Using gene-set association test and epistasis analysis, this research achieved higher statistical power with potentially high accuracy, and detected significant genes and gene networks that influence flowering time in barley.
Keywords: Barley, epistasis, flowering time, gene-set association analysis, GWAS, heritability, phenology, next-generation sequencing, target enrichment, target capture
Abstract
Single-marker genome-wide association studies (GWAS) have successfully detected associations between single nucleotide polymorphisms (SNPs) and agronomic traits such as flowering time and grain yield in barley. However, the analysis of individual SNPs can only account for a small proportion of genetic variation, and can only provide limited knowledge on gene network interactions. Gene-based GWAS approaches provide enormous opportunity both to combine genetic information and to examine interactions among genetic variants. Here, we revisited a previously published phenotypic and genotypic data set of 895 barley varieties grown in two years at four different field locations in Australia. We employed statistical models to examine gene–phenotype associations, as well as two-way epistasis analyses to increase the capability to find novel genes that have significant roles in controlling flowering time in barley. Genetic associations were tested between flowering time and corresponding genotypes of 174 putative flowering time-related genes. Gene–phenotype association analysis detected 113 genes associated with flowering time in barley, demonstrating the unprecedented power of gene-based analysis. Subsequent two-way epistasis analysis revealed 19 pairs of gene×gene interactions involved in controlling flowering time. Our study demonstrates that gene-based association approaches can provide higher capacity for future crop improvement to increase crop performance and adaptation to different environments.
Introduction
Barley (Hordeum vulgare L.) is one the most important cereal crops in the world and is cultivated both in highly productive agricultural regions and in marginal environments prone to adverse conditions (Baum et al., 2007). As a particularly resilient crop compared with other cereals such as wheat and rice, barley has the ability to adapt to biotic and abiotic stresses, holding a great deal of potential to increase production in marginal areas to sustain food security (Tester and Langridge, 2011). It is vital that barley flowers within a particular time window in a given environment to maximize yield, while minimizing exposure to frost, heat, and drought stress during the growing season (Maurer et al., 2015). It is also known that genes controlling phenology including flowering time (FT) overlap with many grain yield-related genes (Hill et al., 2019a). Sharma et al. (2018) identified a total of 96 quantitative trait loci (QTLs) mapped for grain yield in a nested association mapping population, the majority of which also co-localized with known genes controlling FT.
Harnessing the power of genomic tools to manipulate FT for barley improvement is of considerable importance to meet the food and feed demands of the future. Understanding the genetic basis of FT including the interactions between different FT genes has the potential to considerably enhance genetic improvement and future barley breeding. Insights gained from model plants such as Arabidopsis thaliana made it possible to explore the function of gene orthologues and related pathways in barley, but not all genes and gene networks discovered in A. thaliana are conserved across the plant kingdom. For example, monocot-specific genes and gene networks, including species-specific flowering gene networks in rice, have been reported (Xue et al., 2008; Matsubara et al., 2011).
Rapid advancements in genome sequencing technologies including reduced representation sequencing approaches, combined with high-throughput genotyping and the availability of a high-quality reference genome, now allow for an unprecedented view into complex genetic architectures in barley (Waugh et al., 2009; Huang et al., 2011; Mayer et al., 2012; Mascher et al., 2017; Sharma et al., 2018; Hill et al., 2019a). Genome-wide association studies (GWAS) have emerged as powerful tools for identifying genetic variants associated with crop plant phenotypes (Pasam et al., 2012; Yano et al., 2016; Fang et al., 2017). Commonly used single-marker GWAS approaches test each single nucleotide polymorphism (SNP) individually for the association with a trait, which has delivered considerable insight into the genetic control of traits (Yang et al., 2014). However, only the most significant SNPs in the genome are taken into account with the single-marker approach, thus can often explain only a small proportion of the genetic variation. In fact, single SNP variants explained <10% of phenotypic variation for the majority of complex phenotypes (Manolio et al., 2009). Moreover, single SNP analyses consider only the effect of individual SNP and often examine additive models only, while most quantitative traits are polygenic and thus also determined by gene×gene interactions (epistasis).
Epistasis is known to play a crucial role in regulation of many complex traits in plants, animals, and humans (Doust et al., 2014; Phillips, 2008). Different theoretical frameworks and statistical methodologies for epistasis analysis have been developed to improve the detection of genes responsible for complex human diseases (as reviewed in Wei et al., 2014). However, models that take multiple SNP markers into account are still not widely adopted and have only recently been applied to plants, including crops, to identify novel candidate genes and gene networks controlling complex agronomic traits. For example, FT is a crucial yet complex trait of interest in barley and other agronomically important crops (Hill and Li, 2016); several studies have reported gene×gene interactions affecting FT in different plant species (Caicedo et al., 2004; Durand et al., 2012; Maurer et al., 2015). Mathew et al. (2018) observed genomic regions with main or higher order epistatic effects overlapping with known candidate genes that were reported previously in barley and closely related species for FT. In sorghum, it is known that Maturity locus 1 (Ma1) represses expression of the floral activator Early heading date 1 (Ehd1), which activates FT to produce florigen for floral induction (Rooney and Aydin, 1999). Li et al. (2018) revealed a significant interaction between the QTL harbouring Ma1 and the QTL harbouring FT through epistasis analysis. The reported gene×gene interactions are consistent with the networking system proposed for the control of the timing of flowering (Blázquez, 2000; Valverde et al., 2004; Imaizumi and Kay, 2006).
To overcome these limitations, gene-set analysis (GSA) has emerged as a more powerful approach than single SNP analysis (Nam et al., 2010). GSA has several advantages. First, GSA can aggregate effects of many SNPs with weak associations. Although individual SNPs may show little or no effect, their interactions may have a non-linear effect if an unbiased analysis for interactions within combinations of SNPs is performed (Wang et al., 2012; Mooney and Wilmot, 2015; Pers, 2016). Secondly, GSA takes allelic heterogeneity into consideration (i.e. different SNPs within a gene linking to a similar phenotype) which is usually not possible in the single SNP GWAS test (Zöllner and Pritchard, 2005; Guan and Stephens, 2011; Jiang et al., 2018). Thirdly, GSA could capture local epistatic interactions between SNPs within a gene and therefore potentially increase prediction accuracies (Zhang et al., 2014; Jiang et al., 2018). As FT is believed to be controlled by a complex interacting gene network probably influenced by the effects of sets of genes (Hill and Li, 2016, and reference therein), testing associations between a phenotype and the cumulative effect of genes may identify more functionally relevant candidate genes with higher accuracy than single SNP GWAS.
Here, we revisited previously published data sets: (i) phenotypic data of 895 barley varieties grown over two years in four different field locations with varying seasonal temperature and rainfall conditions in Western Australia’s South West; and (ii) genotypic data obtained from the targeted resequencing of 174 putative phenology-related genes and gene orthologues (Hill et al., 2019a, b). Building on the previous study, here we aimed to achieve higher statistical power to detect significant genes and gene networks that influence FT in barley by expanding single SNP GWAS analysis to gene-based analysis and epistasis analysis. By taking only SNPs detected within gene-coding regions of putative FT-related genes into account, we first re-calculated the narrow-sense SNP-based heritability of awn emergence as an equivalent to FT (Alqudah and Schnurbusch, 2017). We then re-assessed the association of individual SNPs and FT by standardizing and averaging FT across multiple locations and experimental years. We further grouped SNPs from the same genes into distinct gene sets and tested the association of each gene set with FT. Finally, we identified interacting SNP pairs using a two-way epistasis analysis and determined an expanded and improved gene interaction network which regulates FT in barley.
Materials and methods
Plant material, phenotypic data, and genes enriched in SNPs
Plant material, phenotypic data, and phenology gene-enriched genetic variants were previously reported in detail in Hill et al. (2019a, b). Briefly, 952 barley accessions from 41 countries in Europe, Asia, North and South America, Africa, and Australia were initially selected to represent the global diversity for phenology genes in barley. These accessions represent the entire spectrum of cultivated barley, including two- and six-row genotypes, and winter and spring growth habits. These accessions were grown in 2015 and 2016 at four locations in Western Australia which significantly differ in rainfall and temperature during the growing season. Awn emergence, defined as the number of days from sowing to the first awn emergence above the flag leaf (Z49) (Zadoks et al., 1974), was recorded as an equivalent to FT (Alqudah and Schnurbusch, 2017). A total of 2758 SNPs were enriched from 174 putative genes that are related to phenology and the development of meristem and inflorescences. Full details of field experiments, targeted resequencing of phenology genes, and SNP discovery and filtering were provided in Hill et al. (2019a, b).
Data preparation
The original measurement of days to Z49 for each accession was transformed to standardized FT (FTD) separately for each growing environment and year using the formula:
| (1) |
We then averaged FTD across four locations and two years for each barley variety to minimize the random effect, while not shrinking the genetic effects (Piepho et al., 2008).
Barley accessions or SNP loci with >10% missing data were excluded from analysis. For the remaining missing SNP data in the data set, we inspected each missing datum individually and replaced the missing data manually with the most likely allelic combinations with consideration of linkage equilibrium and allelic state of the individual in other SNP loci. After the filtering, 895 barley accessions and 2758 SNPs remained for heritability estimation and GWAS analysis.
