Abstract
We analyzed humoral immune responses to nonhuman leukocyte antigen (HLA) after cardiac transplantation to identify antibodies associated with allograft rejection. Protein microarray identified 366 non-HLA antibodies (>1.5 fold, P < .5) from a discovery cohort of HLA antibody–negative, endothelial cell crossmatch–positive sera obtained from 12 cardiac allograft recipients at the time of biopsy-proven rejection. From these, 19 plasma membrane proteins and 10 autoantigens identified from gene ontology analysis were combined with 48 proteins identified through literature search to generate a multiplex bead array. Longitudinal sera from a multicenter cohort of adult cardiac allograft recipients (samples: n = 477 no rejection; n = 69 rejection) identified 18 non-HLA antibodies associated with rejection (P < .1) including 4 newly identified non-HLA antigenic targets (DEXI, EMCN, LPHN1, and SSB). CART analysis showed 5/18 non-HLA antibodies distinguished rejection vs nonrejection. Antibodies to 4/18 non-HLA antigens synergize with HLA donor-specific antibodies and significantly increase the odds of rejection (P < .1). The non-HLA panel was validated using an independent adult cardiac transplant cohort (n = 21 no rejection; n = 42 rejection, >1R) with an area under the curve of 0.87 (P < .05) with 92.86% sensitivity and 66.67% specificity. We conclude that multiplex bead array assessment of non-HLA antibodies identifies cardiac transplant recipients at risk of rejection.
Keywords: autoantibody, autoantigen, clinical research/practice, heart transplantation/cardiology, histocompatibility, immunogenetics, microarray/protein array, organ transplantation in general, rejection, translational research/science
1 |. INTRODUCTION
Long-term outcomes remain largely unchanged and graft loss due to chronic rejection is a significant problem.1–3 Donor-specific HLA antibodies (DSAs) contribute to antibody-mediated rejection (AMR)4–7 and acute cell-mediated rejection (ACR)8–10 after cardiac transplant. However, a number of heart transplant patients present with AMR in the absence of DSAs,11 suggesting antibodies against non-HLA antigens may be associated with an increased risk of AMR. This is further supported by reports that antibodies to vimenten,12–14 MHC class I polypeptide-related sequence A (MICA),15,16 angiotensin II receptor type 1,17,18 and endothelial cell (EC)-specific antibodies19,20 are associated with AMR and cardiac allograft vasculopathy (CAV). Additionally, antibodies to non-HLA antigens have been associated with poor outcomes in other organs.21,22
ECs are the first point of contact between the allograft and the recipient’s immune system and therefore a source of non-HLA antigens that can stimulate a humoral immune response. The endothelial cell crossmatch (ECXM) using primary human aortic ECs (HAECs) and the XM One assay have been shown to identify patient sera containing antibodies against ECs.20,23 However, the utility of cell-based assays is limited, as they do not identify the antigen that binds the non-HLA antibody. Consequently, our understanding of the breadth of non-HLA antigens that elicit a humoral response leading to poor allograft outcomes is hampered by our inability to detect and characterize non-HLA antibodies.
We sought to identify and validate non-HLA antibodies associated with cardiac allograft rejection. We screened a set of ECXM+ sera from cardiac allograft recipients diagnosed with rejection, in the absence of HLA or MICA DSAs20 using protein microarrays containing full-length human protein antigens. Non-HLA antibody targets associated with EC plasma membrane and autoantigens were classified using a bioinformatics and gene ontologic approach. These, with non-HLA antigens identified through literature search were conjugated to a multiplex bead array for high throughput validation using multicenter and single-center cohorts of adult cardiac allograft recipients. Using this proteomics approach, we report the discovery and validation of a novel panel of non-HLA antigens associated with cardiac allograft rejection.
2 |. MATERIALS AND METHODS
2.1 |. Ethics statement
The UCLA institutional review board (12–000656 and 11–000577) approved the use of serum and endomyocardial biopsy (EMB) samples used in this study and all research participants gave written informed consent.
2.2 |. Study populations and sample collection
2.2.1 |. Discovery cohort
The discovery cohort was identified from cardiac allograft recipients transplanted at UCLA between 2001 and 2005 that tested positive by ECXM posttransplant and were negative for HLA DSA and MICA antibodies (n = 12; Table 1). ECXM were performed as previously described.20 HLA antibodies were identified using a Single Antigen Luminex assay (One Lambda). Demographics are listed in Table 1. Rejection was diagnosed by EMB according to the International Society for Heart and Lung Transplantation (ISHLT) criteria24 and as previously reported.25
TABLE 1.
Discovery cohort demographics (rejection samples)
| n = 12 | |
|---|---|
| Age (y), mean ± SD | 44 ± 19 |
| Sex (male), n (%) | 9 (75) |
| Race | |
| White, n (%) | 7 (58) |
| Black, n (%) | 1 (8) |
| Asian, n (%) | 2 (17) |
| Undisclosed, n (%) | 2 (17) |
| End-stage disease, n (%) | |
| Idiopathic cardiomyopathy | 8 (58) |
| Coronary artery disease | 2 (17) |
| Congenital heart disease | 2 (8) |
| Retransplants, n (%) | 0 (0) |
| Mechanical circulatory support, n (%) | 4 (33) |
| HLA donor-specific antibodies, n (%) | 0 (0) |
| Biopsy time from transplant, mean (d), range (d) | 67 (20–222) |
| Antibody-mediated rejection, n (%) | 6 (50) |
| CD68+ | 6 (50) |
| C4d+ | 4 (33) |
| Endothelial cell swelling | 3 (25) |
| Acute cell-mediated rejection 1R | 9 (75) |
| Serum time from biopsy, mean (d), range (d) | 14 (0–76) |
2.2.2 |. Multicenter cohort
The multicenter cohort was a sub-study of the phase 3 clinical trial sponsored by Novartis (www.ClinicalTrials.gov NCT00300274).26 For this sub-study, we obtained IRB approval from 9 participating centers including the University of Puerto Rico; Cleveland Clinic; Drexel College of Medicine; Intermountain Medical Center; Medical University of South Carolina; Methodist Hospital Research Institution; Tufts Medical Center; University of California, Los Angeles; and Washington University. This study included 115 subjects with 546 available posttransplant sera (average of 4.7 samples tested per patient, range of 1–9 samples per patient), protocol EMB collected at 3, 6, 12, and/or 24 months and for cause graded for rejection according to the 1990 ISHLT criteria.26,27 Sera were tested for non-HLA antibodies by Luminex and analyzed for association with rejection using EMB obtained within −21/+2 day (n = 477 no rejection; n = 69 rejection, ≥grade 1B). The median in days from sera collection to EMB for all samples was 0 (P < .39). Posttransplant, 39 patients were maintained on triple-drug immunosuppression (tacrolimus, mycophenolate mofetil, and corticosteroids) and 76 patients received tacrolimus, everolimus, and corticosteroids.
2.2.3 |. Single-center cohort
The single-center cohort consisted of 63 heart transplant recipients transplanted at UCLA between 2009–2016 for which a serum sample with a paired biopsy were available. Rejection (n = 42, ACR > 1R) was scored according to the ISHLT criteria.24 The median in days from sera collection to EMB for samples was 0 (P < .26). Posttransplant, patients were maintained on triple-drug immunosuppression.
