Abstract
Traditional annotation of protein-encoding genes relied on assumptions, such as one open reading frame (ORF) encodes one protein and minimal lengths for translated proteins. With the serendipitous discoveries of translated ORFs encoded upstream and downstream of annotated ORFs, from alternative start sites nested within annotated ORFs and from RNAs previously considered noncoding, it is becoming clear that these initial assumptions are incorrect. The findings have led to the realization that genetic information is more densely coded and that the proteome is more complex than previously anticipated. As such, interest in the identification and characterization of the previously ignored ‘dark proteome’ is increasing, though we note that research in eukaryotes and bacteria has largely progressed in isolation. To bridge this gap and illustrate exciting findings emerging from studies of the dark proteome, we highlight recent advances in both eukaryotic and bacterial cells. We discuss progress in the detection of alternative ORFs as well as in the understanding of functions and the regulation of their expression and posit questions for future work.
INTRODUCTION
Most genome annotation is based on assumptions about what is being translated in the cell. For example, minimal length cutoffs commonly are utilized to prevent spurious annotation of open reading frames (ORFs). Additionally, smaller ORFs nested within larger ORFs generally are not annotated. However, it is becoming clear that these initial assumptions are incorrect. With the serendipitous discoveries of additional translated ORFs upstream and downstream of annotated ORFs, protein variants expressed from alternative start codons both in-frame and out-of-frame of annotated ORFs, and functional small proteins encoded by RNAs previously considered noncoding, such as long noncoding RNAs (lncRNAs) and circular RNAs (circRNAs) (1–6), we now know that many previously ignored ORFs are translated as regulatory mechanisms or to give bioactive proteins (Figure 1). These discoveries are revealing that dense gene architectures previously associated with viruses and bacteriophages may be present in all organisms. The findings are also uncovering new ways proteome complexity is generated, beyond the known mechanisms of mRNA editing, alternative splicing, ribosomal frameshifting, stop codon readthrough or use of non-canonical amino acids. Aspects of the discovery and functions of these ignored ORFs have previously been reviewed (7–10). Here we seek to provide an overview of recent advances, compare and contrast what is known in eukaryotic and bacterial cells, and point out unanswered questions.
Figure 1.
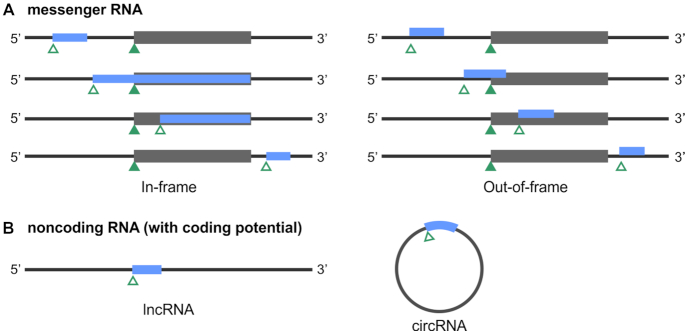
Architecture of alt-ORFs discovered on messenger RNAs (A) and sORFs encoded on transcripts denoted ‘noncoding’ RNAs (B). In (A), the organization of the alt-ORFs (blue) relative to the main ORFs (dark gray) is shown for mature mRNAs with alt-ORFs in the same frame (left), which can result in N-terminal extensions or deletions, and out-of-frame (right) relative to the annotated main-ORF. Annotated start codons for the main-ORF are marked with solid green triangles and alternative start codons are marked with empty green triangles.
We note that nomenclature to denote these newly discovered ORFs has been inconsistent. For instance, regulatory ORFs upstream of annotated ORFs are termed upstream ORFs ‘uORFs’ or upstream conserved coding regions ‘uCCs’ in eukaryotes and ‘leader peptides’ in bacteria. ORFs overlapping annotated ORFs have been denoted ‘alt-ORFs’ in some species, and the gene products of previously unannotated small ORFs ‘sORFs’ have been referred to as ‘small proteins’, ‘micropeptides’ and ‘miniproteins’, as just a few examples. The field would benefit from adoption of a unified terminology. In this review, we denote the ORF encoding the largest, previously annotated protein in one region of a transcript the ‘main-ORF’ and all other ORFs derived from alternative start codons in the same region as ‘alt-ORFs’ (Figure 1). We refer to previously unannotated ORFs of short length (corresponding to the only coding sequence for a particular region of a transcript) as ‘sORFs’, and regulatory upstream ORFs as ‘uORFs’ for both eukaryotes and bacteria. The protein products of sORFs and uORFs will be called ‘small proteins’. A few exceptions are examples where we keep the original designations for consistency.
In thinking about these previously ignored ORFs, it is worth considering some fundamental differences in the translation of eukaryotic and bacterial mRNAs. While eukaryotic mRNAs usually are mono-cistronic, many, but not all, bacterial transcripts are poly-cistronic and encode several proteins. While transcription and translation occur in separate compartments in eukaryotic cells, they can be coupled in bacteria. Additionally, while splicing of mRNAs is common in most eukaryotic species, it is rare in bacteria. The majority of eukaryotic mRNAs are translated by a so-called ‘cap-dependent’ mechanism involving 5′-cap recognition by the eIF4F complex, which associates with eIF3 and the 40S ribosomal subunit to form, with additional factors such as tRNA-bound eIF2, the 43S preinitiation complex (11). The 43S scans through the 5′ untranslated region (UTR) until it encounters an initiation codon. It is commonly assumed that the first AUG codon, which the scanning ribosome encounters, serves as the start site for translation. However, one or more potential start codons for the initiation of uORFs can exist upstream of the main start codon. Likewise, AUG codons immediately downstream of the main start codon can potentially serve as initiators. In contrast, in bacteria, a Shine-Dalgarno or ribosome binding sequence in the 5′ leader usually helps direct the ribosome to the appropriate start codon. Nevertheless ribosomes can also initiate translation at noncanonical sites, including at the 5′ end of ‘leaderless’ transcripts (12). The differences in eukaryotic and bacterial biology result in divergent mechanisms for alt-ORF-mediated translation regulation and for expression control.
IDENTIFICATION AND DETECTION
As more proteins encoded by previously unannotated ORFs were found by chance, it became clear that these represented overlooked translated regions. Thus, systematic approaches for the identification of these ORFs were developed. There are three main approaches: bioinformatic prediction, experimental detection by mass spectrometry, and extrapolation that a sequence is translated based on ribosome occupancy. As identification of alt-ORFs has been reviewed comprehensively (13,14), we only provide a summary with a focus on recent advancements.
Bioinformatic prediction
Genome annotation initially relied on start codon position and ORF length to differentiate between a protein-encoding ORF from an ORF arising from chance occurrence (15–17). The inclusion of additional information such as the assessment of the conservation of codon usage and/or nucleotide frequencies within ORFs and the conservation in closely related species has improved the bioinformatic identification of alt-ORFs in eukaryotes [reviewed in (14)] and bacteria (15,18,19). For example, work with bacteria led to the development of a bioinformatic program, RanSEPs, which is a random forest‐based computational approach that scores sORFs based on coding potential in a species-specific manner (5). Another recent bioinformatic study analyzed existing metagenomic sequencing datasets from the human microbiome to identify thousands of putative sORFs (20). Pioneering studies in mammals revealed more than 12 000 potential regulatory uORFs based on sequence analysis (21). This number may even be low since uORF sequences usually are less conserved than canonical protein-coding ORFs. While regulatory uORFs (leader peptides) have been observed and characterized in bacteria, to our knowledge there have been no systematic computational studies aimed at identification of bacterial uORFs. Given that little is known about nested, internal ORFs, to date there also are no algorithms aimed at detecting this type of gene in any species. Overall, despite significant efforts to improve bioinfomatic prediction, the computational methods are usually subjected to a high rates of false positives. Many of these new ORF annotation programs also rely on previously identified alt-ORFs for training and validation. Thus, their ability to identify alt-ORFs is limited to genes similar to the ones that have already been found. Experimental validation of many of the predictions also is missing. Nevertheless, these programs are valuable tools for improved genome annotation and to provide candidates for future validation.
Mass spectrometry
Mass spectrometry has been adapted for detecting previously unannotated proteins in organisms from all kingdoms. Rather than comparing spectrum matches to databases of previously annotated proteins, the mass spectrometric data must be compared to custom databases generated with all possible translations of a genome. Standard mass spectrometry requires proteins to be supported by multiple spectrum matches. However, small proteins often will have only one putative spectrum match, which may be difficult to detect among all other signals. Thus, biochemical enrichment, less stringent cutoffs or other specific approaches need to be taken to address these limitations. For example, peptidomics approaches inhibiting proteolysis and using electrostatic repulsion hydrophilic interaction chromatography to separate peptides prior to HPLC-MS/MS identified 90 new proteins in human cells, many matching proteins encoded by alt-ORFs (22). This method also was used in Escherichia coli to identify stress-induced small proteins (23,24). A significant drawback of mass spectrometry is that fragmentation of proteins prior to detection means that the true start codon of a gene must be inferred. In bacteria, translation is initiated with N-formylated methionine tRNA, which is deformylated shortly after translation begins. Inhibition of the deformylase and enrichment for formylated N-terminal peptides allows detection of translation initiation start codons by a process termed ‘N-terminomics’ (25). This approach was used to globally map the translation initiation sites of Listeria monocytogenes, revealing 6 putative sORFs and 19 putative alt-ORFs with translation initiation sites internal to an annotated ORF (26). Future advances in the isolation of the small protein fractions and mass spectrometric detection undoubtedly will have a large impact on the field.
Ribosome profiling
Thus far, the most effective identification of translation initiation sites for alt-ORFs has been the mapping of actively translated mRNAs by a technique alternately known as ribosome profiling, Ribo-seq or ribosome foot-printing. The actively translated regions of mRNA bound by ribosomes are protected from nucleolytic digestion, whereupon sequencing of the protected regions gives a snapshot of which sequences are translated. Ribosome profiling was first developed in eukaryotes (27), where strong start and stop codon peaks as well as clear 3 nt periodicity can be observed allowing unambiguous identification of the reading frame. Consequently, ribosome profiling has successfully mapped thousands of ORFs in mammals (28,29), fungi (30) and plants (2,31).
