Abstract
Budding yeast treated with hydroxyurea (HU) activate the S phase checkpoint kinase Rad53, which prevents DNA replication forks from undergoing aberrant structural transitions and nuclease processing. Rad53 is also required to prevent premature extension of the mitotic spindle that assembles during a HU-extended S phase. Here we present evidence that checkpoint restraint of spindle extension is directly coupled to Rad53 control of replication fork stability. In budding yeast, centromeres are flanked by replication origins that fire in early S phase. Mutations affecting the Zn2+-finger of Dbf4, an origin activator, preferentially reduce centromere-proximal origin firing in HU, corresponding with suppression of rad53 spindle extension. Inactivating Exo1 nuclease or displacing centromeres from origins provides a similar suppression. Conversely, short-circuiting Rad53 targeting of Dbf4, Sld3, and Dun1, substrates contributing to fork stability, induces spindle extension. These results reveal spindle extension in HU-treated rad53 mutants is a consequence of replication fork catastrophes at centromeres. When such catastrophes occur, centromeres become susceptible to nucleases, disrupting kinetochore function and spindle force balancing mechanisms. At the same time, our data indicate centromere duplication is not required to stabilize S phase spindle structure, leading us to propose a model for how monopolar kinetochore-spindle attachments may contribute to spindle force balance in HU.
INTRODUCTION
The S phase checkpoint is a conserved signal transduction pathway enabling eukaryotic cells to tolerate DNA replication stress. In budding yeast, checkpoint signaling is initiated when impediments to DNA synthesis cause single-stranded DNA (ssDNA) to accumulate at replication forks, leading to activation of the central checkpoint kinase Mec1 (reviewed in Pardo et al., 2017). Mec1 then acts through the mediator Mrc1 to phosphorylate and activate the effector kinase Rad53 (Sanchez et al., 1996; Osborn and Elledge, 2003). In broad terms, Rad53 controls two seemingly distinct forms of regulation within the S phase checkpoint. First, Rad53 mediates responses that, collectively, allow DNA synthesis to continue despite perturbations to replication forks. These responses include delaying the temporal program of replication origin (ORI) firing (Santocanale and Diffley, 1998; Shirahige et al., 1998), activating the protein kinase Dun1 to upregulate ribonucleotide reductase (RNR; Zhou and Elledge, 1993; Zhao and Rothstein, 2002), and directly regulating the stability and exonucleolytic susceptibility of DNA replication forks (Lopes et al., 2001; Sogo et al., 2002; Katou et al., 2003; Cotta-Ramusino et al., 2005; Bermejo et al., 2011; Rossi et al., 2015; Colosio et al., 2016). Failure to execute these forms of DNA replication control has severe consequences. rad53 mutants treated with the RNR inhibitor hydroxyurea (HU) activate ORIs throughout the genome, greatly increasing the number of forks experiencing replication challenge (Feng et al., 2006). Many of these forks undergo reversal or collapse, corresponding with DNA damage and accumulation of ssDNA (Lopes et al., 2001; Sogo et al., 2002; Feng et al., 2006, 2011).
A second role for Rad53 is to maintain the capacity for accurate chromosome segregation once impediments to DNA replication have been resolved. Budding yeast undergo a closed mitosis, and commitment to spindle pole body (SPB) duplication and intranuclear spindle assembly occurs as cells initiate S phase (Hartwell, 1976). In rich media, S phase is typically complete before SPBs separate and spindle assembly begins. However, when DNA replication is extended by HU, this relative timing is altered such that S phase checkpoint-proficient cells arrest mitotic progression having assembled an ∼1 to 2-μm bipolar spindle (Byers and Goetsch, 1974; referred to here as the S phase spindle). In contrast, S phase checkpoint mutants undergo premature spindle extension in HU (Allen et al., 1994; Weinert et al., 1994; Navas et al., 1995; Alcasabas et al., 2001), but otherwise remain blocked in the cell cycle, eventually completing bulk genome duplication (Desany et al., 1998; Feng et al., 2009). rad53 mutants recovering from HU ultimately fail to biorient sister chromatids on the spindle, indicating a profound defect in their ability to resume chromosome segregation (Feng et al., 2009).
While a molecular understanding of Rad53 control of DNA replication has progressed, checkpoint mechanisms enforcing the block to spindle extension have remained less defined. A clarifying realization was that spindle extension in HU-treated rad53 mutants does not reflect premature anaphase entry (Krishnan et al., 2004; Bachant et al., 2005). In yeast, the metaphase to anaphase transition is controlled through proteolysis of the anaphase inhibitor Pds1 (Cohen-Fix et al., 1996). Pds1 degradation triggers sister chromatid disjunction, after which spindles extend to 8–10 μm (Ciosk et al., 1998; Jensen et al., 2001; Severin et al., 2001b; Khmelinskii et al., 2009; Lianga et al., 2018). In contrast, rad53 spindles extend only partially in HU (3–7 μm), displaying cycles of extension, breakage, and collapse (Bachant et al., 2005) and spindle extension occurs even though Pds1 is stabilized (Feng et al., 2009; Palou et al., 2017). Moreover, Pds1 is not required for short spindle arrest in HU (Yamamoto et al., 1996). A careful kinetic analysis showed HU-treated pds1 mutants do eventually exhibit premature spindle extension, but only after two-thirds of the genome has been duplicated (Clarke et al., 2001). In comparison, spindles extend as soon as they are formed in mec1 and rad53 mutants. These observations suggest that delayed chromosome segregation at the S phase checkpoint involves two sequential responses. The first is a rapidly acting, Pds1-independent response that maintains the structural stability and/or appropriate length regulation of the S phase spindle. The second response is Pds1-dependent and becomes operational after substantial genome duplication has been achieved.
Prior work in our labs has focused on the mechanism of the rapidly acting block to spindle extension in HU. Spindle length regulation reflects a balance between microtubule (MT) motors that extend the central spindle and a counteracting, inward-directed, force (Saunders et al., 1997). In budding yeast, considerable evidence suggests this inward force is generated through amphitelic attachment of sister kinetochores (Ks) and the tensile properties of the C-loop, an elastic pericentromeric (CEN) domain (Bouck and Bloom, 2007; Yeh et al., 2008; Stephens et al., 2011; Nannas et al., 2014). Notably, all 16 yeast CENs are flanked by ORIs that fire in HU-treated cells (Raghuraman et al., 2001). In previous work, we therefore proposed that one role for Rad53 was to ensure replication forks successfully traversed CENs in HU, allowing replicating chromosomes to achieve amphitelic attachment (Bachant et al., 2005). Four observations supported this model. First, mutations affecting the essential K proteins Ndc10, Mif2, and Ndc80 and Ask1—components of K subcomplexes linking CEN chromatin to the MT interface—all induced spindle extension in HU (see also Ma et al., 2007; Liu et al., 2008). Second, dicentric chromosomes rescued rad53 spindle extension. Third, Ipl1/Aurora B, which is required to efficiently orient Ks to both spindle poles (Tanaka et al., 2002), displayed HU spindle extension. Fourth, minichromosomes where CENs are juxtaposed to ORIs blocked rad53 spindle extension in HU. Our interpretation was that these minichromosomes allowed a threshold number of CENs to be duplicated prior to nucleotide exhaustion, compensating for loss of Rad53.
A prediction stemming from the above observations is that Rad53 should prevent HU spindle extension through the same checkpoint substrates as DNA replication fork control. Here we provide evidence this is true, as loss of Rad53 regulation of ORI firing and RNR is sufficient to induce spindle extension. HU-treated rad53 mutants show a loss of both CEN DNA and K integrity, providing an explanation for how DNA replication and spindle control are integrated within the checkpoint. A second issue concerns whether CEN duplication is required to offset S phase spindle extension. Remarkably, we find CEN duplication can lag bipolar spindle assembly, raising the question as to why K attachments are required to stabilize spindle length during an extended S phase. We address this by proposing an S phase spindle structure in which monotelic attachments between immobilized S phase CENs and the spindle generate an inward-acting force that offsets spindle extension.
RESULTS
Rad53 regulation of ORI firing is coupled to spindle extension in HU
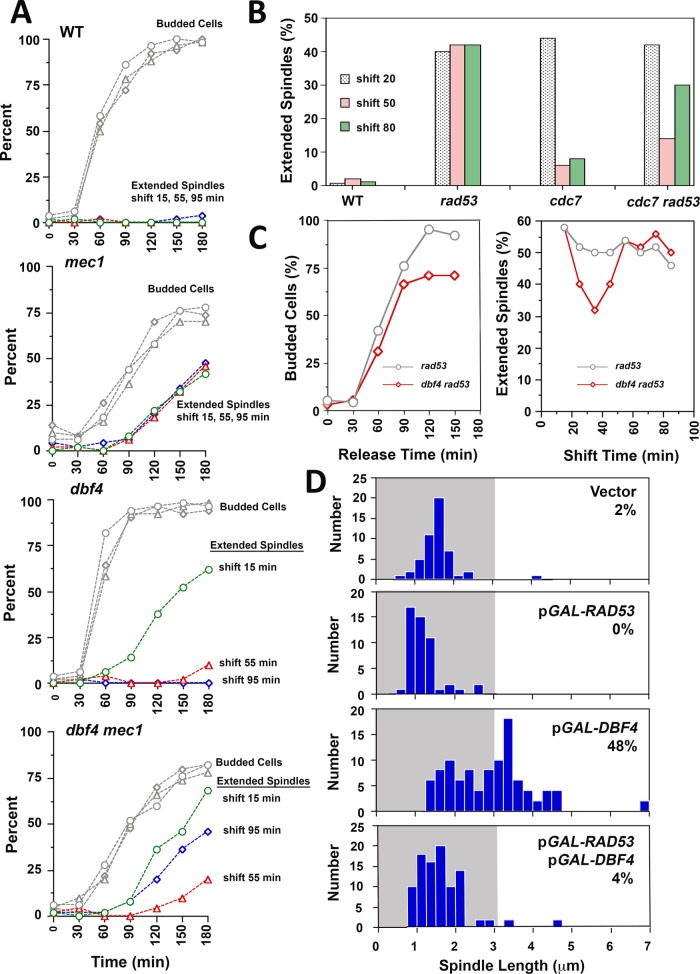
As set out in the Introduction, we initially hypothesized that failure of forks to traverse CENs in HU was the proximal cause of rad53 spindle extension. Interestingly, an inability to duplicate CENs is also thought to be responsible for the reductional anaphase phenotype of replication initiation-defective cdc6, cdc7, and dbf4 mutant strains (Piatti et al., 1995; Warsi et al., 2008). Dbf4 is a regulatory subunit for the Cdc7 kinase (Dbf4-dependent kinase, DDK), an essential and limiting activator of ORI firing (Jackson et al., 1993; Bousset and Diffley, 1998; Mantiero et al., 2011); Cdc6 is a replication initiation protein that localizes to presumptive ORIs as part of the prereplication complex (pre-RC; Cocker et al., 1996). Notably, the role of Rad53 in delaying ORI firing in HU (Rad53-checked ORIs) has been shown to be important in stabilizing replication forks from early firing (unchecked) ORIs, presumably by minimizing competition between forks for limiting factors such as dNTPs (Poli et al., 2012; Zhong et al., 2013; Morafraile et al., 2015). We therefore considered whether the inability to fire ORIs in ddk mutants and unrestrained ORI firing in HU-treated rad53 mutants might lead to spindle extension for the same reason—namely, a failure in CEN duplication. An experimental approach to test this was suggested by the observation that cdc7-1 temperature-sensitive mutants delay firing of some Rad53-checked ORIs when released from a HU block at a nonpermissive temperature (Boussett and Diffley, 1998). On the basis of these considerations, we conducted a series of temperature shift experiments in HU using conditional alleles affecting the DDK in combination with rad53. The rationale was that, by shifting rad53 ddk mutants to a ddk nonpermissive temperature at particular windows in S-phase, it might be possible to reduce ORI firing, including firing of some ORIs that would otherwise be activated due to loss of Rad53. This could, in turn, potentially mitigate fork destabilization, allowing CENs to be duplicated and suppressing spindle extension in ddk rad53 double mutants. Accordingly, dbf4-1 mec1-21, cdc7-1 rad53-21, and dbf4-1 rad53-21 double mutants were released from G1 into media containing 200 mM HU at a permissive temperature of 25°C. Starting at 15 min postrelease, cultures were shifted to a nonpermissive temperature at different times to inactivate the DDK. We observed there was a window 30–55 min postrelease where shifting mec1 ddk and rad53 ddk mutants resulted in a two- to threefold reduction in cells displaying spindle extension (Figure 1, A–C). One explanation for this window is that few ORIs fire in mec1 ddk and rad53 ddk mutants when the DDK is inactivated before 30 min, leading to a reductional anaphase. Inactivating the DDK after 55 min, on the other hand, is too late to prevent ORI firing, leading to spindle extension in a manner similar to mec1 and rad53 controls. Extending these results, we found that overproduction of Dbf4, which advances ORI firing, including ORIs that are normally checked in HU (Mantiero et al., 2011; Tanaka et al., 2011), was sufficient to force HU spindle extension in checkpoint-proficient cells. Cooverproduction of Rad53, which binds to and phosphorylates Dbf4 in HU (Duncker et al., 2002; Chen et al., 2013; Matthews et al., 2014; Almawi et al., 2016), rescued this defect (Figure 1D).