2.3 |. Protein microarray
Serum antibodies were profiled with a human protein microarray as previously described.23,28 The InVitrogen Human ProtoArray v5.0 were analyzed after probing with 12 rejection+ ECXM+ sera, and 6 rejection− ECXM− sera to serve as technical controls. The slides were scanned using a GenePix 4100A fluorescence microarray scanner and GenePix Pro 6.0 software (Molecular Devices). Array normalization and analysis was performed using Prospector 2.0 software (Life Technologies) as previously described.23 Gene ontologic analysis of the 366 antigens was assessed by DAVID gene ontology (https://david.ncifcrf.gov) to identify enriched biological themes as previously described.29
2.4 |. Multiplex non-HLA antibody panel (Luminex)
The non-HLA multiplex bead panel, engineered and provided by Immucor, Inc, included 67 non-HLA antigens (Table S1) conjugated to polystyrene beads. Forty µL of antigen-coated beads were incubated with 10 µL of serum for 30 minutes. After washing beads were stained with 50 µL of phycoerythrin (PE) conjugated goat anti-human IgG diluted 1:10 in buffer and incubated in the dark on a shaking platform for 30 minutes. Antibody binding was reported as the median fluorescence intensity (MFI) of IgG binding on the Luminex 100 (Luminex).
A receiver operating characteristic curve (ROC) analysis was performed to determine the optimal cutoff (using Youden’s Index and the value nearest to [0, 1]) as well as a 1000 MFI cutoff for each of the 18 significant non-HLA antibodies. Both ‘Youden’s Index’ and ‘nearest’ approaches lead to similar ranges (192–961 MFI) of empiric optimal cutoffs per bead (data not shown). However, a positive threshold of MFI > 1000 was chosen for analyses (1) to ensure confidence and fewer false positives, (2) areas under the curve per the ROC analysis was comparable across all 3 methods, and (3) our prior experience with Luminex-based solid phase antibody detection methods suggested using the higher threshold.30 Non-HLA antibodies were assessed in sera isolated from 13 healthy controls who provided informed written consent and compared to the patient samples from the multicenter cohort by 1-way ANOVA (Figure S1).
2.5 |. RNA-sequencing data analysis
Publically available RNA-sequencing data30 were obtained (https://www.proteinatlas.org/) for cardiac, kidney, lung, and liver tissues and human umbilical vein endothelial cells (HUVECs). RNA-sequencing results are reported as number of transcripts per kilobase million (TPM) and protein expression is defined as >1.0.
2.6 |. Histopathology and immunohistochemistry
IHC conducted on EMB (n = 6, 3 rejection and 3 no rejection) from the multicenter cohort that were positive for non-HLA antibodies (either Sjogren syndrome antigen B [SSB] or endomucin [EMCN] antibodies). IHC was performed by the Translational Pathology Core Laboratory at UCLA following a standardized protocol31 with anti-SSB (Abcam, ab75927) and anti-endomucin (Novus Bio, NBP1–92150). Slides were reviewed by a single cardiopathologist and imaged with a Nikon 90i microscope with NIS Elements software version 3.0 (Nikon).
2.7 |. Statistics
For testing differences in the demographic data, t and Fisher exact tests were used for continuous variables and categorical variables, respectively. Odds ratios associating non-HLA antibodies with cardiac allograft rejection were determined to be significantly greater than 1 by Fisher exact tests with P < .1. A P < .1 threshold for significance was set following standard practice for variable selection32 and to prevent exclusion of antibodies that may interact with each other. All tests were 2-tailed, and confidence intervals (95%) were constructed assuming the estimates of standard error are asymptotically normal. To adjust for confounding due to the presence of HLA DSA, a random effects logistic model was used to account for multiple factors generated per patient. Wald tests were used to compare odds ratios. An ROC analysis was performed via a logistic regression model for all 18 non-HLA antibodies on patients with and without HLA DSAs. All preceding analyses were performed using Stata Statistical Software (StataCorp. 2015; Release 14).
In cluster analysis, ordering of the non-HLA markers, circle size and illustrative boxes were based on pairwise correlation coefficients. The analysis was performed using hierarchal clustering with complete linkage and Euclidean distance in the R Corrplot application (https://github.com/taiyun/corrplot). The rpart function in the R library (the R software package version 3.4.0, http://www.r-project.org/) was used to perform a recursive partitioning regression analysis and build a classification tree. In order to avoid over-fitting and to select a parsimonious set of predictor variables, the maximum depth of any node of the final tree (the root node counted as depth 0) was set to 3, and the minimum number of observations that must exist in a node, in order for a split to be attempted was set to 5. All other parameters were set to their default values.
3 |. RESULTS
3.1 |. Discovery of non-HLA antibodies associated with cardiac allograft rejection by protein microarray analysis
A discovery cohort contained sera from 12 cardiac transplant recipients without HLA or MICA DSA, but with biopsy-proven rejection and with positive reactivity in the ECXM (median of 117 MCS; range: 62–529 MCS) (Table 1). An additional 6 sera that were rejection negative, ECXM negative, and MICA DSA negative were used as controls.
The 18 sera (12 rejection+/ECXM+ and 6 rejection−/ECXM− controls) were hybridized to protein microarrays. Bioinformatic analysis of the microarrays identified 366 antigens with increased fluorescence intensity (>1.5-fold; P < .05) in sera isolated from rejection+/ECXM+ patients compared to rejection−/ECXM− controls (Figure 1).
FIGURE 1.

Non-HLA antibody discovery and generation of a high throughput multiplex bead set. Non-HLA antibodies associated with cardiac allograft rejection were identified by protein microarray analysis (9000 full length proteins) using sera from 12 heart transplant patients with biopsy proven rejection in the absence of HLA donor specific antibodies (DSA), but with positive endothelial cell crossmatch (ECXM) (n = 12). 366 proteins binding non-HLA antigens were identified (>1.5 fold increase, P < .05) compared to control sera from rejection negative, DSA negative, and ECXM negative heart transplant patients (n = 6). Of the 366, gene ontology analysis identified 22 plasma membrane proteins and 10 autoantigens. Nineteen of these, with an additional 48 proteins identified by literature search as associated with solid organ rejection, were conjugated to polystyrene beads to generate a multiplex panel of non-HLA proteins for downstream high throughput testing of non-HLA antibodies
3.2 |. Development of a high throughput multiplex bead array displaying non-HLA antigens
Next, gene ontologic analysis of the 366 non-HLA antigens was performed to identify those that represented potential target antigens expressed on the EC surface that may have contributed to a positive ECXM. From this analysis, 19 plasma membrane antigens and 10 autoantigens were expressed and conjugated to Luminex beads for high throughput testing (Table S1, Antigens 1–19). An additional 48 non-HLA antigens identified from published literature as correlating to rejection (9 cardiac, 30 renal, 8 lung, and 1 liver) were also included in the bead set (Figure 1; Table S1, Antigens 20–67).
3.3 |. Identification of non-HLA antibodies associated with cardiac allograft rejection
The multiplex bead array was used to screen sera samples isolated from heart transplant patients enrolled in a multicenter clinical trial.26 Patients in the rejection group experienced at least 1 histologically proven rejection episode (≥grade 1B). The demographics of this cohort are listed in Table 2. There were no significant differences between patient groups with respect to age, race, sex, end-stage disease, induction therapy, presence of mechanical circulatory support, or patient/donor HLA-A, -B, -DR mismatch (Table 2).
TABLE 2.
