Abstract
Background
Nicotinamide phosphoribosyltransferase (NAMPT), also known as pre-B-cell colony-enhancing factor (PBEF) or visfatin, has been reported to be a crucial factor involved in tumor metabolism, angiogenesis and cell apoptosis. However, its definite roles in patients with malignant cancer remain unclear.
Methods
Three online databases PubMed, Embase and Web of Science were looked through comprehensively for eligible articles, published before November, 2018. Pooled hazard ratios (HRs) with 95% confidence intervals (CIs) of overall survival (OS) or disease-free survival time or recurrence-free survival (DFS/RFS) were calculated to determine the associations between NAMPT expression and cancer prognosis.
Results
A total of ten eligible studies were finally enrolled for this analysis. Our results indicated that elevated NAMPT expression was associated with poor OS in breast cancer by both univariate and multivariate analysis (pooled HR =3.23, 95% CI: 1.93–5.41, I2=21.1%, P=0.283; pooled HR =3.34, 95% CI: 2.13–5.22, I2=0.0%, P=0.791; respectively) and in gastric cancer by univariate analysis (pooled HR =2.47, 95% CI: 1.07–5.73, I2=91.1%, P=0.001). Moreover, high expression of NAMPT was also related to poor DFS/RFS in breast cancer by univariate and multivariate analysis (pooled HR =3.85, 95% CI: 2.59–5.71, I2=0.0%, P=0.700; pooled HR =3.43, 95% CI: 2.36–4.99, I2=0.0%, P=0.737; separately). Similar results could be found in urothelial carcinoma (pooled HR =3.14, 95% CI: 1.73–5.71, I2=47.8%, P=0.166; pooled HR =3.06, 95% CI: 1.57–5.98, I2=0.0%, P=0.860). Besides, the translational level of NAMPT was also validated by UALCAN and the Human Protein Atlas database [immunohistochemistry (IHC)].
Conclusions
Our results shed light on that NAMPT might be an oncogenic factor in breast cancer, gastric cancer and urothelial carcinoma.
Keywords: Nicotinamide phosphoribosyltransferase (NAMPT), pre-B-cell colony-enhancing factor (PBEF), visfatin, cancer, meta-analysis
Introduction
As the most concerned health topic worldwide, cancer has been reported to be one of the main life limiting factors in the UK countries (1), the secondary life-threatening factor in the United States (2) and the first cause of death in China (3). In both developed and developing countries, it has also been an important burden restricting the social and economic development (4). At present, medical workers and researchers around the world are dedicating their lives to improve the quality of life in patients with malignant tumors and to prolong their survival time. However, the prognoses of many cancer patients remain poor. Hence, there is an urgent need to explore the possible mechanism affecting the prognosis of these patients.
Nicotinamide phosphoribosyltransferase, or simply called “NAMPT” for short, was regarded as an important rate-limiting enzyme involved in nicotinamide adenine dinucleotide (NAD+) biosynthesis. Moreover, both extracellular and intracellular NAMPT could control the conversation of nicotinamide into nicotinamide mononucleotide (NMN) (5), which was necessary for cancer cell growth, embracing transcription, DNA damage repair, cell-cycle regulation and some other metabolic processes (6). At the very beginning, NAMPT was also known as the pre-B-cell colony-enhancing factor (PBEF), a kind of cytokine, playing a vital role in B cell maturation and in inflammation (7,8). In addition to its enzymatic and cytokine-like functions, NAMPT was also known as visfatin for its high expression in visceral fat, having the insulin-mimetic function. However, the subsequent researches denied this role for uncertain data (9,10).
Recently, NAMPT had become a high-profile topic for its function involved in the progression of many malignant tumors, such as colorectal, ovarian, breast, gastric, prostate cancers and so on (11). NAMPT could be a valuable tumor biomarker and a strong predictor for the survival of cancer patients (12). Tumor cells seemed to be more susceptible to damage by NAMPT inhibitors than normal cells and the inhibition of NAMPT was expected to be a novel antitumor therapeutic strategy (13). However, few articles had analyzed the relationships between NAMPT and tumor prognosis. Its exact mechanism still had many controversies. Therefore, we conducted this meta-analysis to investigate the correlations between NAMPT expression and various cancer prognoses and also discussed the feasibility of using NAMPT as a biomarker.
Methods
Search strategy
This study was conducted in strict accordance with the PRISMA (preferred reporting items for systematic reviews and meta-analyses) guidelines (14). Three online databases PubMed, Embase and Web of Science were looked through systematically and comprehensively for eligible articles exploring the relationship between NAMPT/PBEF/visfatin and cancer prognosis, published before November, 2018. Search terms were as follows: (“NAMPT” or “VF” or “PBEF” or “PBEF1” or “VISFATIN” or “1110035O14Rik”) and (“tumor” or “tumour” or “neoplasm” or “cancer” or “carcinoma”) and (“prognosis” or “survival”). Additionally, we reviewed relevant systematic reviews and meta-analyses to identify articles that were not included in our literature search.
Inclusion and exclusion criteria
Criteria taken to screen for eligible studies were generalized as the following: (I) The subjects were patients with malignant tumors. (II) Focusing on association of NAMPT/PBEF/visfatin expression and cancer prognosis. (III) There was a contrast in survival between high NAMPT/PBEF/visfatin expression and low NAMPT/PBEF/visfatin expression. (IV) Adjusted hazard ratios (HRs) and associated 95% confidence intervals (CIs) were absolutely needed. Only peer-reviewed studies were included and papers reporting a case or conference abstract were excluded. Next, we evaluated the source of data in the included studies to avoid duplicates. If study populations were found to overlap, only data from the study with the most complete information were used. All titles, abstracts and full texts were independently reviewed by two authors. When we encountered an article not certain whether it met the inclusion criteria, another investigator would join in to make an appropriate decision.
Quality assessment
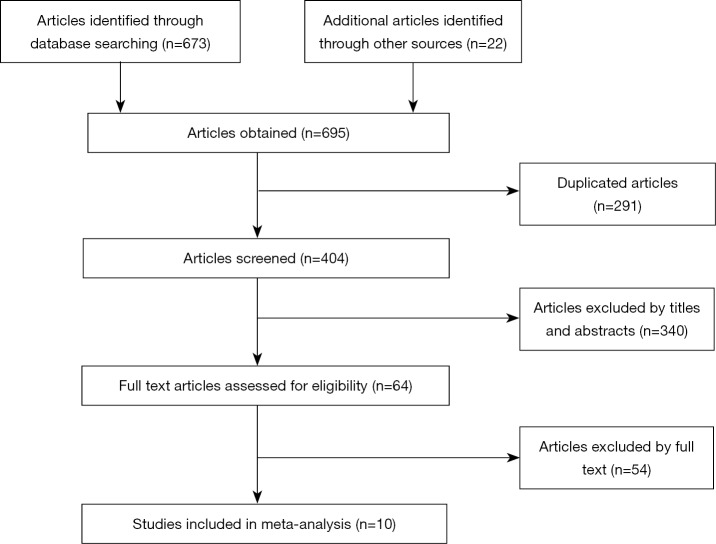
The quality of each included study was strictly evaluated according to a critical review checklist that the Meta-analysis of Observational Studies in Epidemiology (MOOSE) group of the Dutch Cochrane Center has proposed (15). The quality assessment focus on the below information in each study: the study design; the study population and country of origin; the assessment of reported outcomes; assay method applied for NAMPT/PBEF/visfatin testing; the NAMPT/PBEF/visfatin cut-off value; the detected tumor type and the information of pathology; and period of follow-up. We also performed Newcastle-Ottawa Scale (NOS) and sensitivity analyses to ensure the quality of this study. The process of the study selection was presented in Figure S1.