FIGURE 1:
DDK temperature-sensitive alleles reduce spindle extension in S phase checkpoint mutants. (A) WT (Y300), mec1-21 (AY201), dbf4-1 (JBY999), and mec1-21 dbf4-1 (JBY927) strains were released from G1 at 25°C in 200 mM HU media. At 15, 55, or 95 min after G1 release, cultures were shifted to a nonpermissive temperature of 34°C. At times indicated on the x-axis, aliquots were processed for α-tubulin immunofluorescence and DAPI staining. Spindle extension was reduced in mec1-21 dbf4-1 cells shifted at 55 min. (B) WT (Y300), rad53-21 (Y301), cdc7-1 (DES956), and rad53-21 cdc7-1 (DES960) were released into 200 mM HU media as in A and were shifted to 35°C at 20, 50, and 80 min. After a total of 180 min following G1 release, cells were processed for α-tubulin immunofluorescence and DAPI. As with mec1-21 dbf4-1 cells, rad53-21 cdc7-1 cells displayed reduced spindle extension when shifted at 50 min. (C) To more closely bracket the window for suppression of spindle extension, rad53-21 (Y301) and rad53-21 dbf4-1 (JBY1002) strains were released from G1 into 200 mM HU at 25°C and then split into parallel cultures, which were then shifted to 34°C at the indicated times on the x-axis of the right-hand graph (shift time). Aliquots were maintained at 25°C to monitor cell budding (left graph). Spindle extension was evaluated in temperature-shifted samples at 150 min post-G1 release using DAPI and α-tubulin immunofluorescence (right graph). Maximal suppression of spindle extension was observed when rad53-21 dbf4-1 cells were shifted at 40 min, corresponding with bud emergence and S phase entry. (D) SPC42-GFP cells (JBY1129) were transformed with vector (JBY1285), a low copy plasmid expressing RAD53 under control of the inducible GAL promoter (pCEN HIS3 GAL-RAD53, JBY1286), a high copy pGAL-DBF4 plasmid (p2μm URA3 GAL-DBF4; JBY1287), or cotransformed with both plasmids (JBY1288). Transformants were arrested in G1 for 3.5 h in galactose media (YPGAL) to induce RAD53 and/or DBF4. Cells were then released into YPGAL containing 200 mM HU. After 3.5 h, spindle extension was evaluated using Spc42-GFP.
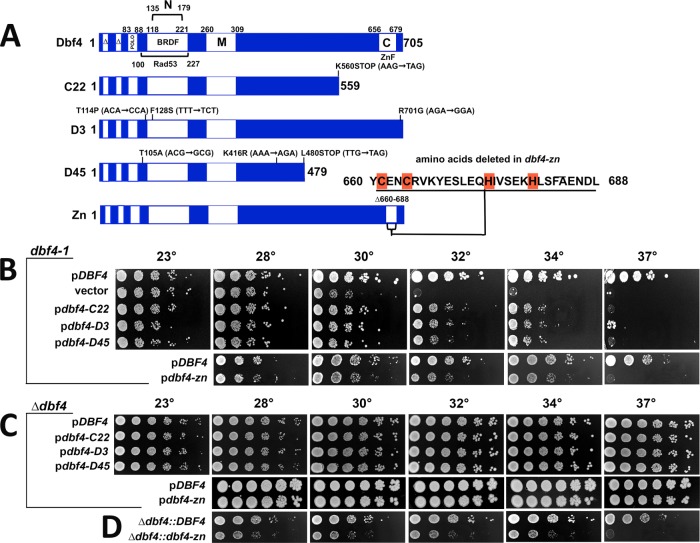
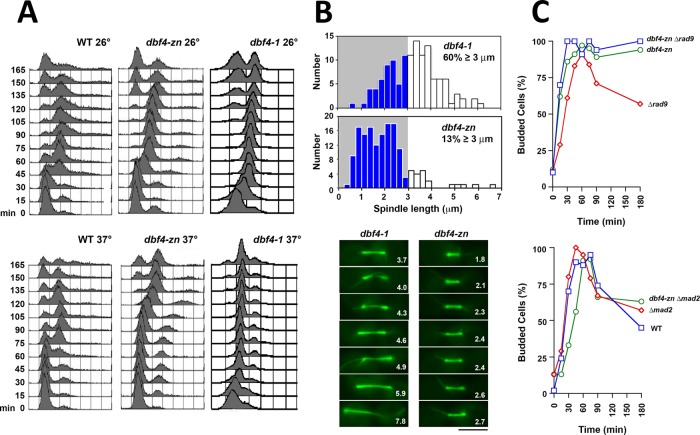
To clarify the epistasis between rad53 and the dbf4 mutations with respect to HU spindle extension, we screened a mutagenized plasmid library for pdbf4 (“p” designates plasmid-based expression) clones that partially suppressed the temperature sensitivity of dbf4-1 and did not exhibit a reductional anaphase. Our presumption was such clones would encode hypomorphic pdbf4 alleles that would reduce ORI firing overall, while still supporting sufficient firing of CEN flanking ORIs to replicate CENs. Three pdbf4 clones that satisfied these criteria were obtained (Khalil et al., 2007). All three harbored nonsense or missense mutations affecting the C-terminus of Dbf4 (collectively referred to as pdbf4-C; Figure 2A). The Dbf4 C-terminus contains a C2H2 Zn2+ finger domain embedded within a 41 amino acid region known as motif-C (Masai and Arai, 2000). Motif-C forms one of two Dbf4 contact surfaces with Cdc7, and, in yeast, it has been shown that the Zn2+ finger plays a nonessential role in stimulating DDK activity (Harkins et al., 2009; Jones et al., 2010; Hughes et al., 2012). We therefore constructed an additional allele, dbf4-zn, which deletes Dbf4 residues 660–688 involved in forming the Zn2+ finger (Figure 2A). Analysis of these dbf4 alleles in the absence of HU indicated, first, similar to pdbf4-C, pdbf4-zn only partially complemented dbf4-1 when expressed from a low copy plasmid (Figure 2B). Second, in agreement with previous work (Harkins et al., 2009; Jones et al., 2010), dbf4-zn exhibited a temperature-sensitive growth defect and slow progression through S phase when integrated at the endogenous locus (Figures 2D and 3A). Third, strains expressing integrated dbf4-zn arrested as budded cells with short preanaphase spindles at a nonpermissive temperature of 37°C (Figure 3B). Fourth, as with dbf4-1 (Warsi et al., 2008), dbf4-zn cell cycle arrest was dependent on the spindle assembly checkpoint (SAC; Figure 3C). Fifth, a surprising finding was that pdbf4-C or pdbf4-zn expressed over a deletion of DBF4 grew at temperatures up to 37°C (Figure 2C). Thus, while dbf4-1 cells transformed with pdbf4-C or pdbf4-zn were temperature sensitive, dbf4-∆ cells transformed with pdbf4-C or pdbf4-zn were not. It is likely that dbf4-1 has a partial dominant negative activity toward pdbf4-C and pdbf4-zn and that the low-copy plasmids used in these experiments lead to increased pdbf4 expression compared with the endogenous locus.
FIGURE 2:
Characterization of dbf4-C and dbf4-zn. Experiments in this figure are without HU treatment. (A) Dbf4 domains and mutant alleles. C22, D3, and D45 are PCR mutagenized alleles, while dbf4-zn was constructed using recombinant techniques. Base pair changes that alter amino acid coding in dbf4-C alleles are indicated; Zn2+ finger amino acids deleted in dbf4-zn are also shown. The diagram also illustrates amino acid boundaries for domains involved in cell cycle proteolysis (∆); Cdc5 (Polo) and Rad53 binding; motifs N, M, and C; and the BRCT-related BFDF domain. (B) Complementation of dbf4-1; 10-fold serial dilutions of a dbf4-1 strain (JBY997) transformed with low copy pCEN ARS DBF4 (JJY059; JJY164), vector (JJY016), pdbf4-C22 (JJY017), pdbf4-D3 (JJY060), pdbf4-D45 (JJY061), and pdbf4-zn (JJY165) plasmids were stamped onto selective media at indicated temperatures. Whereas DBF4 complements dbf4-1 to 37°C, dbf4-C and dbf4-zn alleles partially complement to 34°C. (C) Complementation of dbf4-∆. A dbf4-∆ strain harboring pURA3 DBF4 was transformed with pCEN ARS DBF4 (JJY037, JJY166), pdbf4-C22 (JJY032), pdbf4-D3 (JJY033), pdbf4-D45 (JJY044), and pdbf4-zn (JJY167). The transformants were cured of the covering DBF4 URA3 plasmid on 5′-FOA; 10-fold serial dilutions were stamped onto selective media at indicated temperatures. In this case, low copy dbf4-C and dbf4-zn plasmids complement dbf4-∆ growth to 37°C. (D) Integrated dbf4-zn. Constructs expressing DBF4 (JJY046) or dbf4-zn (JJY076) were integrated immediately upstream of a precise deletion of the DBF4 open reading frame; 10-fold serial dilutions were incubated at indicated temperatures revealing a dbf4-zn temperature sensitive growth phenotype.
FIGURE 3:
dbf4-zn does not undergo a reductional anaphase. Experiments in this figure are without HU treatment. (A) FACs. WT (CRY1), dbf4-1 (JBY999), or dbf4-zn expressed from the native locus (JJY076) strains were arrested in G1, released at 26°C or 37°C, and analyzed by FACs. dbf4-zn shows an ∼15 min S phase delay compared with WT at 26°C and an ∼45–60 min delay at 37°C. (B) Top graphs: a dbf4-1 SPC42-GFP strain (JBY1392) and a strain expressing SPC42-GFP and integrated dbf4-zn (JJY045) were released from G1 at 37°C. After 2 h, spindle length distributions were determined using Spc42-GFP. Bottom micrographs: dbf4-1 (JBY2323) and integrated dbf4-zn (JBY2324) cells expressing GFP-TUB1 were released from G1 at 37°C. After 2 h spindles were visualized at 37°C. Numbers, spindle length in μm; bar, 4 μm. Whereas dbf4-1 exhibits reductional anaphase spindle extension, dbf4-zn arrests with normal length preanaphase spindles. (C) The dbf4-zn cell cycle arrest. Top graph: rad9-∆ (JBY186), integrated dbf4-zn (JJY076) and integrated dbf4-zn rad9-∆ (JJY080) double mutants were released from G1 at 37°C, and budding was used to evaluate cell cycle progression. Bottom graph: WT (CRY1), mad2-∆ (JBY546), and integrated dbf4-zn mad2-∆ (JJY102) strains were analyzed similarly. As with dbf4-1, dbf4-zn cell cycle arrest is dependent on Mad2 but not Rad9.
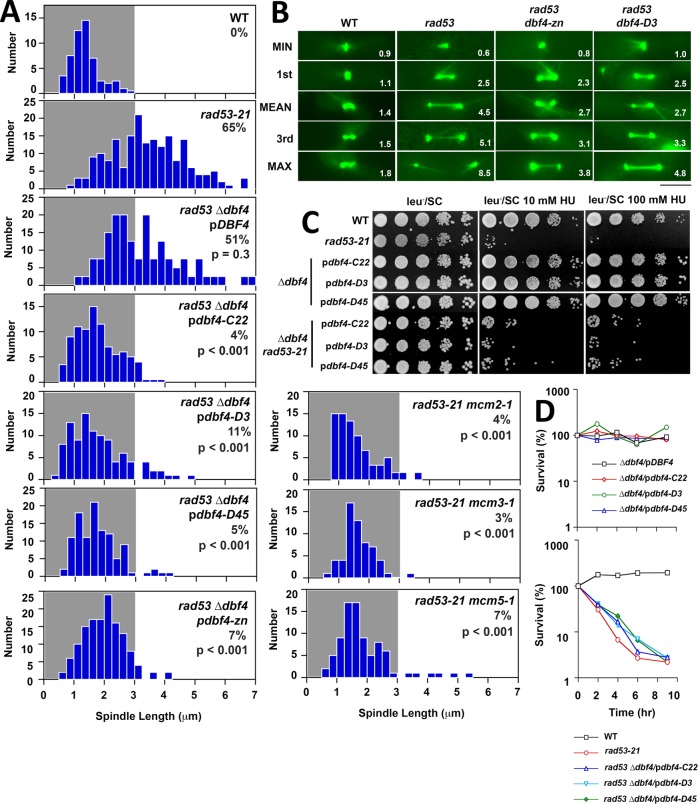
Using pdbf4-C and pdbf4-zn, we reexamined ddk suppression of rad53 spindle extension in HU without having to circumvent the reductional anaphase phenotype associated with dbf4-1. rad53 can display synthetic lethality with ddk mutations (Dohrmann and Sclafani, 2006), and spores combining rad53-21 with integrated dbf4-zn were inviable (unpublished data). However, we readily isolated rad53-21 dbf4-∆ segregants containing pdbf4-C or pdbf4-zn; such transformants were therefore used in subsequent experiments. We found that our three pdbf4-C alleles, as well as pdbf4-zn, provided a robust block to rad53 spindle extension in HU, lowering the percentage of cells with spindles ≥3 μm—our threshold for an extended spindle (Bachant et al., 2005)—from ∼50% in rad53-21 dbf4-∆/pDBF4 strains to 4–11% in rad53-21 dbf4-∆/pdbf4-C and rad53-21 dbf4-∆/pdbf4-zn double mutants (Figure 4, A and B). Suppression of spindle extension was not accompanied by amelioration of rad53-21 HU sensitivity (Figure 4C), or HU recovery (Figure 4D). The essential role of the DDK in ORI firing is to phosphorylate MCM proteins to assemble the CMG replicative helicase (Hardy et al., 1997; Sheu and Stillman, 2010). If pdbf4 alleles suppress spindle extension by reducing MCM activity, mcm mutations should behave similarly. This proved to be the case, as the percentage of cells with spindles ≥3 μm was reduced from 65% in rad53-21 to 5% in HU-treated mcm2-1 rad53-21, mcm3-1 rad53-21, and mcm5-1 rad53-21 mutants (Figure 4A).
