Abstract
G protein mutations are common in uveal melanomas, and the vast majority target amino acid residue Q209 in either GNAQ or GNA11. The GNAQ R183Q mutation is found in a small fraction of uveal melanomas. We report a patient with an unusual presentation of uveal melanoma arising at an early age in the setting of congenital skin and ocular surface melanosis. A 34-year-old Hispanic female with congenital bilateral nevus of Ota and ocular surface melanosis presented with progressive loss of visual acuity and was found to have a juxtapapillary uveal melanoma. She was treated with brachytherapy, but the tumor relapsed. She underwent enucleation that revealed mixed spindle and epithelioid uveal melanoma cells with no extraocular or lymphovascular spread. Next-generation sequencing performed on DNA isolated from the enucleation specimen identified a GNAQ R183Q mutation and a PMS1 truncation mutation. Cytogenetic profiling revealed no monosomy 3. These findings raise the possibility that uveal melanomas bearing G protein R183 mutations may have distinct clinicopathologic profiles compared to those with Q209 mutations. Furthermore, this is the first reported case of a mutation in the mismatch repair gene PMS1 associated with uveal melanoma.
Keywords: Uveal melanoma, Nevus of Ota, Ocular melanosis, GNAQ/GNA11, Mismatch repair
Established Facts
Ocular melanosis and nevus of Ota are associated with uveal melanoma.
GNAQ/GNA11 mutations, most commonly in Q209, are found in uveal melanomas and nevus of Ota.
Mutations in mismatch repair genes have been rarely found in uveal melanomas.
Novel Insights
We identified a patient with oculodermal/ocular melanoses and uveal melanoma carrying a rare GNAQ R183Q mutation.
We discovered a novel mutation in the mismatch repair gene PMS1 in this uveal melanoma by genomic profiling using next-generation sequencing.
Introduction
Uveal melanomas most commonly arise in aged, light-complexioned individuals. Uveal melanomas cause mortality by hematogenous metastasis typically to the liver. Histopathologic features of uveal melanomas associated with poor prognosis include epithelioid morphology, intratumoral vascular “loops,” and tumor size [1, 2]. Retrospective molecular analyses of uveal melanomas have identified genomic sequence and structural mutations with prognostic and therapeutic significance. Loss of chromosome 3 (monosomy 3) correlates with significantly worse prognosis compared to uveal melanomas retaining both copies of chromosome 3 (disomy 3) [2]. Mutations in BAP1, GNAQ, GNA11, and SF3B1 are associated with uveal melanomas [3, 4, 5, 6]. Interestingly, mutations in BRAF and NRAS are not seen in uveal melanomas but are common in dermal melanomas [7, 8].
GNAQ and GNA11 are paralogous genes that encode guanine nucleotide-binding protein alpha (α subunits, and together with β and γ subunits, form heterotrimeric G proteins that regulate intracellular signal transduction pathways). The glutamine (Q) 209 residue of GNAQ or GNA11 is frequently mutated in uveal melanomas (76.7% of primary uveal melanomas and 78.2% of metastatic uveal melanomas) [3, 6]. The arginine (R) 183 residue of GNAQ or GNA11 is mutated in a much smaller fraction of uveal melanomas (4.9% of primary uveal melanomas and 11.8% of metastatic uveal melanomas) [3, 6]. In biochemical studies, the Q209L mutation of GNAQ results in significantly stronger G protein activity compared to the R183C mutation [9]. In immunocompromised mice, GNA11 Q209L-transduced melanocytes potently formed tumors after injection while GNA11 R183C-transduced melanocytes formed significantly slower-growing tumors or no tumors or all [3]. These studies have identified the oncogenic effects of GNAQ and GNA11 mutations found in uveal melanomas and suggest that the R183 mutation is a less potent oncogenic alteration than the Q209 mutation. Clinicopathologic differences, if any, between uveal melanoma patients carrying Q209 versus R183 mutations have not been reported.