Figure S1.
Flow diagram of study selection process.
Data extraction
Two investigators independently extracted all data from eligible studies, including first author, year of publication, case nationality, age of patients with cancer (mean/median, range), the study design, tumor type, pathological information of the tumor, cut-off value, maximum months of follow-up, number of patients with NAMPT/PBEF/visfatin overexpression and low NAMPT/PBEF/visfatin expression, detected sample, the quantitative method for NAMPT/PBEF/visfatin expression, and HRs for survival outcome reported [overall survival (OS) and disease/recurrence/progression-free survival (DFS/RFS/PFS)] as well as their 95% CI and P values. In three studies, survival outcomes were only available in Kaplan-Meier curves. We extracted data from survival curves to deduce manually the overall HRs and corresponding 95% CIs by previously described methods (16,17). Once ambiguous data were found, three investigators would review them in detail. All of the aforementioned information was presented in Table 1 and Table 2.
Table 1. Main characteristics of studies included in the meta-analysis.
| First author, year | Case nationality | Dominant ethnicity | Media or mean age (years) | Study design | Malignant disease | Main type of pathology | Detected sample | Assay method | Survival analysis | Source of HR | Maximum months of follow-up | NOS scores |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pan et al., 2019 | China | Asian | NM | R | Colorectal cancer | AdenoCa | Tissue | IHC | OS | SC | 48 | 6 |
| Ke et al., 2015 | China | Asian | 64 | R | Urothelial carcinoma | TCC | Tissue | IHC | RFS | Reported | 145 | 8 |
| Aneta et al., 2017 | Poland | European | 54.6 | R | Endometrial cancer | AdenoCa | Serum | Immunoassay | RFS | SC | NM | 8 |
| Hung et al., 2016 | China | Asian | NM | R | Breast cancer | AdenoCa | Serum | Immunoassay | OS | Reported | 120 | 7 |
| DFS | ||||||||||||
| Lee et al., 2011 | China | Asian | NM | R | Breast cancer | AdenoCa | Tissue | IHC | DFS | Reported | 60 | 6 |
| OS | ||||||||||||
| Li et al., 2014 | China | Asian | NM | R | Breast cancer | AdenoCa | Serum | Immunoassay | DFS | Reported | NM | 6 |
| OS | ||||||||||||
| Lu et al., 2014 | China | Asian | NM | R | Gastric cancer | AdenoCa | Serum | Immunoassay | OS | Reported | 60 | 7 |
| Zhou et al., 2018 | China | Asian | 47 | R | Breast cancer | AdenoCa | Tissue | IHC | OS | Reported | NM | 7 |
| DFS | ||||||||||||
| Zhang et al., 2014 | China | Asian | NM | R | Urothelial carcinoma | TCC | Serum | ELISA | RFS | Reported | 60 | 7 |
| Li et al., 2012 | China | Asian | 58 | R | Gastric cancer | AdenoCa | Tissue | IHC | OS | SC | 96 | 7 |
HR, hazard ratio; NOS, Newcastle-Ottawa Scale; NM, not mentioned; R, retrospective; AdenoCa, adenocarcinoma; IHC, immunohistochemistry; OS, overall survival; SC, survival curve; TCC, transitional cell carcinoma; DFS, disease-free survival; RFS, progression-free survival.
Table 2. HRs and 95% CIs of patient survival or cancer progression relating to NAMPT/PBEF/visfatin expression in eligible studies.
| First author, publication year | Assay method | Cutoff point | Case number | Multivariate analysis | Univariate analysis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OS | DFS/RFS/PFS | OS | DFS/RFS/PFS | |||||||||||||
| High expression | Low expression | HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | |||||||
| Pan et al., 2018 | IHC | NM | 17 | 18 | 2.28 (1.09–4.76) | 0.023 | NM | NM | NM | NM | NM | NM | ||||
| Ke et al., 2015 | IHC | >50% of tumor cells stained | 60 | 45 | NM | NM | 3.22 (1.34–7.76) | 0.009 | NM | NM | 4.70 (2.06–10.76) | <0.001 | ||||
| Aneta et al., 2017 | Immunoassay | ≥ median | NM | NM | NM | NM | 0.56 (0.23–1.37) | 0.00002 | NM | NM | NM | NM | ||||
| Hung et al., 2016 | Immunoassay | ≥33.75 ng/mL | 176 | 82 | 3.55 (1.22–10.36) | 0.02 | 3.54 (1.48–8.47) | <0.001 | 5.67 (2.01–15.98) | 0.001 | NM | NM | ||||
| Lee et al., 2011 | IHC | Scores 3 and 4 | 50 | 55 | 6.20 (1.18–32.50) | 0.031 | 4.11 (1.15–14.70) | 0.03 | 5.05 (1.42–17.98) | 0.012 | 5.73 (1.92–17.14) | 0.002 | ||||
| Li et al., 2014 | Immunoassay | >65.6 ng/mL | NM | NM | 2.916 (1.684–5.038) | 0.003 | 3.695 (2.288–5.968) | 0.008 | 3.006 (1.741–5.193) | <0.001 | 3.780 (2.343–6.101) | <0.001 | ||||
| Lu et al., 2014 | Immunoassay | >78.4 ng/mL | 110 | 152 | NM | NM | NM | NM | 3.771 (2.735–5.195) | <0.001 | NM | NM | ||||
| Zhou et al., 2018 | IHC | Scores ≥3 | 40 | 43 | 5.00 (1.12–25) | 0.034 | 1.89 (0.63–5.88) | 0.255 | 1.34 (0.41–4.35) | 0.628 | 3.13 (1.27–7.73) | 0.013 | ||||
| Zhang et al., 2014 | ELISA | >14.74 ng/mL | 30 | 29 | 3.02 (0.39–23.37) | 0.291 | 2.85 (1.01–8.06) | 0.048 | 1.46 (0.24–8.73) | 0.679 | 2.02 (0.85–4.79) | 0.11 | ||||
| Li et al., 2012 | IHC | ≥ median | NM | NM | NM | NM | NM | NM | 1.60 (1.09–2.35) | <0.05 | NM | NM | ||||
HR, hazard ratio; CI, confidence interval; NAMPT, nicotinamide phosphoribosyltransferase; PBEF, pre-B-cell colony-enhancing factor; OS, overall survival; DFS, disease-free survival; RFS, recurrence-free survival; PFS, progression-free survival; IHC, immunohistochemistry; NM, not mentioned.
Statistical analysis
We evaluated the heterogeneity of the included literature using I2=100%*(Q−df)/Q. If P<0.10 or I2>50%, a random-effect model (DerSimonian-Laird method) was adopted. If P>0.10 or I2<50%, a fixed-effect model (Mantel-Haenszel method) was applied (18). Besides, subgroup analyses were used to explore the source of heterogeneity. We evaluated the risk of publication bias across studies by Begg’s funnel plot and Egger’s linear regression test (19). The pooled HR was considered insignificant if the CI overlaps 1. And if HR >1, it meant that cancer patients with high expression of NAMPT/PBEF/visfatin had poor survival. We also did a sensitivity analysis and carefully tested the P value. P<0.05 indicated the difference was statistically significant. In addition, all analyses were performed by the statistical software StataSE 12.0 (StataCorp LP, Texas, USA), and Microsoft Excel (V.2019, Microsoft Corporation, Washington, USA).