FIGURE 4:
pdbf4 C, pdbf4-zn and mcm alleles suppress rad53 spindle extension in HU. (A) WT (JBY1129), rad53-21 (JBY1274), rad53-21 dbf4-∆/pDBF4 (JJY184), rad53-21 dbf4-∆/pdbf4-C22 (JJY028), rad53-21 dbf4-∆/pdbf4-D3 (JJY029), rad53-21 dbf4-∆/pdbf4-D45 (JJY030), rad53-21 dbf4-∆/pdbf4-zn (JJY182), rad53-21 mcm2-1 (JJY112), rad53-21 mcm3-1 (JJY117), and rad53-21 mcm5-1 (JJY120) strains, all harboring SPC42-GFP, were released from G1 into 200 mM HU at 30°C. After 90 min spindle lengths were measured in fixed cells. Percentages of cells with spindles ≥3 μm are displayed, along with the results of two-tailed t tests comparing each mutant to the rad53-21 distribution. (B) WT (JBY1129), rad53-21 (JBY1274), rad53-21 dbf4-∆/pdbf4-zn (rad53 dbf4-zn on the figure; JJY182), and rad53-21 dbf4-∆/pdbf4-D3 (rad53 dbf4-D3 on the figure; JJY029) strains were transformed with pGFP-TUB1 to visualize MTs. Cells were released from G1 into 200 mM HU at 30°C. Starting at 90 min, spindles were imaged and measured (values at the bottom right of each panel) in 50 live cells. Spindles corresponding to minimum, maximum, mean, first, and third quadrant measurements are shown. Bar, 4 μm. (C) Tenfold serial dilutions of WT (JBY1129), rad53-21 (JBY1274), dbf4-∆/pDBF4 (JJY037), dbf4-∆/pdbf4-C22 (JJY032), dbf4-∆/pdbf4-D3 (JJY033), and dbf4-∆/pdbf4-D45 (JJY044) strains, along with rad53-21 dbf4-∆ transformants described in A were stamped on indicated media at 30°C to evaluate HU sensitivity. (D) Asynchronous cultures of all the strains described in C were shifted into media containing 200 mM HU at 30°C. At the indicated times, plating efficiency was determined to evaluate HU recovery.
Dbf4, Sld3 and Dun1 are Rad53 substrates in blocking spindle extension
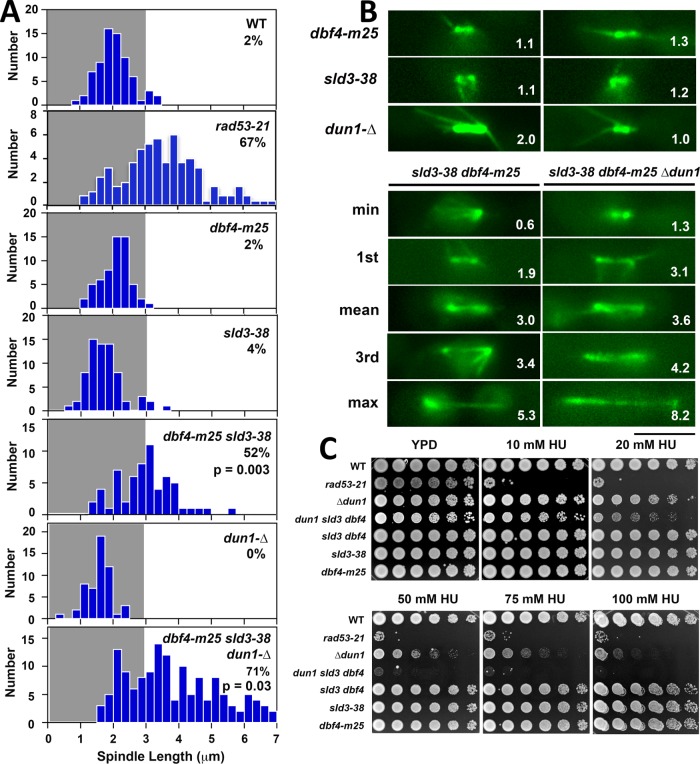
The results presented so far suggest that Rad53 delay of ORI firing in HU might be important in restraining spindle extension. This delay is mediated through Rad53 inhibitory phosphorylation of Dbf4 and another CMG activator, Sld3. A dbf4-m25 sld3-38A mutant that is resistant to Rad53 phosphorylation exhibits firing of checked ORIs even though Rad53 is activated normally (Lopez-Mosqueda et al., 2010; Zegerman and Diffley, 2010). In three experiments we found that, on average, ∼50% of dbf4-m25 sld3-38A cells (56%, 52%, 42%) released into HU displayed spindles ≥3 um compared with 63% for rad53-21 controls (67%, 67%, 54%; Figure 5, A and B). While the frequency of dbf4-m25 sld3-38A spindle extension is similar to rad53-21, the average length of dbf4-m25 sld3-38A spindles was slightly, but significantly, reduced (mean 3.0 μm compared with 3.5 μm for rad53-21, p < 0.001), and the fraction of spindles >4 μm was noticeably diminished (Supplemental Figure S1). Importantly, Dun1 is another Rad53 substrate that contributes to fork stability in HU by expanding dNTP pools (Poli et al., 2012; Morafraile et al., 2015). We therefore examined the effect of ablating both Dun1 and the block to ORI firing (see Figure 10A, red X’s on pathways 1 and 2, later in this article). An average of 72% of dun1-∆ dbf4-m25 sld3-38A triple mutants (79%, 71%, 66%) exhibited spindle extension in HU (Figure 5, A and B), with a distribution of spindle lengths that was largely similar to rad53-21 (Supplemental Figure S1). Thus, short-circuiting Rad53 regulation of Dbf4, Sld3 and Dun1 synergizes to phenocopy rad53 spindle extension in HU. Consistent with previous observations indicating DNA replication control is an essential function of the S phase checkpoint (Desany et al., 1998), dun1-∆ dbf4-m25 sld3-38A mutants also exhibited a synergistic increase in HU sensitivity approaching that of rad53-21 (Figure 5C).
FIGURE 5:
Dbf4 and Sld3 are Rad53 substrates controlling the block to spindle extension. (A) WT (CRY1), rad53-21 (Y301), dbf4-m25 (YJLO157), sld3-38A (YJLO156), dbf4-m25 sld3-38A (YJLO155, JBY2334), dun1-∆ (MY26), and dbf4-m25 sld3-38A dun1-∆ (JJY141, JJY144) strains were arrested in G1 and released into 200 mM HU at 30°C. After 90 min, samples were processed for α-tubulin immunofluorescence, and spindle lengths were measured. The percentage of cells with spindles ≥3 μm is shown, along with p values (two-tailed t test) comparing the sld3-38A dbf4-m25 and sld3-38A dbf4-m25 dun1-∆ distributions to rad53-21. (B) Strains in A were transformed with pGFP-TUB1, arrested in G1, released into 200 mM HU at 30°C, and spindles were imaged and measured in live cells. Representative spindles are shown for dbf4-m25, sld3-38A, and dun1-∆ single mutants, along with spindles corresponding to minimum, maximum, mean, first, and third quadrant measurements from the sld3-38A dbf4-m25 and sld3-38A dbf4-m25 dun1-∆ populations. Numbers in panels, spindle length in μm; bar, 4 μm. (C) Tenfold serial dilutions of strains in A were stamped onto indicated media to evaluate HU sensitivity.
FIGURE 10:
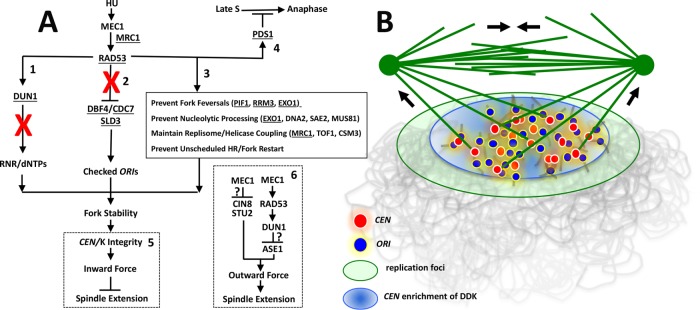
Organization and mechanism of the S phase checkpoint block to spindle extension. (A) Checkpoint organization. Six effector pathways relevant to this work are depicted. Rad53 substrates are underlined. See main text for relevant citations. (1) Rad53 activation of Dun1 controls expansion of dNTP pools through transcriptional and posttranslational regulation of RNR. (2) Rad53 inhibition of Dbf4 and Sld3 blocks firing of checked ORIs. (3) Rad53 acts directly at replication forks (box) to maintain fork structure. Pathways 1–3 work synergistically to ensure fork progression during replication stress. (4) Rad53 controls a Pds1-dependent cell cycle arrest response that becomes operative in late S phase. (5) We propose the role of Rad53 in stabilizing forks ensures early-replicating CEN regions are not subjected to nucleolytic degradation, preserving CEN/K integrity and chromosome attachment to the spindle. These attachments generate inward force to restrain spindle extension. (6) Checkpoint regulators also appear to work through additional pathways to down-regulate outward-directed spindle force. Question marks indicate how the checkpoint mediates these responses is not yet clear. Red Xs in 1 and 2 indicate responses crippled downstream of Rad53 in a dun1-∆ dbf4-m25 sld3-38A mutant. (B) Proposed S phase spindle structure. DDK activation of ORI firing leads to incorporation of CEN proximal ORIs (blue) and CENs (red) into replication foci (green oval). DDK enrichment at the CEN ORI cluster is also depicted (light blue oval). A central assumption is that replicating CENs associated with these structures are partially immobilized, forming a cluster of spindle attachment sites capable of resisting K-MT pulling force (force arrows directed toward SPBs). A distribution of such attachments to both spindle poles could offset spindle extension (force arrows in central spindle) at a characteristic S phase spindle length. Such a spindle assembly intermediate may transiently form during conditions that perturb the relative timing of SPB separation and spindle assembly with completion of S phase. In the illustration, the spindle is depicted as “reaching down” to connect to Ks assembled on immobilized CENs. This is a diagrammatic convenience, as the spindle actually extends through the nucleoplasm and would therefore be emmeshed within the replicating chromatin environment.
dbf4-zn reduces ORI firing in rad53 mutants
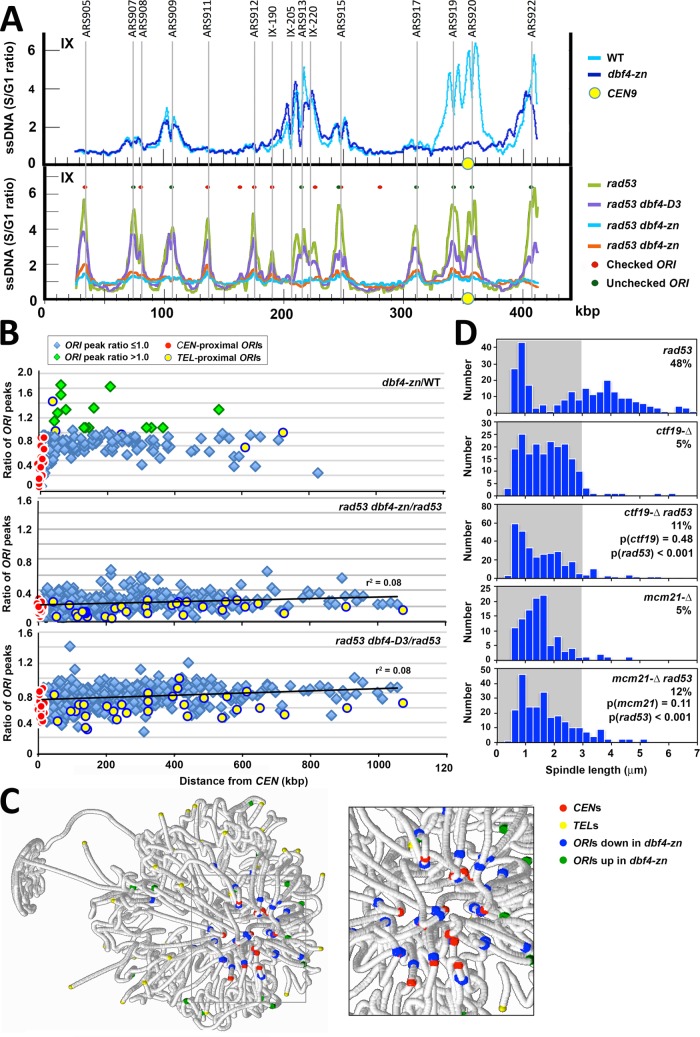
Our initial rationale for how spindle extension might be suppressed in HU-treated dbf4-zn rad53 double mutants made two testable predictions. First, we predicted dbf4-zn rad53 double mutants should exhibit reduced utilization of some, or (given the hypomorphic nature of dbf4-zn) perhaps all, ORIs in HU compared with rad53 single mutants. Second, reduced ORI utilization should correspond with improved duplication of CENs. To examine these predictions, ssDNA hybridization to genome microarrays was used to analyze ORI firing in HU (Feng et al., 2007). In a first experiment, dbf4-∆ cells harboring pDBF4 or pdbf4-zn were released from G1 into 200 mM HU at 30°C. After 60 min, ssDNA was isolated and processed for replication profiling. A total of 177 ORIs activated in dbf4-∆/pDBF4 and dbf4-∆/pdbf4-zn strains was detected (Figure 6A and Supplemental Figure S2). To quantify the effect of pdbf4-zn on ORI utilization in HU, the area under each ssDNA ORI curve (ORI AUC) in the data sets was used to calculate an ORI AUC ratio comparing dbf4-∆/pdbf4-zn to dbf4-∆/pDBF4. The mean pdbf4-zn/pDBF4 ORI AUC ratio for all 177 ORIs was 0.74, an ∼25% reduction (Supplemental Figure S4A). Strikingly, many ORIs in the vicinity of CENs displayed a substantially stronger reduction in HU (Figure 6, A and B; Supplemental Figures S2 and S4A). The set of 32 CEN-flanking ORIs (Supplemental Figure S4B) showed the greatest down-regulation, with a mean pdbf4-zn/pDBF4 ORI AUC ratio of 0.36 (red in Figure 6B, Supplemental Figure S4A; Supplemental Tables S1 and S2, p < 0.0001). Not all CEN-flanking ORIs were affected equivalently (Supplemental Table S2), however, and some ORIs that did not immediately flank a CEN were also reduced (Supplemental Figure S2 and Supplemental Table S3). As a second treatment, we queried the data sets to detect ORI peaks whose amplitudes were altered by three standard deviations from the median difference between HU-treated dbf4-∆/pdbf4-zn and dbf4-∆/pDBF4 (Supplemental Table S3). Thirty-two strongly reduced ORIs were identified, including 20 CEN flanking ORIs. Interesting, 19 differentially up-regulated ORIs were also observed (Supplemental Table S3); these tended to be less efficiently utilized ORIs (Supplemental Figure S4A). To visualize a spatial basis for these patterns, positions for the 32 down-regulated ORIs and 19 up-regulated ORIs were mapped onto a spatial model of chromosome organization in the yeast nucleus (Duan et al., 2010). Down-regulated ORIs (blue) were closely associated with the CEN cluster (red), while up-regulated ORIs (green) tended to reside just beyond the periphery of the CEN cluster (Figure 6C).
