Abstract
Aim.
Whole-genome duplication (polyploidy) can influence the biogeography and ecology of plants that differ in ploidy level (cytotype). Here, we address how two consequences of plant polyploidy (parapatry of cytotypes and altered species interactions) shape the biogeography of herbivorous insects.
Location.
Warm deserts of North America.
Taxa
Gall midges (Asphondylia auripila group, Diptera: Cecidomyiidae) that attack three parapatric cytotypes of creosote bush (Larrea tridentata, Zygophyllaceae).
Methods.
We surveyed Asphondylia species diversity at 177 sites across a 2300-km extent. After noting a correspondence between the distributions of eight Asphondylia species and L. tridentata cytotypes, we fine-mapped Asphondylia species range limits with transects spanning cytotype contact zones. We then tested whether plant-insect interactions and/or abiotic factors explain this coincidence by (1) comparing attack rates and gall midge communities on alternative cytotypes in a narrow zone of sympatry and (2) using species distribution models (SDMs) to determine if climatically suitable habitat for each midge species extended beyond cytotype contact zones
Results.
The range limits of 6/17 Asphondylia species (including two novel putative species confirmed with COI sequencing) perfectly coincided with the contact zone of diploid and tetraploid L. tridentata. One midge species was restricted to diploid host plants while five were restricted to tetraploid and hexaploid host plants. Where diploid and tetraploid L. tridentata are sympatric, cytotype-restricted midge species more frequently attacked their typical host and Asphondylia community structure differed markedly between cytotypes. SDMs predicted that distributions of cytotype-restricted midge species were not constrained by climatic conditions near cytotype contact zones.
Main conclusions.
Contact zones between plant cytotypes are dispersal barriers for many Asphondylia species due to plant-insect interactions. The distribution of L. tridentata cytotypes therefore shapes herbivore species ranges and herbivore community structure across North American deserts. Our results demonstrate that polyploidy in plants can affect the biogeography of ecological communities.
Keywords: Asphondylia, Cecidomyiidae, creosote bush, gall midge, herbivory, Larrea tridentata, North American deserts, polyploidy, species interactions, species distribution models
INTRODUCTION
Autopolyploidy (whole genome duplication without hybridization) is common in vascular plants and can influence plant ecology and evolution (Soltis et al., 2007; Parisod et al., 2010; Ramsey & Ramsey, 2014; Barker et al., 2015, 2016; Van de Peer et al., 2017). Related plants of different ploidy level (cytotype) are often reproductively isolated, which can limit local coexistence through reproductive interference (Levin, 1975) and drive rapid speciation (Coyne & Orr, 2004; Soltis et al., 2007). Cytotypes may also differ in a suite of traits including size, growth form, phenology, water use, cold-hardiness, and secondary metabolism (Levin, 1983; Ramsey & Schemske, 2002) due to genome doubling per se (e.g., Stebbins, 1949) and/or subsequent evolutionary divergence (e.g., Ramsey, 2011). Accordingly, abiotic niches differ between cytotypes in some plant taxa (McIntyre, 2012; Thompson et al., 2014). However, broadly overlapping climatic niches in other taxa indicate that cytotypes may be in direct competition (Laport et al., 2013; Glennon et al., 2014). The joint effects of niche differentiation and population-level processes (reproductive interference and competition) likely underlie a common biogeographic consequence of polyploidy: cytotypes of many taxa have parapatric or allopatric distributions (Lewis, 1980).
The phenotypic effects of polyploidy can also influence interactions with other species, including herbivorous insects (Thompson et al., 1997, 2004; Segraves & Anneberg, 2016). In some systems, rates of herbivory are mediated by insect abundance in a cytotype’s preferred microclimate (Arvanitis et al., 2007; Richardson & Hanks, 2011). Other insect herbivores consistently attack one host cytotype over others (Thompson et al., 1997; Nuismer & Thompson, 2001; Halverson et al., 2008a) or specialize exclusively on a single cytotype (Arvanitis et al., 2010). The herbivores that discriminate most strongly between host plant cytotypes tend to have highly specialized and intimate interactions with their host plants (e.g., gall-makers and other internal-feeding insects), which may make them especially sensitive to phenotypic differences among plant cytotypes (Segraves & Anneberg, 2016).
Given that polyploidy affects plant biogeography and species interactions, its influence on the biogeography of closely-associated species is a notable gap in our understanding of polyploidy’s ecological significance. Thompson and colleagues (1997) hypothesized that the geographic distribution of favorable plant cytotypes may constrain the distribution of herbivore species. The distribution of Greya politella moths on Heuchera grossulariifolia did not support this hypothesis (Thompson et al., 1997). H. grossulariifolia comprises cytotypes with broadly overlapping distributions (Thompson et al., 1997; Segraves et al., 1999), as do most plants for which the effect of polyploidy on species interactions has been studied (Mandáková & Münzbergová, 2006; Arvanitis et al., 2007; Halverson et al., 2008b; Kao, 2008a) (but see Münzbergová et al., 2015). Plants with sympatric cytotypes allow for natural experiments to parse the effect of cytotype from those of geography and environment, but any effect of plant-insect interactions is not necessarily expected to shape the geographic range of herbivore species. Instead, we hypothesized that plants with parapatric cytotypes could constrain herbivore distributions if cytotype contact zones serve as biotically-mediated dispersal barriers.
To address this hypothesis, we tested if plant cytotype variation shapes the biogeography of herbivores of creosote bush (Larrea tridentata (DC.) Coville, Zygophyllaceae), a long-lived and dominant shrub in the warm deserts of North America. Larrea tridentata is a classical polyploid series comprising diploid (2n = 2x = 26), autotetraploid (2n = 4x = 52), and autohexaploid (2n = 6x = 78) cytotypes that roughly assort among the Chihuahuan, Sonoran, and Mojave Deserts (Barbour, 1969; Yang, 1970). Contact zones between L. tridentata cytotypes are well characterized in multiple areas, including sites where cytotypes are sympatric (Laport et al., 2012; Laport & Minckley, 2013; Laport & Ramsey, 2015). These resources enable comparisons with the distributions of insect herbivores and direct tests for the effect of cytotype on species interactions. Diploid and tetraploid cytotypes meet in a short and well-defined contact zone in southeastern Arizona, while a sinuous contact zone between tetraploids and hexaploids extends from central Arizona to southern California (Laport et al., 2012). Established tetraploid populations are hypothesized to have a single origin (Laport et al., 2012). The precise timing of polyploidy events is unknown, but some of the oldest macrofossils of L. tridentata from packrat (Neotoma spp.) middens were inferred to be tetraploid (25,600 cal. y.b.p.; Cole, 1986; Hunter et al., 2001). Hexaploid populations are hypothesized to have arisen from tetraploids in the Holocene (Hunter et al., 2001; Holmgren et al., 2014) and may have multiple origins (Laport et al., 2012). Macrofossil evidence suggests that contemporary cytotype distributions were established in the past 5,000 years during expansion from glacial refugia (Hunter et al., 2001).
The creosote gall midges (Asphondylia auripila group, Diptera: Cecidomyiidae) are an adaptive radiation of 15 species that attack leaves, stems, buds, and flowers of L. tridentata (Gagné & Waring, 1990). Remarkably, the A. auripila group diversified without host plant switching (Joy & Crespi, 2007) and comprises the majority of the gall-forming insect community on L. tridentata. As with most gall midges (Gagné, 1989; Tokuda, 2012), the life cycles of A. auripila group are intimately associated with their host plant (Gagné & Waring, 1990). The small (2–5 mm) and short-lived adults emerge from their natal galls in one or two phenological windows per year. Many gall midges are weak fliers and actively disperse only short distances (i.e., a few meters, Highland, 1964), but wind-borne transport can enable passive dispersal over many kilometers (Yukawa et al., 2003; Yukawa & Rohfritsch, 2005; Miao et al., 2013). After mating, females deposit eggs and spores of a symbiotic fungus (Tokuda, 2012) into a species-specific host plant organ (Gagné & Waring, 1990). Larvae then induce the formation of an enclosed gall in which they develop to maturity. Although the ecology of the A. auripila group has been carefully studied at select sites (Waring & Price, 1989, 1990; Huggins, 2008), the distributions of A. auripila group species have not been previously characterized.
Our goal here was to test whether interactions with L. tridentata cytotypes constrained the distributions of individual Asphondylia species and shaped broader patterns of Asphondylia diversity across North American deserts. We had three specific aims: 1) determine the geographic distributions of species in the A. auripila group, 2) identify concordance between the range limits of Asphondylia species and L. tridentata cytotypes, 3) evaluate whether concordance is due to species interactions or confounding abiotic factors. In doing so, we link genome-scale mutational processes in plants to broad geographic patterns of herbivore diversity.