Nevertheless, standard ribosome profiling predictions have some limitations for predicting translated alt-ORFs. The quality of ribosome profiling data and the prediction model strongly affect the number of alt-ORFs identified (2). Furthermore, given that many are short, the mapping of alt-ORFs is more strongly affected by fluctuations in periodicity. Alternative splicing in 5′ UTRs can disrupt codon periodicity as can overlapping uORFs (32). To overcome these problems, several strategies have been developed to directly capture translation initiation sites by stalling initiating ribosomes (TIS-seq) (3,33,34). Translation inhibitors such as harringtonine and lactimidomycin preferentially arrest the initiating ribosomes at the start codon (3,34). The elongating ribosomes are subsequently run off (GTI-seq) (33) or depleted by another translation inhibitor such as puromycin (QTI-seq) (34), and the enriched sequences protected by initiating ribosomes are subjected to high-throughput sequencing. QTI-seq revealed that ∼50% of mouse mRNAs contain at least one uORF occupied by ribosomes (34). Combined with standard ribosome profiling, these modified approaches substantially improve the sensitivity and specificity of alt-ORF mapping.
In contrast to eukaryotes, ribosome profiling in bacteria has not resulted in clear codon resolution; the signatures are less strong and more variable. Recently however, the use of the ribosome-stalling antibiotics tetracycline (35), retapamulin (1) and Onc112 (36) for ribosome profiling of E. coli has allowed identification of novel alt-ORFs. The treatment with tetracycline revealed 28 genes with multiple start codons and 312 sORFs <50 amino acids long (35). Treatment with retapamulin led to the identification of 124 internal translation initiation sites, 42 in-frame and 78 out-of-frame to the annotated gene (1). Analysis of datasets from both retapamulin and Onc112-treated cells led to the identification of 68 putative sORFs; of the 41 tested for expression by chromosomal tagging, 38 were shown to be translated (36). The treatment with ribosome inhibitors has been a powerful approach, but again some caution is warranted. The accumulation of ribosomes on start codons as well as the elevated levels of free ribosomes that are the result of some treatments can lead to spurious ribosome binding to nonoptimal start codons (37).
The results of these studies underscore the extent to which genome annotation has underestimated the translated proteome. However, given the caveats of each detection method, we stress the importance of additional validation of protein production. Furthermore, given the observation that alt-ORF translation can change based on media type and stress conditions [reviewed in (9,38)], proteomic- and transcriptomic-based studies are limited by the growth condition in which the cells were sampled. It is likely that as detection approaches improve and additional growth conditions are tested, even more previously unannotated proteins will be identified.
ALT-ORF TRANSLATION AS A REGULATORY FUNCTION
The translation of an ORF can itself be a regulatory event. For most studied examples, this is due to translation of an ORF upstream of or overlapping the main-ORF (uORF or leader peptide), an event that can have both positive and negative consequences (Figure 2). However, the translation of internal ORFs or downstream ORFs also can impact synthesis of the protein encoded by the main-ORF.
Figure 2.
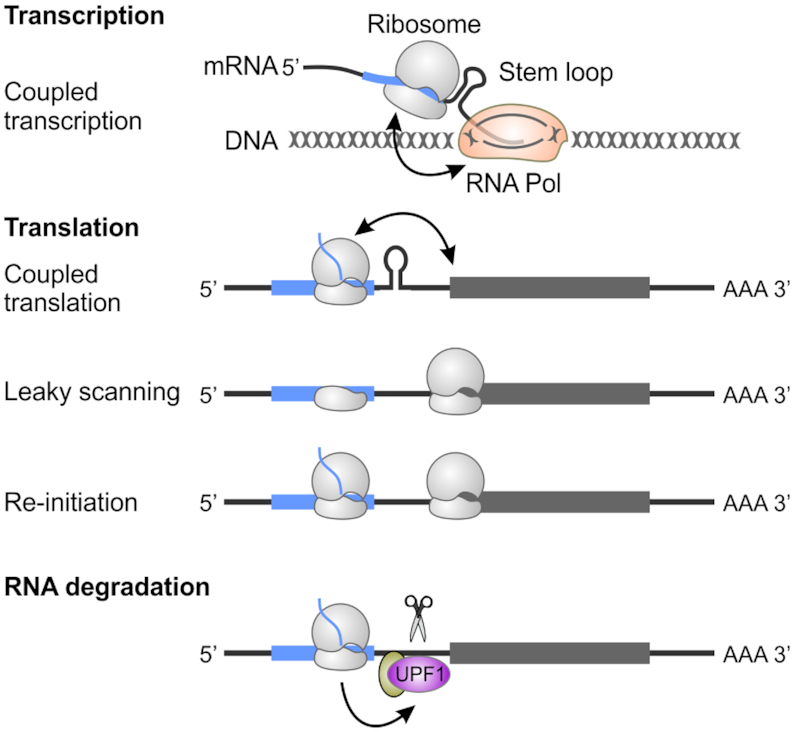
Regulatory roles of alt-ORFs in gene expression. The translation of alt-ORFs has wide-ranging impacts on gene expression. In bacteria, due to coupled transcription-translation, uORF translation may alter downstream RNA structures that impact transcription. A typical example is the trp operon. Uni-directional mRNA translation also can result in a direct effect of uORF translation on downstream main-ORF translation. Possible mechanisms include altering mRNA structures coupled to downstream translation, as well as regulation via leaky scanning and translation re-initiation. uORF translation also can affect mRNA stability throughout the recruitment of components of mRNA surveillance pathways such as UPF1.
Regulation of main-ORF translation initiation in eukaryotes
About 50% of mammalian transcripts contain at least one potential uORF (33). The uORFs were hypothesized to primarily suppress the translation of main-ORFs. This notion has been supported by reporter assays and mutational studies (30,39). Translation inhibition can be achieved by uORF translation blocking scanning 43S ribosomes or sequestering 80S ribosomes. Consequently, uORFs may act as a physical barrier to prevent ribosomal access to the downstream main-ORF (40–42). Translational arrest also can be induced by interaction between the uORF-encoded polypeptides and translating ribosomes (43,44). Additionally, the presence of non-optimal codons slows down the translating ribosomes, as was found for the Cx41 transcript in Xenopus laevis (45) and ATB2/AtbZIP11 in plants (46).
A few eukaryotic uORFs stimulate the translation of mRNAs encoding stress responsive proteins. The best characterized examples are GCN4 in yeast and ATF4 in mammals. The 5′ UTR of ATF4 contains two uORFs: one near the 5′ terminus and the other overlapping the main-ORF but in a different reading frame (47). During normal growth, the eIF2-GTP-tRNAiMet ternary complex is abundant and ribosomes translate the first uORF. The small subunit remains associated with the mRNA, resumes scanning and rapidly reacquires the readily available ternary complex, thus allowing it to reinitate translation at the second uORF. Termination at uORF2 prevents initiation of the main-ORF due to the overlap in sequence. Under stress conditions that trigger eIF2α phosphorylation, the scanning ribosomes take longer to acquire a ternary complex after reinitiating downstream of the first uORF. As a result, more ribosomes bypass the second uORF and become available to translate the downstream main-ORF. It is perplexing to find that uORFs play both stimulatory and inhibitory roles in the translation of main-ORFs. Molecular mechanisms controlling leaky scanning (start codon skipping) versus reinitiation remain incompletely understood. uORF number, length, position and other features might be critical for the overall regulatory effects, and the study of uORFs is an important area for future work given how much remains to be learned.
Control of mRNA stability in eukaryotes
Nonsense-mediated decay (NMD) is an RNA quality control mechanism that eliminates mRNAs with premature translation termination codons (48,49). If the uORF stop codon is recognized as premature, it can trigger NMD, thereby decreasing transcript abundance and protein synthesis. Indeed, transcriptome-wide analysis of NMD in yeast, plant and human cells supports the idea that uORF-containing transcripts are more likely to be targeted by NMD (50–53). Intriguingly, uORFs in plants can trigger NMD in a size-dependent manner (54). The uORFs encoding proteins of >50 amino acids are sensitive to NMD, whereas short uORFs fail to activate NMD response. In yeast, the uORF in the CPA1 mRNA encodes an arginine attenuator peptide. When arginine is abundant, translation of the arginine attenuator peptide causes ribosomal pausing at the uORF termination codon, repressing CPA1 translation (40) and leading to NMD. Nevertheless, not all the uORF-containing mRNAs are subjected to NMD (42,55). For example, although GCN4 contains four uORFs, it is resistant to NMD due to the presence of a stabilizer element (56,57). By contrast, ATF4 contains two uORFs and appears to be targeted by NMD (58,59). The mechanistic correlations between NMD and uORF translation remain to be determined.
Regulation of transcription and translation initiation in bacteria
The tryptophan operon of E. coli is the classic bacterial example of translation of a uORF (traditionally referred to as a leader peptide in bacteria, but perhaps better designated uORF to unify nomenclature moving forward) affecting transcription of the downstream main-ORF. The 5′ UTR of the trp operon encodes a 14 amino acid ORF, trpL, with two codons for tryptophan. Downstream of the tryptophan codons is an RNA sequence that can fold into either a transcription terminator or anti-terminator structure. When the activated tryptophan tRNAs are abundant, the ribosome is able to translate trpL and the terminator structure forms, attenuating transcription. However, when the ribosome pauses on the codons due to a lack of charged tRNA, this stalling allows formation of the anti-terminator and permits transcription of the entire operon. Examples of similar attenuation mechanisms have been documented in other amino acid biosynthetic operons and in other bacteria [reviewed in (60,61)]. Transcription attenuation influenced by uORF translation also has been described for genes impacting other aspects of metabolism such as pyrL regulating pyrimidine biosynthesis (62) and mgtL regulating the expression of the magnesium transporter MgtA (63).
Like in eukaryotes, bacterial uORFs also regulate translation of a downstream gene. The uORF-encoded protein can stall the ribosome due to interactions between the nascent peptides and the ribosome exit channel, resulting in the formation of a secondary structure that frees the ribosome binding site of the downstream gene for translation initiation. For example, the short uORFs upstream of chloramphenicol resistance cat and erythromycin resistance erm genes encode proteins that arrest the ribosome allowing translation of the downstream antibiotic resistance gene [reviewed in (64,65)]. Translation of the uORF can affect translation of downstream genes via translational coupling. This effect is seen in operons, where the rate of translation of downstream (often overlapping) ORFs on the same polycistronic mRNA is affected by translation of the upstream ORF [reviewed in (66)]. The translation of the uORF can also affect binding of regulatory proteins, indirectly impacting the translation of the main-ORF. This mechanism is illustrated by the iraD gene in E. coli (67). The CsrA RNA-binding protein was shown to modulate translation of the iraD mRNA, but this regulation was found to occur via binding to the ribosome binding site of an upstream ORF27 (also designated idlP), which overlaps the start codon of iraD (67). The recent search for sORFs in E. coli identified four uORFs whose translation affects expression of the downstream main-ORF: translation of pssL and yoaL enhanced levels of the main-ORFs pssA and yoeE, while translation of baxL and argL reduced levels of the main-ORFs baxA and argF (36). The regulatory mechanisms for these newly identified uORFs, and likely many others, have yet to be characterized.
