Abstract
The discovery of how to utilize CRISPR (clustered, regularly interspaced, short, palindromic repeats)-Cas (CRISPR-associated) systems for genome modification has accelerated development of the field of genome editing, especially in large animals such as pigs. The low efficiency of somatic cell nuclear transfer (SCNT) is now becoming a major obstacle in the production of genome-edited animals via cell-mediated approaches and improving efficacy of this technique is crucial. In this study, we propose a few simple modifications to a zona-free SCNT protocol that are effective to produce numerous high-quality blastocysts. To refine the SCNT protocol we modified the following steps/factors: 1) culture medium for SCNT embryos, 2) chemical treatment to prevent precocious activation of the manipulated/reconstructed oocytes and 3) donor cell serum starvation treatment. Although changes in each of these steps only resulted in small improvements, the combination of all modifications altogether significantly enhanced developmental competence of SCNT embryos. Our modified method yielded approximately three times greater blastocyst formation rates. Moreover, resulting blastocysts had roughly twice as many cells as compared to blastocysts produced by the conventional SCNT method. With these significant in vitro improvements, our refined SCNT method is potentially suited for use in the production of genome edited pigs.
Keywords: Donor cells, Embryonic development competence, Pigs, Somatic cell nuclear transfer
Once mysterious, CRISPR (clustered, regularly interspaced, short, palindromic repeats)-Cas (CRISPR-associated) systems were first studied more than three decades ago [1]. However, not until recently has the function and purpose of these prokaryote adaptive immune systems been revealed. These discoveries paved the way for the exploration of how CRISPR can be utilized for genome modification [2,3,4]. Only a few years since the first reports showing that these systems could be repurposed and used as genome engineering tools, CRISPR technologies have been now widely exploited to generate genetic modifications in various species, including large animals such as pigs, highlighting the rapid evolution of this field. Applications of CRISPR/Cas9 to modify pig genomes includes growing human organs in immunodeficient pathogen-free pigs [5, 6] or increasing disease resistance in pigs such as production of porcine reproductive and respiratory syndrome (PRRS)-resistant pigs [7, 8]. With the rapid evolution of the field of genome editing, the low efficiency of somatic cell nuclear transfer (SCNT) is now becoming a major obstacle in the production of genome-edited pigs and improving the efficacy of this technique becomes crucial. Despite countless efforts since the birth of the first somatic cell cloned pigs in 2000 [9, 10] the development of pig SCNT embryos is still not optimal, especially when compared to some other livestock species. Simple modifications to SCNT protocols typically do not lead to meaningful improvement, whereas rather complicated procedures or expensive treatments are often required to gain significant achievements [11, 12]. In this study, we propose refinements to a simple and zona-free SCNT protocol that are effective to produce numerous high-quality blastocysts.
We firstly compared the development to blastocysts of SCNT embryos cultured in the two widely used in vitro culture (IVC) systems, namely North Carolina State University (NCSU) or Porcine Zygote Medium (PZM) – Porcine Blastocyst Medium (PBM). Sequential PZM and PBM media are chemically defined media developed by Yoshioka et al. [13, 14]. Based on the observation that glucose consumption occurs after the 8-cell stage [15], in both NCSU and PZM-PBM systems, embryos were firstly cultured in glucose-free media (IVC-PyrLac or PZM) before transfer to glucose supplemented media (namely IVC-Glu or PBM, for each respective system). Glucose-free IVC media contains pyruvate and lactate as substitute energy substrates. In our previous study, we found that in vitro fertilized embryos cultured in NCSU37 or PZM-PBM systems developed to blastocysts at similar rates. However, the PZM-PBM system supported parthenogenetic activated embryo development better than the NCSU system (data not shown). Since SCNT embryos are also artificially activated, we examined whether the PZM-PBM system would be more suitable for this type of embryo. Although not statistically significant, the blastocyst rate and cell number were both greater in SCNT embryos cultured in PZM3 and then PBM compared to NCSU (Table 1). In addition, we also cultured SCNT embryos in PZM3 for 7 days with a fresh medium change on Day 5. In this case, no glucose was supplied during the entire IVC period. However, porcine embryos still developed to blastocysts at the typical rate (Table 1). The presence of both essential and non-essential amino acids in PZM3 medium might compensate for the lack of glucose during later stages of embryo development. The same fibroblast cell line from an adult Western crossbred pig (Landrace × Large White × Duroc) was used as donor cells and were serum-starved in cell culture medium supplemented with 0.5% fetal bovine serum (FBS), for 5–7 days in this series of experiments. We then used the PZM3-PBM system for subsequent experiments.
