Abstract
In recent years our understanding of amyloid structure has been revolutionised by innovations in cryo-electron microscopy, electron diffraction and solid-state NMR. These techniques have yielded high-resolution structures of fibrils isolated from patients with neurodegenerative disease, as well as those formed from amyloidogenic proteins in vitro. The results not only show the expected cross-β amyloid structure, but also reveal that the amyloid fold is unexpectedly diverse and complex. Here, we discuss this diversity, highlighting dynamic regions, ligand binding motifs, cavities, non-protein components, and structural polymorphism. Collectively, these variations combine to allow the generic amyloid fold to be realised in three dimensions in different ways, and this diversity may be related to the roles of fibrils in disease.
Introduction
Amyloid is a conformational state that can be achieved by most (if not all) proteins [1••]. Protein sequences harbor the information necessary to enable them to fold into their native, functional 3D structures [2] or, for intrinsically disordered proteins (IDP), to remain dynamically unstructured [3]. However, proteins also contain sequences capable of forming an alternative structure(s) known as the ‘amyloid fold’ [4]. Upon cellular or physical stress, by a mechanism that is kinetically complex [5] and difficult to characterise structurally [6], one or more of these ‘amyloid-prone regions (APR)’, can rearrange to form β-strands, which stack in layers oriented perpendicular to the fibril’s long axis to generate the ‘amyloid fold’ (Figure 1a) [1••]. These β-strands and their interconnecting loops constitute the ‘amyloid core’. The repeating nature of amyloid cores, involving extensive mainchain hydrogen bonding between adjacent β-strands within the stacked layers (Figure 1b1), and close interdigitation of sidechains (Figure 1b2), results in a fibril structure that is enormously stable both thermodynamically [7] and mechanically [8]. Indeed this stability can far exceed that of the original native fold of the protein, highlighting the physico-chemical knife-edge of cellular life because of the metastable nature of their proteomes [7]. Fascinatingly, despite their high stability, fibrils are dynamic, with monomers and/or oligomers dissociating from their ends [9•,10], while the surface of the fibrils can act as a potent site for secondary nucleation, catalysing the formation of oligomers and new fibrils [11•,12].
Figure 1. Updating the amyloid fold.
(a) Schematic of the cross-β fold viewed from (1) side and (2) cross-section, representing the stacking of molecular layers perpendicular to the long axes of the amyloid fibrils. (b) Detail of the structural elements observed in amyloid cores highlighting (1) in-register parallel β-sheets [30•], (2) ‘dry’ steric zippers [29], (3) sidechain-mainchain loop hydrogen bonding [37], (4) polar zippers [26•], (5) buried salt-bridges [36••], (6) structured solvent molecules [27••], (7) polar and apolar channels (green and purple arrows, respectively) [36••], (8) cofactors (blue density) [27••] and (9) multi-molecular layer interactions between central layer ‘L’ (blue) and the adjacent layers above and below (gold) [35].
The molecular mechanism(s) by which IDPs and initially folded amyloidogenic precursors rearrange into an amyloid core structure and stack into molecular layers is not well understood. However, it is generally accepted that this feat is accomplished via the formation of transient non-native monomeric and oligomeric species [6,13]. The transient and dynamic nature of such species has limited the characterisation of their structures and our understanding of the molecular basis of the cytotoxicity often associated to amyloid formation [14,15].
Here we review recent advances in our understanding of the amyloid fold. We describe the interactions that create the fibril core, as well as less well-ordered and dynamic regions of the amyloid fold. We also discuss differences between fibril structures formed in vitro and in vivo, and how structural polymorphism may rationalise disease phenotype. Finally, we highlight the need to combine information from multiple structural, biophysical, and cellular techniques, including information gained from in vitro and in vivo analyses, to understand amyloid formation and disease.
Updating our understanding of the amyloid fold
One sequence, many structures
The first high resolution structural information about amyloid came from fibrils assembled from synthetic peptides [16–19]. These structures revealed the first atomic resolution details of the stacking of β-strands in the cross-β architecture, which had been visualised at lower resolution some 50 years before [20]. The first structures of amyloid fibrils from intact proteins at atomic/near-atomic resolution were obtained from fibrils assembled in vitro of the amyloi-dogenic IDPs Aβ40, Aβ42, amylin and α-synuclein using solid state NMR (ssNMR) [21–25]. The recent revolution in cryo-EM has now expanded this repertoire to fibrils formed in vitro or extracted ex vivo from β2-microglobulin (β2m), mouse and human serum Amyloid A (AA), Light Chain (LC) amyloid from two human variants, and the IDPs Tau, α-synuclein and Aβ42, [26•,27••,28•,29,30•,31–33, 34••,35,36••,37]. These results show that the same protein sequence can adopt different amyloid structures, leading to more fibril structures than sequences (45 amyloid fibril structures from full proteins and IDPs are currently deposited in the pdb). For α-synuclein, four distinct polymorphs have been solved by cryo-EM [31,32,33,34••] and one structure of a single protofibril by ssNMR [23]. For Tau three different neurodegenerative diseases result in fibrils with distinct, disease-specific structures, all of which are different from the structures formed in vitro in the presence of heparin [26•,27••,28•,38]. Whether this difference between in vitro and in vivo fibril structures is found for other amyloid proteins remains an important, open question.
