Abstract
Eukaryotic genomes are hierarchically organized into protein-DNA assemblies for compaction into the nucleus. Nucleosomes, with the (H3-H4)2 tetrasome as a likely intermediate, are highly dynamic in nature by way of several different mechanisms. We have recently shown that tetrasomes spontaneously change the direction of their DNA wrapping between left- and right-handed conformations, which may prevent torque buildup in chromatin during active transcription or replication. DNA sequence has been shown to strongly affect nucleosome positioning throughout chromatin. It is not known, however, whether DNA sequence also impacts the dynamic properties of tetrasomes. To address this question, we examined tetrasomes assembled on a high-affinity DNA sequence using freely orbiting magnetic tweezers. In this context, we also studied the effects of mono- and divalent salts on the flipping dynamics. We found that neither DNA sequence nor altered buffer conditions affect overall tetrasome structure. In contrast, tetrasomes bound to high-affinity DNA sequences showed significantly altered flipping kinetics, predominantly via a reduction in the lifetime of the canonical state of left-handed wrapping. Increased mono- and divalent salt concentrations counteracted this behavior. Thus, our study indicates that high-affinity DNA sequences impact not only the positioning of the nucleosome but that they also endow the subnucleosomal tetrasome with enhanced conformational plasticity. This may provide a means to prevent histone loss upon exposure to torsional stress, thereby contributing to the integrity of chromatin at high-affinity sites.
Significance
Canonical (H3-H4)2 tetrasomes possess high conformational flexibility as evidenced by their spontaneous flipping between states of left- and right-handed DNA wrapping. Here, we show that these conformational dynamics of tetrasomes cannot be described by a fixed set of rates over all conditions. Instead, an accurate description of their behavior must take into account details of their loading, in particular, the underlying DNA sequence. In vivo, differences in tetrasome flexibility could be regulated by modifications of the histone core or the tetrasomal DNA and as such constitute an intriguing, potentially adjustable mechanism for chromatin to accommodate the torsional stress generated by processes such as transcription and replication.
Introduction
The nucleosome is the basic complex of chromatin in eukaryotic cells (1, 2, 3). It comprises 147 basepairs (bp) of DNA wrapped around a disk-shaped assembly of eight histone proteins in ∼1.7 turns (4, 5, 6). The histone octamer consists of two copies of two types of heterodimers, one of which is formed by histones H3 and H4 and the other by histones H2A and H2B (7, 8). During nucleosome formation, first the two H3-H4 dimers assemble onto the DNA to form a tetrasome, after which the binding of two H2A/H2B dimers completes the full nucleosome (9). The resulting compaction upon DNA wrapping limits the accessibility of the genome for essential cellular processes, such as transcription, replication, and repair. Hence, the positioning and stability of nucleosomes play a key role in gene regulation and cellular function.
Recent research has provided significant new insights into the structure and especially the dynamics of nucleosomes (10). For example, studies using single-molecule techniques have revealed the intrinsically dynamic nature of nucleosomes in the form of “breathing” (i.e., the transient un- and rewrapping of the nucleosomal DNA ends) (11, 12, 13, 14). Nucleosomes were also found to “gap” by transiently opening and closing the two turns of nucleosomal DNA along the direction of the superhelical axis (15). The structure, stability, and dynamics of nucleosomes have furthermore been shown to be extensively regulated by post-translational modifications (16), chaperones (17), and ATP-dependent remodelers (18). Furthermore, nucleosome composition and dynamics can be altered by forces and torques generated by molecular motors that process the genome (19, 20). Such external influences, as well as changes in the ambient conditions, can cause nucleosomes to reorganize into different (sub)structures (21). Specifically, tetrasomes, which wrap ∼80 bp of DNA around the (H3-H4)2 tetramer (Fig. 1 A), have been observed as stable intermediates in several studies (22, 23, 24, 25, 26, 27, 28). Similar to nucleosomes, tetrasomes predominantly adopt a left-handed DNA wrapping, but interestingly, right-handed DNA wrapping has also been identified (29, 30, 31, 32, 33, 34). Recently, we showed that tetrasomes can spontaneously switch between these two conformations without unbinding from the DNA (35, 36, 37) (Fig. 2 A). This conformational flexibility could mitigate the buildup of torsional stress in chromatin during transcription or replication and thereby prevent the eviction of histones from the DNA.
Figure 1.
Real-time assembly of single tetrasomes onto DNA with a 601 sequence. (A) Shown is the top view on ∼80 bp of DNA (dark gray) bound to a tetrameric protein core consisting of the histones H3 (orange) and H4 (cyan). This image was created by modifying the structural data of the Drosophila nucleosome from the Protein Data Bank (PDB) with the PDB identification code 2PYO (82) using the PyMOL Molecular Graphics System, Version 1.8 (Schrödinger, New York, NY). (B) Experimental assay based on FOMT is shown (49). A single DNA construct (black) containing a 601 sequence (red) at its center (DNAw/601) is attached to a coverslip (light blue) at one end and tethered to a superparamagnetic bead (light brown) at the other end. Above the flow cell, a cylindrically shaped permanent magnet (dark blue/red), with its axis precisely aligned with the DNA tether, exerts a constant force (F) on the bead while allowing its free rotation in the (x,y) plane (indicated by the black circular arrow). This enables the direct measurement of the DNA molecule’s length z and linking number Θ, which upon the assembly of a tetrasome (orange), are changed by Δz and ΔΘ (indicated by the red straight and circular arrows, respectively). Tetrasome assembly is induced by flushing in either (H3.1-H4)2 tetramers alone or preincubated histone/NAP1 complexes. Nonmagnetic beads attached to the flow cell surface serve as reference for drift correction. This figure is adapted from (37) (https://doi.org/10.1063/1.5009100), with the permission of AIP Publishing (Melville, NY). (C) Shown are partial time traces of the length z (in nm, top panel) and the linking number Θ (in turns, bottom panel) of a DNAw/601 molecule before and upon the assembly of a (H3.1-H4)2 tetrasome. The formation of a tetrasome simultaneously decreased both quantities in the form of a step identified using a step-finder algorithm (red lines) (Materials and Methods). About 60 s after assembly, free proteins were flushed out with measurement buffer (orange arrows) to prevent further histone binding. In this particular experiment, a tetrasome was assembled by flushing in NAP1/histone complexes (green arrows) in buffer A (Table 1). A corresponding time trace of a DNAw/601 molecule upon the assembly of a tetrasome from histone tetramers only is shown in Fig. S4. (D) Shown is a histogram of the changes in DNA length upon assembly and disassembly Δz(dis)ass (in nm, top panel) of single (H3.1-H4)2 tetrasomes (N = 27) and in DNA linking number upon assembly and disassembly ΔΘ(dis)ass (in turns, bottom panel) of single (H3.1-H4)2 tetrasomes (N = 27) in all buffers (Table 1), shown together with the mean spatial resolution corresponding to the average 1 SD (16 nm and 0.5 turns; green lines) from all experiments. Data beyond the resolution limits (hatched area) were excluded from the determination of the mean change Δz(dis)ass = 23 ± 5 nm (n = 25) and ΔΘ(dis)ass = 0.9 ± 0.2 turns (n = 26). To see this figure in color, go online.