Table 1. Development to blastocysts of somatic cell nuclear transfer (SCNT) oocytes cultured in different in vitro culture (IVC) media systems.
| No. of embryos | No. of blastocysts (%) | Cell no. in blastocysts | |
|---|---|---|---|
| NCSU | 136 | 11 (8.1 ± 3.8) | 35.5 ± 4.5 |
| PZM3 | 136 | 13 (9.6 ± 3.3) | 37.9 ± 5.1 |
| PZM3 + PBM | 136 | 21 (15.4 ± 6.2) | 39.4 ± 6.8 |
NCSU, North Carolina State University; PZM, porcine zygote medium; PBM, porcine blastocyst medium. Three replications were performed.
A major issue in pig SCNT, from our perspective, is the precocious activation of the manipulated or reconstructed oocytes. Pig oocytes are prone to activation by both physical and chemical determinants, especially when they are arrested in metaphase II (M-II) due to the prolonged SCNT procedure. It has been reported that cells arrested or delayed at metaphase more likely to undergo cohesion fatigue, where sister chromatids separate asynchronously, while cells remain in mitosis [16]. Cohesion fatigue contributes to chromosome instability leading to the increased incidence of aneuploidy [16]. In an attempt to prevent precocious activation, we investigated several approaches including shortening the time oocytes were maintained at M-II stage, reducing Ca2+ concentration during the cell fusion step, and keeping manipulated/reconstructed oocytes in medium supplemented with cytochalasin B to prevent extrusion of a pseudo polar body. In our in vitro maturation (IVM) system for pig oocytes, dibutyryl cyclic AMP (dbcAMP), a reversible inhibitor of meiotic resumption, is added during the first half of IVM to prevent germinal vesicle breakdown (GVBD) and synchronize the subsequent progression of oocyte maturation [17]. Oocytes quickly undergo GVBD and progress to M-II about 16–18 h after release from dbcAMP. In our study, we shortened the duration of the second half of IVM by 4 h, from 22 h to 18 h, by increasing the duration of the first half of IVM. In addition to minimizing precocious activation and cohesion fatigue, reducing the duration oocytes are maintained at M-II stage might practically improve the enucleation rate with “blind enucleation methods” since the metaphase plate and first polar body tend to be in close proximity in matured oocytes that have just reached the M-II stage. In “blind enucleation” methods, M-II plate is not visualized by UV and the position of M-II plate is presumed adjacent to the polar body. Although not significant, the enucleation rate was indeed increased by 8% in our study (data not shown). We also lowered the Ca2+ concentration to one tenth in the fusion medium, since this cation also triggers oocyte activation following electrical stimulation in sufficiently aged oocytes. We found that the reduction of Ca2+ concentration, from 0.1 mM to 0.01 mM, significantly decreased oocyte activation rate but did not interfere with cell fusion in preliminary experiments with non-manipulated oocytes. From those observations, we performed SCNT whereby oocytes were maintained at M-II stage for 18 h, then fused with single cells in low-Ca2+ fusion medium before incubation in medium supplemented with cytochalasin after fusion. Using this method, we observed significant increases in both cleavage and blastocyst rates, as well as the cell number in blastocysts (Table 2). The same fibroblast cell line from an adult Western crossbred pig was used as donor cells in this series of experiments. We therefore included these modifications to minimize the occurrence and effects of oocyte precocious activation in our SCNT procedure for subsequent experiments.
Table 2. Effect of precocious activation prevention on development to blastocysts of somatic cell nuclear transfer (SCNT) oocytes.
| No. of embryos | No. of cleaved embryos (%) | No. of blastocysts (%) | Cell no. in blastocysts | |
|---|---|---|---|---|
| Control | 164 | 148 (90.2 ± 2.8) a | 19 (11.6 ± 0.8) a | 49.1 ± 2.8 a |
| Precocious activation prevention | 289 | 283 (97.9 ± 1.1) b | 68 (23.5 ± 6.8) b | 65.0 ± 3.0 b |
Precocious activation prevention: SCNT runs in which oocytes were maintained at metaphase II (M-II) stage for 18 h, then fused with single cells in low-Ca2+ fusion medium, before incubated in medium supplemented with cytochalasin throughout the SCNT after fusion steps. Three replications were performed. a, b Superscript letters denote significant difference (P < 0.05) in the same column.