A convoluted amyloid fold
The structures of amyloid fibrils solved by ssNMR [21–24] and cryo-EM [26•,27••,28•,29,30•,31–33,34••,35,36••,37] have revealed that a wide variety of interactions can stabilise an amyloid core. While all of these structures have canonical cross-β amyloid folds, their structures are more complicated and diverse than originally anticipated. For amyloid precursors that are initially folded, assembly into the amyloid core requires wholesale rearrangement of the polypeptide chain and sometimes reassignment of the secondary structure elements [36••]. As expected, extensive backbone hydrogen bonding between β-strands is observed in these new fibril structures (Figure 1b1), but the topologies of the structural elements that comprise the amyloid core is more complex than those seen in fibrils formed from short peptide fragments [16,18,39,40]. The β-sheets are not, in general, formed from homotypic ‘dry steric zipper’ interactions in which two copies of the same sequence form sidechain-sidechain interactions between the β-sheets [18,41]. Such zippers are observed in fibrils formed from intact proteins, but in the interfaces formed between protofilaments (Figure 1b2) [28•,29,32]. The current list of interactions and structural motifs known to stabilise the amyloid core (Figure 1b) also includes sidechain-mainchain and sidechain-sidechain hydrogen bonding from loops that interconnect β-strands (Figure 1b3), tight interdigitation of polar and charged side chains (named here ‘polar zippers’) (Figure 1b4), buried salt bridges (Figure 1b5), interactions with solvent (Figure 1b6) [27••], and both polar [36••] and apolar [27••,31,36••] internal channels (Figure 1b7). In the latter case, an un-assigned electron density inside the apolar channel of tau filaments isolated from patiens that suffer chronic traumatic encephalopathy suggests that non-proteinaceous aliphatic molecules may participate in this amyloid core (Figure 1b8) [27••]. Finally, a given molecular layer (L), may make interactions with the layer above (L + 1), below (L − 1), oreven beyondits immediate neighbours (L + 2, L − 2, etc). Such interactions also stabilise the amyloid core (Figure 1b9) [35].
Different forms of fibril polymorphism
The biological relevance of amyloid polymorphism has been extensively documented for several amyloid diseases [1••]. Fibrils created from the same precursor have been shown to display different structures in different diseases [26•,27••,28•], different seeding characteristics [42], different rates of spread [43], and distinct patterns of neuropathology [44,45]. Cryo-EM has a major advantage for structural characterisation of fibril polymorphs since each structure can (in principle) be determined independently for each co-populating polymorph, as long as sufficient images of each type can be obtained. Polymorphism can take different forms. Firstly (type 1), it can involve different packing arrangements of the same protofibril, as was observed for amyloidogenic peptides of transthyretin (TTR) [40] and immunoglobulin LC λ-1 (Figure 2a) [46], as well as for entire proteins such as β2m (Figure 2b) [30•]. In some cases, this type of polymorphism can involve subtle changes in the contact angle or arrangement of interactions between protofilaments, exemplified by the difference in Paired Helical Filaments (PHF) and Straight Filaments (SF) of Tau fibrils analysed from Alzheimer’s patients (Figure 2c) [28•], and in early models of fibril structures of Aβ40 observed by ssNMR (Figure 2d) [19,22]. In other cases, this polymorphism can involve fibrils comprised of different numbers of protofilaments, such as in the narrow (NPF) and wide (WPF) filaments observed in Tau fibrils from Pick’s disease (Figure 2e) [26•]. A second type of polymorphism (type 2) can occur when a common structure is adopted by one region of a protein sequence, while different structures are adopted by other regions. This is observed in the ‘Rod’ and ‘Twister’ polymorphs of α-synuclein fibrils (Figure 2f) [32] and in Serum Amyloid A (AA) fibrils [35] (Figure 2g). A third type of polymorphism (type 3) combines types 1 (different packing of protofilaments) and type 2 (partial common fold) and has been observed for Aβ42 in structures elucidated by cryo-EM [29] and ssNMR [24]. These structures exhibit a common fold (the ‘S motif’) that packs through different interfaces and with different structures for the N-terminal domain (Figure 2h). Other polymorphs of Aβ42 have also been observed in which different numbers of protofilaments with different twists are involved [47], but it is not yet known which molecular interactions create these different polymorphs. The fourth (type 4), and most drastic, kind of polymorphism occurs when both the protofilament structure and packing interactions vary, as observed in polymorphs of Tau fibrils formed in vitro in the presence of heparin (Figure 2i) [38], and in fibrils formed from fragments of TDP43 in vitro, in which three polymorphs are observed for the same sequence segment (Figure 2j) [48]. At least for Tau, samples from 17 patients that suffered from two variants of the same disease possessed fibrils with a similar fold, suggesting that a common fibril structure could be associated with a particular disease [49]. Yet, in all of the above cases less abundant polymorphs could also be present, albeit in too low a number to enable structure determination.
Figure 2. The many faces of an amyloid.
Fibril polymorphs observed for (a) peptide fragments of LC λ [46] and for the full-length proteins (b) β2m [30•], (c) Tau in AD [28•], (d) Aβ40 [19,22], (e) Tau in Pick’s Disease [26•], (f) α-synuclein [32], (g) mouse and human serum AA [35], (h) Aβ42 [24,29], (i) Tau in presence of heparin [38] and (j) segments of TDP43 [48]. These fibrils were isolated ex vivo (c,e,g) or formed in vitro (a,b,d,f,h,i,j). Illustrations in (a) and (b) are reproduced with permission [30•,46].
