containing the AATT sequence in the stem region was used for comparison (Supplementary Figure S8b). The SPR sensorgrams were fitted to a 2-to-1 binding model characterized by two equilibrium association constants (Ka1 and Ka2). The b-GGCG DNA exhibits a higher Ka2 (1.18 × 106 M−1) value than Ka1 (2.08 × 105 M−1) with the value of the Ka2/Ka1 ratio being approximately five indicating that the binding of two ActiD molecules to the b-GGCG DNA exhibit cooperative binding mode. The b-AATT showed no response in RU values, indicating that the ActiD is unable to bind to the AATT hairpin duplex.
Binding of ActiD forces a DNA rearrangement and right-handed Z DNA formation of the G:G mismatched DNA duplex
To understand the structural basis of the binding of ActiD to G:G mismatched DNA duplex, we crystallized the d(TTGGCGAA) sequence with a GC selective DNA intercalator, ActiD (Figure 3A). The crystals diffract to 1.52 Å resolution in the tetragonal space group I4122. The asymmetric unit contains one strand of the octamer palindromic DNA sequence, d(TTGGCGAA), bound by one ActiD molecule. In the complex structure, the phenoxazone rings of ActiD (pink) and ActiD* (cyan) intercalate into the G3:A7*/C5:G6* and G6:C5*/A7:G3* base pairs, respectively, through the minor groove side (Figure 3B) consistent with the Job titration analysis. Additionally, the two bulky peptide rings expand the minor groove width at the central (G3:A7*)/(C5:G6*) step in the ActiD–d(TTGGCGAA)2 complex and force the two backbones to unwind to generate additional space. The 5′ guanosine (G4) in the central GGCG core is flipped out of the duplex DNA upon drug binding, and the guanine base becomes perpendicular to the long axis of the flanking G:C base pairs (Figure 3B). The 5′ cytosine (C5*) that was originally paired with G4 forms a base pair with the 3′ guanosine (G6) from the destabilised G:G mispair. Another guanine from the G:G mismatch (G3) forms a mismatched base pair with the adenosine (A7*) from the complementary strand of d(TTGGCGAA). The G3:A7 mismatch observed in the current structure also adopts the more common and favoured type of anti–syn purine:purine mismatch conformation (Figure 3C). The helix axis of the top half is bent with an angle of c.a. 61° away from that of the bottom half. The bending angle and the backbone distortion are far greater than those observed in the previous structure of the drug–DNA complex.
Figure 3.
DNA rearrangement and right-handed Z DNA formation in the G:G mismatched DNA duplex forced by actinomycin D. (A) Chemical structure of actinomycin D (ActiD). (B) Biological assembly and schematic representation of the ActiD–d(TTGGCGAA)2 complex viewed from the front and a side-view. The bases are numbered from T1 to A8; the bases in the asymmetric unit are numbered with an asterisk (*) sign. The DNA backbone is coloured blue, adenine in green, thymine in yellow, cytosine in blue and guanine in red. (C) Refined (2Fo-Fc) difference Fourier electron density map showing A:G and G:A mismatched base pairs of the ActiD–d(TTGGCGAA)2 complex. (D) Comparison of the central core of the d(TTGGCGAA)2 DNA backbone with the d(CGCGCG)2 Z-DNA structure (PDB ID: 2DCG). (E) Twist and Roll angle parameter comparison for DNA backbones of the ActiD–d(TTGGCGAA)2 complex and the d(CGCGCG)2 Z-DNA. Values for perfect A- and B-DNA helices are also shown for comparison.
The binding of ActiD to the central -GGCG- causes considerable changes in DNA geometry compared to canonical A- or B-DNA (Supplementary Figure S9 and Table S4). The flipped-out G4 and G4* have torsion angles (ζ) of 107°, which differ by 289° and 265°, respectively, from those observed in A- and B-DNA. The central C5 and G6*, which adopt anti (C5) and syn (G6*) conformations, exhibit greater propeller twist angles and low stretch distances. The DNA helical twist at the ActiD intercalation site at the GpC step is 59°, which indicates that this step is overwound whereas the helix is unwound at the central CpG step with a low twist angle of 3°. Notably, the duplex again shows over winding at the second ActiD binding site (twist angle 49°). The large positive roll angle at the central CpG step accompanied by a low twist angle induce a sharp bend at the central step of the DNA helix toward the major groove. Upon comparing the opposite twist characteristics and zig-zag-like backbone shape of the DNA duplex with those of the left-handed Z DNA structure (49), we propose that the present ActiD–DNA complex structure may be categorized as a right-handed Z-DNA structure (Figure 3D and E). This zig-zag structure may relieve the steric conflicts between the inwardly pointing bulky peptide rings associated with the two ActiD molecules and absorb the torsional stress caused by over-winding at the GpC step caused by ActiD binding. Moreover, the G:A mismatched base pairs also have a larger opening angle along with low shear and the central G:C base pairs has a smaller opening angle with high shear (Supplementary Figure S9).