Patient demographics
| Multicenter adult |
Single-center adult |
|||||
|---|---|---|---|---|---|---|
| Patients | No rejection n = 59 | Rejection n = 56 | P-value | No rejection n = 21 | Rejection n = 42 | P-value |
| Age (y), mean ± SD | 52 ± 11 | 51 ± 11 | .43 | 50 ± 14 | 50 ± 12 | .91 |
| Sex (male), n (%) | 47 (80) | 46 (82) | .82 | 15 (71) | 29 (69) | .99 |
| Race | ||||||
| White, n (%) | 37 (63) | 46 (82) | .04 | 12 (57) | 25 (60) | .06 |
| End-stage disease, n (%) | ||||||
| Idiopathic cardiomyopathy | 25 (42) | 28 (50) | .27 | 7 (33) | 15 (36) | .99 |
| Coronary artery disease | 12 (20) | 14 (25) | 0 | 1 (2) | ||
| Congenital heart disease | 0 (0) | 1 (2) | 2 (10) | 3 (7) | ||
| Myocarditis | 0 (0) | 1 (2) | 0 | 1 (2) | ||
| Valvular heart disease | 1 (2) | 0 (0) | 0 | 0 | ||
| Other etiology | 21 (36) | 12 (21) | 12 (57) | 22 (52) | ||
| Induction therapy, n (%) | 15 (25) | 19 (34) | .41 | 5 (24) | 11 (26) | .73 |
| Mechanical circulatory support, n (%) | 14 (24) | 15 (27) | .08 | 9 (43) | 11 (26) | .25 |
| HLA-A, -B, -DR mismatches ≥3, n/total (%) | 53/55 (96) | 45/48 (94) | .66 | 20/21 (95) | 42/42 (100) | .44 |
Five hundred forty-six serum samples from 115 recipients (n = 477 no rejection; n = 69 rejection, ≥grade 1B) collected longitudinally with paired EMB were tested for non-HLA antibodies by multiplex analysis. The distribution of rejection grades is listed in Table 3. Eighteen non-HLA antibodies were significantly associated with rejection (Figure 2A, grade ≥1B, odds ratio >1, 95% CI, P < .1). These included 4 non-HLA antibodies that target antigens newly described as being associated with allograft rejection including EMCN, dexamethasone-induced transcript (DEXI), latrophilin 1 (LPHN1) and SSB. Seven non-HLA antibodies to cytoskeletal proteins were also identified including: myosin, vimentin (VIM), actin, keratin 8 (KRT8), keratin 18 (KRT18), tubulin33 (TUBα1b), and agrin34 (AGRN). Non-HLA antibodies to 3 enzymes were also identified: enolase 1 (ENO1), peroxisomal trans-2-enoyl-coA reductase35 (PECR), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH).36 Additionally, non-HLA antibodies were identified to bind the cytokines, IL-21 and CXCL11, as well as the kinases protein kinase C37 (PRKCH) and Fms related tyrosine kinase 3 ligand (FLT3LG) (Table 4). To further characterize the 18 non-HLA antibodies significantly associated with rejection (Figure 2A) in healthy controls and patient samples, box plots showing the distribution of the MFI (y-axis) are shown in Figure S1. Within each trio from left to right, the first box shows the healthy controls (n = 13). The next, shows the no rejection samples from the multicenter cohort (n = 477) and the last box shows the rejection samples (n = 69) for each of the 18 non-HLA antibodies (x-axis). Significant differences (1-way ANOVA, P < .05) among the MFI distributions occurred between healthy controls, no rejection samples, and rejection samples for the 18 non-HLA antibodies identified to be significantly associated with rejection except for PECR, CXCL11, and FLT3LG.
TABLE 3.
Multicenter cohort biopsy results (N = 546)
| n (%) | |
|---|---|
| No rejection | 298 (54.6) |
| Acute cell-mediated rejection (ACR) 1A | 179 (32.8) |
| ACR 1B | 50 (9.2) |
| ACR 2 | 11 (2.0) |
| ACR 3A | 8 (1.5) |
| ACR 4 | 0 (0.0) |
| Antibody-mediated rejection | 0 (0.0)a |
| Mixed rejection | 0 (0.0) |
AMR was likely underreported in the multicenter cohort using the earlier 1990 ISHLT scoring28 criteria.
FIGURE 2.
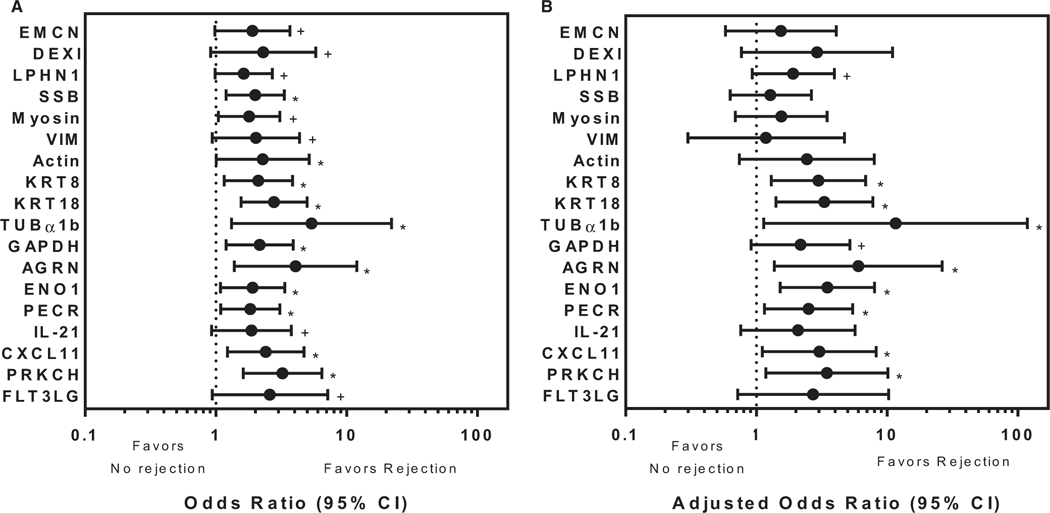
Non-HLA antibodies significantly associated with cardiac allograft rejection. High throughput multiplex bead analysis was used to identify non-HLA antibodies in sera from a multicenter adult cardiac transplant cohort. A, Antibodies to 18 non-HLA antigens (y-axis) were identified to be significantly associated with rejection with an odds ratio >1 (x-axis; n = 69 rejection sera acute-cellular rejection [ACR] ≥ 1b, n = 477 nonrejection sera). Four of these, endomucin, dexamethasone-induced transcript, latrophilin 1 and Sjogren syndrome antigen B, are newly described in relationship to transplant rejection. Bars represent the 95% confidence interval (CI). The odds ratios for antibody binding to the remaining non-HLA antigens on the multiplex panel that were not significantly different from 1 are not shown. +Denotes P < .1, *Denotes P < .05. B, Odds ratios (x-axis) of the 18 non-HLA antibodies identified in A were adjusted to remove the effects of HLA donor specific antibodies using a random effects logistic regression (n = 26 rejection sera ACR ≥ 1b, n = 106 nonrejection sera). Antibodies to 10/18 non-HLA antigens are significantly associated with rejection with odds ratio >1. Bars represent the 95% CI. The odds ratios for antibody binding to the remaining 49 non-HLA antigens not significantly different from 1 are not shown. +Denotes P < .1, *Denotes P < .05
TABLE 4.