Regulation of main ORF translation by internal ORF translation in bacteria
Translation from an internal alternative start codon can affect translation of the main-ORF in bacteria. For example, mutation of the out-of-frame AUG start codon of a 12 amino acid ORF internal to the E. coli sfsA gene resulted in loss of expression of the sfsA main-ORF (1). The smallest possible open reading frame is a start codon followed by a stop codon, which have been referred to as ‘minimal ORFs’ or ‘start-stops’. Ribosome profiling revealed out-of-frame internal minimal ORFs in 13 E. coli genes (1). Mutation of the start codon of a minimal ORF in one gene, yecJ, resulted in an increase in translation of the main-ORF, suggesting that these minimal ORFs also can modulate translation of the main-ORF (1). How commonly internal alt-ORFs regulate translation elongation in bacteria, whether the same regulation occurs in eukaryotic cells, and the exact mechanisms of regulation are currently unknown.
INTRINSIC FUNCTIONS AS PROTEINS
In addition to regulatory roles of uORF and alt-ORF translation mentioned above, the translational products of some these ORFs have cellular functions. For instance, small proteins encoded by uORFs in fruit flies exert critical functions in development (68), and the uORF-encoded AtCDC26 is part of the plant anaphase promoting complex/cyclosome (APC/C), regulating the accumulation of APC/C target proteins (69). Nevertheless, only a small fraction of uORF-encoding proteins are evolutionarily conserved and the codon usage in uORFs generally is similar to the random triplet frequency found in 5′ UTRs (28,70). These observations suggest that most uORFs are selected not to encode bioactive proteins, but to regulate main-ORF translation. An alt-ORF in-frame with and upstream of main-ORF can encode a signal sequence that relocalizes the main-ORF encoded protein (71,72). For example, PTEN is primarily localized to the cytoplasm and the nucleus, where it acts as a tumor suppressor by negatively regulating PI3K/AKT-mediated cell survival and proliferation (72). eIF2A-mediated translation of an alt-ORF leads to an N-terminally extended form of PTEN, which is found in mitochondria and is required for mitochondrial structure and function. Other alt-ORFs, in contrast, may just be translated spuriously. Detailed mutational studies are needed to differentiate between eukaryotic uORFs and altORFs that produce functional proteins and uORFs that simply provide upstream initiation and termination sites or alt-ORFs corresponding to spurious translation.
In bacteria, independent functions have not been demonstrated for uORFs (leader peptides), but may exist in some cases. In contrast, internal alt-ORFs have been shown to have functions both related and unrelated to the main-ORF [reviewed in (13)]. As one example, ribosome profiling identified an in-frame alt-ORF in the C-terminal region of the E. coli ArcB sensor kinase coding region, resulting in the production of just the C-terminus of ArcB (ArcB-C) (1). ArcB is a part of the two-component regulatory system ArcAB that controls responses to changes in oxygen (73). Under microaerobic conditions, wild-type E. coli outcompeted the E. coli strain in which the acrB-C start codon was mutated, indicating that the AcrB-C alt-ORF is important for survival in low oxygen environments (1). Whether production of AcrB-C changes in response to aeration or other stress conditions, how translation of this alt-ORF is regulated and how AcrB-C acts remain to be identified. The frequency of alt-ORF translation indicated by ribosome profiling suggests there is much to be learned about the physiological roles of bacterial alt-ORFs.
In addition to functional proteins encoded by uORFs and alt-ORFs, another major group of previously overlooked proteins are those encoded by sORFs found on independent ‘noncoding’ transcripts such as lncRNAs and circRNAs or in operons with other main-ORFs. Only a limited number of sORF-encoded proteins have been characterized but already it is clear that they have a broad range of functions [reviewed in (8,13,38,74)]. However, a few general categories observed for small membrane proteins are coming into focus (Figure 3).
Figure 3.
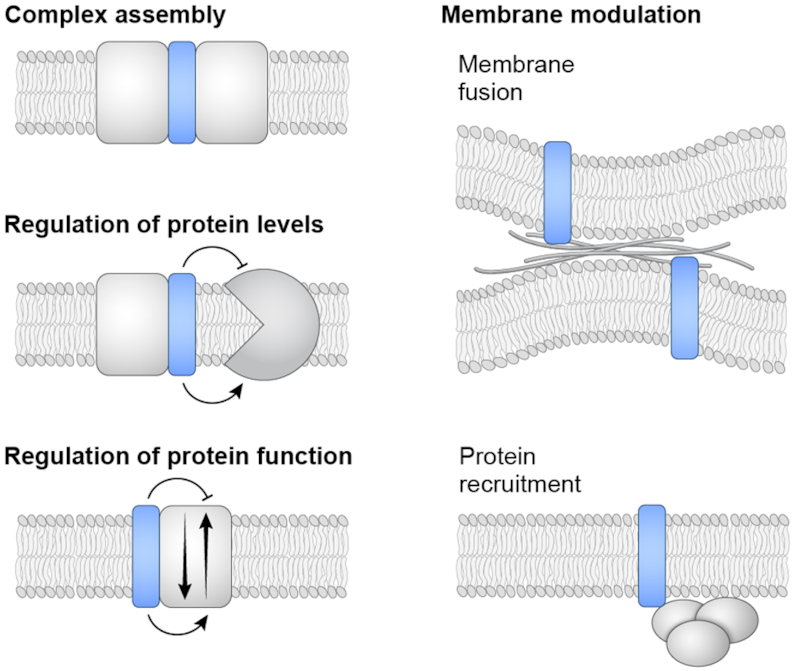
Biological functions of sORF products. The general functions of several small membrane proteins are illustrated. The small proteins can be subunits of a larger protein complexes (such as the CydX, CcoQ and CcoM protein components of cytochrome oxidase complexes in bacteria), affect protein stability (such as the low-magnesium induced MgtS protein that stabilizes the E. coli MgtA magnesium importer) or function (such as the Sarcolipin, Phospholamban, Myoregulin and DWORF modulators of the mammalian CIRCA calcium pump) that mediates membrane reorganization (such as the Minion-Myomerger-Myomixer small protein in skeletal muscle cells) and recruit complexes to cell membranes (such as the Prli42 and SpoVM proteins in L. monocytogenes and B. subtilis, respectively).
Components of larger protein complexes
A number of small proteins have been found as part of larger protein complexes, often in the membrane. Among the best characterized examples are the small proteins associated with photosystems I and II as well as cytochrome oxidases [reviewed in (75)]. Some of the associated small proteins were found as unexpected density in structural studies of these complexes (76). CydX, a small transmembrane α-helical protein associated with the E. coli cytochrome bd-I oxidase, is required for the function of this complex (77–79). However, very little is known about what CydX and other small proteins found in similar complexes are doing. One can imagine that they might be required for complex assembly as reported for the CcoQ protein (80) or for stability as suggested for the CcoM protein (81), both associated with a cytochrome c oxidases in Pseudomonas stutzeri.
Interestingly, CydX homologs have a wide diversity of sequences and a range of protein lengths. Furthermore, CydX activity withstands extensive mutagenesis and highly divergent CydX homologs can complement the E. coli cydX mutant (82,83). This heterogeneity makes predictions of small protein homologs across species difficult, particularly if genomic context is not conserved. The observations also suggest that evolution could occur rapidly in this type of small protein, as mutations in residues other than a few specific binding determinants are well-tolerated.
Modulating membrane protein activity and levels
In addition to assisting in the assembly or function of a membrane protein complex, small proteins have been found to act as regulators of membrane proteins. In eukaryotes, the small single-transmembrane α-helical proteins Sarcolipin (84), Phospholamban (85), Sarcolamban (86), Myoregulin (87) and DWORF (88,89) have been shown, in different species, to positively or negatively control the activity of the SERCA calcium pump. Sarcolipin, Phospholamban, Sarcolamban and Myoregulin share very few conserved amino acids, but structural analyses indicate that they bind SERCA in the same transmembrane groove with a similar orientation.
In E. coli, the small membrane protein AcrZ binds AcrB, the inner membrane resistance-nodulation-division (RND) family component of the AcrAB-TolC multidrug efflux pump (90). AcrZ modulates the pump substrate specificity, as loss of AcrZ results in increased sensitivity to a subset of the antibiotics transported by AcrB (90). AcrZ binds AcrB in a hydrophobic groove conserved in the transmembrane domain of RND pump proteins (91). While RND pumps with this conserved groove are found broadly across bacterial classes, AcrZ is only conserved in enterobacterial species, raising the question whether other small proteins of dissimilar sequence bind and regulate RND pumps in species lacking AcrZ.
Several bacterial small proteins have also been found to affect the activity or levels of membrane-embedded transporters or signaling proteins related to magnesium homeostasis. In E coli, the small proteins MgtS and MgrB are induced by PhoPQ, a two-component system activated by signals such as low magnesium (92). MgtS expression increases intracellular magnesium by binding two membrane transporters, the magnesium importer MgtA (93) and the phosphate-cation symporter PitA (94). MgtS appears to prevent MgtA degradation by a membrane protease while modulating PitA activity. The determinants governing the specific MgtS interaction with two different transporters are not fully understood. Conversely, in Salmonella enterica, the PhoPQ-regulated MgtR small protein enhances membrane protease degradation of MgtC (95) and MgtA (96). Furthermore, the MgrB small protein interacts with the sensor kinase PhoQ itself to repress autophosphorylation, resulting in a negative feedback loop to control expression of the PhoPQ regulon (97,98). The example small proteins described here fine-tune the cellular responses to environmental changes. Since many small proteins are induced under stress conditions or are tissue or species specific, they could have arisen as a rapidly evolving means to regulate protein activity. It is likely that many more examples of these modulators will be discovered as additional small proteins are characterized.
Modulating the membrane by membrane fusion and protein recruitment
Small proteins also act at the membrane to affect fusion and protein recruitment. The small membrane protein Minion-Myomerger-Myomixer mediates membrane fusion during skeletal muscle development and regeneration (99–101). The mechanism of action is unknown, but co-immunoprecipitation showed high enrichment of cytoskeletal proteins, suggesting that this small protein is involved in coordinating cytoskeletal reorganization events involved in fusion. In human and mouse skeletal muscle cells, the lncRNA LINC00961 produces the SPAR protein (102). SPAR is localized to the late endosomal/lysosomal membranes where it associates with the v-ATPase complex via conserved residues in its N-terminal transmembrane domain (102). SPAR enhances v-ATPase association with the Ragulator-Rag complex, which prevents Ragulator recruitment of mTORC1 to the membrane, resulting in decreased mTORC1 activation (102). During muscle injury, a reduction in SPAR levels results in enhanced mTORC1 signaling and increased muscle repair (102).
