Abstract
Synthetic modified RNA (modRNA) is a novel vector for gene transfer to the heart and other organs. modRNA can mediate strong, transient protein expression with minimal induction of the innate immune response and risk for genome integration. modRNA is already being used in several human clinical trials, and its use in basic and translational science is growing. Due to the complexity of preparing modRNA and the high cost of its reagents, there is a need for an improved, cost-efficient protocol to make modRNA. Here we show that changing the ratio between anti-reverse cap analog (ARCA) and N1-methyl-pseudouridine (N1mΨ), favoring ARCA over N1mΨ, significantly increases the yield per reaction, improves modRNA translation, and reduces its immunogenicity in vitro. This protocol will make modRNA preparation more accessible and financially affordable for basic and translational research.
Keywords: in vitro mRNA synthesis, modified mRNA, gene therapy
Introduction
Gene therapy treats disease by using genetic material to change or correct gene expression to cure disease. The concept of gene therapy emerged in the early 1970s as a consequence of both increased understanding of the role of genes in human disease and the development of genetic engineering techniques.1 First attempts to treat human patients with exogenous genes were conducted in the 1980s and 1990s; however, safety issues hampered their progress.2, 3 Extensive research in recent years has yielded better and safer gene delivery strategies that reduce vector immunogenicity; consequently, the gene therapy field is flourishing again, with exciting successes in treating rare genetic diseases, including a first US Food and Drug Administration (FDA)-approved gene therapy for inherited retinal dystrophy,4 and thousands of gene therapy clinical trials worldwide.5 In pursuit of improved treatments for cardiovascular disease, including complications of myocardial infarction (MI) and heart failure (HF), extensive gene therapy preclinical research resulted in numerous clinical trials aiming to promote angiogenesis6 and improve calcium homeostasis.7 Although proving safety, all cardiac gene therapy trials failed to demonstrate significant therapeutic efficacy.7 It has been suggested that inefficient gene transfer that resulted in poor expression of the target gene is a potential reason for these neutral results.7
Gene transfer can be achieved by using viral vectors including adenovirus, retrovirus, adeno-associated virus (AAV), lentivirus, vaccinia virus, poxvirus, and herpes simplex virus or non-viral vectors including naked plasmid DNA (pDNA) or lipofection.5 Modified RNA (modRNA) is a novel vector for gene transfer to both dividing and non-dividing mammalian cells that mediate fast, robust, transient expression of proteins in the targeted cells or tissue.8 Pre-clinical attempts to use mRNA as a gene transfer vector showed successful expression of the encoded proteins9 and improved symptoms in a rodent model of diabetes insipidus.10 Yet the high immunogenicity of exogenous mRNA11, 12, 13, 14, 15 and high susceptibility to degradation by RNase have hampered progress in developing mRNA as a vector for treating humans.16, 17 Since the first description of a biologically active and translatable in vitro transcription (IVT) product by Krieg and Melton18 in 1984, however, immense technological advances have led to synthetic mRNA’s emergence as a promising vector for gene delivery.19 Two important milestones in developing IVT technology turned synthetic mRNA from a scientific tool to a potential platform for gene therapy. First, pioneering work by Karikó et al.20 demonstrated that incorporating chemically modified nucleotides in the synthetic mRNA results in a more stable and less immunogenic mRNA that mediates rapid, high expression of the encoded protein.21, 22 Second, the discovery of a stable cap analog, anti-reverse cap analog (ARCA), increases synthetic mRNA’s stability and translatability.23
We and others have recently shown that modRNA can be used to express reporter proteins in rodent and porcine myocardium,8, 24, 25 and that functional genes can be used to promote angiogenesis,26 cardiomyocyte proliferation,27 and cardiac cell survival.28 Previously, we described a detailed protocol for synthesizing modRNA capped with ARCA 3′-O-Me-m7G(5′)ppp(5′)G and substituting 100% cytidine (C) with 5-methylcytidine (m5C) and 100% uridine (U) with pseudouridine (Ψ),29 or substituting 100% uridine with N1-methyl-pseudouridine (N1mΨ), which has been shown to be superior over the combination of Ψ and m5C.8, 30 Here we describe an optimized protocol that indicates that a change in ARCA and N1mΨ ratios leads to cost-effective modRNA with higher protein expression and lower immunogenicity in vitro.