Non-HLA antibodies significantly associated with cardiac allograft rejection
| Gene symbol | Gene name | Cellular localization | Originally described in | Reference |
|---|---|---|---|---|
| EMCN | Endomucin | Plasma membrane and cytosol | Heart | a |
| DEXI | Dexamethasone induced transcript | Plasma membrane and cytosol | Heart | a |
| LPHN1 | Latrophilin 1 | Plasma membrane | Heart | a |
| SSB | Sjogren syndrome antigen B (autoantigen La) | Nucleus | Heart | a |
| Myosin | Myosin | Cytoskeleton | Heart | 46,47 |
| VIM | Vimentin | Cytoskeleton | Heart | 12 |
| Actin | Actin | Cytoskeleton | Heart | 48 |
| KRT8 | Cytokeratin 8 | Cytoskeleton | Heart | 48 |
| KRT18 | Cytokeratin 18 | Cytoskeleton | Heart | 48 |
| TUBα1b | Tubulin, α1b | Cytoskeleton | Heart | 33 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | Plasma membrane and cytosol | Heart | 36 |
| AGRN | Agrin | Plasma membrane and cytosol | Kidney | 34 |
| ENO1 | Alpha-enolase | Plasma membrane and cytosol | Kidney | 39 |
| PECR | Peroxisomal trans-2-enoyl-CoA reductase | Peroxisomes | Kidney | 35 |
| IL-21 | Integrin 21 | Secreted | Kidney | 51 |
| CXCL11 | Chemokine (C-X-C motif) ligand 11 | Secreted | Kidney | 51 |
| PRKC | Protein kinase C | Secreted | Kidney | 37 |
| FLT3LG | Fms-like tyrosine kinase-3 ligand | Intracellular and plasma membrane | Kidney | 23 |
Newly described here.
The odds ratios were adjusted to remove the confounder of HLA DSAs (Figure 2B, grade ≥1B, odds ratio >1, 95% CI, P < .1; HLA DSA+ samples n = 26 [37.6%] in rejection and n = 106 [22.2%] in nonrejection samples). Applying this criterion, 10 of 18 non-HLA antibodies remained significantly associated with cardiac allograft rejection (95% CI, P < .1). The odds ratios for antibody binding to the remaining non-HLA antigens on the multiplex panel were not significantly different (not shown).
To determine if the number of non-HLA antibodies is increased in rejection sera we tallied the number of non-HLA antibody specificities per sample and plotted it as a percentage of the total number of samples. The median number of non-HLA antibodies per sample was significantly increased (P < .001) in sera collected at the time of rejection (3, interquartile range 1–5) compared to nonrejection samples (1, interquartile range 1–3; Figure 3). As shown, the distribution of the number of antibodies per sample shifts to the right in rejection samples.
FIGURE 3.

Heart allograft rejection is associated with an increased number of non-HLA antibodies. The number of non-HLA antibodies identified by multiplex analysis is significantly increased (P < .001) in sera obtained at the time of rejection in comparison to nonrejection. The median number of non-HLA antibodies identified from rejection samples (n = 69) is 3 (interquartile range: 1–5) in comparison to 1 (interquartile range: 1–3) in nonrejection samples (n = 477). Horizontal bar indicates interquartile range. Black circle indicates median
3.4 |. Identification of non-HLA antibodies that independently associate with and classify cardiac allograft rejection
A correlation matrix analysis evaluated clustering of non-HLA antibodies associated with rejection. Non-HLA antibodies significantly associated with cardiac allograft rejection selectively cluster into 9 groups (Figure 4, red boxes). The size of the dot/color correlates to the association between the non-HLA antibodies in rejection samples. Three of 4 newly identified non-HLA antibodies do not cluster with any other non-HLA antibodies—SSB, EMCN, and LPHN1—and are independently associated with cardiac allograft rejection. AGRN, KRT18, ENO1, and GAPDH are found independently. In comparison, non-HLA antibodies targeting the 11 remaining antigens are found in clusters and, therefore, are not independently informative.
FIGURE 4.

Correlation matrix analysis showing hierarchical clustering of non-HLA antibodies in cardiac allograft rejection sera. The matrix describes correlation of non-HLA antibodies found in sera of rejection patients. Non-HLA antibodies associated with cardiac allograft rejection (n = 69/546) selectively cluster into 9 groups (red boxes). The coefficient of correlation (scale bar at right) is indicated by the dot color and size and describes the tendency that the non-HLA antibodies will be identified independently or together in the sera of rejection patients. Dark blue and large dots along the diagonal represent a marker’s perfect correlation with itself. Correlation to other non-HLA antibodies is evaluated off the diagonal and indicated by the dot color and size (smaller dot size and lighter color indicates lower correlation; blue color indicates positive correlation). Three of the 4 non-HLA antibodies that are newly identified in this study (Sjogren syndrome antigen B, endomucin [EMCN], and latrophilin 1) are independently associated with rejection and do not group with other non-HLA antibodies. In comparison, non-HLA antibodies to 11 other antibodies cluster together and are not considered independently informative. At left, solid boxes indicate non-HLA antigens newly identified in this study; antigens identified in the CART analysis (shown in Figure 5) are indicated by dotted line boxes. EMCN is boxed in both a solid and dotted line designating it as newly identified, and identified in the CART analysis
To supplement the individual odds ratios, CART explored potential interactions and synergies of non-HLA antibodies differentiating nonrejection from rejection samples (Figure 5). Five non-HLA antibodies (KRT18, GAPDH, AGRN, ENO1, and EMCN) including 1 newly described EMCN were informative classifiers of rejection in the CART analysis at a range of strengths (800–4900 MFI, values rounded to the nearest 100). The root node, KRT18, that includes all 546 sera, 13% of which are rejection samples (69/546 = 0.13) splits at an MFI > 1500 into child nodes. As the algorithm progresses from the root node to terminal nodes at the bottom of the decision tree, rejection and nonrejection are classified. The algorithm accurately identifies nonrejection in 86% of the 546 sera analyzed (Figure 5, far left). The 5 non-HLA targets identified as nodes in the CART analysis are also independently associated with rejection (Figure 4, identified by dotted line boxes). The other 13 non-HLA antibodies identified to be significant in Figure 2A were included in the CART analysis but trimmed from the analysis because they did not add additional discrimination power.
FIGURE 5.

Classification algorithm identifying non-HLA antibodies that predict cardiac allograft rejection. Classification and regression tree (CART) analysis showing a binary decision tree that assesses the classification of rejection based on non-HLA median fluorescence intensity (MFI) in sera isolated from cardiac allograft transplant patients (n = 546 sera). The algorithm selects MFI values which optimize the differentiation of the sera into rejection and nonrejection categories at each node in the tree. For display, selected MFI’s were rounded to the nearest 100. The root node, KRT18, that includes all 546 sera, 13% of which are rejection samples (69/546 = 0.13) splits at an MFI > 1500 into child nodes. As the algorithm progresses to terminal nodes, 86% (n = 467) of nonrejection samples are correctly identified (far left). Sera samples with KRT18 MFI > 1500 strongly correlate to cardiac allograft rejection (far right). KRT18 appears in the model twice as nodes are split recursively in decision tree construction. Scale bar indicates association with rejection with lighter terminal node boxes correlating to nonrejection
3.5 |. Non-HLA antibodies synergize with HLA DSAs in cardiac allograft rejection
To determine if HLA DSAs and non-HLA antibodies are synergistic in cardiac rejection, we used logistic regression (Figure 6, n = 546 samples). The odds ratio of cardiac allograft rejection in the presence of HLA DSA alone is 2.3, and 14/18 non-HLA antibodies initially found to be significantly associated with cardiac allograft rejection retained significance in comparison to those samples without any antibodies (HLA DSA or non-HLA antibodies, P < .1). Comparison of the effect of HLA DSA alone or HLA DSA and any of the non-HLA antibodies identified as associated with cardiac allograft rejection indicates SSB, Myosin, VIM or PRKCH synergistically interact with HLA DSA to significantly increase the odds of rejection (Figure 6, brackets, P < .1).
FIGURE 6.