There are also bacterial examples of small protein-mediated recruitment of protein complexes to the membrane. The stressosome in Gram-positive bacteria is a large multi-protein cytoplasmic complex comprised of the RsbR, RsbS and RsbT proteins that sense environmental signals to activate the general stress response sigma factor (103). The Prli42 small protein of the pathogen L. monocytogenes was found to co-immunoprecipitate with components of the stressosome (26). Prli42 inserts C-terminally into the cytoplasmic face of the inner membrane and binds RbsR via residues in the N-terminus to tether the stressosome to the inner membrane. Prli42 is upregulated when L. monocytogenes grow in blood, and cells lacking the small protein are more sensitive to oxidative stress and show decreased survival in macrophages. Thus, it appears that Prli42 is required for activation of the stressosome-response pathway. The recruitment of a protein complex is reminiscent of the function of B. subtilis SpoVM, which is amphipathic helix that embeds horizontally into the membrane at sites of convex curvature and recruits the spore cortex component SpoIVA to the membrane of the nascent spore (104,105). These examples illustrate how small membrane-associated proteins play regulatory roles by recruiting larger protein complexes.
REGULATION OF ALT-ORF AND sORF EXPRESSION
Expression of alt-ORFs and sORFs themselves is often extensively regulated at the transcriptional and post-transcriptional levels, though the impact of various regulatory mechanisms can differ. For instance, alt-ORFs with non-canonical start codon such as CUG or UUG may be more highly dependent on the activity of translation initiation factors or the dynamics of the scanning complex.
Modulation by transcription or alternate splicing
In eukaryotes, alternative splicing or altered transcriptional initiation is quite common at different development stages. By generating isoforms with different 5′ UTR lengths, it is possible to create new ORFs or abrogate pre-existing ORFs from certain transcripts. During meiotic differentiation of budding yeast, translational reprogramming occurs via a switch between mRNA isoforms that differ in uORF content (106). This feature is also exploited by Drosophila melanogaster during development (68). In Arabidopsis thaliana, alternative transcription to eliminate uORFs is induced after exposure to blue light (52). Intriguingly, Caenorhabditis elegans uses trans-splicing to remove upstream AUG codons in native 5′ UTRs in order enhance the translation efficiency of a subset of mRNAs (107). Producing transcript isoforms probably is the most straightforward way to achieve alt-ORF diversity without altering the translation machinery.
In bacteria, transcription of the mRNAs encoding the small proteins also has been shown to be strongly regulated. As already mentioned, in E. coli, the transcription of the mgtS and mgrB mRNAs is induced by the response regulator PhoP in low magnesium. Likewise, the transcription of the mRNA encoding AcrZ is induced by the same toxic compounds that activate transcription of the acrB mRNA. Global analysis of small protein levels in E. coli showed that many are highest under specific environmental conditions (108), most likely regulated at the level of transcription.
Modulation by translation
In eukaryotes, the translation of alt-ORFs can change in response to different developmental stages, environmental cues and stress conditions. For instance, ribosome profiling showed an increase of uORF occupancy under many stress conditions such as starvation, oxidative stress, heat shock and proteotoxic stress (47,109,110). The mechanisms of this dynamic regulation are beginning to be understood [reviewed in (111,112)] and we summarize a few well-characterized pathways below (Figure 4).
Figure 4.
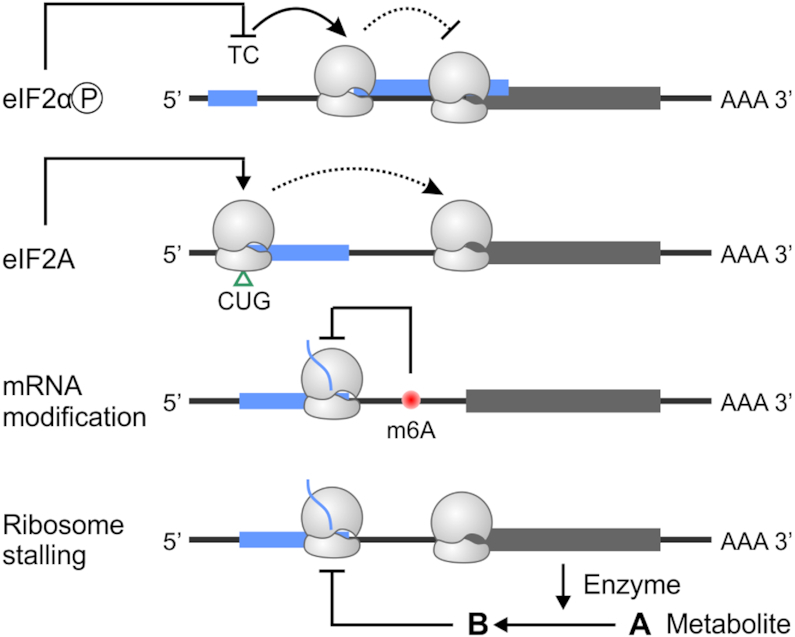
Regulation of alt-ORF translation in eukaryotes. Translation of alt-ORFs is modulated by both cis-sequence elements and trans-acting factors. Stress signaling pathways lead to phosphorylation of eIF2α, which triggers translation re-initiation at a second uORF by affecting the eIF2-GTP-tRNAiMet ternary complex (TC) availability. This can block translation of the downstream main-ORF. In response to certain stress conditions, eIF2A promotes non-AUG translation (CUG shown here), which appears to stimulate main ORF translation. mRNA modification in the form of m6A may influence alternative start codon selection by slowing ribosome scanning. Additionally, translation of some uORFs is sensitive to intracellular metabolites whose levels are controlled by enzymes encoded by the downstream main-ORF.
It has been well-established that the stringency of start codon recognition can be strengthened or weakened by cis elements and trans-acting factors. In higher eukaryotes, the optimal context for the initiator AUG triplet is a purine at position -3 and a guanine at position +4 (113). The presence of mRNA secondary structures at or near the start codon influences the recognition efficiency (114,115). In addition to these cis sequence elements, the stringency of start codon selection is subject to regulation by nearly all initiation factors, including eIF1, eIF1A and eIF3 (116–120). Impediments to 43S scanning by secondary structures or RNA-binding proteins enhances the selection of non-canonical start codons that otherwise would be skipped. Inefficient recognition of an initiator codon results in a portion of 43S ribosomes continuing to scan and initiating at a downstream site, in a process known as leaky scanning.
In response to stress conditions, phosphorylation of eIF2α reduces the abundance of eIF2-GTP-tRNAiMet ternary complex. This reduction leads to a global attenuation of protein synthesis. However, stress genes such as ATF4, GADD34 and CHOP are translationally maintained or even induced by a re-initiation mechanism that relies on uORFs. Similarly, UV-induced DNA damage triggers selective translation of mRNAs containing uORFs in the 5′ UTR (121), indicating that this regulatory mechanism is broadly prevalent under stress conditions. In mammalian cells, a gene encoding a mitochondrial ribosome protein MRPL18 undergoes alternative translation initiation in response to heat shock stress (122). The selection of a downstream CUG codon gives rise a truncated MRPL18 that is cytoplasmic. Intriguingly, cytoplasmic MRPL18 incorporates into the 80S ribosome in a stress-dependent manner, amplifying the impact of alternative translation.
eIF2A is an alternative tRNA carrier that appears to be responsible for initiation events from non-AUG codons such as CUG. The BiP transcript encodes an endoplasmic reticulum (ER) chaperone protein whose translation during ER stress relies on two uORFs with non-AUG codons (–190 UUG and –61 CUG). In response to ER stress, upregulated eIF2A initiates translation from these non-AUG uORFs, which promotes the downstream translation of the BiP main-ORF (123). As another example in yeast, the levels of Ded1p, a DEAD-box RNA helicase, are reduced during meiosis (124). Ded1p catalyzes the forward scanning of ribosomes by interacting with the scanning ribosome at the mRNA entry channel and unwinding mRNA secondary structures. Reduced levels of Ded1p lead to alternative initiation from near-cognate start codons immediately upstream of RNA secondary structures (118). These results suggest that activation of alt-ORF translation is regulated by modulating the ribosome scanning process.
Polyamines such as spermidine are essential for a wide range of cellular functions, and key enzymes in polyamine biogenesis are tightly regulated at the level of translation. The mammalian AZIN1 gene encoding an antizyme inhibitor, a regulator of polyamine synthesis, contains a uORF essential for polyamine-mediated translational repression of this gene (125). A recent study reported that polyamine competitively inhibits elongation factor eIF5A, causing the pausing of elongating ribosomes on a conserved Pro-Pro-Trp (PPW) motif in the uORF (126). The queuing of scanning ribosomes behind a paused ribosome promotes initiation at upstream weak start sites, thereby reducing AZIN1 translation. Likewise, translation of S-adenosylmethionine decarboxylase is sensitive to intracellular polyamine levels via uORF translation (127). In this case, a Met-Ala-Gly-Asp-Ile-Ser (MAGDIS) hexapeptide stalls ribosomes at the uORF stop codon and the duration of arrest depends on the concentration of polyamines. It is clear that there is extensive communication between alt-ORF and main-ORF translation to achieve polyamine homeostasis.
In bacteria, the expression of alt-ORFs can also be regulated at the post-transcriptional level. This is illustrated by genome-wide mapping of regulatory small RNA (sRNA) targets in E. coli, which showed that many small protein-coding mRNAs are associated with sRNAs, suggesting sRNA-mediated post-transcriptional regulation (128). In the intriguing case of the gndA alt-ORF encoded out of frame within the gndP main-ORF (6-phosphogluconate dehydrogenase) in E. coli (23), GndA protein levels were found to be elevated upon heat shock, while there is no evidence that gndP mRNA levels increase under this condition.
Modulation by RNA modification
Nucleotide modifications have been observed in many types of RNA. Some of the RNA modifications serve as sentinels for stress conditions, while others directly affect the decoding process of translation. In eukaryotic mRNAs, different types of methylation have been documented. One abundant and conserved mRNA modification is N6-methyladenine (m6A) (129–132). The abundance of m6A has been estimated to be, on average, 3–5 residues per mRNA in human cells. Importantly, the m6A modification is dynamic and can be reprogrammed under different conditions. For instance, yeast cells have low levels of m6A modification during regular mitotic growth, but 50% of all mRNAs contain m6A sites during meiosis (133). In addition, the m6A landscape changes in response to various stimuli (134,135). Intriguingly, m6A in 5′ UTRs affects the ribosome scanning process, thereby influencing uORF translation (136). In the ATF4 mRNA, the stress-induced removal of m6A from uORF2 facilitates uORF2 bypass, thereby enhancing ATF4 translation. How exactly m6A affects the scanning process remains unclear.