Estimation of narrow-sense SNP-based heritability
A genome-based restricted maximum likelihood method (GREML-LDMS) was used to estimate the heritability of FT using all filtered SNPs. GREML-LDMS corrects linkage disequilibrium biases in the estimated SNP-based heritability (Yang et al., 2015). To calculate narrow-sense heritability from SNP data, h2SNP, we first computed linkage disequilibrium (LD) scores between SNPs with the block size of 100 kb using the computer software package GCTA (Yang et al., 2011). We used the GREML (a function within GCTA) to estimate the proportion of variance in a phenotype explained by all SNPs (i.e. the SNP-based heritability), following an LD score regression approach as detailed in Yang et al. (2015). h2SNP was estimated both with and without additional data descriptors (growth habit, row type, and origin of the barley accessions) fitted as fixed effects.
Genome-wide association analysis
We used a linear mixed model (LMM) for GWAS analysis as implemented in the Factored Spectrally Transformed Linear Mixed Models (FaST-LMM) package to perform single SNP, gene-set GWAS, and epistasis analysis (Lippert et al., 2011; Listgarten et al., 2012; Widmer et al., 2014). GWAS are often confounded by population substructure and sample relatedness. LMMs are a powerful and established tool for studying genotype–phenotype relationships. LMMs can capture confounders (e.g. population substructure and family relatedness) of GWAS simultaneously, without requiring prior knowledge of whether the confounders are present or not (Lippert et al., 2011). Its computational efficiency also makes it feasible for an exhaustive search for gene×gene interactions (Lippert et al., 2013; Widmer et al., 2014).
For GWAS analysis, we calculated the first five principal eigenvectors from principal components analysis (PCA) using GCTA (Yang et al., 2011) and subsequently included them as covariates in the model as fixed effects for association analysis. GWAS analysis was conducted using the Python-based program FaST-LMM (Listgarten et al., 2012) following the developers’ instructions (available from http://microsoftgenomics.github.io/FaST-LMM/). Genetic data were formatted into the binary Plink ped input file format (*.bed, *.bim, and *.fam) using Plink 2.0 (Chang et al., 2015). For single SNP association analysis, we used the average FTD (see Equation 1) of each filtered barley accession as the phenotypic data, all filtered SNPs as genetic data, and the first five principal eigenvectors from the PCA as the covariate. For GSA, we first grouped the SNPs into 174 gene sets with each set of SNPs corresponding to one gene (each gene set had an average of 18 SNPs ranging from 1 to 167). The algorithm as employed in FaST-LMM uses two random effects—one to capture the confounder’s effect and the other to reflect the set association signal—to correct for confounder, and uncovers signal not recoverable by single-SNP GWAS analysis (Listgarten et al., 2013). For epistasis testing, one SNP (the first polymorphic SNP locus) was taken from each gene, as such a filtering approach significantly reduces the required statistical power for multiple testing. The GWAS analysis was then used to test whether pairs of SNPs taken together explain a higher proportion of variance than the sum of the individual effects of each SNP analysed separately (Widmer et al., 2014).
Because the SNPs were enriched from putative genes that were reported to be associated with FT in barley, A. thaliana, and other cereal crops, we adopted a less stringent threshold than Bonferroni correction to define the significance in GWAS. We instead used the Holm’s sequential Bonferroni correction (Holm, 1979) with a significance threshold at P<0.05 to determine significant SNPs, gene sets, and SNP pairs with epistatic interaction. Sequential Bonferroni correction is an adjusted Bonferroni correction depending on rank to maximize the statistical power in GWAS whilst being stringent. ANOVA was implemented using SPSS (Statistical Package for the Social Sciences, SPSS Inc., Chicago, Il, USA) software, and P<0.05 was used as the statistically significant threshold.
Regulatory connections between flowering genes
Interacting network of flowering genes was constructed using STRING, a database of known and predicted gene–gene (protein–protein) interactions (Szklarczyk et al., 2017). In STRING, each protein–protein interaction is assigned a score, as an indicator of confidence of a true interaction. A score of 0.7 was used to assign high confidence when retaining the interaction. Connections between the networks of each key gene were achieved by connecting the overlapping genes and epistatic interactions as revealed in the epistatic analysis.
Results
Flowering time and environmental influence
All 895 barley accessions were grown across multienvironment field trials, conducted over four geographical locations and two years in Western Australia. Significant phenotypic differences of agronomic and phenological traits measured were present for the set of barley genotypes grown in the field at different geographical locations in WA in the 2015 and 2016 growing seasons (Table 1; with more details in Hill et al., 2019a). Average time to flowering for the 895 accessions ranged from 65 d to 85 d, with median time from 72 d to 111 d, after sowing across the trial environments. The range in FTs for all accessions evaluated varied from 42–94 d to 63–136 d across the environments. Geraldton in the North of WA is characterized by a hot and dry environment with a short growing season, with the lowest median number of days to Z49 recorded for any environment (72 d), with a range of 46–89 d recorded in 2015. The trial environments at Esperance (ESP) in Southern WA have a longer, wetter, and cooler growing season, and thus recorded the longest maximum days to Z49 (146 d) in 2016.
Table 1.
Locations and experimental years, major climatic factors, and flowering time mean (days to Z49)
| Location (year) | Tmin–Tmax (°C) | Tmean (°C) | Rainfall (mm) | Global solar radiation (MJ m−2) | Growth period (d) | Days to Z49 median (range) |
|---|---|---|---|---|---|---|
| Geraldton (2015) | 2–40 | 17.3 | 189.8 | 18.03 | 182 | 72 (46–89) |
| Geraldton (2016) | 3–41 | 15.1 | 355.4 | 17.50 | 210 | 80 (44–91) |
| Katanning (2015) | 4–36 | 14.0 | 550.0 | 14.63 | 208 | 105 (69–132) |
| Katanning (2016) | -3–38 | 12.9 | 256.2 | 16.87 | 244 | 105 (75–131) |
| Esperance (2015) | 1–41 | 14.7 | 318.2 | 12.35 | 203 | 104 (60–136) |
| Esperance (2016) | 3–37 | 13.6 | 343.6 | 12.86 | 201 | 110 (72–146) |
| Merredin (2016) | -1–37 | 14.1 | 181.4 | 16.39 | 191 | 111 (80–131) |
Tmin/Tmax/Tmean: minimum/maximum/mean temperature during the growing season. Environmental data were taken over 200 d since the sowing date during the growth period for comparisons. Modified from Hill et al. (2019a)
The average range to Z49 between the earliest and the latest flowering types was 59 d, showing the considerable genetic difference in controlling the switch from vegetative growth to reproduction among the tested barley accessions. Variation of FT within barley accessions in different environments is strongly influenced by average temperature during the growth period. Growing season average temperature explained 62.9% (P=0.0001) of variance in FT across environments of four locations and two years (Fig. 1), while minimum/maximum temperature, global solar radiation, and rainfall during the growth period had no significant influence on FT (P>0.05). The trial environments received an optimum rainfall throughout the two growing seasons at all four locations.
Fig. 1.
Mean temperature influencing flowering time (days to Z49) in barley in seven experimental environmental sets across four locations in two years. r and p represent correlation coefficient and probability, respectively, assuming a linear relationship between flowering and temperature. Whiskers are standard deviations.
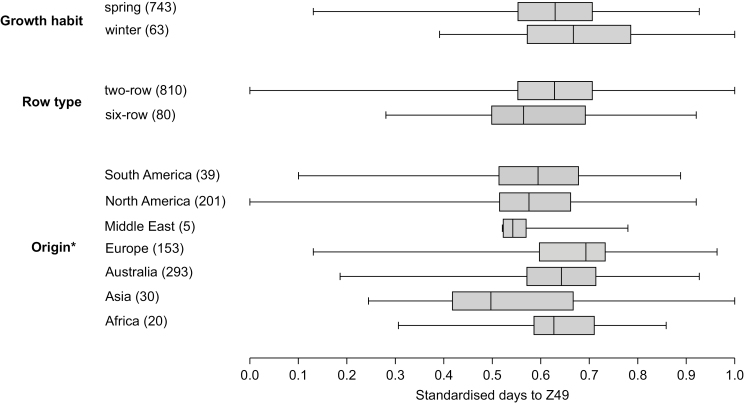
Barley accessions with contrasting growth habit (spring or winter type) had similar standardized days to Z49 (FTD), as did the barley accessions with different row type (P>0.05). However, average FTD of barley accessions with different origin was significantly different (ANOVA P<0.0001) (Fig. 2). Barley accessions with different origin also had unequal variances in FTD (ANOVA, F=9.117, df=44.2, P<0.0001).
Fig. 2.
Phenology of barley accessions with contrasting growth habits, row types, and geographic origin of accessions. Asterisk indicates significant difference in ANOVA. Numbers in parentheses indicate the number of samples, and only the samples positively identified were included.
Gene-set GWAS analysis of flowering time
After filtering (<10% missingness) and pruning to only SNPs located within gene-coding regions of the 174 targeted phenology genes, 895 barley accessions and 2758 SNP markers were retained. Genetic variation of the 2758 SNPs across the barley accessions were not structured by row type, nor by growth habit, nor by geographic origin, confirming previous findings (Hill et al., 2019a, b). Narrow-sense heritability as estimated from all SNP (h2SNP) was estimated at 0.395±0.048. Specifying the origin of each barley line in the analysis as a fixed effect increased h2SNP to 0.503±0.056, while including growth habit or row type as fixed effects did not improve the estimation of heritability. Average temperature, as one of the most significant environmental factors, explained 3.8% of the variance in FTD between barley accessions which was a small yet significant (P<0.001) amount.