FIGURE 6:
ORI firing in HU-treated pdbf4-zn and rad53 pdbf4-zn mutants. dbf4-∆/pDBF4 (WT on figure; JJY108), dbf4-∆/pdbf4-zn (dbf4-zn on the figure; JJY181), rad53-21 dbf4-∆/pDBF4 (rad53 on the figure; JJY023), rad53-21 dbf4-∆/pdbf4-zn (rad53 dbf4-zn on the figure; JJY182), and rad53-21 dbf4-∆/pdbf4-D3 (rad53 dbf4-D3 on the figure; JJY029) strains were arrested in G1. The cultures were split with aliquots either released into 200 mM HU at 30°C or retained at the G1 block. After 60 min, ssDNA replication intermediates were isolated from G1 and HU-treated samples and hybridized to microarrays. The ratio of S phase (HU) to G1 hybridization values (S/G1 ratio) was determined at each array position. (A) Replication profiles for chromosome IX showing reduced firing of CEN-flanking ARS919 and ARS920 in pdbf4-zn (top profile) and more uniformly reduced ORI utilization in rad53-21 pdbf4-zn and rad53-21 pdbf4-D3 (bottom profile). (B) ORI AUCs were calculated for 177 ORI peaks in pDBF4 (WT) and pdbf4-zn strains treated with HU and 403 ORI peaks in rad53-21 pDBF4 (rad53) and rad53 pdbf4 strains treated with HU. The ratios of the pdbf4-zn to WT ORI AUCs, the rad53 pdbf4-zn to rad53 ORI AUCs, and the rad53 pdbf4-D3 to rad53 ORI AUCs (ratio of ORI peaks, y-axis) were plotted as a function of ORI distance to the CEN. CEN flanking ORIs, red circles; TEL proximal ORIs, yellow circles; ORIs with ORI AUC ratios ≥1.0, green diamonds. Regression lines are shown for rad53 plots. (C) pdbf4-zn ORIs decreased (blue) or increased (green) by ≥3 SD from the median difference between pdbf4-zn and pDBF4 peak amplitude values are highlighted (RasMol) on a spatial map of the yeast nucleus (see also Supplemental Table S3). Positions of CENs (red) and TELs (yellow) are also shown. Down-regulated ORIs in HU-treated pdbf4-zn cells tend to be located in close proximity to the CEN/SPB cluster, while up-regulated ORIs are often located somewhat more distally (inset). (D) The rad53-21 (JBY2274), ctf19-∆ (JBY2250), ctf19-∆ rad53-21 (JBY2251), mcm21-∆ (JBY2327), and mcm21-∆ rad53-21 (JBY2330) SPC42-GFP strains were released from G1 into 200 mM HU at 30°C. Spindle lengths were measured after 90 min. Percentages of cells with spindles ≥3 μm are shown, along with p values (two-tailed t test) for indicated comparisons.
We next proceeded to examine ssDNA replication profiles for 403 ORIs detected in HU-treated rad53-21 dbf4-∆ mutants transformed with pDBF4, pdbf4-zn, or pdbf4-D3 (a pdbf4-C allele harboring R701G, Figure 2A). Utilization of all 403 ORIs was substantially reduced in rad53-21 dbf4-∆/pdbf4-zn compared with rad53-21 dbf4-∆/pDBF4, with a mean ORI AUC ratio of 0.26 (Figure 6, A and B; Supplemental Figures S2, S3, and S4A). The set of 32 CEN-flanking ORIs, however, still showed a preferential reduction (ORI AUC ratio 0.15, red in Figure 6B and Supplemental Figure S4A, p < 0.0001). The rad53-21 dbf4-∆/pdbf4-D3 HU data set trended similarly, although ORI diminishment was not as pronounced, with an ORI AUC ratio of 0.72 for all 403 ORIs and 0.61 for CEN-flanking ORIs (Figure 6, A and B; Supplemental Figures S3 and S4A, p < 0.0001 for both comparisons). Overall, these results reveal that the Zn2+ finger of Dbf4 makes a complex contribution to ORI firing in HU. When the S phase checkpoint is active, this contribution is strongly manifested in the vicinity of CENs. When the S phase checkpoint is inactive, however, global ORI firing become more reliant on the Zn2+ finger. In terms of our first prediction, the HU replication profiles of rad53-21 dbf4-∆/pdbf4-zn mutants clearly show reduced ORI utilization in HU compared with rad53-21 dbf4-∆/pDBF4-consistent with our expectations. With respect to our second prediction, however, the pattern of ORI firing does not necessarily support the idea that CEN duplication is required for suppression of spindle extension in HU. This is because the exact population of ORIs whose firing would be expected to give rise to CEN duplication was the most strongly reduced in rad53-21 dbf4-∆/pdbf4-zn. In fact, a different interpretation of the rad53-21 dbf4-∆/pdbf4-zn data set is that preventing, rather than promoting, replication in the vicinity of CENs might correspond with rescue of spindle extension in HU-treated rad53-21 dbf4-∆/pdbf4-zn mutants.
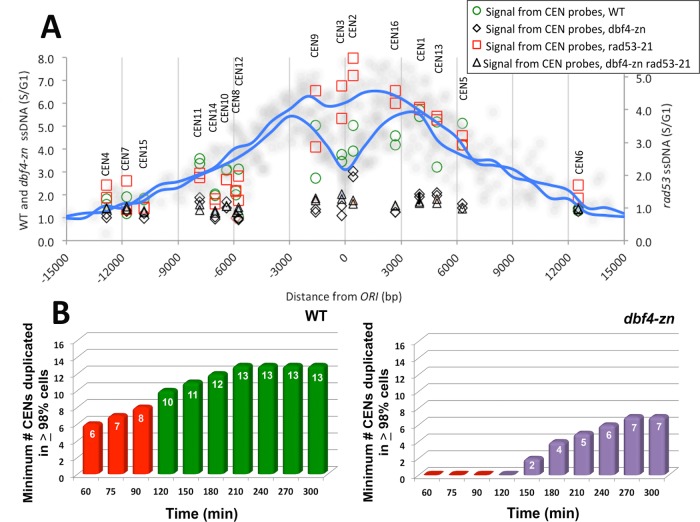
In light of the realization that avoiding replication fork destabilization in proximity to CENs might underlie suppression of spindle extension in HU-treated rad53 mutants, several additional analyses were performed. First, using the 16 ORIs closest to each CEN (Supplemental Figure S4B; Supplemental Tables S1 and S2), we generated composite HU replication profiles for dbf4-∆/pDBF4 cells and rad53-21 dbf4-∆/pDBF4 mutants (Figure 7A). The dbf4-∆/pDBF4 cells display a characteristic split peak profile indicative of bidirectional replication fork movement. The rad53-21 dbf4-∆/pDBF4 mutants, in contrast, tend to display a single ssDNA peak whose maximum amplitude is centered over the ORI, reflecting accumulation of aberrant ssDNA. ssDNA values for CDEI, II, and III CEN DNA elements from the dbf4-∆/pDBF4, dbf4-∆/pdbf4-zn, rad53-21 dbf4-∆/pDBF4, and rad53-21 dbf4-∆/pdbf4-zn data sets were superimposed on these HU replication composites. The rad53-21 dbf4-∆/pDBF4 showed a comparative increase in ssDNA at CENs 2, 3, 9, and 16, the four closest CENs to an ORI (Figure 7A). In contrast, only baseline levels of CEN ssDNA were observed in dbf4-∆/pdbf4-zn and rad53-21 dbf4-∆/pdbf4-zn. Second, we used the dbf4-∆/pDBF4 and dbf4-∆/pdbf4-zn composite profiles to perform a statistical analysis of CEN duplication in HU (Supplemental Results). We found that between 60 and 90 min, ≥98% of HU-treated dbf4-∆/pDBF4 cells are predicted to have duplicated a minimum of 6–8 CENs (Figure 7B, Supplemental Figure S5B, and Supplemental Tables S4 and S5). In contrast, during the same period ∼10% of dbf4-∆/pdbf4-zn cells have not duplicated any CEN. Third, ctf19-∆ strains defective for the Ctf19/COMA K complex fail to recruit the DDK to Ks and, like dbf4-∆/pdbf4-zn, exhibit reduced firing of CEN-proximal ORIs (Natsume et al., 2013; Hinshaw et al., 2017). We therefore tested whether loss of Ctf19 behaved similarly to pdbf4-zn in suppressing rad53 spindle extension in HU. Unlike our previous results with strains defective for essential K proteins (Bachant et al., 2005), ctf19-∆ and mcm21-∆ mutants arrested with short spindles in HU, and spindle extension in rad53-21 ctf19-∆ and rad53-21 mcm21-∆ strains was reduced to 11% (p < 0.001) and 12% (p < 0.001) compared with 48% for rad53-21 controls (Figure 6D).
FIGURE 7:
Analysis of CEN ssDNA and CEN duplication in HU. (A) Using the HU replication profiles described in Figure 6, the closest ORI to each CEN was identified (see also Supplemental Figure S4B; Supplemental Tables S1 and S2). S/G1 ssDNA values spanning these 16 ORIs were determined at 1000-base-pair intervals from the ORI center. The average value at each position was used to plot a composite replication profile for the dbf4-∆/pDBF4 (WT on the y-axis) and rad53-21 dbf4-∆/pDBF4 (rad53 on the y-axis) data sets (blue lines). Data points from each of the 16 individual profiles are also shown (soft gray circles). The split peak characteristic of the WT composite reflects the average extent of bidirectional replication fork movement, while the rad53 composite appears as a more uniform peak due to accumulation of aberrant ssDNA. Genome arrays used in these experiments have two positions overlapping each CEN DNA element. G1/S ssDNA values for these positions were extracted from the dbf4-∆/pDBF4 (WT, green circles), dbf4-∆/pdbf4-zn (dbf4-zn; black diamonds), rad53-21 dbf4-∆/pDBF4 (rad53-21, red squares), and rad53-21 dbf4-∆/pdbf4-zn (dbf4-zn rad53-21; black triangles) data sets and superimposed on the composite profiles to compare the extent of CEN ssDNA in each strain. The relative position of each CEN from the center of the composite is shown at the top of the graph. (B) Statistical projections for CEN duplication in HU were computed as described in the Supplemental Results. The simulation spans a 60–300 min period following G1 release into 200 mM HU. The contributions of 45 CEN-proximal ORIs to CEN duplication are included. Graphs display the minimum number of CENs predicted to be duplicated in ≥ 98% of the cell population at the indicated times for dbf4-∆/pDBF4 (WT) and dbf4-∆/pdbf4-zn (dbf4-zn). Red columns (60–90 min) denote the period in which the spindle forms in HU. Extending the simulation allows the kinetics and maximal extent of CEN duplication to be evaluated.