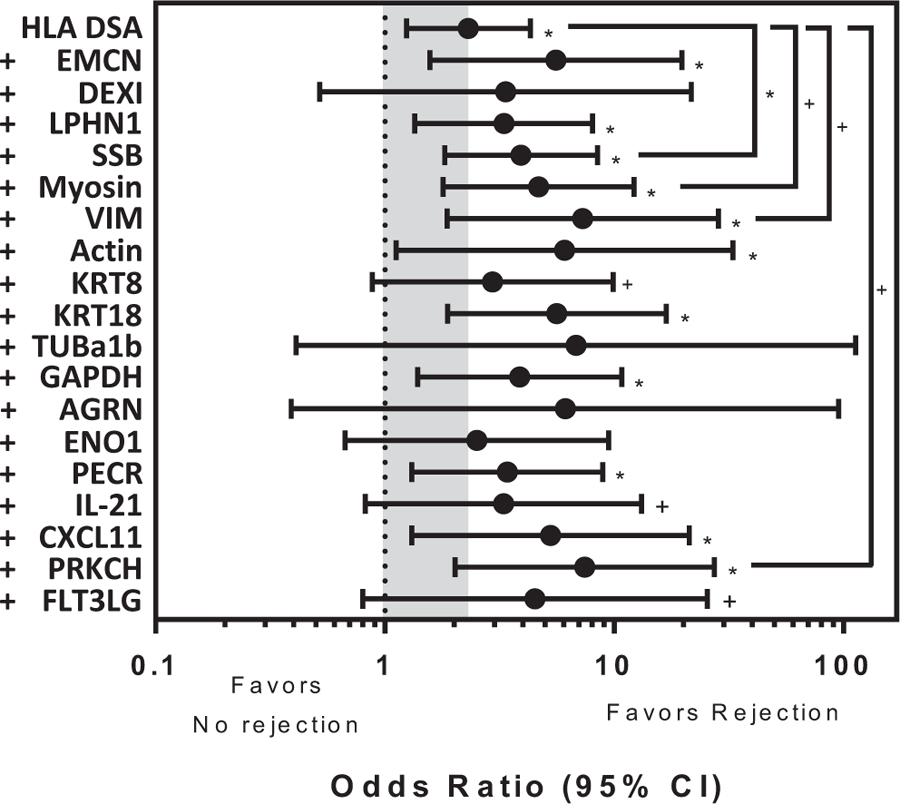
Synergy of non-HLA antibodies and HLA donor specific antibodies (DSA) in cardiac allograft rejection. Logistic regression models (using 546 samples) showing the odds ratio (x-axis) of HLA DSA alone or HLA DSA plus each non-HLA antibody (y-axis) in comparison to no antibodies. The gray shaded box highlights the significant association of cardiac allograft rejection when HLA DSA alone is present (HLA DSA, odds ratio = 2.31, P < .05). The odds ratio of 14 of the 18 non-HLA antibodies initially identified as significantly associated with cardiac allograft rejection retain significance in this model (*P < .05, +P < .1). When conditions are compared, the odds ratio of HLA DSA with non-HLA antibody raised against Sjogren syndrome antigen B, Myosin, vimentin and PRKCH with cardiac allograft rejection are significantly increased in comparison to HLA DSA alone suggesting synergistic interactions between the 2 types of antibodies (brackets, *P < .05, +P < .1). Bars represent the 95% confidence interval (CI)
3.6 |. Validation in an independent single-center cohort
We next sought to validate the 18 non-HLA antibodies (log values of MFI) to classify rejection using an independent single-center cohort (n = 63). The demographics of this cohort are listed in Table 2. There were no significant differences between patient groups with respect to age, sex, race, end-stage disease, induction therapy, and presence of pretransplant mechanical circulatory support or patient/donor HLA-A, -B, -DR mismatch. Patients in the rejection group had rejection24 of grade 1R or higher (Table 5). ROC shows that the set of 18 non-HLA antibodies associated with cardiac allograft rejection significantly differentiated the 42 patients with rejection (area under the curve [AUC] = 0.87, P < .05 with 92.86% sensitivity and 66.67% specificity) (Figure 7A). After removing the 17 patients with HLA DSA (Figure 7B), ROC analysis identified an AUC of 0.92 and sensitivity and specificity of 88.89% and 68.42%, respectively (P < .05) in the independent cohort of cardiac transplant recipients (n = 46 patients without HLA DSA, 19 with no rejection and 27 with rejection).
TABLE 5.
Single-center cohort biopsy results (N = 63)
| n (%) | |
|---|---|
| No rejection | 21 (33.3) |
| Acute cell-mediated rejection (ACR) 1R | 31 (49.2) |
| ACR 2R | 2 (3.2) |
| ACR 3R | 2 (3.2) |
| Antibody-mediated rejection | 5 (7.9) |
| Mixed rejection | 2 (3.2) |
FIGURE 7.
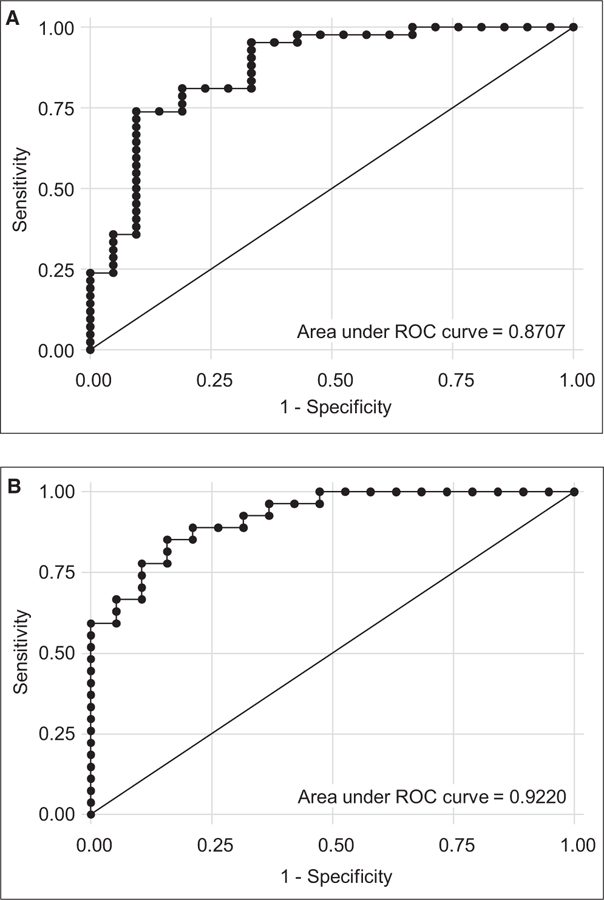
Validation of the panel of 18 non-HLA antibodies for the prediction of cardiac allograft rejection using an independent single-center adult cardiac transplant cohort. A, ROC analysis identified an area under the curve (AUC) of 0.87 and sensitivity and specificity of 92.86% and 66.67%, respectively (P < .05) in an independent cohort of cardiac transplant recipients (n = 63 patients, 21 with no rejection and 42 with rejection). B, After removing those patients with HLA donor-specific antibodies (DSAs), ROC analysis identified an AUC of 0.92 and sensitivity and specificity of 88.89% and 68.42%, respectively (P < .05) in the independent cohort of cardiac transplant recipients (n = 46 patients without HLA DSAs, 19 with no rejection and 27 with rejection)
3.7 |. Tissue expression of non-HLA antigens associated with cardiac allograft rejection
To confirm that the non-HLA antigens associated with cardiac allograft rejection are expressed in vivo, publicly available RNA-seq data30 from cardiac, kidney, lung, and liver tissues as well as HUVECs was assessed (Figure 8A). Transcripts for all of the non-HLA antigens were found to be expressed in cardiac tissue (TPM > 1), as well as additional tissues with the exception of myosin which is specific for cardiac tissue. IHC was performed to identify the tissue localization of SSB and EMCN in EMB tissues from cardiac allograft rejection patients. Representative images of biopsies from 3 patients in the multicenter cohort show SSB is a nuclear protein in the EC microvasculature. EMCN is expressed in the membrane and cytoplasm of cardiomyocytes and in the microvasculature (Figure 8B). A similar expression for SSB and EMCN was seen in biopsies without rejection (data not shown).