METHODS
(a). Diversity and distributions of Asphondylia auripila group
We surveyed Asphondylia diversity on L. tridentata at 177 sites across the Chihuahuan, Sonoran, and Mojave Deserts between March 2015 and August 2016 (Fig. 1). At most sites we haphazardly collected branch segments from 5–10 plants, transported them to the laboratory, and a single investigator (TKO) searched them for galls with the naked eye. Two sites near Tucson, AZ were surveyed more intensively (up to 15 plants / site), and three sites were represented by opportunistic collections from a single plant (Appendix S6). We sorted galls into morphotypes and identified the causative Asphondylia species using the original species descriptions (Gagné & Waring, 1990). Gall morphology is the most reliable diagnostic character for species of the A. auripila group (Gagné & Waring, 1990) and corresponds to species identity (Joy & Crespi, 2007).
Figure 1.
Map of sampling sites. (a) 177 total sites surveyed in this study. Shading shows the approximate distribution of diploid (2x), tetraploid (4x), and hexaploid (6x) L. tridentata (after Laport and Ramsey, 2015). L. tridentata cytotypes in the Lower Colorado River Valley and Baja California Peninsula have not been directly determined by flow cytometry. Arrowheads indicate transect locations. (b) Four contact zone transects. Symbols indicate creosote bush cytotype: white squares = 2x, gray circles = 4x, dark gray triangles = 6x. (c) Site of diploid–tetraploid sympatry within San Pedro transect showing location of 72 surveyed plants.
(b). Contact zone transects
To evaluate concordance between the range limits of Asphondylia species and L. tridentata cytotypes, we sampled along four transects spanning cytotype contact zones (Fig 1). We used two diploid–tetraploid transects (Gila and San Pedro) and two tetraploid–hexaploid transects (Salton and Bend) previously established by Laport and colleagues (Laport et al., 2012, 2016; Laport & Ramsey, 2015). Each transect includes 4–6 sites with permanently marked plants of known cytotype as determined by flow cytometry. In most transects the transition between parapatric cytotypes has been delimited to within ≤ 8 km (except Gila transect: 19 km). We did not sample marked plants directly to protect ongoing research. Instead, we sampled plants found among marked plants (< 5 m away) or at sites near those of Laport and colleagues. Additional details are provided in Appendix S1.
In all transects, sampling sites were ≥ 10 m from the nearest paved road and embedded in regionally typical vegetation. We collected ≥ 10 branch segments (30 cm long) from throughout the canopy of 10 focal plants per site. All plants were separated by at least 5 m to reduce the chance of sampling from the same clone and limit spatial autocorrelation among samples. Collections were stored in airtight containers under cool conditions for up to 24 hours. We then clipped stems into 5 cm-long segments, thoroughly mixed them, and subsampled 50 g fresh mass. A single investigator (TKO) exhaustively searched subsamples for galls with the naked eye. It was often impossible to differentiate senesced galls of the current season from those of previous seasons, which may remain on the plant for months. We included all galls in analyses described below, and results may therefore integrate patterns of gall midge diversity over several generations.
We tested whether the prevalence of Asphondylia species differed between cytotypes in each transect using binomial generalized linear mixed models (GLMMs) with site as a random effect. We evaluated the significance of model terms with Wald F-test and applied a 5% false discovery rate (FDR) correction to P-values to account for multiple testing. We only fit models for 13 species found on > 5 plants in a given transect. Analyses were performed in the R Environment for Statistical Computing v. 3.4.2 (R Core Team, 2017) with packages ‘lme4’ (Bates et al., 2015) and ‘car’ (Fox & Weisberg, 2011).
(c). Diploid–tetraploid sympatry
To isolate the effect of cytotype from confounding factors, we next tested whether gall midge attack rates differed between sympatric diploid and tetraploid L. tridentata in the San Pedro River Valley (Fig. 1). Only a single such site has been characterized along the diploid–tetraploid contact zone. We surveyed galls on 72 permanently-marked plants of known cytotype (36 diploid, 36 tetraploid; Laport & Ramsey, 2015; RGL, unpublished data) near the peak of spring gall maturation in March–April 2016. To protect ongoing research on focal plants, we used a non-destructive field survey. A single investigator (TKO) inspected each plant for eight minutes under a standardized search routine and recorded all galls encountered. Although most plants were observed while blind to cytotype, morphological differences between diploids and tetraploids at this site (Laport & Ramsey, 2015) precluded a truly blind experiment. We measured mean diameter for 58/72 focal plants.
We used binomial generalized linear models (GLMs) to test whether the prevalence of Asphondylia galls differed between sympatric diploid and tetraploid hosts. We constructed GLMs for 13 Asphondylia species found on > 5 plants and assessed significance as for GLMMs. Initial models included cytotype, plant diameter, and their interaction as independent variables. Plant diameter did not differ between cytotypes (t-test, P > 0.2), so we present models with cytotype as the only independent variable. The inferred effect of cytotype from GLMs including or excluding plant diameter were generally consistent (Table S4.1). Analyses were performed with base functions in R and the ‘car’ package (Fox & Weisberg, 2011).
In addition to single-species analyses, we compared overall Asphondylia community structure between sympatric diploid and tetraploid L. tridentata. This analysis integrates over all Asphondylia species to test the hypothesis that interactions between Asphondylia species differ between L. tridentata cytotypes. We applied square-root and Wisconsin double-standardizations to gall abundance data (Bray & Curtis, 1957) and calculated Bray-Curtis dissimilarities among Asphondylia communities on individual plants. We analyzed only plants with ≥ 10 galls (N = 54). We then visualized variation among communities with non-metric multidimensional scaling (NMDS) ordinations and tested if community structure differed among cytotypes with permutational MANOVA (PERMANOVA) (Anderson, 2001). Analyses were implemented with the R packages ‘MASS’ (Venables & Ripley, 2002) and ‘vegan’ (Oksanen et al., 2017).
(d). Species distribution models
As a complement to our field observations of Asphondylia distributions and abundance, we used species distribution models (SDMs) to assess whether abiotic factors or plant–insect interactions set the range limits of Asphondylia species. Our approach followed Anderson et al. (2002), who compared the predicted extent of climatically suitable habitat for parapatric spiny pocket mice (Heteromys) species to their observed distributions. Because H. anomalus was absent from climatically suitable areas that were occupied by H. australis, the authors concluded that competition set the range limit of H. anomalus. We conducted an analogous comparison for Asphondylia species with range limits near cytotype contact zones. If climatically suitable Asphondylia habitat extended beyond the cytotype contact zone, we would conclude that plant-insect interactions limit Asphondylia distributions. If suitable habitat was instead confined to the observed species distribution, we would conclude that abiotic factors are sufficient to explain the location of Asphondylia species range limits.
We built species distribution models (SDMs) using MaxEnt (Phillips et al., 2004). Models were fit using WorldClim v. 2.0 BioClim variables at 2.5 arc-minute resolution (Fick & Hijmans, 2017) and implemented with the R packages ‘dismo’ (Hijmans et al., 2017), ‘ENMeval’ (Muscarella et al., 2014), ‘maxnet’ (Phillips, 2017), ‘MASS’ (Venables & Ripley, 2002), ‘raster’ (Hijmans, 2016), and ‘spThin’ (Aiello-Lammens et al., 2015). Expanded methods are reported in Appendix S2.
We selected eight uncorrelated BioClim variables (∣r∣ < 0.7; Dormann et al., 2013) for modeling: Bio2 (mean diurnal temperature range), Bio 4 (temperature seasonality), Bio10 (mean temperature of warmest quarter), Bio11 (mean temperature of coldest quarter), Bio15 (precipitation seasonality), Bio16 (precipitation of wettest quarter), Bio17 (precipitation of driest quarter), and Bio19 (precipitation of coldest quarter). We combined our collection data with published surveys of the creosote gall midge community (Werner & Olsen, 1973; Waring & Price, 1989; Gagné & Waring, 1990; Schowalter et al., 1999; Huggins, 2008).
Sampling bias can substantially affect Maxent model predictions (Kramer-Schadt et al., 2013), so we applied and compared two methods to account for bias in our dataset: spatial thinning of occurrence records (Pearson et al., 2007; Aiello-Lammens et al., 2015) and biased background sampling (Dudík et al., 2005). Spatially thinned analyses only included occurrences separated by ≥ 25 km, resulting in relatively uniform sampling across the range of each species. We randomly drew 10,000 background points from a species-specific background region (see below). The second set of SDMs used biased background sampling to match the bias in occurrence records. We modeled geographic variation in sampling intensity by estimating the 2-dimensional kernel density of our 177 sampling sites, then drew 10,000 background points with probability defined by local kernel density. To avoid model overfitting (Kremen et al., 2008), background regions for each species were defined as (1) within 100 km of the minimum convex polygon fit to occurrence records, and (2) within 100 km of the range of L. tridentata.