Another factor that we investigated in this study was serum starvation of the donor cells used for SCNT. The standard serum starvation protocol for commonly used somatic cells is to culture sub-confluent cells in low FBS culture medium, normally DMEM supplemented with 0.5% FBS, for 5–7 days. The purpose of this treatment is to obtain a high proportion of diploid (G1/G0) cells at the time of nuclear transfer, otherwise re-replication of previously replicated DNA will occur by the end of the first cell cycle in the SCNT embryos, and the DNA ploidy of daughter cells will be incorrect [18]. Experiments on transgenic and non-transgenic bovine fetal fibroblast cells suggested that it may be necessary to coordinate donor cell type and cell cycle stage to maximize overall cloning efficiency [19]. In our previous experiments with porcine donor cells, a slightly greater percentage of cells were at G1/G0 phases after serum starvation with 0% FBS for a shorter time, normally 2 days, compared to the standard serum starvation method (unpublished data). Following these observations, we assessed developmental competence, in terms of rate and quality of resulting blastocysts, in SCNT embryos derived from cells that were serum starved using different methods: low (0.5%) serum for 5 days or no (0%) serum for 2 days. In this series of experiments, we used both cumulus cells and adult fibroblasts from Western crossbred pigs as donor cells. It should also be noted that fresh cumulus cells were used soon after removal from in vitro matured oocytes, since the proportion of G1/G0 cells with fresh cumulus cells was similar compared to low-serum starved cultured cells [20]. Although the proportion of cleaved embryos derived from no-serum starved cumulus cells was higher than that derived from fresh cumulus cells, the blastocyst formation rates and cell numbers in blastocysts were similar between the two treatment groups (Table 3). Although insignificant, increased blastocyst rates were observed in the no-serum starvation group using fibroblast donor cells compared with low-serum starvation group (Table 3). No-serum starvation was then used for subsequent experiments, because of the convenience of this method and resulting development.
Table 3. Developmental competence of somatic cell nuclear transfer (SCNT) embryos derived from cells serum-starved by different methods.
| No. of embryos | No. of cleaved embryos (%) | No. of blastocysts (%) | Cell no. in blastocysts | ||
|---|---|---|---|---|---|
| Fibroblasts | No-serum | 175 | 170 (97.1 ± 1.4) | 56 (32.0 ± 7.7) | 86.8 ± 5.9 |
| Low-serum | 188 | 184 (97.9 ± 1.2) | 33 (17.6 ± 7.9) | 79.9 ± 5.2 | |
| Cumulus cells | No-serum | 102 | 99 (96.0 ± 3.5) a | 21 (20.6 ± 3.5) | 60.9 ± 5.6 |
| Fresh (no starvation) | 102 | 90 (88.2 ± 1.4) b | 24 (23.5 ± 3.5) | 50.0 ± 8.6 | |
No-serum: cells were serum-starved in fetal bovine serum (FBS)-free DMEM for 2 days before used for nuclear transfer. Low-serum: cells were serum-starved in 0.5% FBS DMEM for 5 days before used for nuclear transfer. Three replications were performed. a, b Superscript letters denote significant difference in the same column.
Finally, we assessed development to blastocyst of SCNT embryos using different donor cell types from two breeds. No-serum starved adult fibroblasts from a wild boar or from a Western crossbred pig, or cumulus cells from Western crossbred pigs were used in this series of experiments. Blastocyst rates were significantly increased when fibroblasts from adult wild boar and Western crossbred pigs were used as donor cells (Table 4, Fig. 1). The cell number in blastocysts derived from fibroblasts from adult Western crossbred pig was significantly greater than those derived from either fibroblasts from wild boars or cumulus cells. Nevertheless, these results suggested that our method may be suitable for a range of fibroblasts. Although we obtained high quality blastocysts at an acceptable rate with cumulus cells, development appeared less than for fibroblasts from the adult Western crossbred pig examined. It should be noted that, in all the experiments in this present study, oocytes were pre-selected prior to SCNT according to our sucrose treatment method which has proven to be an effective method to select oocytes with highly developmental competence [21].
Table 4. Developmental competence of somatic cell nuclear transfer (SCNT) embryos derived from different types of donor cells.
| No. of embryos | No. of cleaved embryos (%) | No. of blastocysts (%) | Cell no. in blastocysts | |
|---|---|---|---|---|
| FWB | 1120 | 1098 (98.0 ± 0.2) | 442 (39.5 ± 5.1) a | 59.4 ± 4.0 a |
| FLWD | 101 | 99 (98.0 ± 2.0) | 35 (34.7 ± 6.6) a | 72.0 ± 4.3 b |
| Cumulus | 119 | 114 (95.8 ± 3.4) | 25 (21.0 ± 1.3) b | 59.0 ± 4.9 a |
FWB, fibroblasts from wild boar; FLWD, fibroblasts from Western crossbred pig; Cumulus: cumulus cells from Western crossbred pigs. Three (FLWD and cumulus cells) or eight (FWB cells) replications were performed. a, b Superscript letters denote significant difference in the same column.