What drives fibril diversity is unclear. It could result from the intrinsic properties of the polypeptide sequence; the presence or absence of post-translational modifications; interaction with cofactors or cellular components; the nature of the environment (pH, ionic strength etc), or the cell type in which amyloid is formed. That interaction with cofactors can modulate the abundance of polymorphs has been shown for Tau, with heparin inducing structurally heterogeneous fibrils, while RNA induces structurally homogeneous fibrils [50••]. Sequence variation can also contribute to polymorphism. For example, fibrils generated from the 3R isoform of Tau in Pick’s disease (NPF and WPF, Figure 2e) [26•] are different to those generated by the 4R isoform in Alzheimer’s disease (HPF and SF, Figure 2c) [28•].
Dynamic regions are integral to the amyloid fold
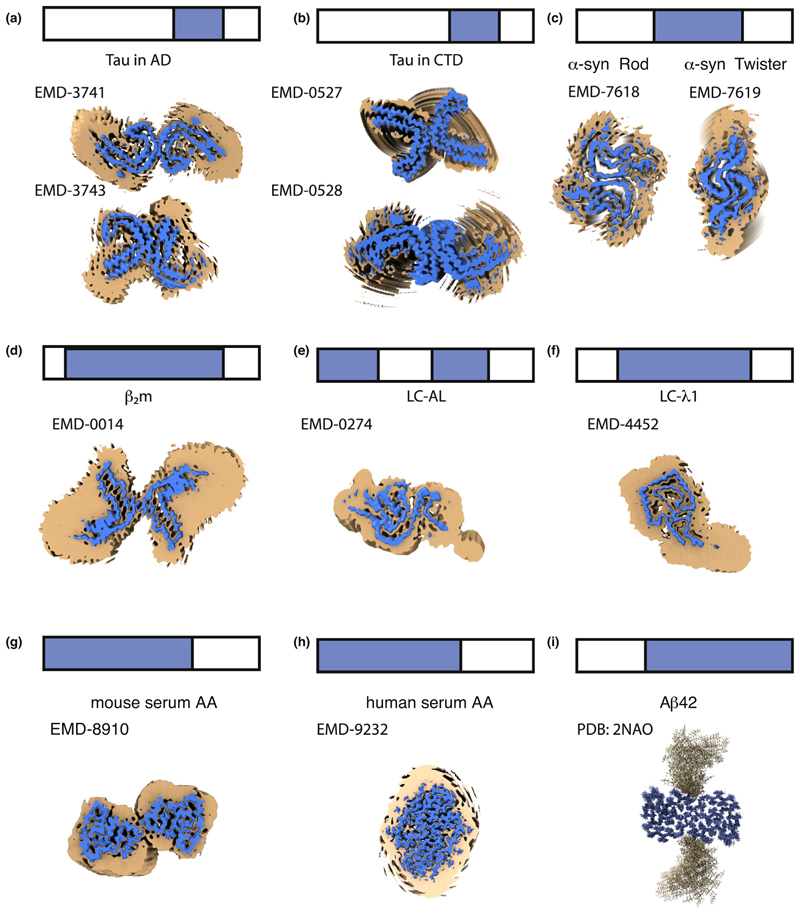
Another remarkable characteristic of the amyloid fibril structures determined recently is that relatively short regions of a protein sequence form the amyloid core [26•,27••,28•,30•,31,32,33,35,36••,37], with the remaining segments exhibiting high structural variability (Figure 3). Disordered regions map to the termini (Figure 3a−i) [23,24,26•,27••,28•,32], and to internal loops/segments of the polypeptide chain (Figure 3e) [37]. For example, the amyloid core of Tau fibrils involves between 72 and 94 of its 441 residues, with the number of ordered amino acids depending on the tauopathy (Figure 3a−b) [26•,27••]. For α-synuclein, 40−59 of its 140 residues form the amyloid core depending on the polymorph (Figure 3c) [32]. Similar observations have been made for β2m (63 out of 100 residues [30•] (Figure 3d), antibody LC case 1 (77 out of 111 residues [37] (Figure 3e)), or case 2 (91 out of 118 residues [36••] (Figure 3f)), SAA from mouse (69 out of 83 residues), and SAA from humans (53 out of 67 residues) [35] (Figure 3g and h, respectively). We refer to these as ‘Locally Disordered Regions’ (LDRs) to signify their localised high structural variability. LDRs have also been described in fibrils of Aβ42, where the N-terminal 14−15 residues, that coincide with the least amyloidogenic regions, are not involved in the amyloid core (Figure 3i) [24]. Similarly, the N-terminal 10 residues of β2m are highly dynamic, with the succeeding 10 residues less so (but not organised into the amyloid core) [30•]. Finally, for the structure of α-synuclein fibrils determined using ssNMR, three regions (1-24, 55-62 and 97-140) lack assignment [23] and this is usually interpreted as signifying dynamic behaviour.
Figure 3. Amyloid is more than a rigid core.
LDRs are shown on the structures of amyloid fibrils of full-length proteins formed in vitro or isolated ex vivo. The top of each panel shows a schematic of the full-length sequence (bar) with the sequence involved in forming the ordered amyloid core in blue and LDRs in white. The image on panels (a−h) corresponds to an orthogonal view down the fibril axis of the reported density maps contoured at two different levels. The regions of localised disorder are shown as broad/noisy density (orange) surrounding the amyloid core density (blue) for (a) Tau in Alzheimer’s disease [28•], (b) Tau in CTE [27••], (c) two polymorphs of α-synuclein [32], (d) β2m [30•], (e) IGLV6-57 derived LC amyloid [37], (f) GLV1-44 derived LC amyloid [36••], (g) mouse AA and (h) human AA [35]. Panel (i) shows the structure of Aβ42 fibrils determined by ssNMR [24], where regions of disorder are modeled as loops that point away from the core. The EMD code of each map used is indicated on each panel. The blue maps are countered to the recommended level indicated for the deposited maps. The orange maps are 5 Å low-pass filtered of the deposited map countered to 1.75 RMS using ChimeraX [72].