Structural details of the ActiD binding sites
Figure 4A and B shows a close-up view of the ActiD–DNA complex at the drug intercalation site. The two halves of the complex exhibit global similarities albeit with local differences. In the complex, the two drug molecules ActiD and ActiD* maintain a pseudo 2-fold symmetry in the same orientation, which is different than a previously reported ActiD–DNA complex, in which the two ActiDs occur in opposite orientations (42). The phenoxazone ring of ActiD (and ActiD*) is intercalated at the G3:A7*/C5:G6* base pairs (and A7:G3*/G6:C5* base pairs) from the minor groove side by pushing out G4 (and G4*). The high G:C preference of ActiD binding can even extrude nucleotides located between a 5′-G and a 3′-C. There are extensive stacking interactions between the phenoxazone ring and the guanine bases from both sides of the ring, which were also observed in earlier X-ray (42) and nuclear magnetic resonance (NMR) (50) structures (Figure 4C). The flipped-out G4 and G4* sites, which are not disordered, form a perpendicular Π-ring/C-H interaction with the chromophore groups 4-CH3 and 6-CH3 in ActiD* and ActiD, along with a single hydrogen bond between G4:C5 and G4*:C5*, respectively (Figure 4D). In the ActiD–DNA complex, G3 and A7* form a non-canonical anti–syn purine:purine mismatch base pair. The central C5:G6* and G6:C5* base pairs are standard Watson–Crick ones. There are short intermolecular hydrogen bonds between A7*-N1 and O4′ with O3 and N2 of PXZ at a distance of 3.2 Å. In addition, hydrogen-bonded interactions are present between G3-N2 and PXZ-O1, along with G3*-N2 and PXZ-O1′ (3.1 Å). There are also strong intermolecular hydrogen bonds between G3-N3 and the NH of Thr7 and G6-N3 Thr1 of the two ActiD moieties (Figure 4E).
Figure 4.
Skeletal models showing the antibiotic binding sites in the actinomycin D (ActiD)–DNA complex. Close-up view of the ActiD-TTGGC part (ActiD shown as ball-and-stick and DNA as skeletal representation). The two phenoxazone rings (PXZ) are intercalated individually into the (G3pC5)-(G6*pA7*) step in (A) and the (G3*pC5*)-(G6pA7) step in (B), respectively. (C–E) Detailed conformation showing the stacking interactions in the ActiD–DNA complex at various base-pair steps of the refined structure. Hydrogen bonding is marked by black dotted lines. The phenoxazone ring of ActiD (yellow) and ActiD* (green) is intercalated at the A7:G3*/G6:C5* base pairs from the minor groove side by pushing out G4 (and G4*) in (C). The stacking interactions between the phenoxazone ring and C5*:G6/ A7:G3* base pairs is shown in (D). The flipped-out G4 form a single hydrogen bond between G4:C5 and G4*:C5*. The intermolecular hydrogen bonds between G3-N3 and the NH of Thr7 and G6-N3 Thr1 of the two ActiD moieties are shown in (E).
Water-cluster, triplet-stranded base pair formation and Na+-mediated hydrogen bonding affect the stabilization of ActiD and the right-handed Z DNA complex
A cluster of six linked water molecules was observed in the ActiD–DNA complex structure (Figure 5A). Four of these waters mediate the intra-strand and two mediate the inter-strand interactions. This cluster appears to play a key structural role in stabilizing the kinked DNA structure with the flipped out guanines. The N2 atoms of the flipped out guanines (G4 and G4*) are linked to each other through two water-mediated interactions, W38 and W38*. The guanine O6-G4 (O6-G4*) is linked to the N7-G3 (N7-G3*) through water (W13/W13*)-mediated hydrogen bonding with distances of 2.7 and 2.8 Å. On the other side, N2-G4 (N2-G4*) interacts with O6-G6 (O6-G6*) through the water (W14/W14*) at distances of 2.9 and 2.8 Å. Analysis of the high-resolution crystal structure of the ActiD–DNA complex reveals that water molecules are positioned in order to hold and stabilize the entire kinked DNA backbone structure toward the major groove side.
Figure 5.

Water-cluster, triplet-stranded base pair formation and Na+-mediated hydrogen bonding stabilizing the ActiD–DNA complex structure. (A) Water-mediated hydrogen bonding of the flipped out guanines, stabilizing the overall structure of the ActiD–DNA complex. (B) Coordination of the metal ions in the ActiD–DNA complex. The 2Fo-Fc electron density is contoured at 1.0 σ. The coordinated metal ions and water molecules appear clearly in the refined structures between the symmetry units. Coordination and hydrogen bonds are shown by dashed lines. The hydrated Na+ ion, with ∼50% occupancy in the complex, coordinates with two symmetry-related actinomycin residues, having square-planar antiprismatic geometry. The terminal bases form a (T:A):T triplet with the symmetry-related strands. In a triplet, adenine forming a Watson–Crick base pair and a Hoogsteen base pair with thymine.