Fig. 1.
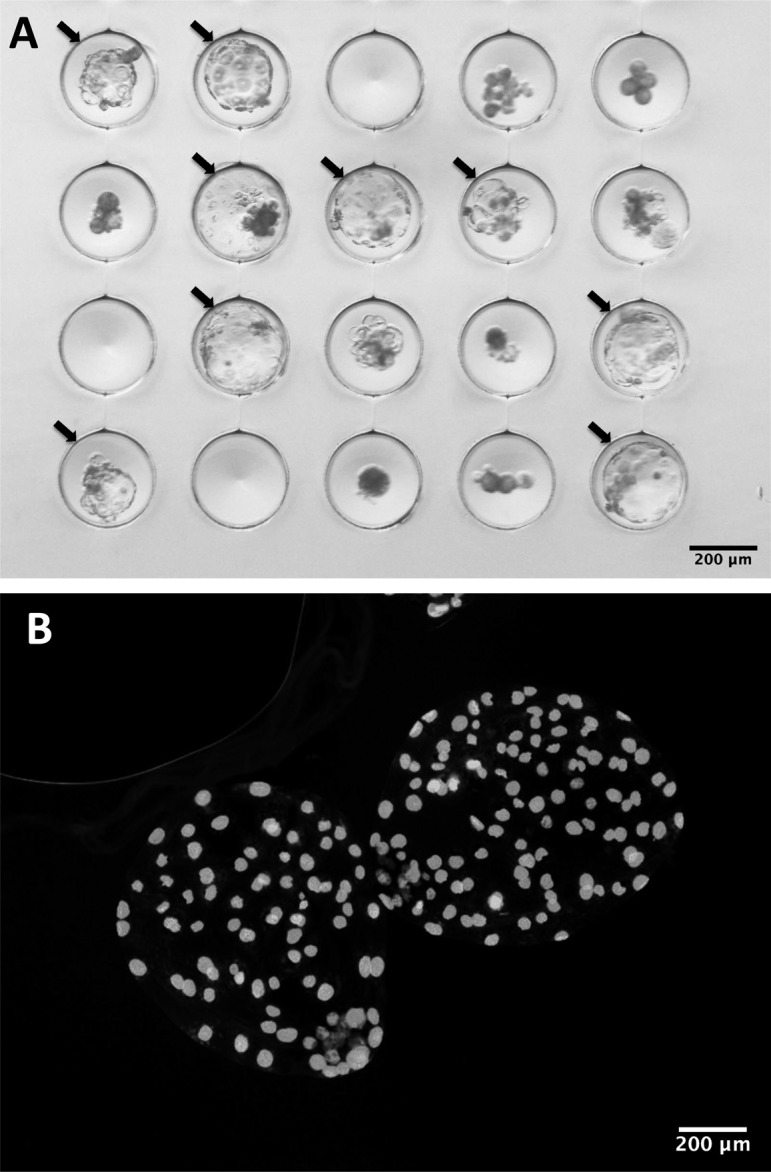
Day 7 somatic cell nuclear transfer (SCNT) blastocysts derived from adult fibroblasts from a wild boar produced according to the optimized protocol in culture (A) or after staining with DAPI (B). Arrows indicate blastocysts.
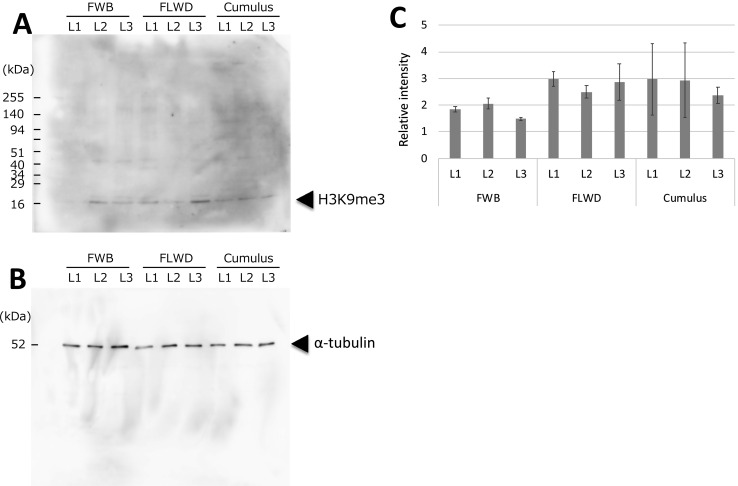
Histone modifications play an important role in gene regulation and cell linage [22]. Tri-methylation of lysine 9 on histone 3 (H3K9me3) is a repressive mark found abundantly at heterochromatic regions and is implicated in maintaining silencing imprinting related gene repression [23]. H3K9me3 of donor cell genome is considered a major epigenetic barrier in efficient reprogramming by SCNT and reduction of H3K9me3 levels improved in vitro development into cloned embryos [22, 24]. We therefore measured H3K9me3 level in the donor cells and examined whether H3K9me3 level contributes to the difference in developmental competence of SCNT embryos derived from different donor cells serum-starved with different methods. Western blotting analysis of H3K9me3 in adult fibroblasts from wild boar or from Western crossbred pig, or in cumulus cells from Western crossbred pigs, revealed no notable differences (Fig. 2A–C). This suggests that differential reprograming between cumulus and fibroblast cells at this chromatin mark was unlikely to be the main reason for the differing developmental competence with these two cell types.