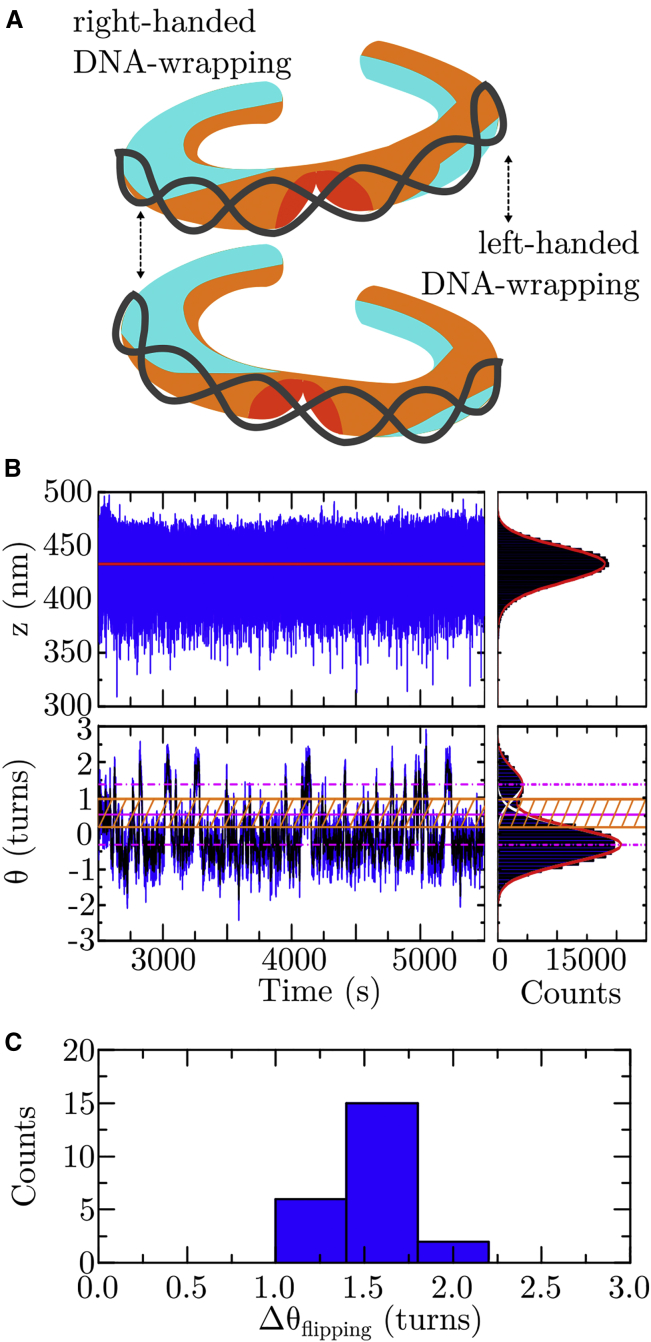
Figure 2.
The structural dynamics of single tetrasomes on DNA with a 601 sequence. (A) Representation of the two tetrasome conformations with the DNA wrapped in either a left-handed or a right-handed superhelix. Tetrasomes were observed to spontaneously flip between these two states (35, 36, 37). This figure is adapted from (37) (https://doi.org/10.1063/1.5009100), with the permission of AIP Publishing. (B) Shown are partial time traces of the length z (in nm, top panel) and the linking number Θ (in turns, bottom panel) of a DNA molecule after the assembly of a (H3.1-H4)2 tetrasome in buffer A (Table 1) upon flushing in histone/NAP1 complexes. As indicated from the fit by the step-finder algorithm to the time trace (red line, left panel) and the fit of a mirrored γ function (red line in histogram plot, right panel) to the skewed data, the DNA length remains constant. The DNA linking number spontaneously fluctuates (i.e., “flips”) between two states identified by fitting two Gaussian functions (white lines in histogram plot, right panel) underlying the full profile (red line in histogram plot, right panel). The two states correspond to a prevalent left-handed and a less adopted right-handed conformation of DNA wrapping, with the respective mean linking numbers Θleft = −0.31 ± 0.01 turns and Θright = +1.38 ± 0.06 turns (dashed-dotted magenta lines, 95% confidence level for estimated values). Because of drift, the mean value for Θleft obtained here is offset from the average change in DNA linking number upon tetrasome dis-/assembly (Fig. 1D, bottom panel). The structural dynamics were quantified in terms of the dwell times in the two states based on a threshold zone (hatched orange area) that is bounded by 1 SD from each mean value (orange solid lines) about their average (solid magenta line) (Materials and Methods). A corresponding partial time trace of a DNAw/601 molecule upon the assembly of a tetrasome from histone tetramers only is shown in Fig. S7. (C) Shown is a histogram of the change in DNA linking number ΔΘflipping (in turns) upon flipping of single (H3.1-H4)2 tetrasomes in their handedness of DNA wrapping in all buffers (Table 1). The data yield a mean value of ΔΘflipping = 1.6 ± 0.2 turns (N = 23). The individual values are provided in Table S9. To see this figure in color, go online.
DNA sequence has been suggested to play an important role in determining the structural and dynamic properties of nucleosomes (38). The formation of a nucleosome is significantly affected by the ability of the underlying DNA to bend sharply (39, 40). This feature has not only been examined in numerous in vitro studies (41), but it has also been observed at a global level in vivo (42). For example, in yeast, it was shown that ∼50% of the in vivo nucleosome positions can be attributed to sequence-specific features. In particular, high-affinity nucleosome binding sequences were found to be enriched in intergenic regions where an integral chromatin structure is important, whereas low affinity sequences were most dominant in highly transcribed regions, such as ribosomal RNA and transfer RNA genes (42). High-affinity sequences were also detected in strongly regulated genes, and it was proposed that such sequences may favor rapid reassembly of nucleosomes promoting repression of these genes under unfavorable conditions (42).
Although the structure and dynamics of high-affinity nucleosomes have also been studied extensively by in vitro single-molecule assays (43, 44, 45, 46, 47, 48), an interesting open question is whether association of histones with high-affinity DNA sequences affects the dynamics of subnucleosomal structures, such as (H3-H4)2 tetrasomes, which can arise in the course of transcription or other torque-generating processes. In this work, we have addressed this question by a single-molecule approach. Using freely orbiting magnetic tweezers (FOMT) (49), we systematically investigated the structure and handedness dynamics of single (H3.1-H4)2 tetrasomes assembled onto the well-characterized high-affinity Widom 601 nucleosome binding sequence (50). In addition, we have examined the contribution of different buffers commonly employed in chromatin studies (51, 52, 53) to the dynamics of high-affinity tetrasomes. Our results revealed intriguing effects of DNA sequence on handedness flipping dynamics that can be counteracted by mono- and divalent salts. Thus, our findings provide conceptual evidence for a model in which chromatin regions containing high-affinity nucleosomes may be better protected than other regions from complete histone loss during torque-generating processes, such as transcription or replication, because of inherently increased handedness flipping dynamics of (H3-H4)2 tetrasomes.
Materials and Methods
Preparation of DNA molecules
Tetrasome assembly was performed on linear double-stranded DNA fragments of 1.96 kilo-bp length containing a single 601 sequence of 147 bp at their center. The two ends of the DNA fragments were ligated to digoxigenin-coated (Roche Diagnostics, Basel, Switzerland) or biotin-coated (Roche Diagnostics) double-stranded fragments (handle) of 643-bp length, respectively, for immobilization and tethering. The schematics, sequence, and preparation of the DNA molecules are described in Fig. S1 and Table S1. Further details on the handles have been described in (37).
Expression and purification of proteins
The recombinant canonical Drosophila histones H3.1-H4 and nucleosome assembly protein 1 (NAP1) chaperones were expressed and purified as described in (35, 37). Whereas in vivo NAP1 has been identified as a chaperone for histones H2A and H2B (54), in vitro, it has been found to also interact with histones H3 and H4 and prevent histone aggregation (35, 36, 37, 55, 56, 57, 58, 59, 60).