We also observed π–π stacking, triplet-stranded base pairing, and metal ion-mediated hydrogen bonding, which stabilizes the crystal packing of the complex. The adenine (A8) at the end of the sequences is stacked on the adenine (A8) from the symmetry-related strand (Figure 5B). The symmetry-related strands can also interact through the triplet hydrogen bonding between the (A:T):T from three independent strands in an asymmetric unit. The first A:T base pair follows standard Watson–Crick base pairing whereas another T:A base pair in a triplet is Hoogsteen-type. Moreover, we also observed monovalent metal ions that stabilize the crystal packing of the complex. A single Na+ cation was found to interact with two symmetry-related thymines in each strand as well as three water molecules with tri-coordination geometry.
DISCUSSION
The presence of mismatches usually causes local distortions in DNA duplex structure, and accumulation of mismatches may eventually alter the structural topology of DNA (11,51). Failure to correct for mismatched base pairs eventually leads to abnormal functioning of the cell and is the dominant mechanism behind a significant number of genetic defects and cancers (2,4,52). Out of the eight known mismatches, the G:G mismatch has been reported to be profoundly destabilizing of DNA duplexes but has been rarely characterized structurally by X-ray crystallography (23,53) and NMR (54,55). G:G mismatches have been reported to be present in G-quadruplexes, purine–purine DNA triplexes and RNA stem bulge structures associated with various disease states (56–58). Moreover, neurological disorders such as fragile X syndrome are also correlated with the expansion of (CGG)/(CCG)n trinucleotide repeats containing many contiguous GpC pairs flanked by mismatched G:G base pairs (59,60). Thus, because of its structure and biological implications, the G:G base pair has been of considerable interest for studies at the interface of structural biology. In the current study, CoII(Chro)2 has been shown to recognize GpC sites through groove binding and stabilize the overall d(TTGGCGAA)2 duplex, which has enabled us to analyse the mismatch polymorphism of G:G mismatches in DNA duplexes. Previous studies have shown that anti–syn conformations are more common in G:G mismatch structures (Supplementary Table S5), in which the guanine bases tend to form hydrogen bonds in a Hoogsteen-type manner (61–64). We now confirm the existence of both G(syn):G(syn) and G(anti):G(syn) arrangements in the folded backbone structures of d(TTGGCGAA)2 DNA duplexes. The anti–syn geometry for the G6:G11 mismatch in the duplexes is well suited to optimize the stability of the mismatch, with G11 in a syn conformation engaging in Hoogsteen-edge pairing with G6 to reduce the steric clashes between the mismatched residues. Alternatively, the G3:G14 mismatch pair shows considerable conformational heterogeneity, adopting syn–syn, anti–syn or syn–syn-like geometry in the three complexes. The G3:G14 in CPX1 adopts a syn–syn conformation, with van der Waals interaction and amide-π stacking interactions between G3 and G4 ring to stabilize the syn–syn geometry (Figure 6A). In spite of the weak nature of such interactions, similar ones have been reported in several nucleic acid structures including Z-DNA and RNA structures (65). For instance, the geometrical arrangement of the oxygen atom of the sugar unit and the π orbitals of the guanine rings provides stability to Z-DNA structures (66). A single hydrogen-bonded anti–syn arrangement is observed in G3:G14 of CPX2, where the stability to the more distant anti–syn base pairing is compensated via the water-mediated interaction between the mismatched bases (Figure 6B). Previous studies also showed that two mismatches with larger opening angles including T:T and C:T can be reinforced by water-mediated pairing to enhance the overall structural stability of the ligand–DNA interactions (61,67). Notably, in CPX3, G3:G14 has the most unusual base pairing, in which the G3 adopts a syn like conformation based on the unusual ‘χ’ value of 147° yet G3 exhibits a C1′-exo pucker most usually associated with syn geometry (Figure 6C). Previous NMR studies proposed the conformational exchange between anti and syn base pairs in G:G mismatch structures owing to the flipping of the guanine base in DNA alone structures (54,55). Cognet et al. also noted that the mismatched guanine residue can exhibit a conformation intermediate between that expected for anti or syn types (68). Thus, it would be interesting in the future to study the pathways for conformational interchange amongst the polymorphic G:G mismatches.
Figure 6.
Close-up views of G3:G14 mismatched pairs in the CoII-(Chro)2–d(TTGGCGAA)2 structure showing the base-pair geometry, stacking interactions and hydrogen bonding patterns. (A) The G3(syn):G14(syn) pair in CPX1 is stabilized by amide–Π stacking interactions (shown in red-dashed lines) between N1 of the G3 and G4 ring and the van der Waals interactions between the oxygen of the C6 carbonyl group of the G3 and G14 bases (indicated by green-dashed lines). (B) Water-mediated hydrogen bonds between the N2 atom of G3(anti) and N7 of G14(syn) bases are represented as black dotted lines in CPX2, where the water is shown as a cyan sphere. (C) Distorted geometry between the G3(anti):G14(syn) shows that the syn–syn-like characteristic is stabilized by a single hydrogen bond between the N1 of G3 and the oxygen of the C6 carbonyl group in CPX3.