In addition to some uveal melanomas, GNAQ R183 mutations are also found in other neoplasms. The abnormal vascular malformations in the Sturge-Weber syndrome and port-wine stains bear somatic GNAQ R183 mutations [10]. GNAQ R183 mutations have also been identified in phakomatosis pigmentovascularis, sporadic syndromes characterized by concomitant congenital melanocytic and vascular proliferations that can lead to neurologic and ophthalmologic complications [11]. The Q209 mutation in GNAQ/GNA11 has not been found in these syndromes. These diseases highlight additional clinical phenotypes specific to the R183 mutation. Here, we describe a patient bearing a GNAQ R183Q mutation with ocular surface melanosis, nevus of Ota, and uveal melanoma.
Case Report
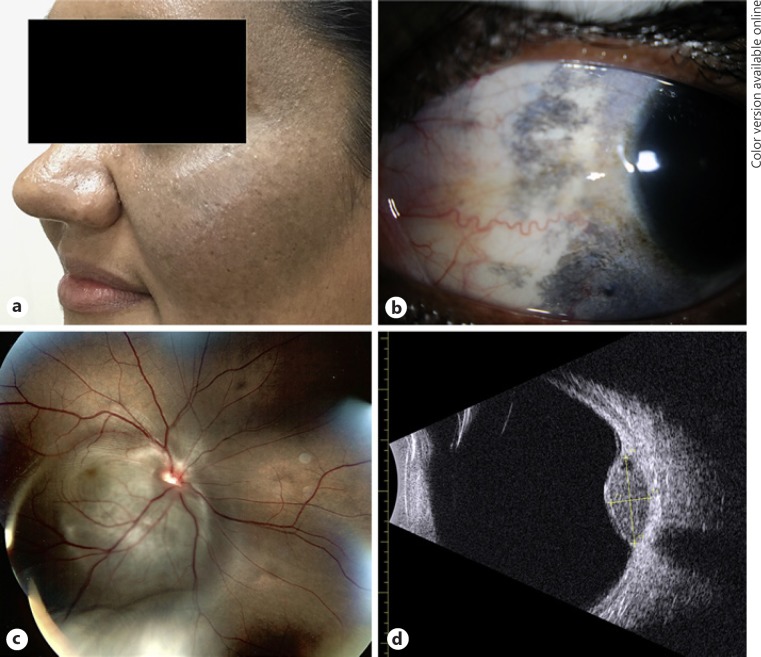
A 34-year-old Hispanic female was referred for evaluation of loss of visual acuity with an intraocular mass. Over the previous year, the patient experienced gradual deterioration of vision in the right eye. The patient's ocular history was significant for bilateral nevus of Ota and ocular surface melanosis (Fig. 1a, b). The patient had no other significant medical history. The patient reported no additional birthmarks. On examination, visual acuity was 20/20 in the left eye and 20/400 in the right eye. Fundus examination of the right eye showed a raised pigmented juxtapupillary mass (Fig. 1c), and B-scan ultrasonography revealed a discrete mushroom-shaped lesion (9.5 × 8.1 × 4.1 mm) arising from the choroid abutting the optic nerve head (Fig. 1d). The patient underwent a trans pars plana biopsy of the tumor with a 25-G vitrectomy instrument that confirmed a diagnosis of uveal melanoma (Fig. 2a, b). She was treated by brachytherapy with a notched 125I plaque, but the melanoma relapsed with a tumor arising from the optic nerve head 3 years later (2017). CT scans of the chest, abdomen, and pelvis were negative for metastasis at the time of enucleation.
Fig. 1.
a External photograph demonstrating periocular dermal hyperpigmentation typical of nevus of Ota. b External photograph demonstrating episcleral pigmentation typical of ocular melanocytosis. c Color fundus image showing a large juxtapapillary choroidal lesion at the time of diagnosis. d B-scan ultrasound of the lesion measuring 9.5 × 8.1 × 4.1 mm.