Results
modRNA Yield and Quality
Currently, there are two strategies for mRNA capping in an IVT reaction: (1) post-transcriptional capping and (2) chemical capping. In the post-transcriptional approach, the vaccinia virus capping enzyme is used to cap the enzyme after the mRNA synthesis is completed.31, 32 Although effective, this procedure is time-consuming and expensive. In chemical capping, a cap analog is added directly to the IVT reaction, after which the cap structure is incorporated at the 5′ end of the mRNA. To achieve a high percentage of capped mRNA using this method, the ratio between the concentrations of the cap analog and the guanosine triphosphate (GTP) in the reaction are kept high. For IVT reactions, the ratio of ARCA to GTP should always be 1:3.7. In order to reduce the synthetic mRNA’s immunogenicity and increase its translatability, we replace 100% U with N1mΨ. Previously, for the IVT reaction, we used a composition of 5 nM ARCA, 1.35 mM GTP, 7.5 mM N1mΨTP, and 7.5 mM MTP (composition 1 in Table 1, hereafter termed ARCA 5 protocol). To achieve our aim of increasing the translation capacity and lowering the cost of modRNA production, we reduce the amount of template DNA by 85% (Figure S1). We then explored different nucleotide compositions and desalting methods for making and cleaning the modRNA (Table 1). Out of the various nucleotide stoichiometries tested, composition 5, comprising higher concentrations of ARCA, GTP, ATP, and cytidine triphosphate (CTP) and less N1mΨTP, another costly element in modRNA production, resulted in the greatest modRNA yield compared with the previously used protocol (ARCA 5 protocol), keeping the amount of T7 RNA Polymerase consistent (Table 1). To desalt the modRNA in the first cleaning process, we compared the MEGAclear RNA purification kit (used previously) with the more cost-efficient Amicon filter. We found that the nuclear GFP (nGFP) modRNA yield using the Amicon filter was equivalent to the amount generated by the MEGAclear RNA purification kit. Thus, we established that the most cost-effective protocol for IVT reaction uses nucleotide concentrations of 10 mM ARCA, 2.7 mM GTP, 8.1 mM ATP, 8.1 mM CTP, and 2.7 mM N1mΨTP followed by desalting and nucleotide clearing using the Amicon filter (this protocol will now be referred to as the ARCA 10 protocol).
Table 1.
Nucleotide Composition and Cleaning Methods
| Nucleotide Composition 1 (ARCA 5) | Nucleotide Composition 2 | Nucleotide Composition 3 | Nucleotide Composition 4 | Nucleotide Composition 5 (ARCA 10) | Nucleotide Composition 6 | |
|---|---|---|---|---|---|---|
| ARCA | 5 mM | 10 mM | 10 mM | 10 mM | 10 mM | 7.5 mM |
| GTP | 1.35 mM | 2.7 mM | 2.7 mM | 2.7 mM | 2.7 mM | 2.25 mM |
| ATP | 7.5 mM | 8.1 mM | 15 mM | 15 mM | 8.1 mM | 10 mM |
| CTP | 7.5 mM | 2.7 mM | 15 mM | 15 mM | 8.1 mM | 10 mM |
| N1mΨTP | 7.5 mM | 2.7 mM | 15 mM | 2.7 mM | 2.7 mM | 2.25 mM |
| Total | 28.85 mM | 26.2 mM | 57.7 mM | 45.4 mM | 31.6 mM | 32 mM |
| Desalting with MEGAclear | ||||||
| Total mg nGFP modRNA per 1-mL reaction | 1.33 | 3.12 | 1.19 | 2.13 | 2.94 | 1.93 |
| % of ARCA 5 protocol | 100 | 235 | 89 | 160 | 221 | 145 |
| Desalting with Amicon Filter | ||||||
| Total mg nGFP modRNA per 1-mL reaction | 2.12 | 1.04 | 1.92 | 2.94 | 1.74 | |
| % of ARCA 5 protocol | 159 | 78 | 144 | 221 | 131 | |
Summary of nucleotide composition and desalting (first cleansing) methods used in this study and their effects on the final modRNA yield. For modRNA integrity, see Figure 1.