LDRs are important in amyloid formation and in disease. For example, they can kinetically define the amyloid structures that result from aggregation [51]. Fibrils, including their LDRs, are also known to be involved in engaging with cellular components that regulate the health of the cell, including molecular chaperones [52], other proteins that contain IDRs or IDPs [53], components of the extracellular matrix [54•], biological membranes of different type [55] or other cellular components [56]. These dynamic regions must not be over-looked, despite the fact that they are difficult, if not impossible, to structurally characterise using cryo-EM or ssNMR. Single-molecule FRET, hydrogen/deuterium exchange, oxidative labelling and cross-linking methods offer exciting possibilities to characterise these regions and their interactions in vitro and in vivo in the future.
Left or right-handed, parallel or anti-parallel?
Contrary to the canonical right-handed β-sheets observed in globular proteins, amyloid fibrils can adopt right-handed or left-handed β-sheets, with a switch between handedness requiring only subtle differences in the β-strand φ/ψ angles [57]. For example, mouse and human AA amyloid have opposite chirality despite having 78% sequence identity [35]. While anti-parallel β-strands were observed in amyloidogenic fragments using X-ray crystallography [18,41], ssNMR [16] and X-ray fibre diffraction [58], amyloid fibrils formed from longer precursors commonly adopt a parallel in-register structure (Figure 1a and b1) [1••]. In these structures each molecular layer ‘L’ deviates from planarity, which allows intermolecular interactions beyond the immediate neighbouring layers ‘L + 1’ and ‘L-1’ [29,35]. The number of molecules that can interact in this mode can span up to 10 molecular layers, as observed in human AA amyloid (Figure 1b9) [35]. Non-planarity of the layers also confers a subtle polarity to the fibrils because it generates structural differences between the two fibril ends.
Amyloid structures: beyond protein
The interactions between amyloid fibrils and cofactors has been long studied, with the list of ligands including nucleic acids [50••,59], lipids [60], metal ions [61], glycosaminoglycans [62], glycoproteins [63] and others (reviewed in Ref. [54•]). The consequences of these interactions include modulation of fibril growth kinetics and fibril stability [62,64], changes in amyloid-associated cytotoxicity [65] and altered seeding efficiency [50••]. The recent elucidation of the structure of Tau fibrils extracted from patients with Chronic Traumatic Encephalopaty (CTE) have provided the first atomic-resolution information showing that co-factors can be an integral part of the amyloid core (Figure 1b8) [27••].
The amyloid fold in vivo
Recent developments in Cryo-Electron Tomography (CryoET) have started to reveal the organisation of amyloid fibrils in situ and how fibrils can disturb cellular processes [66,67•]. Using kinetic experiments in vitro amyloid fibril formation can be explained as a combination of elementary mechanisms including primary/secondary nucleation, elongation and fragmentation [68••]. This has allowed the identification of small molecules and antibodies/chaperones that target specific steps in amyloid assembly [69,70]. Importantly, the finding that the same compounds are active in vitro and in vivo, validates the utility of in vitro observations for understanding amyloid disease. In the same way, fluorescent oligothiophene conjugates designed in vitro have also been used to analyse amyloid in situ and shown to be able to differentiate conformational variants of Ab plaques in patients with different subtypes of Alzheimer’s disease [71].
Although extraordinary progress has been made in our understanding of amyloid in vivo, the resolution currently possible by cryo-ET (~3 to 10 nm) does not enable fibril structure details to be visualised within cells. Hence, the full diversity and complexity of the amyloid fold in vivo are yet to be revealed. To complete the picture (Figure 4) we need to improve the resolution of fibril structures in vitro and in situ, and to employ different techniques, in combination, to inform on different aspects of the amyloid fold, including the functionally important dynamic regions discussed above. We also need to remember that oligomers also play a key role in amyloid disease [14,15], yet structurally characterising these species remains a hugely challenging task in vitro, and is not possible currently in vivo.
Figure 4.
A combination of techniques is required to understand the amyloid fibril structure and its cellular consequences. However, the picture is still incomplete. The missing aspects will be achieved through biochemical, biophysical and cellular investigation. Only by an integrative approach in which in vitro, ex vivo, in situ and in vivo approaches are combined can we hope to achieve the structural, cellular and mechanistic understanding required to fully understand the amyloid structure and to inspire biomedical progress.
Summary and outlook
Recent discoveries about the structure of amyloid fibrils have shifted our understanding of the amyloid fold from an initially simple set of structural elements, to a complex architecture in which apolar, polar, ionic and hydrogen bonding interactions, together with solvent and co-factor binding, structured cores and locally disordered regions, build the fibril. Cofactors and post-translational modifications can also have profound effects on the structure, kinetic and thermodynamic properties of amyloid fibrils and their cellular effects. By integrating methods able to interrogate the structured and dynamic regions of amyloid, and exploiting the powers of cryo-EM/ET to determine amyloid structures in vitro, ex vivo and in situ, we will soon have a better understanding of the amyloid fold as a whole and how this amazingly diverse, but stable structure, affects cells. Rather than a generic and simple amyloid fold, there is a remarkable array of amyloid structures, each of which may uniquely contribute to the generation of cellular dysfunction and disease.