Fig. 2.
a, b Histopathology of the vitrectomy biopsy shows malignant, epithelioid, pigmented melanocytes at low (a) and high magnifications (b). c Histopathology of the uveal melanoma with overlying retina. d Higher magnification reveals uveal melanoma associated with ocular melanosis of choroid and sclera.
Histopathological analysis showed a heavily pigmented uveal melanoma with mixed epithelioid and spindle-shaped morphology (Fig. 2c–f). Vascular loops were identified within the tumor. Extraocular extension was seen. Melanosis was noted in the uninvolved choroid and sclera (Fig. 2f). FISH analysis of the enucleation slides revealed no monosomy 3 in the uveal melanoma. Tumor DNA was extracted from a vitreous biopsy specimen and a formalin-fixed enucleation specimen for molecular studies. Estimated malignant cell concentrations from the sequencing samples were determined by manually counting the number of total cells and malignant cells within the histology specimens. 80 and 30% of estimated malignant cell concentrations were determined for the vitreous biopsy and enucleation specimen, respectively. Next-generation sequencing of a panel of 397 genes identified a GNAQ c.548G>A alteration resulting in the p.R183Q protein change in both samples with a variant allele fraction of 70 and 15%, respectively (Table 1). Two additional genetic variants were also identified: PMS1 c.1888C>T (p.R630*) at a 47 and 43% variant allele fraction in each specimen, respectively, and MITF c.440C>T (p.T147M) at a 47 and 46% variant allele fraction, respectively (Table 1). No BAP-1 mutations were identified. Additionally, an SF3B1 c.1874G>A (p.R625H) mutation was observed in the vitreous biopsy specimen at 40% allele fraction (Table 1). Chromosomal microarray analysis of the tumor DNA was also performed using the Affymetrix OncoScan platform (Santa Clara) and revealed female chromosome complement with no chromosomal deletions or duplications (data not shown).
Table 1.
Summary of genomic alterations detected
| Biopsy specimen |
Estimated malignant cells (80%) |
||
|---|---|---|---|
| gene/transcript | coding variant | protein change | variant allele fraction, % |
| Clinically significant variants detected | |||
| SF3B1 | |||
| NM_012433 | c.1874G>A | p.R625H | 40 |
| GNAQ | |||
| NM_002072 | c.548G>A | p.R183Q | 70 |
| Variants of uncertain clinical significance detected | |||
| PMS1 | |||
| NM_000534 | c.1888C>T | p.R630* | 47 |
| MITF | |||
| NM_000248 | c.440C>T | p.T147M | 47 |
| HNF1A | |||
| NM_000545 | c.1624-1G>T | splice site exon 9 | 43 |
| Enucleation specimen |
Estimated malignant cells (30%) |
||
|---|---|---|---|
| gene/transcript | coding variant | protein change | variant allele fraction, % |
| Clinically significant variants detected | |||
| GNAQ | |||
| NM_002072 | c.548G>A | p.R183Q | 15 |
| Variants of uncertain clinical significance detected | |||
| PMS1 | |||
| NM_000534 | c.1888C>T | p.R630* | 43 |
| MITF | |||
| NM_000248 | c.440C>T | p.T147M | 44 |
Indicated a truncation mutation.
The patient was stable following surgery. Follow-up CT scans of the brain, chest, and abdomen revealed no metastasis of the uveal melanoma in the 18 months since enucleation.
Discussion and Conclusion
Ocular melanosis (melanosis confined to the conjunctiva and uvea) and oculodermal melanosis (ocular melanosis with cutaneous involvement) are congenital conditions characterized by abundant melanocytes that present as uveal hyperpigmentation, conjunctival melanosis, and nevus of Ota. Ocular or oculodermal melanoses are well-recognized risk factors for the development of uveal melanoma in Caucasian populations [12, 13]. Singh et al. [13] reported a 1 in 400 lifetime risk of developing uveal melanoma in patients with ocular or oculodermal melanoses, compared to a 1 in 13,000 lifetime risk in the general population. The median age at diagnosis of uveal melanoma was similar between both groups (60.5 compared to 62.5 years with or without melanosis, respectively) [13, 14]. However, ocular or oculodermal melanoses double the risk of metastatic uveal melanoma [14]. In other ethnic groups (i.e., Spanish patients), ocular and oculodermal melanoses were also found to be significant risk factors for uveal melanoma [15]. The increased risk for uveal melanoma with ocular or oculodermal melanosis has been proposed to arise from the increased numbers of melanocytes in these affected individuals [14]. However, the precise molecular and genetic pathomechanisms that enhance melanocyte transformation and metastasis are unclear. Furthermore, it is unclear if uveal melanomas arising in individuals with ocular or oculodermal melanosis share the same molecular and genetic alterations found in uveal melanomas arising in the absence of melanosis.