Exo1 is a determinant of CEN/K integrity in HU-treated rad53 mutants
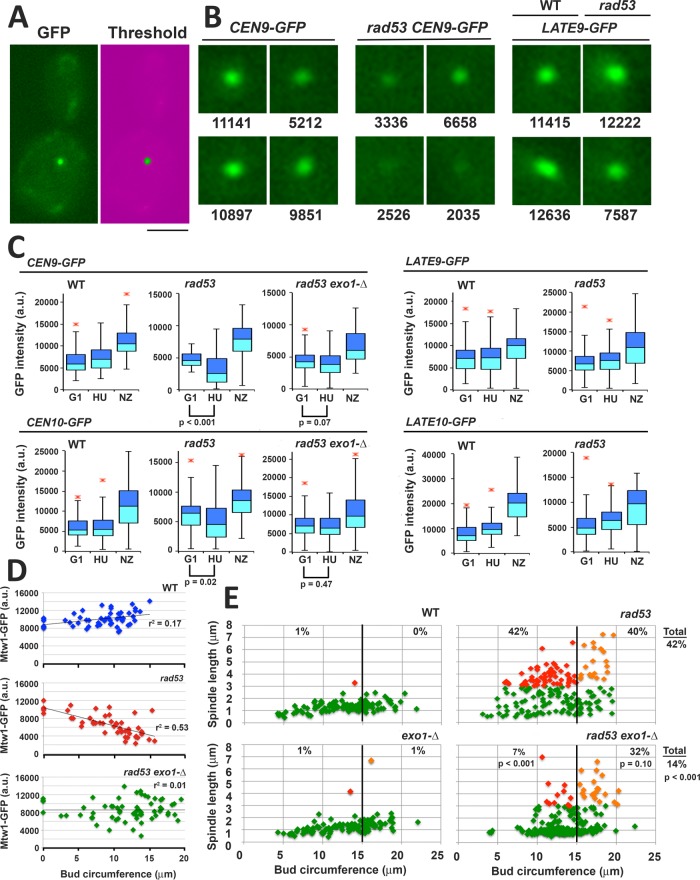
The exonuclease Exo1 is responsible for much of the aberrant ssDNA that is generated at replication forks in HU-treated rad53 mutants (Cotta-Ramusino et al., 2005; Feng et al., 2006). During this study, we noticed GFP chromosome tags in the vicinity of CENs often exhibited reduced fluorescence in rad53 mutants exposed to HU (Figure 8B). To determine whether this loss of signal might be a result of Exo1 activity, LacO arrays were integrated adjacent to the CEN on chromosomes 2 (CEN2-GFP), 9 (CEN9-GFP), and 10 (CEN10-GFP) and visualized using LacI-GFP (Supplemental Figures S6 and S7). We also targeted LacO arrays to regions on chromosomes 9 and 10 ∼20 kbp distant from ORIs in wild type (WT) and rad53 cells (LATE9-GFP and LATE10-GFP; Supplemental Figures S6 and S7); these regions are unlikely to be duplicated during the initial phase of HU treatment. WT, rad53-21, and rad53-21 exo1-∆ strains harboring CEN-GFP and LATE-GFP were released into HU, or into media containing nocodazole (NZ). For all tags, we observed the anticipated increase in fluorescence in NZ, reflecting duplication of the tags (Figure 8C). In contrast, for WT cells, median tag intensity only increased slightly in HU. In HU-treated rad53 cells, CEN2-GFP (p < 0.001), CEN9-GFP (p < 0.001), and CEN10-GFP (p = 0.02) all showed significant decreases in fluorescence (Figure 8C and Supplemental Figure S8A). Such a decrease was not observed in rad53 exo1-∆ CEN9-GFP, rad53 exo1-∆ CEN10-GFP, rad53 LATE9-GFP, or rad53 LATE10-GFP strains. Thus, reduced tag intensity in HU-treated rad53 cells is influenced by both Exo1 and tag distance from an ORI. To examine whether rad53 mutants might also display an Exo1-mediated disruption of K integrity in HU, WT, rad53-21, and rad53-21 exo1-∆ strains harboring the K protein Mtw1-GFP were released into media containing both HU and NZ. Bud circumference was measured as a proxy for elapsed time in S phase. Whereas HU-treated WT cells showed a gradual increase in Mtw1-GFP signal as bud size increased, Mtw1-GFP intensity decreased in rad53 cells (Figure 8D, p < 0.001). This decrease was partially ameliorated in rad53 exo1-∆, which showed an intermediate distribution of Mtw1-GFP intensities.
FIGURE 8:
Exo1 is a determinant of CEN/K integrity and spindle extension in HU-treated rad53 mutants. Strains harboring the indicated GFP chromosome tags were released from G1 into media containing 200 mM HU or with 15 μg/ml NZ at 30°C: CEN9-GFP (WT, JBY2283; rad53-21, JBY2295; exo1-∆ rad53-21, JBY2299); CEN10-GFP (WT, JBY2297; rad53-21, JBY2298; exo1-∆ rad53-21, JBY2301); LATE9-GFP (WT, JBY2289; rad53-21, JBY2290); LATE10-GFP (WT, JBY2291; rad53-21, JBY2293). Cells were processed for microscopy after 90 min. (A) Example of low-end mask to threshold GFP signal. Bar, 4 μm. (B) Representative CEN9-GFP and LATE9-GFP foci in HU-treated cells, with corresponding intensity values (arbitrary units). (C) Box plots of GFP tag intensities in G1, HU, and NZ arrested cells. At least 50 cells were analyzed for G1 and NZ samples; 100 cells were analyzed for HU samples. The p values (two-tailed t test) are provided in cases where the signal intensity of the HU sample is reduced compared with the G1 sample. (D) WT (JBY2252), rad53-21 (JBY2253), and exo1-∆ rad53-21 (JBY2264) MTW1-GFP strains were released from G1 into media containing both HU and NZ at 30°C. NZ was included so that Ks were not dispersed by spindle extension, facilitating quantification of Mtw1-GFP. After 90 min, live cell mounts were analyzed for Mtw1-GFP intensity. Bud circumference was measured as a proxy for time post-G1 release. Cells that did not leave the G1 block were scored to provide a signal baseline. Regression lines and fit estimates are indicated. (E) WT (JBY1129), rad53-21 (JBY1274), exo1-∆ (JBY2246), and exo1-∆ rad53-21 (JBY2303) SPC42-GFP strains were treated with HU as in A and evaluated for bud circumference and spindle length. Color coding: cells with spindles <3 μm, green; cells with spindles ≥3 μm and bud circumferences <15 μm (small to medium budded cells), red; cells with spindles ≥3 μm and buds ≥15 μm (medium to large budded cells), orange. The percentages of small/medium and medium/large budded cells with extended spindles are shown on the corresponding regions of the graphs. The total percentage rad53 and rad53 exo1 cells with extended spindles is shown on the right-hand side of the graphs. The p values (two-tailed t tests) compare differences in spindle extension between the rad53 and rad53 exo1 data sets.
We then examined spindle length in HU-treated WT, exo1-∆, rad53-21, and rad53-21 exo1-∆ strains, again using bud circumference as a metric for elapsed time in S phase. For HU-treated rad53 mutants, spindle extension is observed shortly after spindle poles separate (Bachant et al., 2005). Consistent with this, 42% of rad53 cells with a bud circumference of ≤15 μm (small- to medium-budded cells) exhibited extended spindles (red diamonds, Figure 8E), and 40% of cells with buds >15 μm (medium- to large-budded cells) showed extended spindles (orange diamonds). In comparison, only 7% of small- to medium-budded rad53 exo1-∆ exhibited spindle extension (sixfold decrease, p < 0.001), while 32% of medium- to large-budded cells showed extended spindles (1.25-fold decrease, p = 0.10). Thus, loss of Exo1 significantly delays rad53 spindle extension in HU.
Artificial Ks at sites distant from ORIs delay rad53 spindle extension in HU
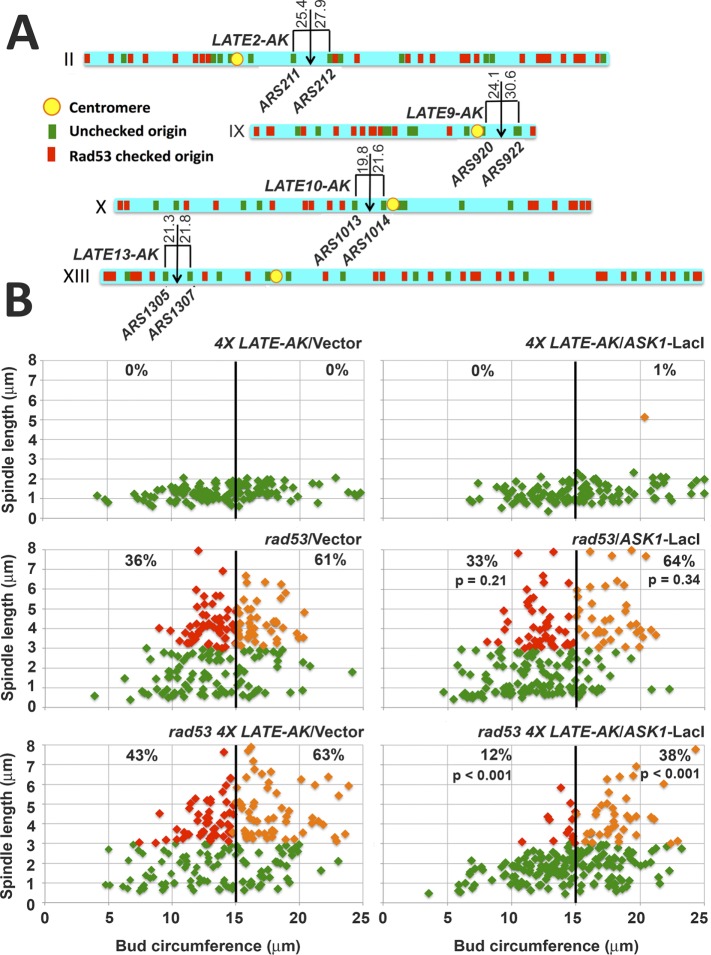
In light of our observations, we sought to devise an experiment to test whether transferring CENs from early to late replicating genome regions would suffice to bypass the role of Rad53 in restraining spindle extension in HU. Our approach was based on the finding that assembly of an artificial K (AK) can be directed by integrating bacterial operator sequences at desired locations, followed by production of the outer K proteins Ask1 or Dam1 fused to the corresponding bacterial repressor (Kiermaier et al., 2009; Lacefield et al., 2009). LacO-based AK targeting constructs were integrated at sites on chromosomes 2, 9, 10, and 13 ∼20 kbp distant from unchecked ORIs that fire in both HU-treated WT cells and rad53 mutants (Figure 9A; Supplemental Figures S6, S7, and S8B). Cells were then transformed with a plasmid encoding Ask1 fused to LacI to induce AK assembly (Lacefield et al., 2009). WT and rad53 transformants harboring all four AK insertions (4× LATE-AK), transformed with vector or pASK1-LacI, were released into HU. After 90 min, spindle lengths were measured and correlated with bud circumference. We observed that activation of the four AKs imposed a reproducible and significant (p < 0.001 for three experiments) reduction in rad53 spindle extension in HU (Figure 9B and Supplemental Figure S8C). In particular, small- to medium-budded rad53 4X LATE-AK cells expressing ASK1-LacI exhibited an average of 10% spindle extension compared with an average of 40% for vector controls (fourfold reduction, red diamonds, Figure 9B and Supplemental Figure S8C). These observations suggest that a limited number of AKs, inserted into sites that are unlikely to replicate in HU during the period of spindle assembly, can significantly offset spindle extension in HU-treated rad53 mutants.
FIGURE 9:
AKs at late replicating sites offset rad53 spindle extension in HU. (A) AK insertion sites on chromosomes 2, 9, 10, and 13 are illustrated by downward arrows. Distances to flanking ORIs (kbp), as well as positions of unchecked ORIs (green), checked ORIs (red) and CENs (yellow) are indicated. (B) RAD53 SPC42-GFP (JBY2271) and rad53-21 SP42-GFP (JBY2273) strains harboring all four LATE-AK insertions (4X LATE-AK) were transformed with a vector control or a plasmid expressing ASK1-LacI to activate synthetic K activity. A rad53-21 SPC42-GFP strain (JBY1274) lacking 4X LATE-AK was similarly transformed. Small colonies emerging on transformation plates were resuspended in liquid media, arrested in G1, and released into 200 mM HU. After 90 min, cells were analyzed for bud circumference and spindle length. Color scheme for graphs is as described for Figure 8E. The percentages of small/medium and medium/large budded cells with extended spindles are shown in the corresponding regions of the graphs. The p values (two-tailed t tests) compare spindle extension in rad53/pASK1-LacI and rad53 4X LATE-AK/pASK1-LacI transformed samples with corresponding vector controls.
DISCUSSION
Three principal findings arise from this study. First, the C-motif of Dbf4 plays a novel role in the K-directed pathway specifying early firing of CEN-proximal ORIs. Second, our data indicate rad53 spindle extension in HU is a consequence of replication fork catastrophes in proximity to CENs. This has a bearing on how Rad53 effector functions are organized within the S phase checkpoint. Third, while K-MT attachments are required to prevent S phase spindle extension, CEN duplication, and thus the capacity for amphitelic K attachments, does not appear to be essential. This finding necessitates a partial revision of our previous model (Bachant et al., 2005) for how spindle extension is restrained at the S phase checkpoint.
The Dbf4 Zn 2+ finger/C-motif stimulates firing of CEN proximal ORIs
In the K-directed pathway, Dbf4 localizes to the Ctf19/COMA complex and is then transferred to adjacent ORIs, leading to accumulation of limiting replication factors (Natsume et al., 2013; Hinshaw et al., 2017). Possible roles for the Dbf4 C-motif are therefore to bind Ctf19/COMA to facilitate DDK recruitment to the K, or to bind components of pre-RCs to facilitate ORI transfer. In particular, a dbf4-zn defect in the transfer step might increase local DDK availability to regions that are peripheral to the CEN ORI cluster, consistent with the increased utilization of some adjacent, less efficient ORIs observed in our study. A different possibility is that the C-motif facilitates firing of CEN-proximal ORIs by stimulating DDK activation of MCM (Harkins et al., 2009; Jones et al., 2010). Positively charged residues flanking the Dbf4 C-motif have been proposed to mediate recognition of priming phosphorylation events (Hughes et al., 2012), and both Mec1 and S phase forms of Cdk1 have been implicated in phosphorylating chromatin-bound MCM to facilitate DDK targeting (Nougaréde et al., 2000; Randell et al., 2010) Speculatively, the R701G mutation in the pdbf4-D3 allele, which has a less severe effect on ORI firing compared with pdbf4-zn, could impair such targeting interactions. We note that the Dbf4 C-terminus has previously been implicated in controlling a different group of ORIs where early firing is conferred through binding of the Forkhead (Fkh) transcription factors Fkh1 and Fkh2 (Knott et al., 2012; Fang et al., 2017). This suggests the C-motif is acting through at least two pathways to specify early ORI firing timing.