UALCAN and The Human Protein Atlas database
UALCAN (http://ualcan.path.uab.edu/index.html) is a user-friendly, interactive web resource for analyzing cancer transcriptome data (20). It is used to provide graphs and plots that describe gene expression and correlative patient survival information. The translational-level validation of NAMPT was carried out by using The Human Protein Atlas database (https://www.proteinatlas.org/) (21). These results from UALCAN and The Human Protein Atlas database were utilized to confirm the reliability of our meta-analysis at the translational level.
Results
Search results and Study characteristics
A total of 673 records were identified by searching PubMed, Embase and Web of Science. After removing duplicates (n=291), 404 records were retained for further review. By reviewing the literature titles and abstracts, we preliminarily removed 340 literatures, including 5 review and 3 meta-analyses. Among the remaining 64 records, 10 eligible were ultimately enrolled after reviewing full text articles (22-31) (Figure S1).
Tables 1,2 showed the main characters of the ten enrolled studies. All studies were published between 2011 and 2018. There were seven articles covering OS, seven articles covering RFS or DFS. Of all ten studies, 9 were Asian and the rest were European. Study designs of these ten literatures included were all retrospective studies. Colorectal cancer, urothelial cancer, endometrial cancer, breast cancer and gastric cancer were involved in these researches. Among them, there were four articles on breast cancer, two on gastric cancer and two on urothelial cancer, one on endometrial epithelial cancer and one on colorectal cancer. As for pathological types, adenocarcinoma (adenoCA) and transitional cell carcinoma (TCC) were included. To determine the expression of NAMPT/PBEF/visfatin, five studies examined tissue samples by immunohistochemistry (IHC), and other studies examined serum samples via ELISA or other immunoassays.
OS associated with NAMPT/PBEF/visfatin expression based on univariate analysis
A total of seven articles contributed to the univariate analysis of OS. On account of the significant heterogeneity (P=0.013, I2=62.8%), a random-effects model was adopted. The results showed that the high expression of NAMPT/PBEF/visfatin could indicate poor survival prognosis of patients with malignant tumors (pooled HR =2.75, 95% CI: 1.79–4.23; Figure 1A). We did further subgroup analysis to find the source of heterogeneity. In detected sample subgroup analysis, high expression of NAMPT/PBEF/visfatin seemed as a predictor of poor OS in serum (pooled HR =3.60, 95% CI: 2.76–4.69) as well as in tissue (pooled HR =1.90, 95% CI: 1.03–3.50). Subgroup analysis based on tissue (P=0.215, I2=34.9%) and serum samples (P=0.531, I2=0.0%) showed low heterogeneity (Figure 1B). In addition, we also performed subgroup analysis based on tumor types. The results showed that the high expression of NAMPT/PBEF/visfatin had a negative effect on the survival rate of patients with breast cancer (pooled HR =3.23, 95% CI: 1.93–5.41) and gastric cancer (pooled HR =2.47, 95% CI: 1.07–5.73). Subgroup analysis of gastric cancer patients showed significant heterogeneity (P=0.001, I2=91.1%), while the heterogeneity in breast cancer subgroup was low relatively (P=0.283, I2=21.1%) (Figure 1C). In the remaining reports, the results of other tumors overlapped with the invalid line with no significant difference.
Figure 1.
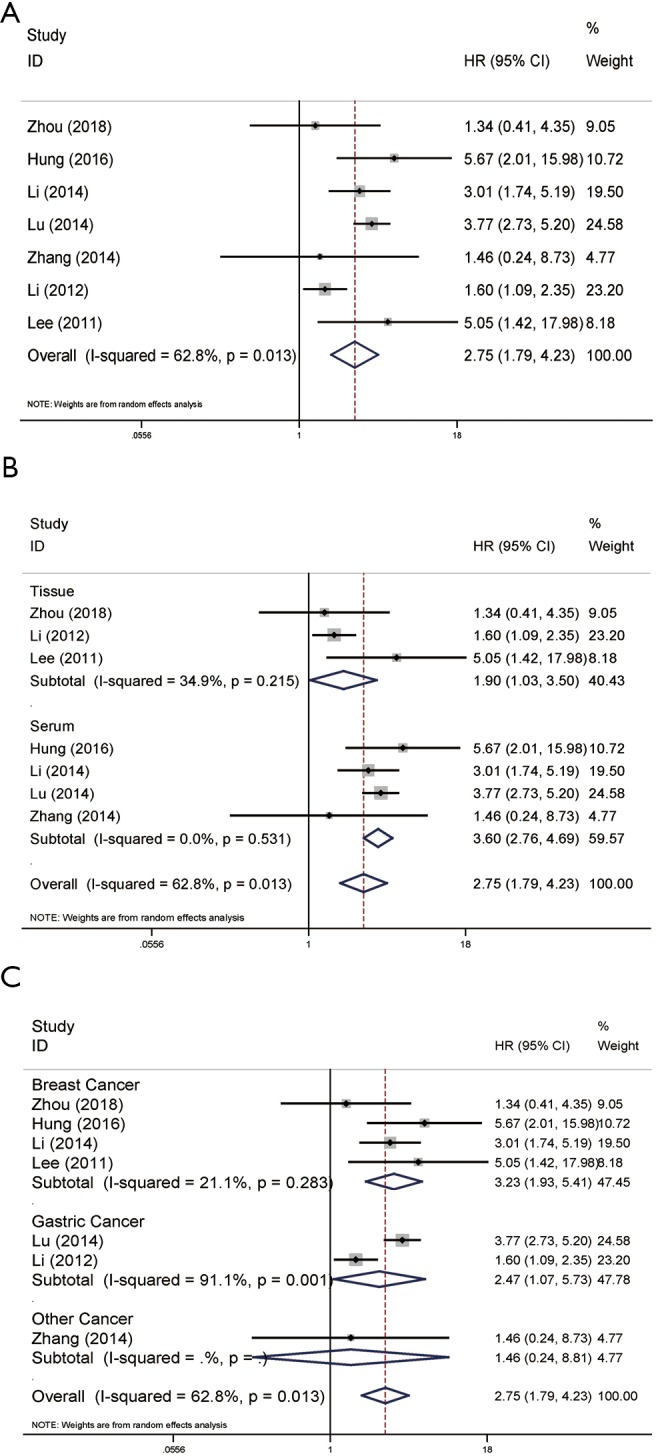
Forest plots of OS in association with NAMPT/PBEF/visfatin expression in various cancers based on univariate analysis. (A) The overall group; (B) the subgroup analysis of detected samples; (C) the subgroup analysis of cancer types. HR, hazard ratio; CI, confidence interval; OS, overall survival; NAMPT, nicotinamide phosphoribosyltransferase; PBEF, pre-B-cell colony-enhancing factor.
OS associated with NAMPT/PBEF/visfatin expression based on multivariate analysis
A total of six articles contributed to the multivariate analysis of the OS. We used a fixed-effects model due to the low heterogeneity (P=0.877, I2=0.0%) showed by these articles. The pooled HR (3.01, 95% CI: 2.07–4.39; Figure 2A) demonstrated that high NAMPT/PBEF/visfatin play an unfavorable role of OS in malignant tumor patients. Through subgroup analysis, we found consistent results in tissue samples (pooled HR =2.97, 95% CI: 1.60–5.50) as well as serum samples (pooled HR =3.04, 95% CI: 1.89–4.88; Figure 2B). Cancer type subgroup analysis was also conducted and showed the negative effect of high NAMPT/PBEF/visfatin expression in breast cancer (pooled HR =3.34, 95% CI: 2.13–5.22; Figure 2C).