FIGURE 8.

Non-HLA antigenic targets are expressed in human tissues and endothelial cells. A, mRNA expression of the 18 non-HLA antibody antigens associated with cardiac allograft rejection from publically available data of the 18 non-HLA antibodies in cardiac, kidney, lung, and liver tissue as well as HUVEC. B, Expression of Sjogren syndrome antigen B (SSB) and endomucin (EMCN) on cardiac biopsies. Immunohistochemistry of biopsies taken at the time of rejection shows expression of SSB and EMCN proteins on cardiac endothelium. Micrographs are representative of biopsies tested from 3 cardiac transplant recipients from the multicenter cohort. SSB is a nuclear protein expressed in endothelial cell (EC) microvasculature. EMCN is expressed in the membrane and cytoplasm of some EC in the microvasculature and in all cardio myocytes
4 |. DISCUSSION
We used a proteomics approach to identify antibodies to 18 non-HLA antigens as significantly associated with cardiac allograft rejection. Four of 18 targets (EMCN, DEXI, LAPHN, and SSB) are newly described as being associated with cardiac transplant rejection. Transcripts for each of the non-HLA targets discovered are expressed in cardiac, liver, kidney, and lung tissues, with the exception of cardiac myosin, which is exclusive to cardiac tissue. We further confirmed, that 2/4 novel targets (SSB and EMCN) are expressed in EMB.
Three of 4 newly described targets (EMCN, LAPHN, and SSB) are found independent of other non-HLA antibodies, indicating they are independent markers of rejection. Further, the absence of EMCN antibodies was associated with nonrejection in the CART analysis designed to explore synergistic effects. The ability of the 18-non-HLA antigen panel to classify rejection was validated using an independent single-center cohort of adult cardiac transplant recipients. ROC analysis showed the panel correctly classifies rejection 87% of the time (AUC = 0.87, P < .05).
We observed the median number of non-HLA antibodies in rejection sera was significantly higher than in nonrejection samples (3, interquartile range 1–5 vs 1, interquartile range 1–3; P < .001). Allograft damage due to rejection may therefore stimulate antibody production to self-antigens. Non-HLA antibodies may also function to mediate rejection. We also identified non-HLA antibodies present in nonrejection samples suggesting association with underlying disease etiologies. While we do not yet know whether these antibodies are directly mediating injury or reflecting an inflammatory process, they may be clinically useful to identify patients at risk of rejection or poor outcomes.
A model of HLA DSA and non-HLA antibodies showed synergies with rejection. The odds of rejection were significantly increased when antibodies to SSB, myosin, VIM, or PRKCH were present with HLA DSAs, in comparison to samples with HLA DSAs alone. Whether this reflects a 2-hit mechanism of injury or HLA mediated remodeling of the actin cytoskeleton remains to be determined.
The 4 novel non-HLA antigen targets include EMCN, DEXI, LPHN1, and SSB. EMCN is a mucin-like sialoglycoprotein that inhibits assembly of focal adhesions. It is highly expressed in HAEC and microvascular EC.42 DEXI is a protein of unknown function implicated in autoimmune disease43 and immunoglobulin A deficiency.44 LPHN1 is a G protein–coupled receptor with roles in cell adhesion and signal transduction.45 SSB antibodies are found in patients with systemic lupus erythematosus (SLE) and Sjogren’s syndrome. Mothers with SSB antibodies are at an increased risk for neonatal lupus associated with fetal congenital heart block.
Additionally, we found 14 non-HLA autoantibodies that were previously reported in cardiac allograft rejection including myosin, vimentin, actin, KRT8, KRT18, tubulin, and GAPDH. While autoantibodies to agrin, ENO1, PECR, IL-21, CXCL11, PRKCH, and FLT3LG were previously associated with renal allograft rejection. Our findings are consistent with studies showing vimentin and myosin autoantibodies are associated with acute rejection.14,46 Anti-vimentin and myosin antibodies are also associated with development of CAV in heart transplant.12,47 Cardiac allograft rejection was also associated with anti-cytoskeletal EC antibodies to actin, keratin, and tubulin.48 Joosten et al34 identified agrin autoantibodies in 44% of patients with transplant glomerulopathy after kidney transplant. Antibodies to GAPDH were previously associated with dilated cardiomyopathy36 and SLE.49 Furthermore, GAPDH has been found in exosomes50 and at increased concentrations of cell free DNA in rheumatoid arthritis.38 ENO1 antibodies have been associated with membranous nephropathy in kidney transplant.39 Antibodies to PECR are associated with transplant glomerulopathy in kidney transplant. Antibodies to the chemokine CXCL11 and cytokine IL-21 have been associated with chronic renal allograft injury.52 Additionally, the transcription level of CXCL11 has been shown to be increased in patients with AMR by the molecular microscope40; therefore, antibodies to CXCL11 may be generated in response to the increased expression of CXCL11 produced during rejection. PRKCH is a calcium-independent serine/threonine protein kinase involved in the differentiation of pre–B cell receptors and regulation of epithelial cell tight junction and is involved in proliferation and apoptosis in cancer models. FLT3LG antibodies were associated with AMR in kidney allografts.23
Limitations of this study include the retrospective nature and largely white population of the studied cohorts and hence the need to confirm these findings in a larger, multicenter prospective study. Additionally, the multicenter cohort and single-center cohorts were graded according to different ISHLT criteria, 199027 and 2004,24 respectively, according to how the data were originally published or scored clinically. Thus, to compare the results between cohorts, we included ≥ACR 1R rejection episodes to validate the panel of 18 non-HLA antigen targets to predict rejection in the single-center cohort. Additionally, the diagnosis of AMR was likely underreported in the multicenter cohort using the earlier 1990 ISHLT scoring27 criteria and the histologic evidence for these non-HLA antibodies has not been clearly defined.
The CART was used to identify synergies and potential interactions among the 18 significant non-HLA antibodies that were identified in the odds ratio analysis. We acknowledge the limitation of considering the selected MFI values as definitive due to the low number of rejection sera positive for each non-HLA antibody.
A positive threshold of MFI > 1000 was used for the odds ratio analyses performed in this study. However, this may have resulted in lower sensitivity, but higher specificity in calling rejection. Going forward, the positive threshold for non-HLA antibodies will have to be established by individual labs until enough data can be generated by the field to support a common cutoff similar to the process that was used for HLA single antigen. The protein microarray expressed in a baculovirus system, may not be posttranslationally modified in the same way as proteins expressed in human cells although, they provide some posttranslational modifications.41 In addition, certain non-HLA antibodies such as AT1R could not be conjugated to Luminex beads to be included in the panel. We acknowledge that non-HLA protein antigens excluded during our discovery strategy may be biologically relevant and merit additional investigation.