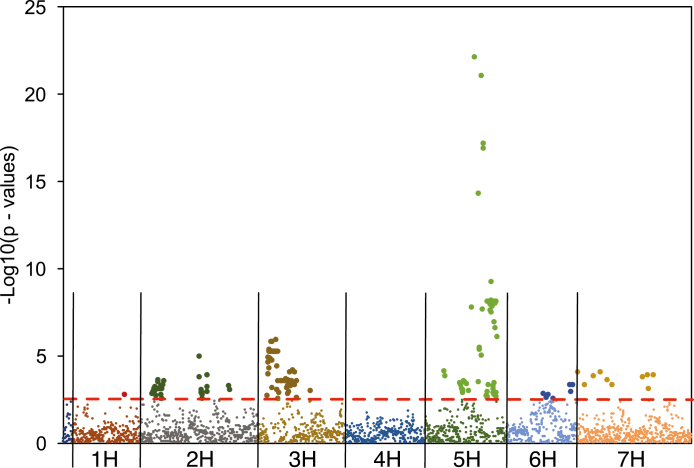
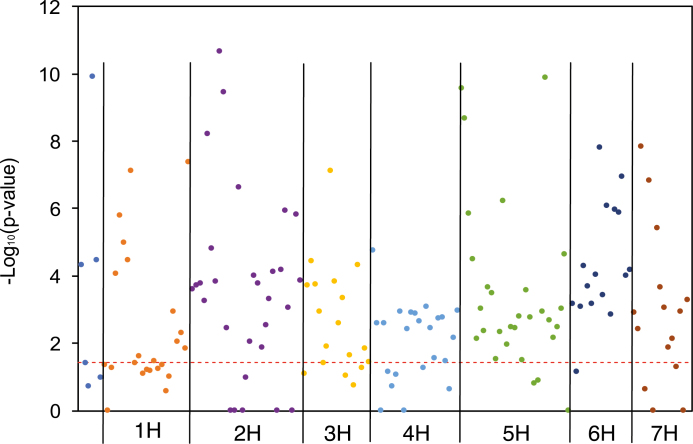
Using the sequential Bonferroni correction and significance threshold of P<0.05, GWAS analysis was performed using FaST-LMM (Listgarten et al., 2012), and 170 SNP loci were found to be associated with FTD across all environments (see the Materials and methods). Systematic biases in GWAS were low, indicated by a λ GC, the genomic inflation factor, close to 1.1 (Winkler et al., 2014) (Supplementary Fig. S1 at JXB online). These SNPs were located within the gene-coding regions of 32 genes on six chromosomes, with no significant SNPs detected on chromosomes 4H (Fig. 3). The subsequent GSA revealed 113 gene sets, corresponding to 113 putative genes, among the 174 genes that were previously shown as flowering-related genes in cereal crop species or in A. thaliana (Hill et al., 2019a, b), associated with FTD in the barley accessions. Those significant genes are located on all seven chromosomes (Fig. 4) and from all flowering pathways (Table 2): photoperiod and circadian clock (34 genes), meristem response and development (27 genes), gibberellin signalling and metabolism (19 genes), grain development (15 genes), vernalization regulation (14 genes), and light perception and signalling (10 genes). Among the 170 significant SNPs as detected in single SNP GWAS, 167 SNPs and the 29 corresponding genes they belong to were also detected as part of gene sets to be significantly associated with FTD (Table 2).
Fig. 3.
Manhattan plot of single SNP GWAS showing significant SNPs that are associated with flowering time in barley accessions. Significant SNPs are shown with larger symbols above the red dashed line as the significance threshold. Significance was determined by sequential Bonferroni correction at P<0.05.
Fig. 4.
Manhattan plot of gene-set GWAS showing significant genes that are associated with phenology in the barley accessions. Significant genes are shown above the red dashed line as the significance threshold as determined by sequential Bonferroni correction at P<0.05.
Table 2.
Genes, their annotation, and associated flowering pathways in barley, as revealed to be significantly associated with flowering time through gene-set analysis
| Putative gene name | Annotation (Hv_IBSC_PGSB_r1_HighConf) | Gene ID (Hv_IBSC_PGSB_r1 _HighConf) | Flowering pathway |
|---|---|---|---|
| HvADA2 | Transcriptional adapter 2 | HORVU5Hr1G095400 | Vernalization |
| HvAGL1 | MADS-box transcription factor TaAGL1 | HORVU6Hr1G002330 | Vernalization and autonomous pathways |
| HvAGL32 | MADS-box transcription factor 31 | HORVU2Hr1G098930 | Meristem response and development |
| HvAGLG1 | MADS-box transcription factor 34 | HORVU5Hr1G095710 | Meristem response and development |
| HvAP2 | AP2-like ethylene-responsive transcription factor | HORVU2Hr1G113880 | Meristem response and development |
| HvARF2 | auxin response factor 2 | HORVU3Hr1G096510 | Grain size and reproductive development |
| HvBB | E3 ubiquitin ligase BIG BROTHER | HORVU4Hr1G055690 | Grain development |
| HvBM1 | MADS-box transcription factor 47 | HORVU4Hr1G077850 | Meristem response and development |
| HvBM16 | MADS-box transcription factor 16 | HORVU7Hr1G091210 | Meristem response and development |
| HvBM3 | MADS-box transcription factor 18 | HORVU0Hr1G003020 | Meristem response and development |
| HvBM5 (HvVRN-H1) | MADS-box transcription factor 14 | HORVU5Hr1G095630 | Vernalization |
| HvBM8 | MADS-box transcription factor 15 | HORVU2Hr1G063800 | Meristem response and development |
| HvBM9 | MADS-box transcription factor 7 | HORVU7Hr1G054220 | Meristem response and development |
| HvCBF10A | ethylene-responsive element binding factor 13 | HORVU5Hr1G080430 | Vernalization |
| HvCBF14 | Ethylene-responsive element binding factor 14 | HORVU5Hr1G080350 | Vernalization |
| HvCBF2A | Dehydration-responsive element-binding protein 1B | HORVU5Hr1G080310 | Vernalization |
| HvCBF3 | C-repeat-binding factor 4 | HORVU5Hr1G080420 | Vernalization |
| HvCBF4A | Dehydration-responsive element-binding protein 1B | HORVU5Hr1G080300 | Vernalization |
| HvCBF6 | C-repeat-binding factor 4 | HORVU5Hr1G080450 | Vernalization |
| HvCBF8A | C-repeat binding factor 3-like protein | HORVU2Hr1G041090 | Vernalization |
| HvCBF9 | Dehydration-responsive element-binding protein 1B | HORVU5Hr1G080230 | Vernalization |
| HvCCA1 | circadian clock-associated 1 | HORVU7Hr1G070870 | Photoperiod and circadian clock |
| HvCDF1 | DOF zinc finger protein 1 | HORVU2Hr1G017290 | Photoperiod and circadian clock |
| HvCEN | Protein TERMINAL FLOWER 1 | HORVU2Hr1G072750 | Meristem response and development |
| HvCIGARP | GRAS family transcription factor | HORVU2Hr1G043780 | Gibberellin signalling and metabolism |
| HvCIGARP-2 | SCARECROW-like 1 | HORVU3Hr1G091250 | Gibberellin signalling and metabolism |
| HvCK2B | casein kinase II beta subunit 4 | HORVU1Hr1G055250 | Photoperiod and circadian clock |
| HvCKX | Cytokinin dehydrogenase 2 | HORVU3Hr1G027460 | Grain development |
| HvCMF4 | CCT motif family protein | HORVU4Hr1G084020 | Photoperiod and circadian clock |
| HvCMF6b | Zinc finger protein CONSTANS-LIKE 4 | HORVU1Hr1G095410 | Photoperiod and circadian clock |
| HvCO11 | Zinc finger protein CONSTANS-LIKE 16 | HORVU6Hr1G073170 | Photoperiod and circadian clock |
| HvCO2 | receptor kinase 3 | HORVU6Hr1G072620 | Photoperiod and circadian clock |
| HvCO8 | CONSTANS-like 5 | HORVU7Hr1G027560 | Photoperiod and circadian clock |
| HvCOP1 | Erect panicle 2 protein | HORVU2Hr1G031030 | Grain development |
| HvCry1a | cryptochrome 1 | HORVU6Hr1G049950 | Light perception and signalling |
| HvCry2 | cryptochrome 2 | HORVU6Hr1G058740 | Light perception and signalling |
| HvCYP1 | Cytochrome P450 superfamily protein | HORVU2Hr1G081650 | Grain development |
| HvDRF1 | Ethylene-responsive transcription factor 4 | HORVU1Hr1G060490 | Meristem response and development |
| HvDRF2 | Ethylene-responsive transcription factor 4 | HORVU6Hr1G050500 | Meristem response and development |
| HvEFS | Histone-lysine N-methyltransferase 2A | HORVU2Hr1G000940 | Photoperiod and circadian clock |
| HvELF3 | Early flowering 3 | HORVU1Hr1G094980 | Photoperiod and circadian clock |
| HvELF4-like4 | ELF4-like 4 | HORVU5Hr1G060000 | Photoperiod and circadian clock |
| HvELF7 | RNA polymerase II-associated factor 1 homolog | HORVU3Hr1G001430 | Photoperiod and circadian clock |
| HvFCA | FCA-A1 | HORVU5Hr1G050820 | Photoperiod and circadian clock |
| HvFD | Lysine-specific histone demethylase 1 homolog 3 | HORVU2Hr1G096300 | Meristem response and development |
| HvFT1 | FLOWERING LOCUS T 1 | HORVU7Hr1G024610 | Photoperiod and circadian clock |
| HvFT2 | Protein FLOWERING LOCUS T | HORVU3Hr1G027590 | Photoperiod and circadian clock |
| HvFT3 | Protein FLOWERING LOCUS T | HORVU1Hr1G076420 | Photoperiod and circadian clock |
| HvFT5 | Protein FLOWERING LOCUS T | HORVU4Hr1G090390 | Photoperiod and circadian clock |
| HvFTL5 | Protein FLOWERING LOCUS T | HORVU2Hr1G084540 | Photoperiod and circadian clock |
| HvGA20ox1 | gibberellin 20 oxidase 1 | HORVU5Hr1G124120 | Gibberellin signalling and metabolism |
| HvGA20ox2 | gibberellin 20-oxidase 2 | HORVU3Hr1G090980 | Gibberellin signalling and metabolism |