Acknowledgements
We thank the Ranson and Radford laboratories especially members of our amyloid team for many helpful discussions and comments. We acknowledge with thanks funding from the Wellcome Trust (204963) and the ERC ((FP7/2007-2013)/ERC grant agreement no.⁰32240.
Footnotes
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Iadanza MG, Jackson MP, Hewitt EW, Ranson NA, Radford SE. A new era for understanding amyloid structures and disease. Nat Rev Mol Cell Biol. 2018;19:755–773. doi: 10.1038/s41580-018-0060-8. [Comprehensive review on the history of discovery, fibrillation and the relationship of fibril structure and disease] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowman GR, Voelz VA, Pande VS. Taming the complexity of protein folding. Curr Opin Struct Biol. 2011;21:4–11. doi: 10.1016/j.sbi.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babu MM, van der Lee R, de Groot NS, Gsponer J. Intrinsically disordered proteins: regulation and disease. Curr Opin Struct Biol. 2011;21:432–440. doi: 10.1016/j.sbi.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 5.Knowles TP, Waudby CA, Devlin GL, Cohen SI, Aguzzi A, Vendruscolo M, Terentjev EM, Welland ME, Dobson CM. An analytical solution to the kinetics of breakable filament assembly. Science. 2009;326:1533–1537. doi: 10.1126/science.1178250. [DOI] [PubMed] [Google Scholar]
- 6.Karamanos TK, Kalverda AP, Thompson GS, Radford SE. Mechanisms of amyloid formation revealed by solution NMR. Prog Nucl Magn Reson Spectrosc. 2015;88–89:86–104. doi: 10.1016/j.pnmrs.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baldwin AJ, Knowles TP, Tartaglia GG, Fitzpatrick AW, Devlin GL, Shammas SL, Waudby CA, Mossuto MF, Meehan S, Gras SL, et al. Metastability of native proteins and the phenomenon of amyloid formation. J Am Chem Soc. 2011;133:14160–14163. doi: 10.1021/ja2017703. [DOI] [PubMed] [Google Scholar]
- 8.Knowles TP, Fitzpatrick AW, Meehan S, Mott HR, Vendruscolo M, Dobson CM, Welland ME. Role of intermolecular forces in defining material properties of protein nanofibrils. Science. 2007;318:1900–1903. doi: 10.1126/science.1150057. [DOI] [PubMed] [Google Scholar]
- 9.Hoshino M. Fibril formation from the amyloid-beta peptide is governed by a dynamic equilibrium involving association and dissociation of the monomer. Biophys Rev. 2017;9:9–16. doi: 10.1007/s12551-016-0217-7. [Review that argues fibril formation and stability as a dynamic, fine balance between association and dissociation of soluble oligomers at the tips of amyloid fibrils] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tipping KW, Karamanos TK, Jakhria T, Iadanza MG, Goodchild SC, Tuma R, Ranson NA, Hewitt EW, Radford SE. pH-induced molecular shedding drives the formation of amyloid fibril-derived oligomers. Proc Natl Acad Sci U S A. 2015;112:5691–5696. doi: 10.1073/pnas.1423174112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bunce SJ, Wang Y, Stewart KL, Ashcroft AE, Radford SE, Hall CK, Wilson AJ. Molecular insights into the surface-catalyzed secondary nucleation of amyloid-beta40 (Abeta40) by the peptide fragment Abeta16-22. Sci Adv. 2019;5:eaav8216. doi: 10.1126/sciadv.aav8216. [Clear example of secondary nucleation catalysing amyloid formation. By combining in vitro experiments and molecular dynamic simulations, the authors generate a real-time molecular movie that depicts the secondary nucleation process] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tornquist M, Michaels TCT, Sanagavarapu K, Yang X, Meisl G, Cohen SIA, Knowles TPJ, Linse S. Secondary nucleation in amyloid formation. Chem Commun (Camb) 2018;54:8667–8684. doi: 10.1039/c8cc02204f. [DOI] [PubMed] [Google Scholar]
- 13.Birol M, Kumar S, Rhoades E, Miranker AD. Conformational switching within dynamic oligomers underpins toxic gain-of-function by diabetes-associated amyloid. Nat Commun. 2018;9:1312. doi: 10.1038/s41467-018-03651-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 15.Lee SJ, Nam E, Lee HJ, Savelieff MG, Lim MH. Towards an understanding of amyloid-beta oligomers: characterization, toxicity mechanisms, and inhibitors. Chem Soc Rev. 2017;46:310–323. doi: 10.1039/c6cs00731g. [DOI] [PubMed] [Google Scholar]
- 16.Balbach JJ, Ishii Y, Antzutkin ON, Leapman RD, Rizzo NW, Dyda F, Reed J, Tycko R. Amyloid fibril formation by A beta 16-22, a seven-residue fragment of the Alzheimer’s beta-amyloid peptide, and structural characterization by solid state NMR. Biochemistry. 2000;39:13748–13759. doi: 10.1021/bi0011330. [DOI] [PubMed] [Google Scholar]
- 17.Blake C, Serpell L. Synchrotron X-ray studies suggest that the core of the transthyretin amyloid fibril is a continuous beta-sheet helix. Structure. 1996;4:989–998. doi: 10.1016/s0969-2126(96)00104-9. [DOI] [PubMed] [Google Scholar]
- 18.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model for Alzheimer’s beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci U S A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geddes AJ, Parker KD, Atkins ED, Beighton E. “Cross-beta” conformation in proteins. J Mol Biol. 1968;32:343–358. doi: 10.1016/0022-2836(68)90014-4. [DOI] [PubMed] [Google Scholar]