Monosomy 3 is a significant cytogenetic risk factor for aggressive uveal melanomas with poor prognoses [2]. The BAP1 tumor suppressor gene is found on chromosome 3p21.1 and encodes a deubiquitinase enzyme [4]. Somatic inactivating mutations of BAP1 are frequently found in metastatic uveal melanomas [4, 16]. Biallelic inactivation of BAP1 through loss of one copy of chromosome 3 combined with somatic mutation of BAP1 on the remaining chromosome 3 is an important step in uveal melanoma metastasis. BAP1 mutations are also found in Spitzoid dermal melanocytic lesions, mesothelioma, clear cell renal cell carcinoma, and other tumors [16]. Germline BAP1 mutations have also been reported in families with hereditary cancer syndromes [16]. GNAQ and GNA11 activating mutations at amino acid residue Q209, and to a lesser extent R183, are found in uveal melanomas, nevus of Ota, and dermal blue nevi [3, 6]. Other genes frequently affected by somatic mutations in uveal melanomas include the SF3B1 splicing factor and the EIF2AX translation initiation factor [5, 17].
Our case highlights a uveal melanoma arising in a young Hispanic woman with ocular and oculodermal melanosis, where genetic sequencing identified both previously known as well as novel mutations in the tumor. A GNAQ c.548G>A (p.R183Q) variant was detected at 70% allelic fraction in the tumor biopsy specimen obtained during vitrectomy (estimated 80% malignant cells) and 15% allelic fraction in the enucleation specimen (estimated 30% malignant cells), consistent with a somatic origin of this mutation in the tumor cells. A PMS1 c.1888C>T mutation, which introduces a premature stop codon in the protein, was also detected at 47% allelic fraction in the vitreous biopsy specimen (estimated 80% malignant cells) and 43% allelic fraction in the enucleation specimen (estimated 30% malignant cells), possibly indicating a germline or early neural crest origin for this mutation. PMS1 encodes a mismatch repair protein [18]. Germline mutations in mismatch repair genes such as PMS1, PMS2, MSH2, MSH3, MSH6, MLH1, and MLH3 cause hereditary nonpolyposis colorectal cancer [19], and somatic mutations in mismatch repair genes are common in sporadic colorectal cancers [20]. Loss of PMS1 has been identified in dermal melanomas but not uveal melanoma [21, 22, 23]. Microsatellite instability and mutations in mismatch repair genes MSH2 and MLH3 are infrequently found in uveal melanomas [24, 25, 26]. To our knowledge, this is the first report of a uveal melanoma associated with a mutation in the PMS1 mismatch repair gene. This specific PMS1 mutation, p.R630*, has not been experimentally tested, and it is considered a variant of uncertain clinical significance. However, it is likely a loss-of-function mutation because premature stop codons typically initiate nonsense-mediated mRNA decay that degrades the mutant transcript. Furthermore, any surviving transcript will produce a truncated and likely malfunctional destabilized protein. A mechanism linking PMS1 and GNAQ to the development of uveal melanoma has not been shown, but we speculate that defects in genomic stability (arising from loss of PMS1-mediated mismatch repair) may have led to the secondary somatic mutations found in GNAQ. Interestingly, molecular markers portending poor prognoses, BAP1 mutations and monosomy 3, were not present in this patient's melanoma, even though prior reports indicated uveal melanomas associated with ocular/oculodermal melanoses were more aggressive [14]. Furthermore, the histopathology showed some spindle cell morphology in this patient's uveal melanoma tumor and suggests a more favorable outcome.
