Candida auris is a recently emerged pathogenic yeast that within a few years after its initial description has spread all over the globe. C. auris is a major concern for human health, because it can cause life-threatening systemic infections, is easily transmissible, and is difficult to eradicate from hospital environments. Furthermore, C. auris is highly drug resistant, especially against the widely used antifungal drug fluconazole. Mutations in the drug target and high activity of efflux pumps are associated with azole resistance, but it is not known how drug resistance genes are regulated in C. auris. We have investigated the potential role of several candidate transcriptional regulators in the intrinsic fluconazole resistance of C. auris and identified a transcription factor that contributes to the high resistance to fluconazole and voriconazole of two C. auris strains from different genetic clades, thereby providing insight into the molecular basis of drug resistance of this medically important yeast.
KEYWORDS: Candida auris, fluconazole resistance, transcription factor
ABSTRACT
The recently emerged pathogenic yeast Candida auris is a major concern for human health, because it is easily transmissible, difficult to eradicate from hospitals, and highly drug resistant. Most C. auris isolates are resistant to the widely used antifungal drug fluconazole due to mutations in the target enzyme Erg11 and high activity of efflux pumps, such as Cdr1. In the well-studied, distantly related yeast Candida albicans, overexpression of drug efflux pumps also is a major mechanism of acquired fluconazole resistance and caused by gain-of-function mutations in the zinc cluster transcription factors Mrr1 and Tac1. In this study, we investigated a possible involvement of related transcription factors in efflux pump expression and fluconazole resistance of C. auris. The C. auris genome contains three genes encoding Mrr1 homologs and two genes encoding Tac1 homologs, and we generated deletion mutants lacking these genes in two fluconazole-resistant strains from clade III and clade IV. Deletion of TAC1b decreased the resistance to fluconazole and voriconazole in both strain backgrounds, demonstrating that the encoded transcription factor contributes to azole resistance in C. auris strains from different clades. CDR1 expression was not or only minimally affected in the mutants, indicating that Tac1b can confer increased azole resistance by a CDR1-independent mechanism.
IMPORTANCE Candida auris is a recently emerged pathogenic yeast that within a few years after its initial description has spread all over the globe. C. auris is a major concern for human health, because it can cause life-threatening systemic infections, is easily transmissible, and is difficult to eradicate from hospital environments. Furthermore, C. auris is highly drug resistant, especially against the widely used antifungal drug fluconazole. Mutations in the drug target and high activity of efflux pumps are associated with azole resistance, but it is not known how drug resistance genes are regulated in C. auris. We have investigated the potential role of several candidate transcriptional regulators in the intrinsic fluconazole resistance of C. auris and identified a transcription factor that contributes to the high resistance to fluconazole and voriconazole of two C. auris strains from different genetic clades, thereby providing insight into the molecular basis of drug resistance of this medically important yeast.
INTRODUCTION
Candida auris is a recently emerged pathogenic Candida species that was described for the first time only a decade ago (1) but has since been isolated with increasing frequency from all over the world (2). Four genetically distinct clades of C. auris have been identified and named according to their geographic origin: South Asian (clade I), East Asian (clade II), South African (clade III), and South American (clade IV) (3). Within each clade, isolates are genetically nearly identical, but strains from different clades differ by tens of thousands of single nucleotide polymorphisms. C. auris is a major health concern, because it persists on skin and hospital surfaces despite disinfection measures, is easily transmitted, and has already caused nosocomial outbreaks (4). Except for strains from clade II, which have been mainly isolated from the ear canal of patients, C. auris can cause systemic infections with high mortality rates. This problem is aggravated by the fact that C. auris is a highly drug-resistant species, and resistance to all classes of antifungal drugs that are available to treat such infections has been reported. Resistance to the most widely used antifungal drug, fluconazole, is especially prominent, with >90% of all isolates being resistant, such that C. auris can be considered an intrinsically fluconazole-resistant species that requires alternative drugs for treatment. Fluconazole resistance is frequently associated with mutations in the target enzyme Erg11, a well-known azole resistance mechanism in other fungal pathogens (3, 5–7). However, ERG11 mutations alone cannot explain the very high fluconazole resistance levels of many C. auris strains, and high activity of drug efflux pumps has been suggested as an additional mechanism (8).
In the well-studied, distantly related pathogenic yeast Candida albicans, which is normally susceptible to fluconazole but can acquire resistance to the drug under selective pressure, three efflux pumps are known to contribute to azole resistance, the ABC transporters Cdr1 and Cdr2 and the major facilitator Mdr1 (9). Constitutive overexpression of the encoding genes is caused by gain-of-function (GOF) mutations in the zinc cluster transcription factors Tac1 and Mrr1, respectively, and is a major mechanism of acquired azole resistance in C. albicans (10–14). A CDR1 homolog has been identified in C. auris and shown by targeted gene deletion to mediate azole resistance (15, 16). How the transcription of CDR1 is regulated in C. auris and how high expression levels resulting in drug resistance are achieved in this species are currently unknown. We hypothesized that homologs of Tac1 and Mrr1 might control expression of CDR1 and other potential drug efflux pump-encoding genes also in C. auris and be constitutively active to ensure the high fluconazole resistance of many strains. We therefore identified TAC1 and MRR1 homologs in the C. auris genome and investigated their possible involvement in azole resistance.
RESULTS
Identification of MRR1 and TAC1 homologs in C. auris.
Highly complete genome assemblies have recently been published for four C. auris isolates representing each clade of this species (17). Among these isolates, B11243 (clade IV) exhibited the highest level of fluconazole resistance (>256 μg/ml), which cannot be explained solely by the Y132F mutation found in Erg11 of this strain. We therefore chose isolate B11243 to investigate a possible role of Mrr1 and Tac1 homologs in fluconazole resistance of C. auris. A BLAST search of the B11243 genome sequence with the Mrr1 and Tac1 protein sequences of C. albicans strain SC5314 identified three and two predicted proteins, respectively, for which a reciprocal BLAST search yielded Mrr1 and Tac1 as best hits. For simplicity, we designated the corresponding genes MRR1a (PSK78296.1), MRR1b (PSK79149.1), MRR1c (PSK77655.1), TAC1a (PSK79380.1), and TAC1b (PSK79381.1) in the present study. MRR1a encodes a protein of 1,133 amino acids that has 35.4% identity and 54% similarity over its entire length to CaMrr1. MRR1b encodes a protein of 1,059 amino acids with 28.8% identity and 47.1% similarity to CaMrr1. MRR1c encodes a protein of 851 amino acids with 25.3% identity and 40.5% similarity to CaMrr1. Of the two Tac1 homologs, Tac1a (805 amino acids) has 29% identity and 46.4% similarity to CaTac1, while Tac1b (863 amino acids) has 26.7% identity and 45.4% similarity to CaTac1. TAC1a and TAC1b are located in tandem in the genome of C. auris, suggesting that they arose by gene duplication; however, the encoded proteins display only low similarity to each other (24.4% identity and 43.6% similarity). All five predicted proteins contain the consensus motif CX2CX6CX5–12CX2CX6–8C in their N-terminal region, as is typical for zinc cluster transcription factors (18).