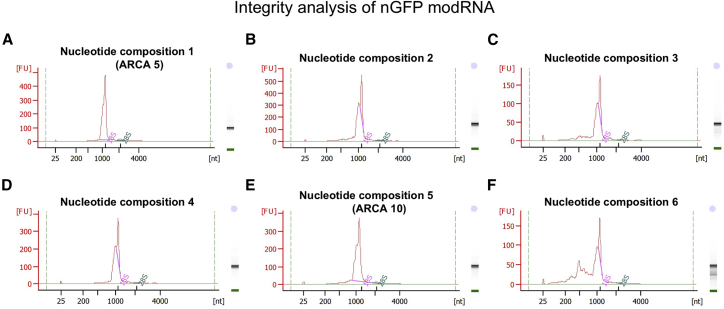
We also evaluated modRNA quality using a bioanalyzer. We found no differences in the integrity of the modRNA generated using the ARCA 10 protocol compared with that produced via the ARCA 5 protocol (Figures 1A and 1B). Interestingly, when using different nucleotide compositions in the reaction, we observed high levels of uncompleted IVT products (Figure 1F).
Figure 1.
Effect of Nucleotide Composition in IVT Reaction on modRNA Integrity
(A–F) modRNA integrity was evaluated using a bioanalyzer (Tap Station 2200) of compositions 1 (A), 2 (B), 3 (C), 4 (D), 5 (E), and 6 (F).
Translational Capacity of modRNA In Vitro
Next, we analyzed the translational capacity of modRNA generated with the ARCA 10 protocol by transfecting several human cell lines and primary cardiac cells isolated from neonatal rat hearts and measuring the expression levels of two different reporter genes: nGFP and firefly luciferase (Luc) (Figure 2). We observed significantly increased nGFP expression in HeLa cells (data not shown), primary rat cardiac cells, and neonatal rat cardiomyocytes (nrCM) transfected with nGFP modRNA generated with the ARCA 10 protocol compared with the ARCA 5 protocol (Figures 2C and 2D). Similar results were observed in primary rat cardiac cells, nrCM, human umbilical vein endothelial cells (HUVECs), HeLa cells, and HEK293 cells when transfected with Luc modRNA (Figures 2E–2H). We did not observe any difference in cell death levels post transfection in vitro as measured by DAPI or propidium iodide (PI)-positive cells (data not shown).
Figure 2.
Effect of Nucleotide Composition in IVT Reaction on Gene Expression in Human Cell Lines and Rat Cardiac Cells
(A) GFP expression in rat cardiac cells 18 h post transfection with or without nGFP modRNA generated with the ARCA 5 protocol and ARCA 10 protocol. (B) Representative image of Luc activity level in neonatal rat cardiomyocytes 24 h post transfection with or without Luc modRNA generated using the ARCA 5 protocol and ARCA 10 protocol. (C and D) Quantification of GFP expression in neonatal rat cardiac cells (C) or cardiomyocytes (D) 18 h post transfection with nGFP modRNA generated using the ARCA 5 protocol and ARCA 10 protocol. (E–H) Quantification of Luc activity level in HUVECs (E), rat neonatal cardiomyocytes (F), HeLa (G), or Hek293 (H) cells 18 h post transfection with or without Luc modRNA generated using the ARCA 5 protocol and ARCA 10 protocol. One-way ANOVA, Tukey’s multiple comparison test; ***p < 0.001, **p < 0.01, *p < 0.05. n = 5 (A and B); n = 4 (C–F).