Figure 2.
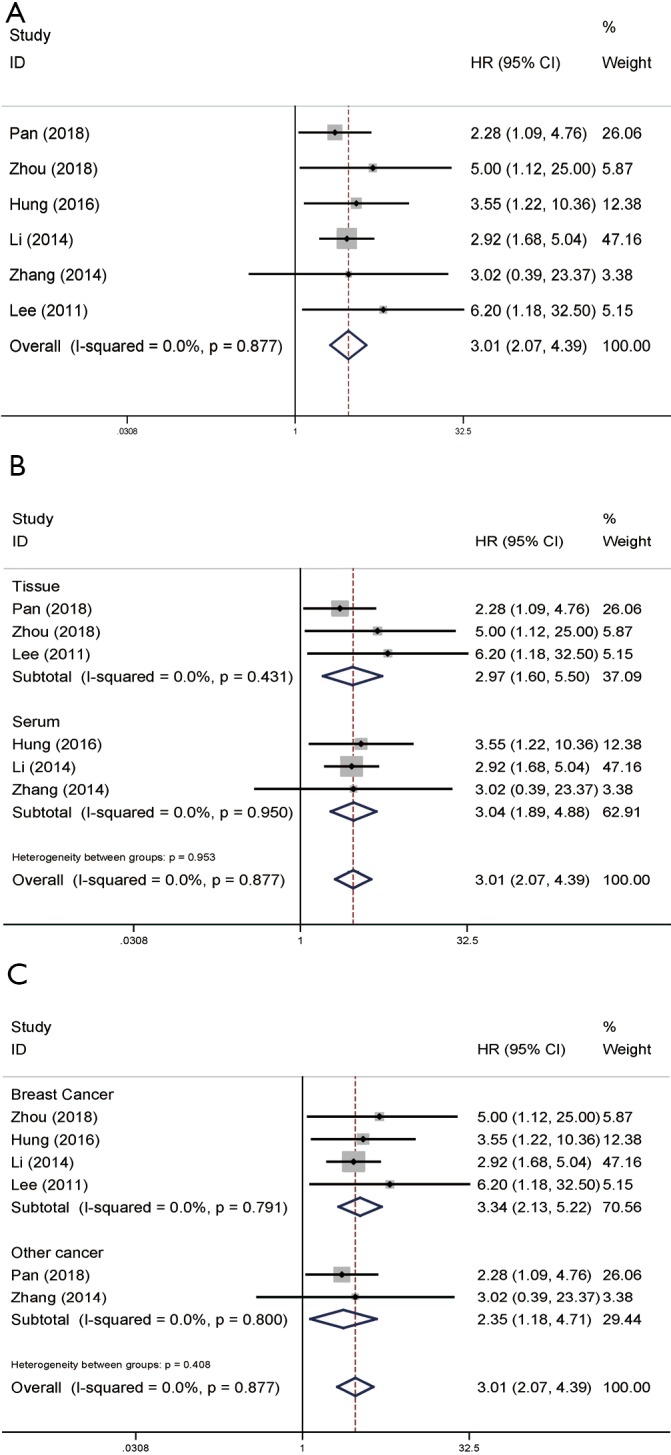
Forest plots of OS in association with NAMPT/PBEF/visfatin expression in various cancers based on multivariate analysis. (A) The overall group; (B) the subgroup analysis of detected samples; (C) the subgroup analysis of cancer types. HR, hazard ratio; CI, confidence interval; OS, overall survival; NAMPT, nicotinamide phosphoribosyltransferase; PBEF, pre-B-cell colony-enhancing factor.
DFS/RFS associated with NAMPT/PBEF/visfatin expression based on univariate analysis
The calculated individual and pooled HRs for DFS/RFS based on univariate analysis were presented specifically in Figure 3. The pooled HR (3.62, 95% CI: 2.60–5.03) showed that high NAMPT/PBEF/visfatin expression may be an unfavorable predictor of progression in patients with cancer. Because of the low heterogeneity (P=0.568, I2=0.0%; Figure 3A), a fixed-effects model was adopted. We further did subgroup analysis based on detected samples and cancer types, the results are consistent. In the tissue subgroup, the HR of the tissue group was 4.28 (95% CI: 2.51–7.28; Figure 3B). The HR of the serum group was 3.26 (95% CI: 2.15–4.96; Figure 3B). In the tumor type subgroup, the HR of the breast cancer group was 3.85 (95% CI: 2.59–5.71; Figure 3C), and that of the urothelial carcinoma was 3.14 (95% CI: 1.73–5.71; Figure 3C).
Figure 3.
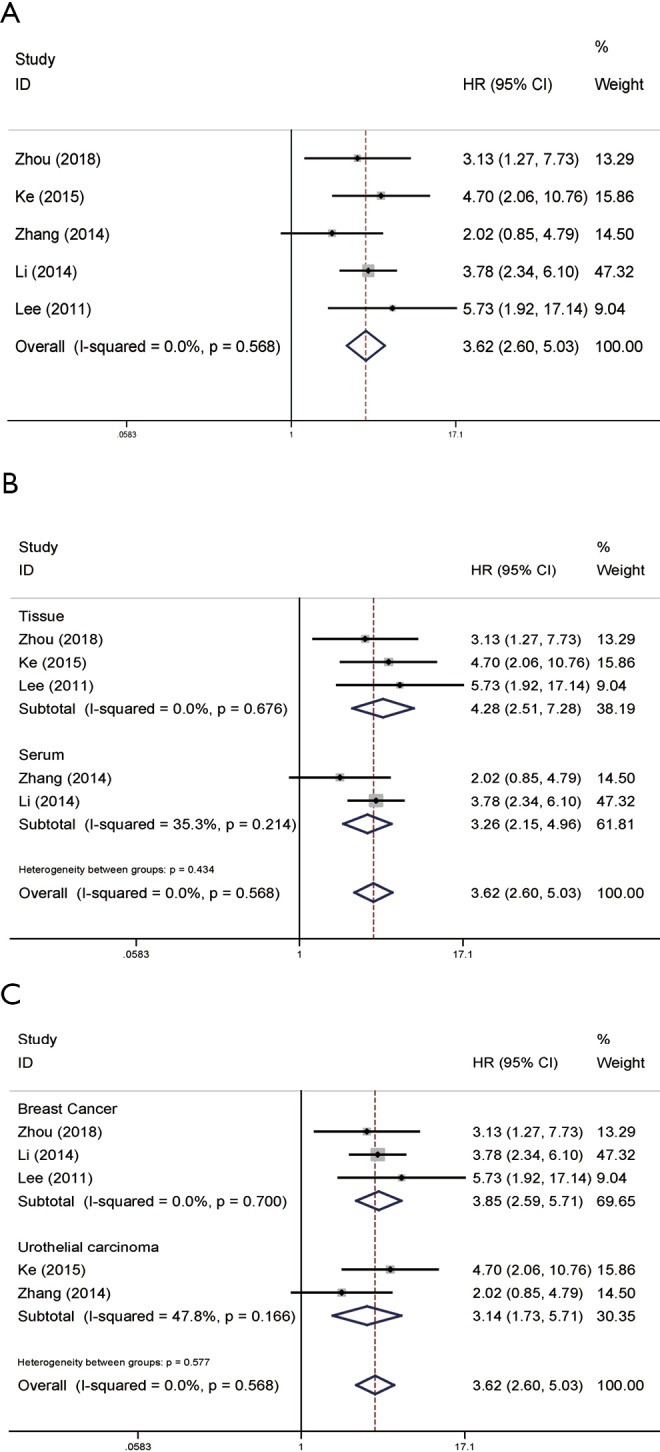
Forest plots of DFS/RFS in association with NAMPT/PBEF/visfatin expression in various cancers based on univariate analysis. (A) The overall group; (B) the subgroup analysis of detected samples; (C) the subgroup analysis of cancer types. HR, hazard ratio; CI, confidence interval; DFS, disease-free survival; RFS, recurrence-free survival; NAMPT, nicotinamide phosphoribosyltransferase; PBEF, pre-B-cell colony-enhancing factor.