Preparation of histones and tetrasome assembly
Individual tetrasomes were assembled and monitored in real time using magnetic tweezers (see below) in three buffers with varying compositions of core components employed in previous studies (35, 36, 37, 51, 52, 53) (Table 1). To allow for direct comparison to our previous study (35), the protein samples were prepared similarly by incubating 51 nM of an equimolar solution of H3.1-H4 histones—either without or with 192 nM NAP1—on ice for 30 min. The incubation buffers were identical to the measurement buffers (Table 1), except for buffer A. In that case, the incubation buffer contained 0.25% (w/v) polyethylene glycol (PEG), 0.25% (w/v) polyvinyl alcohol (PVA), and 0.1% (w/v) bovine serum albumin (BSA), as in our previous study (35). The incubated protein solutions were then diluted by at most 1:5000, and 100 μL of the diluted solution was flushed into the flow cell to assemble a single tetrasome. About 60 s after assembly, 100–300 μL of measurement buffer was flushed through the flow cell to remove free proteins and to prevent further assembly of tetrasomes.
Table 1.
Composition of the Measurement Buffers Used in This Work
| Measurement buffers | Buffer Components |
|---|---|
| A [Standard] | 25 mM HEPES-KOH (pH 7.5) (Sigma-Aldrich; St. Louis, MO) |
| 0.1 mM EDTA (Sigma-Aldrich) | |
| 50 mM KCl (Merck, Darmstadt, Germany) | |
| 0.01% (w/v) BSA (Sigma-Aldrich) | |
| 0.025% (w/v) PEG (Sigma-Aldrich) | |
| 0.025% (w/v) PVA (Sigma-Aldrich) | |
| B | 10 mM HEPES (pH 7.5) 100 mM KCl 0.2% (w/v) BSA |
| 0.1% (w/v) Tween-20 (Sigma-Aldrich) 10 mM NaN3 (Merck) | |
| C | 10 mM HEPES (pH 7.5) 100 mM KCl 0.2% (w/v) BSA |
| 0.1% (w/v) Tween-20 10 mM NaN3 2 mM MgCl2 |
The details of the sample and flow cell preparation for magnetic tweezers experiments have been described in (37). Where the concentrations and volumes of DNA and magnetic bead solutions used in this work differ, this is stated in Table S2. Five (out of total Nexp, total = 20) experiments were performed by resuming the measurement on a tetrasome that did not disassemble during a preceding experiment. Two such experiments involved an exchange between buffers B and C (Table 1) with 300–500 μL of the respective measurement buffer. In one of these two experiments, a measurement was resumed in buffer C on a tetrasome that had been assembled before in buffer B, and in the other, the reverse procedure was employed.
Magnetic tweezers instrumentation
The assembly and dynamics of an individual tetrasome was monitored for up to 10 h by directly measuring the length and angular position (i.e., the linking number) of a single DNA molecule in each experiment using FOMT (49) (Fig. 1 B). The magnetic tweezers hardware is detailed in (37). All experiments were performed at an exerted force of 0.6–0.7 pN and at room temperature (22°C).
Data analysis
The analysis of the acquired data was performed using custom-written scripts with built-in functions in MATLAB (The MathWorks, Natick, MA). An improved version of the custom-written step-finder algorithm described in (61) was used to detect stepwise changes in the time traces of the DNA length and DNA linking number. Subsequently, these fits were analyzed to identify simultaneous steps in both DNA length and DNA linking number within a time window of 19.1 s (see below), resulting from the assembly or disassembly of a tetrasome (Fig. 1 C; Fig. S4). With this approach, 48% (n = 13) of all such steps (N = 27) were automatically identified, whereas the remaining 52% (n = 14) were corrected manually to better match the data. The retrieved sizes of these simultaneous changes in DNA length (N = 27) and DNA linking number (N = 27) form the basic quantities that characterize tetrasome structure (Tables S3 and S4). Step sizes were also compared to the mean spatial resolution, determined by averaging the SDs of the time trace profiles in all experiments (Fig. 1 D; Fig. S5).
The dynamics of tetrasome structure in terms of handedness flipping (Fig. 2 A) was separately analyzed in the parts of the time traces with a stable baseline in DNA length and DNA linking number (Fig. 2 B; Fig. S7). The change in DNA linking number upon flipping was determined by fitting a double Gaussian function to the DNA linking number data (N = 23; Figs. 2 B and S7; Table S9) and calculating the difference between the mean values. The probability of a tetrasome to occupy one of the two states was calculated from the relative ratio of the corresponding peak areas of the two Gaussian fits (Table S11).
The handedness dynamics of single, flipping (H3.1-H4)2 tetrasomes was further analyzed in terms of the times spent in the left- or right-handed state using another custom-written algorithm based on (62). A threshold zone was defined by 1 SD from the mean values of the two Gaussian distributions about their average (Fig. 2 B; Fig. S7). The details of the dwell-time analysis have been described in (37). Here, the corresponding DNA linking number data of the time traces of sufficiently long duration (>1600 s) were smoothed by filtering over N = 330 (N = 1910) points, corresponding to a time average of τshort = 3.3 s (τlong = 19.1 s). These timescales were obtained from autocorrelation analysis of the bead’s angular fluctuations, as described in (37, 49). (Fig. S2). The mean dwell time in each state was then determined by combining all data sets obtained in identical buffer conditions and fitting these to an exponential function (Figs. 3, A–C and S8, A–C; Tables 2 and S12). For direct comparison, we also performed dwell-time analysis with the same settings on the partial time traces (N = 6) of one of our earlier experiments published in (35). In that experiment, we note that three tetrasomes were assembled, whereas mainly a single tetrasome was flipping between the two conformations. Because a second tetrasome only flipped with a relative occurrence of 2.8% and 0.7% in two of the partial time traces, all dwell times in the right-handed state were assigned to one flipping tetrasome, yielding a mean dwell time of τD,right = 14 (+/−1) s (N = 161). Because tetrasomes flip independently of one another, the mean dwell time of a single tetrasome in the left-handed state was obtained by multiplying the mean dwell time from the exponential fit by three, yielding τD,left = 183 (+15/–12) s (N = 159).
Figure 3.
Kinetics and energetics of single tetrasomes on DNA with a 601 sequence. (A) Shown are histograms of the dwell times of a single NAP1-loaded (H3.1-H4)2 tetrasome on a DNAw/601 molecule in the left- (left panel) and right-handed conformation (right panel) in buffer A (Tables 1 and 2). Exponential fits (red lines) yielded a mean dwell time of τD,left = 37 ± 2 s (N = 371) and τD,right = 28 + 2/−1 s (N = 373), respectively (68% confidence level for estimated values). (B) is the same as in (A) but now in buffer B (Tables 1 and 2). Exponential fits (red lines) yielded a mean dwell time of τD,left = 62 ± 3 s (N = 559) and τD,right = 33 ± 1 s (N = 564), respectively (68% confidence level for estimated values). (C) Shown are histograms of the dwell times of a single NAP1-loaded (H3.1-H4)2 tetrasome on a DNAw/601 molecule in the left-handed (top panel) and right-handed conformation (bottom panel) in buffer C (Tables 1 and 2). Exponential fits (red lines) yielded a mean dwell time of τD,left = 141 + 11/−10 s (N = 187) and τD,right = 30 ± 2 s (N = 188), respectively (68% confidence level for estimated values). All data in (A)–(C) were obtained by dwell-time analysis of the DNA linking number time traces filtered by averaging over 3.3 s (N = 330) (Materials and Methods). (D) Schematic energy diagrams of single NAP1-loaded tetrasomes on DNAw/601 in all buffer conditions are based on the dwell-time values and the probabilities obtained from the linking number distributions (Tables 2, S11, and S13–S15). The solid black lines illustrate the energy levels in buffer A (Table 1) for a NAP1-loaded (H3.1-H4)2 tetrasome on DNArandom, whereas the solid blue lines depict the energetics for a NAP1-loaded (H3.1-H4)2 tetrasome on DNAw/601. The green and orange dashed lines show the energy levels for a NAP1-loaded (H3.1-H4)2 tetrasome on DNAw/601 in buffers B and C, respectively. The free energy differences (ΔE) between the left- and right-handed conformations of tetrasomes on DNAw/601, with the respective energies Eleft and Eright, are considerably decreased compared to tetrasomes loaded onto DNArandom (Table 2). The heights of the energy barriers ΔG∗left and ΔG∗right are estimated from the rates kl->r and kr->l, respectively. For tetrasomes on DNAw/601 relative to DNArandom in buffer A, ΔG∗left decreases strongly, whereas ΔG∗right is largely unchanged. In the presence of mono- and divalent salts in buffers B and C, tetrasomes on DNAw/601 become considerably longer lived in the left-handed conformation (i.e., ΔG∗left is higher) compared with tetrasomes on DNAw/601 in buffer A, whereas the right-handed state (hence ΔG∗right) remains essentially unaltered. To see this figure in color, go online.