The polymorphism and local instabilities in the mismatches may act as hotspots for small molecules to recognize the mismatch sites. Barton's group has shown that rhodium metalloinsertors can recognize DNA mismatches by extruding the mismatched base pairs into the major groove to cause moderate structural perturbations in the DNA structures (47,69–70). The present study demonstrates that the non-metallo drug ActiD can tightly bind to G:G mismatched DNA duplexes through large scale structural rearrangements, resulting in a right-handed Z-DNA-type structure with a guanine flipping out and a marked DNA kink with a 5′-GC/GA-5′ intercalation site (Figure 7A). The kink associated with minor groove complex formation has been previously observed in the structures of the complexes formed between small molecules such as antibiotics and ruthenium-based compounds and their DNA target sites. In these kink angles induced by these ligands are different for each binding mode. Cardin and coworkers characterized induction of substantial kinks of ∼51° and 49° in duplex DNA by λ–[Ru (TAP)2(dppz)]2+ and λ–[Ru(phen)2(dppz)]2+ complexes, respectively (17,71) (Figure 7B and C). In both cases, the chromophores TAP and phen caused a single-step sharp kink with greater stacking between the purine residues through semi-intercalation binding at the GG step, resulting in an overall curved appearance for the bound duplex. In comparison, Wu et al. reported that the bis-intercalator echinomycin preferentially binds to consecutive CpG steps separated by a T:T mismatch, causing a smooth bend (roll angle c.a. −3°), caused by the stacking interactions starting at the thymines of the sheared T:T base pair through the quinoxaline ring of echinomycin and ending at the guanines of the flanking G:C base pairs (67) (Figure 7D). The kinked DNA structure induced by anticancer drugs such as cisplatin and oxaliplatin has been found to play a significant role in the cytotoxic actions of these compounds (72,73). This specific structure might be unrecognizable to DNA repair enzymes and thus facilitate cell death by modulating processes such as replication and transcription.
Figure 7.
Comparison of DNA bending in different sequence context associated with ligand binding. The marked kink induced by (A). ActiD in the d(TTGGCGAA)2 complex (B). λ-[Ru-(TAP)2(dppz)]2+ in the d(TCGGCGCCGA)2 complex (C). λ–[Ru(phen)2(dppz)]2+ in the d(CCGGTACCGG)2 complex and (D) echinomycin in the d(ACGTCGT)2 complex, at the single-step in various DNA duplexes. (E) The sharp bend associated with the ActiD ligand bound to the neighbouring GpC sites flanking a G:G mismatch in the d(ATGCGGCAT)2 duplex is due to the sum of roll angles of three different steps at the intercalation site. The DNA bending angles are measured in terms of the respective roll angles (°) at the kinked step. The lower figure shows a schematic representation of ligand-induced DNA bending due to different intercalators in each duplex. The DNA bases are coloured as adenine-green, thymine-pink, cytosine-yellow, and guanine-blue. The respective PDB IDs for each structure are given in bold, red letters.
Previous studies have shown that the GpC site is the preferred intercalation site for ActiD when flanked with A:T, T:T, or G:G pairs (15,42,50). ActiD bound to the neighbouring GpC sites flanking a G:G mismatch has also been reported to cause a bend of c.a. 41° toward the major groove as a result of crowding between the two neighbouring cyclic peptides of ActiD at two overlapping GpC sites, with the two GpC sites flanking a G:G mismatch showing over- and unwinding at each ActiD binding site (42) (Figure 7E). The unusually large twist angle of −43° at the central base pair generates a left-handed helix structure similar to Z-DNA (Supplementary Figure S10a and b). In the present structure of the ActiD–TTGGCGAA complex, we demonstrate for the first time the consequences of direct binding of a widely-studied intercalator to the G:G mismatch with cooperative binding mode, revealing that a symmetric mode of binding results in sequence rearrangements to form a 5′-GC/GA-5′ intercalation site rather than the classic 5′-GC/CG-5′ site, together with a homopurine G:A mismatch (Supplementary Figure S10c). The marked kink is associated with significant over-winding at the ActiD binding sites (twist angles >40°) and abrupt unwinding at the centre (3° twist). The large change in twist angles at the central G4-C5-G6 residues also causes the DNA backbone to bend c.a. 90° twice around the central C5 and follow a zigzag path. Nevertheless, despite these differences, there are parallels between these observed modes of DNA binding of ActiD to the G:G mismatched DNA duplexes. Irrespective of whether the ActiD binds to a GpC site flanked by G:G mismatches or directly intercalates into the G:G mispair, the binding of ActiD will follow two general features: (i) the G:G mismatch will break to cause overall structural displacements resulting in new base pairs; and (ii) adjacent guanines to the G:G mismatches will invariably flip out in order to form a new GpC step in the re-arranged duplex (Supplementary Figure S11).