| HvGA20ox2-2 | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | HORVU1Hr1G070710 | Gibberellin signalling and metabolism |
| HvGA20ox2-2 | 1-aminocyclopropane-1-carboxylate oxidase 1 | HORVU2Hr1G114980 | Gibberellin signalling and metabolism |
| HvGA20ox2-3 | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | HORVU4Hr1G013840 | Gibberellin signalling and metabolism |
| HvGA20ox3 | gibberellin 20 oxidase 2 | HORVU3Hr1G089980 | Gibberellin signalling and metabolism |
| HvGA2betadiox7 | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | HORVU3Hr1G117870 | Gibberellin signalling and metabolism |
| HvGA3ox1 | gibberellin 3-oxidase 1 | HORVU2Hr1G118350 | Gibberellin signalling and metabolism |
| HvGA3ox2 | gibberellin 3-oxidase 2 | HORVU3Hr1G022840 | Gibberellin signalling and metabolism |
| HvGARMP | Scarecrow-like transcription factor PAT1 | HORVU4Hr1G071670 | Gibberellin signalling and metabolism |
| HvGID1 | Gibberellin receptor GID1 | HORVU1Hr1G060810 | Gibberellin signalling and metabolism |
| HvGID1L2-3 | alpha/beta-Hydrolases superfamily protein | HORVU5Hr1G068140 | Gibberellin signalling and metabolism |
| HvGID1L2-4 | alpha/beta-Hydrolases superfamily protein | HORVU5Hr1G069040 | Gibberellin signalling and metabolism |
| HvGID1L2-5 | alpha/beta-Hydrolases superfamily protein | HORVU5Hr1G098770 | Gibberellin signalling and metabolism |
| HvGID1L2-8 | Acetylesterase | HORVU4Hr1G015550 | Gibberellin signalling and metabolism |
| HvGRP7a | Histone-lysine N-methyltransferase | HORVU4Hr1G003060 | Photoperiod and circadian clock |
| HvGW7 | unknown function | HORVU2Hr1G032710 | Grain development |
| HvHYL | alpha/beta-Hydrolases superfamily protein | HORVU0Hr1G004410 | Gibberellin signalling and metabolism |
| HvLFY1 | Floricaula/leafy homolog | HORVU2Hr1G102590 | Meristem response and development |
| HvLNG1 | unknown function | HORVU2Hr1G063820 | Grain development |
| HvLUX1 | Two-component response regulator ARR1 | HORVU3Hr1G114970 | Circadian clock |
| HvMADS25-2 | MADS-box transcription factor 25 | HORVU7Hr1G023940 | Meristem response and development |
| HvMADS25-3 | MADS-box transcription factor 25 | HORVU7Hr1G024000 | Meristem response and development |
| HvMADS26 | MADS-box transcription factor 26 | HORVU7Hr1G076310 | Meristem response and development |
| HvMADS68 | MADS-box transcription factor family protein | HORVU4Hr1G032440 | Meristem response and development |
| HvMADS75 | MADS-box transcription factor family protein | HORVU5Hr1G110470 | Meristem response and development |
| HvNAT | Acyl-CoA N-acyltransferases (NAT) superfamily protein | HORVU7Hr1G113480 | Grain development |
| HvNHL | NHL domain-containing protein | HORVU6Hr1G045970 | Grain development |
| HvPAF | Phytochrome A-associated F-box protein | HORVU1Hr1G058630 | Light perception and signalling |
| HvPFT1 | Mediator of RNA polymerase II transcription subunit 25 | HORVU5Hr1G054650 | Light perception and signalling |
| HvPhyA | phytochrome A | HORVU4Hr1G008610 | Light perception and signalling |
| HvPhyB | phytochrome B | HORVU4Hr1G053400 | Light perception and signalling |
| HvPhyC | phytochrome C | HORVU5Hr1G095530 | Light perception and signalling |
| HvPI | MADS-box transcription factor 4 | HORVU1Hr1G063620 | Meristem response and development |
| HvPI-2 | MADS-box transcription factor 2 | HORVU3Hr1G091000 | Meristem response and development |
| HvPIF4 | Transcription factor EB | HORVU5Hr1G011780 | Light perception and signalling |
| HvPPD-H1 | pseudo-response regulator 7 | HORVU2Hr1G013400 | Photoperiod and circadian clock |
| HvPRR59 | Two-component response regulator-like APRR5 | HORVU4Hr1G021010 | Photoperiod and circadian clock |
| HvPRR73 | pseudo-response regulator 7 | HORVU4Hr1G057550 | Photoperiod and circadian clock |
| HvPRR95 | Two-component response regulator-like PRR95 | HORVU5Hr1G081620 | Photoperiod and circadian clock |
| HvRLPK | Leucine-rich receptor-like protein kinase family protein | HORVU4Hr1G079040 | Grain development |
| HvRNG | Protein SIP5 | HORVU6Hr1G044080 | Grain development |
| HvSCPL33 | Carboxypeptidase Y homolog A | HORVU3Hr1G033550 | Grain development |
| HvSHP1 | MADS-box transcription factor 13 | HORVU1Hr1G023620 | Meristem response and development |
| HvSP1 | Protein NRT1/ PTR FAMILY 4.3 | HORVU4Hr1G015640 | Grain development |
| HvSPL11 | squamosa promoter binding protein-like 2 | HORVU6Hr1G031450 | Photoperiod and circadian clock |
| HvSPL12 | squamosa promoter-binding-like protein 3 | HORVU6Hr1G019700 | Photoperiod and circadian clock |
| HvSPL14 | squamosa promoter-binding-like protein 17 | HORVU0Hr1G020810 | Photoperiod and circadian clock |
| HvSPL3 | squamosa promoter binding protein-like 8 | HORVU6Hr1G030490 | Meristem response and development |
| HvSS1 | strictosidine synthase-like 3 | HORVU5Hr1G091230 | Grain development |
| HvSTK | MADS-box transcription factor 21 | HORVU1Hr1G064150 | Meristem response and development |
| HvTEM1 | AP2/B3 transcription factor family protein | HORVU3Hr1G010100 | Photoperiod and circadian clock |
| HvTFL1 | Protein TERMINAL FLOWER 1 | HORVU5Hr1G042230 | Meristem response and development |
| HvTOC1 | Two-component response regulator-like PRR1 | HORVU6Hr1G057630 | Photoperiod and circadian clock |
| HvTT16 | MADS-box transcription factor 29 | HORVU6Hr1G032220 | Meristem response and development |
| HvTUBA3 | tubulin alpha-4 chain | HORVU4Hr1G009520 | Meristem response and development |
| HvVEL1 | Protein VERNALIZATION INSENSITIVE 3 | HORVU6Hr1G022770 | Vernalization |
| HvVIN3 | Protein VERNALIZATION INSENSITIVE 3 | HORVU7Hr1G099250 | Vernalization |
| HvWPSRLK | Mitochondrial transcription termination factor family protein | HORVU2Hr1G061060 | Grain development |
| HvWRKY61 | WRKY DNA-binding protein 3 | HORVU5Hr1G028340 | Grain development |
| HvZCCTc | Zinc finger protein CONSTANS-LIKE 4 | HORVU1Hr1G056120 | Vernalization |
| HvZTLa | Kelch repeat-containing F-box family protein | HORVU7Hr1G099010 | Photoperiod and circadian clock |
| HvZTLb | Adagio-like protein 1 | HORVU6Hr1G022330 | Photoperiod and circadian clock |
| a HvAG1 | MADS-box transcription factor 3 | HORVU3Hr1G026650 | Meristem response and development |
| a HvCK2A | Protein kinase superfamily protein | HORVU0Hr1G030500 | Photoperiod and circadian clock |
| a HvPAP2 | Auxin-responsive protein IAA17 | HORVU3Hr1G031460 | Light perception and signalling |
| b HvBM7 | MADS-box transcription factor 1 | HORVU4Hr1G067680 | Meristem response and development |
| b HvCO1 | B-Box-type zinc finger transcription factor | HORVU7Hr1G043030 | Photoperiod and circadian clock |
| b HvCry1b | cryptochrome 1 | HORVU2Hr1G079220 | Light perception and signalling |
| b HvEDL2 | EID1-like F-box protein 2 | HORVU2Hr1G034270 | Photoperiod and circadian clock |
| b HvGA2ox3 | gibberellin 2-oxidase | HORVU3Hr1G072810 | Gibberellin signalling and metabolism |
Significance was determined by sequential Bonferroni correction (P<0.05). The detailed list with chromosome position is in table S1 in Hill et al. (2019a). Annotation and Gene ID follows Hv_IBSC_PGSB_r1_HighConf.
a Significant only in single SNP GWAS analysis.
b Significant only in epistasis analysis.