We tuned Maxent models by fitting 40 alternative combinations of feature classes and regularization multipliers and then selected the preferred model using the sample size-corrected Akaike information criterion (AICc) (Warren & Seifert, 2011). We fit final SDMs with Maxent v. 3.4 (Phillips et al., 2017) using optimized model settings for each species. We predicted habitat suitability across North America and estimated model performance using geographically-structured 4-fold cross-validation (Radosavljevic & Anderson, 2014; “block” method in ‘ENMeval’). We converted probabilistic predictions of habitat suitability to binary predictions with the commonly-used 10% omission threshold (Pearson et al., 2007).
(e). Evaluating SDM transferability
Our approach to test whether L. tridentata cytotypes determine the range limits of Asphondylia species assumes that SDMs can predict habitat suitability in unoccupied areas (i.e., that models are transferable across space). However, an array of methodological and biological factors can reduce model transferability (reviewed in Petitpierre et al., 2017). We addressed methodological challenges of inferring transferable SDMs using a limited number of uncorrelated, biologically meaningful predictors, as suggested by Petitpierre et al. (2017). Another persistent challenge is non-analog conditions between training and prediction regions (Fitzpatrick & Hargrove, 2009). The diploid–tetraploid contact zone coincides with the ecotone of the Chihuahuan and Sonoran Deserts, and the contrasting temperature and precipitation regimes in each desert region (MacMahon & Wagner, 1985) may limit SDM transferability.
We therefore evaluated SDM transferability across the study extent to identify potential limitations of our hypothesis testing framework. First, we sub-setted the occurrences of nine widespread species to include records only from the range of diploid L. tridentata (“diploid sites”) or the range of polyploid L. tridentata (“polyploid sites”). These sub-setted records mimicked the observed distributions of some Asphondylia species (see Results). For each species, we then trained SDMs on either diploid or polyploid sites using model tuning and sampling bias correction as described above. We applied a 10% omission threshold calculated from training data (e.g., polyploid sites) and calculated omission rates for occurrences on the other side of the contact zone (e.g., diploid sites). Omission rates greater than the expected 10% would indicate limited model transferability across desert regions. Finally, we compared habitat suitability predictions from models trained on complete vs. sub-setted occurrence records. By visualizing the agreement between these predictions across North American deserts, we identified where models trained on one region could be successfully transferred across the diploid–tetraploid contact zone and where they could not.
(f). Molecular phylogenetics
Three gall morphotypes identified in this study did not match any species descriptions. We collected gall midges from two of these galls (“acuminata” and “hirsuta”; informal names explained in Appendix S3) and used molecular phylogenetics to test the hypothesis that they correspond to novel Asphondylia species. Detailed molecular methods and specimen information are reported in Appendix S3. Briefly, we sequenced fragments of cytochrome c oxidase subunit I (COI) from novel gall morphotypes (“acuminata”, N = 8; “hirsuta”, N = 5) and combined them with a previously ungenotyped member of the group (A. discalis, N = 1) and published sequences representing the other 14 described species in the A. auripila group (Joy & Crespi, 2007). Following initial phylogenetic analyses, we sequenced more exemplars of A. florea (N = 10), A. apicata, (N = 3), A. rosetta (N = 3) and A. silicula, (N = 4) to provide additional phylogenetic context. Our final dataset comprised 34 new COI sequences and 34 sequences from the literature representing all 15 described species in the A. auripila group, midges, two novel gall morphotypes, and seven outgroups (Table S3.2). We aligned sequences with muscle (Edgar, 2004) using default parameters, trimmed sequences to a uniform length (432 bp), and estimated a maximum likelihood phylogeny with PhyML v. 3 (Guindon et al., 2010) under a GTR+G model. Branch support was estimated with 200 bootstrap replicates.
RESULTS
(a). Diversity of the A. auripila group
We found 18 gall morphotypes from 177 total sites across a 2,300-km extent (Appendix S5). Among these were galls of all 15 species and three gall morphotypes that did not match any species description (Fig. S3.1). A large (10 mm-long) spade-shaped bud gall (“acuminata”) was widespread in the Chihuahuan Desert. A smaller (2.5–3 mm), oblong gall found on the abaxial leaf surface (“hirsuta”) resembled the gall of A. silicula but was densely covered in long trichomes. The “hirsuta” morphotype was also present in the Chihuahuan Desert but at lower frequency than “acuminata” galls. A third undescribed morphotype strongly resembled the gall of A. silicula but was attached to the adaxial leaf margin rather than the abaxial surface (“adaxial silicula”). The “adaxial silicula” galls were found in all contact zone transects, but because we did not record the position of A. silicula galls at most sites, the full distribution of this morphotype is unknown.
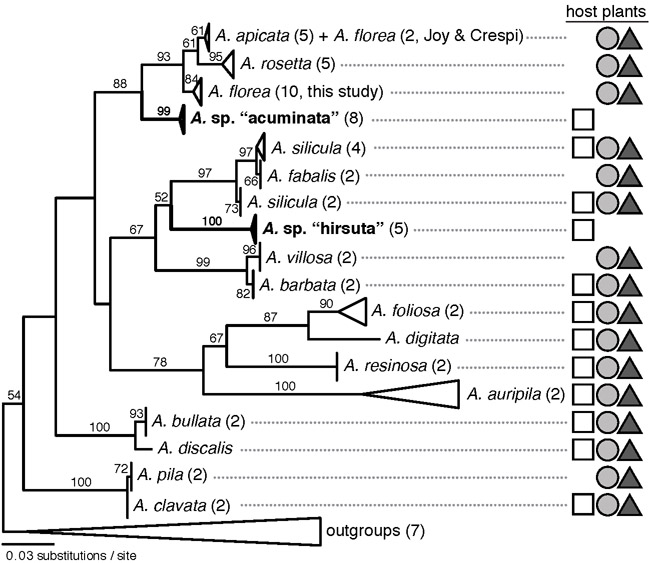
The COI phylogeny indicated that gall midges collected from “acuminata” and “hirsuta” galls represent distinct mitochondrial lineages in the A. auripila group (Fig. 2). A. sp. “acuminata” are monophyletic (bootstrap support [BS] = 99) and found as sister to A. rosetta (A. apicata + A. florea) with moderate support (BS = 88). A. sp. “hirsuta” was strongly supported as monophyletic (BS = 100), but relationships with other Asphondylia species were poorly resolved. We considered A. sp. “acuminata” and A. sp. “hirsuta” to be novel undescribed species based on the distinctness of their gall morphology, their phylogenetic placement, and the extent of mitochondrial sequence divergence from sister lineages (uncorrected divergence: A. sp. “acuminata” ≥ 4.3%, A. sp. “hirsuta” ≥ 7.7%).
Figure 2.
Maximum likelihood phylogeny of Asphondylia auripila group inferred from cytochrome c oxidase subunit I (COI) sequences. Two novel putative species (A. sp. “acuminata” and A. sp. “hirsuta”) are indicated in bold. Number of samples per species given in parentheses. Bootstrap support values ≥ 50% are indicated above branches. Scale bar represents number of substitutions per site. Host plant use shown with symbols at right: white squares = 2x, gray circles = 4x, dark gray triangles = 6x.
We did not design our phylogenetic analyses to re-evaluate relationships within the A. auripila group or uncover cryptic diversity. Nevertheless, we note that A. florea specimens from across Arizona and California formed a clade (BS = 84) that was strongly supported as sister to the clade of A. rosetta (A. apicata + A. florea) sequenced by Joy & Crespi (2007) (BS = 93; Fig. 2). We also identified a divergent lineage of A. silicula from the southern Chihuahuan desert (represented by samples MX D8S4P2G1 and MX D7S2P2G1, Fig. S3.2) that renders the species paraphyletic with respect to A. fabalis. This lineage co-occurs with other A. silicula (MX D8S4P3G1) that are closely related to populations in the Sonoran Desert (A. silicula J&C1, Fig. S3.2). Additionally, we found that A. discalis (not sequenced by Joy & Crespi (2007)) is sister to A. bullata with strong support (BS = 100, Fig. S3.2).