Fig. 2.
Western blotting analysis of tri-methylation of lysine 9 on histone 3 (H3K9me3) in fibroblasts from wild boar (FWB), in fibroblasts from Western crossbred pig (FLWD), and in cumulus cells from Western crossbred pigs. Cells were serum starved in serum-free DMEM for 2 days (L1), in 0.5% FBS DMEM for 5 days (L2) or were not serum starved (L3).
Embryo transfer in pigs is normally performed using one or sometimes two-cell embryos. In this case, the presence of a zona pellucida is important to prevent embryos from adhering together. The main reason for transferring embryos at this early stage is due to lower quality blastocysts produced with IVC, especially following SCNT. Embryos produced using our SCNT method are zona-free and thus, not suitable to transfer at an early stage, but should be transferred at the blastocyst stage instead. However, using our method, we could produce a high number of good-quality blastocysts that could be transferred to recipients at this stage. Numerous reports have shown that blastocyst transfer results in healthy piglets [13, 22].
In conclusion, in this present study, we propose a simple and zona-free SCNT protocol that efficiently promotes development of cloned pig embryos. In this protocol, fibroblasts, which is no-serum starved for 2 days, were used as donor cells. The donor cells were then fused with zona-free enucleated oocytes, which were maintained at M-II stage for 18 h, in low-Ca2+ fusion medium. Reconstructed oocytes were incubated in medium supplemented with cytochalasin throughout the SCNT after fusion steps before cultured in PZM3 for 5 days and PBM for 2 days. Embryos produced according to this method are of high quality and suitable for transfer to surrogate recipients at the blastocyst stage. This refined SCNT method is expected to contribute to the rapid development of genome editing research in pigs.
Methods
Oocyte collection and IVM
Porcine ovaries were collected from pre-pubertal crossbred gilts (Landrace × Large White) at a local abattoir and transported to the laboratory in Dulbecco’s phosphate buffered saline (PBS; Nissui Pharmaceutical, Tokyo, Japan) at 35oC within 1 h. Cumulus-oocyte complexes (COCs) were aspirated from 3–6 mm follicles, and cultured in groups of 40 to 50 in 500 μL of modified NCSU-37 medium [26] without oil overlay according to Kikuchi et al. [25] in 4-well dishes (Nunclon Multidishes, Nalge Nunc International, Roskilde, Denmark) for either 22 or 26 h. The IVM medium was modified by adding 10% (v/v) porcine follicular fluid, 0.6 mM cysteine (Sigma, St. Louis, MO, USA), 50 mM β-mercaptoethanol (Axon Medchem, Groningen, Netherlands), 1 mM dbcAMP (Sigma), 10 IU/mL eCG (Serotropin; ASKA Pharmaceutical, Tokyo, Japan), and 10 IU/mL hCG (Puberogen; Novartis Animal Health, Tokyo, Japan). The COCs were then transferred to IVM medium without dbcAMP and hormones and cultured for another 22 or 18 h. IVM was performed in 5% CO2 and 5% O2 at 38.5oC.
Donor cell preparation
Cumulus cells from Western crossbred pigs and adult fibroblasts from either a Western crossbred pig or a wild boar were used as donor cells. Fresh cumulus cells were removed from COCs following IVM and then washed with PBS before used as donor cells for nuclear transfer on the same day. After reaching confluence, cultured cumulus cells or adult fibroblasts were serum starved in FBS-free DMEM for 2 days or in 0.5% FBS DMEM for 5 days before used for nuclear transfer.