In rad53-21 ∆dbf4/pdbf4-zn mutants, activation of all ORIs in HU—both unchecked ORIs that fire in ∆dbf4/pdbf4-zn and rad53-21 ∆dbf4/pDBF4 mutants and checked ORIs activated in rad53-21 ∆dbf4/pDBF4—is greatly reduced. The basis for this synergism is unclear. It is possible that in HU-treated rad53-21 ∆dbf4/pdbf4-zn mutants, overall DDK activity is reduced below a threshold necessary to fire ∼400 ORIs throughout the genome, leading to a diminishment at all ORIs. Alternatively, Rad53 may function in parallel with the DDK in distinct aspects of ORI firing, with loss of both activities producing a synthetic phenotype (Dohrmann and Sclafani, 2006; Holzen and Sclafani, 2010).
The block to spindle extension in HU is coupled to Rad53 regulation of replication fork stability
As shown in Figure 10A, our findings support an organization of Rad53 effector functions within the S phase checkpoint such that the initial, Pds1-independent, block to spindle extension is coupled to regulation of DNA replication fork stability. First, mutations affecting the DDK (pdbf4-zn, pdbf4-C alleles), MCM (mcm2-1, mcm3-1, mcm5-1), or K components that specify CEN ORI firing (ctf19-∆, mcm21-∆) rescue rad53 spindle extension in HU. Second, overproduction of Dbf4, which forces some checked ORIs to fire in HU (Tanaka et al., 2011), induces spindle extension. Third, Dbf4 and Sld3 are Rad53 substrates that mediate the block to spindle extension, a role that also involves Dun1. Fourth, loss of Exo1 ameliorates CEN/K integrity in HU-treated rad53 mutants and significantly delays spindle extension. Thus, a sequence of events to explain spindle extension in HU is that, in WT cells and rad53 mutants, unchecked ORIs, including CEN-flanking ORIs, fire normally. In rad53 cells, however, unrestrained ORI firing, combined with failure to up-regulate RNR and misregulation of fork modifying activities, results in pathological fork structures that allow nuclease access to replication forks (Figure 10A, pathways 1–3; Chen et al., 2007; Naylor et al., 2009; Lopez-Mosqueda et al., 2010; Zegerman and Diffley, 2010; Hu et al., 2012; Rossi et al., 2015; Chappidi et al., 2019), leading to a loss of CEN DNA integrity in a manner proportional to CEN distance from ORIs (Xed CENs in Supplemental Figure S9). Based on our analysis of Mtw1-GFP, loss of CEN integrity likely corresponds with a cis-acting defect in K assembly, which we argue is a proximal cause of spindle extension in HU. As mentioned in the Introduction, Rad53 also enforces a later-acting, Pds1-dependent, cell cycle arrest response in HU-treated cells (Figure 10A, pathway 4). This response becomes operative once HU replication stress has been largely circumvented, and likely significantly overlaps Rad53 activity in the DNA damage checkpoint (Gardner et al., 1999; Sanchez et al., 1999; Agarwal et al., 2003), involving stabilization of Pds1, modulation of Cdk1, and regulation of the mitotic exit network (Stueland et al., 1993; Clarke et al., 2001; Palou et al., 2015; Zhou et al., 2016; Zhang et al., 2017).
If spindle extension in HU-treated rad53 mutants is, in essence, a byproduct of defective DNA replication control, a significant issue concerns whether restraint of spindle extension can be considered a specific checkpoint response that contributes to tolerating DNA replication stress. Insight into this comes from a consideration of mrc1 mutants, which display an identical spindle extension phenotype in HU to rad53 strains (Alcasabas et al., 2001). A key difference, however, is that within 30 min HU-treated mrc1 mutants activate Rad53 due to rerouting the checkpoint signal through the DNA damage checkpoint mediator Rad9 (Alcasabas et al., 2001; Osborn and Elledge, 2003). mrc1 mutants survive HU challenge to a much greater extent than rad53 mutants (Alcasabas et al., 2001) and are able to reform normal preanaphase spindles and biorient duplicated CENs during HU recovery (J.B., unpublished observation). In contrast, rad53 strains fail in biorientation when HU is removed, a defect that persists despite eventual recovery of CEN and bulk genome duplication (Feng et al., 2009). These considerations suggest premature spindle extension in HU is not, in and of itself, necessarily an irrecoverable or lethal checkpoint defect. Rather, our data suggest that spindle extension in HU can be viewed as an early manifestation of a replication-associated perturbation to CEN function. If the underlying CEN defect is not ameliorated, it ultimately leads to terminal chromosome attachment and/or alignment errors.
S phase spindle structure
Two models have been proposed to explain how Rad53 prevents spindle extension in HU (Figure 10A, pathways 5 and 6). In one, checkpoint signaling down-regulates at least three spindle proteins (Cin8, Stu2, and Ase1; Krishnan et al., 2004; McKnight et al., 2014) that produce or contribute to MT-sliding forces that extend the central spindle (Hoyt et al., 1992; Severin et al., 2001a; Schuyler et al., 2003). Cin8 and Stu2 abundance was found to decline in HU, associated with reduced Cin8 and Stu2 stability and reduced STU2 transcript accumulation (Krishnan et al., 2004). This depletion required Mec1, suggesting a role for the checkpoint in limiting spindle extension. In a different study, it was found that HU-treated cells preferentially utilize an intragenic transcription start site within ASE1, resulting in an Ask1 isoform lacking MT bundling activity (McKnight et al., 2014). This regulation required Rad53, again suggesting checkpoint dampening of spindle extension. Regulation of gene expression through the Mec1-Rad53-Dun1 axis is complex, involving transcriptional and posttranscriptional mechanisms (Jaehnig et al., 2013; Corcoles-Saez et al., 2018; Gay et al., 2018), and specific checkpoint mechanisms controlling Cin8, Stu2, and Ase1 remain to be defined (Figure 10A, question marks in pathway 6). Rad53 inhibition of Ndd1 has been shown to down-regulate G2/M-expressed genes, including ASE1 (Edenberg et al., 2014; Yelamanchi et al., 2014); whether this is related to control of ASE1 intragenic transcription is unclear.
In comparison, the model arising from our previous work sought to explain the requirement for the K in stabilizing spindle structure in HU, leading us to propose that precocious amphitelic K attachments prevented spindle extension during an extended S phase (Bachant et al., 2005). Based on the statistical modeling presented here, at the time HU-treated rad53 cells exhibit spindle extension, ≥98% of WT cells will have duplicated at least 4–8 CENs. As proposed, biorientation of these duplicated CENs, likely in the context of ongoing replication rather than the C-loop, could contribute to spindle force balance. However, dbf4-∆/pdbf4-zn mutants assemble short spindles in HU even though reduced firing of CEN-proximal ORIs greatly delays CEN duplication. Moreover, in the case of rad53 mutants, the proximity of CENs to early ORIs clearly becomes a liability. Reducing ORI firing in the vicinity of CENs, or, conversely, placing AKs at late replicating sites, corresponds with rescue of spindle extension. In comparison, ddk mutants that are globally defective in ORI firing exhibit a reductional anaphase in HU. Thus, initiating DNA synthesis and CEN/K integrity, but not CEN duplication, are minimal preconditions to restrict HU spindle extension.
A revised model (Figure 10B and Supplemental Figure S9) to accommodate these findings arises from observations that the movement of interphase chromosomes becomes constrained in S phase (Heun et al., 2001), corresponding with coalescence of ORIs into large replication assemblages (Kitamura et al., 2006; Natsume et al., 2013). During this time, Ks remain coupled to MTs, detaching for 1–2 min as they spool through replication foci (Kitamura et al., 2007). On detachment, CENs recoil away from SPBs, and, following duplication and reattachment, are pulled back. These dynamics raise the possibility that replicating CENs are immobilized in a manner that provides resistance to K-MT pulling. Thus, as envisioned in Figure 10B and Supplemental Figure S9, as interdigitating MTs within the forming spindle push SPBs apart, a distribution of monopolar K-MT attachments to both poles could limit extension when S phase is slowed in HU, establishing a distinct force regime/length regulation setpoint for the early S phase spindle. Achieving this distribution would likely require Ipl1, consistent with the role for Ipl1 in preventing HU spindle extension we documented previously (Bachant et al., 2005). In addition to preventing fork catastrophes, a role for Rad53 could also be to stabilize the replication structures proposed to immobilize CENs (Meister et al., 2007). Furthermore, as resistance to extension may be limited, complementary checkpoint responses dampening extension could also be involved. As S phase continued in HU, spindle stability would become increasingly reliant on amphitelic K attachments, the tensile properties of the C-loop, and a checkpoint block to Pds1 proteolysis (Supplemental Figure S9). To our knowledge this is a novel proposal, with implications for budding yeast spindle and karyotype evolution. It suggests that the spindle is a remarkably flexible structure, where tensegrity principles can be accommodated through diverse mechanisms.
MATERIALS AND METHODS
Yeast strains and culture
All Saccharomyces cerevisiae strains were derived from the W303-related CRY1 strain (Allen et al., 1994) and are listed in Supplemental Table S6. A list of episomal and integrating plasmids used in this study is provided in Supplemental Table S7. Cells were cultured in standard formulations of yeast extract/peptone/dextrose (YPD) and synthetic complete minimal (SC) media, with 2% glucose or 2% galactose as a carbon source. For G1 synchronization, α-factor (Bio-Synthesis Corp.) was used at 10 μg/ml. For liquid media, HU was used at 200 mM and NZ was used at 15 μg/ml in YPD. HU and NZ were purchased from Sigma-Aldrich. The 5-fluoroorotic acid (5-FOA) was purchased from Biovectra/Fisher and used at 1 mg/ml. G418 was purchased from Mediatech/Fisher and used at 200 μg/ml in YPD.
Microscopy
Cultures for microscopy were supplemented with 50 μg/ml adenine to quench autofluorescence. For visualizing spindles by α-tubulin immunofluorescence, cells were fixed 4 h in 3.7% formaldehyde; permeabilized in 70% ethanol; spheroplasted for 1 h in 1.2 M sorbitol, 100 mM KPO4 (pH 7.5), and 50 μg/ml Zymolyase 100T at 37°C; and stained using a 1:50 dilution of YOL1/34 (Accurate Scientific) and a 1:100 dilution of FITC–conjugated α-rat antibodies (Sigma). 4’,6-Diamidino-2-phenylindole (DAPI) staining was performed using Vecta-Shield (Vector Laboratories) containing 10 μg/ml DAPI. To visualize GFP chromosome tags and Spc42-GFP, cells were fixed in 1% formaldehyde for 1.5 min and washed into phosphate-buffered saline. Mtw1-GFP and Tub1-GFP were visualized as live mounts. Cells were visualized on either Nikon E-800 or Nikon Eclipse 80i microscopes equipped with florescence optics and 100× (Plan Apo, 1.40 NA) or 60× (Plan Apo, 1.40 NA) objectives. Spindle length and bud circumference measurements were performed using the MetaMorph (Molecular Devices) suite of software tools. GFP chromosome tag and Mtw1-GFP intensity measurements were performed by first capturing images of specimens. A low-end mask was then applied using MetaMorph image processing features. The mask was adjusted until only the chromosome tag to be measured remained above the low-end threshold value. The fluorescent tag was then defined as a region, and the integrated intensity value of the region was used as the intensity measurement.
Flow cytometry
DNA content of 95% ethanol-fixed yeast samples was analyzed by prodium idodide staining, followed by flow cytometry using a Becton Dickinson FACSort instrument as previously described (Schober-Ditmore and Bachant, 2000).
ssDNA isolation, hybridization, and analysis
Genome-wide replication profiling by ssDNA labeling was performed as previously described (Peng et al., 2014). Briefly, cells were released from a G1 block (G1 control sample) into media containing 200 mM HU for 60 min (S phase sample). Cells from the G1 control and S phase samples were embedded in agarose plugs for spheroplasting, followed by Klenow (3′-5′ exo-)-assisted ssDNA labeling without denaturation of the template DNA. The labeling was done such that the DNA from the G1 and S samples were differentially decorated with Cy5– and Cy3–conjugated dUTPs, respectively, which were then pooled and cohybridized onto the Agilent 4 × 44 K ChIP-to-chip yeast microarray (G4493A). The ratio of fluorescence intensity from the S sample to that from the G1 sample was calculated and normalized as a “ratio of ssDNA” as previously described (Peng et al., 2014).
Superimposition of insertion sites onto the yeast 3C nuclear model
The coordinates from a three-dimensional model of the yeast genome (Duan et al., 2010) were downloaded from the W. Noble lab website (https://noble.gs.washington.edu/proj/yeast-architecture/sup.html). The coordinates for unchecked origins, checked origins, and K-insertions were transformed from the original atom designation, “C,” to “P,” “S,” and “H,” respectively, in the modified visualization script. Visualization was performed with the Rasmol software (windows version Raswin 2.7.5.1, www.openrasmol.org/OpenRasMol.html).
Construction and expression of dbf4 mutant alleles
The dbf4-C22, dbf4-D3, and dbf4-D45 alleles were isolated by screening a PCR mutagenized plasmid library for clones that could partially suppress the temperature sensitivity of dbf4-1 mutants (Khalil et al., 2007). The DBF4 insert in these constructs was PCR amplified using JB.83 and JB.84 and included 623 base pairs of upstream promoter sequence and 146 base pairs downstream of the DBF4 open reading frame:
JB.83: 5′-AGCTCCATGGCATTTTACTTCTCGCAGTACACCG-3′
JB.84: 5′-AGCTGTTAACAGTCAATAGCAAGAAAGTAACAAGGG-3′
Before screening the dbf4 library, in vivo Cre/lox recombination (Khalil et al., 2007) was used to batch fuse library clones with pHY314, a pCEN ARS TRP1 lox yeast episomal vector. The pdbf4-C22, pdbf4-D3, and pdbf4-D45 clones were subsequently mobilized to other plasmids, including the pCEN ARS LEU2 lox(dbf4)lox constructs used in a number of experiments described in the Results, through in vitro Cre/lox recombination (Liu et al., 1998) with target plasmid pJBN242 (pCEN ARS LEU2 lox(kanMX)lox).