(b). Concordance of Asphondylia and L. tridentata range limits
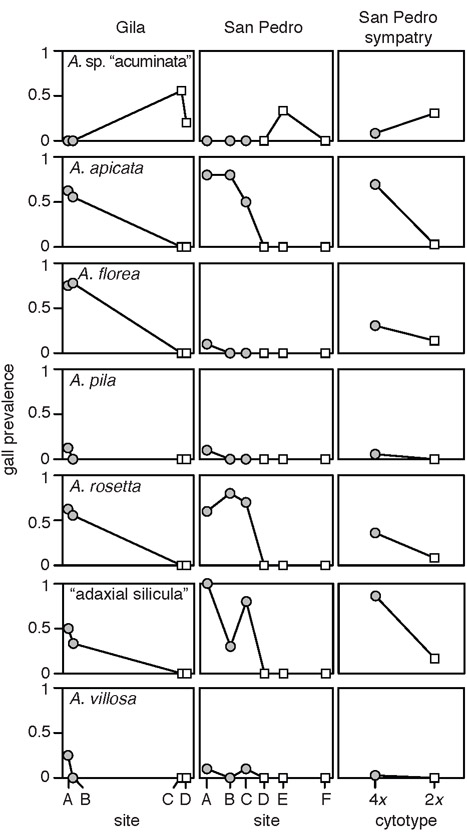
Nine of 17 Asphondylia species were found across portions of all the Chihuahuan, Sonoran, and Mojave Deserts and therefore attack all L. tridentata cytotypes (Appendix S5). No species had range margins near the tetraploid–hexaploid contact zone. The other eight species were initially found only in the Chihuahuan Desert (on diploid hosts) or in the Sonoran and Mojave Deserts (on polyploid hosts). We found six such species in diploid–tetraploid transects, and each occurred on a single host cytotype. A. sp. “acuminata” was found in diploid sites while A. apicata, A. florea, A. pila (= “A. pilosa” of Gagne & Warning (1990) and Joy & Crespi (2007); see Gagné & Jaschof (2014)), A. rosetta, and A. villosa were found in tetraploid sites (Fig. 3). “Adaxial silicula” galls were also found only in tetraploid sites. These species included all four bud-galling species, two leaf-galling species, and one leaf-galling morphotype (Table S5.1).
Figure 3.
The range limits of six Asphondylia species and the adaxial silicula gall morphotype are concordant with the contact zone between diploid (white square) and tetraploid (gray circle) L. tridentata. For each species or morphotype, the first two columns show gall prevalence (proportion of plants with a gall) in contact zone transects. Relative position along the transect is shown on the x-axis. The third column shows gall prevalence on each plant cytotype where they naturally occur in sympatry.
Concordance between Asphondylia range limits and the diploid–tetraploid contact zone is especially striking for A. apicata, A. rosetta, and “adaxial silicula” galls. Each was absent from diploid sites in the San Pedro transect but found at frequencies ≥ 0.5 in a tetraploid site < 8 km away (Fig. 3). We found similarly striking trends in the prevalence of A. apicata, A. florea, A. rosetta, and A. silicula in the Gila transect, although diploid and tetraploid sites were more distant than in the San Pedro transect. All six species and the “adaxial silicula” gall morphotype were also found in the sympatric site within the San Pedro transect. The range limits of these species were thus perfectly concordant with those of their typical host plant cytotype, a pattern we describe as “cytotype-restricted.” Note that Joy & Crespi (2007) reported A. florea 65 km east of the diploid–tetraploid contact zone, within the range of diploid L. tridentata. In light of our phylogeny, this likely represents a different species from the A. florea we consider here.
(c). Plant-insect interactions differ between L. tridentata cytotypes
All cytotype-restricted species attacked diploid and tetraploid hosts at different rates in sympatry. A. pila and A. villosa were rare in the sympatric site and found only on their typical tetraploid hosts (N = 2 and N = 1, respectively). A. sp. “acuminata”, A. apicata, A. rosetta, and “adaxial silicula” galls were more prevalent on their typical host cytotype (binomial GLM, P < 0.05; Table 1). A. florea was also more than twice as prevalent on its typical host (11/36 tetraploids vs. 5/36 diploids), but this was only a trend (P > 0.1). Mean gall intensity (gall abundance on plants with ≥ 1 gall) for these four species and the “adaxial silicula” morphotype was higher on the species’ typical host plant cytotype (1.3—3.3× higher). However, we had limited power to statistically evaluate differences between cytotypes because atypical cytotypes were rarely attacked.
Table 1.
Summary of binomial generalized linear models and Wald F-tests for differences in prevalence of Asphondylia species between sympatric diploid (2x) and tetraploid (4x) L. tridentata. We fit models only for species found on ≥ 5 plants.
| cytotype-restricted? | species | N 2x | N 4x | b (± s.e.)a | O.R. (95% CI)a | F1,70b | Pb | |
|---|---|---|---|---|---|---|---|---|
| yes | A. sp. "acuminata" | 11 | 3 | −1.58 (± 0.70) | 0.21 (0.04 – 0.74) | 5.8 | 0.035 | |
| yes | A. apicata | 1 | 25 | 4.38 (± 1.08) | 79.5 (14.3 – 1504) | 39.6 | <0.001 | |
| yes | A. florea | 5 | 11 | 1 (± 0.60) | 2.73 (0.87 – 9.62) | 2.87 | 0.137 | |
| yes | A. pila | 0 | 2 | - | - | - | - | |
| yes | A. rosetta | 3 | 13 | 1.83 (± 0.70) | 6.22 (1.77 – 29.39) | 8.3 | 0.014 | |
| yes | “adaxial silicula” morphotype | 6 | 31 | 3.43 (± 0.66) | 31 (9.3 – 125.8) | 37.24 | <0.001 | |
| yes | A. villosa | 0 | 1 | - | - | - | - | |
| no | A. auripila | 4 | 6 | 0.47 (± 0.69) | 1.6 (0.42 – 6.78) | 0.45 | 0.544 | |
| no | A. barbata | 9 | 3 | −1.30 (± 0.72) | 0.27 (0.06 – 1.02) | 3.64 | 0.099 | |
| no | A. bullata | 15 | 15 | 0 (± 0.48) | 1 (0.39 – 2.56) | 0 | 1.000 | |
| no | A. clavata | 17 | 5 | −1.71 (± 0.59) | 0.18 (0.05 – 0.54) | 9.55 | 0.009 | |
| no | A. digitata | 6 | 2 | −1.22 (± 0.85) | 0.29 (0.04 – 1.39) | 2.28 | 0.176 | |
| no | A. discalis | 3 | 28 | 3.65 (± 0.72) | 38.5 (10.6 – 193) | 38.53 | <0.001 | |
| no | A. foliosa | 0 | 2 | - | - | - | - | |
| no | A. resinosa | 14 | 17 | 0.34 (± 0.48) | 1.41 (0.55 – 3.63) | 0.5 | 0.544 | |
| no | A. silicula (typical) | 35 | 28 | −2.30 (± 1.09) | 0.10 (0.01 – 0.59) | 6.78 | 0.024 |
= Coefficients from GLM: b: estimated effect of cytotype, O.R.: odds ratio that galls of an Asphondylia species were found on a tetraploid rather than diploid plant. We report the inverse of the O.R. for A. sp. “acuminata” in the main text, which is the O.R. that this species was found on a diploid plant (its typical host) rather than a tetraploid.
= Results of Wald F-tests. P-values reflect 5% false discovery rate correction.
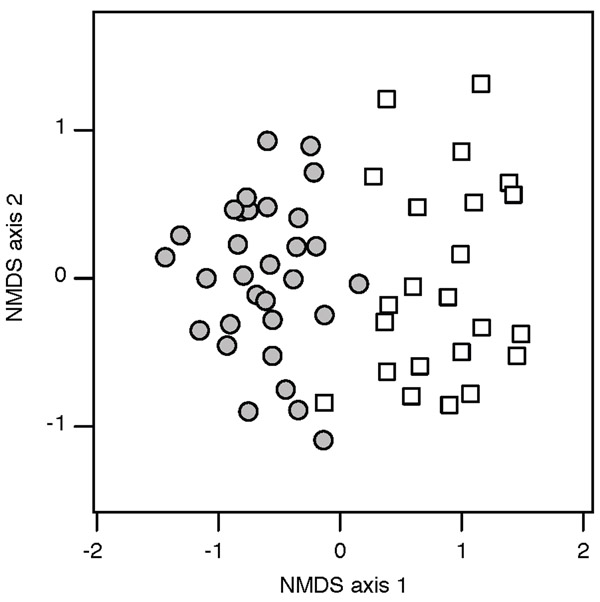
Asphondylia community structure differed markedly between sympatric diploid and tetraploid L. tridentata (Fig. 4, PERMANOVA R2 = 0.33, P < 0.001). This is consistent with the tendency of cytotype-restricted species to attack their typical host cytotype. However, the correlation of Asphondylia community structure and host cytotype was nearly as strong when cytotype-restricted species were removed from the dataset (R2 = 0.28, P < 0.001). This indicates that some widespread Asphondylia species also attacked diploid and tetraploid hosts at different rates in sympatry. A. discalis was more prevalent on tetraploids in the sympatric site while A. clavata, A. digitata, and A. silicula were more prevalent on diploids (GLM, P < 0.05) (Table 1). The prevalence of A. discalis was also higher in tetraploid sites than diploid sites within the San Pedro transect (diploid = 0.07, tetraploid = 0.66; GLMM, P < 0.001), although not in the Gila transect. By contrast, A. clavata, A. digitata, and A. silicula did not show consistent bias in prevalence in the San Pedro or Gila transects (Table S1.2).