Epistatic effects of genes associated with flowering time
Two-way (interaction of two SNPs) epistasis analysis revealed 19 pairs of SNPs (sequential Bonferroni corrected P<0.05), among the overall 30 276 pairs between each of the phenology genes studied here, interacting to influence FT. Depending on the combination of SNPs in their allelic state, 12 pairs significantly promoted earlier flowering (–8 d), and seven pairs were linked with later flowering (+10 d) when compared with average FT (Table 3). A homozygote at an alternative state (‘GG’ versus ‘TT’ in the reference genome) in HvELF7 (an RNA polymerase II-associated factor 1 homologue gene) interacted with six other SNPs promoting earlier flowering, while the HvGA2ox3 (a gibberellin 2-oxidase gene) homozygote at an alternative state (‘AA’ versus ‘GG’ in the reference genome) interacts with other genes to delay flowering in barley (Fig. 5). For example, cultivar ‘UWA2Rsel9506’ which has genotype ‘GG’ in HvELF7 tends to flower earlier when HvCO1 (a zinc finger protein CONSTANS-LIKE gene) has genotype ‘GG’ across all experimental locations. Seven accessions (‘07T741’, ‘B559’, ‘B751’, ‘Han 85-222’, ‘I92-562’, ‘ICB104039’, and ‘Lao Wu Hu Xu Mai’) with HvGA2ox having genotype ‘AA’ and HvCKX (a cytokinin dehydrogenase gene) having genotype ‘GG’ usually flower later across our trials. When homozygous in an alternative state (‘GG’ versus ‘CC’ in the reference genome), HvPhyB (a phytochrome B gene) interacts with two other genes (HvNHL, an NHL domain-containing protein gene, and HvTOC, a two-component response regulator-like PRR1 gene) to promote early flowering, while when in the heterozygous state, this gene interacts with other genes (HvSPL3, a squamosa promoter-binding protein-like gene) to promote late flowering. Eight out of the 13 genes revealed to have epistatic interactions were also significant in the SNP-set GWAS analysis, while the remaining five were defined as insignificant both in the single SNP and gene-set GWAS analyses (Table 2).
Table 3.
SNP–SNP interaction in determining flowering time in barley as revealed by epistasis analysis
| Gene_1 | FTD | Gene_2 | FTD | Gene interaction | FTD |
|---|---|---|---|---|---|
| Gene interactions to promote early flowering | |||||
| HvCBF8A (CC) | 0.64±0.12 | HvELF7 (GG) | 0.52±0.09 | CC–GG | 0.48±0.09 |
| HvCO1 (GG) | 0.63±0.12 | HvELF7 (GG) | 0.52±0.09 | GG–GG | 0.48±0.10 |
| HvCry1b (TT) | 0.64±0.12 | HvELF7 (GG) | 0.52±0.09 | TT–GG | 0.48±0.09 |
| HvBM7 (CC) | 0.64±0.12 | HvELF7 (GG) | 0.52±0.09 | CC–GG | 0.48±0.09 |
| HvPhyB (CC) | 0.65±0.12 | HvELF7 (GG) | 0.52±0.09 | CC–GG | 0.48±0.09 |
| HvFT1 (CC) | 0.64±0.13 | HvELF7 (GG) | 0.52±0.09 | CC–GG | 0.48±0.09 |
| HvCK2B (GG) | 0.64±0.12 | HvCO1 (CC) | 0.64±0.12 | GG–CC | 0.55±0.13 |
| HvCO1 (GG) | 0.63±0.12 | HvZCCTc (CC) | 0.60±0.13 | GG–CC | 0.58±0.11 |
| HvPhyB (GG) | 0.60±0.13 | HvNHL (GG) | 0.61±0.12 | GG–GG | 0.56±0.08 |
| HvPhyB (GG) | 0.60±0.13 | HvTOC1 (TT) | 0.62±0.13 | GG–TT | 0.56±0.08 |
| HvPhyA (AA) | 0.61±0.13 | HvZTLa (GG) | 0.64±0.12 | AA–GG | 0.57±0.13 |
| HvCBF8A (TT) | 0.59±0.12 | HvEDL2 (TT) | 0.64±0.13 | TT–TT | 0.58±0.10 |
| Gene interactions to delay flowering | |||||
| HvPhyA (GG) | 0.64±0.12 | HvZTLb (GG) | 0.65±0.13 | GG–GG | 0.66±0.11 |
| HvPhyB (CC) | 0.64±0.12 | HvSPL3 (CC) | 0.61±0.12 | CC–CC | 0.69±0.11 |
| HvSLN1 (CC) | 0.67±0.12 | HvCO8 (TT) | 0.70±0.16 | CC–TT | 0.74±0.14 |
| HvCKX (CC) | 0.70±0.15 | HvGA2ox3 (AA) | 0.69±0.15 | CC–AA | 0.77±0.15 |
| HvFT2 (GG) | 0.70±0.16 | HvGA2ox3 (AA) | 0.69±0.15 | GG–AA | 0.77±0.15 |
| HvFT2 (GG) | 0.70±0.16 | HvCBF6 (TT) | 0.69±0.15 | GG–TT | 0.77±0.15 |
| HvCBF6 (TT) | 0.69±0.15 | HvCKX (CC) | 0.70±0.15 | TT–CC | 0.77±0.15 |
Flowering time (days to Z49) was standardized to 0–1 as FTD (see the Materials and methods). Letters in parentheses indicate the genotype of the first SNP of the gene. FTD is presented as mean ±SD. Note that the average FTD across all samples was 0.64±0.12
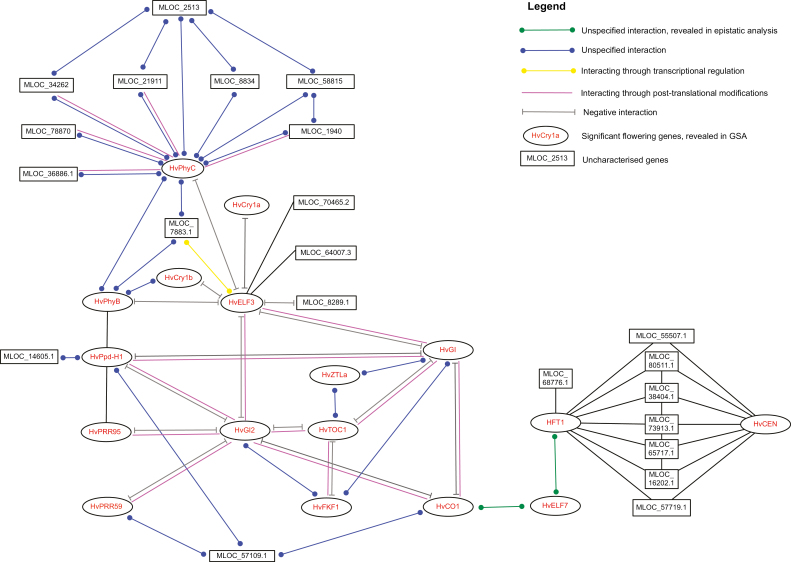
Fig. 5.
Significant flowering genes and their regulatory connections in barley (Hordeum vulgare L.). Putative gene name and gene IDs were from Ensembl Plants Hordeum vulgare Genome assembly 082214v1 that was archived in STRING (Szklarczyk et al., 2017). The interactions, including type and effects, were based direct (physical) and indirect (functional) associations from computational prediction and knowledge transfer between organisms, as implemented in STRING (Szklarczyk et al., 2017).
Gene interaction network in regulation of flowering time
Using key genes involved in flowering regulation in barley as recorded in the comprehensive protein–protein interaction database ‘STRING’ and also including additional candidate genes as revealed in our gene–gene interactions (epistatic interaction) analysis, we constructed a complex gene regulatory network (Fig. 5). The network involved 18 genes that were identified as significant in the above gene-based associated analysis. These genes are known to have roles in light signalling (e.g. HvPhyC), photoperiod response (HvPpD-H1), circadian clock (HvELF3), and development of the inflorescence meristem (HvCEN). Twenty-one genes were uncharacterized in the Hordeum vulgare genome assembly 082214v1.
Discussion
We have previously identified 429 functional alleles within the coding regions of 95 genes associated with FT in barley using single-marker GWAS (Hill et al., 2019a). In this study, by expanding to GSA and epistasis analysis, we achieved higher statistical power, and with potentially high accuracy, to detect significant genes and gene networks that influence FT in barley. We have identified 121 genes that have been associated with FT in barley, including 26 that have not been described in barley in previous research. All 121 genes have been previously described in dicot A. thaliana, and monocot cereal crops (e.g. rice, maize, and sorghum), indicating that many of the flowering genes are conserved across angiosperms including dicots and monocots (Blümel et al., 2015). FT genes involved in the photoperiod, vernalization, circadian clock, and gibberellin biosynthesis pathways were previously studied in barley (Turner et al., 2005; Wang et al., 2010; Maurer et al., 2015; Mathew et al., 2018). Our GSA detected essential genes involved in the key flowering pathways and confirmed that these genes were indeed controlling the FT in the barley accessions with a broad geographic origin. We note that our SNPs have been enriched from putative flowering genes; it is highly likely that there are additional genes, and gene interactions between flowering genes and other genes that may not directly be involved in flowering, influencing flowering in barley. Further research into the genetic mechanism of flowering in barley should expand to include genome-wide genetic variants.