DFS/RFS associated with NAMPT/PBEF/visfatin expression based on multivariate analysis
As displayed in Figure 4, it provides the result of seven literature, exploring the association between NAMPT/PBEF/visfatin expression and DFS/RFS based on multivariate analysis. The pooled HR was 2.47 (95% CI: 1.46–4.18) with slightly hard heterogeneity (P=0.021, I2=59.8%) (Figure 4A). Thus, a random-effects model was performed. In detected samples subgroup, the results of tissue group indicated that high NAMPT/PBEF/visfatin expression was negatively correlated with the prognosis of cancer patients (pooled HR =2.91, 95% CI: 1.58–5.34; Figure 4B), while the result of Serum group overlaps the invalid line (Figure 4B). In subgroup analysis of tumor types, results for breast and urothelial carcinoma were consistent with the overall results (pooled HR =3.43, 95% CI: 2.36–4.99; pooled HR =3.06, 95% CI: 1.57–5.98; Figure 4C).
Figure 4.
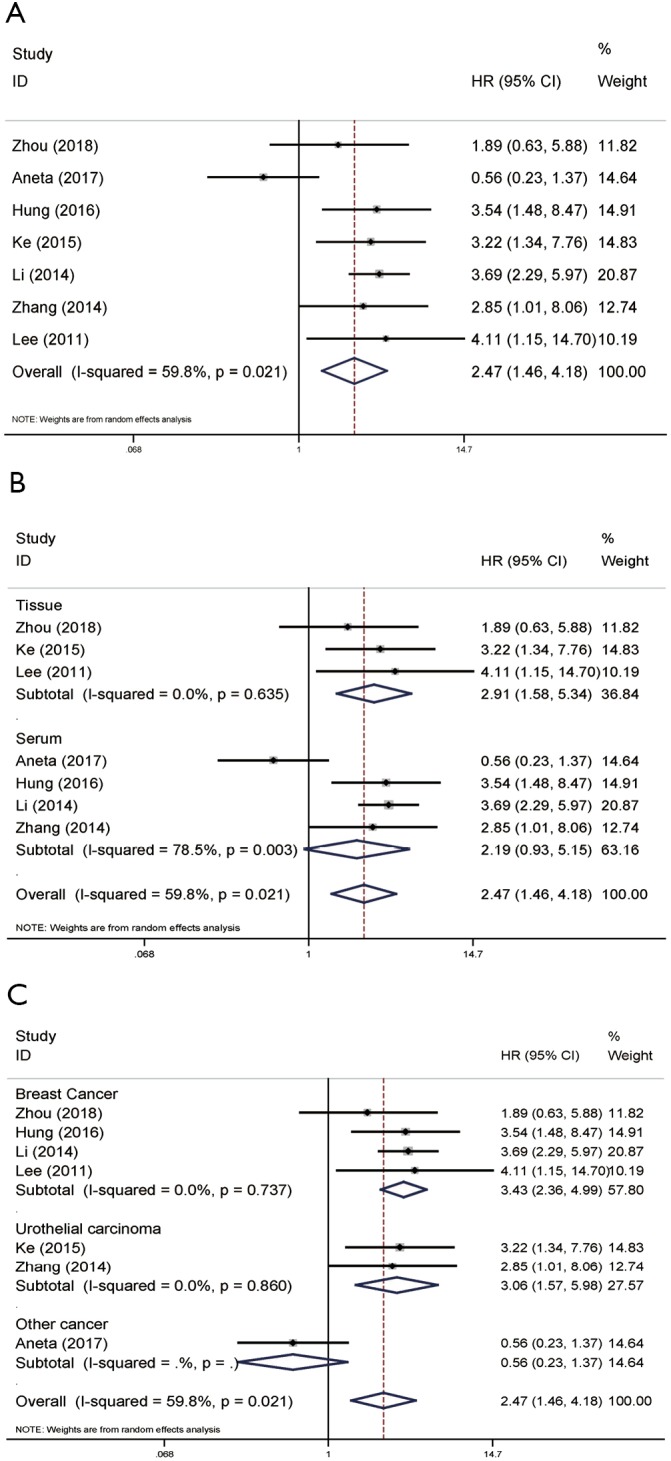
Forest plots of DFS/RFS in association with NAMPT/PBEF/visfatin expression in various cancers based on multivariate analysis. (A) The overall group; (B) the subgroup analysis of detected samples; (C) the subgroup analysis of cancer types. HR, hazard ratio; CI, confidence interval; DFS, disease-free survival; RFS, recurrence-free survival; NAMPT, nicotinamide phosphoribosyltransferase; PBEF, pre-B-cell colony-enhancing factor.
Sensitivity analysis and publication bias
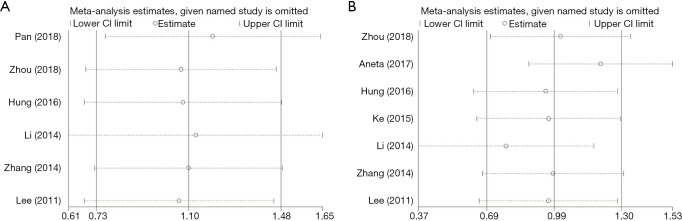
A sensitivity analysis was conducted by Stata12.0 software to judge whether our results on OS and DFS/RFS are reliable. We mainly conducted a sensitivity analysis on OS and DFS/RFS studies. To determine the stability of this study, we removed the included literature one by one and observed the changes in the HR with 95% CIs. The results of our analysis showed that our research is quite reliable (Figure S2).
Figure S2.
Sensitivity analysis of each included study. (A) OS for individual studies by multivariate analysis; (B) DFS/RFS for individual studies by multivariate analysis. CI, confidence interval; OS, overall survival; DFS, disease-free survival; RFS, recurrence-free survival.
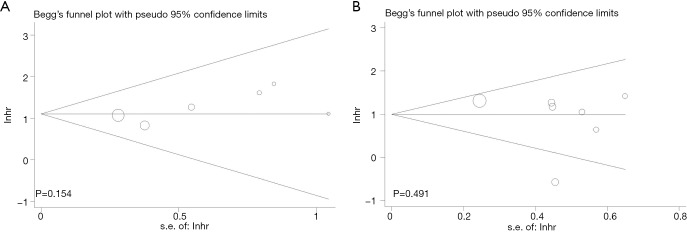
In order to determine the publication bias that may exist in this meta-analysis, Begg’s funnel and the Egger’s test were used. Meanwhile, funnel plots were presented in Figure S3. In the comprehensive analysis of OS or DFS/RFS, the P values of Begg’s test and the P values of Egger’s test were all above 0.05, indicating no publication bias in this study.
Figure S3.