Figure 4.
Non-metric multidimensional scaling (NMDS) ordination of Asphondylia communities on sympatric diploid (white square) and tetraploid (gray circle) plants. Cytotype was a significant predictor of community structure (PERMANOVA, R2 = 0.33, P < 0.001)
In general, gall prevalence was similar on both cytotypes at the tetraploid–hexaploid contact zone. However, one species (A. foliosa) was more prevalent on tetraploids in the Salton transect (GLMM, P < 0.05, Table S1.2). We also detected differences in prevalence for several other species (A. auripila, A. florea in the Salton transect, A. discalis in the Bend transect), but these were not statistically different after FDR correction.
(d). Abiotic limits to Asphondylia species ranges
Accounting for sampling bias using a biased background yielded SDMs with visual fits that were generally better or comparable to those using spatial thinning. For simplicity we discuss only the results using biased background sampling, but we note where spatially thinned models differ.
Tuned SDMs for all cytotype-restricted species predicted suitable abiotic habitat beyond their observed range limit at the diploid–tetraploid contact zone (Fig. 5). Nearly the full range of L. tridentata was predicted to be suitable for A. acuminata, which is found only in the Chihuahuan Desert. Suitability predictions for A. apicata, A. florea, and A. rosetta included most of the Sonoran and Mojave Deserts and regions of unoccupied habitat in the Chihuahuan Desert. Unoccupied Chihuahuan Desert habitat was contiguous with the observed distribution of A. rosetta, but was interspersed with larger unsuitable regions for A. apicata and A. florea. The A. villosa SDM based on biased background selection produced the biologically unrealistic prediction of suitable habitat in the coastal plains of Texas and Tamaulipas. Bias may have been more difficult to correct in this case due to limited occurrence records (N = 23) that were spatially clustered. The SDM based on spatial thinning resembled those of A. apicata, A. florea, and A. rosetta, although suitable habitat in the Chihuahuan Desert was smaller than in those species. The SDM for A. pila least resembled other cytotype-restricted species. Regions beyond the northern limit of L. tridentata were predicted to be suitable for A. pila, as were isolated patches of the southeastern Chihuahuan Desert. Further details of model parameters, evaluation, and performance are reported in Appendix S2.
Figure 5.
Species records and habitat suitability predictions for six cytotype-restricted species. Dark gray areas show suitable habitat when applying a 10% omission rate threshold. All species have predicted suitable habitat beyond their observed range limits. For species found on polyploid L. tridentata (all but A. sp. “acuminata”), tests of model transferability (Appendix S2) suggest species distribution models may under-predict the extent of suitable habitat in the Chihuahuan Desert. Predictions for all species except A. villosa are based on models fit using biased background sampling; A. villosa predictions are based upon predictions from spatially thinned models.
Transferability experiments for nine widespread Asphondylia species revealed that models trained with polyploid sites systematically under-predicted suitable habitat in the Chihuahuan Desert, suggesting abiotically suitable habitat is more widespread than indicated by SDM predictions. Omission rate of diploid sites was well above the expected 10% for most species (up to 71%, Table 2), although A. clavata and A. digitata were exceptions to this trend. Under-prediction was even more pronounced in spatially thinned datasets (omission rates: 51–77%, Table S2.5). Notably, SDMs trained on polyploid sites predicted relatively small and non-contiguous regions of suitable habitat in the western Chihuahuan Desert (Fig. S2.1), similar to those observed in models for cytotype-restricted species (A. apicata, A. florea, A. rosetta, and A. villosa). By contrast, models trained only on occurrences from diploid sites tended to predict the distribution in the Sonoran and Mojave Deserts well (Table 2). The species with the least transferrable models (A. auripila, A. bullata, and A. resinosa) all have very limited distributions and / or sparse occurrence records confined to the Northern Chihuahuan Desert (Fig. S5.1). Species with broader ranges in the Chihuahuan Desert had omission rates near or below the expected 10% (range: 0–12%), indicating good model transferability. Results were qualitatively similar for spatially thinned models.
Table 2.
Results of SDM transferability experiments with models employing biased background sampling. SDMs were trained on occurrences from the range of diploid or polyploid L. tridentata, then tested with occurrences from the opposite dataset. Columns report mean omission rates when applying a 10% omission threshold calculated from the training dataset.
| Species | Training: polyploid Test: diploid |
Training: diploid Test: polyploid |
|---|---|---|
| A. auripila | 67% | 38% |
| A. barbata | 43% | 3.3% |
| A. bullata | 48% | 57% |
| A. clavata | 12% | 2.8% |
| A. digitata | 0% | 8.3% |
| A. discalis | 67% | 5.2% |
| A. foliosa | 71% | 12% |
| A. resinosa | 52% | 45% |
| A. silicula | 66% | 5.9% |
DISCUSSION
There is an emerging consensus that cytotype variation is an important dimension of plant biodiversity (Soltis et al., 2007; Ramsey & Ramsey, 2014; Barker et al., 2015; Laport & Ng, 2017), and growing interest in how polyploidy affects interactions with other species (Segraves & Anneberg, 2016). Only one study has formally tested whether host plant polyploidy influences the biogeography of herbivorous insects (Thompson et al., 1997). Here, we tested if cytotype variation in creosote bush (L. tridentata) affects interactions with specialized herbivores and shapes herbivore biogeography.
The observed range limits of six Asphondylia species and an additional gall morphotype coincided perfectly with the contact zone of diploid and tetraploid L. tridentata. Although it is likely that cytotype-restricted species disperse beyond their observed distributions, they must occur at only low densities: among > 5,800 galls identified from the San Pedro and Gila transects, cytotype-restricted species were only found on atypical host cytotypes where diploids and tetraploids are sympatric. Galls of all cytotype-restricted species were more prevalent on their typical host where diploid and tetraploid L. tridentata naturally co-occur, and overall Asphondylia community structure differed markedly between sympatric cytotypes. In contrast, we found only modest differentiation in Asphondylia galling between tetraploid and hexaploid L. tridentata. The same pool of Asphondylia species attacked tetraploids and hexaploids, and while several species tended to attack these cytotypes at different rates near contact zones, prevalence differed statistically for only one species in one transect. Although our findings are generally consistent across contact zone surveys, we note that these results would be more compelling with further replication.
Species distribution models (SDMs) indicated that abiotic factors are unlikely to determine the range limits of most cytotype-restricted species. Suitable habitat for A. sp. “acuminata” was predicted across the full range of L. tridentata, while A. apicata, A. florea, A. rosetta were predicted to have large and nearly contiguous regions of suitable, but unoccupied, habitat in the Chihuahuan Desert. SDM transferability experiments demonstrated that models trained on occurrences from the range of polyploid creosote bush systematically underestimate suitable habitat in the northwest Chihuahuan Desert. Therefore, predicted distributions for A. apicata, A. florea, and A. rosetta on diploid L. tridentata are likely conservative. Both A. pila and A. villosa were predicted to have disjunct distributions separated by a broad region of unsuitable habitat in the western and central Chihuahuan Desert. It is unclear whether this reflects true abiotic tolerances or methodological bias, so we cannot exclude the possibility that abiotic and biotic factors jointly limit the distribution of these species.
It is possible that competition between Asphondylia species contributes to the parapatric distribution of some cytotype-restricted species (Sexton et al., 2009), although we have limited data to test this hypothesis. This is most plausible among bud-galling species, which include A. sp. “acuminata” on diploid hosts and A. apicata, A. florea, and A. rosetta on polyploids. We consider this unlikely because commonly find all three polyploid–restricted species on the same plant, some plants host all four species where diploid and tetraploid L. tridentata are sympatric, and many buds remain ungalled.
The Larrea–Asphondylia system demonstrates that extreme specialization on a host plant cytotype (as seen in Dasineura cardaminis on Cardamine pratensis, Arvanitis et al. 2010) is not required for polyploidy to shape insect biogeography. Asphondylia herbivory was biased between sympatric L. tridentata cytotypes, but less so than insect herbivory on Solidago altissima (Halverson et al., 2008a), Heuchera grossulariifolia (Nuismer & Thompson, 2001), and Arnica cordifolia (Kao, 2008b). In the A. auripila group, a host plant with parapatric cytotypes and modest differences in herbivore preference or performance appear sufficient to constrain herbivore distributions. Because cytotype parapatry is common among autopolyploid plants (Lewis, 1980), our results may be generalizable to other systems. For example, the sister lineage to the A. auripila group attacks a geographically segregated complex of Atriplex canescens cytotypes (2x – 20x) (Hawkins et al., 1986; Sanderson & Stutz, 1994) and is a compelling system for future study.