- 21.Luca S, Yau WM, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: constraints from solid-state NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer’s beta-amyloid fibrils. Proc Natl Acad Sci U S A. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuttle MD, Comellas G, Nieuwkoop AJ, Covell DJ, Berthold DA, Kloepper KD, Courtney JM, Kim JK, Barclay AM, Kendall A, et al. Solid-state NMR structure of a pathogenic fibril of full-length human alpha-synuclein. Nat Struct Mol Biol. 2016;23:409–415. doi: 10.1038/nsmb.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walti MA, Ravotti F, Arai H, Glabe CG, Wall JS, Bockmann A, Guntert P, Meier BH, Riek R. Atomic-resolution structure of a disease-relevant Abeta(1-42) amyloid fibril. Proc Natl Acad Sci U S A. 2016;113:E4976–4984. doi: 10.1073/pnas.1600749113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colvin MT, Silvers R, Frohm B, Su Y, Linse S, Griffin RG. High resolution structural characterization of Abeta42 amyloid fibrils by magic angle spinning NMR. J Am Chem Soc. 2015;137:7509–7518. doi: 10.1021/jacs.5b03997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falcon B, Zhang W, Murzin AG, Murshudov G, Garringer HJ, Vidal R, Crowther RA, Ghetti B, Scheres SHW, Goedert M. Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature. 2018;561:137–140. doi: 10.1038/s41586-018-0454-y. [Structure of Tau amyloid fibrils from patients with Pick’s disease is completely different from those observed in other tauopathies, which sustain the hypothesis that different amyloid folds are associated to different disease] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falcon B, Zivanov J, Zhang W, Murzin AG, Garringer HJ, Vidal R, Crowther RA, Newell KL, Ghetti B, Goedert M, et al. Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature. 2019;568:420–423. doi: 10.1038/s41586-019-1026-5. [Tau amyloid fibrils from patients with Chronic Traumatic Encephalopathy contain cofactor molecules into the amyloid core. It is likely that the cofactor alters kinetically or thermodynamically the formation of these fibrils] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fitzpatrick AWP, Falcon B, He S, Murzin AG, Murshudov G, Garringer HJ, Crowther RA, Ghetti B, Goedert M, Scheres SHW. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature. 2017;547:185–190. doi: 10.1038/nature23002. [Structure of Tau amyloid fibrils from patients with Alzheimer’s illustrates the molecular differences between two characteristic polymorphs and the presence of unstructured regions associated to the fibril core] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gremer L, Scholzel D, Schenk C, Reinartz E, Labahn J, Ravelli RBG, Tusche M, Lopez-Iglesias C, Hoyer W, Heise H, et al. Fibril structure of amyloid-beta(1-42) by cryo-electron microscopy. Science. 2017;358:116–119. doi: 10.1126/science.aao2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iadanza MG, Silvers R, Boardman J, Smith HI, Karamanos TK, Debelouchina GT, Su Y, Griffin RG, Ranson NA, Radford SE. The structure of a beta2-microglobulin fibril suggests a molecular basis for its amyloid polymorphism. Nat Commun. 2018;9:4517. doi: 10.1038/s41467-018-06761-6. [Different association between the same fibril units supports the amyloid polymorphism observed for β2m in vitro fibrillation] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guerrero-Ferreira R, Taylor NM, Mona D, Ringler P, Lauer ME, Riek R, Britschgi M, Stahlberg H. Cryo-EM structure of alpha-synuclein fibrils. eLife. 2018;7 doi: 10.7554/eLife.36402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li B, Ge P, Murray KA, Sheth P, Zhang M, Nair G, Sawaya MR, Shin WS, Boyer DR, Ye S, et al. Cryo-EM of full-length alpha-synuclein reveals fibril polymorphs with a common structural kernel. Nat Commun. 2018;9:3609. doi: 10.1038/s41467-018-05971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Zhao C, Luo F, Liu Z, Gui X, Luo Z, Zhang X, Li D, Liu C, Li X. Amyloid fibril structure of alpha-synuclein determined by cryo-electron microscopy. Cell Res. 2018;28:897–903. doi: 10.1038/s41422-018-0075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ni X, McGlinchey RP, Jiang J, Lee JC. Structural insights into alpha-synuclein fibril polymorphism: effects of Parkinson’s disease-related C-terminal truncations. J Mol Biol. 2019;431:3913–3919. doi: 10.1016/j.jmb.2019.07.001. [Truncation experiments demonstrate that regions that do not participate in the amyloid core define the structure of polymorphs observed for a-synuclein fibrillation in vitro] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liberta F, Loerch S, Rennegarbe M, Schierhorn A, Westermark P, Westermark GT, Hazenberg BPC, Grigorieff N, Fandrich M, Schmidt M. Cryo-EM fibril structures from systemic AA amyloidosis reveal the species complementarity of pathological amyloids. Nat Commun. 2019;10:1104. doi: 10.1038/s41467-019-09033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radamaker L, Lin YH, Annamalai K, Huhn S, Hegenbart U, Schonland SO, Fritz G, Schmidt M, Fandrich M. Cryo-EM structure of a light chain-derived amyloid fibril from a patient with systemic AL amyloidosis. Nat Commun. 