Table 2.
Summary of Quantified Properties for NAP1-Loaded Tetrasomes
| Quantity | (H3.1-H4)2 Tetrasomes on DNArandom | (H3.1-H4)2 Tetrasomes on DNAw/601 |
|---|---|---|
| Δz(dis)ass (nm) | 24 (±3)a | 23 (±5)b |
| Δθ(dis)ass (turns) | 0.73 (±0.05)a | 0.9 (±0.2)b |
| Δθflipping (turns) | 1.7 (±0.1)a | 1.6 (±0.2)b |
| τD,left (s) with time average in filtering of 3.3 s [N = 330] | 183 (+15/−12)c [A] | 37 (±2)d [A] |
| 62 (±3)d [B] | ||
| 141 (+11/−10)d [C] | ||
| τD,left (s) with time average in filtering of 19.1 s [N = 1910] | 417 (+60/−48)c [A] | 67 (+5/−4)d [A] |
| 121 (+8/−7)d [B] | ||
| 231 (+24/−20)d [C] | ||
| τD,right (s) with time average in filtering of 3.3 s [N = 330] | 14 (±1) [A] | 28 (+2/−1)d [A] |
| 33 (±1)d [B] | ||
| 30 (±2)d [C] | ||
| τD,right (s) with time average in filtering of 19.1 s [N = 1910] | 34 (+5/−4)d [A] | 48 (+4/−3)d [A] |
| 64 (±4)d [B] | ||
| 44 (+5/−4)d [C] | ||
| τD,left/τD,right with time average in filtering of 3.3 s [N = 330] | 13 (±1)c [A] | 1 (±0)c [A] |
| 2 (±0)c [B] | ||
| 4 (±0)c [C] | ||
| τD,left/τD,right with time average in filtering of 19.1 s [N = 1910] | 12 (±3)c [A] | 1 (±0)c [A] |
| 2 (±0)c [B] | ||
| 5 (±1)c [C] | ||
| ΔEdwell-times (kBT) | 2.6 (±0.1)c [A] | 0.3 (±0.1)c [A] |
| 0.6 (±0.1)c [B] | ||
| 1.5 (±0.1)c [C] | ||
| ΔEpeak areas (kBT) | – | 0.9 (±0.7)c [A] |
| 1.4 (±0.8)c [B] | ||
| 1.7 (±—–) [C] |
Specific buffers employed are indicated within brackets. The results for spontaneously loaded tetrasomes are provided in Table S12.
Values taken from (35).
Values obtained for spontaneously loaded tetrasomes on DNAw/601 are included for improved statistics.
Errors calculated by error propagation.
Errors correspond to 68% confidence interval for estimated values from exponential fits.
Further details of the data analysis have been described in (37). The significance of the similarity between the structural quantities obtained in the different conditions was assessed by respective unpaired two-sample t-tests (Tables S5–S8 and S10). The results for the dwell times obtained in the different conditions were checked for similarity by respective Wilcoxon rank-sum tests (Tables S13–S15). Finally, we note that the times measured in this study are an upper boundary due to the finite bead response time. The errors reported for the mean values obtained in this study correspond to 1 SD from the underlying distributions, unless indicated otherwise. The errors of calculated quantities were computed by error propagation.
Construction of energy diagrams
To construct the schematic energy diagrams of the left- and right-handed states of tetrasomes (Fig. 3 D; Document S1. Supporting Results and Discussion, Figs. S1–S8, Tables S1–S15, Document S2. Article plus Supporting Material), we have computed the free energy difference between them and estimated the height of the barrier separating them. The free energy difference can be calculated from the ratio of the probabilities pleft = p and pright = 1−p for a tetrasome to be in the left- and right-handed conformations, respectively, using ΔEpeak areas = −kBTln((1−p)/p) (see Data analysis above, and Table S11). It can alternatively be computed from the ratio of the dwell times according to ΔEdwell-times = −kBTln(τD,right/τD,left). The obtained results are summarized in Table 2 and Table S12. Although the results from both approaches agree within error, we consider the energy differences calculated from the dwell times as more reliable because of the larger sample size. To estimate the height of the energy barrier ΔG∗ relative to the left- and right-handed states of tetrasomes, we compute ΔG∗ = −kBTln(k/k0), where the transition rates kl->r and kr->l equal the inverse of the dwell times τD,left and τD,right, respectively, and the transition rate between the two states at zero force k0 is assumed to be symmetric and estimated at ∼107 s−1 (37).
Results
DNA sequence and buffer conditions do not affect the structural properties of tetrasomes
To assess whether high-affinity DNA sequences impact the formation and structure of tetrasomes, we loaded (H3.1-H4)2 tetramers onto individual DNA molecules containing a single centrally positioned high-affinity 601 sequence (DNAw/601) known for its strong nucleosome-positioning capability (50) with the help of the histone chaperone NAP1. Using FOMT (49), tetrasome assembly was recorded in real time by tracking the length z and linking number Θ of the DNA. In this setting, a single immobilized DNA molecule is tethered to a magnetic bead that is subject to constant force applied by a cylindrical permanent magnet, allowing its free rotation in the (x,y) plane (Fig. 1 B). This force was set to 0.6–0.7 pN for direct comparison to our previous study of (H3.1-H4)2 tetrasomes assembled by histone chaperone NAP1 on DNA molecules of random sequence (DNArandom) (35).
As in our previous studies with DNArandom (35, 36, 37), flushing in preincubated histone/NAP1 complexes (nexp, subset = 9 out of total Nexp, total = 20) induced a simultaneous step-like change in both the length z and linking number Θ of DNAw/601, indicating the assembly of a single tetrasome (Fig. 1 C). The mean values of these changes (Table S3) are in excellent agreement with those previously obtained for NAP1-loaded (H3.1-H4)2 tetrasomes on DNArandom (35). Also in agreement with previous observations (35, 36, 37) is the lack of observable interactions between NAP1 chaperones and DNA molecules in the absence of histones (Fig. S3). To probe the effects of ambient conditions on tetrasome structure and dynamics, we performed the experiments in three different buffers employed in prior studies (35, 36, 37, 51, 52, 53) that mainly differed in the concentrations of mono- and divalent salts (Table 1) and obtained highly similar results (Table S3). We also flushed in (H3.1-H4)2 tetramers in the absence of NAP1 (nexp,subset= 11). Thereby, DNAw/601 also exhibited a simultaneous step-like change in both measured quantities (Fig. S4). The mean values of the changes in length z and linking number Θ (Table S3) correspond excellently to those we obtained for (H3.1-H4)2 tetrasomes loaded by NAP1 onto DNAw/601 or onto DNArandom (35). The similarity of these results, which were furthermore independent of buffer conditions, indicate the formation of proper complexes in all cases. This was unexpected because we never observed tetrasome formation in the absence of NAP1 in previous studies using random DNA sequences (35, 36, 37). Thus, the specific features of the high-affinity 601 sequence are capable of inducing spontaneous chaperone-independent assembly of histone tetramers.