Several plausible mechanisms may underlie specialized interactions between Asphondylia species and host plant cytotypes. Traits that differ among L. tridentata cytotypes such as volatile organic compounds (Bohnstedt & Mabry, 1979) and organ size and proportions (Laport & Ramsey, 2015) affect host plant preference in other gall midges (Kanno & Harris, 2000; Hall et al., 2012). Alternatively, cytotype-restricted species may perform better on typical host cytotypes due to differences in plant defense. Although the major constituent of L. tridentata resin (nordihydroguaiaretic acid, NDGA) is a defense against many chewing herbivores (Mabry et al., 1977; Rhoades, 1977), internal-feeding Asphondylia spp. larvae are unlikely to encounter biologically meaningful levels of this compound. Moreover, NDGA levels are comparable between diploids and tetraploids (Zuravnsky, 2014). We are unaware of any studies investigating inducible defenses in L. tridentata, which are more likely to target internal parasites such as gall midges (e.g., Williams & Whitham, 1986; Harris et al., 2003).
Whether whole-genome duplication directly alters species interactions or potentiates their subsequent evolution is an unresolved question (Segraves & Anneberg, 2016). The limited evidence from other systems is mixed. Eurosta solidaginis flies tend to attack independently-derived tetraploid plants over alternative cytotypes, suggesting that a trait directly affected by genome duplication per se might mediate plant-insect interactions (Halverson et al., 2008a). By contrast, although Greya politella moths prefer tetraploid Heuchera grossulariifolia, this is not due to pre-existing bias and likely evolved after polyploidization (Janz & Thompson, 2002). Comparisons between contact zone transects suggest an indirect effect of polyploidy in the Larrea–Asphondylia system. We found that while the prevalence of many Asphondylia species differed between adjacent diploids and tetraploid populations, few species attacked tetraploids and hexaploids at different rates. We hypothesize that this difference is due to 1) the more ancient divergence between tetraploids and diploids and 2) periods of allopatry between diploid and tetraploid L. tridentata during Pleistocene glacial cycles. However, we cannot rule out the possibility that phenotypic differences accompanying transitions from diploidy to tetraploidy present a steeper adaptive gradient for Asphondylia than transitions from tetraploidy to hexaploidy (Ramsey & Schemske, 2002; Madlung, 2013; Laport & Ramsey, 2015)
Joy & Crespi (2007) found that the A. auripila group diversified without host plant switching, a remarkable pattern among herbivorous insects. Our results suggest at least some diversification in this group may be associated with switching among L. tridentata cytotypes. For example, the diploid–restricted species A. sp. “acuminata” shares a common ancestor with the clade comprising A. rosetta, A. apicata, and A. florea, all of which are restricted to polyploid L. tridentata (Fig. 2). Robustly testing the hypothesis that speciation coincides with host cytotype switching will require greater phylogenetic resolution and elucidation of cryptic species in the A. auripila group.
Overall, our results are consistent with the hypothesis of Thompson and colleagues (1997) that polyploidy, by altering plant traits, can affect plant-insect interactions and impact herbivore distributions. The distribution of L. tridentata cytotypes indirectly contributes to the disparity in A. auripila group diversity between the Chihuahuan vs. Sonoran and Mojave Deserts (max 11 species / site vs. max 15 species / site, Fig S5.2). Given the strong influence of plant polyploidy on many species interactions (Segraves & Anneberg, 2016), genomic variation at the primary trophic level may affect the biogeography and composition of ecological communities. Future research on the community-wide effects of plant polyploidy (Laport & Ng, 2017; Segraves, 2017) should explicitly test for such biogeographic linkages.
Supplementary Material
ACKNOWLEDGMENTS
We thank NM Alexandre, AE Baniaga, CL Kivarkis, SM Lambert, KM O’Connor, UO Garcia-Vazquez, and KM Yule for field assistance. Collections in Mexico were made under SEMERNAT permit FAUT-0062, and we thank TR van Devender, AL Reina, and HU Brailovsky for assistance with permitting. Funding for this project was provided by the Society for the Study of Evolution Rosemary Grant Award, Society for Integrative and Comparative Biology Grant-In-Aid of Research, and National Science Foundation Graduate Research Fellowship to TKO, a grant from the National Science Foundation to RGL (DEB-1556371), and grants from the National Science Foundation (DEB-1405966) and National Institute of General Medical Sciences of the National Institutes of Health to NKW (R35GM119816).
Biographies
Timothy (Tim) O’Connor employs the methods of biogeography, evolutionary ecology, and population genomics to understand how species interactions generate biodiversity. His dissertation research addresses a variety of evolutionary questions using creosote bush and its herbivorous insect community as a model system.
Robert Laport is a plant evolutionary ecologist interested in the processes generating patterns of biodiversity. His research focuses on the population-level interplay between genome duplication (polyploidy), ecological processes, and speciation. Additional details can be found at: https://robertlaport.com.
Noah Whiteman studies the evolution of plant–herbivore and host–parasite interactions with a focus on the genomic basis of adaptation. He also has an interest in the assembly of host-parasite communities. His dissertation research was on the phylogeography of the Galápagos hawk (Buteo galapagoensis) and its parasite community. His research group’s website can be found at: https://noahwhiteman.org.
Footnotes
Data Accessibility.
COI sequences have been deposited in GenBank (accession numbers: MK058365-MK058398). Collection records for all species are reported in Appendix S6.
REFERENCES
- Aiello-Lammens ME, Boria RA, Radosavljevic A, Vilela B, & Anderson RP (2015) spThin: An R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography, 38, 541–545. [Google Scholar]
- Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Austral Ecology, 26, 32–46. [Google Scholar]
- Anderson RP, Peterson AT, & Gómez-Laverde M (2002) Using niche-based GIS modeling to test geographic predictions of competitive exclusion and competitive release in South American pocket mice. Oikos, 98, 3–16. [Google Scholar]
- Arvanitis L, Wiklund C, & Ehrlén J (2007) Butterfly seed predation: effects of landscape characteristics, plant ploidy level and population structure. Oecologia, 152, 275–285. [DOI] [PubMed] [Google Scholar]
- Arvanitis L, Wiklund C, Münzbergova Z, Dahlgren JP, & Ehrlén J (2010) Novel antagonistic interactions associated with plant polyploidization influence trait selection and habitat preference. Ecology Letters, 13, 330–337. [DOI] [PubMed] [Google Scholar]
- Barbour M (1969) Patterns of genetic similarity between Larrea divaricata of North and South America. American Midland Naturalist, 81, 54–67. [Google Scholar]
- Barker MS, Arrigo N, Baniaga AE, Li Z, & Levin DA (2015) On the relative abundance of autopolyploids and allopolyploids. New Phytologist, 210, 391–398. [DOI] [PubMed] [Google Scholar]
- Barker MS, Husband BC, & Pires JC (2016) Spreading Winge and flying high: The evolutionary importance of polyploidy after a century of study. American Journal of Botany, 103, 1139–1145. [DOI] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, & Walker S (2015) Fitting linear mixed-effects models using lme4. Journal of Statistical Software, 67, 1–48. [Google Scholar]
- Bohnstedt CF & Mabry TJ (1979) The volatile constituents of the genus Larrea (Zygophyllaceae). Revista Latinoamericana de Química, 10, 128–131. [Google Scholar]
- Bray JR & Curtis JT (1957) An ordination of the upland forest communities of southern Wisconsin. Ecological Monographs, 27, 325–349. [Google Scholar]
- Cole KL (1986) The Lower Colorado River Valley: a Pleistocene desert. Quaternary Research, 25, 392–400. [Google Scholar]
- Coyne JA & Orr HA (2004) Speciation. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Dormann CF, Elith J, Bacher S, Buchmann C, Carl G, Carré G, Marquéz JRG, Gruber B, Lafourcade B, Leitão PJ, Münkemüller T, Mcclean C, Osborne PE, Reineking B, Schröder B, Skidmore AK, Zurell D, & Lautenbach S (2013) Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography, 36, 027–046. [Google Scholar]
- Dudík M, Phillips SJ, & Schapire RE (2005) Correcting sample selection bias in maximum entropy density estimation Advances in Neural Information Processing Systems (ed. by Weiss Y, Schölkopf B, and Platt JC), pp. 323–330. MIT Press, Cambridge, MA. [Google Scholar]
- Edgar RC (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fick SE & Hijmans RJ (2017) WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. International Journal of Climatology, 37, 4302–4315. [Google Scholar]
- Fitzpatrick MC & Hargrove WW (2009) The projection of species distribution models and the problem of non-analog climate. Biodiversity and Conservation, 18, 2255–2261. [Google Scholar]