Begg’s funnel plots of the publication bias. (A) OS for individual studies by multivariate analysis; (B) DFS/RFS for individual studies by multivariate analysis. OS, overall survival; DFS, disease-free survival; RFS, recurrence-free survival.
Validation of NAMPT/PBEF/visfatin in the translational level
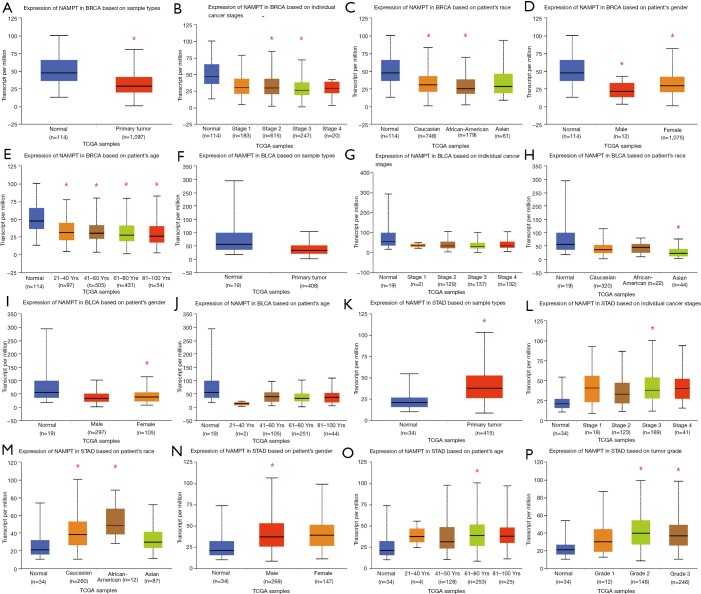
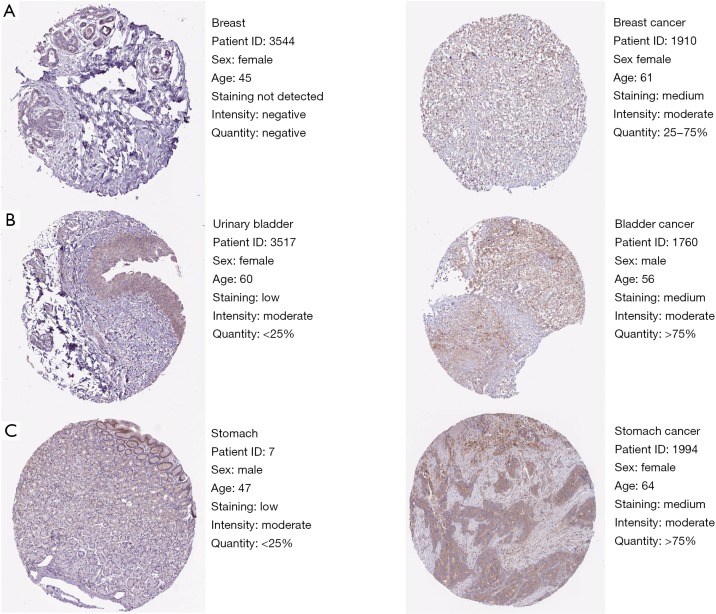
As presented in Figure 5A,B,C,D,E, it showed that in breast cancer patients, NAMPT/PBEF/visfatin expression differs between sample types, tumor stages, patient races, patient genders, and patient age. Expressions of NAMPT in bladder cancer based on sample types, tumor stages, patient races, patient genders and patient age were presented in Figure 5F,G,H,I,J. Figure 5K,L,M,N,O,P provided the expression of NAMPT in gastric cancer patients. IHC status of the above three tumor tissues was shown in Figure 6, which further verified that NAMPT could be a poor predictor of prognosis.
Figure 5.
Boxplots of NAMPT/PBEF/vifatin expression level based on UALCAN. (A,B,C,D,E) Breast cancer (BRCA); (F,G,H,I,J) bladder cancer (BLCA); (K,L,M,N,O,P) stomach adenocarcinoma((STAD). *, P<0.05. NAMPT, nicotinamide phosphoribosyltransferase; TCGA, The Cancer Genome Atlas; PBEF, pre-B-cell colony-enhancing factor.
Figure 6.
Validation of NAMPT/PBEF/visfatin in the translational level by The Human Protein Atlas database (IHC). The translational expression level of NAMPT/PBEF/visfatin was positively correlated with disease status as they were upregulated in cancer samples. (A) Breast cancer sample (40×); (B) bladder cancer sample (40×); (C) stomach cancer sample (40×). NAMPT, nicotinamide phosphoribosyltransferase; PBEF, pre-B-cell colony-enhancing factor; IHC, immunohistochemistry.
Discussion
Accumulating data had explored the prognostic roles of NAMPT in various malignant tumors, however, the definite relationships between NAMPT and prognosis of patients with malignant tumors remained ambiguous. Our study combined the results of univariate and multivariate analyses to further elucidate their associations. No such study had been reported before. Subgroup analysis, sensitivity analysis and heterogeneity analysis were also carried out. The results of most studies suggested that cancer patients with high NAMPT expression shall have a poor prognosis. The pooled HR of OS is 2.75 (95% CI: 1.79–4.23) based on univariate analysis,while it was 3.01 (95% CI: 2.07–4.39) in multivariate analysis. The pooled HR of DFS/RFS was 3.62 (95% CI: 2.60–5.03) based on univariate analysis, while it was 2.47 (95% CI: 1.46–4.18) in multivariate analysis. Of all these studies, only one literature had the opposite result of the HR <1 (22). Subsequent subgroup analysis and sensitivity analysis were further performed. In subgroup analysis of tumor types, survival of breast cancer, urothelial carcinoma and gastric cancer showed significant statistical significance in both univariate and multivariate analysis. As for breast cancer, urothelial carcinoma and gastric cancer, shorter OS or DFS/RFS could be inferred, if the NAMPT/PBEF/visfatin expression was high. Moreover, no significant publication biases were found, indicating this meta-analysis was relatively stable. In addition, we also implemented a search in UALCAN and The Human Protein Atlas database and the results were consistent with ours, proving that our meta-analysis had certain credibility to some extent.
With the proposal of Warburg effect theory, energy source of tumors was not completely dependent on the tricarboxylic acid cycle, glycolysis had also become an important metabolic pathway (32). Recently, NAD+ had attracted more and more researchers' attention, playing an important coenzyme role not only in cell redox reaction, but also in many signaling pathways including calcium signaling, cellular metabolism, mitochondrial biogenesis, DNA repair and so on (33-35). Normally, NAD+ levels in cells decreased with consumption, but their expression could also be up-regulated in tumor cells, owing to increased energy demand (36,37). Inhibiting the biosynthesis of NAD+ provided a new idea for future anti-tumor researches.