Under the highly diluted conditions required for the controlled assembly of single tetrasomes (Materials and Methods), we also observed disassembly of tetrasomes in 40% (nexp,subset = 8) of all experiments (Nexp,total = 20) within 3364 ± 765 s (mean ± 1 SE; Fig. S6). The absolute mean values of the disassembly-associated changes in DNA length z and linking number Θ (Table S4) are in excellent agreement with those obtained upon assembly of tetrasomes (Table S3). Because disassembly occurred for both NAP1-loaded and spontaneously loaded tetrasomes in all buffer conditions, we can exclude a destabilizing effect of NAP1 and/or specific buffer conditions.
Given the similarity of the absolute changes in DNA length z and linking number Θ in all experiments in different buffer conditions (Nexp, total = 20) (as assessed by respective t-tests, Tables S5–S8), all results were grouped together to yield improved statistics. The absolute changes in DNA length z and DNA linking number Θ upon the assembly and disassembly of single tetrasomes (Ncounts,total = 27) yielded mean values of Δz(dis)ass = 23 ± 5 nm (ncounts,subset= 25) and ΔΘ(dis)ass = 0.9 ± 0.2 turns (ncounts,subset= 26), respectively (Fig. 1 D). The distributions of the changes upon the assembly or disassembly alone are shown in Fig. S5, A and B, respectively. These results are in excellent agreement with values reported in other studies involving tetrasomes (25, 30, 31, 32, 33, 34, 35, 36, 37). Overall, our findings indicate that the high-affinity 601 sequence fully supports the assembly of (H3.1-H4)2 tetramers into proper tetrasome complexes, irrespective of the presence or absence of NAP1 or of buffer conditions.
High-affinity tetrasomes exhibit spontaneous DNA handedness flipping
To investigate the time-dependent behavior of tetrasomes stably bound to DNAw/601, the FOMT measurements were continued for several hours following assembly. Like stably bound tetrasomes on DNArandom (35, 36, 37), high-affinity tetrasomes exhibited spontaneous dynamics in the handedness of their DNA wrapping (Fig. 2 A), irrespective of the buffer conditions. Whereas the length z of the DNA molecules remained constant, the linking number Θ spontaneously fluctuated (or flipped) between two distinct mean values (Fig. 2 B; Fig. S7). Based on the negative and positive signs of these values, we deduce that they correspond to conformations of left- and right-handed DNA wrapping, respectively. As observed with tetrasomes on DNArandom (35, 36, 37), tetrasomes on DNAw/601 most frequently adopted the left-handed conformation. The difference in mean DNA linking number between the two observed states quantifies the induced change in tetrasome structure. The values of this quantity obtained under different experimental conditions, including tetrasomes loaded in the absence of NAP1, are summarized in Table S9. Because of the similarity of their values (as assessed by respective t-tests, Table S10), the results from all experiments were pooled, which yielded a mean change in DNA linking number Θ upon the flipping of ΔΘflipping = 1.6 ± 0.2 turns (Ncounts,total = 23) (Fig. 2 C). These values are in excellent agreement with previous studies (35, 36, 37). Overall, our findings indicate that DNA sequence, the presence or absence of NAP1 during assembly, or buffer conditions do not affect the structural rearrangement of tetrasomes associated with handedness flipping.
High-affinity DNA sequences increase the kinetics of tetrasome handedness flipping
We further examined the handedness dynamics of single tetrasomes in terms of the times spent in each of the two states of DNA wrapping (Materials and Methods). To specifically examine the role of DNA sequence, we focused these studies on NAP1-assembled tetrasomes, comparing the dynamics of NAP1-assembled (H3.1-H4)2 tetramers on either DNAw/601 or DNArandom. The details of tetrasome handedness dynamics upon spontaneous, NAP1-independent assembly can be found in the Supporting Results and Discussion and in Fig. S8. Our results indicate that the underlying DNA sequence significantly alters the kinetics of tetrasome handedness flipping (as assessed by respective Wilcoxon rank-sum tests, Tables S13–S15). This can be deduced from the observation that a tetrasome on DNAw/601 spent an average time of τleft = 37 s in the left-handed state (in standard buffer A, Fig. 3 A, left), which is 4.9 ± 0.5-fold shorter than a tetrasome assembled onto DNArandom in the same buffer (τleft = 183 s, Table 2). The right-handed state was affected to a lesser extent, displaying a lifetime τright = 28 s (in standard buffer A, Fig. 3 A, right), which is 2.0 ± 0.3 times longer than a tetrasome on DNArandom in the same buffer (τright = 14 s, Table 2). These results clearly show that tetrasomes on high-affinity DNA sequences have a substantially enhanced tendency to flip from the left-to the right-handed state compared to tetrasomes on random DNA sequences (Table S13). Because the experimental conditions were otherwise identical, these effects must arise from the 601 sequence in DNAw/601.
We can translate the information from dwell-time measurements into schematic energy diagrams depicting the left- and right-handed states of tetrasomes (Materials and Methods; Fig. 3 D; Table 2). Notably, the free energy differences ΔE between the left- and right-handed conformations of tetrasomes are reduced ∼ninefold on DNAw/601 (ΔE = 0.3 ± 0.1 kBT) relative to DNArandom (ΔE = 2.6 ± 0.1 kBT), reflecting the shift of the equilibrium occupancy toward the right-handed conformation for NAP1-loaded tetrasomes on DNAw/601. This results predominantly from the smaller estimated energy difference between the left-handed state and the transition state (at the top of the barrier) for tetrasomes loaded onto DNAw/601 (ΔG∗left ∼19.7 kBT) relative to DNArandom (ΔG∗left ∼21.3 kBT).
We next examined the flipping dynamics of tetrasomes on DNAw/601 in two different buffer conditions. In buffer B (containing a twofold increased concentration of monovalent salt in the absence of crowding agents, Table 1), tetrasomes dwelled in the left-handed state for τleft = 62 s (Fig. 3 B, left), 1.7 ± 0.1 times longer than in standard buffer A (τleft = 37 s, Fig. 3 A; Table 2, left). The average lifetimes in the right-handed conformation were affected only slightly by the change to buffer B (Fig. 3 B, right; Tables 2, S14, and S15). In buffer C, which contained additional magnesium chloride relative to buffer B (Table 1), a tetrasome dwelled in the left-handed conformation for an even longer time of τleft = 141 s (Fig. 3 C, top), which is 2.3 ± 0.2-fold longer than in buffer B (τleft = 62 s, Fig. 3 B; Table 2, left). Again, the mean dwell time in the right-handed state showed essentially no change (Fig. 3 C, bottom; Fig. 3 B, right).
These findings can also be transferred into schematic energy diagrams (Materials and Methods; Fig. 3 D). Because the predominant effect of increased monovalent salt concentration in buffer B for tetrasomes on DNAw/601 is an increase in the lifetime of the left-handed conformation, the free energy difference between the two states increases to ΔE = 0.6 ± 0.1 kBT (compared to ΔE = 0.3 ± 0.1 kBT in buffer A; Table 2). The estimated barrier height relative to the left-handed state is altered to ΔG∗left ∼20.2 kBT, whereas the barrier height relative to the right-handed state remains essentially unaffected. Similarly, because the higher concentration of divalent ions in buffer C caused a further extension of the lifetime of the left-handed conformation, the free energy difference between the two states correspondingly increases to ΔE = 1.5 ± 0.1 kBT (Table 2). Thus, the estimated barrier height relative to the left-handed state for tetrasomes on DNAw/601 is larger in buffer C (ΔG∗left ∼21.1 kBT) than in buffer B (ΔG∗left ∼20.2 kBT), whereas the barrier height relative to the right-handed state remains essentially unaffected. In summary, higher concentrations of monovalent salt in the absence of crowding agents as well as divalent salts attenuate the handedness flipping dynamics of tetrasomes assembled on the high-affinity Widom 601 sequence by increasing the dwell time in the left-handed state and thus reducing the tendency for tetrasomes to flip to the right-handed state.