Deletion of MRR1 and TAC1 homologs in C. auris strain B11243.
To investigate if the Mrr1 and Tac1 homologs are important for the high fluconazole resistance of C. auris strain B11243, we generated deletion mutants lacking the coding sequences of the corresponding genes. In contrast to C. albicans, C. auris is a haploid species, so that null mutants can be obtained in a single gene replacement step. We constructed deletion cassettes in which the caSAT1 selection marker (19), which confers resistance to nourseothricin and has been used successfully by other researchers for the genetic manipulation of C. auris (16), was flanked by ca. 0.5 kb of the upstream and downstream sequences of the target genes. Nourseothricin-resistant clones that were obtained after transformation with the deletion cassettes were then analyzed by Southern hybridization of genomic DNA that was digested with suitable restriction enzymes (schematics in Fig. 1).
FIG 1.

Schematics of the genomic MRR1a (A), MRR1b (B), MRR1c (C), TAC1a (D), and TAC1b (E) loci in the wild-type strain B11243 (top panels) and after replacement of the coding sequences by the caSAT1 marker via homologous recombination (bottom panels). Diagnostic restriction sites used to analyze transformants are indicated: B, BamHI; N, NdeI; EV, EcoRV; S, SalI; EI, EcoRI. The coding regions of the target genes are indicated by the gray arrows, and the caSAT1 marker is indicated by the red arrows. The gray lines represent the flanking sequences; the thicker parts were cloned in the deletion cassettes and also used as upstream and downstream probes in Southern hybridizations. The predicted sizes of hybridizing fragments in the parental strain and deletion mutants are given. The green bars indicate the parts of the coding sequences that were used as probes to confirm the absence of the target genes in deletion mutants.
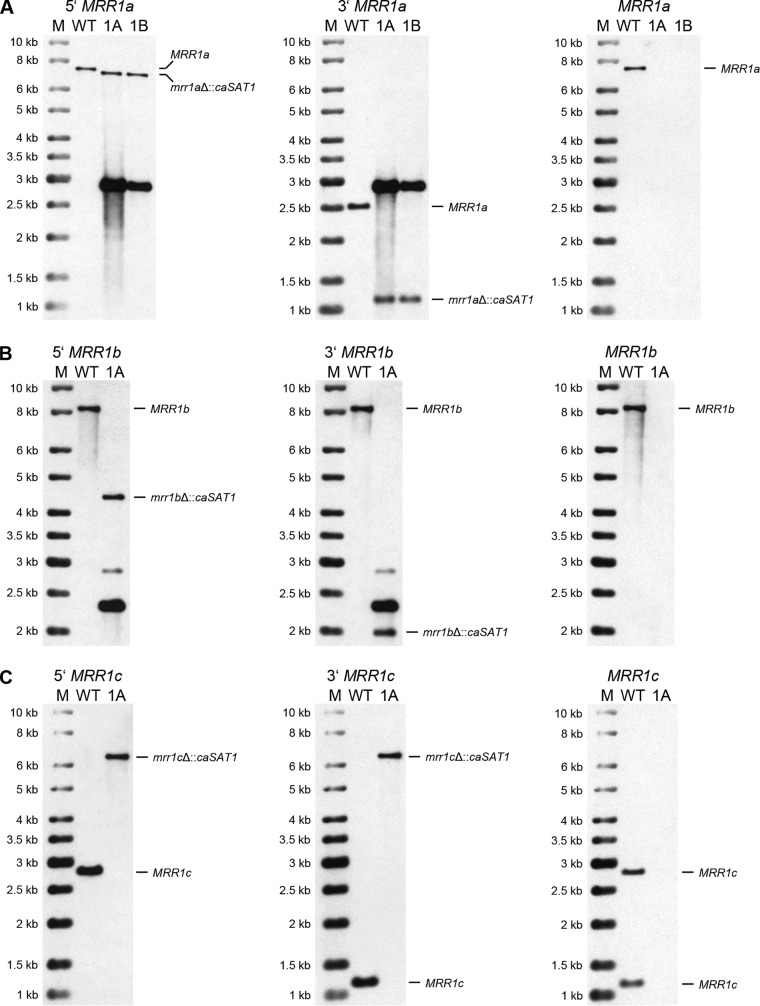
Among 26 tested clones that were transformed with the MRR1a deletion cassette, only four had lost the wild-type BamHI fragments and contained the expected new fragments after hybridization with the upstream and downstream flanking sequences, whereas the other transformants had ectopically integrated the deletion cassette. The correct transformants contained an additional hybridizing fragment of 2.9 kb that produced a strong signal with the probes and corresponded to the size of the MRR1a deletion cassette (which contained a single BamHI site), indicative of tandem integration of multiple copies of the cassette. This hybridizing fragment was also present in most of the clones with ectopic integrations. Rehybridization of the blot with a probe from the MRR1a coding sequence confirmed the deletion of the target gene in the four correct transformants, and two clones from independent sets of transformants, termed AR0931MRR1aM1A and -B (Fig. 2A), were kept for phenotypic analysis (Table 1 lists all strain names and genotypes).
FIG 2.
Southern hybridization analysis of the parental strain B11243 (WT) and the mrr1aΔ (A), mrr1bΔ (B), and mrr1cΔ (C) mutants derived from it. Genomic DNA of the strains was digested with appropriate restriction enzymes as shown in Fig. 1 and hybridized with upstream probes (left panels), downstream probes (middle panels), and open reading frame (ORF) probes (right panels). Molecular size markers (M) are on the left, and the identity of hybridizing fragments is given on the right side of the blots. Names of the mutants are abbreviated; for example, 1A and 1B in panel A indicate the mrr1aΔ mutants AR0931MRR1aM1A and AR0931MRR1aM1B, respectively.