ROLES IN DISEASE
A growing body of evidence shows that alternative translation initiation plays a critical role in the maintenance of cellular homeostasis. In humans, mutations that create or delete alt-ORFs are associated with various physiological abnormalities. Examples include craniofrontonasal syndrome (CFNS) and thrombocythaemia (137). CFNS is a rare X-linked dominant disorder caused by loss-of-function mutations in the EFNB1 transcript. There are at least two CFNS-associated mutations found in the EFNB1 5′ UTR. Either 95T>C or 95T>G abolishes the stop codon of a uORF, resulting in an overlapped, out-of-frame ORF relative to the main-ORF. Another mutation (411C>G) leads to creation of a new uORF. Translation of these abnormal uORFs leads to repressed translation of main-ORF. In the case of thrombocythaemia (138), a mutation in 5′ UTR of thrombopoietin gene causes abnormal translation of the pre-existing uORF, resulting in systemic overexpression of thrombopoietin leading to thrombocythaemia.
A number of neurological disorders are caused by the expansion of nucleotide repeats. Typical examples include Huntington’s disease with CAG repeat expansions in the coding sequence of the Huntingtin gene (139), and fragile X disorders with CGG repeat expansions in the 5′ UTR of the FMR1 gene (140). Importantly, repeat-associated non-AUG (RAN) translation appears to be the etiology of many neurodegenerative diseases. RAN translation occurs in multiple reading frames and appears to initiate within expanded nucleotide repeats, generating toxic polypeptides. Regulatory mechanisms underlying RAN translation remain incompletely understood.
Both genome-wide and gene-specific analyses have provided evidence for the functional and clinical relevance of 5′ UTR mutations in cancer (141–143). Cancer cells exploit multiple mechanisms to modulate main-ORF translation. For example, human epidermal growth factor receptor 2 (HER2, also known as ERBB2) is overexpressed in ∼15–30% of breast cancers (144). The translation of HER2 is normally repressed by uORF translation. In tumor cells, however, the RNA-binding proteins HUR and hnRNPA1 overcome the inhibitory effect of this translation by binding to the 3′ UTR of HER2 (145,146). The deregulation of uORF translation in cancer also can have the opposite effect. The CDKN2A gene encodes two proteins p16INK4a and p14ARF, which act as tumor suppressors by regulating the cell cycle (147,148). In addition to mutations in the main-ORF, a single point mutation in the 5′ UTR of CDKN2A creates a new translation initiation site. Translation of this novel uORF decreases CDKN2A protein levels in hereditary melanoma. The repeated observation of uORF mutations associated with disease implies they play a crucial role in pathogenesis.
De-regulation of uORF translation also can occur in a global manner. A recent study reported that alternative translation initiation is globally elevated in embryonic skin cells from a mouse tumorigenesis model (149). It was hypothesized that the phosphorylation of eIF2α in early stages of tumorigenesis leads to reprogrammed translation via eIF2A-dependent alternative initiation. How differential usage of uORFs genome-wide can drive cancer phenotypes as of yet is unknown.
PERSPECTIVES
As seen from the studies summarized here, the density of information encoded in eukaryotic and bacterial genomes is far greater than previously appreciated, with many more translation events than expected. Translation can serve a regulatory role or result in a functional protein, or possibly both. The tight regulation observed suggests that the translation of these previously ignored regions is important to the cell, a point emphasized by the association of abnormal alt-ORF expression with disease. These studies open up many exciting new avenues of research already mentioned and raise additional questions worth considering.
Standard of evidence
It is unlikely that every possible translated ORF gives rise to a functional protein, and the functional analysis of every predicted translated alt-ORF will take time. This leads to the question of when to annotate a putative ORF. What level of evidence is necessary? Is evidence of translation by ribosome profiling or matches by mass spectrometry sufficient? The presence of ribosome density does not necessarily indicate translation of a protein product. Similarly, a single spectral match by mass spectrometry does not provide concrete evidence for synthesis. Furthermore, translation of an ORF could be translational noise that neither has a regulatory function nor produces a stable protein. We suggest that multiple lines of evidence must be present before a putative ORF is annotated.
Complication of dual-function
For nested alt-ORFs such as the gndA-gndP pair, there are questions about how expression of the two functions encoded by overlapping regions impact each other. Related to this, there are an increasing number of examples where transcripts shown to act as regulatory RNAs also encode small proteins, thus serving a ‘dual-function’ (150). The regulatory RNA and the small protein could either act in the same pathway or have very different consequences. The plant Medicago truncatula regulatory miRNA miR171b encodes the small protein miPEP171, which was shown to increase accumulation of miR171b to enhance lateral root formation (151). In bacteria, the E. coli SgrS regulatory sRNA also encodes the SgrT protein; both SgrS and SgrT regulate the same target, the glucose transporter EIICB (152). The Staphylococcus aureus RNAIII sRNA, which base pairs with multiple mRNAs to regulate virulence, also encodes a δ-hemolysin protein. While δ-hemolysin is important in S. aureus virulence, in this case the regulatory sRNA and the protein it produces do not impact the same target [reviewed in (153,154)]. Very little is known about when and how these dual-function RNAs act as mRNAs versus regulatory RNAs or whether the two activities interfere with each other.
Evolution
Consideration of how alt-ORFs in general, and overlapping functions in particular, evolve also leads to interesting questions. For example, in the case of dual-function RNAs, which activity evolved first? For small, hydrophobic membrane proteins, did these evolve from the duplication of a transmembrane domain? An attractive hypothesis is that alt-ORFs, which are less conserved than main-ORFs (consistent with their regulatory role in gene expression), have evolved more recently. Unlike main-ORFs that primarily employ canonical AUG start codons, alt-ORFs often use near cognate start codons (AUG-like triplets) for initiation. This feature further argues that translation of alt-ORFs is of much lower efficiency when compared to the main-ORF. It was recently shown that in a D. melanogaster population, a considerable fraction of alt-ORFs are beneficial and rapidly fixed under positive selection (68). In addition to mutations, the insertion of transposable elements can generate new alt-ORFs. Notably, an estimated ∼10% human alt-ORFs are derived from transposable elements (155). The evolutionary forces governing alt-ORF generation in all organisms remain to be explored.
Potential for exploitation
Given their broad roles, it is desirable to engineer alt-ORFs for precise control of protein translation (156–158). Indeed, genome editing of endogenous uORFs in plants enabled translational modulation of main-ORFs encoding proteins involved in development or antioxidant biosynthesis (156). Additionally, inserting pathogen-responsive uORFs into transgenes encoding key immune regulators allows plants to gain broad-spectrum disease resistance without compromising plant fitness (158). More recently, efforts have been undertaken to identify small molecule compounds that target uORF translation (159). Since the majority of uORFs rely on near-cognate start codons, targeting non-AUG translation may be a therapeutic strategy.
Along these same lines, once more is known about small protein mechanisms of action, their functions could be exploited for disease treatment. Both stable synthetic analogs of natural small regulatory proteins and molecules that affect small protein binding could have therapeutic value [reviewed for eukaryotes in (13)]. For instance, the inhibition or mimicry of small proteins involved in infection, such as the stressosome-recruiting protein Prli42, or antibiotic resistance, such as the the AcrZ multidrug efflux pump regulator, could be valuable tools for combating infection or multi-drug resistance, particularly in conjunction with other therapies.
CONCLUSION
Going forward, we suggest that addressing the questions raised in this review will be of great interest and importance. This work will provide novel insights into translational regulation and uncover hidden coding potential for the genomes and additional functions for the proteomes of all organisms.
ACKNOWLEDGEMENTS
We thank Nicholas Guydosh and Allen Buskirk for comments on this review. Research in the G.S. lab is supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
FUNDING
National Institutes of Health [R01GM1222814, R21CA227917]; HHMI Faculty Scholar Program [55108556] to S.B.-Q. Funding for open access charge: NICHD [ZIA HD008855-12].
Conflict of interest statement. None declared.