Discussion
A strong component determining the positioning and stability of nucleosomes throughout chromatin is the DNA sequence (63). For instance, nucleosome maps obtained from chromatin assembled in vitro on purified yeast genomic DNA showed a remarkable similarity to nucleosome maps from native chromatin (64). The observation that specific periodic patterns of AA, TT, TA, or GC dinucleotides favor DNA wrapping around the histone core led to the development of algorithms to predict nucleosome positions in different organisms (42, 65, 66). High-affinity nucleosome binding sequences have been found to be enriched at the transcriptional start site, in intragenic locations, or over genes subject to strong regulation (42). In this study, we have investigated to what extent such high-affinity sites affect the structure and dynamics of subnucleosomal (H3-H4)2 tetrasomes. Our previous single-molecule experiments revealed that the left-handed wrapping of DNA around the histone octamer is predominantly static, whereas the absence of H2A-H2B dimers results in highly dynamic flipping of DNA handedness between the left- and right-handed state (35, 36, 37). However, these studies were carried out on tetrasomes assembled onto random DNA sequences, and it remained unclear whether high-affinity sequences, exemplified by the well-characterized Widom 601 sequence (50), would have an impact on the dynamics of tetrasomes. Our assessment of the assembly and structural features of (H3-H4)2 tetrasomes on DNA including the 601 sequence revealed that the key characteristics of assembled tetrasomes were very similar to those assembled onto random DNA (35, 36, 37). Interestingly, however, we observed that the 601 sequence also supported spontaneous, histone chaperone-independent formation of long-lived, stable tetrasomes. As this property strongly distinguishes DNA including the high-affinity 601 sequence from DNA of random sequence, this suggests that proper tetrasome assembly occurs at the 601 sequence under these conditions. Such spontaneous chaperone-independent in situ assembly of histone tetramers may facilitate sample preparation in future single-molecule experiments. In the following, we assume that a similar sequence specificity holds for the chaperoned assembly of tetrasomes onto DNA including the 601 sequence.
In contrast to the overall structural similarity between tetrasomes assembled onto high-affinity versus random DNA sequences, our analyses revealed a striking difference with respect to handedness flipping dynamics. The presence of the 601 sequence considerably decreased the lifetime of the canonical left-handed state of high-affinity tetrasomes, corresponding to a reduction in the free energy ΔE between the left- and right-handed conformations, which results in enhanced flipping frequency.
These results may at first sight seem surprising given the high propensity of the 601 sequence to support nucleosome/tetrasome formation relative to other DNA sequences (67), which presumably derives from a combination of enhanced binding to histones and increased bendability or twistability of the DNA (38, 50). Indeed, our observation that histone tetramers assemble spontaneously onto DNA molecules with a 601 sequence but not onto DNA molecules of random sequence emphasizes the strong affinity of the sequence for histones. Yet what could be the reason for the increased flipping dynamics of high-affinity tetrasomes? A possible explanation of these findings is that the stronger binding of the histone tetramer to the DNA reduces the impact of thermal forces on the histone-DNA contacts, such as possible disruption (in analogy to nucleosome “breathing” (11, 12, 13, 14)), and instead transmits them to the H3-H3 interface. This could in turn enhance the rotation at the H3-H3 interface that is proposed to underlie tetrasome handedness flipping (30, 31, 32, 33, 34, 35, 36, 37). In this model, high-affinity sequences might not only promote energetically favorable histone-DNA interactions but may also endow tetrasomes with an enhanced ability to accommodate torsional stress by flipping between left- and right-handed conformation. This feature may be particularly important during transcription, when tetrasomes can arise from nucleosomes by the loss or removal of H2A/H2B dimers and when torque can be generated by elongating RNA polymerase II (68, 69, 70). Although most transcription-linked torsional stress is removed by topoisomerases (71, 72), high-affinity tetrasomes may provide extra protection against complete histone ejection at crucial locations in the genome by their inherently stronger ability to absorb this stress.
In the presence of higher concentrations of mono- and divalent salt and an absence of synthetic crowding agents, the left-handed states of NAP1-loaded tetrasomes on DNA molecules with a 601 sequence become longer lived. Hence, these conditions decrease the tendency of tetrasomes to adopt the right-handed conformation. We consider a direct influence of the synthetic crowding agents PEG and PVA on tetrasome dynamics to be unlikely because studies show that the influence of PEG (73) or PVA (74) on the kinetics of biological systems is negligible at the low concentrations (below 1%) employed in our standard buffer. Thus, these changes must arise from the higher concentration of salts, which will reduce the strength of electrostatic interactions through screening. Even a twofold increase in salt concentration as employed here can induce small but significant changes in the properties of DNA (e.g., elasticity (75, 76, 77), torsional stiffness (78), and supercoiling (79)) and proteins (80) and, in the case of histones, reduce their affinity for DNA (81). In agreement with the model proposed above, such a generic reduction in affinity could account for the observed increase of the lifetime of the left-handed state for tetrasomes on DNA molecules with a 601 sequence because it would shift the balance between a thermally induced opening of DNA-histone contacts and dimer rotation at the H3-H3 interface toward the former. Upon the further addition of divalent salt, which increases electrostatic screening to an even larger extent, the dwell time in the left-handed state of NAP1-loaded tetrasomes on DNA molecules with a 601 sequence becomes more pronounced, as one might expect by analogy with the above. These findings also illustrate that the interpretation of results obtained using nucleosome-positioning sequences must take into account the specific buffer conditions employed.
In summary, we provide conceptual evidence that the DNA sequence impacts tetrasome handedness flipping dynamics, which may contribute to the mitigation of torsional stress generated by molecular motors without accompanying the eviction of histones.
Author Contributions
O.O., A.L., and N.H.D. designed the research. O.O. performed the research and analyzed the data. O.O., A.L., and N.H.D. discussed the results and wrote the article.
Acknowledgments
The authors thank Artur Kaczmarczyk for insightful discussions, the laboratory of John van Noort for providing the pGEM3Z-mono601 plasmid and several stock solutions, Theo van Laar for preparing the DNA constructs, and Jacob Kerssemakers for providing the improved step-finder algorithm.
The authors also acknowledge financial support from the funding agencies mentioned below. This work was funded from the European Research Council with a Consolidator Grant DynGenome (No:312221) to N.H.D and by the Austrian Science Fund (FWF) P31377–B30 to A.L.
Editor: Massa Shoura.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.07.055.