TABLE 1.
C. auris strains
| Strain | Parent | Genotype | Reference |
|---|---|---|---|
| B11243 (AR#0931) | Clinical isolate | 17 | |
| AR0931MRR1aM1A and B | B11243 | mrr1aΔ::caSAT1a | This study |
| AR0931MRR1bM1A | B11243 | mrr1bΔ::caSAT1b | This study |
| AR0931MRR1cM1A | B11243 | mrr1cΔ::caSAT1a | This study |
| AR0931TAC1aM1A and B | B11243 | tac1aΔ::caSAT1b | This study |
| AR0931TAC1bM1A and B | B11243 | tac1bΔ::caSAT1 | This study |
| AR0931TAC1bM2A | B11243 | tac1bΔ::cauSAT1 | This study |
| B11221 (AR#0383) | Clinical isolate | 17 | |
| AR0383MRR1aM1A | B11221 | mrr1aΔ::caSAT1 | This study |
| AR0383MRR1bM1A | B11221 | mrr1bΔ::caSAT1 | This study |
| AR0383MRR1cM1A | B11221 | mrr1cΔ::caSAT1 | This study |
| AR0383TAC1aM1A and B | B11221 | tac1aΔ::caSAT1 | This study |
| AR0383TAC1bM1A | B11221 | tac1bΔ::caSAT1 | This study |
Strains contain tandem integrations of the deletion cassette.
Strain contains additional ectopic integrations.
Of seven tested clones that were transformed with the MRR1b deletion cassette, only one had lost the wild-type NdeI fragment and contained the expected new fragments after hybridization with the upstream and downstream flanking sequences (Fig. 2B); the other transformants had ectopically integrated multiple copies of the deletion cassette. The positive clone additionally contained a strongly hybridizing 2.3-kb fragment, which could be explained by tandem integration of more than one copy of the deletion cassette. Rehybridization with a probe from the MRR1b coding sequence confirmed its deletion in strain AR0931MRR1bM1A (Fig. 2B).
Among five analyzed transformants with the MRR1c deletion construct, only one had lost the wild-type EcoRV fragments, while the other clones contained ectopic insertions. The size of the single new hybridizing fragment (6.3 kb) indicated that two copies of the deletion cassette were integrated at the endogenous MRR1c locus of the deletion mutant AR0931MRR1cM1A (Fig. 2C).
For TAC1a, we analyzed 36 clones and identified two independent transformants in which the target gene was deleted. In addition to the predicted new SalI fragments, several extra fragments hybridized with the upstream and downstream flanking sequences in these transformants (Fig. 3A), suggestive of additional integration events. Rehybridization with a probe from the TAC1a coding region confirmed the deletion of TAC1a in strains AR0931TAC1aM1A and -B.
FIG 3.
Southern hybridization analysis of the parental strain B11243 (WT) and the tac1aΔ (A) and tac1bΔ (B) mutants derived from it. See Fig. 2 for explanations. The tac1bΔ mutant AR0931TAC1bM2A (2A) has a different hybridization pattern because the cauSAT1 marker contains an internal EcoRI site that is not present in the caSAT1 marker used to generate AR0931TAC1bM1A (1A) and AR0931TAC1bM1B (1B).
Our initial efforts to delete TAC1b in C. auris strain B11243 were unsuccessful, as none of the first 48 analyzed transformants displayed the desired allelic replacement. Since the majority of our transformants in the previous experiments contained multiple and often ectopically integrated copies of the deletion cassettes (see above), we hypothesized that the caSAT1 marker might not be well expressed in C. auris and the nourseothricin concentration used in our experiments (200 μg/ml) selected for clones with multiple copies. We also observed that B11243 and several other tested C. auris strains were more sensitive to nourseothricin than C. albicans and did not grow on plates containing 10 μg/ml of the antibiotic, on which C. albicans can still grow (19). We therefore lowered the nourseothricin concentration in the selection plates and obtained five mutants with the correct hybridization pattern from plates with 50 μg/ml nourseothricin (out of 24 tested transformants) and two additional correct deletion mutants out of 12 tested clones from plates with 100 μg/ml nourseothricin. Two independent tac1bΔ mutants, AR0931TAC1bM1A and -B (Fig. 3B), were kept for phenotypic analysis. In parallel, we replaced the C. albicans ACT1 promoter in the caSAT1 marker by the upstream region of the C. auris ACT1 gene (see Materials and Methods) to improve expression of the so-generated modified nourseothricin resistance marker (termed cauSAT1 to distinguish it from caSAT1). This did not increase the frequency of correct transformants, but we obtained an additional mutant (out of 24 tested transformants) from a plate with 200 μg/ml nourseothricin. In this mutant (AR0931TAC1bM2A), TAC1b was also replaced by a single copy of the marker (Fig. 3B).
TAC1b contributes to azole resistance in C. auris strain B11243.
To investigate if deletion of any of the MRR1 and TAC1 homologs resulted in increased susceptibility of the mutants to fluconazole, we compared the fluconazole MICs for the parental strain B11243 and the deletion mutants. We confirmed the reported high fluconazole resistance of isolate B11243 (MIC, >256 μg/ml). All mutants lacking MRR1a, MRR1b, MRR1c, or TAC1a displayed the same high resistance, indicating that the encoded transcription factors did not detectably contribute to fluconazole resistance in strain B11243. In contrast, the fluconazole MIC for all three independently generated tac1bΔ mutants was reduced to between 128 μg/ml and 256 μg/ml, i.e., at least 2- to 4-fold (Table 2). Strain B11243 is also highly resistant to voriconazole, with a reported MIC of 8 μg/ml (17). In our assays, the MIC of voriconazole for B11243 was 4 μg/ml and no decrease was observed after deletion of MRR1a, MRR1b, MRR1c, and TAC1a (the MIC was even minimally increased for the tac1aΔ mutants). In contrast, the MIC of voriconazole was reduced to between 0.5 μg/ml and 1 μg/ml, i.e., 4- to 8-fold, for all three tac1bΔ mutants of this strain (Table 2). These results demonstrate that Tac1b contributes to fluconazole and voriconazole resistance in strain B11243, although the mutants retained high resistance levels.
TABLE 2.