Immunogenicity of modRNA
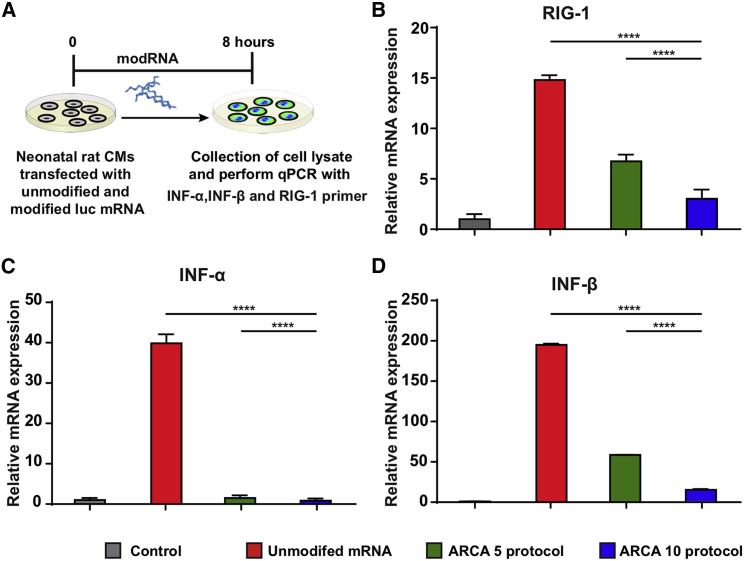
Evaluating immunogenicity is critical to defining the safety profile of mRNA therapeutics and their use in humans. To evaluate the innate immune response in vitro, we measured the expression levels of three innate immune-related genes, retinoic acid-inducible gene I (RIG1), interferon alpha (IFNα), and interferon beta (IFNβ), in nrCM 8 h post transfection with Luc modRNA (made by using either the ARCA 5 protocol or the ARCA 10 protocol) or unmodified mRNA (Figure 3). Importantly, Luc modRNA made via both the ARCA 5 and ARCA 10 protocols has significantly lower levels of RIG1, IFNα, and IFNβ when compared with unmodified mRNA (Figures 3B–3D). However, Luc made using the ARCA 5 protocol was significantly more immunogenic than Luc modRNA made using the ARCA 10 protocol (Figures 3B–3D).
Figure 3.
Immunogenicity of modRNA Compared with Unmodified mRNA
(A) Luc mRNA or modRNA transfected into neonatal rat cardiomyocytes 8 h after transfection cells were collected. qPCR was performed to evaluate the gene expression of innate immune response markers. (B–D) Expression of innate immune response markers: RIG1 (B), IFNα (C), or IFNβ (D) 8 h post transfecting unmodified mRNA, Luc modRNA (ARCA 5 protocol), or Luc modRNA (ARCA 10 protocol). One-way ANOVA, Tukey’s multiple comparison test; ****p < 0.0001. n = 5.
Discussion
Synthetic modRNA is now emerging as a promising vector for gene delivery.19 However, synthesizing modRNA in the high quantities required by pre-clinical and clinical trials is expensive and time-consuming. In light of recent successes in promoting angiogenesis,26 CM proliferation,27 and cardiac cell survival28 in rodents, there in an urgent need to reduce the cost and improve the quality of synthesizing modRNA.
At present, our capping strategy is chemical capping; in this capping method, the IVT reaction includes a cap analog, resulting in a cap structure at the mRNA’s 5′ end. This capping method requires a high ratio of cap analog and GTP concentrations to produce a high percentage of capped mRNA.
This study aimed to optimize the protocol for synthesizing mRNA using ARCA and N1mΨ. Previously published protocols8, 26, 28, 29 maintained the correct ratio between ARCA and GTP by keeping the GTP concentration low (1.35 mM) and the nucleotides high (7.5 mM). In this composition, the GTP becomes the limiting factor in the reaction, leading the reaction to terminate without incorporating the other nucleotides present in excess. The unincorporated nucleotides are later washed away during the cleaning process. Although the ratio between ARCA and GTP must be kept high, we realized that the composition of the other nucleotides in the reaction can be modified, leading us to explore the effect of different nucleotide compositions on the final yield and quality of the modRNA. Interestingly, we found that when doubling the concentration of all nucleotides to increase the final concentration to 57.7 mM, the final yield is lower than the original concentration of 28.85 mM. The optimal final nucleotide concentration in our study was 31.6 mM (ARCA 10 mM, GTP 2.7 mM, CTP 8.1 mM, ATP 8.1 mM, and N1mΨ 2.7 mM). Using this concentration, we were able to increase the final modRNA yield by 290%. Combining this protocol with improved cleaning methods results in modRNA with increases translatability in both human cell lines and primary cardiac cells (Figure 2), as well as reduced immunogenicity (Figure 3). A possible explanation for this result is that optimizing nucleotide composition in the IVT reaction raises the percentage of successfully capped modRNA.