Supporting Material
References
- 1.Kornberg R.D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 2.Olins A.L., Olins D.E. Spheroid chromatin units (v bodies) Science. 1974;183:330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- 3.Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975;4:281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- 4.Richmond T.J., Finch J.T., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984;311:532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- 5.Luger K., Mäder A.W., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 6.Davey C.A., Sargent D.F., Richmond T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 7.Klug A., Rhodes D., Thomas J.O. A low resolution structure for the histone core of the nucleosome. Nature. 1980;287:509–516. doi: 10.1038/287509a0. [DOI] [PubMed] [Google Scholar]
- 8.Arents G., Burlingame R.W., Moudrianakis E.N. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc. Natl. Acad. Sci. USA. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polo S.E., Almouzni G. Chromatin assembly: a basic recipe with various flavours. Curr. Opin. Genet. Dev. 2006;16:104–111. doi: 10.1016/j.gde.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 10.Ordu O., Lusser A., Dekker N.H. Recent insights from in vitro single-molecule studies into nucleosome structure and dynamics. Biophys. Rev. 2016;8(Suppl 1):33–49. doi: 10.1007/s12551-016-0212-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li G., Levitus M., Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 12.Koopmans W.J., Brehm A., van Noort J. Single-pair FRET microscopy reveals mononucleosome dynamics. J. Fluoresc. 2007;17:785–795. doi: 10.1007/s10895-007-0218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyagi A., Ando T., Lyubchenko Y.L. Dynamics of nucleosomes assessed with time-lapse high-speed atomic force microscopy. Biochemistry. 2011;50:7901–7908. doi: 10.1021/bi200946z. [DOI] [PubMed] [Google Scholar]
- 14.Wei S., Falk S.J., Lee T.H. A novel hybrid single molecule approach reveals spontaneous DNA motion in the nucleosome. Nucleic Acids Res. 2015;43:e111. doi: 10.1093/nar/gkv549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ngo T.T., Ha T. Nucleosomes undergo slow spontaneous gaping. Nucleic Acids Res. 2015;43:3964–3971. doi: 10.1093/nar/gkv276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowman G.D., Poirier M.G. Post-translational modifications of histones that influence nucleosome dynamics. Chem. Rev. 2015;115:2274–2295. doi: 10.1021/cr500350x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurard-Levin Z.A., Quivy J.P., Almouzni G. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu. Rev. Biochem. 2014;83:487–517. doi: 10.1146/annurev-biochem-060713-035536. [DOI] [PubMed] [Google Scholar]
- 18.Clapier C.R., Cairns B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 19.Lavelle C., Praly E., Croquette V. Nucleosome-remodelling machines and other molecular motors observed at the single-molecule level. FEBS J. 2011;278:3596–3607. doi: 10.1111/j.1742-4658.2011.08280.x. [DOI] [PubMed] [Google Scholar]
- 20.Lavelle C. Pack, unpack, bend, twist, pull, push: the physical side of gene expression. Curr. Opin. Genet. Dev. 2014;25:74–84. doi: 10.1016/j.gde.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Andrews A.J., Luger K. Nucleosome structure(s) and stability: variations on a theme. Annu. Rev. Biophys. 2011;40:99–117. doi: 10.1146/annurev-biophys-042910-155329. [DOI] [PubMed] [Google Scholar]
- 22.Sollner-Webb B., Camerini-Otero R.D., Felsenfeld G. Chromatin structure as probed by nucleases and proteases: evidence for the central role of histones H3 and H4. Cell. 1976;9:179–193. doi: 10.1016/0092-8674(76)90063-5. [DOI] [PubMed] [Google Scholar]
- 23.Eickbush T.H., Moudrianakis E.N. The histone core complex: an octamer assembled by two sets of protein-protein interactions. Biochemistry. 1978;17:4955–4964. doi: 10.1021/bi00616a016. [DOI] [PubMed] [Google Scholar]
- 24.Dong F., van Holde K.E. Nucleosome positioning is determined by the (H3-H4)2 tetramer. Proc. Natl. Acad. Sci. USA. 1991;88:10596–10600. doi: 10.1073/pnas.88.23.10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brower-Toland B.D., Smith C.L., Wang M.D. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc. Natl. Acad. Sci. USA. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall M.A., Shundrovsky A., Wang M.D. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat. Struct. Mol. Biol. 2009;16:124–129. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Böhm V., Hieb A.R., Langowski J. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 2011;39:3093–3102. doi: 10.1093/nar/gkq1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheinin M.Y., Li M., Wang M.D. Torque modulates nucleosome stability and facilitates H2A/H2B dimer loss. Nat. Commun. 2013;4:2579. doi: 10.1038/ncomms3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson V. Preferential binding of histones H3 and H4 to highly positively coiled DNA. Biochemistry. 1995;34:10607–10619. doi: 10.1021/bi00033a036. [DOI] [PubMed] [Google Scholar]
- 30.Hamiche A., Carot V., Prunell A. Interaction of the histone (H3-H4)2 tetramer of the nucleosome with positively supercoiled DNA minicircles: potential flipping of the protein from a left- to a right-handed superhelical form. Proc. Natl. Acad. Sci. USA. 1996;93:7588–7593. doi: 10.1073/pnas.93.15.7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamiche A., Richard-Foy H. The switch in the helical handedness of the histone (H3-H4)2 tetramer within a nucleoprotein particle requires a reorientation of the H3-H3 interface. J. Biol. Chem. 1998;273:9261–9269. doi: 10.1074/jbc.273.15.9261. [DOI] [PubMed] [Google Scholar]
- 32.Alilat M., Sivolob A., Prunell A. Nucleosome dynamics. Protein and DNA contributions in the chiral transition of the tetrasome, the histone (H3-H4)2 tetramer-DNA particle. J. Mol. Biol. 1999;291:815–841. doi: 10.1006/jmbi.1999.2988. [DOI] [PubMed] [Google Scholar]
- 33.Sivolob A., Prunell A. Nucleosome dynamics V. Ethidium bromide versus histone tails in modulating ethidium bromide-driven tetrasome chiral transition. A fluorescence study of tetrasomes on DNA minicircles. J. Mol. Biol. 2000;295:41–53. doi: 10.1006/jmbi.1999.3301. [DOI] [PubMed] [Google Scholar]
- 34.Sivolob A., De Lucia F., Prunell A. Nucleosome dynamics. VI. Histone tail regulation of tetrasome chiral transition. A relaxation study of tetrasomes on DNA minicircles. J. Mol. Biol. 2000;295:55–69. doi: 10.1006/jmbi.1999.3302. [DOI] [PubMed] [Google Scholar]
- 35.Vlijm R., Lee M., Dekker N.H. Nucleosome assembly dynamics involve spontaneous fluctuations in the handedness of tetrasomes. Cell Rep. 2015;10:216–225. doi: 10.1016/j.celrep.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 36.Vlijm R., Lee M., Dekker C. Comparing the assembly and handedness dynamics of (H3.3–H4)2 tetrasomes to canonical tetrasomes. PLoS One. 2015;10:e0141267. doi: 10.1371/journal.pone.0141267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ordu O., Kremser L., Dekker N.H. Modification of the histone tetramer at the H3-H3 interface impacts tetrasome conformations and dynamics. J. Chem. Phys. 2018;148:123323. doi: 10.1063/1.5009100. [DOI] [PubMed] [Google Scholar]
- 38.Widom J. Role of DNA sequence in nucleosome stability and dynamics. Q. Rev. Biophys. 2001;34:269–324. doi: 10.1017/s0033583501003699. [DOI] [PubMed] [Google Scholar]
- 39.Sekinger E.A., Moqtaderi Z., Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 40.Anderson J.D., Widom J. Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Mol. Cell. Biol. 2001;21:3830–3839. doi: 10.1128/MCB.21.11.3830-3839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trifonov E.N., Nibhani R. Review fifteen years of search for strong nucleosomes. Biopolymers. 2015;103:432–437. doi: 10.1002/bip.22604. [DOI] [PubMed] [Google Scholar]