2019;10:1103. doi: 10.1038/s41467-019-09032-0. [LDRs are not limited to the fibril ends and might contain protein−protein interaction motives, as in the case of LC amyloids from patients with systemic AL amyloidosis] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swuec P, Lavatelli F, Tasaki M, Paissoni C, Rognoni P, Maritan M, Brambilla F, Milani P, Mauri P, Camilloni C, et al. Cryo-EM structure of cardiac amyloid fibrils from an immunoglobulin light chain AL amyloidosis patient. Nat Commun. 2019;10:1269. doi: 10.1038/s41467-019-09133-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W, Falcon B, Murzin AG, Fan J, Crowther RA, Goedert M, Scheres SH. Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer’s and Pick’s diseases. eLife. 2019;8 doi: 10.7554/eLife.43584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antzutkin ON, Leapman RD, Balbach JJ, Tycko R. Supramolecular structural constraints on Alzheimer’s beta-amyloid fibrils from electron microscopy and solid-state nuclear magnetic resonance. Biochemistry. 2002;41:15436–15450. doi: 10.1021/bi0204185. [DOI] [PubMed] [Google Scholar]
- 40.Fitzpatrick AW, Debelouchina GT, Bayro MJ, Clare DK, Caporini MA, Bajaj VS, Jaroniec CP, Wang L, Ladizhansky V, Muller SA, et al. Atomic structure and hierarchical assembly of a cross-beta amyloid fibril. Proc Natl Acad Sci U S A. 2013;110:5468–5473. doi: 10.1073/pnas.1219476110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 42.Yamasaki TR, Holmes BB, Furman JL, Dhavale DD, Su BW, Song ES, Cairns NJ, Kotzbauer PT, Diamond MI. Parkinson’s disease and multiple system atrophy have distinct alpha-synuclein seed characteristics. J Biol Chem. 2019;294:1045–1058. doi: 10.1074/jbc.RA118.004471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibbons GS, Lee VMY, Trojanowski JQ. Mechanisms of cell-to-cell transmission of pathological Tau: a review. JAMA Neurol. 2019;76:101–108. doi: 10.1001/jamaneurol.2018.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiang W, Yau WM, Lu JX, Collinge J, Tycko R. Structural variation in amyloid-beta fibrils from Alzheimer’s disease clinical subtypes. Nature. 2017;541:217–221. doi: 10.1038/nature20814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaquer-Alicea J, Diamond MI. Propagation of protein aggregation in neurodegenerative diseases. Annu Rev Biochem. 2019;88:785–810. doi: 10.1146/annurev-biochem-061516-045049. [DOI] [PubMed] [Google Scholar]
- 46.Close W, Neumann M, Schmidt A, Hora M, Annamalai K, Schmidt M, Reif B, Schmidt V, Grigorieff N, Fandrich M. Physical basis of amyloid fibril polymorphism. Nat Commun. 2018;9:699. doi: 10.1038/s41467-018-03164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meinhardt J, Sachse C, Hortschansky P, Grigorieff N, Fandrich M. Abeta(1-40) fibril polymorphism implies diverse interaction patterns in amyloid fibrils. J Mol Biol. 2009;386:869–877. doi: 10.1016/j.jmb.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao Q, Boyer DR, Sawaya MR, Ge P, Eisenberg DS. Cryo-EM structures of four polymorphic TDP-43 amyloid cores. Nat Struct Mol Biol. 2019;26:619–627. doi: 10.1038/s41594-019-0248-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Falcon B, Zhang W, Schweighauser M, Murzin AG, Vidal R, Garringer HJ, Ghetti B, Scheres SHW, Goedert M. Tau filaments from multiple cases of sporadic and inherited Alzheimer’s disease adopt a common fold. Acta Neuropathol. 2018;136:699–708. doi: 10.1007/s00401-018-1914-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fichou Y, Lin Y, Rauch JN, Vigers M, Zeng Z, Srivastava M, Keller TJ, Freed JH, Kosik KS, Han S. Cofactors are essential constituents of stable and seeding-active tau fibrils. Proc Natl Acad Sci U S A. 2018;115:13234–13239. doi: 10.1073/pnas.1810058115. [Different cofactors give specific structural and functional properties to amyloid fibrils of the same protein, which highlight the role of interactions with the molecular environment in defining amyloid structure] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Simone A, Kitchen C, Kwan AH, Sunde M, Dobson CM, Frenkel D. Intrinsic disorder modulates protein self-assembly and aggregation. Proc Natl Acad Sci U S A. 2012;109:6951–6956. doi: 10.1073/pnas.1118048109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao L, Vecchi G, Vendruscolo M, Korner R, Hayer-Hartl M, Hartl FU. The Hsp70 chaperone system stabilizes a thermo-sensitive subproteome in E. coli. Cell Rep. 2019;28:1335–1345.:e6. doi: 10.1016/j.celrep.2019.06.081. [DOI] [PubMed] [Google Scholar]