MICs of fluconazole and voriconazole for C. auris strains
| Strain | Genotype | MIC (μg/ml) of drug: |
|
|---|---|---|---|
| Fluconazole | Voriconazole | ||
| B11243 (AR#0931) | Wild type | >256 | 4 |
| AR0931MRR1aM1A | mrr1aΔ | >256 | 4 |
| AR0931MRR1aM1B | mrr1aΔ | >256 | 4 |
| AR0931MRR1bM1A | mrr1bΔ | >256 | 4 |
| AR0931MRR1cM1A | mrr1cΔ | >256 | 4 |
| AR0931TAC1aM1A | tac1aΔ | >256 | 4–8 |
| AR0931TAC1aM1B | tac1aΔ | >256 | 4–8 |
| AR0931TAC1bM1A | tac1bΔ | 128–256 | 0.5–1 |
| AR0931TAC1bM1B | tac1bΔ | 128–256 | 0.5–1 |
| AR0931TAC1bM2A | tac1bΔ | 128–256 | 0.5–1 |
| B11221 (AR#0383) | Wild type | 256 | 1–2 |
| AR0383MRR1aM1A | mrr1aΔ | 128 | 0.5–1 |
| AR0383MRR1bM1A | mrr1bΔ | 256 | 1–2 |
| AR0383MRR1cM1A | mrr1cΔ | 256 | 1–2 |
| AR0383TAC1aM1A | tac1aΔ | 256 | 1–2 |
| AR0383TAC1aM1B | tac1aΔ | 256 | 1–2 |
| AR0383TAC1bM1A | tac1bΔ | 128 | 0.5–1 |
Deletion of MRR1 and TAC1 homologs in C. auris strain B11221.
Since the MIC of fluconazole for strain B11243 was above the highest tested concentration (256 μg/ml), a minor contribution of TAC1a and the MRR1 homologs of C. auris to the high fluconazole resistance of this strain might not have been detected in our experiments. We therefore chose an additional fluconazole-resistant C. auris strain (B11221, clade III) with a reported fluconazole MIC of 64 μg/ml (17) to generate a separate set of deletion mutants. B11221 also contains an Erg11 mutation (F126L) that is supposed to contribute to its fluconazole resistance but that cannot fully explain it (17). The Mrr1 and Tac1 homologs of strain B11221 are highly similar to their counterparts in strain B11243, with identities of 98.3% for Mrr1a (PIS54262.1), 97.8 for Mrr1b (PIS53339.1), 99.9% for Mrr1c (PIS50876.1), 99.3 for Tac1a (PIS49946.1), and 97.9% for Tac1b (PIS49945.1). With this strain, we faced the same problem of unspecific integration of the deletion cassettes, which may have been exacerbated by the fact that the flanking sequences were derived from strain B11243 and are not identical to those in strain B11221. Nevertheless, we obtained one mrr1aΔ mutant (AR0383MRR1aM1A) out of 60 tested transformants, two mrr1bΔ mutants out of 36 tested transformants (only one, AR0383MRR1bM1A, was kept, because they were recovered from the same plate), and two independent tac1aΔ mutants (AR0383TAC1aM1A and -B) out of 36 tested transformants after selection with 50 μg/ml nourseothricin. In all these mutants, the target gene was correctly replaced by a single copy of the caSAT1 marker (Fig. 4A and B and Fig. 5A). We did not obtain mrr1cΔ and tac1bΔ mutants with the previously used deletion cassettes (48 clones were tested in each case) and therefore tried a split marker approach in an effort to increase the frequency of homologous recombination (20, 21) (see Materials and Methods). Although this also did not improve the efficiency of gene deletion, we obtained one mutant for each gene (out of 36 tested clones in each case). The mrr1cΔ mutant AR0383MRR1cM1A (Fig. 4C) exhibited the expected hybridization pattern (the EcoRV fragments on which MRR1c is located in strain B11221 are different from those in strain B11243; the 2,718-bp and 4,973-bp fragments hybridizing with the upstream and downstream sequences, respectively, in the wild-type parent were predicted to be replaced by a single 7,053-bp fragment hybridizing with both probes in correct mutants). The EcoRI restriction pattern at the TAC1b locus is also different in B11221 compared to B11243, and the 6,581-bp upstream and the 2,105-bp downstream fragment were predicted to be replaced by a single 8,014-bp fragment after insertion of the caSAT1 marker. In the tac1bΔ mutant AR0383TAC1bM1A, the new fragment that hybridized with both the upstream and downstream probes was slightly smaller than expected (Fig. 5B), indicating that the allelic exchange was unprecise, most likely because of sequence differences in the flanking homology regions. Rehybridization with probes from the coding sequences confirmed the deletion of all five target genes also in the mutants derived from strain B11221 (Fig. 4 and 5).
FIG 4.
Southern hybridization analysis of the parental strain B11221 (WT) and the mrr1aΔ (A), mrr1bΔ (B), and mrr1cΔ (C) mutants derived from it. See Fig. 2 for explanations.
FIG 5.
Southern hybridization analysis of the parental strain B11221 (WT) and the tac1aΔ (A) and tac1bΔ (B) mutants derived from it. See Fig. 2 for explanations.
The fluconazole and voriconazole MICs for strain B11221 were somewhat higher in our assays than previously reported (256 μg/ml instead of 64 μg/ml for fluconazole, and 1 μg/ml to 2 μg/ml instead of 0.5 μg/ml for voriconazole) (17). As for strain B11243, deletion of MRR1b, MRR1c, and TAC1a in strain B11221 did not alter the MICs of the two drugs, but the tac1bΔ mutant, and in this case also the mrr1aΔ mutant, displayed a 2-fold-reduced resistance to fluconazole and voriconazole (Table 2). The reduced voriconazole resistance of the mrr1aΔ and tac1bΔ mutants was also observed in a dilution spot assay on agar plates (Fig. 6). In summary, Tac1b contributes to the azole resistance of C. auris strains from both clade III and clade IV, while a minor contribution of Mrr1a to azole resistance was observed only in the clade III strain B11221.
FIG 6.

Voriconazole sensitivity of strain B11221 and deletion mutants. Serial dilutions of the indicated strains were spotted on SD agar plates without (control) or with voriconazole and grown for 4 days at 37°C.
CDR1 expression in tac1aΔ and tac1bΔ mutants.