- 42.Segal E., Fondufe-Mittendorf Y., Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blosser T.R., Yang J.G., Zhuang X. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009;462:1022–1027. doi: 10.1038/nature08627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gansen A., Valeri A., Seidel C.A. Nucleosome disassembly intermediates characterized by single-molecule FRET. Proc. Natl. Acad. Sci. USA. 2009;106:15308–15313. doi: 10.1073/pnas.0903005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kruithof M., Chien F.T., van Noort J. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat. Struct. Mol. Biol. 2009;16:534–540. doi: 10.1038/nsmb.1590. [DOI] [PubMed] [Google Scholar]
- 46.Hodges C., Bintu L., Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325:626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bintu L., Ishibashi T., Bustamante C. Nucleosomal elements that control the topography of the barrier to transcription. Cell. 2012;151:738–749. doi: 10.1016/j.cell.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ngo T.T., Zhang Q., Ha T. Asymmetric unwrapping of nucleosomes under tension directed by DNA local flexibility. Cell. 2015;160:1135–1144. doi: 10.1016/j.cell.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lipfert J., Wiggin M., Dekker N.H. Freely orbiting magnetic tweezers to directly monitor changes in the twist of nucleic acids. Nat. Commun. 2011;2:439. doi: 10.1038/ncomms1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lowary P.T., Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 51.Meng H., Andresen K., van Noort J. Quantitative analysis of single-molecule force spectroscopy on folded chromatin fibers. Nucleic Acids Res. 2015;43:3578–3590. doi: 10.1093/nar/gkv215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaczmarczyk A., Allahverdi A., van Noort J. Single-molecule force spectroscopy on histone H4 tail-cross-linked chromatin reveals fiber folding. J. Biol. Chem. 2017;292:17506–17513. doi: 10.1074/jbc.M117.791830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaczmarczyk A., Brouwer T.B., van Noort J. Probing chromatin structure with magnetic tweezers. In: Lyubchenko Y.L., editor. Nanoscale Imaging: Methods and Protocols. Springer; 2018. pp. 297–323. [DOI] [PubMed] [Google Scholar]
- 54.Zlatanova J., Seebart C., Tomschik M. Nap1: taking a closer look at a juggler protein of extraordinary skills. FASEB J. 2007;21:1294–1310. doi: 10.1096/fj.06-7199rev. [DOI] [PubMed] [Google Scholar]
- 55.Vlijm R., Smitshuijzen J.S., Dekker C. NAP1-assisted nucleosome assembly on DNA measured in real time by single-molecule magnetic tweezers. PLoS One. 2012;7:e46306. doi: 10.1371/journal.pone.0046306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fujii-Nakata T., Ishimi Y., Kikuchi A. Functional analysis of nucleosome assembly protein, NAP-1. The negatively charged COOH-terminal region is not necessary for the intrinsic assembly activity. J. Biol. Chem. 1992;267:20980–20986. [PubMed] [Google Scholar]
- 57.Nakagawa T., Bulger M., Ito T. Multistep chromatin assembly on supercoiled plasmid DNA by nucleosome assembly protein-1 and ATP-utilizing chromatin assembly and remodeling factor. J. Biol. Chem. 2001;276:27384–27391. doi: 10.1074/jbc.M101331200. [DOI] [PubMed] [Google Scholar]
- 58.Mazurkiewicz J., Kepert J.F., Rippe K. On the mechanism of nucleosome assembly by histone chaperone NAP1. J. Biol. Chem. 2006;281:16462–16472. doi: 10.1074/jbc.M511619200. [DOI] [PubMed] [Google Scholar]
- 59.Andrews A.J., Chen X., Luger K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol. Cell. 2010;37:834–842. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McBryant S.J., Park Y.J., Luger K. Preferential binding of the histone (H3-H4)2 tetramer by NAP1 is mediated by the amino-terminal histone tails. J. Biol. Chem. 2003;278:44574–44583. doi: 10.1074/jbc.M305636200. [DOI] [PubMed] [Google Scholar]
- 61.Kerssemakers J.W., Munteanu E.L., Dogterom M. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- 62.Lipfert J., Skinner G.M., Dekker N.H. Double-stranded RNA under force and torque: similarities to and striking differences from double-stranded DNA. Proc. Natl. Acad. Sci. USA. 2014;111:15408–15413. doi: 10.1073/pnas.1407197111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eslami-Mossallam B., Schiessel H., van Noort J. Nucleosome dynamics: sequence matters. Adv. Colloid Interface Sci. 2016;232:101–113. doi: 10.1016/j.cis.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 64.Kaplan N., Moore I.K., Segal E. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van der Heijden T., van Vugt J.J., van Noort J. Sequence-based prediction of single nucleosome positioning and genome-wide nucleosome occupancy. Proc. Natl. Acad. Sci. USA. 2012;109:E2514–E2522. doi: 10.1073/pnas.1205659109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang C., Pugh B.F. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thåström A., Lowary P.T., Widom J. Measurement of histone-DNA interaction free energy in nucleosomes. Methods. 2004;33:33–44. doi: 10.1016/j.ymeth.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 68.Teves S.S., Henikoff S. Transcription-generated torsional stress destabilizes nucleosomes. Nat. Struct. Mol. Biol. 2014;21:88–94. doi: 10.1038/nsmb.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kouzine F., Gupta A., Levens D. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat. Struct. Mol. Biol. 2013;20:396–403. doi: 10.1038/nsmb.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cole H.A., Ocampo J., Clark D.J. Heavy transcription of yeast genes correlates with differential loss of histone H2B relative to H4 and queued RNA polymerases. Nucleic Acids Res. 2014;42:12512–12522. doi: 10.1093/nar/gku1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baranello L., Wojtowicz D., Levens D. RNA polymerase II regulates topoisomerase 1 activity to favor efficient transcription. Cell. 2016;165:357–371. doi: 10.1016/j.cell.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Levens D., Baranello L., Kouzine F. Controlling gene expression by DNA mechanics: emerging insights and challenges. Biophys. Rev. 2016;8:259–268. doi: 10.1007/s12551-016-0216-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dupuis N.F., Holmstrom E.D., Nesbitt D.J. Molecular-crowding effects on single-molecule RNA folding/unfolding thermodynamics and kinetics. Proc. Natl. Acad. Sci. USA. 2014;111:8464–8469. doi: 10.1073/pnas.1316039111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ariga O., Sano T., Sano Y. Effect of polyvinyl alcohol on kinetic and thermal properties of β-galactosidase. J. Ferment. Bioeng. 1992;74:120–122. [Google Scholar]
- 75.Baumann C.G., Smith S.B., Bustamante C. Ionic effects on the elasticity of single DNA molecules. Proc. Natl. Acad. Sci. USA. 1997;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wenner J.R., Williams M.C., Bloomfield V.A. Salt dependence of the elasticity and overstretching transition of single DNA molecules. Biophys. J. 2002;82:3160–3169. doi: 10.1016/S0006-3495(02)75658-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Manning G.S. The persistence length of DNA is reached from the persistence length of its null isomer through an internal electrostatic stretching force. Biophys. J. 2006;91:3607–3616. doi: 10.1529/biophysj.106.089029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kriegel F., Ermann N., Lipfert J. Probing the salt dependence of the torsional stiffness of DNA by multiplexed magnetic torque tweezers. Nucleic Acids Res. 2017;45:5920–5929. doi: 10.1093/nar/gkx280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schlick T., Li B., Olson W.K. The influence of salt on the structure and energetics of supercoiled DNA. Biophys. J. 1994;67:2146–2166. doi: 10.1016/S0006-3495(94)80732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Damodaran S., Kinsella J.E. The effects of neutral salts on the stability of macromolecules. A new approach using a protein-ligand binding system. J. Biol. Chem. 1981;256:3394–3398. [PubMed] [Google Scholar]
- 81.Rippe K., Mazurkiewicz J., Kepper N. Interactions of histones with DNA: nucleosome assembly, stability and dynamics. In: Dias R., Lindman B., editors. DNA Interactions with Polymers and Surfactants. John Wiley & Sons, Inc.; 2008. pp. 135–172. [Google Scholar]
- 82.Clapier C.R., Chakravarthy S., Müller C.W. Structure of the Drosophila nucleosome core particle highlights evolutionary constraints on the H2A-H2B histone dimer. Proteins. 2008;71:1–7. doi: 10.1002/prot.21720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.