As for NAMPT, it was a key rate-limiting enzyme in the biosynthesis of NAD+ (38,39), which partly explained the elevated NAMPT expressions in patients with malignant tumors. In addition, NAMPT had an important function in promoting angiogenesis via a variety of pathways, related to cancer angiogenesis (40). SIRT6, as one of the NAD+-dependent deacetylases, was crucial in the development of metabolism, cancer and inflammation (41,42). SIRT6 had been reported to have multiple enzyme activities, including deacetylase activity, de-fatty acylase activity, and mono-ADP-ribosyltransferase activity (43-45). A researcher explored the role of SIRT6 in cancer cells and found that the expression of SIRT6 had a negative correlation with NAMPT (46). Subsequent researchers also believed that SIRT6 could regulate the activity of NAMPT and displayed a crucial effect on maintaining the sufficient level of NAD+ in tumors (47). As a hot topic in recent years, SIRT1 was also related to the expression of NAMPT. SIRT1 was well known for its role in tumor metabolism and had the ability to inhibit the activity of oncogene p53 and improve the expression level of oncogene c-MYC (48-50). Recent studies found that in prostate cancer, liver cancer and breast cancer, NAMPT could regulate pi3k-akt, mapk-erk1/2 and p38 signaling pathways and promote the proliferation of tumor cells (5,51,52). By upregulating phosphorylation of GSK-3β, tumor cell apoptosis could be avoided (53-55). NAMPT could also directly or indirectly promote tumor inflammation and help tumors escape immune damage by regulating the expressions of CD50/CD40/CD80, IL-1/IL-6, NF-κB and so on (56-58). The roles of NAMPT in the prognosis of patients with malignant tumors might be associated with these molecular mechanisms and biological activities.
Even though our analysis had rigorously followed the PRISMA guidelines and applied rigorous statistical methods, the conclusions were still limited for the following reasons. Firstly, OS and DFS/RFS analysis had some heterogeneity, which might be caused by the fact that three groups of data were extracted from the survival curve (22,27,29), the source of HR was inconsistent. It was also possible with the difference in patient characteristics including age, race, tumor type and so on. In particular, when it comes to race, 9 of the 10 studies we included were Chinese. This might affect the persuasiveness of our findings. Secondly, the samples included in the study were not sufficient, especially the persuasiveness of gastric cancer and urothelial cancer was not enough. Thirdly, there was no uniform standard for the critical value of NAMPT expression, and the cut-off value of each article was different. This might bias the role of NAMPT in tumor prognosis. Fourthly, since these studies were retrospective and lacked prospective studies, our results might be impacted. Finally, we only analyzed the relationship between the single expression of NAMPT and the prognosis of malignant tumors. Recent studies had shown that the NAMPT expression in tumor tissues was decreased after the use of NAMPT inhibitors, but the tumors could show tolerance (59), which might be related to other pathways of NAD+ biosynthesis, such as the NAPRT pathway. From what had been discussed above, NAMPT/PBEF/visfatin was a potential prognostic tool for patients with malignant tumors. More well-designed studies were warranted to make the impact of NAMPT clear in different cancers. In the future, we shall combine NAMPT and NAPRT to study their relationship with tumorigenesis and development.
Conclusions
In summary, this systematic review and meta-analysis clearly revealed that high expression of NAMPT/PBEF/visfatin was associated with poor prognosis of cancer patients, especially in breast cancer, gastric cancer and urothelial carcinoma, indicating it could serve as a prognostic biomarker for these tumors. More well-designed studies on the relationship between NAMPT/PBEF/visfatin expression and the prognosis of cancer patients were also warranted to provide additional evidence for its future clinical value and the biological mechanism of NAMPT/PBEF/visfatin in tumors was of great research value.
Acknowledgments
We would like to thank the researchers and study participants for their contributions.
Funding: This article was funded by Medical key talent of Jiangsu Province: ZDRCA2016009 and the National Natural Science Foundation of China (grant number: 81871151).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Steel N, Ford JA, Newton JN, et al. Changes in health in the countries of the UK and 150 English Local Authority areas 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 2018;392:1647-61. 10.1016/S0140-6736(18)32207-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 4.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 5.Revollo JR, Grimm AA, Imai S. The regulation of nicotinamide adenine dinucleotide biosynthesis by Nampt/PBEF/visfatin in mammals. Curr Opin Gastroenterol 2007;23:164-70. 10.1097/MOG.0b013e32801b3c8f [DOI] [PubMed] [Google Scholar]
- 6.Chiarugi A, Dölle C, Felici R, et al. The NAD metabolome--a key determinant of cancer cell biology. Nat Rev Cancer 2012;12:741-52. 10.1038/nrc3340 [DOI] [PubMed] [Google Scholar]
- 7.Samal B, Sun Y, Stearns G, et al. Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol Cell Biol 1994;14:1431-7. 10.1128/MCB.14.2.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moschen AR, Gerner RR, Tilg H. Pre-B cell colony enhancing factor/NAMPT/visfatin in inflammation and obesity-related disorders. Curr Pharm Des 2010;16:1913-20. 10.2174/138161210791208947 [DOI] [PubMed] [Google Scholar]
- 9.Fukuhara A, Matsuda M, Nishizawa M, et al. Visfatin: a protein secreted by visceral fat that mimics the effects of insulin. Science 2005;307:426-30. 10.1126/science.1097243 [DOI] [PubMed] [Google Scholar]
- 10.Fukuhara A, Matsuda M, Nishizawa M, et al. Retraction. Science 2007;318:565. 10.1126/science.318.5850.565b [DOI] [PubMed] [Google Scholar]
- 11.Galli U, Travelli C, Massarotti A, et al. Medicinal chemistry of nicotinamide phosphoribosyltransferase (NAMPT) inhibitors. J Med Chem 2013;56:6279-96. 10.1021/jm4001049 [DOI] [PubMed] [Google Scholar]
- 12.Bi TQ, Che XM. Nampt/PBEF/visfatin and cancer. Cancer Biol Ther 2010;10:119-25. 10.4161/cbt.10.2.12581 [DOI] [PubMed] [Google Scholar]
- 13.Hasmann M, Schemainda I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res 2003;63:7436-42. [PubMed] [Google Scholar]
- 14.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 2009;339:b2535. 10.1136/bmj.b2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 2000;283:2008-12. 10.1001/jama.283.15.2008 [DOI] [PubMed] [Google Scholar]
- 16.Williamson PR, Smith CT, Hutton JL, et al. Aggregate data meta-analysis with time-to-event outcomes. Stat Med 2002;21:3337-51. 10.1002/sim.1303 [DOI] [PubMed] [Google Scholar]
- 17.Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007;8:16. 10.1186/1745-6215-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986;7:177-88. 10.1016/0197-2456(86)90046-2 [DOI] [PubMed] [Google Scholar]
- 19.Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629-34. 10.1136/bmj.315.7109.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- 22.Cymbaluk-Ploska A, Chudecka-Glaz A, Pius-Sadowska E, et al. Circulating Serum Level of Visfatin in Patients with Endometrial Cancer. Biomed Res Int 2018;2018:8576179. 10.1155/2018/8576179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hung AC, Lo S, Hou MF, et al. Extracellular Visfatin-Promoted Malignant Behavior in Breast Cancer Is Mediated Through c-Abl and STAT3 Activation. Clin Cancer Res 2016;22:4478-90. 10.1158/1078-0432.CCR-15-2704 [DOI] [PubMed] [Google Scholar]