In this study, we have demonstrated for the first time the existence of G:G mismatch polymorphism through X-ray crystallography. The mismatch adopts a variety of conformations and may act as a hotspot for intercalator binding. Although metallo-insertion binding has been observed as a common mismatch recognition mode for DNA binding molecules with extrusion of the mismatched bases (14,69,74), the structure presented here is the first to demonstrate that a well-studied non-metallo intercalator, ActiD, is equally capable of recognizing mismatches through intercalation accompanied by sequence rearrangement. Several small-molecule ligands have been reported to target mismatch DNA, with potential applications in the diagnosis and therapy of cancer. For instance, Barton et al. proposed the use of rhodium metalloinsertors as the basis for the development of new types of chemotherapeutic agents active against MMR-deficient cancers (75). ActiD has been used in clinical practice for a long time; however, it is highly cytotoxic with many different side effects. The structural understanding from the current study will hopefully help guide the development of future generations of more selective intercalating agents, such as ActiD derivatives, as chemical tools for the interrogation and detection of mismatch-related diseases.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession numbers 6J0H and 6J0I.
Supplementary Material
ACKNOWLEDGEMENTS
We sincerely thank the National Synchrotron Radiation Research Center (Taiwan), and the SPring-8 (Japan) staff for X-ray data collection. We especially thank Dr Masato Yoshimura for his generous help in X-ray data collection at SPring-8, Japan. We are also grateful to the staffs of Technology Commons, College of Life Science, NTU for help with surface plasmon resonance (SPR) experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology, Taiwan [106-2628-M-005-001-MY3 to M.-H.H.]. Funding for open access charge: Ministry of Science and Technology, Taiwan.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jeggo P.A., Pearl L.H., Carr A.M.. DNA repair, genome stability and cancer: a historical perspective. Nat. Rev. Cancer. 2015; 16:35–42. [DOI] [PubMed] [Google Scholar]
- 2. Kunkel T.A., Erie D.A.. Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 2015; 49:291–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gaillard H., Garcia-Muse T., Aguilera A.. Replication stress and cancer. Nat. Rev. Cancer. 2015; 15:276–289. [DOI] [PubMed] [Google Scholar]
- 4. Lord C.J., Ashworth A.. The DNA damage response and cancer therapy. Nature. 2012; 481:287–294. [DOI] [PubMed] [Google Scholar]
- 5. Rossetti G., Dans P.D., Gomez-Pinto I., Ivani I., Gonzalez C., Orozco M.. The structural impact of DNA mismatches. Nucleic Acids Res. 2015; 43:4309–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Raiber E.A., Murat P., Chirgadze D.Y., Beraldi D., Luisi B.F., Balasubramanian S.. 5-Formylcytosine alters the structure of the DNA double helix. Nat. Struct. Mol. Biol. 2015; 22:44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Varani G., McClain W.H.. The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000; 1:18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Faibis V., Cognet J.A., Boulard Y., Sowers L.C., Fazakerley G.V.. Solution structure of two mismatches G.G and I.I in the K-ras gene context by nuclear magnetic resonance and molecular dynamics. Biochemistry. 1996; 35:14452–14464. [DOI] [PubMed] [Google Scholar]
- 9. Ghosh A., Kar R.K., Krishnamoorthy J., Chatterjee S., Bhunia A.. Double GC:GC mismatch in dsDNA enhances local dynamics retaining the DNA footprint: a high-resolution NMR study. Chemmedchem. 2014; 9:2059–2064. [DOI] [PubMed] [Google Scholar]
- 10. Satange R., Chang C.K., Hou M.H.. A survey of recent unusual high-resolution DNA structures provoked by mismatches, repeats and ligand binding. Nucleic Acids Res. 2018; 46:6416–6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Granzhan A., Kotera N., Teulade-Fichou M.P.. Finding needles in a basestack: recognition of mismatched base pairs in DNA by small molecules. Chem. Soc. Rev. 2014; 43:3630–3665. [DOI] [PubMed] [Google Scholar]
- 12. Tseng W.H., Chang C.K., Wu P.C., Hu N.J., Lee G.H., Tzeng C.C., Neidle S., Hou M.H.. Induced-Fit recognition of CCG trinucleotide repeats by a Nickel-Chromomycin complex resulting in Large-Scale DNA deformation. Angew. Chem. Int. Ed. Engl. 2017; 56:8761–8765. [DOI] [PubMed] [Google Scholar]