In C. albicans, Tac1 is not required for basal CDR1 expression levels, but hyperactive forms of Tac1 cause constitutive CDR1 overexpression (12, 22). We therefore tested if the Tac1 homologs Tac1a and Tac1b regulate CDR1 expression in C. auris. Northern hybridization analysis showed that CDR1 transcript levels were slightly reduced in the tac1bΔ mutants of strain B11243 but not in the tac1bΔ mutant derived from strain B11221 and not in any of the tac1aΔ mutants (Fig. 7A). We considered the possibility that Tac1b might upregulate CDR1 expression in response to fluconazole and thereby promote increased drug resistance. However, CDR1 mRNA levels were not increased in wild-type cells in the presence of fluconazole and were not or only minimally affected by the deletion of TAC1a and TAC1b (Fig. 7B). Therefore, the contribution of Tac1b to azole resistance in strains B11243 and B11221 seems to involve other, CDR1-independent mechanisms.
FIG 7.

CDR1 expression in the parental strains B11243 and B11221 and deletion mutants. Strains were grown to log phase without (A) or with (B) fluconazole (FLC) exposure and analyzed by Northern hybridization with CDR1- and ACT1-specific probes. The identities of the mRNAs are indicated. Signals were quantified and normalized to ACT1 signals for each strain. The signals of the wild-type strains grown in the absence of fluconazole were set to 1, and the normalized CDR1 expression values for each strain are given below the blots. The wild-type samples from panel A were included in panel B to compare CDR1 expression levels in the absence (−) and presence (+) of fluconazole (FLC).
DISCUSSION
The efflux pump encoded by CDR1 has recently been found to play a major role in the fluconazole resistance of C. auris. CDR1 deletion decreased the MIC by 4-fold in a fluconazole-susceptible isolate and by 8-fold and 64-fold in two different fluconazole-resistant isolates, all from clade I (15, 16). Since activating mutations in the transcription factor Tac1 are responsible for CDR1 overexpression in C. albicans and a major cause of azole resistance in this species, we hypothesized that a Tac1 homolog might be constitutively active in C. auris and confer fluconazole resistance by promoting high CDR1 expression levels. However, deletion of TAC1a, one of two CaTAC1 homologs in C. auris, in two different strains did not affect fluconazole resistance, and deletion of TAC1b only mildly reduced their high fluconazole MICs. Furthermore, the Tac1 homologs did not or only minimally contribute to CDR1 expression in the presence of fluconazole under the conditions used in our experiments. Other transcription factors may therefore be more important for CDR1 expression in C. auris. We also considered the possibility that TAC1a and TAC1b, which are located in tandem in the C. auris genome, might have partially redundant roles. Unfortunately, our efforts to construct tac1aΔ tac1bΔ double mutants of strains B11243 and B11221 were unsuccessful. It is possible that CDR1 is less critical for azole resistance in the two strains from clades III and IV investigated in our study than in the clade I strains studied previously, in which it may be more strongly expressed, possibly due to activating mutations in Tac1a or Tac1b. Nevertheless, Tac1b contributed to azole resistance in strains B11243 and B11221, even if this did not involve CDR1 upregulation.
Activating mutations in the transcription factor Mrr1 are another common cause of fluconazole resistance in C. albicans, which is partly mediated by the constitutive overexpression of the efflux pump-encoding gene MDR1. C. auris possesses three Mrr1 homologs, but we did not find clear evidence for their involvement in the fluconazole resistance of the two C. auris isolates investigated in our present study. A possible exception is Mrr1a, as MRR1a deletion in strain B11221 resulted in a slightly increased susceptibility to fluconazole and voriconazole. A caveat here is that only one mrr1aΔ mutant was obtained from this parental strain, and MRR1a deletion did not detectably affect azole resistance in strain B11243. We therefore cannot exclude the possibility that the increased azole susceptibility of the mrr1aΔ mutant derived from strain B11221 was caused by an unspecific mutation during the construction of the strain.
Since C. auris is a haploid species, we anticipated that the generation of specific gene deletion mutants would be straightforward, as also inferred from the successful construction of C. auris mutants by other researchers (15, 16, 23–26). Unexpectedly, the vast majority of our transformants had unspecifically inserted the five different gene deletion cassettes at ectopic sites in the genome instead of integrating them at the target locus, suggesting that homologous recombination is much less efficient in C. auris than in C. albicans. Ectopic integration apparently was not such a significant problem in studies by other researchers who generated C. auris gene deletion mutants. Grahl et al. (25) used longer flanking sequences (ca. 1 kb) and a different nourseothricin resistance marker (NAT1) to delete the CAT1 gene in a C. auris clade I strain. Of 10 tested transformants, five contained the desired gene replacement. This was slightly improved (7/10) when CRISPR-Cas9 was used to introduce a double-strand break at the genomic target locus. Day et al. (24) also used the NAT1 marker, but short flanking homology regions (100 bp), to delete the HOG1 gene in a C. auris strain from clade I. Using this strategy, they found that HOG1 was accurately deleted in 30% of the nourseothricin-resistant transformants, suggesting that it is not necessary to include long fragments of sequence homology to achieve targeted integration of a selection marker. Kim et al. (15) applied the CRISPR-Cas9 technology to replace the HSP90 promoter by a doxycycline-repressible promoter and to delete CDR1 in a clade I strain, using a hygromycin resistance marker or the NAT1 marker, respectively, for the selection of transformants. However, they did not report the frequency of specific integration events. Rybak et al. (16) also used a CRISPR-Cas9 system in combination with a replacement cassette containing the caSAT1 selection marker and short (50-bp) flanking homology regions to delete CDR1 and MDR1 in two clade I isolates but did not report the frequency of specific gene replacement. Integration specificity was also not detailed in two very recent studies in which the NAT1 marker and long flanking regions were used for gene deletions in C. auris (23, 26). The reason for the low frequency of specific marker integration into the target locus in our experiments is not evident. Nourseothricin resistance was used for the selection of transformants in all six previous studies, and Rybak et al. (16) also used the same nourseothricin resistance gene (caSAT1) as we did. The use of electroporation for transformation of C. auris also does not seem to be the problem, because electroporation was also used in the studies by Grahl et al., Kim et al., Rybak et al., and Iyer et al. (15, 16, 25, 26). It is possible that homologous recombination is less efficient in strains from clade III and clade IV, used to generate mutants in our present study, as opposed to clade I strains, which were the parents of the mutants generated in the six previous studies (15, 16, 23–26). Alternatively, the frequency of specific marker integration may vary considerably depending on the target locus.