Validation of these non-HLA antigens, assessment of their evolution, and relationship to rejection and other clinical factors including mechanical circulatory devices is needed. Longitudinal analysis including pretransplant time points is needed to determine the role of preformed and/or de novo non-HLA antibodies in rejection and long-term graft outcome. Another question is which of these antibodies are autoantibodies or antibodies to minor antigen differences between the donor/recipient.52 In summary, using a combination of high throughput proteomics and bioinformatics, we identified an informative panel of non-HLA antigens that can identify antibodies associated with cardiac allograft rejection and upon validation could be extended to other solid organ transplants. We envision using the panel to support clinical decision making as part of the protocols for noninvasive monitoring, for biopsy interpretation, and recalcitrant rejection.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported in part by a grant from Immucor, Inc, NIAID R01 AI024819, R01 AI135201 (to Dr Reed), and an NIDDK T32 fellowship (to Dr Butler). The multicenter cohort was a sub-study of the phase 3 clinical trial sponsored by Novartis (www.ClinicalTrials.gov NCT00300274). The authors thank all individuals and centers involved in the collection and maintenance of the multicenter cohort patient sera including: Cleveland Clinic, Drexel, LDS Hospital, Cardiology Methodist, Medical University of South Carolina, Tufts, UCLA, University of Puerto Rico, and Washington University. We also extend our gratitude to the staff of the UCLA Immunogenetics Center for maintenance of the biorepository sera and HLA typing of endothelial cells. Finally, the authors hereby express their thanks for the cooperation of OneLegacy and gratitude to the organ and tissue donors and their families, for giving the gift of life and the gift of knowledge, facilitated by their generous donation.
Funding information
Immucor, Inc.; National Institute of Diabetes and Digestive and Kidney Diseases, Grant/Award Number: T32 DK10468702; National Institute of Allergy and Infectious Diseases, Grant/Award Number: RO1AI024819 and RO1AI35201; Novartis, Grant/Award Number: NCT00300274
Drs Jiang, Balazs, and Bryan Ray are employees of Immucor, Inc. Dr Reed has an investigator-initiated research grant from Immucor, Inc.
Abbreviations:
- ACR
acute cell-mediated rejection
- AGRN
agrin
- AMR
antibody-mediated rejection
- AUC
area under the curve
- CART
Classification and Regression Tree
- CXCL11
C-X-C motif chemokine ligand 11
- DEXI
dexamethasone-induced transcript
- DSA
donor-specific antibody
- EC
endothelial cell
- ECXM
endothelial cell crossmatch
- EMB
endomyocardial biopsy
- EMCN
endomucin
- ENO1
alpha enolase
- FLT3LG
Fms-related tyrosine kinase 3 ligand
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- HAEC
human aortic endothelial cell
- HUVEC
human umbilical vein endothelial cell
- IHC
immunohistochemistry
- IL
interleukin
- ISHLT
International Society for Heart and Lung Transplantation
- KRT18
keratin 18
- KRT8
keratin 8
- LPHN1
latrophilin 1
- MFI
median fluorescence intensity
- MICA
MHC class I polypeptide-related sequence A
- PECR
peroxisomal trans-2-enoyl-CoA reductase
- PRKCH
protein kinase C eta
- ROC
receiver operating characteristic curve
- SLE
systemic lupus erythematosus
- SSB
Sjogren syndrome antigen B
- TUBα1b
tubulin alpha 1 b
- VIM
vimentin
Footnotes
DISCLOSURE
The authors of this manuscript have conflicts of interest to disclose as described by the American Journal of Transplantation.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
REFERENCES
- 1.Tonsho M, Michel S, Ahmed Z, Alessandrini A, Madsen JC. Heart transplantation: challenges facing the field. Cold Spring Harb Perspect Med. 2014;4(5):pii:a015636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stehlik J, Edwards LB, Kucheryavaya AY, et al. The Registry of the International Society for Heart and Lung Transplantation: 29th official adult heart transplant report–2012. J Heart Lung Transplant. 2012;31(10):1052–1064. [DOI] [PubMed] [Google Scholar]
- 3.Colvin-Adams M, Smith JM, Heubner BM, et al. OPTN/SRTR 2013 annual data report: heart. Am J Transplant. 2015;15(Suppl 2):1–28. [DOI] [PubMed] [Google Scholar]
- 4.Tible M, Loupy A, Vernerey D, et al. Pathologic classification of antibody-mediated rejection correlates with donor-specific antibodies and endothelial cell activation. J Heart Lung Transplant. 2013;32(8):769–776. [DOI] [PubMed] [Google Scholar]
- 5.Tait BD, Süsal C, Gebel HM, et al. Consensus guidelines on the testing and clinical management issues associated with HLA and non-HLA antibodies in transplantation. Transplantation. 2013;95(1):19–47. [DOI] [PubMed] [Google Scholar]
- 6.Loupy A, Toquet C, Rouvier P, et al. Late failing heart allografts: pathology of cardiac allograft vasculopathy and association with antibody-mediated rejection. Am J Transplant. 2016;16(1):111–120. [DOI] [PubMed] [Google Scholar]
- 7.Kobashigawa J, Colvin M, Potena L, et al. The management of antibodies in heart transplantation: an ISHLT consensus document. J Heart Lung Transplant. 2018;37(5):537–547. [DOI] [PubMed] [Google Scholar]
- 8.Tambur AR, Pamboukian SV, Costanzo M-R, et al. The presence of HLA-directed antibodies after heart transplantation is associated with poor allograft outcome. Transplantation. 2005;80(8):1019–1025. [DOI] [PubMed] [Google Scholar]
- 9.Starling RC, Stehlik J, Baran DA, et al. Multicenter analysis of immune biomarkers and heart transplant outcomes: results of the clinical trials in Organ Transplantation-05 Study. Am J Transplant. 2016;16(1):121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Filippo S, Girnita A, Webber SA, et al. Impact of ELISA-detected anti-HLA antibodies on pediatric cardiac allograft outcome. Hum Immunol. 2005;66(5):513–518. [DOI] [PubMed] [Google Scholar]
- 11.Clerkin KJ, Farr MA, Restaino SW, et al. Donor-specific anti-HLA antibodies with antibody-mediated rejection and long-term outcomes following heart transplantation. J Heart Lung Transplant. 2017;36(5):540–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jurcevic S, Ainsworth ME, Pomerance A, et al. Antivimentin antibodies are an independent predictor of transplant-associated coronary artery disease after cardiac transplantation. Transplantation. 2001;71(7):886–892. [DOI] [PubMed] [Google Scholar]
- 13.Leong HS, Mahesh BM, Day JR, et al. Vimentin autoantibodies induce platelet activation and formation of platelet-leukocyte conjugates via platelet-activating factor. J Leukoc Biol. 2008;83(2):263–271. [DOI] [PubMed] [Google Scholar]
- 14.Mahesh B, Leong HS, McCormack A, Sarathchandra P, Holder A, Rose ML. Autoantibodies to vimentin cause accelerated rejection of cardiac allografts. Am J Pathol. 2007;170(4):1415–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kauke T, Kaczmarek I, Dick A, et al. Anti-MICA antibodies are related to adverse outcome in heart transplant recipients. J Heart Lung Transplant. 2009;28(4):305–311. [DOI] [PubMed] [Google Scholar]
- 16.Suárez-Álvarez B, López-Vázquez A, Gonzalez MZ, et al. The relationship of anti-MICA antibodies and MICA expression with heart allograft rejection. Am J Transplant. 2007;7(7):1842–1848. [DOI] [PubMed] [Google Scholar]
- 17.Reinsmoen NL, Lai C-H, Heidecke H, et al. Anti-angiotensin type 1 receptor antibodies associated with antibody mediated rejection in donor HLA antibody negative patients. Transplantation. 2010;90(12):1473–1477. [DOI] [PubMed] [Google Scholar]