- 13. Jourdan M., Granzhan A., Guillot R., Dumy P., Teulade-Fichou M.P.. Double threading through DNA: NMR structural study of a bis-naphthalene macrocycle bound to a thymine-thymine mismatch. Nucleic Acids Res. 2012; 40:5115–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zeglis B.M., Pierre V.C., Kaiser J.T., Barton J.K.. A bulky rhodium complex bound to an adenosine-adenosine DNA mismatch: general architecture of the metalloinsertion binding mode. Biochemistry. 2009; 48:4247–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hou M.H., Robinson H., Gao Y.G., Wang A.H.. Crystal structure of actinomycin D bound to the CTG triplet repeat sequences linked to neurological diseases. Nucleic Acids Res. 2002; 30:4910–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hall J.P., Sanchez-Weatherby J., Alberti C., Quimper C.H., O'Sullivan K., Brazier J.A., Winter G., Sorensen T., Kelly J.M., Cardin D.J. et al.. Controlled dehydration of a ruthenium complex-DNA crystal induces reversible DNA kinking. J. Am. Chem. Soc. 2014; 136:17505–17512. [DOI] [PubMed] [Google Scholar]
- 17. Hall J.P., O'Sullivan K., Naseer A., Smith J.A., Kelly J.M., Cardin C.J.. Structure determination of an intercalating ruthenium dipyridophenazine complex which kinks DNA by semiintercalation of a tetraazaphenanthrene ligand. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:17610–17614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang T.Y., Chang C.K., Kao Y.F., Chin C.H., Ni C.W., Hsu H.Y., Hu N.J., Hsieh L.C., Chou S.H., Lee I.R. et al.. Parity-dependent hairpin configurations of repetitive DNA sequence promote slippage associated with DNA expansion. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:9535–9540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carter E.K., Laughlin-Toth S., Dodd T., Wilson W.D., Ivanov I.. Small molecule binders recognize DNA microstructural variations via an induced fit mechanism. Phys. Chem. Chem. Phys. 2019; 21:1841–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Y.W., Satange R., Wu P.C., Jhan C.R., Chang C.K., Chung K.R., Waring M.J., Lin S.W., Hsieh L.C., Hou M.H.. Co(II)(Chromomycin)(2) complex induces a conformational change of CCG repeats from i-Motif to Base-Extruded DNA duplex. Int. J. Mol. Sci. 2018; 19:E2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Da L.T., Yu J.. Base-flipping dynamics from an intrahelical to an extrahelical state exerted by thymine DNA glycosylase during DNA repair process. Nucleic Acids Res. 2018; 46:5410–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yin Y., Yang L., Zheng G., Gu C., Yi C., He C., Gao Y.Q., Zhao X.S.. Dynamics of spontaneous flipping of a mismatched base in DNA duplex. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:8043–8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Skelly J.V., Edwards K.J., Jenkins T.C., Neidle S.. Crystal structure of an oligonucleotide duplex containing G.G base pairs: influence of mispairing on DNA backbone conformation. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:804–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Menéndez N., Nur-e-Alam M., Braña A.F., Rohr J., Salas J.A., Méndez C.. Biosynthesis of the antitumor chromomycin A3 in streptomyces griseus: analysis of the gene cluster and rational design of novel chromomycin analogs. Chem. Biol. 2004; 11:21–32. [DOI] [PubMed] [Google Scholar]
- 25. Murase H., Noguchi T., Sasaki S.. Evaluation of simultaneous binding of Chromomycin A3 to the multiple sites of DNA by the new restriction enzyme assay. Bioorg. Med. Chem. Lett. 2018; 28:1832–1835. [DOI] [PubMed] [Google Scholar]
- 26. Hou M.H., Robinson H., Gao Y.G., Wang A.H.. Crystal structure of the [Mg2+-(chromomycin A3)2]-d(TTGGCCAA)2 complex reveals GGCC binding specificity of the drug dimer chelated by a metal ion. Nucleic Acids Res. 2004; 32:2214–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen Y.-W., Hou M.-H.. The binding of the Co(II) complex of dimeric chromomycin A3 to GC sites with flanking G:G mismatches. J. Inorg. Biochem. 2013; 121:28–36. [DOI] [PubMed] [Google Scholar]
- 28. Hou M.H., Robinson H., Gao Y.G., Wang A.H.J.. Crystal structure of actinomycin D bound to the CTG triplet repeat sequences linked to neurological diseases. Nucleic Acids Res. 2002; 30:4910–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang S.-Y., Lee Y.-L., Lai Y.-H., Chen J.J.W., Wu W.-L., Yuann J.-M.P., Su W.-L., Chuang S.-M., Hou M.-H.. Spermine attenuates the action of the DNA intercalator, actinomycin D, on DNA binding and the inhibition of transcription and DNA replication. PLoS One. 2012; 7:e47101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilson W.D., Jones R.L.. Intercalating drugs: DNA binding and molecular pharmacology. Adv. Pharmacol. Chemother. 1981; 18:177–222. [DOI] [PubMed] [Google Scholar]