Another interesting observation in our experiments was that nourseothricin-resistant transformants often contained multiple copies of the deletion cassette (we note that such multiple integration events would not be detected by the diagnostic PCR methods employed by most researchers to confirm gene replacements). While this might be related to the mechanism used by C. auris to integrate exogenously supplied DNA into the genome, it also suggested that multiple copies of the resistance marker were required for growth on the selection plates. We considered the possibility that the caSAT1 marker was not as efficiently expressed in C. auris as in C. albicans and conferred lower levels of resistance. Yet, after replacement of the CaACT1 promoter in the selection marker by the ACT1 promoter from C. auris, many transformants with the TAC1b deletion cassette still contained multiple integrations. We found that wild-type C. auris strains were more susceptible to nourseothricin than wild-type C. albicans, which may have favored growth of transformants containing more than one copy of the resistance marker immediately after plating on selective medium if the marker was not yet efficiently expressed. We therefore lowered the nourseothricin concentration used for selection of transformants from 200 μg/ml to 50 μg/ml in subsequent experiments. Although we still observed multiple ectopic integration events in many transformants, all correct gene deletion mutants selected with the lower nourseothricin concentration (the tac1bΔ mutants of strain B11243 and all deletion mutants of strain B11221) contained only a single copy of the selection marker instead of the tandem integrations that had occurred in the previously obtained mutants (mrr1aΔ, mrr1bΔ, and mrr1cΔ mutants of strain B11243). In any case, and from a practical perspective, our results argue that a lower nourseothricin concentration is sufficient for the selection of C. auris transformants when using this rather expensive reagent, which considerably reduces the costs of experiments.
In conclusion, our study has provided novel insights into the molecular basis of drug resistance of the recently emerged pathogenic yeast C. auris, which is a major concern for human health. Our work also provides useful information about the genetic manipulation of C. auris that should be valuable for the increasing community of researchers studying this fungus.
MATERIALS AND METHODS
Strains and growth conditions.
The C. auris strains used in this study are listed in Table 1. The clinical isolates B11243 and B11221 were obtained from the Centers for Disease Control and Prevention (CDC AR Bank numbers 0931 and 0383, respectively). All strains were stored as frozen stocks with 17.2% glycerol at −80°C and subcultured on YPD agar plates (10 g yeast extract, 20 g peptone, 20 g glucose, 15 g agar per liter) at 30°C. Strains were routinely grown in YPD liquid medium at 30°C in a shaking incubator.
Plasmid constructions.
To generate deletion constructs for MRR1a, MRR1b, MRR1c, TAC1a, and TAC1b, ca. 0.5 kb of the upstream and downstream regions of these genes was PCR amplified from genomic DNA of strain B11243 with the primers listed in Table 3. The upstream fragments were digested at the KpnI and ApaI sites introduced with primers 1 and 2, and the downstream fragments were digested at the SacII and SacI sites introduced with primers 3 and 4, respectively. The upstream and downstream flanking sequences of each target gene were then cloned together with an ApaI-SacII fragment from plasmid pSAT1, which contains the caSAT1 marker (19), in the KpnI/SacI-digested vector pBluescript II KS, resulting in plasmids pCauMRR1aM1, pCauMRR1bM1, pCauMRR1cM1, pCauTAC1aM1, and pCauTAC1bM1. To obtain a C. auris-adapted SAT1 marker (cauSAT1), a fragment containing 1,059 bp of the upstream region of the C. auris ACT1 gene was amplified from genomic DNA of strain B11243 with primers CauACT1.01 and CauACT1.02. A part of the caSAT1 marker, without the CaACT1 sequences, was amplified with primers SAT9 and SAT10. The PCR products were then used as the templates in a fusion PCR with primers CauACT1.01 and SAT10. The PCR product was digested at the introduced ApaI and SacII sites and substituted for the caSAT1 marker in pCauTAC1bM1, yielding pCauTAC1bM2.
TABLE 3.
Oligonucleotide primers
| Primer | Sequence (5′–3′)a |
|---|---|
| ACT1CauNBF | TCGAGACCTTCAACGTTCCT |
| ACT1CauNBR | ACGCACATCGACATCACATT |
| CauACT1.01 | ATATGGGCCCGAGTAGTAATTTGTAACGGG |
| CauACT1.02 | CCGAAATTTTCATATTGACTTAATTGAATTCTTCG |
| CauMRR1a.01 | TATAGGTACCTCGTGAACTTCATCATTGTCACACGG |
| CauMRR1a.02 | TATAGGGCCCATACCATTATCAAAGTTTTTCTGGGGAG |
| CauMRR1a.03 | TATACCGCGGTAAGTTTCATACTACGTGAATATACATGCG |
| CauMRR1a.04 | TATAGAGCTCGGTACCTATTTGATTACTTAGCGATACGATCTCC |
| CauMRR1a_F | GATAACGCTGCACTCGAACA |
| CauMRR1a_R | AGGGGCCAAAATTGAGTCTT |
| CauMRR1b.01 | TATAGGTACCGAACCGGACAATGTATGCGAACCG |
| CauMRR1b.02 | TATAGGGCCCATTCGCTTTTTGGAGCTTCCGG |
| CauMRR1b.03 | TATACCGCGGATAGCACGGAGTTAGTGACAATTATG |
| CauMRR1b.04 | TATAGAGCTCGGTACCTATCAGAAATAATGGGTATACTGTATCG |
| CauMRR1b_F | TTACCCATTTGTCCCGGTTA |
| CauMRR1b_R | CTCCACCATCATACCCATCC |
| CauMRR1c.01 | TATAGGTACCTTCTATTGGCTGATCTTGAACCTTTGTG |
| CauMRR1c.02 | TATAGGGCCCCATTGGGGTGCTGTTGGTGGAAG |
| CauMRR1c.03 | TATACCGCGGTAAGGGATGCTTCGACCTCTG |
| CauMRR1c.04 | TATAGAGCTCGGTACCGTCTGAAAATTCGAGTTCCTCGG |
| CauMRR1c_F | GCTACTTCCGGCTCTTCCTT |
| CauMRR1c_R | AGGCACGACGAGCTCAGTAT |
| CauTAC1a.01 | TATAGGTACCGCTTGATCACGCCACAGCAACTTCAC |
| CauTAC1a.02 | TATAGGGCCCGCATACGGCACTTCGGCTGC |
| CauTAC1a.03 | TATACCGCGGTCAGCAATTCTAAAAGAGATACTACAATAC |
| CauTAC1a.04 | TATAGAGCTCGGTACCATAGCTTCTTGAGATTCGAATGAG |
| CauTAC1a_F | CACCCCACTCGTACACTCCT |
| CauTAC1a_R | AGTTCATGCACGTTGTCAGC |
| CauTAC1b.01 | TATAGGTACCGATACTACTGCCAGGCTTGACAG |
| CauTAC1b.02 | TATAGGGCCCAGCTTCTTGAGATTCGAATGAGC |
| CauTAC1b.03 | TATACCGCGGTTAACTTTGTAAATAGTATGCTTACCACG |
| CauTAC1b.04 | TATAGAGCTCGGTACCTGTCGAAGACTGTAACAAAGCC |
| CauTAC1b_F | GGCCGATTCATCCTCAACTA |
| CauTAC1b_R | CTGTCCACACGCTCAGAAAA |
| CDR1CauNBF | GCCAGAACCTTCACCAACAT |
| CDR1CauNBR | ACAACCAGAACCAGGACGAC |
| SAT9 | CAATTAAGTCAATATGAAAATTTCGGTGATCCC |
| SAT10 | TATACCGCGGGACCACCTTTGATTGTAAATAG |
| SAT1Nrev1 | ATGAGACTGTGCGCGACTCC |
| SAT1Cfor1 | GTTCGATGTGCACCTATCCG |
Introduced restriction sites are underlined.