- 18.Reinsmoen NL, Lai C-H, Mirocha J, et al. Increased negative impact of donor HLA-specific together with non-HLA-specific antibodies on graft outcome. Transplantation. 2014;97(5):595–601. [DOI] [PubMed] [Google Scholar]
- 19.Dunn MJ, Crisp SJ, Rose ML, Taylor PM, Yacoub MH. Anti-endothelial antibodies and coronary artery disease after cardiac transplantation. Lancet. 1992;339(8809):1566–1570. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Q, Cecka JM, Gjertson DW, et al. HLA and MICA: targets of antibody-mediated rejection in heart transplantation. Transplantation. 2011;91(10):1153–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gunasekaran M, Sharma M, Hachem R, Bremner R, Smith MA, Mohanakumar T. Circulating exosomes with distinct properties during chronic lung allograft rejection. J Immunol. 2018;200(8):2535–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soulez M, Pilon E-A, Dieudé M, et al. The perlecan fragment LG3 is a novel regulator of obliterative remodeling associated with allograft vascular rejection. Circ Res. 2012;110(1):94–104. [DOI] [PubMed] [Google Scholar]
- 23.Jackson AM, Sigdel TK, Delville M, et al. Endothelial cell antibodies associated with novel targets and increased rejection. J Am Soc Nephrol. 2015;26(5):1161–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart S, Winters GL, Fishbein MC, et al. Revision of the 1990 working formulation for the standardization of nomenclature in the diagnosis of heart rejection. J Heart Lung Transplant. 2005;24(11):1710–1720. [DOI] [PubMed] [Google Scholar]
- 25.Michaels PJ, Espejo ML, Kobashigawa J, et al. Humoral rejection in cardiac transplantation: risk factors, hemodynamic consequences and relationship to transplant coronary artery disease. J Heart Lung Transplant. 2003;22(1):58–69. [DOI] [PubMed] [Google Scholar]
- 26.Eisen HJ, Kobashigawa J, Starling RC, et al. Everolimus versus mycophenolate mofetil in heart transplantation: a randomized, multicenter trial. Am J Transplant. 2013;13(5):1203–1216. [DOI] [PubMed] [Google Scholar]
- 27.Billingham ME, Cary NR, Hammond ME, et al. A working formulation for the standardization of nomenclature in the diagnosis of heart and lung rejection: Heart Rejection Study Group. The International Society for Heart Transplantation. J Heart Transplant. 1990;9(6):587–593. [PubMed] [Google Scholar]
- 28.Li L, Wadia P, Chen R, et al. Identifying compartment-specific non-HLA targets after renal transplantation by integrating transcriptome and “antibodyome” measures. Proc Natl Acad Sci USA. 2009;106(11):4148–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uhlen M, Fagerberg L, Hallstrom BM, et al. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- 31.Sosa RA, Zarrinpar A, Rossetti M, et al. Early cytokine signatures of ischemia/reperfusion injury in human orthotopic liver transplantation. JCI Insight. 2016;1(20):e89679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Draper N, Smith H. Applied Regression Analysis. 2nd edn. New York: John Wiley & Sons; 1981. [Google Scholar]
- 33.Hoffman WH, Sharma M, Cihakova D, et al. Cardiac antibody production to self-antigens in children and adolescents during and following the correction of severe diabetic ketoacidosis. Autoimmunity. 2016;49(3):188–196. [DOI] [PubMed] [Google Scholar]
- 34.Joosten SA, Sijpkens YWJ, van Ham V, et al. Antibody response against the glomerular basement membrane protein agrin in patients with transplant glomerulopathy. Am J Transplant. 2005;5(2):383–393. [DOI] [PubMed] [Google Scholar]
- 35.Dinavahi R, George A, Tretin A, et al. Antibodies reactive to non-HLA antigens in transplant glomerulopathy. J Am Soc Nephrol. 2011;22(6):1168–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buse C, Altmann F, Amann B, et al. Discovering novel targets for autoantibodies in dilated cardiomyopathy. Electrophoresis. 2008;29(6):1325–1332. [DOI] [PubMed] [Google Scholar]
- 37.Sutherland SM, Li L, Sigdel TK, et al. Protein microarrays identify antibodies to protein kinase Czeta that are associated with a greater risk of allograft loss in pediatric renal transplant recipients. Kidney Int. 2009;76(12):1277–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong X-Y, von Mühlenen I, Li Y, et al. Increased concentrations of antibody-bound circulatory cell-free DNA in rheumatoid arthritis. Clin Chem. 2007;53(9):1609–1614. [DOI] [PubMed] [Google Scholar]
- 39.Murtas C, Bruschi M, Candiano G, et al. Coexistence of different circulating anti-podocyte antibodies in membranous nephropathy. Clin J Am Soc Nephrol. 2012;7(9):1394–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loupy A, Duong Van Huyen JP, Hidalgo L, et al. Expression profiling for the identification and classification of antibody-mediated heart rejection. Circulation. 2017;135(10):917–935. [DOI] [PubMed] [Google Scholar]
- 41.Murphy CI, Piwnica-Worms H, Grunwald S, Romanow WG, Francis N, Fan HY. Overview of the baculovirus expression system. Curr Protoc Mol Biol. 2004;Chapter 16: Unit 16 19 10.1002/0471142727.mb1609s65. [DOI] [PubMed] [Google Scholar]
- 42.Liu C, Shao Z-M, Zhang L, et al. Human endomucin is an endothelial marker. Biochem Biophys Res Commun. 2001;288(1):129–136. [DOI] [PubMed] [Google Scholar]
- 43.Davison LJ, Wallace C, Cooper JD, et al. Long-range DNA looping and gene expression analyses identify DEXI as an autoimmune disease candidate gene. Hum Mol Genet. 2012;21(2):322–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bronson PG, Chang D, Bhangale T, et al. Common variants at PVT1, ATG13-AMBRA1, AHI1 and CLEC16A are associated with selective IgA deficiency. Nat Genet. 2016;48(11):1425–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silva JP, Ushkaryov YA. The latrophilins, “split-personality” receptors. Adv Exp Med Biol. 2010;706:59–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nath DS, Ilias Basha H, Tiriveedhi V, et al. Characterization of immune responses to cardiac self-antigens myosin and vimentin in human cardiac allograft recipients with antibody-mediated rejection and cardiac allograft vasculopathy. J Heart Lung Transplant. 2010;29(11):1277–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalache S, Dinavahi R, Pinney S, Mehrotra A, Cunningham MW, Heeger PS. Anticardiac myosin immunity and chronic allograft vasculopathy in heart transplant recipients. J Immunol. 2011;187(2):1023–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Álvarez-Márquez A, Aguilera I, Blanco RM, et al. Positive association of anticytoskeletal endothelial cell antibodies and cardiac allograft rejection. Hum Immunol. 2008;69(3):143–148. [DOI] [PubMed] [Google Scholar]
- 49.Sun J, Li X, Zhou H, et al. Anti-GAPDH autoantibody is associated with increased disease activity and intracranial pressure in systemic lupus erythematosus. J Immunol Res. 2019;2019:7430780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jella KK, Yu L, Yue Q, Friedman D, Duke BJ, Alli AA. Exosomal GAPDH from proximal tubule cells regulate ENaC activity. PLoS ONE. 2016;11(11):e0165763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sigdel TK, Li LI, Tran TQ, et al. Non-HLA antibodies to immunogenic epitopes predict the evolution of chronic renal allograft injury. J Am Soc Nephrol. 2012;23(4):750–763. [DOI] [PubMed] [Google Scholar]
- 52.Steers NJ, Li Y, Drace Z, et al. Genomic mismatch at LIMS1 locus and kidney allograft rejection. N Engl J Med. 2019;380(20):1918–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