Statement of Ethics
The authors have no ethical conflicts to disclose. This research has been approved by the UCSD committee on human research.
Disclosure Statement
The authors have no conflicts of interest to declare.
Funding Sources
This work was supported by the NIH NEI P30 P30EY0225789 Vision Research Core.
Author Contributions
C.B.T., K.F., and J.H.L. were responsible for data collection, analysis, and writing/preparation of the manuscript. M.H.G. and J.A.T. were responsible for data collection and analysis.
Acknowledgment
This work was supported by the NIH NEI P30 P30EY0225789 Vision Research Core.
References
- 1.Design and methods of a clinical trial for a rare condition: the Collaborative Ocular Melanoma Study. COMS Report No. 3. Control Clin Trials. 1993 Oct;14((5)):362–91. doi: 10.1016/0197-2456(93)90052-f. [DOI] [PubMed] [Google Scholar]
- 2.Damato B, Duke C, Coupland SE, Hiscott P, Smith PA, Campbell I, et al. Cytogenetics of uveal melanoma: a 7-year clinical experience. Ophthalmology. 2007 Oct;114((10)):1925–31. doi: 10.1016/j.ophtha.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 3.Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010 Dec;363((23)):2191–9. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010 Dec;330((6009)):1410–3. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harbour JW, Roberson ED, Anbunathan H, Onken MD, Worley LA, Bowcock AM. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet. 2013 Feb;45((2)):133–5. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O'Brien JM, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009 Jan;457((7229)):599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruz F, 3rd, Rubin BP, Wilson D, Town A, Schroeder A, Haley A, et al. Absence of BRAF and NRAS mutations in uveal melanoma. Cancer Res. 2003 Sep;63((18)):5761–6. [PubMed] [Google Scholar]
- 8.Weber A, Hengge UR, Urbanik D, Markwart A, Mirmohammadsaegh A, Reichel MB, et al. Absence of mutations of the BRAF gene and constitutive activation of extracellular-regulated kinase in malignant melanomas of the uvea. Lab Invest. 2003 Dec;83((12)):1771–6. doi: 10.1097/01.lab.0000101732.89463.29. [DOI] [PubMed] [Google Scholar]
- 9.Orth JH, Preuss I, Fester I, Schlosser A, Wilson BA, Aktories K. Pasteurella multocida toxin activation of heterotrimeric G proteins by deamidation. Proc Natl Acad Sci USA. 2009 Apr;106((17)):7179–84. doi: 10.1073/pnas.0900160106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shirley MD, Tang H, Gallione CJ, Baugher JD, Frelin LP, Cohen B, et al. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N Engl J Med. 2013 May;368((21)):1971–9. doi: 10.1056/NEJMoa1213507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas AC, Zeng Z, Rivière JB, O'Shaughnessy R, Al-Olabi L, St-Onge J, et al. Mosaic Activating Mutations in GNA11 and GNAQ Are Associated with Phakomatosis Pigmentovascularis and Extensive Dermal Melanocytosis. J Invest Dermatol. 2016 Apr;136((4)):770–8. doi: 10.1016/j.jid.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yanoff M, Zimmerman LE. Histogenesis of malignant melanomas of the uvea. 3. The relationship of congenital ocular melanocytosis and neurofibromatosis in uveal melanomas. Arch Ophthalmol. 1967 Mar;77((3)):331–6. doi: 10.1001/archopht.1967.00980020333007. [DOI] [PubMed] [Google Scholar]