- 31. Gniazdowski M., Denny W.A., Nelson S.M., Czyz M.. Transcription factors as targets for DNA-interacting drugs. Curr. Med. Chem. 2003; 10:909–924. [DOI] [PubMed] [Google Scholar]
- 32. Xu Y., McSally J., Andricioaei I., Al-Hashimi H.M.. Modulation of Hoogsteen dynamics on DNA recognition. Nat. Commun. 2018; 9:1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murphy J.H., Trapane T.L.. Concentration and extinction coefficient determination for oligonucleotides and analogs using a general phosphate analysis. Anal. Biochem. 1996; 240:273–282. [DOI] [PubMed] [Google Scholar]
- 34. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 35. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sheldrick G.M. A short history of SHELX. Acta Crystallogr. A. 2008; 64:112–122. [DOI] [PubMed] [Google Scholar]
- 37. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Parkinson G., Vojtechovsky J., Clowney L., Brunger A.T., Berman H.M.. New parameters for the refinement of nucleic acid-containing structures. Acta Crystallogr. D. Biol. Crystallogr. 1996; 52:57–64. [DOI] [PubMed] [Google Scholar]
- 39. Kudlicki A., Rowicka M., Otwinowski Z.. The crystallographic fast Fourier transform. Recursive symmetry reduction. Acta Crystallogr. A. 2007; 63:465–480. [DOI] [PubMed] [Google Scholar]
- 40. Li S., Olson W.K., Lu X.-J.. Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures. Nucleic Acids Res. 2019; 47:W26–W34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Blanchet C., Pasi M., Zakrzewska K., Lavery R.. CURVES+ web server for analyzing and visualizing the helical, backbone and groove parameters of nucleic acid structures. Nucleic Acids Res. 2011; 39:W68–W73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lo Y.S., Tseng W.H., Chuang C.Y., Hou M.H.. The structural basis of actinomycin D-binding induces nucleotide flipping out, a sharp bend and a left-handed twist in CGG triplet repeats. Nucleic Acids Res. 2013; 41:4284–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sha F., Chen F.-M.. Actinomycin D Binds Strongly to d(CGACGACG) and d(CGTCGTCG). Biophys. J. 2000; 79:2095–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hung H.-C., Liu C.-L., Hsu J.T.A., Horng J.-T., Fang M.-Y., Wu S.-Y., Ueng S.-H., Wang M.-Y., Yaw C.-W., Hou M.-H.. Development of an Anti-Influenza drug screening assay targeting nucleoproteins with tryptophan fluorescence quenching. Anal. Chem. 2012; 84:6391–6399. [DOI] [PubMed] [Google Scholar]
- 45. Neidle S. Neidle S. Chapter 3 DNA structure as observed in fibers and crystals. Principles of Nucleic Acid Structure. 2008; NY: Academic Press; 38–80. [Google Scholar]
- 46. Nakatani K., Hagihara S., Goto Y., Kobori A., Hagihara M., Hayashi G., Kyo M., Nomura M., Mishima M., Kojima C.. Small-molecule ligand induces nucleotide flipping in (CAG)n trinucleotide repeats. Nat. Chem. Biol. 2005; 1:39–43. [DOI] [PubMed] [Google Scholar]
- 47. Pierre V.C., Kaiser J.T., Barton J.K.. Insights into finding a mismatch through the structure of a mispaired DNA bound by a rhodium intercalator. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Olson E.J., Bühlmann P.. Getting more out of a job plot: Determination of reactant to product stoichiometry in cases of displacement reactions and n:n complex formation. J. Org. Chem. 2011; 76:8406–8412. [DOI] [PubMed] [Google Scholar]
- 49. Wang A.H.J., Quigley G.J., Kolpak F.J., Crawford J.L., van Boom J.H., van der Marel G., Rich A.. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979; 282:680–686. [DOI] [PubMed] [Google Scholar]
- 50. Chen H., Liu X., Patel D.J.. DNA bending and unwinding associated with actinomycin D antibiotics bound to partially overlapping sites on DNA. J. Mol. Biol. 1996; 258:457–479. [DOI] [PubMed] [Google Scholar]
- 51. Chakarov S., Petkova R., Russev G.C., Zhelev N.. DNA damage and mutation. Types of DNA damage. Biodiscovery. 2014; 11:e8957. [Google Scholar]
- 52. Gonzalez-Perez A., Sabarinathan R., Lopez-Bigas N.. Local determinants of the mutational landscape of the human genome. Cell. 2019; 177:101–114. [DOI] [PubMed] [Google Scholar]
- 53. Batra V.K., Beard W.A., Pedersen L.C., Wilson S.H.. Structures of DNA polymerase mispaired DNA termini transitioning to Pre-catalytic complexes support an Induced-Fit fidelity mechanism. Structure. 2016; 24:1863–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lane A.N., Peck B.. Conformational flexibility in DNA duplexes containing single G.G mismatches. Eur. J. Biochem. 1995; 230:1073–1087. [DOI] [PubMed] [Google Scholar]
- 55. Borden K.L., Jenkins T.C., Skelly J.V., Brown T., Lane A.N.. Conformational properties of the G.G mismatch in d(CGCGAATTGGCG)2 determined by NMR. Biochemistry. 1992; 31:5411–5422. [DOI] [PubMed] [Google Scholar]