REFERENCES
- 1. Meydan S., Marks J., Klepacki D., Sharma V., Baranov P.V., Firth A.E., Margus T., Kefi A., Vázquez-Laslop N., Mankin A.S.. Retapamulin-assisted ribosome profiling reveals the alternative bacterial proteome. Mol. Cell. 2019; 74:481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hsu P.Y., Calviello L., Wu H.-Y.L., Li F.-W., Rothfels C.J., Ohler U., Benfey P.N.. Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fritsch C., Herrmann A., Nothnagel M., Szafranski K., Huse K., Schumann F., Schreiber S., Platzer M., Krawczak M., Hampe J. et al.. Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res. 2012; 22:2208–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shell S.S., Wang J., Lapierre P., Mir M., Chase M.R., Pyle M.M., Gawande R., Ahmad R., Sarracino D.A., Ioerger T.R.. Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLos Genet. 2015; 11:e1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miravet‐Verde S., Ferrar T., Espadas‐García G., Mazzolini R., Gharrab A., Sabido E., Serrano L., Lluch‐Senar M.. Unraveling the hidden universe of small proteins in bacterial genomes. Mol. Syst. Biol. 2019; 15:e8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ragan C., Goodall G.J., Shirokikh N.E., Preiss T.. Insights into the biogenesis and potential functions of exonic circular RNA. Sci. Rep. 2019; 9:2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Couso J.-P., Patraquim P.. Classification and function of small open reading frames. Nat. Rev. Mol. Cell Biol. 2017; 18:575–589. [DOI] [PubMed] [Google Scholar]
- 8. Duval M., Cossart P.. Small bacterial and phagic proteins: an updated view on a rapidly moving field. Curr. Opin. Microbiol. 2017; 39:81–88. [DOI] [PubMed] [Google Scholar]
- 9. Meydan S., Vázquez-Laslop N., Mankin A.S.. Genes within genes in bacterial genomes. Microbiol. Spectr. 2018; 6:doi:10.1128/microbiolspec.RWR-0020-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ruiz-Orera J., Albà M.M.. Translation of small open reading frames: roles in regulation and evolutionary innovation. Trends Genet. 2019; 35:186–198. [DOI] [PubMed] [Google Scholar]
- 11. Hinnebusch A.G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014; 83:779–812. [DOI] [PubMed] [Google Scholar]
- 12. Laursen B.S., Sørensen H.P., Mortensen K.K., Sperling-Petersen H.U.. Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 2005; 69:101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saghatelian A., Couso J.P.. Discovery and characterization of smORF-encoded bioactive polypeptides. Nat. Chem. Biol. 2015; 11:909–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pueyo J.I., Magny E.G., Couso J.P.. New peptides under the s(ORF)ace of the genome. Trends Biochem. Sci. 2016; 41:665–678. [DOI] [PubMed] [Google Scholar]
- 15. Samayoa J., Yildiz F.H., Karplus K.. Identification of prokaryotic small proteins using a comparative genomic approach. Bioinformatics. 2011; 27:1765–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou P., Silverstein K.A.T., Gao L., Walton J.D., Nallu S., Guhlin J., Young N.D.. Detecting small plant peptides using SPADA (Small Peptide Alignment Discovery Application). BMC Bioinform. 2013; 14:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ladoukakis E., Pereira V., Magny E.G., Eyre-Walker A., Couso J.P.. Hundreds of putatively functional small open reading frames in Drosophila. Genome Biol. 2011; 12:R118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goli B., Nair A.S.. The elusive short gene – an ensemble method for recognition for prokaryotic genome. Biochem. Biophys. Res. Commun. 2012; 422:36–41. [DOI] [PubMed] [Google Scholar]
- 19. Chen S., Zhang C.-y, Song K.. Recognizing short coding sequences of prokaryotic genome using a novel iteratively adaptive sparse partial least squares algorithm. Biol. Direct. 2013; 8:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sberro H., Fremin B.J., Zlitni S., Edfors F., Greenfield N., Snyder M.P., Pavlopoulos G.A., Kyrpides N.C., Bhatt A.S.. Large-scale analyses of human microbiomes reveal thousands of small, novel genes. Cell. 2019; 30781-0:S0092-8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matsui M., Yachie N., Okada Y., Saito R., Tomita M.. Bioinformatic analysis of post-transcriptional regulation by uORF in human and mouse. FEBS Lett. 2007; 581:4184–4188. [DOI] [PubMed] [Google Scholar]
- 22. Slavoff S.A., Mitchell A.J., Schwaid A.G., Cabili M.N., Ma J., Levin J.Z., Karger A.D., Budnik B.A., Rinn J.L., Saghatelian A.. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 2013; 9:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yuan P., D’Lima N.G., Slavoff S.A.. Comparative membrane proteomics reveals a nonannotated E. coli heat shock protein. Biochemistry. 2018; 57:56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. D’Lima N.G., Khitun A., Rosenbloom A.D., Yuan P., Gassaway B.M., Barber K.W., Rinehart J., Slavoff S.A.. Comparative proteomics enables identification of nonannotated cold shock proteins in E. coli. J. Proteome Res. 2017; 16:3722–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bienvenut W.V., Giglione C., Meinnel T.. Proteome-wide analysis of the amino terminal status of Escherichia coli proteins at the steady-state and upon deformylation inhibition. Proteomics. 2015; 15:2503–2518. [DOI] [PubMed] [Google Scholar]
- 26. Impens F., Rolhion N., Radoshevich L., Bécavin C., Duval M., Mellin J., García del Portillo F., Pucciarelli M.G., Williams A.H., Cossart P.. N-terminomics identifies Prli42 as a membrane miniprotein conserved in Firmicutes and critical for stressosome activation in Listeria monocytogenes. Nat. Microbiol. 2017; 2:17005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ingolia N.T., Ghaemmaghami S., Newman J.R.S., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Johnstone T.G., Bazzini A.A., Giraldez A.J.. Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J. 2016; 35:706–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chew G.L., Pauli A., Schier A.F.. Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat. Commun. 2016; 7:11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Spealman P., Naik A.W., May G.E., Kuersten S., Freeberg L., Murphy R.F., McManus J.. Conserved non-AUG uORFs revealed by a novel regression analysis of ribosome profiling data. Genome Res. 2018; 28:214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lei L., Shi J., Chen J., Zhang M., Sun S., Xie S., Li X., Zeng B., Peng L., Hauck A. et al.. Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress. Plant J. 2015; 84:1206–1218. [DOI] [PubMed] [Google Scholar]
- 32. Lim C.S., SJ T.W., Kleffmann T., Brown C.M.. The exon-intron gene structure upstream of the initiation codon predicts translation efficiency. Nucleic Acids Res. 2018; 46:4575–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee S., Liu B., Lee S., Huang S.X., Shen B., Qian S.B.. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E2424–E2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gao X., Wan J., Liu B., Ma M., Shen B., Qian S.-B.. Quantitative profiling of initiating ribosomes in vivo. Nat. Methods. 2015; 12:147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nakahigashi K., Takai Y., Kimura M., Abe N., Nakayashiki T., Shiwa Y., Yoshikawa H., Wanner B.L., Ishihama Y., Mori H.. Comprehensive identification of translation start sites by tetracycline-inhibited ribosome profiling. DNA Res. 2016; 23:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weaver J., Mohammad F., Buskirk A.R., Storz G.. Identifying small proteins by ribosome profiling with stalled initiation complexes. mBio. 2019; 10:e02819-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Santos D.A., Shi L., Tu B.P., Weissman J.S.. Cycloheximide can distort measurements of mRNA levels and translation efficiency. Nucleic Acids Res. 2019; 47:4974–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Khitun A., Ness T.J., Slavoff S.A.. Small open reading frames and cellular stress responses. Mol. Omics. 2019; 15:108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Occhi G., Regazzo D., Trivellin G., Boaretto F., Ciato D., Bobisse S., Ferasin S., Cetani F., Pardi E., Korbonits M.. A novel mutation in the upstream open reading frame of the CDKN1B gene causes a MEN4 phenotype. PLos Genet. 2013; 9:e1003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gaba A., Jacobson A., Sachs M.S.. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol. Cell. 2005; 20:449–460. [DOI] [PubMed] [Google Scholar]
- 41. Young S.K., Palam L.R., Wu C., Sachs M.S., Wek R.C.. Ribosome elongation stall directs gene-specific translation in the integrated stress response. J. Biol. Chem. 2016; 291:6546–6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ribone P.A., Capella M., Arce A.L., Chan R.L.. A uORF represses the transcription factor AtHB1 in aerial tissues to avoid a deleterious phenotype. Plant Physiol. 2017; 175:1238–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wethmar K. The regulatory potential of upstream open reading frames in eukaryotic gene expression. Wiley Interdiscip. Rev. RNA. 2014; 5:765–778. [DOI] [PubMed] [Google Scholar]
- 44. Wilson D.N., Arenz S., Beckmann R.. Translation regulation via nascent polypeptide-mediated ribosome stalling. Curr. Opin. Struct. Biol. 2016; 37:123–133. [DOI] [PubMed] [Google Scholar]
- 45. Meijer H.A. Ribosomes stalling on uORF1 in the Xenopus Cx41 5′ UTR inhibit downstream translation initiation. Nucleic Acids Res. 2003; 31:3174–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wiese A., Elzinga N., Wobbes B., Smeekens S.. A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell. 2004; 16:1717–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vattem K.M., Wek R.C.. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lykke-Andersen S., Jensen T.H.. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 2015; 16:665–677. [DOI] [PubMed] [Google Scholar]
- 49. Jaffrey S.R., Wilkinson M.F.. Nonsense-mediated RNA decay in the brain: emerging modulator of neural development and disease. Nat. Rev. Neurosci. 2018; 19:715–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Colombo M., Karousis E.D., Bourquin J., Bruggmann R., Muhlemann O.. Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways. RNA. 2017; 23:189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Celik A., Baker R., He F., Jacobson A.. High-resolution profiling of NMD targets in yeast reveals translational fidelity as a basis for substrate selection. RNA. 2017; 23:735–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kurihara Y., Makita Y., Kawashima M., Fujita T., Iwasaki S., Matsui M.. Transcripts from downstream alternative transcription start sites evade uORF-mediated inhibition of gene expression in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:7831–7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Saul H., Elharrar E., Gaash R., Eliaz D., Valenci M., Akua T., Avramov M., Frankel N., Berezin I., Gottlieb D. et al.. The upstream open reading frame of the Arabidopsis AtMHX gene has a strong impact on transcript accumulation through the nonsense-mediated mRNA decay pathway. Plant J. 2009; 60:1031–1042. [DOI] [PubMed] [Google Scholar]
- 54. Nyiko T., Sonkoly B., Merai Z., Benkovics A.H., Silhavy D.. Plant upstream ORFs can trigger nonsense-mediated mRNA decay in a size-dependent manner. Plant Mol. Biol. 2009; 71:367–378. [DOI] [PubMed] [Google Scholar]
- 55. Tanaka M., Sotta N., Yamazumi Y., Yamashita Y., Miwa K., Murota K., Chiba Y., Hirai M.Y., Akiyama T., Onouchi H. et al.. The minimum open reading frame, AUG-Stop, induces boron-dependent ribosome stalling and mRNA degradation. Plant Cell. 2016; 28:2830–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ruiz-Echevarría M.J., Peltz S.W.. The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell. 2000; 101:741–751. [DOI] [PubMed] [Google Scholar]