The dbf4-zn allele was constructed by cleaving pJJ019 (pURA3 DBF4) at unique BspE1 and SwaI sites within the DBF4 open reading frame. The BspEI overhang was made flush with T4 DNA polymerase and ligated to the blunt end of SwaI via an intramolecular reaction. The resulting plasmid, pJJ022, contains an 86-base-pair in-frame deletion of the zinc finger region. The dbf4-zn was subsequently transferred to other yeast episomal vectors via standard subcloning procedures. The dbf4-zn was targeted for integration by cleaving pJJ022 (and pJJ019 as a DBF4 control) at a unique PspXI site within the promoter. The linearized fragments were looped in through homologous recombination at the corresponding promoter site in a ∆dbf4::kanMX6 strain, placing dbf4-zn (and DBF4) in the normal 5′ regulatory context of the endogenous locus.
Construction of AK and chromosomal GFP tags
Regions on chromosomes 2, 9, 10, and 13 were selected for AK insertion based on these sites being predicted to be maximally distant from unchecked ORIs that fire in both WT and rad53 mutant cells treated with HU (Supplemental Figures S6 and S7). Assembly of the four AKs was performed by targeting eight copies of the Lac operator sequence (LacO8X) as previously described (Lacefield et al., 2009). In each of the four LATE-AK constructs, PCR-amplified fragments (described below) corresponding to the identified insertion sites were cloned into pJBN291, a pLEU2-LacO8X integrating plasmid using SacI and XhoI sites incorporated into the PCR primers. Restriction enzyme sites used to linearize the LacO8X plasmids for targeting, as well as the structure of each recombinant, are diagrammed in Supplemental Figure S7. Additional information regarding sequences or PCR strategies used for verifying accurate insertion of each construct is available on request.
LATE2-AK.
A 742-base-pair fragment of chromosome 2 corresponding to base pairs 351419–352184 was amplified using JB.305 and JB.306 and cloned into pJBN291, yielding pJBN316, the final targeting plasmid:
JB.305: 5′-GTCTCTCGAGAATTGATCTATGTTGTAGCTGC-3′
JB.306: 5′-AAAGAGCTCTACTCATTATCGAGAACATATGGC-3′
LATE9-AK.
A 664-base-pair fragment of chromosome 9 corresponding to base pairs 380966–381606 was amplified using JB.307 and JB.308. After cloning into pJBN291 to yield pJBN317, AhdI/BssHII (blunt/blunt) and SacI/SacI junctions were used to subclone the targeting insert and LacO sequences into pAK047, yielding pJBN322, the final targeting plasmid. This second cloning step incorporates a kanMX marker for G418 resistance:
JB.307: 5′-TTTAGAGCTCGTAATATAAACTTCTCATATGGC-3′
JB.308: 5′-CTGAAGACTCGAGCTGAGTGAGCCG-3′
LATE10-AK.
A 548-base-pair fragment of chromosome 10 corresponding to base pairs 395468–395995 was amplified using JB.309 and JB.310. After cloning into pJBN291 to yield pJBN318, AhdI/AhdI and Sac/Sac junctions were used to subclone the targeting insert and LacO sequences into pJBN196, yielding the final targeting plasmid pJBN323. The second cloning step incorporates yeast ADE2 as a selectable marker:
JB.309: 5′-CTGACTCGAGGCATTTCAAGGATCAAAAATGCC-3′
JB.310: 5′-ACGTGAGCTCGATTTGGAAGCGCACTACAAGCG-3′
LATE13-AK.
A 578-base-pair fragment of chromosome 13 corresponding to base pairs 115353–115930 was amplified using JB.311 and JB.312. After cloning into pJBN291 to yield pJBN319, Sal/Xho and Sac/Sac junctions were used to subclone the targeting insert and LacO sequences into pRS406 (Sikorski and Hieter, 1989), yielding the final targeting plasmid pJBN324. The second cloning step incorporates yeast URA3 as a selectable marker:
JB.311: 5′-TCCACTCGAGAATTCAACCCTGAAGATCTTCCC-3′
JB.312: 5′-ACCAGAGCTCGTCAATTAGCAAGAATAGTTGCC-3′
To activate AK function, JB.303 and JB.304 were used to amplify an 899-base-pair region of the ASK1 gene. The PCR primers were positioned to remove the ASK1 start and stop codons, replacing them with unique XhoI and EcoRI sites, respectively. The ASK1 PCR fragment was then cloned into the XhoI and EcoRI sites of pAFS135 (Straight et al., 1997) to yield pJBN320, creating an in frame C-terminal fusion between ASK1 and the DNA binding domain of LacI. In pJBN320, the ASK1-LacI fusion is expressed under control of the HIS3 promoter, and a nuclear localization sequence (NLS) is additionally fused in frame at the 3′ end of the LacI sequence. A ScaI/XbaI fragment from pJBN320, containing entire (HIS3 promoter-ASK1-LacI-NLS) element was cloned into pRS413 (Sikorski and Hieter, 1989), yielding pJBN321, a low-copy episomal vector for expression of the ASK1-LacI-NLS fusion. pJBN321 fully complemented the growth defect associated with ask1-2 and ask1-3 temperature-sensitive mutants, indicating the ASK1 fusion encoded by pJBN321 is capable of providing ASK1 essential function:
JB.303: 5′-GACACAACTCGAGATGGATTCTGCAAGCAAAGAGG-3′
JB.304: 5′-GATCGAATTCTCTATTCGTAGAAAAATGAATGATGG-3′
LATE9-GFP and LATE10-GFP.
To create more robust GFP visualization of LATE9-GFP and LATE10-GFP regions than was provided by 8XLacO insertions, pAFS59 (Straight et al., 1997), a LEU2-marked integrating plasmid containing 256 copies of the LacO sequence, was transformed into strains harboring LATE9-AK-LacO8X-kanMX and LATE10-AK LacO8X-ADE2 insertions. Leu+ transformants were visually screened for robust GFP foci, and, for selected LATE9-GFP and LATE10-GFP transformants, the expected cosegregation of the linked kanMX-LEU2 and ADE2-LEU2 markers was verified by tetrad analysis. For all LacO chromosome tags, foci were visualized using an integrated GFP-LacI fusion construct under control of the HIS3 promoter as previously described (Straight et al., 1997).
CEN2-GFP.
A 916-base-pair fragment of chromosome 2 corresponding to base pairs 240348–241260 was amplified using JB.297 and JB.298. SacI and XhoI sites included on the primers were used to clone the PCR fragment into the corresponding sites of pJBN164, yielding pJBN334. As depicted in Supplemental Figure S7, pJBN334 was used to target LacO256X-LEU2 sequences 2552 base pairs to the right side of the SCG midpoint of CEN2:
JB.297: 5′-CCAGCTCGAGGATTAACGTGTTCTTCCATAGCC-3′
JB.298: 5′-TACCGAGCTCAACGGAAATAAATCCTCCATCCG-3′
CEN9-GFP.
A 770-base-pair fragment of chromosome 9 corresponding to base pairs 353395–354212 was amplified using JB.299 and JB.300. SacI and XhoI sites included on the primers were used to clone the PCR fragment into pJBN164, yielding pJBN335. pJBN335 was used to target LacO256X-LEU2 sequences 1884 base pairs to the left side of the SCG midpoint of CEN9 (Supplemental Figure S7):
JB.299: 5′-GAAGCTCGAGCAATCGACCGTGATCTTCTACCG-3′
JB.300: 5′-CATAGAGCTCAAGGGTATCTCTGATAGTATCGG-3′
CEN10-GFP.
A 711-base-pair fragment of chromosome 10 corresponding to base pairs 438082–438793 was amplified using JB.301 and JB.302. SacI and XhoI sites on the primers were used to clone the PCR fragment into pJBN164, yielding pJBN336. pJBN336 was used to target LacO256X-LEU2 1992 base pairs to the right side of the midpoint of CEN10 (Supplemental Figure S7):
JB.301: 5′-TGAACTCGAGCTATAAGATAACATCGGTTACGG-3′
JB.302: 5′-CAAAACACTGTTTCTTCAAGAGCTCATCGC-3′
Supplementary Material
Acknowledgments
Portions of this work were supported by a grant to J.B. from the California Cancer Research Coordinating Committee, a National Science Foundation award to C.N., and a National Institutes of Health Pathway to Independence Award (5R00GM08137805) to W.F. We thank D. Clarke for providing critical comments on an early draft of the manuscript and O. Aparicio for useful discussions. Thanks also go to S. Elledge, J. Lechner, A. Murray, B. Sclafani, P. Sorger, B. Stillman, and D. Toczyski for strains and reagents. This work is dedicated to the memory of Tim Ngo.
Abbreviations used:
- AK
artificial K
- AUC
area under the curve
- CEN
centromeric
- DDK
Dbf4-dependent kinase
- Fkh
Forkhead
- HU
hydroxyurea
- K
kinetochore
- MT
microtubule
- NLS
nuclear localization sequence
- NZ
nocodazole
- ORI
replication origin
- pre-RC
prereplication complex
- RNR
ribonucleotide reductase
- SAC
spindle assembly checkpoint
- SC
synthetic complete
- SPB
spindle pole body
- ssDNA
single-stranded DNA
- WT
wild type
- YPD
yeast extract/peptone/dextrose.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www .molbiolcell.org/cgi/doi/10.1091/mbc.E19-03-0156) on September 11, 2019.
REFERENCES
- Agarwal R, Tang Z, Yu H, Cohen-Fix O. (2003). Two distinct pathways for inhibiting pds1 ubiquitination in response to DNA damage. J Biol Chem , 45027–45033. [DOI] [PubMed] [Google Scholar]
- Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ. (2001). Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol , 958–965. [DOI] [PubMed] [Google Scholar]
- Allen JB, Zhou Z, Siede W, Friedberg EC, Elledge SJ. (1994). The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev , 2401–2415. [DOI] [PubMed] [Google Scholar]
- Almawi AW, Matthews LA, Larasati , Myrox P, Boulton S, Lai C, Moraes T, Melacini G, Ghirlando R, Duncker BP, et al. (2016). “AND” logic gates at work: crystal structure of Rad53 bound to Dbf4 and Cdc7. Sci Rep , 34237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachant J, Jessen SR, Kavanaugh SE, Fielding CS. (2005). The yeast S phase checkpoint enables replicating chromosomes to bi-orient and restrain spindle extension during S phase distress. J Cell Biol , 999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Jossen R, Colosio A, Frattini C, Carotenuto W, Cocito A, Doksani Y, Klein H, Gómez-González B, et al. (2011). The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell , 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck DC, Bloom K. (2007). Pericentric chromatin is an elastic component of the mitotic spindle. Curr Biol , 741–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousset K, Diffley JFX. (1998). The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev , 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. (1974). Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb Symp Quant Biol , 123–131. [DOI] [PubMed] [Google Scholar]
- Chappidi N, De Gregorio G, Ferrari S. (2019). Replication stress-induced Exo1 phosphorylation is mediated by Rad53/Pph3 and Exo1 nuclear localization is controlled by 14-3-3 proteins. Cell Div , 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Smolka MB, Zhou H. (2007). Mechanism of Dun1 activation by Rad53 phosphorylation in Saccharomyces cerevisiae. J Biol Chem , 986–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-C, Kenworthy J, Gabrielse C, Hanni C, Zegerman P, Weinreich M. (2013). DNA replication checkpoint signaling depends on a Rad53-Dbf4 N-terminal interaction in Saccharomyces cerevisiae. Genetics , 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. (1998). An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell , 1067–1076. [DOI] [PubMed] [Google Scholar]
- Clarke DJ, Segal M, Jensen S, Reed SI. (2001). Mec1p regulates Pds1p levels in S phase: complex coordination of DNA replication and mitosis. Nat Cell Biol , 619–627. [DOI] [PubMed] [Google Scholar]
- Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JF. (1996). An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature , 180–182. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. (1996). Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev , 3081–3093. [DOI] [PubMed] [Google Scholar]
- Colosio A, Frattini C, Pellicanò G, Villa-Hernández S, Bermejo R. (2016). Nucleolytic processing of aberrant replication intermediates by an Exo1-Dna2-Sae2 axis counteracts fork collapse-driven chromosome instability. Nucleic Acids Res , 10676–10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoles-Saez I, Dong K, Johnson AL, Waskiewicz E, Costanzo M, Boone C, Cha RS. (2018). Essential function of Mec1, the budding yeast ATM/ATR checkpoint-response kinase, in protein homeostasis. Dev Cell , 495–503.e2. [DOI] [PubMed] [Google Scholar]
- Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M. (2005). Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell , 153–159. [DOI] [PubMed] [Google Scholar]
- Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. (1998). Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev , 2956–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann PR, Sclafani RA. (2006). Novel role for checkpoint Rad53 protein kinase in the initiation of chromosomal DNA replication in Saccharomyces cerevisiae. Genetics , 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. (2010). A three-dimensional model of the yeast genome. Nature , 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncker BP, Shimada K, Tsai-Pflugfelder M, Pasero P, Gasser SM. (2002). An N-terminal domain of Dbf4p mediates interaction with both origin recognition complex (ORC) and Rad53p and can deregulate late origin firing. Proc Natl Acad Sci USA , 16087–16092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg ER, Vashisht A, Benanti JA, Wohlschlegel J, Toczyski DP. (2014). Rad53 downregulates mitotic gene transcription by inhibiting the transcriptional activator Ndd1. Mol Cell Biol , 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D, Lengronne A, Shi D, Forey R, Skrzypczak M, Ginalski K, Yan C, Wang X, Cao Q, Pasero P, et al. (2017). Dbf4 recruitment by forkhead transcription factors defines an upstream rate-limiting step in determining origin firing timing. Genes Dev , 2405–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Bachant J, Collingwood D, Raghuraman MK, Brewer BJ. (2009). Centromere replication timing determines different forms of genomic instability in saccharomyces cerevisiae checkpoint mutants during replication stress. Genetics , 1249–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Collingwood D, Boeck ME, Fox LA, Alvino GM, Fangman WL, Raghuraman MK, Brewer BJ. (2006). Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat Cell Biol , 148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Di Rienzi SC, Raghuraman MK, Brewer BJ. (2011). Replication stress-induced chromosome breakage is correlated with replication fork progression and is preceded by single-stranded DNA formation. G3 (Bethesda) , 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Raghuraman MK, Brewer BJ. (2007). Mapping yeast origins of replication via single-stranded DNA detection. Methods , 151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R, Putnam CW, Weinert T. (1999). RAD53, DUN1 and PDS1 define two parallel G2/M checkpoint pathways in budding yeast. EMBO J , 3173–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay S, Piccini D, Bruhn C, Ricciardi S, Soffientini P, Carotenuto W, Biffo S, Foiani M. (2018). A Mad2-mediated translational regulatory mechanism promoting S-phase cyclin synthesis controls origin firing and survival to replication stress. Mol Cell , 628–638.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CFJ, Dryga O, Seematter S, Pahl PMB, Sclafani RA. (1997). mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc Natl Acad Sci USA , 3151–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkins V, Gabrielse C, Haste L, Weinreich M. (2009). Budding yeast Dbf4 sequences required for Cdc7 kinase activation and identification of a functional relationship between the Dbf4 and Rev1 BRCT domains. Genetics , 1269–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH. (1976). Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J Mol Biol , 803–817. [DOI] [PubMed] [Google Scholar]