- Fox J & Weisberg S (2011) An R Companion to Applied Regression. Sage, Thousand Oaks, CA. [Google Scholar]
- Gagné R & Waring G (1990) The Asphondylia (Cecidomyiidae: Diptera) of creosote bush (Larrea tridentata) in North America. Proceedings of the Entomological Society of Washington, 92, 649–671. [Google Scholar]
- Gagné RJ (1989) The Plant-Feeding Gall Midges of North America. Cornell University Press, Ithaca, NY. [Google Scholar]
- Gagné RJ & Jaschof M (2014) A Catalog of the Cecidomyiidae (Diptera) of the World. 3rd Edition. Digital Version 2 Available from https://www.ars.usda.gov/ARSUserFiles/80420580/Gagne_2014_World_Cecidomyiidae_Catalog_3rd_Edition.pdf. [Google Scholar]
- Glennon KL, Ritchie ME, & Segraves KA (2014) Evidence for shared broad-scale climatic niches of diploid and polyploid plants. Ecology Letters, 17, 574–582. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, & Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology, 59, 307–321. [DOI] [PubMed] [Google Scholar]
- Hall DR, Amarawardana L, Cross JV, Francke W, Boddum T, & Hillbur Y (2012) The chemical ecology of Cecidomyiid midges (Diptera: Cecidomyiidae). Journal of Chemical Ecology, 38, 2–22. [DOI] [PubMed] [Google Scholar]
- Halverson K, Heard SB, Nason JD, & Stireman JO (2008a) Differential attack on diploid, tetraploid, and hexaploid Solidago altissima L. by five insect gallmakers. Oecologia, 154, 755–761. [DOI] [PubMed] [Google Scholar]
- Halverson K, Heard SB, Nason JD, & Stireman JO (2008b) Origins, distribution, and local co-occurrence of polyploid cytotypes in Solidago altissima (Asteraceae). American Journal of Botany, 95, 50–58. [DOI] [PubMed] [Google Scholar]
- Harris MO, Stuart JJ, Mohan M, Nair S, Lamb RJ, & Rohfritsch O (2003) Grasses and gall midges: plant defense and insect adaptation. Annual Review of Entomology, 48, 549–577. [DOI] [PubMed] [Google Scholar]
- Hawkins BA, Goeden RD, & Gagné RJ (1986) Ecology and taxonomy of the Asphondylia spp. (Diptera: Cecidomyiidae) forming galls on Atriplex spp. (Chenopodiaceae) in southern California. Entomography, 4, 55–107. [Google Scholar]
- Highland HA (1964) Life history of Asphondylia ilicicola (Diptera: Cecidomyiidae), a pest of American holly. Journal of Economic Entomology, 57, 81–83. [Google Scholar]
- Hijmans RJ (2016) raster: Geographic data analysis and modeling. R package version 2.5–8. Available from https://CRAN.R-project.org/package=raster.
- Hijmans RJ, Phillips S, Leathwick J, & Elith J (2017) dismo: species distribution modeling. R package version 1.1–4. Available from https://CRAN.R-project.org/package=dismo.
- Holmgren C. a., Betancourt JL, Peñalba MC, Delgadillo J, Zuravnsky K, Hunter KL, Rylander KA, & Weiss JL (2014) Evidence against a Pleistocene desert refugium in the Lower Colorado River Basin. Journal of Biogeography, 41, 1769–1780. [Google Scholar]
- Huggins TR (2008) Gall morphology and the effects of host plant water status on the Asphondylia auripila group on Larrea tridentata in the Mojave Desert, Granite Mountains, California (Unpublished doctoral dissertation). University of California Los Angeles, Los Angeles, CA. [Google Scholar]
- Hunter KL, Betancourt JL, Riddle BR, Van Devender TR, Cole KL, & Spaulding WG (2001) Ploidy race distributions since the Last Glacial Maximum in the North American desert shrub, Larrea tridentata. Global Ecology and Biogeography, 10, 521–533. [Google Scholar]
- Janz N & Thompson JN (2002) Plant polyploidy and host expansion in an insect herbivore. Oecologia, 130, 570–575. [DOI] [PubMed] [Google Scholar]
- Joy JB & Crespi BJ (2007) Adaptive radiation of gall-inducing insects within a single host-plant species. Evolution, 61, 784–95. [DOI] [PubMed] [Google Scholar]
- Kanno H & Harris MO (2000) Physical features of grass leaves influence the placement of eggs within the plant by the Hessian fly. Entomologia Experimentalis et Applicata, 96, 69–80. [Google Scholar]
- Kao RH (2008a) Origins and widespread distribution of co-existing polyploids in Arnica cordifolia (Asteraceae). Annals of Botany, 101, 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao RH (2008b) Implications of polyploidy in the host plant of a dipteran seed parasite. Western North American Naturalist, 68, 225–230. [Google Scholar]
- Kramer-Schadt S, Niedballa J, Pilgrim JD, Schröder B, Lindenborn J, Reinfelder V, Stillfried M, Heckmann I, Scharf AK, Augeri DM, Cheyne SM, Hearn AJ, Ross J, Macdonald DW, Mathai J, Eaton J, Marshall AJ, Semiadi G, Rustam R, Bernard H, Alfred R, Samejima H, Duckworth JW, Breitenmoser-Wuersten C, Belant JL, Hofer H, & Wilting A (2013) The importance of correcting for sampling bias in MaxEnt species distribution models. Diversity and Distributions, 19, 1366–1379. [Google Scholar]
- Kremen C, Cameron A, Moilanen A, Phillips SJ, Thomas CD, Beentje H, Dransfield J, Fisher BL, Glaw F, Good TC, Harper GJ, Hijmans RJ, Lees DC, Louis E, Nussbaum RA, Raxworthy CJ, Razafimpahanana A, Schatz GE, Vences M, Vieites DR, Wright PC, & Zjhra ML (2008) Aligning conservation priorities across taxa in Madagascar with high-resolution planning tools. Science, 320, 222–226. [DOI] [PubMed] [Google Scholar]
- Laport R & Minckley R (2013) Cytogeography of Larrea tridentata at the Chihuahuan–Sonoran Desert ecotone. USDA Forest Service Proceedings, 218–224. [Google Scholar]
- Laport RG, Hatem L, Minckley RL, & Ramsey J (2013) Ecological niche modeling implicates climatic adaptation, competitive exclusion, and niche conservatism among Larrea tridentata cytotypes in North American deserts. The Journal of the Torrey Botanical Society, 140, 349–363. [Google Scholar]
- Laport RG, Minckley RL, & Ramsey J (2012) Phylogeny and cytogeography of the North American creosote bush (Larrea tridentata, Zygophyllaceae). Systematic Botany, 37, 153–164. [Google Scholar]
- Laport RG, Minckley RL, & Ramsey J (2016) Ecological distributions, phenological isolation, and genetic structure in sympatric and parapatric populations of the Larrea tridentata polyploid complex. American Journal of Botany, 103, 1358–1374. [DOI] [PubMed] [Google Scholar]
- Laport RG & Ng J (2017) Out of one, many: The biodiversity considerations of polyploidy. American Journal of Botany, 104, 1119–1121. [DOI] [PubMed] [Google Scholar]
- Laport RG & Ramsey J (2015) Morphometric analysis of the North American creosote bush (Larrea tridentata, Zygophyllaceae) and the microspatial distribution of its chromosome races. Plant Systematics and Evolution, 301, 1581–1599. [Google Scholar]
- Levin DA (1975) Minority cytotype exclusion in local plant populations. Taxon, 24, 35–43. [Google Scholar]
- Levin DA (1983) Polyploidy and novelty in flowering plants. The American Naturalist, 122, 1–25. [Google Scholar]
- Lewis WH (1980) Polyploidy in species populations Polyploidy: Biological Relevance (ed. by Lewis WH), pp. 103–144. Plenum Press, New York, NY. [Google Scholar]
- Mabry TJ, DiFeo DR Jr, Sakakibara M, Bohnstedt CF Jr, & Siegler D (1977) The natural products chemistry of Larrea Creosote Bush: Biology and Chemistry of Larrea in New World Deserts (ed. by Mabry TJ, Hunziker AJH, and DiFeo DR Jr), pp. 115–134. Dowden, Hutchinson & Ross Inc, Stroudsburg, PA. [Google Scholar]
- MacMahon JA & Wagner FH (1985) Mojave, Sonoran and Chihuahuan Deserts of North America Ecosystems of the World, Vol. 12A (ed. by Evanri M, Noy-Meir I, and Goodall D), pp. 105–198. Elsevier; Amsterdam. [Google Scholar]
- Madlung A (2013) Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity, 110, 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T & Münzbergová Z (2006) Distribution and ecology of cytotypes of the Aster amellus aggregates in the Czech Republic. Annals of Botany, 98, 845–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre PJ (2012) Polyploidy associated with altered and broader ecological niches in the Claytonia perfoliata (Portulacaceae) species complex. American Journal of Botany, 99, 655–662. [DOI] [PubMed] [Google Scholar]
- Miao J, Wu YQ, Gong ZJ, He YZ, Duan Y, & Jiang YL (2013) Long-distance wind-borne dispersal of Sitodiplosis mosellana Géhin (Diptera: Cecidomyiidae) in Northern China. Journal of Insect Behavior, 26, 120–129. [Google Scholar]
- Münzbergová Z, Skuhrovec J, & Maršík P (2015) Large differences in the composition of herbivore communities and seed damage in diploid and autotetraploid plant species. Biological Journal of the Linnean Society, 115, 270–287. [Google Scholar]
- Muscarella R, Galante PJ, Soley-Guardia M, Boria RA, Kass JM, Uriarte M, & Anderson RP (2014) ENMeval: An R package for conducting spatially independent evaluations and estimating optimal model complexity for Maxent ecological niche models. Methods in Ecology and Evolution, 5, 1198–1205. [Google Scholar]
- Nuismer SL & Thompson JN (2001) Plant polyploidy and non-uniform effects on insect herbivores. Proceedings of the Royal Society B: Biological Sciences, 268, 1937–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, & Wagner H (2017) vegan: community ecology package. R package version 2.4–4. Available from https://CRAN.R-project.org/package=vegan.