- 13.Singh AD, De Potter P, Fijal BA, Shields CL, Shields JA, Elston RC. Lifetime prevalence of uveal melanoma in white patients with oculo(dermal) melanocytosis. Ophthalmology. 1998 Jan;105((1)):195–8. doi: 10.1016/s0161-6420(98)92205-9. [DOI] [PubMed] [Google Scholar]
- 14.Shields CL, Kaliki S, Livesey M, Walker B, Garoon R, Bucci M, et al. Association of ocular and oculodermal melanocytosis with the rate of uveal melanoma metastasis: analysis of 7872 consecutive eyes. JAMA Ophthalmol. 2013 Aug;131((8)):993–1003. doi: 10.1001/jamaophthalmol.2013.129. [DOI] [PubMed] [Google Scholar]
- 15.Carreño E, Saornil MA, Garcia-Alvarez C, Lopez-Lara F, De Frutos-Baraja JM, Almaraz A. Prevalence of ocular and oculodermal melanocytosis in Spanish population with uveal melanoma. Eye (Lond) 2012 Jan;26((1)):159–62. doi: 10.1038/eye.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murali R, Wiesner T, Scolyer RA. Tumours associated with BAP1 mutations. Pathology. 2013 Feb;45((2)):116–26. doi: 10.1097/PAT.0b013e32835d0efb. [DOI] [PubMed] [Google Scholar]
- 17.Martin M, Maßhöfer L, Temming P, Rahmann S, Metz C, Bornfeld N, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013 Aug;45((8)):933–6. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prolla TA, Pang Q, Alani E, Kolodner RD, Liskay RM. MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science. 1994 Aug;265((5175)):1091–3. doi: 10.1126/science.8066446. [DOI] [PubMed] [Google Scholar]
- 19.Papadopoulos N, Lindblom A. Molecular basis of HNPCC: mutations of MMR genes. Hum Mutat. 1997;10((2)):89–99. doi: 10.1002/(SICI)1098-1004(1997)10:2<89::AID-HUMU1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 20.Li SK, Martin A. Mismatch Repair and Colon Cancer: Mechanisms and Therapies Explored. Trends Mol Med. 2016 Apr;22((4)):274–89. doi: 10.1016/j.molmed.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Korabiowska M, Dengler H, Kellner S, Stachura J, Schauer A. Decreased expression of MLH1, MSH2, PMS1 and PMS2 in pigmented lesions indicates accumulation of failed DNA repair along with malignant transformation and tumour progression. Oncol Rep. 1997 May-Jun;4((3)):653–5. doi: 10.3892/or.4.3.653. [DOI] [PubMed] [Google Scholar]
- 22.Korabiowska M, Brinck U, Dengler H, Stachura J, Schauer A, Droese M. Analysis of the DNA mismatch repair proteins expression in malignant melanomas. Anticancer Res. 2000 Nov-Dec;20(6B):4499–505. [PubMed] [Google Scholar]
- 23.Korabiowska M, Brinck U, Kotthaus I, Berger H, Droese M. Comparative study of the expression of DNA mismatch repair genes, the adenomatous polyposis coli gene and growth arrest DNA damage genes in melanoma recurrences and metastases. Melanoma Res. 2000 Dec;10((6)):537–44. doi: 10.1097/00008390-200012000-00005. [DOI] [PubMed] [Google Scholar]
- 24.Cross NA, Murray AK, Rennie IG, Ganesh A, Sisley K. Instability of microsatellites is an infrequent event in uveal melanoma. Melanoma Res. 2003 Oct;13((5)):435–40. doi: 10.1097/00008390-200310000-00001. [DOI] [PubMed] [Google Scholar]
- 25.Hajkova N, Hojny J, Nemejcova K, Dundr P, Ulrych J, Jirsova K, et al. Germline mutation in the TP53 gene in uveal melanoma. Sci Rep. 2018 May;8((1)):7618. doi: 10.1038/s41598-018-26040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hussein MR, Haemel AK, Albert DM, Wood GS. Microsatellite instability and alterations of mismatch repair protein expression in choroidal melanomas. Arch Ophthalmol. 2005 Dec;123((12)):1705–11. doi: 10.1001/archopht.123.12.1705. [DOI] [PubMed] [Google Scholar]