Here we developed an improved protocol for large-scale production of modRNA. By increasing the final nucleotide concentrations while reducing the N1mΨ molarity and using the same amount of T7 RNA polymerase, we established a new method that is more cost-effective than the previously published approach.29 Our novel protocol generated improved modRNA with higher translatability and lower immunogenicity in vitro, and will allow basic and translational research labs to overcome the high cost of modRNA preparation and attain more effective modRNA for their research.
Materials and Methods
Synthesizing modRNA
In brief, clean PCR products generated with plasmid templates purchased from GenScript were used as the template for mRNA. modRNAs were generated by transcription in vitro with a customized ribonucleoside blend of ARCA; 30-O-Me-m7G(50) ppp(50)G (catalog no. [cat. #] N-1081; Trilink Biotechnologies); GTP (cat. #am1334-5; Life Technologies); ATP (cat. #am1334-5; Life Technologies); cytidine triphosphate (cat. #am1334-5; Life Technologies), and N1-methylpseudouridine-50-triphosphate (cat. #N-1081; Trilink Biotechnologies). The mRNA was purified with the MEGAclear kit (cat. #AM1908; Life Technologies) according to the manufacturer’s instructions or using Amicon Ultra-4 Centrifugal Filter Unit 4 mL,10 kDa (cat. #UFC801024; MilliporeSigma) and treated with Antarctic Phosphatase (cat. #M0289L; NEB). It was then re-purified with the MEGAclear kit. The mRNA was quantified using a NanoDrop spectrometer (Thermo Scientific), precipitated with ethanol and ammonium acetate, and re-suspended in 10 mM Tris-HCl and 1 mM EDTA. The open reading frame for the modRNA used is listed in Table S1. This protocol has been described in detail elsewhere.29
Desalting and Nucleotide Clearing
Amicon Ultra-4 Centrifugal Filter Unit 4 mL, 10 kDa (cat. #UFC801024; MilliporeSigma) was washed with 4 mL nuclease-free water two times. A total of 400 μL IVT reaction was loaded to the filter and diluted with 3.5 mL nuclease-free water. The filter was centrifuged at 3,200 × g for 20 min until the volume reduces to 100 μL. A total of 3.9 mL water was added to the filter and centrifuged at 3,200 × g for 20 min two times. The purified modRNA was incubated at 70°C for 10 min (to inactivate T7).
Neonatal Rat Cardiac Cells Isolation
Neonatal rat ventricular CMs were isolated from 3- to 4-day-old Sprague-Dawley rats (Jackson ImmunoResearch Laboratories) by multiple rounds of digestion with 0.1% collagenase II (Invitrogen) in PBS. After each digestion, the supernatant was collected in horse serum (Invitrogen). The total cell suspension was centrifuged at 300 × g for 5 min. The supernatants were discarded and cells were resuspended in DMEM (GIBCO) supplemented with 0.1 mM ascorbic acid (Sigma), 0.5% insulin-transferrin-selenium (100×), penicillin (100 U/mL), and streptomycin (100 mg/mL). Cells were plated in plastic culture dishes for 90 min until most of the non-myocytes were attached to the dish while the myocytes remained in suspension. Myocytes were then used to seed 24-well plates at a density of 1 × 105 cells/well. nrCMs were incubated for 48 h in DMEM supplemented with 5% horse serum before transfection with modRNAs. Twenty-four hours post isolation, the non-myocytes were used to seed 24-well plates at a density of 1 × 105 cells/well. The non-myocytes were transfected with modRNA 24 h post plating.
In Vitro Transfection with modRNA
Using a 24-well plate, we complexed 2.5 μg/well mRNA with Lipofectamine RNAiMAX Transfection Reagent (cat. #13778030; Life Technologies) and used the resulting complex to transfect either neonatal rat cardiac cells or human cell lines according to the manufacturer’s instructions.
Assessing Number of nGFP-Expressing Cells
Eighteen hours post transfection, cells were imaged using a Zeiss Slide Scanner Axio Scan. Quantification of nGFP-positive cells was performed using ImageJ software.