Strain constructions.
C. auris strains were transformed by electroporation (19) with the gel-purified inserts (KpnI-KpnI fragments) from plasmids pCauMRR1aM1, pCauMRR1bM1, pCauMRR1cM1, pCauTAC1aM1, pCauTAC1bM1, and pCauTAC1bM2. Transformants were initially selected on YPD plates containing 200 μg/ml nourseothricin, but lower concentrations (50 μg/ml and 100 μg/ml) were used in later experiments. For the split-marker approach, the deletion cassettes contained in pCauMRR1cM1 and pCauTAC1bM1 were amplified as two overlapping fragments using primers 1 and SAT1Nrev1 for the 5′ part and primers 4 and SAT1Cfor1 for the 3′ part. The two fragments of each deletion cassette were then used for electroporation. In this case, a functional nourseothricin resistance marker can be regenerated only when the two fragments are joined by recombination between the overlapping sequences in the transformed cells, which might also increase the frequency of integration at the target locus by homologous recombination with the flanking sequences (21). Genomic integration of the deletion cassettes and absence of the target genes were tested by Southern hybridization using the upstream (amplified with primers 1 and 2) and downstream (amplified with primers 3 and 4) flanking sequences as well as parts of the coding sequences (amplified with primers F and R) as probes.
Isolation of genomic DNA and Southern hybridization.
Genomic DNA from C. auris strains was isolated as described previously for C. albicans (19). The DNA was digested with appropriate restriction enzymes, separated on a 1% agarose gel, transferred by vacuum blotting onto a nylon membrane, and fixed by UV cross-linking. Southern hybridization with enhanced chemiluminescence-labeled probes was performed with the Amersham ECL direct nucleic acid labeling and detection system (GE Healthcare UK Limited, Little Chalfont, Buckinghamshire, United Kingdom) according to the instructions of the manufacturer. A molecular size marker was included in the probes to facilitate size determination of the hybridizing genomic DNA fragments. ECL signals were captured by exposing the membranes to Hyperfilm (GE Healthcare) and digitized with an HP ScanJet 8300 (HP Inc., Palo Alto, CA).
Northern hybridization analysis.
Overnight cultures of the strains were diluted to an optical density at 600 nm (OD600) of 0.4 in fresh YPD medium and grown for 4 h at 30°C. In a separate experiment, to compare CDR1 expression levels in the various strains in the presence of fluconazole, 50 μg/ml of fluconazole was added to the cultures after 3 h, followed by further incubation for 1 h. Total RNA was extracted using a Quick-RNA fungal/bacterial miniprep kit (Zymo Research, Irvine, CA) according to the manufacturer’s instructions. RNA samples were separated on a 1.2% agarose gel, transferred by capillary blotting onto a nylon membrane, and fixed by UV cross-linking. The blots were simultaneously hybridized with digoxigenin-labeled probes for CauCDR1 (positions +157 to +615 in the CDR1 coding sequence, amplified with primers CDR1CauNBF and CDR1CauNBR) and CauACT1 (positions +374 to +873 in the ACT1 coding sequence, amplified with primers ACT1CauNBF and ACT1CauNBR). Bound probe was detected with a peroxidase-labeled antidigoxigenin alkaline phosphatase (AP)-conjugate (Roche, Basel, Switzerland). Signals were generated using CSPD (Roche, Basel, Switzerland) as the substrate and captured with the ImageQuant LAS 4000 imaging system (GE Healthcare). Signal intensities were quantified using the image analysis software Fiji (27).
Azole susceptibility tests.
The azole susceptibilities of the strains were determined by a previously described broth microdilution method (28), with slight modifications. Three to five 2-day-old colonies from a YPD agar plate were suspended in 2 ml of an 0.9% NaCl solution, and 4 μl of the suspension was mixed with 2 ml 2× SD-CSM medium (13.4 g yeast nitrogen base with ammonium sulfate [YNB; MP Biomedicals, Illkirch, France], 40 g glucose, 1.58 g complete supplement medium [CSM; MP Biomedicals]). Stock solutions of fluconazole and voriconazole (Sigma GmbH, Deisenhofen, Germany) were made in water and dimethyl sulfoxide (DMSO), respectively, and 2-fold dilution series were prepared in water, starting from initial concentrations of 512 μg/ml (fluconazole) and 64 μg/ml (voriconazole). One hundred microliters of each drug solution was then mixed with 100 μl of the cell suspension in a 96-well microtiter plate, and the plates were incubated for 48 h at 37°C. The MIC was defined as the drug concentration that abolished or drastically reduced visible growth compared to a drug-free control. For dilution spot assays, YPD overnight cultures of the strains were diluted to an OD600 of 2.0. Tenfold dilutions from 100 to 10−5 were prepared in a 96-well microtiter plate, and ca. 5 μl of the cell suspensions was transferred with a replicator onto SD agar plates without or with 2.5 μg/ml voriconazole. Plates were incubated for 4 days at 37°C and photographed.