- Parisod C, Holderegger R, & Brochmann C (2010) Evolutionary consequences of autopolyploidy. New Phytologist, 186, 5–17. [DOI] [PubMed] [Google Scholar]
- Pearson RG, Raxworthy CJ, Nakamura M, & Peterson AT (2007) Predicting species distributions from small numbers of occurrence records: A test case using cryptic geckos in Madagascar. Journal of Biogeography, 34, 102–117. [Google Scholar]
- Van de Peer Y, Mizrachi E, & Marchal K (2017) The evolutionary significance of polyploidy. Nature Reviews Genetics, 18, 411–424. [DOI] [PubMed] [Google Scholar]
- Petitpierre B, Broennimann O, Kueffer C, Daehler C, & Guisan A (2017) Selecting predictors to maximize the transferability of species distribution models: lessons from cross-continental plant invasions. Global Ecology and Biogeography, 26, 275–287. [Google Scholar]
- Phillips S (2017) maxnet: fitting ‘Maxent’ species distribution models with ‘glmnet.’ R package version 0.1.2. Available from https://CRAN.R-project.org/package=maxnet.
- Phillips SJ, Anderson RP, Dudík M, Schapire RE, & Blair ME (2017) Opening the black box: an open-source release of Maxent. Ecography, 40, 887–893. [Google Scholar]
- Phillips SJ, Dudík M, & Schapire RE (2004) A maximum entropy approach to species distribution modeling. 21st International Conference on Machine Learning, Banff, Canada, 655–662. [Google Scholar]
- R Core Team. (2018). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; Retrieved from http://www.R-project.org/. [Google Scholar]
- Radosavljevic A & Anderson RP (2014) Making better Maxent models of species distributions: complexity, overfitting and evaluation. Journal of Biogeography, 41, 629–643. [Google Scholar]
- Ramsey J (2011) Polyploidy and ecological adaptation in wild yarrow. Proceedings of the National Academy of Sciences of the United States of America, 108, 7096–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J & Ramsey TS (2014) Ecological studies of polyploidy in the 100 years following its discovery. Philosophical Transactions of the Royal Society B: Biological Sciences, 369, 1–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J & Schemske DW (2002) Neopolyploidy in flowering plants. Annual Review of Ecology and Systematics, 33, 589–639. [Google Scholar]
- Rhoades DF (1977) The antiherbivore chemistry of Larrea Creosote Bush: Biology and Chemistry of Larrea in New World Deserts (ed. by Mabry TJ, Hunziker AJH, and DiFeo DR Jr), pp. 135–175. Dowden, Hutchinson & Ross, Stroudsburg, Pennsylvania. [Google Scholar]
- Richardson ML & Hanks LM (2011) Differences in spatial distribution, morphology, and communities of herbivorous insects among three cytotypes of Solidago altissima (Asteraceae). American Journal of Botany, 98, 1595–1601. [DOI] [PubMed] [Google Scholar]
- Sanderson SC & Stutz HC (1994) High chromosome numbers in Mojavean and Sonoran Desert Atriplex canescens (Chenopodiaceae). American Journal of Botany, 81, 1045–1053. [Google Scholar]
- Schowalter TD, Lightfoot DC, & Whitford WG (1999) Diversity of arthropod responses to host-plant water stress in a desert ecosystem in southern New Mexico. The American Midland Naturalist, 142, 281–290. [Google Scholar]
- Segraves KA (2017) The effects of genome duplications in a community context. New Phytologist, 215, 57–69. [DOI] [PubMed] [Google Scholar]
- Segraves KA & Anneberg TJ (2016) Species interactions and plant polyploidy. American Journal of Botany, 103, 1326–1335. [DOI] [PubMed] [Google Scholar]
- Segraves KA, Thompson JN, Soltis PS, & Soltis DE (1999) Multiple origins of polyploidy and the geographic structure of Heuchera grossulariifolia. Molecular Ecology, 8, 253–262. [Google Scholar]
- Sexton JP, Mcintyre PJ, Angert AL, & Rice KJ (2009) Evolution and ecology of species range limits. Annual Review of Ecology & Systematics, 40, 415–436. [Google Scholar]
- Soltis DE, Soltis PS, Schemske DW, Hancock JF, John N, Husband BC, Judd WS, Hancock JF, Soltis DE, Soltis PS, Schemske DW, Thompson JN, Husband BC, & Judd WS (2007) Autopolyploidy in angiosperms: have we grossly underestimated the number of species? Taxon, 56, 13–30. [Google Scholar]
- Stebbins GL (1949) The evolutinoary significance of natural and artificial polyploids in the family Graminae. Hereditas, 35, 461–485. [Google Scholar]
- Thompson JN, Nuismer SL, & Merg K (2004) Plant polyploidy and the evolutionary ecology of plant/animal interactions. Biological Journal of the Linnean Society, 82, 511–519. [Google Scholar]
- Thompson JN, Segraves KA, Cunningham BM, Althoff DM, & Wagner D (1997) American Naturalist. 150, 730–743. [DOI] [PubMed] [Google Scholar]
- Thompson KA, Husband BC, & Maherali H (2014) Climatic niche differences between diploid and tetraploid cytotypes of Chamerion angustifolium (Onagraceae). American Journal of Botany, 101, 1868–1875. [DOI] [PubMed] [Google Scholar]
- Tokuda M (2012) Biology of Asphondyliini (Diptera: Cecidomyiidae). Entomological Science, 15, 361–383. [Google Scholar]
- Venables WN & Ripley BD (2002) Modern Applied Statistics with S. Springer, New York. [Google Scholar]
- Waring G & Price P (1989) Parasitoid pressure and the radiation of a gallforming group (Cecidomyiidae: Asphondylia spp.) on creosote bush (Larrea tridentata). Oecologia, 293–299. [DOI] [PubMed] [Google Scholar]
- Waring G & Price P (1990) Plant water stress and gall formation (Cecidomyiidae: Asphondylia spp.) on creosote bush. Ecological Entomology, 87–95. [Google Scholar]
- Warren DL & Seifert S (2011) Ecological niche modeling in Maxent: the importance of model complexity and the performance of model selection criteria. Ecological Applications, 21, 335–342. [DOI] [PubMed] [Google Scholar]
- Werner F & Olsen A (1973) Consumption of Larrea by Chewing Insects. US International Biological Program, Desert Biome, Logan, UT. [Google Scholar]
- Williams AG & Whitham TG (1986) Premature leaf abscission: an induced plant defense against gall aphids. Ecology, 67, 1619–1627. [Google Scholar]
- Yang T (1970) Major chromosome races of Larrea divaricata in North America. Journal of the Arizona Academy of Science, 6, 41–45. [Google Scholar]
- Yukawa J & Rohfritsch O (2005) Biology and ecology of gall-inducing Cecidomyiidae (Diptera: Cecidomyiidae; ). Biology, Ecology, and Evolution of Gall-Inducing Arthropods. (ed. by Raman A, Schaefer CW, and Withers TM), pp. 273–304. Science Publishers, Enfield, NH. [Google Scholar]
- Yukawa J, Uechi N, Horikiri M, & Tuda M (2003) Description of the soybean pod gall midge, Asphondylia yushimai sp n. (Diptera: Cecidomyiidae), a major pest of soybean and findings of host alternation. Bulletin of Entomological Research, 93, 73–86. [DOI] [PubMed] [Google Scholar]
- Zuravnsky KN (2014) Understanding the roles of polyploidy and the environment on nordihydroguaiaretic variation in Larrea tridentata (Unpublished master’s thesis). Salisbury University, Salisbury, MD. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.