Detection of Luciferase Activity
Bioluminescence of the transfected cells was measured 18 h post transfection. Each unit of Luc signal represents p/s/cm2/sr × 106. Luciferin (cat. #L9504; Sigma) (1.5 mg/mL culture media) was added to cell culture 10 s before imaging. Cells were imaged using an IVIS100 charge-coupled device imaging system every 20 s until the Luc signal reached a plateau. Imaging data were analyzed and quantified with Living Image software.
qRT-PCR
Total RNA was reverse transcribed with Superscript III reverse transcriptase (cat. #LS18080044; Invitrogen), according to the manufacturer’s instructions. qRT-PCR analyses were performed on a Mastercycler Realplex 4 Sequence Detector (Eppendorf) with the HotStart-IT SYBR Green qPCR master mix (×2) (cat. #75762; Affymetrix). Data were normalized relative to GAPDH. Fold-changes in gene expression were determined by the delta-delta-Ct (ddCT) method. The PCR primer sequences used are listed in Table S2.
Statistical Analyses
Statistical analyses were performed with GraphPad Prism software. Values are reported as means ± SD. Two-tailed Student’s t tests (*p < 0.05 considered significant) or one-way ANOVA with Bonferroni correction (*p < 0.05 considered significant) were used for comparisons between groups.
Author Contributions
Y.H. designed most of the experiments, performed experiments, analyzed most of the data, and wrote the manuscript. N.S. performed most of the experiments, analyzed data, and revised the manuscript. E.Y. performed experiments. M.T.K.S. prepared modRNAs and performed experiments. K.K. revised the manuscript. E.E. performed CM isolation method. L.Z. designed experiments, analyzed data, and revised the manuscript.
Acknowledgments
This work was funded by a cardiology start-up grant awarded to the Zangi laboratory and also by NIH grant R01 HL142768-01.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2019.07.006.
Supplemental Information
References
- 1.Friedmann T., Roblin R. Gene therapy for human genetic disease? Science. 1972;175:949–955. doi: 10.1126/science.175.4025.949. [DOI] [PubMed] [Google Scholar]
- 2.Walters L. DIANE Publishing; 1984. Human Gene Therapy. [Google Scholar]
- 3.Somia N., Verma I.M. Gene therapy: trials and tribulations. Nat. Rev. Genet. 2000;1:91–99. doi: 10.1038/35038533. [DOI] [PubMed] [Google Scholar]
- 4.Smalley E. First AAV gene therapy poised for landmark approval. Nat. Biotechnol. 2017;35:998–999. doi: 10.1038/nbt1117-998. [DOI] [PubMed] [Google Scholar]
- 5.Ginn S.L., Amaya A.K., Alexander I.E., Edelstein M., Abedi M.R. Gene therapy clinical trials worldwide to 2017: An update. J. Gene Med. 2018;20:e3015. doi: 10.1002/jgm.3015. [DOI] [PubMed] [Google Scholar]
- 6.Gupta R., Tongers J., Losordo D.W. Human studies of angiogenic gene therapy. Circ. Res. 2009;105:724–736. doi: 10.1161/CIRCRESAHA.109.200386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishikawa K., Weber T., Hajjar R.J. Human Cardiac Gene Therapy. Circ. Res. 2018;123:601–613. doi: 10.1161/CIRCRESAHA.118.311587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sultana N., Magadum A., Hadas Y., Kondrat J., Singh N., Youssef E., Calderon D., Chepurko E., Dubois N., Hajjar R.J., Zangi L. Optimizing Cardiac Delivery of Modified mRNA. Mol. Ther. 2017;25:1306–1315. doi: 10.1016/j.ymthe.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 10.Jirikowski G.F., Sanna P.P., Maciejewski-Lenoir D., Bloom F.E. Reversal of diabetes insipidus in Brattleboro rats: intrahypothalamic injection of vasopressin mRNA. Science. 1992;255:996–998. doi: 10.1126/science.1546298. [DOI] [PubMed] [Google Scholar]
- 11.Diebold S.S., Massacrier C., Akira S., Paturel C., Morel Y., Reis e Sousa C. Nucleic acid agonists for Toll-like receptor 7 are defined by the presence of uridine ribonucleotides. Eur. J. Immunol. 2006;36:3256–3267. doi: 10.1002/eji.200636617. [DOI] [PubMed] [Google Scholar]