SCNT
After IVM, cumulus cells were removed from the COCs by gentle pipetting after treatment with 0.1% (w/v) hyaluronidase (Sigma) for 2–5 min, and the denuded oocytes were then washed three times in Medium 199 (with Hanks’ balanced salts; Sigma) supplemented with 10% (v/v) FBS (Gibco, Life Technologies, Grand Island, NY, USA), 20 mM HEPES (Dojindo Laboratories, Kumamoto, Japan), antibiotics (100 units/ml penicillin G potassium (Sigma), and 0.1 mg/ml streptomycin sulfate (Sigma)), with pH adjusted to 7.4. After washing, oocytes selected for the presence of the polar body were treated with 0.2 M sucrose in Medium 199 supplemented with HEPES and 5% FBS (524 mOsm/l) for 5 min. Oocytes that maintained a round shape following sucrose treatment were selected for SCNT. After treatment with sucrose, the oocytes were washed three times in Medium 199 with HEPES plus 5% FBS to remove sucrose completely. SCNT was performed according to Oback and Wells [27] with modifications for pig. Briefly, oocytes with a first polar body were enucleated in HEPES buffered Medium 199 supplemented with 20% FBS by the “squeezing method” according to Akagi et al. [28]. Enucleated oocytes were then treated with 0.5% pronase to remove the zona pellucida. Each resulting zona-free cytoplast was attached to a single donor cell possessing a smooth and clear membrane by incubation in 300 µg/ml phytohemagglutinin (Sigma) for 15 min. The cytoplast-cell couplets were orientated between a pair of parallel electrodes, 1 mm apart, with the assistance of alternating current in fusion medium comprising 0.28 M mannitol, 0.01 mM or 0.1 mM CaCl2, 0.1 mM MgSO4, 0.5 mM HEPES and 0.1 mg/ml bovine serum albumin (BSA; Fraction V, Sigma). A single direct current (DC) pulse of 2 kV/cm was then applied for 20 µsec for cytoplast-cell fusion using an electro cell fusion generator (LF101; Nepa-Gene, Japan). Cytoplast-cell couplets were kept in HEPES buffered Medium 199 supplemented with 20% FBS, either with or without 5 µg/mL cytochalasin B for around 30 min before evaluating fusion. After approximately 2–3 h, fused couplets were activated with a single DC pulse of 1.5 kV/cm for 99 µsec by the electro cell fusion generator in medium comprising 0.28 M mannitol, 0.1 mM CaCl2, 0.1 mM MgSO4, 0.5 mM HEPES and 0.1 mg/ml BSA, followed by incubation in PZM3 medium (Research Institute for the Functional Peptides, Yamagata, Japan) supplemented with 5 µg/ml cytochalasin B for 3 h.
IVC
Reconstructed oocytes were cultured individually in microwell dishes (DNP, Tokyo, Japan) at 39°C in humidified 5% CO2, 5% O2 and 90% N2. Two IVC systems were used in this study: NCSU-37 and PZM-PBM. In the NCSU-37 system, embryos were cultured in IVC-PyrLac from Day 0 to Day 2, and then in IVC-Glu until Day 7 according to Kikuchi et al. [25]. Day 0 was designated the day of SCNT. In the PZM-PBM system, embryos were cultured either in PZM3 for 7 days with a fresh medium change on Day 5, or in PZM3 for 5 days and then in PBM (Research Institute for the Functional Peptides) for another 2 days.
Western blotting
Cells were washed twice with PBS and cell numbers adjusted equally between samples. Histone proteins were isolated from the cells according to Sidoli et al. [29]. The supernatants containing cytosolic fraction were also used for internal control. The samples were resolved on 4–15% precast polyacrylamide gels (#4561084, Bio-Rad, Hercules, CA, USA), transferred onto a polyvinylidene fluoride membrane (Immobilon-P, Millipore, Burlington, MA, USA) and blocked for 1 h at room temperature with 5% skim milk (BD Biosciences, San Jose, CA, USA) in TBST buffer (100 mM Tris-HCl, pH 7.5, 50 mM NaCl, 0.1% Tween-20; blocking buffer). The membranes were probed with anti-trimethyl-histone H3 (Lys9) (1:500, #07-442, Millipore) or anti-α-Tubulin (1:2,000, #2125, Cell Signaling Technology, Danvers, MA, USA) antibodies in blocking buffer overnight at 4°C. After washing with TBST, the membranes were incubated with anti-rabbit IgG (1:2,000, #7074, Cell Signaling Technology) in blocking buffer for 1 h at room temperature. The membranes were washed with TBST and processed using Chemi-Lumi One Ultra (Nacalai Tesque, Kyoto, Japan). Immunoblots were visualized using ImageQuant LAS 500 (GE Healthcare, Chicago, IL, USA). The signal intensity of H3K9me3 and α-Tubulin were measured using the software ImageQuant TL8.1 (GE Healthcare). The signal intensity of H3K9me3 was normalized to that of α-Tubulin using ImageQuant TL8.1.