Our gene-set association analysis detected key photoperiod response genes controlling FT. The photoperiod response gene Photoperiod 1 (Ppd-H1), located at chromosome 2H, is a pseudoresponse regulator gene. This gene has previously been identified as one essential gene for providing adaptation to photoperiod in barley by flowering induction under long days (Turner et al., 2005). It is known that the Ppd-H1 dominant allele induces early flowering in wild and winter barley varieties, while recessive ppd-H1 delays flowering in spring barleys (Turner et al., 2005; Jones et al., 2008). The second photoperiod gene Ppd-H2, also known as HvFT3 in barley, located on chromosome 1H, was shown to regulate FT under short days (Börner et al., 2002; Wang et al., 2010). GSA identified HvCEN as a significant flowering gene, corroborating the report from Comadran et al. (2012). TFL1, the homologue of HvCEN, is a key regulator of FT by controlling the development of the inflorescence meristem in A. thaliana (Hanano and Goto, 2011). HvCEN and associated QTLs were also reported to be associated with components of grain yield traits in barley (Comadran et al., 2012; Pasam and Sharma, 2014; Sharma et al., 2018). Saade et al. (2016) reported that the HvCEN locus promoted early FT, and resulted in higher grain yield, under salt stress conditions.
Among the three light receptor phytochrome genes—HvPhyA, HvPhyB, and HvPhyC—identified as associated with FT in our GSA, HvPhyC has previously been reported as an essential component in photoperiodic flowering in barley (Faure et al., 2012; Nishida et al., 2013; Pankin et al., 2014; Hill et al., 2019a). As phytochromes are involved in plants’ ability to intercept and translate light signals, they play a crucial role in modulating and regulating growth and development (Mathews, 2010). The HvPHYC gene was reported to interact with several other photoperiod response genes under different photoperiods (Pankin et al., 2014). Meanwhile, existing evidence suggests that variation at the HvPHYC locus has no pleiotropic effects on important agronomic traits and starch pasting properties (Nishida et al., 2013; Pankin et al., 2014). As such, Ibrahim et al. (2018) suggested that HvPHYC can be used effectively in barley breeding programmes to manipulate FT for yield improvement for varieties in stressful growing conditions.
Circadian clock-controlled mechanisms enable plants to measure changes of photoperiod as a cue for seasonal changes in their environment and therefore control developmental transitions, such as from vegetative growth to initiating flowering (Shim et al., 2017). Previous reports identified HvELF3 as one of the key genes affecting the circadian clock (Faure et al., 2012; Zakhrabekova et al., 2012), which was also confirmed in this study. The HvELF3 locus regulates flowering under the influence of photoperiod (Boden et al., 2014). In A. thaliana, it is known that ELF3, LUX, and ELF4 form a protein complex, termed the evening complex (EC). This complex represses the expression of PRR9 and LUX (two core circadian components in A. thaliana) through binding to LUX-binding sites (reviewed in Shim et al., 2017). Huang et al. (2016) recently reported that the PhyB–ELF3 complex forms one of the signalling hubs that connects red light signalling with the circadian clock. It is not clear whether the circadian clock-controlled mechanisms involving ELF3, ELF4, LUX, PRR9, and PhyB operate in the same way in barley as in A. thaliana. However, HvELF3, HvELF4, HvLUX, HvPRR9, and HvPhyB were all identified as significant in controlling FT in our gene-set test.
It is known that the early flowering of some barley genotypes is closely linked to gibberellin biosynthesis (Boden et al., 2014). We identified 19 genes related to gibberellin biosynthesis [e.g. HvGA20ox1 (GA20 oxidase 1)] as significant flowering genes in the barley accessions we investigated. Our findings corroborate with the notion that gibberellin is an important signal in flower development in barley. In A. thaliana, paclobutrazol—a gibberellin biosynthesis inhibitor—significantly reduces the long hypocotyl and petiole phenotypes of Arabidopsis elf3 mutants (Filo et al., 2015). As discussed above, ELF3 is a key gene in a tripartite transcriptional complex, the EC. Filo et al. (2015) further suggested that the role of the EC in the regulation of gibberellin biosynthesis and flowering in dicots is shared with monocots and is a highly conserved mechanism for growth control. As such, mechanisms of the circadian clock-controlled pathway linking regulation of gibberellin biosynthesis and flowering induction, as reported in A. thaliana, may provide a useful template for exploring clock-controlled mechanisms in barley.
Fourteen genes that were reportedly involved in the vernalization pathway have been identified in the GSA. The interaction of Vrn-H1, Vrn-H2, and Vrn-H3 has been reported as an important mechanism controlling flowering in response to vernalization in barley (von Zitzewitz et al., 2005). HvBM5 (equivalent to HvVrn-H1), a MADS-box transcription factor gene, was identified as a significant flowering gene, and was also previously reported to promote the transition from the vegetative to the reproductive phase (Hemming et al., 2008). In the interaction, HvVrn-H1 represses the expression of Vrn-H2 (a zinc-finger CONSTANS); in turn, that represses Vrn-H3 in regulating flowering as the response to vernalization (Yan et al., 2003, 2004). Vrn-H3 (equivalent to HvFT1) in barley was thought to be a central integrator of different FT pathways (Yan et al., 2006). Yan et al. (2006) also reported that the Vrn-H3 gene in both barley and wheat is responsible for natural allelic variation in vernalization requirement. Five FT genes (HvFT1, HvFT2, HvFT4, HvFT5, and HvFTL5) were identified as significantly influencing FT in this study. These genes were observed to play different roles in their response to photoperiod, while HvFT1 has an essential role in the transition from the vegetative growth to reproductive stage (Alqudah et al., 2014).
We identified 22 genes involving 19 two-way epistatic interactions in either promoting early flowering or delaying flowering. Epistatic interactions have previously been reported in barley. Yan et al. (2004, 2006) have previously reported significant two-way epistasis between vernalization genes VRN-H1 (syn. HvBM5) and VRN-H3 (syn. HvFT1), and between Vrn-H1 and Vrn-H2, to play an essential role in FT regulation in barley. Griffiths et al. (2003) postulated that FT genes HvGI, Vrn-H2, Vrn-H1, and HvCO1 could be involved in two-way epistatic interactions. Cuesta-Marcos et al. (2010) proposed that Vrn-H1 (HvBM5), Vrn-H2, Vrn-H3 (HvFT1), and Vrn4 could interact to determine vernalization sensitivity in barley. A few of the epistatic interactions revealed in this study could be linked to previously reported interaction in barley or A. thaliana. For example, the interaction of homozygous HvFT2 and HvGA2ox3 delayed flowering in our study, which is consistent with the previous report by Filo et al. (2015). Our results also demonstrate the extensive epistatic interactions controlling the FT between genes involved in response to photoperiod, circadian clock pathway genes, response to vernalization, and gibberellin biosynthesis (Fig. 5). HvELF7, a homologue of the RNA polymerase II-associated factor 1 gene, is notable. This gene interacted with six other genes involved with photoperiod and vernalization to induce flowering up to 10 d earlier. Its effects were consistent across our experimental locations and years, implying that its role is probably independent of environmental impacts.
HvCO1 is another key gene identified in this study. HvCO1 and HvCO8 were involved in four epistatic interactions in influencing flowering. It is known that CONSTANS (CO) plays a crucial role in the photoperiodic regulation of flowering in A. thaliana (Kim et al., 2008). At least eight homologues of CO-like genes (HvCO1–HvCO8) were identified in barley, but their roles in controlling the FT pathway are not clear (Griffiths et al., 2003; Cockram et al., 2012). Our findings for HvCO1 and other genes involved in photoperiodic regulation and vernalization could provide some testing hypothesis of the role of CO in the regulation of flowering in barley.
Interestingly, HvCO1 was involved in epistatic interactions promoting early flowering, while HvCO8 interacting with HvSLN1 delayed flowering in the studied barley accessions, implying the possible different roles that different homologues of CO-like genes may play in the regulation of FT in barley. The broad epistatic interactions in the regulation of FT in barley as revealed in our study suggest the presence of other functional networks of genes involved in controlling FT. Based on the fact that more genes and their interactions were identified as important in regulating FT in barley, this study added more details to the gene regulatory network that Hill and Li (2016) proposed. Our results on epistatic interaction and proposed gene regulatory networks could provide further insight to refine the current model of the regulatory network controlling flowering in barley and other cereal crops (e.g. Woods et al., 2017), while further studies, such as with knock-out accessions, may validate the observed interaction effects and regulatory network.