- 57. Toma K.G., Rebbapragada I., Durand S., Lykke-Andersen J.. Identification of elements in human long 3′ UTRs that inhibit nonsense-mediated decay. RNA. 2015; 21:887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Park Y., Reyna-Neyra A., Philippe L., Thoreen C.C.. mTORC1 balances cellular amino acid supply with demand for protein synthesis through post-transcriptional control of ATF4. Cell Rep. 2017; 19:1083–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Keeling K.M., Wang D., Dai Y., Murugesan S., Chenna B., Clark J., Belakhov V., Kandasamy J., Velu S.E., Baasov T.. Attenuation of nonsense-mediated mRNA decay enhances in vivo nonsense suppression. PLoS One. 2013; 8:e60478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kolter R., Yanofsky C.. Attenuation in amino acid biosynthetic operons. Annu. Rev. Genet. 1982; 16:113–134. [DOI] [PubMed] [Google Scholar]
- 61. Gollnick P., Babitzke P.. Transcription attenuation. Biochim. Biophys. Acta Gene Struct. Expr. 2002; 1577:240–250. [DOI] [PubMed] [Google Scholar]
- 62. Levin H.L., Schachman H.K.. Regulation of aspartate transcarbamoylase synthesis in Escherichia coli: analysis of deletion mutations in the promoter region of the pyrBI operon. Proc. Natl. Acad. Sci. U.S.A. 1985; 82:4643–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Park S.-Y., Cromie M.J., Lee E.-J., Groisman E.A.. A bacterial mRNA leader that employs different mechanisms to sense disparate intracellular signals. Cell. 2010; 142:737–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ramu H., Mankin A., Vazquez-Laslop N.. Programmed drug-dependent ribosome stalling. Mol. Microbiol. 2009; 71:811–824. [DOI] [PubMed] [Google Scholar]
- 65. Ito K., Chiba S.. Arrest peptides: cis-acting modulators of translation. Annu. Rev. Biochem. 2013; 82:171–202. [DOI] [PubMed] [Google Scholar]
- 66. McCarthy J.E.G., Gualerzi C.. Translational control of prokaryotic gene expression. Trends Genet. 1990; 6:78–85. [DOI] [PubMed] [Google Scholar]
- 67. Park H., McGibbon L.C., Potts A.H., Yakhnin H., Romeo T., Babitzke P.. Translational repression of the RpoS antiadapter IraD by CsrA is mediated via translational coupling to a short upstream open reading frame. mBio. 2017; 8:e01355-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang H., Dou S., He F., Luo J., Wei L., Lu J.. Genome-wide maps of ribosomal occupancy provide insights into adaptive evolution and regulatory roles of uORFs during Drosophila development. PLoS Biol. 2018; 16:e2003903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lorenzo-Orts L., Witthoeft J., Deforges J., Martinez J., Loubery S., Placzek A., Poirier Y., Hothorn L.A., Jaillais Y., Hothorn M.. Concerted expression of a cell cycle regulator and a metabolic enzyme from a bicistronic transcript in plants. Nat. Plants. 2019; 5:184–193. [DOI] [PubMed] [Google Scholar]
- 70. Fields A.P., Rodriguez E.H., Jovanovic M., Stern-Ginossar N., Haas B.J., Mertins P., Raychowdhury R., Hacohen N., Carr S.A., Ingolia N.T. et al.. A regression-based analysis of ribosome-profiling data reveals a conserved complexity to mammalian translation. Mol. Cell. 2015; 60:816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Outten C.E., Culotta V.C.. Alternative start sites in the Saccharomyces cerevisiae GLR1 gene are responsible for mitochondrial and cytosolic isoforms of glutathione reductase. J. Biol. Chem. 2004; 279:7785–7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liang H., He S., Yang J., Jia X., Wang P., Chen X., Zhang Z., Zou X., McNutt M.A., Shen W.H. et al.. PTENα, a PTEN isoform translated through alternative initiation, regulates mitochondrial function and energy metabolism. Cell Metab. 2014; 19:836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Iuchi S., Cameron D.C., Lin E.C.. A second global regulator gene (arcB) mediating repression of enzymes in aerobic pathways of Escherichia coli. J. Bacteriol. 1989; 171:868–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Plaza S., Menschaert G., Payre F.. In search of lost small peptides. Annu. Rev. Cell Dev. Biol. 2017; 33:391–416. [DOI] [PubMed] [Google Scholar]
- 75. Storz G., Wolf Y.I., Ramamurthi K.S.. Small proteins can no longer be ignored. Annu. Rev. Biochem. 2014; 83:753–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Safarian S., Rajendran C., Müller H., Preu J., Langer J.D., Ovchinnikov S., Hirose T., Kusumoto T., Sakamoto J., Michel H.. Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases. Science. 2016; 352:583–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sun Y.-H., de Jong M.F., den Hartigh A.B., Roux C.M., Rolán H.G., Tsolis R.M.. The small protein CydX is required for function of cytochrome bd oxidase in Brucella abortus. Front. Cell Infect. Microbiol. 2012; 2:47–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. VanOrsdel C.E., Bhatt S., Allen R.J., Brenner E.P., Hobson J.J., Jamil A., Haynes B.M., Genson A.M., Hemm M.R.. The Escherichia coli CydX protein is a member of the CydAB cytochrome bd oxidase complex and is required for cytochrome bd oxidase activity. J. Bacteriol. 2013; 195:3640–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chen H., Luo Q., Yin J., Gao T., Gao H.. Evidence for the requirement of CydX in function but not assembly of the cytochrome bd oxidase in Shewanella oneidensis. Biochim. Biophys. Acta Gene Subj. 2015; 1850:318–328. [DOI] [PubMed] [Google Scholar]
- 80. Kohlstaedt M., Buschmann S., Langer J.D., Xie H., Michel H.. Subunit CcoQ is involved in the assembly of the Cbb3-type cytochrome c oxidases from Pseudomonas stutzeri ZoBell but not required for their activity. Biochim. Biophys. Acta Bioenerg. 2017; 1858:231–238. [DOI] [PubMed] [Google Scholar]
- 81. Kohlstaedt M., Buschmann S., Xie H., Resemann A., Warkentin E., Langer J.D., Michel H.. Identification and characterization of the novel subunit ccoM in the cbb3-cytochrome c oxidase from Pseudomonas stutzeri ZoBell. mBio. 2016; 7:e01921-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Allen R.J., Brenner E.P., VanOrsdel C.E., Hobson J.J., Hearn D.J., Hemm M.R.. Conservation analysis of the CydX protein yields insights into small protein identification and evolution. BMC Genom. 2014; 15:946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hobson J.J., Gallegos A.S., Atha B.W. III, Kelly J.P., Lein C.D., VanOrsdel C.E., Weldon J.E., Hemm M.R.. Investigation of amino acid specificity in the CydX small protein shows sequence plasticity at the functional level. PLoS One. 2018; 13:e0198699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tada M., Kirchberger M.A., Katz A.M.. Phosphorylation of a 22,000-dalton component of the cardiac sarcoplasmic reticulum by adenosine 3′: 5′-monophosphate-dependent protein kinase. J. Biol. Chem. 1975; 250:2640–2647. [PubMed] [Google Scholar]
- 85. Wawrzynow A., Theibert J.L., Murphy C., Jona I., Martonosi A., Collins J.H.. Sarcolipin, the “proteolipid” of skeletal muscle sarcoplasmic reticulum, is a unique, amphipathic, 31-residue peptide. Arch. Biochem. Biophys. 1992; 298:620–623. [DOI] [PubMed] [Google Scholar]
- 86. Magny E.G., Pueyo J.I., Pearl F.M., Cespedes M.A., Niven J.E., Bishop S.A., Couso J.P.. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science. 2013; 341:1116–1120. [DOI] [PubMed] [Google Scholar]
- 87. Anderson D.M., Anderson K.M., Chang C.-L., Makarewich C.A., Nelson B.R., McAnally J.R., Kasaragod P., Shelton J.M., Liou J., Bassel-Duby R. et al.. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015; 160:595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Nelson B.R., Makarewich C.A., Anderson D.M., Winders B.R., Troupes C.D., Wu F., Reese A.L., McAnally J.R., Chen X., Kavalali E.T. et al.. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016; 351:271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Payre F., Desplan C.. Small peptides control heart activity. Science. 2016; 351:226–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hobbs E.C., Yin X., Paul B.J., Astarita J.L., Storz G.. Conserved small protein associates with the multidrug efflux pump AcrB and differentially affects antibiotic resistance. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:16696–16701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Du D., Wang Z., James N.R., Voss J.E., Klimont E., Ohene-Agyei T., Venter H., Chiu W., Luisi B.F.. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014; 509:512–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Moon K., Gottesman S.. A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol. Microbiol. 2009; 74:1314–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wang H., Yin X., Wu Orr M., Dambach M., Curtis R., Storz G.. Increasing intracellular magnesium levels with the 31-amino acid MgtS protein. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:5689–5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yin X., Wu Orr M., Wang H., Hobbs E.C., Shabalina S.A., Storz G.. The small protein MgtS and small RNA MgrR modulate the PitA phosphate symporter to boost intracellular magnesium levels. Mol. Microbiol. 2019; 111:131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Alix E., Blanc-Potard A.-B.. Peptide-assisted degradation of the Salmonella MgtC virulence factor. EMBO J. 2008; 27:546–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Choi E., Lee K.-Y., Shin D.. The MgtR regulatory peptide negatively controls expression of the MgtA Mg2+ transporter in Salmonella enterica serovar Typhimurium. Biochem. Biophys. Res. Commun. 2012; 417:318–323. [DOI] [PubMed] [Google Scholar]
- 97. Lippa A.M., Goulian M.. Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLoS Genet. 2009; 5:e1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Salazar M.E., Podgornaia A.I., Laub M.T.. The small membrane protein MgrB regulates PhoQ bifunctionality to control PhoP target gene expression dynamics. Mol. Microbiol. 2016; 102:430–445. [DOI] [PubMed] [Google Scholar]
- 99. Zhang Q., Vashisht A.A., O’Rourke J., Corbel S.Y., Moran R., Romero A., Miraglia L., Zhang J., Durrant E., Schmedt C. et al.. The microprotein Minion controls cell fusion and muscle formation. Nat. Commun. 2017; 8:15664–15664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Quinn M.E., Goh Q., Kurosaka M., Gamage D.G., Petrany M.J., Prasad V., Millay D.P.. Myomerger induces fusion of non-fusogenic cells and is required for skeletal muscle development. Nat. Commun. 2017; 8:15665–15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Bi P., Ramirez-Martinez A., Li H., Cannavino J., McAnally J.R., Shelton J.M., Sánchez-Ortiz E., Bassel-Duby R., Olson E.N.. Control of muscle formation by the fusogenic micropeptide myomixer. Science. 2017; 356:323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Tajbakhsh S. lncRNA-encoded polypeptide SPAR(s) with mTORC1 to regulate skeletal muscle regeneration. Cell Stem Cell. 2017; 20:428–430. [DOI] [PubMed] [Google Scholar]
- 103. Pané-Farré J., Q.M.B. Lewis R.J., Marles-Wright J.. Structure and function of the stressosome signalling hub. Subcell. Biochem. 2017; 83:1–41. [DOI] [PubMed] [Google Scholar]
- 104. Ramamurthi K.S., Clapham K.R., Losick R.. Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol. Microbiol. 2006; 62:1547–1557. [DOI] [PubMed] [Google Scholar]