- Heun P, Laroche T, Shimada K, Furrer P, Gasser SM. (2001). Chromosome dynamics in the yeast interphase nucleus. Science , 2181–2186. [DOI] [PubMed] [Google Scholar]
- Hinshaw SM, Makrantoni V, Harrison SC, Marston AL. (2017). The Kinetochore Receptor for the Cohesin Loading Complex. Cell , 72–84.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzen TM, Sclafani RA. (2010). Genetic interaction of RAD53 protein kinase with histones is important for DNA replication. Cell Cycle , 4735–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. (1992). Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol , 109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Sun L, Shen F, Chen Y, Hua Y, Liu Y, Zhang M, Hu Y, Wang Q, Xu W, et al. (2012). The intra-S phase checkpoint targets Dna2 to prevent stalled replication forks from reversing. Cell , 1221–1232. [DOI] [PubMed] [Google Scholar]
- Hughes S, Elustondo F, Di Fonzo A, Leroux FG, Wong AC, Snijders AP, Matthews SJ, Cherepanov P. (2012). Crystal structure of human CDC7 kinase in complex with its activator DBF4. Nat Struct Mol Biol , 1101–1107. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Pahl PM, Harrison K, Rosamond J, Sclafani RA. (1993). Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol Cell Biol , 2899–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaehnig EJ, Kuo D, Hombauer H, Ideker TG, Kolodner RD. (2013). Checkpoint kinases regulate a global network of transcription factors in response to DNA damage. Cell Rep , 174–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S, Segal M, Clarke DJ, Reed SI. (2001). A novel role of the budding yeast separin Esp1 in anaphase spindle elongation: evidence that proper spindle association of Esp1 is regulated by Pds1. J Cell Biol , 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DR, Prasad AA, Chan PK, Duncker BP. (2010). The Dbf4 motif C zinc finger promotes DNA replication and mediates resistance to genotoxic stress. Cell Cycle , 2018–2026. [DOI] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K. (2003). S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature , 1078–1083. [DOI] [PubMed] [Google Scholar]
- Khalil A-M, Julius JA, Bachant J. (2007). One step construction of PCR mutagenized libraries for genetic analysis by recombination cloning. Nucleic Acids Res , e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A, Roostalu J, Roque H, Antony C, Schiebel E. (2009). Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation. Dev Cell , 244–256. [DOI] [PubMed] [Google Scholar]
- Kiermaier E, Woehrer S, Peng Y, Mechtler K, Westermann S. (2009). A Dam1-based artificial kinetochore is sufficient to promote chromosome segregation in budding yeast. Nat Cell Biol , 1109–1115. [DOI] [PubMed] [Google Scholar]
- Kitamura E, Blow JJ, Tanaka TU. (2006). Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell , 1297–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E, Tanaka K, Kitamura Y, Tanaka TU. (2007). Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev , 3319–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott SRV, Peace JM, Ostrow AZ, Gan Y, Rex AE, Viggiani CJ, Tavaré S, Aparicio OM. (2012). Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell , 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Nirantar S, Crasta K, Cheng AYH, Surana U. (2004). DNA replication checkpoint prevents precocious chromosome segregation by regulating spindle behavior. Mol Cell , 687–700. [DOI] [PubMed] [Google Scholar]
- Lacefield S, Lau DTC, Murray AW. (2009). Recruiting a microtubule-binding complex to DNA directs chromosome segregation in budding yeast. Nat Cell Biol , 1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianga N, Doré C, Kennedy EK, Yeh E, Williams EC, Fortinez CM, Wang A, Bloom KS, Rudner AD. (2018). Cdk1 phosphorylation of Esp1/Separase functions with PP2A and Slk19 to regulate pericentric Cohesin and anaphase onset. PLoS Genet , a1007029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Liang F, Jin F, Wang Y. (2008). The coordination of centromere replication, spindle formation, and kinetochore-microtubule interaction in budding yeast. PLoS Genet , e1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Li MZ, Leibham D, Cortez D, Elledge SJ. (1998). The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr Biol , 1300–1309. [DOI] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M. (2001). The DNA replication checkpoint response stabilizes stalled replication forks. Nature , 557–561. [DOI] [PubMed] [Google Scholar]
- Lopez-Mosqueda J, Maas NL, Jonsson ZO, DeFazio Eli LG, Wohlschlegel J, Toczyski DP. (2010). Damage-induced phosphorylation of Sld3 is important to block late origin firing. Nature , 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, McQueen J, Cuschieri L, Vogel J, Measday V. (2007). Spc24 and Stu2 promote spindle integrity when DNA replication is stalled. Mol Biol Cell , 2805–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantiero D, Mackenzie A, Donaldson A, Zegerman P. (2011). Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J , 4805–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Arai K. (2000). Dbf4 motifs: conserved motifs in activation subunits for Cdc7 kinases essential for S-phase. Biochem Biophys Res Commun , 228–232. [DOI] [PubMed] [Google Scholar]
- Matthews LA, Selvaratnam R, Jones DR, Akimoto M, McConkey BJ, Melacini G, Duncker BP, Guarné A. (2014). A novel non-canonical forkhead-associated (FHA) domain-binding interface mediates the interaction between Rad53 and Dbf4 proteins. J Biol Chem , 2589–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight K, Liu H, Wang Y. (2014). Replicative stress induces intragenic transcription of the ASE1 gene that negatively regulates Ase1 activity. Curr Biol , 1101–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P, Taddei A, Ponti A, Baldacci G, Gasser SM. (2007). Replication foci dynamics: replication patterns are modulated by S-phase checkpoint kinases in fission yeast. EMBO J , 1315–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morafraile EC, Diffley JFX, Tercero JA, Segurado M. (2015). Checkpoint-dependent RNR induction promotes fork restart after replicative stress. Sci Rep , 7886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannas NJ, O’Toole ET, Winey M, Murray AW. (2014). Chromosomal attachments set length and microtubule number in the Saccharomyces cerevisiae mitotic spindle. Mol Biol Cell , 4034–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume T, Müller CA, Katou Y, Retkute R, Gierliński M, Araki H, Blow JJ, Shirahige K, Nieduszynski CA, Tanaka TU. (2013). Kinetochores coordinate pericentromeric cohesion and early DNA replication by Cdc7-Dbf4 kinase recruitment. Mol Cell , 661–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas TA, Zhou Z, Elledge SJ. (1995). DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell , 29–39. [DOI] [PubMed] [Google Scholar]
- Naylor ML, Li J, Osborn AJ, Elledge SJ. (2009). Mrc1 phosphorylation in response to DNA replication stress is required for Mec1 accumulation at the stalled fork. Proc Natl Acad Sci USA , 12765–12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nougarède R, Della Seta F, Zarzov P, Schwob E. (2000). Hierarchy of S-phase-promoting factors: yeast Dbf4-Cdc7 kinase requires prior S-phase cyclin-dependent kinase activation. Mol Cell Biol , 3795–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn AJ, Elledge SJ. (2003). Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev , 1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palou R, Palou G, Quintana DG. (2017). A role for the spindle assembly checkpoint in the DNA damage response. Curr Genet , 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palou G, Palou R, Zeng F, Vashisht AA, Wohlschlegel JA, Quintana DG. (2015). Three different pathways prevent chromosome segregation in the presence of DNA damage or replication stress in budding yeast. PLoS Genet , e1005468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo B, Crabbé L, Pasero P. (2017). Signaling pathways of replication stress in yeast. FEMS Yeast Res , 10.1093/femsyr/fow101. [DOI] [PubMed] [Google Scholar]
- Peng J, Raghuraman MK, Feng W. (2014). Analysis of ssDNA gaps and DSBs in genetically unstable yeast cultures. Methods Mol Biol , 501–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S, Lengauer C, Nasmyth K. (1995). Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a “reductional” anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J , 3788–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli J, Tsaponina O, Crabbé L, Keszthelyi A, Pantesco V, Chabes A, Lengronne A, Pasero P. (2012). dNTP pools determine fork progression and origin usage under replication stress. EMBO J , 883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL. (2001). Replication dynamics of the yeast genome. Science , 115–121. [DOI] [PubMed] [Google Scholar]
- Randell JCW, Fan A, Chan C, Francis LI, Heller RC, Galani K, Bell SP. (2010). Mec1 is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7. Mol Cell , 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi SE, Ajazi A, Carotenuto W, Foiani M, Giannattasio M. (2015). Rad53-mediated regulation of Rrm3 and Pif1 DNA helicases contributes to prevention of aberrant fork transitions under replication stress. Cell Rep , 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Bachant J, Wang H, Hu F, Liu D, Tetzlaff M, Elledge SJ. (1999). Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science , 1166–1171. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ. (1996). Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science , 357–360. [DOI] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF. (1998). A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature , 615–618. [DOI] [PubMed] [Google Scholar]
- Saunders W, Lengyel V, Hoyt MA. (1997). Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol Biol Cell , 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober-Ditmore W, Bachant J. (2000). Yeast DNA flow cytometry. In: In Living Color: Protocols in Flow Cytometry and Cell Sorting, ed. Diamond RA, DeMaggio, New York S: Springer Science & Business Media, 455–460. [Google Scholar]
- Schuyler SC, Liu JY, Pellman D. (2003). The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J Cell Biol , 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin F, Habermann B, Huffaker T, Hyman T. (2001a). Stu2 promotes mitotic spindle elongation in anaphase. J Cell Biol , 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin F, Hyman AA, Piatti S. (2001b). Correct spindle elongation at the metaphase/anaphase transition is an APC-dependent event in budding yeast. J Cell Biol , 711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu Y-J, Stillman B. (2010). The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature , 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. (1998). Regulation of DNA-replication origins during cell-cycle progression. Nature , 618–621. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics , 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M. (2002). Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science , 599–602. [DOI] [PubMed] [Google Scholar]
- Stephens AD, Haase J, Vicci L, Taylor RM, 2nd, Bloom K. (2011). Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J Cell Biol , 1167–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, Murray AW. (1997). Mitosis in living budding yeast: anaphase A but no metaphase plate. Science , 574–578. [DOI] [PubMed] [Google Scholar]
- Stueland CS, Lew DJ, Cismowski MJ, Reed SI. (1993). Full activation of p34CDC28 histone H1 kinase activity is unable to promote entry into mitosis in checkpoint-arrested cells of the yeast Saccharomyces cerevisiae. Mol Cell Biol , 3744–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H. (2011). Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr Biol , 2055–2063. [DOI] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJR, Nasmyth K. (2002). Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell , 317–329. [DOI] [PubMed] [Google Scholar]
- Warsi TH, Navarro MS, Bachant J. (2008). DNA topoisomerase II is a determinant of the tensile properties of yeast centromeric chromatin and the tension checkpoint. Mol Biol Cell , 4421–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert TA, Kiser GL, Hartwell LH. (1994). Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev , 652–665. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Guacci V, Koshland D. (1996). Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s). J Cell Biol , 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS. (2008). Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol , 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelamanchi SK, Veis J, Anrather D, Klug H, Ammerer G. (2014). Genotoxic stress prevents Ndd1-dependent transcriptional activation of G2/M-specific genes in Saccharomyces cerevisiae. Mol Cell Biol , 711–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JFX. (2010). Checkpoint dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature , 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ren P, Vashisht AA, Wohlschlegel JA, Quintana DG, Zeng F. (2017). Cdk1-interacting protein Cip1 is regulated by the S phase checkpoint in response to genotoxic stress. Genes Cells , 850–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Rothstein R. (2002). The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc Natl Acad Sci USA , 3746–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Nellimoottil T, Peace JM, Knott SRV, Villwock SK, Yee JM, Jancuska JM, Rege S, Tecklenburg M, Sclafani RA, et al. (2013). The level of origin firing inversely affects the rate of replication fork progression. J Cell Biol , 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Elia AEH, Naylor ML, Dephoure N, Ballif BA, Goel G, Xu Q, Ng A, Chou DM, Xavier RJ, et al. (2016). Profiling DNA damage-induced phosphorylation in budding yeast reveals diverse signaling networks. Proc Natl Acad Sci USA , E3667–E3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Elledge SJ. (1993). DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell , 1119–1127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.