The main environmental factors that influence FT include the ambient temperature and day length. In sorghum, temperature explained 69.4% of the variation in average FT in different environments (Li et al., 2018). Similarly, our study found that 62.9% of the variation in FT of a barley line was due to variation in average temperature in the growing locations. Gene and environment interactions explained 3.85% (P<0.001) of variance for FT. This figure, although much less than that by the average temperature, was found to be highly significant. FT in barley is highly heritable. Broad-sense heritability of FT was estimated at 88% in wild barley (Herzig et al., 2018). In maize, the genetic architecture of FT is predominantly determined by small additive loci with few environmental interactions, and FT is also highly heritable (h2 >0.85) (Buckler et al., 2009). Our estimate of heritability from 2758 SNPs was 0.503 if the origin of the experimental accessions was included as a fixed effect, while it was only 0.395 if the origin was not specified. Previously, we reported that peak SNPs at the identified loci explained 31–78% of the phenotypic variance for phenology in different environments (Hill et al., 2019a).
Both our current and previous estimates of heritability seem to be low, which could be explained by four aspects. (i) There may be more genes that are essential parts of the network regulating FT in barley yet to be captured in our study. For example, Bouché et al. (2016) curated a database containing 306 genes that were reported to have functions and interactions within the flowering pathways in A. thaliana, while we analysed 174 putative genes. (ii) Causal SNPs related to FT could be located far from the known gene in its regulatory regions; therefore, SNP enrichment based on genes could fail to capture the effect. (iii) The epistatic effect could be more extensive because of the existence of a complex regulation network in controlling flowering. (iv) Broader sampling to include samples from broader genetic background and origin could be required. Future research that builds on the insights generated from this study, and with the aim of finding the missing heritability and the genes that are important in regulating FT in barley, will help to decipher the genetic mechanism of flowering regulation, and therefore facilitate barley breeding programmes to increase performance and grain yield under optimal cultivation conditions as well as under stress.
GWAS has been a powerful tool to connect genomic variation (SNPs) to complex phenotype, while pinpointing the actual genes underlying biology is still not straightforward. Our previous research (Hill et al., 2019a) demonstrated that targeted enrichment of SNPs from function-related genes combined with GWAS could provide great opportunities to associate DNA variations with complex phenotypes in plants. In this study, we further demonstrated that GSA could provide higher power to detect genetic association than the analysis of SNPs individually. We suggest that GSA is particularly useful for dissecting the genetic determinants of complex traits such as FT, as it is likely that many SNPs with small effects contribute to these complex traits, while their effects are difficult to detect when testing SNPs individually (Holmans, 2010). Our research also shows that the incorporation of analysis of gene interaction and gene-set GWAS offers great promise in the characterization of the biological pathway of genetic determination of complex traits. It should be noted that, despite the power to connect sequence diversity to complex traits, GSA has its limits. First, GWAS analysis so far revealed that most of the significant SNPs fall within the category of non-protein coding, and many are a distance away from the known gene (Maurano et al., 2012); it is not clear how far the flanking sequencing of each gene should be included in the mapping of SNPs to a gene set (Fridley and Biernacka, 2011). Further, as with the single SNP GWAS analysis, GSA reveals the genetic changes to be correlated with a particular phenotype; this does not mean that genes identified by the studies control the phenotype, which needs to be tested in controlled experiments.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Q–Q plots showing the low level of systematic biases in genome-wide association study (GWAS) results.
Acknowledgements
The project is supported by the Grain Research and Development Corporation (GRDC)—UMU00050. The authors declare no conflicts of interest.
Author contributions
CL, DM, and PT conceived the project; CH and ZX generated the genotypic data; TA, CL, and SW generated the phenotypic data; TH performed the data analysis; KC performed spatial analysis, and TH and CH completed the manuscript with inputs from CL, PT, TA, and the other co-authors.
References
- Alqudah AM, Schnurbusch T. 2017. Heading date is not flowering time in spring barley. Frontiers in Plant Science 8, 896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alqudah AM, Sharma R, Pasam RK, Graner A, Kilian B, Schnurbusch T. 2014. Genetic dissection of photoperiod response based on GWAS of pre-anthesis phase duration in spring barley. PLoS One 9, e113120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M, von Korff M, Guo P, Lakew B, Udupa SM, Sayed H, Choumane W, Grando S, Ceccarelli S. 2007. Molecular approaches and breeding strategies for drought tolerance in barley. In: Varshney RK, Tuberosa R, eds. Genomic assisted crop improvement, vol. 2, genomics applications in crops. Dordrecht, The Netherlands: Springer, 51–79. [Google Scholar]
- Blázquez MA, Weigel D. 2000. Integration of floral inductive signals in Arabidopsis. Nature 404, 889–892. [DOI] [PubMed] [Google Scholar]
- Blümel M, Dally N, Jung C. 2015. Flowering time regulation in crops—what did we learn from Arabidopsis? Current Opinion in Biotechnology 32, 121–129. [DOI] [PubMed] [Google Scholar]
- Boden SA, Weiss D, Ross JJ, Davies NW, Trevaskis B, Chandler PM, Swain SM. 2014. EARLY FLOWERING3 regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression. The Plant Cell 26, 1557–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner A, Buck‐Sorlin G, Hayes P, Malyshev S, Korzun V. 2002. Molecular mapping of major genes and quantitative trait loci determining flowering time in response to photoperiod in barley. Plant Breeding 121, 129–132. [Google Scholar]
- Bouché F, Lobet G, Tocquin P, Périlleux C. 2016. FLOR-ID: an interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Research 44, D1167–D1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachi B, Morris GP, Borevitz JO. 2011. Genome-wide association studies in plants: the missing heritability is in the field. Genome Biology 12, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, et al. 2009. The genetic architecture of maize flowering time. Science 325, 714–718. [DOI] [PubMed] [Google Scholar]
- Caicedo AL, Stinchcombe JR, Olsen KM, Schmitt J, Purugganan MD. 2004. Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proceedings of the National Academy of Science, USA 101, 15670–15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockram J, Thiel T, Steuernagel B, Stein N, Taudien S, Bailey PC, O’Sullivan DM. 2012. Genome dynamics explain the evolution of flowering time CCT domain gene families in the Poaceae. PLoS One 7, e45307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comadran J, Kilian B, Russell J, et al. 2012. Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nature Genetics 44, 1388–1392. [DOI] [PubMed] [Google Scholar]
- Cuesta-Marcos A, Szucs P, Close TJ, Filichkin T, Muehlbauer GJ, Smith KP, Hayes PM. 2010. Genome-wide SNPs and re-sequencing of growth habit and inflorescence genes in barley: implications for association mapping in germplasm arrays varying in size and structure. BMC Genomics 11, 707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doust AN, Lukens L, Olsen KM, Mauro-Herrera M, Meyer A, Rogers K. 2014. Beyond the single gene, how epistasis and gene-by-environment effects influence crop domestication. Proceedings of the National Academy of Sciences, USA 111, 6178–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand E, Bouchet S, Bertin P, Ressayre A, Jamin P, Charcosset A, Dillmann C, Tenaillon MI. 2012. Flowering time in maize: linkage and epistasis at a major effect locus. Genetics 190, 1547–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Wang Q, Hu Y, et al. 2017. Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nature Genetics 49, 1089–1098. [DOI] [PubMed] [Google Scholar]
- Faure S, Turner AS, Gruszka D, Christodoulou V, Davis SJ, von Korff M, Laurie DA. 2012. Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proceedings of the National Academy of Sciences, USA 109, 8328–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filo J, Wu A, Eliason E, Richardson T, Thines BC, Harmon FG. 2015. Gibberellin driven growth in elf3 mutants requires PIF4 and PIF5. Plant Signaling & Behavior 10, e992707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridley BL, Biernacka JM. 2011. Gene set analysis of SNP data: benefits, challenges, and future directions. European Journal of Human Genetics 19, 837–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S, Dunford RP, Coupland G, Laurie DA. 2003. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiology 131, 1855–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Stephens M. 2011. Bayesian variable selection regression for genome-wide association studies, and other large-scale problems. Annals of Applied Statistics 5, 1780–1815. [Google Scholar]
- Hanano S, Goto K. 2011. Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. The Plant Cell 23, 3172–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming MN, Peacock WJ, Dennis ES, Trevaskis B. 2008. Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiology 147, 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig P, Maurer A, Draba V, Sharma R, Draicchio F, Bull H, Milne L, Thomas WTB, Flavell AJ, Pillen K. 2018. Contrasting genetic regulation of plant development in wild barley grown in two European environments revealed by nested association mapping. Journal of Experimental Botany 69, 1517–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CB, Angessa TT, McFawn LA, et al. 2019a. Hybridisation-based target enrichment of phenology genes to dissect the genetic basis of yield and adaptation in barley. Plant Biotechnology Journal 17, 932–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CB, Li C. 2016. Genetic architecture of flowering phenology in cereals and opportunities for crop improvement. Frontiers in Plant Science 7, 1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CB, Wong D, Tibbits J, Forrest K, Hayden M, Zhang XQ, Westcott S, Angessa TT, Li C. 2019b. Targeted enrichment by solution-based hybrid capture to identify genetic sequence variants in barley. Scientific Data 6, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm S. 1979. A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics 6, 65–70. [Google Scholar]