- 12.Heil F., Hemmi H., Hochrein H., Ampenberger F., Kirschning C., Akira S., Lipford G., Wagner H., Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 13.Diebold S.S., Kaisho T., Hemmi H., Akira S., Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 14.Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 15.Pichlmair A., Schulz O., Tan C.P., Rehwinkel J., Kato H., Takeuchi O., Akira S., Way M., Schiavo G., Reis e Sousa C. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dyer K.D., Rosenberg H.F. The RNase a superfamily: generation of diversity and innate host defense. Mol. Divers. 2006;10:585–597. doi: 10.1007/s11030-006-9028-2. [DOI] [PubMed] [Google Scholar]
- 17.Rigby R.E., Rehwinkel J. RNA degradation in antiviral immunity and autoimmunity. Trends Immunol. 2015;36:179–188. doi: 10.1016/j.it.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krieg P.A., Melton D.A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984;12:7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hadas Y., Katz M.G., Bridges C.R., Zangi L. Modified mRNA as a therapeutic tool to induce cardiac regeneration in ischemic heart disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2017;9:e1367. doi: 10.1002/wsbm.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Karikó K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jemielity J., Fowler T., Zuberek J., Stepinski J., Lewdorowicz M., Niedzwiecka A., Stolarski R., Darzynkiewicz E., Rhoads R.E. Novel “anti-reverse” cap analogs with superior translational properties. RNA. 2003;9:1108–1122. doi: 10.1261/rna.5430403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh R.D., Hillestad M.L., Livia C., Li M., Alekseev A.E., Witt T.A., Stalboerger P.G., Yamada S., Terzic A., Behfar A. M3RNA drives targeted gene delivery in acute myocardial infarction. Tissue Eng. Part A. 2019;25:145–158. doi: 10.1089/ten.tea.2017.0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turnbull I.C., Eltoukhy A.A., Anderson D.G., Costa K.D. Lipidoid mRNA Nanoparticles for Myocardial Delivery in Rodents. Methods Mol. Biol. 2017;1521:153–166. doi: 10.1007/978-1-4939-6588-5_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zangi L., Lui K.O., von Gise A., Ma Q., Ebina W., Ptaszek L.M., Später D., Xu H., Tabebordbar M., Gorbatov R. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013;31:898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magadum A., Singh N., Kurian A.A., Sharkar M.T.K., Chepurko E., Zangi L. Ablation of a Single N-Glycosylation Site in Human FSTL 1 Induces Cardiomyocyte Proliferation and Cardiac Regeneration. Mol. Ther. Nucleic Acids. 2018;13:133–143. doi: 10.1016/j.omtn.2018.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang C.L., Leblond A.L., Turner E.C., Kumar A.H., Martin K., Whelan D., O’Sullivan D.M., Caplice N.M. Synthetic chemically modified mrna-based delivery of cytoprotective factor promotes early cardiomyocyte survival post-acute myocardial infarction. Mol. Pharm. 2015;12:991–996. doi: 10.1021/mp5006239. [DOI] [PubMed] [Google Scholar]
- 29.Kondrat J., Sultana N., Zangi L. Synthesis of Modified mRNA for Myocardial Delivery. Methods Mol. Biol. 2017;1521:127–138. doi: 10.1007/978-1-4939-6588-5_8. [DOI] [PubMed] [Google Scholar]
- 30.Svitkin Y.V., Cheng Y.M., Chakraborty T., Presnyak V., John M., Sonenberg N. N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 2017;45:6023–6036. doi: 10.1093/nar/gkx135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuchs A.L., Neu A., Sprangers R. A general method for rapid and cost-efficient large-scale production of 5′ capped RNA. RNA. 2016;22:1454–1466. doi: 10.1261/rna.056614.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paterson B.M., Rosenberg M. Efficient translation of prokaryotic mRNAs in a eukaryotic cell-free system requires addition of a cap structure. Nature. 1979;279:692–696. doi: 10.1038/279692a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.