- 24.Ke HL, Lin HH, Li WM, et al. High visfatin expression predicts poor prognosis of upper tract urothelial carcinoma patients. Am J Cancer Res 2015;5:2447-54. [PMC free article] [PubMed] [Google Scholar]
- 25.Lee YC, Yang YH, Su JH, et al. High visfatin expression in breast cancer tissue is associated with poor survival. Cancer Epidemiol Biomarkers Prev 2011;20:1892-901. 10.1158/1055-9965.EPI-11-0399 [DOI] [PubMed] [Google Scholar]
- 26.Li XY, Tang SH, Zhou XC, et al. Preoperative serum visfatin levels and prognosis of breast cancer among Chinese women. Peptides 2014;51:86-90. 10.1016/j.peptides.2013.11.010 [DOI] [PubMed] [Google Scholar]
- 27.Long HL, Che XM, Bi TQ, et al. The expression of nicotinamide phosphoribosyl transferase and vascular endothelial growth factor-A in gastric carcinoma and their clinical significance. Zhonghua Wai Ke Za Zhi 2012;50:839-42. [PubMed] [Google Scholar]
- 28.Lu GW, Wang QJ, Xia MM, et al. Elevated plasma visfatin levels correlate with poor prognosis of gastric cancer patients. Peptides 2014;58:60-4. 10.1016/j.peptides.2014.05.016 [DOI] [PubMed] [Google Scholar]
- 29.Pan JH, Zhou H, Zhu SB, et al. Nicotinamide phosphoribosyl transferase regulates cell growth via the Sirt1/P53 signaling pathway and is a prognosis marker in colorectal cancer. J Cell Physiol 2019;234:4385-95. 10.1002/jcp.27228 [DOI] [PubMed] [Google Scholar]
- 30.Zhang K, Zhou B, Zhang P, et al. Prognostic value of serum nicotinamide phosphoribosyltransferase in patients with bladder cancer. Croat Med J 2014;55:507-13. 10.3325/cmj.2014.55.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou SJ, Bi TQ, Qin CX, et al. Expression of NAMPT is associated with breast invasive ductal carcinoma development and prognosis. Oncol Lett 2018;15:6648-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 2012;21:297-308. 10.1016/j.ccr.2012.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corda D, Di Girolamo M. Functional aspects of protein mono-ADP-ribosylation. EMBO J 2003;22:1953-8. 10.1093/emboj/cdg209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee HC. Physiological functions of cyclic ADP-ribose and NAADP as calcium messengers. Annu Rev Pharmacol Toxicol 2001;41:317-45. 10.1146/annurev.pharmtox.41.1.317 [DOI] [PubMed] [Google Scholar]
- 35.Ménissier de Murcia J, Ricoul M, Tartier L, et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J 2003;22:2255-63. 10.1093/emboj/cdg206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshino J, Mills KF, Yoon MJ, et al. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab 2011;14:528-36. 10.1016/j.cmet.2011.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajman L, Chwalek K, Sinclair DA. Therapeutic Potential of NAD-Boosting Molecules: The In Vivo Evidence. Cell Metab 2018;27:529-47. 10.1016/j.cmet.2018.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rongvaux A, Shea RJ, Mulks MH, et al. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur J Immunol 2002;32:3225-34. [DOI] [PubMed] [Google Scholar]
- 39.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem 2004;279:50754-63. 10.1074/jbc.M408388200 [DOI] [PubMed] [Google Scholar]
- 40.Garten A, Petzold S, Korner A, et al. Nampt: linking NAD biology, metabolism and cancer. Trends Endocrinol Metab 2009;20:130-8. 10.1016/j.tem.2008.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kugel S, Mostoslavsky R. Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem Sci 2014;39:72-81. 10.1016/j.tibs.2013.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lerrer B, Gertler AA, Cohen HY. The complex role of SIRT6 in carcinogenesis. Carcinogenesis 2016;37:108-18. 10.1093/carcin/bgv167 [DOI] [PubMed] [Google Scholar]
- 43.Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem 2013;288:31350-6. 10.1074/jbc.C113.511261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liszt G, Ford E, Kurtev M, et al. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem 2005;280:21313-20. 10.1074/jbc.M413296200 [DOI] [PubMed] [Google Scholar]
- 45.Mao Z, Hine C, Tian X, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science 2011;332:1443-6. 10.1126/science.1202723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sociali G, Grozio A, Caffa I, et al. SIRT6 deacetylase activity regulates NAMPT activity and NAD(P)(H) pools in cancer cells. FASEB J 2019;33:3704-17. 10.1096/fj.201800321R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sampath D, Zabka TS, Misner DL, et al. Inhibition of nicotinamide phosphoribosyltransferase (NAMPT) as a therapeutic strategy in cancer. Pharmacol Ther 2015;151:16-31. 10.1016/j.pharmthera.2015.02.004 [DOI] [PubMed] [Google Scholar]
- 48.Menssen A, Hydbring P, Kapelle K, et al. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc Natl Acad Sci U S A 2012;109:E187-96. 10.1073/pnas.1105304109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ong ALC, Ramasamy TS. Role of Sirtuin1-p53 regulatory axis in aging, cancer and cellular reprogramming. Ageing Res Rev 2018;43:64-80. 10.1016/j.arr.2018.02.004 [DOI] [PubMed] [Google Scholar]
- 50.Knight JR, Milner J. SIRT1, metabolism and cancer. Curr Opin Oncol 2012;24:68-75. 10.1097/CCO.0b013e32834d813b [DOI] [PubMed] [Google Scholar]
- 51.Patel ST, Mistry T, Brown JE, et al. A novel role for the adipokine visfatin/pre-B cell colony-enhancing factor 1 in prostate carcinogenesis. Peptides 2010;31:51-7. 10.1016/j.peptides.2009.10.001 [DOI] [PubMed] [Google Scholar]
- 52.Kim JG, Kim EO, Jeong BR, et al. Visfatin stimulates proliferation of MCF-7 human breast cancer cells. Mol Cells 2010;30:341-5. 10.1007/s10059-010-0124-x [DOI] [PubMed] [Google Scholar]
- 53.Ninomiya S, Shimizu M, Imai K, et al. Possible role of visfatin in hepatoma progression and the effects of branched-chain amino acids on visfatin-induced proliferation in human hepatoma cells. Cancer Prev Res (Phila) 2011;4:2092-100. 10.1158/1940-6207.CAPR-11-0340 [DOI] [PubMed] [Google Scholar]
- 54.Luo J. Glycogen synthase kinase 3beta (GSK3beta) in tumorigenesis and cancer chemotherapy. Cancer Lett 2009;273:194-200. 10.1016/j.canlet.2008.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Desbois-Mouthon C, Blivet-Van Eggelpoël MJ, Beurel E, et al. Dysregulation of glycogen synthase kinase-3beta signaling in hepatocellular carcinoma cells. Hepatology 2002;36:1528-36. 10.1002/hep.1840360630 [DOI] [PubMed] [Google Scholar]
- 56.Bauer L, Venz S, Junker H, et al. Nicotinamide phosphoribosyltransferase and prostaglandin H2 synthase 2 are up-regulated in human pancreatic adenocarcinoma cells after stimulation with interleukin-1. Int J Oncol 2009;35:97-107. [DOI] [PubMed] [Google Scholar]
- 57.Kim SR, Bae YH, Bae SK, et al. Visfatin enhances ICAM-1 and VCAM-1 expression through ROS-dependent NF-kappaB activation in endothelial cells. Biochim Biophys Acta 2008;1783:886-95. 10.1016/j.bbamcr.2008.01.004 [DOI] [PubMed] [Google Scholar]
- 58.Moschen AR, Kaser A, Enrich B, et al. Visfatin, an adipocytokine with proinflammatory and immunomodulating properties. J Immunol 2007;178:1748-58. 10.4049/jimmunol.178.3.1748 [DOI] [PubMed] [Google Scholar]
- 59.Piacente F, Caffa I, Ravera S, et al. Nicotinic Acid Phosphoribosyltransferase Regulates Cancer Cell Metabolism, Susceptibility to NAMPT Inhibitors, and DNA Repair. Cancer Res 2017;77:3857-69. 10.1158/0008-5472.CAN-16-3079 [DOI] [PubMed] [Google Scholar]