- 105. Ebmeier S.E., Tan I.S., Clapham K.R., Ramamurthi K.S.. Small proteins link coat and cortex assembly during sporulation in Bacillus subtilis. Mol. Microbiol. 2012; 84:682–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Rojas-Duran M.F., Gilbert W.V.. Alternative transcription start site selection leads to large differences in translation activity in yeast. RNA. 2012; 18:2299–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Yang Y.-F., Zhang X., Ma X., Zhao T., Sun Q., Huan Q., Wu S., Du Z., Qian W.. Trans-splicing enhances translational efficiency in C. elegans. Genome Res. 2017; 27:1525–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Hemm M.R., Paul B.J., Miranda-Ríos J., Zhang A., Soltanzad N., Storz G.. Small stress response proteins in Escherichia coli: proteins missed by classical proteomic studies. J. Bacteriol. 2010; 192:46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Gerashchenko M.V., Lobanov A.V., Gladyshev V.N.. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:17394–17399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zhu X., Thalor S.K., Takahashi Y., Berberich T., Kusano T.. An inhibitory effect of the sequence‐conserved upstream open‐reading frame on the translation of the main open‐reading frame of HsfB1 transcripts in Arabidopsis. Plant Cell Environ. 2012; 35:2014–2030. [DOI] [PubMed] [Google Scholar]
- 111. Wek R.C. Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb. Perspect. Biol. 2018; 10:a032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Liu B., Qian S.-B.. Translational reprogramming in cellular stress response. Wiley Interdiscip. Rev. RNA. 2014; 5:301–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986; 44:283–292. [DOI] [PubMed] [Google Scholar]
- 114. Gu W., Zhou T., Wilke C.O.. A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput. Biol. 2010; 6:e1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Mortimer S.A., Kidwell M.A., Doudna J.A.. Insights into RNA structure and function from genome-wide studies. Nat. Rev. Genet. 2014; 15:469–479. [DOI] [PubMed] [Google Scholar]
- 116. Fijalkowska D., Verbruggen S., Ndah E., Jonckheere V., Menschaert G., Van Damme P.. eIF1 modulates the recognition of suboptimal translation initiation sites and steers gene expression via uORFs. Nucleic Acids Res. 2017; 45:7997–8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kim T.H., Kim B.H., Yahalom A., Chamovitz D.A., von Arnim A.G.. Translational regulation via 5′ mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit eIF3h. Plant Cell. 2004; 16:3341–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Roy B., Vaughn J.N., Kim B.H., Zhou F., Gilchrist M.A., Von Arnim A.G.. The h subunit of eIF3 promotes reinitiation competence during translation of mRNAs harboring upstream open reading frames. RNA. 2010; 16:748–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Schepetilnikov M., Dimitrova M., Mancera-Martinez E., Geldreich A., Keller M., Ryabova L.A.. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 2013; 32:1087–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Hinnebusch A.G. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem. Sci. 2017; 42:589–611. [DOI] [PubMed] [Google Scholar]
- 121. Powley I.R., Kondrashov A., Young L.A., Dobbyn H.C., Hill K., Cannell I.G., Stoneley M., Kong Y.-W., Cotes J.A., Smith G.C.M. et al.. Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes Dev. 2009; 23:1207–1220. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 122. Zhang X., Gao X., Coots R.A., Conn C.S., Liu B., Qian S.-B.. Translational control of the cytosolic stress response by mitochondrial ribosomal protein L18. Nat. Struct. Mol. Biol. 2015; 22:404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Starck S.R., Tsai J.C., Chen K., Shodiya M., Wang L., Yahiro K., Martins-Green M., Shastri N., Walter P.. Translation from the 5′ untranslated region shapes the integrated stress response. Science. 2016; 351:aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Guenther U.-P., Weinberg D.E., Zubradt M.M., Tedeschi F.A., Stawicki B.N., Zagore L.L., Brar G.A., Licatalosi D.D., Bartel D.P., Weissman J.S.. The helicase Ded1p controls use of near-cognate translation initiation codons in 5′ UTRs. Nature. 2018; 559:130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Dever T.E., Ivanov I.P.. Roles of polyamines in translation. J. Biol. Chem. 2018; 293:18719–18729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ivanov I.P., Shin B.-S., Loughran G., Tzani I., Young-Baird S.K., Cao C., Atkins J.F., Dever T.E.. Polyamine control of translation elongation regulates start site selection on antizyme inhibitor mRNA via ribosome queuing. Mol. Cell. 2018; 70:254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Hanfrey C., Franceschetti M., Mayer M.J., Illingworth C., Michael A.J.. Abrogation of upstream open reading frame-mediated translational control of a plant S-adenosylmethionine decarboxylase results in polyamine disruption and growth perturbations. J. Biol. Chem. 2002; 277:44131–44139. [DOI] [PubMed] [Google Scholar]
- 128. Melamed S., Peer A., Faigenbaum-Romm R., Gatt Y.E., Reiss N., Bar A., Altuvia Y., Argaman L., Margalit H.. Global mapping of small RNA-target interactions in bacteria. Mol. Cell. 2016; 63:884–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Frye M., Harada B.T., Behm M., He C.. RNA modifications modulate gene expression during development. Science. 2018; 361:1346–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Roundtree I.A., Evans M.E., Pan T., He C.. Dynamic RNA modifications in gene expression regulation. Cell. 2017; 169:1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Fu Y., Dominissini D., Rechavi G., He C.. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014; 15:293–306. [DOI] [PubMed] [Google Scholar]
- 132. Helm M., Motorin Y.. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 2017; 18:275–291. [DOI] [PubMed] [Google Scholar]
- 133. Schwartz S., Agarwala S.D., Mumbach M.R., Jovanovic M., Mertins P., Shishkin A., Tabach Y., Mikkelsen T.S., Satija R., Ruvkun G.. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013; 155:1409–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Anders M., Chelysheva I., Goebel I., Trenkner T., Zhou J., Mao Y., Verzini S., Qian S.-B., Ignatova Z.. Dynamic m6A methylation facilitates mRNA triaging to stress granules. Life Sci. Alliance. 2018; 1:e201800113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Zhou J., Wan J., Gao X., Zhang X., Jaffrey S.R., Qian S.-B.. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature. 2015; 526:591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Zhou J., Wan J., Shu X.E., Mao Y., Liu X.-M., Yuan X., Zhang X., Hess M.E., Brüning J.C., Qian S.-B.. N6-methyladenosine guides mRNA alternative translation during integrated stress response. Mol. Cell. 2018; 69:636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Romanelli Tavares V.L., Kague E., Musso C.M., Alegria T.G.P., Freitas R.S., Bertola D.R., Twigg S.R.F., Passos-Bueno M.R.. Craniofrontonasal syndrome caused by introduction of a novel uATG in the 5′ UTR of EFNB1. Mol. Syndromol. 2019; 10:40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Wiestner A., Schlemper R.J., van der Maas A.P., Skoda R.C.. An activating splice donor mutation in the thrombopoietin gene causes hereditary thrombocythaemia. Nat. Genet. 1998; 18:49–52. [DOI] [PubMed] [Google Scholar]
- 139. Zühlke C., Rless O., Bockel B., Lange H., Thies U.. Mitotic stability and meiotic variability of the (CAG)n repeat in the Huntington disease gene. Hum. Mol. Genet. 1993; 2:2063–2067. [DOI] [PubMed] [Google Scholar]
- 140. Nolin S.L., Brown W.T., Glicksman A., Houck J.G.E., Gargano A.D., Sullivan A., Biancalana V., Bröndum-Nielsen K., Hjalgrim H., Holinski-Feder E. et al.. Expansion of the fragile X CGG repeat in females with premutation or intermediate alleles. Am. J. Hum. Genet. 2003; 72:454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Schulz J., Mah N., Neuenschwander M., Kischka T., Ratei R., Schlag P.M., Castanos-Velez E., Fichtner I., Tunn P.U., Denkert C. et al.. Loss-of-function uORF mutations in human malignancies. Sci. Rep. 2018; 8:2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Calvo S.E., Pagliarini D.J., Mootha V.K.. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:7507–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Barbosa C., Peixeiro I., Romão L.. Gene expression regulation by upstream open reading frames and human disease. PLos Genet. 2013; 9:e1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Iqbal N., Iqbal N.. Human epidermal growth factor receptor 2 (HER2) in cancers: overexpression and therapeutic implications. Biochem. Mol. Biol. Int. 2014; 2014:852748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Mehta A., Trotta C.R., Peltz S.W.. Derepression of the Her-2 uORF is mediated by a novel post-transcriptional control mechanism in cancer cells. Genes Dev. 2006; 20:939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Child S.J., Miller M.K., Geballe A.P.. Translational control by an upstream open reading frame in the HER-2/neu transcript. J. Biol. Chem. 1999; 274:24335–24341. [DOI] [PubMed] [Google Scholar]
- 147. Zhao R., Choi B.Y., Lee M.-H., Bode A.M., Dong Z.. Implications of genetic and epigenetic alterations of CDKN2A (p16(INK4a)) in cancer. EBioMedicine. 2016; 8:30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Liu L., Dilworth D., Gao L., Monzon J., Summers A., Lassam N., Hogg D.. Mutation of the CDKN2A 5′ UTR creates an aberrant initiation codon and predisposes to melanoma. Nat. Genet. 1999; 21:128–132. [DOI] [PubMed] [Google Scholar]
- 149. Sendoel A., Dunn J.G., Rodriguez E.H., Naik S., Gomez N.C., Hurwitz B., Levorse J., Dill B.D., Schramek D., Molina H. et al.. Translation from unconventional 5′ start sites drives tumour initiation. Nature. 2017; 541:494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Vanderpool C.K., Balasubramanian D., Lloyd C.R.. Dual-function RNA regulators in bacteria. Biochimie. 2011; 93:1943–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Lauressergues D., Couzigou J.-M., Clemente H.S., Martinez Y., Dunand C., Bécard G., Combier J.-P.. Primary transcripts of microRNAs encode regulatory peptides. Nature. 2015; 520:90–93. [DOI] [PubMed] [Google Scholar]
- 152. Lloyd C.R., Park S., Fei J., Vanderpool C.K.. The small protein SgrT controls transport activity of the glucose-specific phosphotransferase system. J. Bacteriol. 2017; 199:e00869-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Gimpel M., Brantl S.. Dual-function small regulatory RNAs in bacteria. Mol. Microbiol. 2017; 103:387–397. [DOI] [PubMed] [Google Scholar]
- 154. Raina M., King A., Bianco C., Vanderpool C.K.. Dual-function RNAs. Microbiol. Spectr. 2018; 6:doi:10.1128/microbiolspec.RWR-0032-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Kitano S., Kurasawa H., Aizawa Y.. Transposable elements shape the human proteome landscape via formation of cis-acting upstream open reading frames. Genes Cells. 2018; 23:274–284. [DOI] [PubMed] [Google Scholar]
- 156. Zhang H., Si X., Ji X., Fan R., Liu J., Chen K., Wang D., Gao C.. Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol. 2018; 36:894–898. [DOI] [PubMed] [Google Scholar]
- 157. Liang X.H., Shen W., Sun H., Migawa M.T., Vickers T.A., Crooke S.T.. Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nat. Biotechnol. 2016; 34:875–880. [DOI] [PubMed] [Google Scholar]
- 158. Xu G., Yuan M., Ai C., Liu L., Zhuang E., Karapetyan S., Wang S., Dong X.. uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature. 2017; 545:491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Rabouw H.H., Langereis M.A., Anand A.A., Visser L.J., de Groot R.J., Walter P., van Kuppeveld F.J.M.. Small molecule ISRIB suppresses the integrated stress response within a defined window of activation. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:2097–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]