- 56. Kettani A., Kumar A.R., Patel D.J.. Solution structure of a DNA quadruplex containing the fragile X syndrome triplet repeat. J. Mol. Biol. 1995; 254:638–656. [DOI] [PubMed] [Google Scholar]
- 57. Macaya R.F., Gilbert D.E., Malek S., Sinsheimer J.S., Feigon J.. Structure and stability of X.G.C mismatches in the third strand of intramolecular triplexes. Science. 1991; 254:270–274. [DOI] [PubMed] [Google Scholar]
- 58. Zhang Y., Roland C., Sagui C.. Structure and dynamics of DNA and RNA double helices obtained from the GGGGCC and CCCCGG hexanucleotide repeats that are the hallmark of C9FTD/ALS diseases. ACS Chem. Neurosci. 2017; 8:578–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McMurray C.T. Mechanisms of trinucleotide repeat instability during human development. Nat. Rev. Genet. 2010; 11:786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yu S., Pritchard M., Kremer E., Lynch M., Nancarrow J., Baker E., Holman K., Mulley J.C., Warren S.T., Schlessinger D. et al.. Fragile X genotype characterized by an unstable region of DNA. Science. 1991; 252:1179–1181. [DOI] [PubMed] [Google Scholar]
- 61. Johnson S.J., Beese L.S.. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004; 116:803–816. [DOI] [PubMed] [Google Scholar]
- 62. Natrajan G., Lamers M.H., Enzlin J.H., Winterwerp H.H., Perrakis A., Sixma T.K.. Structures of Escherichia coli DNA mismatch repair enzyme MutS in complex with different mismatches: a common recognition mode for diverse substrates. Nucleic Acids Res. 2003; 31:4814–4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Thiviyanathan V., Somasunderam A., Hazra T.K., Mitra S., Gorenstein D.G.. Solution structure of a DNA duplex containing 8-hydroxy-2′-deoxyguanosine opposite deoxyguanosine. J. Mol. Biol. 2003; 325:433–442. [DOI] [PubMed] [Google Scholar]
- 64. Pan F., Man V.H., Roland C., Sagui C.. Structure and dynamics of DNA and RNA double helices obtained from the CCG and GGC trinucleotide repeats. J. Phys. Chem. B. 2018; 122:4491–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Singh S.K., Das A.. The n → π* interaction: a rapidly emerging non-covalent interaction. PCCP. 2015; 17:9596–9612. [DOI] [PubMed] [Google Scholar]
- 66. Bancroft D., Williams L.D., Rich A., Egli M.. The low-temperature crystal structure of the pure-spermine form of Z-DNA reveals binding of a spermine molecule in the minor groove. Biochemistry. 1994; 33:1073–1086. [DOI] [PubMed] [Google Scholar]
- 67. Wu P.C., Tzeng S.L., Chang C.K., Kao Y.F., Waring M.J., Hou M.H.. Cooperative recognition of T:T mismatch by echinomycin causes structural distortions in DNA duplex. Nucleic Acids Res. 2018; 46:7396–7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cognet J.A., Gabarro-Arpa J., Le Bret M., van der Marel G.A., van Boom J.H., Fazakerley G.V.. Solution conformation of an oligonucleotide containing a G.G mismatch determined by nuclear magnetic resonance and molecular mechanics. Nucleic Acids Res. 1991; 19:6771–6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cordier C., Pierre V.C., Barton J.K.. Insertion of a bulky rhodium complex into a DNA cytosine-cytosine mismatch: an NMR solution study. J. Am. Chem. Soc. 2007; 129:12287–12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Boyle K.M., Barton J.K.. Targeting DNA mismatches with rhodium metalloinsertors. Inorg. Chim. Acta. 2016; 452:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Niyazi H., Hall J.P., O'Sullivan K., Winter G., Sorensen T., Kelly J.M., Cardin C.J.. Crystal structures of Lambda-[Ru(phen)(2)dppz](2)(+) with oligonucleotides containing TA/TA and AT/AT steps show two intercalation modes. Nat. Chem. 2012; 4:621–628. [DOI] [PubMed] [Google Scholar]
- 72. Ohndorf U.-M., Rould M.A., He Q., Pabo C.O., Lippard S.J.. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature. 1999; 399:708–712. [DOI] [PubMed] [Google Scholar]
- 73. Spingler B., Whittington D.A., Lippard S.J.. 2.4 Å crystal structure of an oxaliplatin 1,2-d(GpG) intrastrand Cross-Link in a DNA dodecamer duplex. Inorg. Chem. 2001; 40:5596–5602. [DOI] [PubMed] [Google Scholar]
- 74. Song H., Kaiser J.T., Barton J.K.. Crystal structure of Delta-[Ru(bpy)(2)dppz](2)(+) bound to mismatched DNA reveals side-by-side metalloinsertion and intercalation. Nat. Chem. 2012; 4:615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Komor A.C., Schneider C.J., Weidmann A.G., Barton J.K.. Cell-Selective biological activity of rhodium metalloinsertors correlates with subcellular localization. J. Am. Chem. Soc. 2012; 134:19223–19233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession numbers 6J0H and 6J0I.