ACKNOWLEDGMENTS
This study was funded by the Deutsche Forschungsgemeinschaft (DFG grant MO 846/7). Publication of the work was supported by the Open Access Publication Fund of the University of Würzburg.
REFERENCES
- 1.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 2.Rhodes J, Fisher MC. 2019. Global epidemiology of emerging Candida auris. Curr Opin Microbiol 52:84–89. doi: 10.1016/j.mib.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chowdhary A, Sharma C, Meis JF. 2017. Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog 13:e1006290. doi: 10.1371/journal.ppat.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chowdhary A, Prakash A, Sharma C, Kordalewska M, Kumar A, Sarma S, Tarai B, Singh A, Upadhyaya G, Upadhyay S, Yadav P, Singh PK, Khillan V, Sachdeva N, Perlin DS, Meis JF. 2018. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother 73:891–899. doi: 10.1093/jac/dkx480. [DOI] [PubMed] [Google Scholar]
- 6.Healey KR, Kordalewska M, Jimenez Ortigosa C, Singh A, Berrio I, Chowdhary A, Perlin DS. 2018. Limited ERG11 mutations identified in isolates of Candida auris directly contribute to reduced azole susceptibility. Antimicrob Agents Chemother 62:e01427-18. doi: 10.1128/AAC.01427-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon YJ, Shin JH, Byun SA, Choi MJ, Won EJ, Lee D, Lee SY, Chun S, Lee JH, Choi HJ, Kee SJ, Kim SH, Shin MG. 2019. Candida auris clinical isolates from South Korea: identification, antifungal susceptibility, and genotyping. J Clin Microbiol 57:e01624-18. doi: 10.1128/JCM.01624-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-Ami R, Berman J, Novikov A, Bash E, Shachor-Meyouhas Y, Zakin S, Maor Y, Tarabia J, Schechner V, Adler A, Finn T. 2017. Multidrug-resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg Infect Dis 23:195–203. doi: 10.3201/eid2302.161486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morschhäuser J. 2016. The development of fluconazole resistance in Candida albicans—an example of microevolution of a fungal pathogen. J Microbiol 54:192–201. doi: 10.1007/s12275-016-5628-4. [DOI] [PubMed] [Google Scholar]
- 10.Coste A, Selmecki A, Forche A, Diogo D, Bougnoux M-E, d’Enfert C, Berman J, Sanglard D. 2007. Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot Cell 6:1889–1904. doi: 10.1128/EC.00151-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coste A, Turner V, Ischer F, Morschhäuser J, Forche A, Selmecki A, Berman J, Bille J, Sanglard D. 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172:2139–2156. doi: 10.1534/genetics.105.054767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coste AT, Karababa M, Ischer F, Bille J, Sanglard D. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot Cell 3:1639–1652. doi: 10.1128/EC.3.6.1639-1652.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunkel N, Blass J, Rogers PD, Morschhäuser J. 2008. Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol Microbiol 69:827–840. doi: 10.1111/j.1365-2958.2008.06309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morschhäuser J, Barker KS, Liu TT, Blaß-Warmuth J, Homayouni R, Rogers PD. 2007. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog 3:e164. doi: 10.1371/journal.ppat.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SH, Iyer KR, Pardeshi L, Munoz JF, Robbins N, Cuomo CA, Wong KH, Cowen LE. 2019. Genetic analysis of Candida auris implicates Hsp90 in morphogenesis and azole tolerance and Cdr1 in azole resistance. mBio 10:e00346-19. doi: 10.1128/mBio.00346-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rybak JM, Doorley LA, Nishimoto AT, Barker KS, Palmer GE, Rogers PD. 2019. Abrogation of triazole resistance upon deletion of CDR1 in a clinical isolate of Candida auris. Antimicrob Agents Chemother 63:e00057-19. doi: 10.1128/AAC.00057-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munoz JF, Gade L, Chow NA, Loparev VN, Juieng P, Berkow EL, Farrer RA, Litvintseva AP, Cuomo CA. 2018. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun 9:5346. doi: 10.1038/s41467-018-07779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacPherson S, Larochelle M, Turcotte B. 2006. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev 70:583–604. doi: 10.1128/MMBR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reuss O, Vik A, Kolter R, Morschhäuser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 20.Fairhead C, Llorente B, Denis F, Soler M, Dujon B. 1996. New vectors for combinatorial deletions in yeast chromosomes and for gap-repair cloning using ‘split-marker’ recombination. Yeast 12:1439–1457. doi:. [DOI] [PubMed] [Google Scholar]
- 21.Fu J, Hettler E, Wickes BL. 2006. Split marker transformation increases homologous integration frequency in Cryptococcus neoformans. Fungal Genet Biol 43:200–212. doi: 10.1016/j.fgb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 22.Schillig R, Morschhäuser J. 2013. Analysis of a fungus-specific transcription factor family, the Candida albicans zinc cluster proteins, by artificial activation. Mol Microbiol 89:1003–1017. doi: 10.1111/mmi.12327. [DOI] [PubMed] [Google Scholar]
- 23.Bravo Ruiz G, Ross ZK, Gow NAR, Lorenz A. 2020. Pseudohyphal growth of the emerging pathogen Candida auris is triggered by genotoxic stress through the S phase checkpoint. mSphere 5:e00151-20. doi: 10.1128/mSphere.00151-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Day AM, McNiff MM, da Silva Dantas A, Gow NAR, Quinn J. 2018. Hog1 regulates stress tolerance and virulence in the emerging fungal pathogen Candida auris. mSphere 3:e00506-18. doi: 10.1128/mSphere.00506-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grahl N, Demers EG, Crocker AW, Hogan DA. 2017. Use of RNA-protein complexes for genome editing in non-albicans Candida species. mSphere 2:e00218-17. doi: 10.1128/mSphere.00218-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iyer KR, Whitesell L, Porco JA Jr, Henkel T, Brown LE, Robbins N, Cowen LE. 2020. Translation inhibition by rocaglates activates a species-specific cell death program in the emerging fungal pathogen Candida auris. mBio 11:e03329-19. doi: 10.1128/mBio.03329-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruhnke M, Eigler A, Tennagen I, Geiseler B, Engelmann E, Trautmann M. 1994. Emergence of fluconazole-resistant strains of Candida albicans in patients with recurrent oropharyngeal candidosis and human immunodeficiency virus infection. J Clin Microbiol 32:2092–2098. doi: 10.1128/JCM.32.9.2092-2098.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]