Evaluation of total cell number in blastocysts
To evaluate the total cell number in embryos, Day 7 blastocysts were stained using Vectashield mounting medium with DAPI (Vector Laboratories, CA, USA). Blastocysts were washed in PBS and fixed for 15 min in 4% paraformaldehyde. The blastocysts were then washed twice in PBS supplemented with 0.3% polyvinylpyrrolidone followed by staining with DAPI for 5 min. The blastocysts were gently flattened under a coverslip, so that the cells appeared in a similar focal plane. Total cell numbers were counted using an epi-fluorescent microscope (Nikon Diaphot 200, Nikon, Tokyo, Japan).
Statistical analysis
The data on cleavage and blastocyst rates, as well as mean cell number in blastocysts, were analyzed by one-way ANOVA followed by Bonferroni correction by using the Stata/SE 15.0 software package (StataCorp., College Station, TX, USA). A value of P < 0.05 was considered statistically significant. All data were expressed as mean ± SEM values.
Acknowledgments
We would like to thank Drs Hayashi Satoshi, Fuchimoto Daiichiro, and Suzuki Shunichi for supplying fibroblasts from Western crossbred pigs and wild boars. We would like to thank Ms. Nagai for technical assistance. This study was partly supported by a JSPS – MBIE/RSNZ joint bilateral research project (DW, KK and TQ D-N).
References
- 1.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 1987; 169: 5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pennisi E. The CRISPR craze. Science 2013; 341: 833–836. [DOI] [PubMed] [Google Scholar]
- 3.Barrangou R, May AP. Unraveling the potential of CRISPR-Cas9 for gene therapy. Expert Opin Biol Ther 2015; 15: 311–314. [DOI] [PubMed] [Google Scholar]
- 4.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014; 346: 1258096. [DOI] [PubMed] [Google Scholar]
- 5.Kang JT, Cho B, Ryu J, Ray C, Lee EJ, Yun YJ, Ahn S, Lee J, Ji DY, Jue N, Clark-Deener S, Lee K, Park KW. Biallelic modification of IL2RG leads to severe combined immunodeficiency in pigs. Reprod Biol Endocrinol 2016; 14: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lei S, Ryu J, Wen K, Twitchell E, Bui T, Ramesh A, Weiss M, Li G, Samuel H, Clark-Deener S, Jiang X, Lee K, Yuan L. Increased and prolonged human norovirus infection in RAG2/IL2RG deficient gnotobiotic pigs with severe combined immunodeficiency. Sci Rep 2016; 6: 25222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitworth KM, Rowland RRR, Ewen CL, Trible BR, Kerrigan MA, Cino-Ozuna AG, Samuel MS, Lightner JE, McLaren DG, Mileham AJ, Wells KD, Prather RS. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat Biotechnol 2016; 34: 20–22. [DOI] [PubMed] [Google Scholar]
- 8.Burkard C, Lillico SG, Reid E, Jackson B, Mileham AJ, Ait-Ali T, Whitelaw CB, Archibald AL. Precision engineering for PRRSV resistance in pigs: Macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog 2017; 13: e1006206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Onishi A, Iwamoto M, Akita T, Mikawa S, Takeda K, Awata T, Hanada H, Perry AC. Pig cloning by microinjection of fetal fibroblast nuclei. Science 2000; 289: 1188–1190. [DOI] [PubMed] [Google Scholar]
- 10.Polejaeva IA, Chen SH, Vaught TD, Page RL, Mullins J, Ball S, Dai Y, Boone J, Walker S, Ayares DL, Colman A, Campbell KH. Cloned pigs produced by nuclear transfer from adult somatic cells. Nature 2000; 407: 86–90. [DOI] [PubMed] [Google Scholar]
- 11.Tao J, Zhang Y, Zuo X, Hong R, Li H, Liu X, Huang W, Cao Z, Zhang Y. DOT1L inhibitor improves early development of porcine somatic cell nuclear transfer embryos. PLoS One 2017; 12: e0179436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruan D, Peng J, Wang X, Ouyang Z, Zou Q, Yang Y, Chen F, Ge W, Wu H, Liu Z, Zhao Y, Zhao B, Zhang Q, Lai C, Fan N, Zhou Z, Liu Q, Li N, Jin Q, Shi H, Xie J, Song H, Yang X, Chen J, Wang K, Li X, Lai L. XIST derepression in active X chromosome hinders pig somatic cell nuclear transfer. Stem Cell Reports 2018; 10: 494–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshioka K, Suzuki C, Tanaka A, Anas IM, Iwamura S. Birth of piglets derived from porcine zygotes cultured in a chemically defined medium. Biol Reprod 2002; 66: 112–119. [DOI] [PubMed] [Google Scholar]