- 53.Hosp F, Gutierrez-Angel S, Schaefer MH, Cox J, Meissner F, Hipp MS, Hartl FU, Klein R, Dudanova I, Mann M. Spatiotemporal proteomic profiling of Huntington’s disease inclusions reveals widespread loss of protein function. Cell Rep. 2017;21:2291–2303. doi: 10.1016/j.celrep.2017.10.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stewart KL, Radford SE. Amyloid plaques beyond Abeta: a survey of the diverse modulators of amyloid aggregation. Biophys Rev. 2017;9:405–419. doi: 10.1007/s12551-017-0271-9. [A comprehensive guide thoroughly described interactions of amyloid fibrils and cofactors] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fusco G, Pape T, Stephens AD, Mahou P, Costa AR, Kaminski CF, Kaminski Schierle GS, Vendruscolo M, Veglia G, Dobson CM, et al. Structural basis of synaptic vesicle assembly promoted by alpha-synuclein. Nat Commun. 2016;7:12563. doi: 10.1038/ncomms12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frottin F, Schueder F, Tiwary S, Gupta R, Korner R, Schlichthaerle T, Cox J, Jungmann R, Hartl FU, Hipp MS. The nucleolus functions as a phase-separated protein quality control compartment. Science. 2019;365:342–347. doi: 10.1126/science.aaw9157. [DOI] [PubMed] [Google Scholar]
- 57.Chothia C. Conformation of twisted beta-pleated sheets in proteins. J Mol Biol. 1973;75:295–302. doi: 10.1016/0022-2836(73)90022-3. [DOI] [PubMed] [Google Scholar]
- 58.Serpell LC, Blake CC, Fraser PE. Molecular structure of a fibrillar Alzheimer’s A beta fragment. Biochemistry. 2000;39:13269–13275. doi: 10.1021/bi000637v. [DOI] [PubMed] [Google Scholar]
- 59.Dinkel PD, Holden MR, Matin N, Margittai M. RNA binds to Tau fibrils and sustains template-assisted growth. Biochemistry. 2015;54:4731–4740. doi: 10.1021/acs.biochem.5b00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malishev R, Kolusheva S, Jelinek R. Vesicle-based assays to study membrane interactions of amyloid peptides. Methods Mol Biol. 2019;1873:39–51. doi: 10.1007/978-1-4939-8820-4_3. [DOI] [PubMed] [Google Scholar]
- 61.Viles JH. Metal ions and amyloid fiber formation in neurodegenerative diseases. Copper, zinc and iron in Alzheimer’s, Parkinson’s and prion diseases. Coord Chem Rev. 2012;256:2271–2284. [Google Scholar]
- 62.Stewart KL, Hughes E, Yates EA, Middleton DA, Radford SE. Molecular origins of the compatibility between glycosaminoglycans and Abeta40 amyloid fibrils. J Mol Biol. 2017;429:2449–2462. doi: 10.1016/j.jmb.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kanekiyo T, Xu H, Bu G. ApoE and Abeta in Alzheimer’s disease: accidental encounters or partners? Neuron. 2014;81:740–754. doi: 10.1016/j.neuron.2014.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valle-Delgado JJ, Alfonso-Prieto M, de Groot NS, Ventura S, Samitier J, Rovira C, Fernandez-Busquets X. Modulation of Abeta42 fibrillogenesis by glycosaminoglycan structure. FASEB J. 2010;24:4250–4261. doi: 10.1096/fj.09-153551. [DOI] [PubMed] [Google Scholar]
- 65.Iannuzzi C, Irace G, Sirangelo I. The effect of glycosaminoglycans (GAGs) on amyloid aggregation and toxicity. Molecules. 2015;20:2510–2528. doi: 10.3390/molecules20022510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bauerlein FJB, Saha I, Mishra A, Kalemanov M, Martinez-Sanchez A, Klein R, Dudanova I, Hipp MS, Hartl FU, Baumeister W, et al. In situ architecture and cellular interactions of polyQ inclusions. Cell. 2017;171:179–187.:e10. doi: 10.1016/j.cell.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 67.Guo Q, Lehmer C, Martinez-Sanchez A, Rudack T, Beck F, Hartmann H, Perez-Berlanga M, Frottin F, Hipp MS, Hartl FU, et al. In situ structure of neuronal C9orf72 poly-GA aggregates reveals proteasome recruitment. Cell. 2018;172:696–705.:e12. doi: 10.1016/j.cell.2017.12.030. [In situ characterisation of amyloid fibrillation inside the cell illustrates the many ways by which amyloid fibrils affect cellular homeostasis] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Michaels TCT, Saric A, Habchi J, Chia S, Meisl G, Vendruscolo M, Dobson CM, Knowles TPJ. Chemical kinetics for bridging molecular mechanisms and macroscopic measurements of amyloid fibril formation. Annu Rev Phys Chem. 2018;69:273–298. doi: 10.1146/annurev-physchem-050317-021322. [An excellent review on the kinetic steps of fibril formation including primary and secondary nucleation, fragmentation and elongation that provides a general formalism to understand and analyse amyloid fibrillation] [DOI] [PubMed] [Google Scholar]
- 69.Aprile FA, Sormanni P, Perni M, Arosio P, Linse S, Knowles TPJ, Dobson CM, Vendruscolo M. Selective targeting of primary and secondary nucleation pathways in Abeta42 aggregation using a rational antibody scanning method. Sci Adv. 2017;3:e1700488. doi: 10.1126/sciadv.1700488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Limbocker R, Chia S, Ruggeri FS, Perni M, Cascella R, Heller GT, Meisl G, Mannini B, Habchi J, Michaels TCT, et al. Trodusquemine enhances Abeta42 aggregation but suppresses its toxicity by displacing oligomers from cell membranes. Nat Commun. 2019;10:225. doi: 10.1038/s41467-018-07699-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rasmussen J, Mahler J, Beschorner N, Kaeser SA, Hasler LM, Baumann F, Nystrom S, Portelius E, Blennow K, Lashley T, et al. Amyloid polymorphisms constitute distinct clouds of conformational variants in different etiological subtypes of Alzheimer’s disease. Proc Natl Acad Sci U S A. 2017;114:13018–13023. doi: 10.1073/pnas.1713215114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]