- Holmans P. 2010. Statistical methods for pathway analysis of genome-wide data for association with complex genetic traits. Advances in Genetics 72, 141–179. [DOI] [PubMed] [Google Scholar]
- Huang H, Chanda P, Alonso A, Bader JS, Arking DE. 2011. Gene-based tests of association. PLoS Genetics 7, e1002177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Yoo CY, Bindbeutel R, Goldsworthy J, Tielking A, Alvarez S, Naldrett MJ, Evans BS, Chen M, Nusinow DA. 2016. PCH1 integrates circadian and light-signaling pathways to control photoperiod-responsive growth in Arabidopsis. eLife 5, e13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim A, Harrison M, Meinke H, Fan Y, Johnson P, Zhou M. 2018. A regulator of early flowering in barley (Hordeum vulgare L.). PLoS One 13, e0200722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA. 2006. Photoperiodic control of flowering: not only by coincidence. Trends in Plant Science 11, 550–558. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Schmidt RH, Reif JC. 2018. Haplotype-based genome-wide prediction models exploit local epistatic interactions among markers. Genes, Genomes, Genetics 16, g3–300548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones H, Leigh FJ, Mackay I, Bower MA, Smith LM, Charles MP, Jones G, Jones MK, Brown TA, Powell W. 2008. Population-based resequencing reveals that the flowering time adaptation of cultivated barley originated east of the Fertile Crescent. Molecular Biology and Evolution 25, 2211–2219. [DOI] [PubMed] [Google Scholar]
- Kim SY, Yu X, Michaels SD. 2008. Regulation of CONSTANS and FLOWERING LOCUS T expression in response to changing light quality. Plant Physiology 148, 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Guo T, Mu Q, Li X, Yu J. 2018. Genomic and environmental determinants and their interplay underlying phenotypic plasticity. Proceedings of the National Academy of Sciences, USA 115, 6679–6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert C, Listgarten J, Davidson RI, Baxter S, Poon H, Poong H, Kadie CM, Heckerman D. 2013. An exhaustive epistatic SNP association analysis on expanded Wellcome Trust data. Scientific Reports 3, 1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D. 2011. FaST linear mixed models for genome-wide association studies. Nature Methods 8, 833–835. [DOI] [PubMed] [Google Scholar]
- Listgarten J, Lippert C, Kadie CM, Davidson RI, Eskin E, Heckerman D. 2012. Improved linear mixed models for genome-wide association studies. Nature Methods 9, 525–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listgarten J, Lippert C, Kang EY, Xiang J, Kadie CM, Heckerman D. 2013. A powerful and efficient set test for genetic markers that handles confounders. Bioinformatics 29, 1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, et al. 2009. Finding the missing heritability of complex diseases. Nature 461, 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M, Gundlach H, Himmelbach A, et al. 2017. A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433. [DOI] [PubMed] [Google Scholar]
- Mathew B, Léon J, Sannemann W, Sillanpää MJ. 2018. Detection of epistasis for flowering time using Bayesian multilocus estimation in a barley MAGIC population. Genetics 208, 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews S. 2010. Evolutionary studies illuminate the structural–functional model of plant phytochromes. The Plant Cell 22, 4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K, Yamanouchi U, Nonoue Y, Sugimoto K, Wang ZX, Minobe Y, Yano M. 2011. Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. The Plant Journal 66, 603–612. [DOI] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, et al. 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer A, Draba V, Jiang Y, Schnaithmann F, Sharma R, Schumann E, Kilian B, Reif JC, Pillen K. 2015. Modelling the genetic architecture of flowering time control in barley through nested association mapping. BMC Genomics 16, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KFX, Waugh R, Brown JWS, et al. 2012. A physical, genetic and functional sequence assembly of the barley genome. Nature 491, 711–716. [DOI] [PubMed] [Google Scholar]
- Mooney MA, Wilmot B. 2015. Gene set analysis: a step-by-step guide. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics 168, 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam D, Kim J, Kim SY, Kim S. 2010. GSA-SNP: a general approach for gene set analysis of polymorphisms. Nucleic Acids Research 38, W749–W754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H, Ishihara D, Ishii M, et al. 2013. Phytochrome C is a key factor controlling long-day flowering in barley. Plant Physiology 163, 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankin A, Campoli C, Dong X, et al. 2014. Mapping-by-sequencing identifies HvPHYTOCHROME C as a candidate gene for the early maturity 5 locus modulating the circadian clock and photoperiodic flowering in barley. Genetics 198, 383–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasam RK, Sharma R. 2014. Association mapping, a new paradigm for dissection of complex traits in crops. In: Kavi Kashor PB, Bandopadhyay R, Suravajhala P, eds. Agricultural bioinformatics. New Delhi: Springer India, 1–20. [Google Scholar]
- Pasam RK, Sharma R, Malosetti M, van Eeuwijk FA, Haseneyer G, Kilian B, Graner A. 2012. Genome-wide association studies for agronomical traits in a world wide spring barley collection. BMC Plant Biology 12, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pers TH. 2016. Gene set analysis for interpreting genetic studies. Human Molecular Genetics 25, R133–R140. [DOI] [PubMed] [Google Scholar]
- Phillips PC. 2008. Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems. Nature Reviews. Genetics 9, 855–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepho HP, Möhring J, Melchinger AE, Büchse A. 2008. BLUP for phenotypic selection in plant breeding and variety testing. Euphytica 161, 209–228. [Google Scholar]
- Rooney W, Aydin S. 1999. Genetic control of a photoperiod-sensitive response in Sorghum bicolor (L.) Moench. Crop Science 39, 397–400. [Google Scholar]
- Saade S, Maurer A, Shahid M, Oakey H, Schmöckel SM, Negrão S, Pillen K, Tester M. 2016. Yield-related salinity tolerance traits identified in a nested association mapping (NAM) population of wild barley. Scientific Reports 6, 32586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Draicchio F, Bull H, Herzig P, Maurer A, Pillen K, Thomas WTB, Flavell AJ. 2018. Genome-wide association of yield traits in a nested association mapping population of barley reveals new gene diversity for future breeding. Journal of Experimental Botany 69, 3811–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JS, Kubota A, Imaizumi T. 2017. Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiology 173, 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Morris JH, Cook H, et al. 2017. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Research 18, D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tester M, Langridge P. 2011. Breeding technologies to increase. Science 818, 818–822. [DOI] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. 2005. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310, 1031–1034. [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. 2004. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006. [DOI] [PubMed] [Google Scholar]
- von Zitzewitz J, Szucs P, Dubcovsky J, Yan L, Francia E, Pecchioni N, Casas A, Chen TH, Hayes PM, Skinner JS. 2005. Molecular and structural characterization of barley vernalization genes. Plant Molecular Biology 59, 449–467. [DOI] [PubMed] [Google Scholar]
- Wang JM, Yang JM, McNeil DL, Zhou MX. 2010. Identification and molecular mapping of a dwarfing gene in barley (Hordeum vulgare L.) and its correlation with other agronomic traits. Euphytica 175, 331–342. [Google Scholar]
- Wang X, Morris NJ, Schaid DJ, Elston RC. 2012. Power of single- vs. multi-marker tests of association. Genetic Epidemiology 36, 480–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh R, Jannink JL, Muehlbauer GJ, Ramsay L. 2009. The emergence of whole genome association scans in barley. Current Opinion in Plant Biology 12, 218–222. [DOI] [PubMed] [Google Scholar]
- Wei WH, Hemani G, Haley CS. 2014. Detecting epistasis in human complex traits. Nature Reviews. Genetics 15, 722–733. [DOI] [PubMed] [Google Scholar]
- Widmer C, Lippert C, Weissbrod O, Fusi N, Kadie C, Davidson R, Listgarten J, Heckerman D. 2014. Further improvements to linear mixed models for genome-wide association studies. Scientific Reports 4, 6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler TW, Day FR, Croteau-Chonka DC, et al. ; Genetic Investigation of Anthropometric Traits (GIANT) Consortium. 2014. Quality control and conduct of genome-wide association meta-analyses. Nature Protocols 9, 1192–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods DP, Bednarek R, Bouché F, Gordon SP, Vogel JP, Garvin DF, Amasino RM. 2017. Genetic architecture of flowering-time variation in Brachypodium distachyon. Plant Physiology 173, 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, et al. 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature Genetics 40, 761–767. [DOI] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. 2006. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proceedings of the National Academy of Sciences, USA 103, 19581–19586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J. 2004. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303, 1640–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Bakshi A, Zhu Z, et al. ; LifeLines Cohort Study. 2015. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nature Genetics 47, 1114–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, et al. 2010. Common SNPs explain a large proportion of the heritability for human height. Nature Genetics 42, 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. 2011. GCTA: a tool for genome-wide complex trait analysis. American Journal of Human Genetics 88, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Guo Z, Huang C, et al. 2014. Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nature Communications 5, 5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Yamamoto E, Aya K, et al. 2016. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nature Genetics 48, 927–934. [DOI] [PubMed] [Google Scholar]
- Zadoks JC, Chang TT, Konzak CF. 1974. A decimal code for the growth stages of cereals. Weed Research 14, 415–421. [Google Scholar]
- Zakhrabekova S, Gough SP, Braumann I, et al. 2012. Induced mutations in circadian clock regulator Mat-a facilitated short-season adaptation and range extension in cultivated barley. Proceedings of the National Academy of Sciences, USA 109, 4326–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang W, Valdar W. 2014. Bayesian modeling of haplotype effects in multiparent populations. Genetics 198, 139–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zöllner S, Pritchard JK. 2005. Coalescent-based association mapping and fine mapping of complex trait loci. Genetics 169, 1071–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.