- 14.Mito T, Yoshioka K, Yamashita S, Suzuki C, Noguchi M, Hoshi H. Glucose and glycine synergistically enhance the in vitro development of porcine blastocysts in a chemically defined medium. Reprod Fertil Dev 2012; 24: 443–450. [DOI] [PubMed] [Google Scholar]
- 15.Sturmey RG, Leese HJ. Energy metabolism in pig oocytes and early embryos. Reproduction 2003; 126: 197–204. [DOI] [PubMed] [Google Scholar]
- 16.Daum JR, Potapova TA, Sivakumar S, Daniel JJ, Flynn JN, Rankin S, Gorbsky GJ. Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Curr Biol 2011; 21: 1018–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Somfai T, Kikuchi K, Onishi A, Iwamoto M, Fuchimoto D, Papp AB, Sato E, Nagai T. Meiotic arrest maintained by cAMP during the initiation of maturation enhances meiotic potential and developmental competence and reduces polyspermy of IVM/IVF porcine oocytes. Zygote 2003; 11: 199–206. [DOI] [PubMed] [Google Scholar]
- 18.Campbell KHS, Alberio R. Reprogramming the genome: role of the cell cycle. Reprod Suppl 2003; 61: 477–494. [PubMed] [Google Scholar]
- 19.Wells DN, Laible G, Tucker FC, Miller AL, Oliver JE, Xiang T, Forsyth JT, Berg MC, Cockrem K, L’Huillier PJ, Tervit HR, Oback B. Coordination between donor cell type and cell cycle stage improves nuclear cloning efficiency in cattle. Theriogenology 2003; 59: 45–59. [DOI] [PubMed] [Google Scholar]
- 20.Akagi S, Takahashi S, Adachi N, Hasegawa K, Sugawara T, Tozuka Y, Yamamoto E, Shimizu M, Izaike Y. In vitro and in vivo developmental potential of nuclear transfer embryos using bovine cumulus cells prepared in four different conditions. Cloning Stem Cells 2003; 5: 101–108. [DOI] [PubMed] [Google Scholar]
- 21.Dang-Nguyen TQ, Nguyen HT, Somfai T, Wells D, Men NT, Viet-Linh N, Noguchi J, Kaneko H, Kikuchi K, Nagai T. Sucrose assists selection of high-quality oocytes in pigs. Anim Sci J 2018; 89: 880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antony J, Oback F, Chamley LW, Oback B, Laible G. Transient JMJD2B-mediated reduction of H3K9me3 levels improves reprogramming of embryonic stem cells into cloned embryos. Mol Cell Biol 2013; 33: 974–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008; 453: 948–951. [DOI] [PubMed] [Google Scholar]
- 24.Matoba S, Liu Y, Lu F, Iwabuchi KA, Shen L, Inoue A, Zhang Y. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell 2014; 159: 884–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kikuchi K, Onishi A, Kashiwazaki N, Iwamoto M, Noguchi J, Kaneko H, Akita T, Nagai T. Successful piglet production after transfer of blastocysts produced by a modified in vitro system. Biol Reprod 2002; 66: 1033–1041. [DOI] [PubMed] [Google Scholar]
- 26.Petters RM, Wells KD. Culture of pig embryos. J Reprod Fertil Suppl 1993; 48: 61–73. [PubMed] [Google Scholar]
- 27.Oback B, Wells DN. Cloning cattle. Cloning Stem Cells 2003; 5: 243–256. [DOI] [PubMed] [Google Scholar]
- 28.Akagi S, Kaneyama K, Adachi N, Tsuneishi B, Matsukawa K, Watanabe S, Kubo M, Takahashi S. Bovine nuclear transfer using fresh cumulus cell nuclei and in vivo- or in vitro-matured cytoplasts. Cloning Stem Cells 2008; 10: 173–180. [DOI] [PubMed] [Google Scholar]
- 29.Sidoli S, Bhanu NV, Karch KR, Wang X, Garcia BA. Complete workflow for analysis of histone post-translational modifications using bottom-up mass spectrometry: from histone extraction to data analysis. J Vis Exp 2016; 111. [DOI] [PMC free article] [PubMed] [Google Scholar]