Abstract
Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous pollutants that are metabolized to carcinogenic dihydrodiol epoxides (PAHDE) by cytochrome P450 1B1 (CYP1B1). This metabolism occurs in bone marrow (BM) mesenchymal stem cells (MSC), which sustain hematopoietic stem and progenitor cells (HSPC). In BM, CYP1B1-mediated metabolism of 7, 12-dimethylbenz[a]anthracene (DMBA) suppresses HSPC colony formation within 6 h, whereas benzo(a)pyrene (BP) generates protective cytokines. MSC, enriched from adherent BM cells, yielded the bone marrow stromal, BMS2, cell line. These cells express elevated basal CYP1B1 that scarcely responds to Ah receptor (AhR) inducers. BMS2 cells exhibit extensive transcriptome overlap with leptin receptor positive mesenchymal stem cells (Lepr+ MSC) that control the hematopoietic niche. The overlap includes CYP1B1 and the expression of HSPC regulatory factors (Ebf3, Cxcl12, Kitl, Csf1 and Gas6). MSC are large, adherent fibroblasts that sequester small HSPC and macrophage in the BM niche (Graphic abstract). High basal CYP1B1 expression in BMS2 cells derives from interactions between the Ah-receptor enhancer and proximal promoter SP1 complexes, boosted by autocrine signaling. PAH effects on BMS2 cells model Lepr+MSC niche activity. CYP1B1 metabolizes DMBA to PAHDE, producing p53-mediated mRNA increases, long after the in vivo HSPC suppression. Faster, direct p53 effects, favored by stem cells, remain possible PAHDE targets. However, HSPC regulatory factors remained unresponsive. BP is less toxic in BMS2 cells, but, in BM, CYP1A1 metabolism stimulates macrophage cytokines (Il1b > Tnfa> Ifng) within 6 h. Although absent from BMS2 and Lepr+MSC, their receptors are highly expressed. The impact of this cytokine signaling in MSC remains to be determined.
Keywords: CYP1B1, BMS2 cells, Lepr+MSC, Bone marrow vascular niche, Hematopoietic stem and progenitor cells, Mesenchymal stem cells
Graphic abstract
HSPC, in vivo, are controlled in the BM vascular niche by Lepr+MSC. The mouse BMS2 line models Lepr+MSC regulation, with parallel CYP1B1, HSPC support factors (Cxcl12, Kitl, Ebf3) and cytokine receptor expression. PAHs can produce opposing effects. Rapid suppression of HSPC, effected by CYP1B1-mediated PAH metabolism to PAHDE in MSC (blue), is marked in BMS2 cells by p53-mediated gene activations (DMBA> > BP). Protection arises from in vivo BP metabolism by CYP1A1 that stimulates cytokines from local macrophage (Il1b, Tnf, Ifng), each with corresponding Lepr+MSC receptors. PAH quinones are the probable CYP1A1-mediated protectants.
Highlights
-
•
BMS2 and Lepr+MSC cells co-express CYP1B1 and 12 functional niche activity markers.
-
•
CYP1B1 mRNA in BMS2 cells depends on activation of SP1 coupled to an AhR enhancer unit.
-
•
DMBA metabolism by CYP1B1 activates p53 gene targets in BMS2 cells far more than BP.
-
•
HSPC suppression by CYP1B1 generation of PAHDE requires rapid, non-genomic targets.
-
•
BMS2 and Lepr+MSC share receptors activated by BP stimulation of macrophage cytokines.
1. Introduction
Polycyclic aromatic hydrocarbons (PAHs) are major health risk factors through the association of smoking with lung cancer and their contributions to multiple adverse effects of vehicle air particulates (Bostrom et al., 2002; Castano-Vinyals et al., 2004; Layshock et al., 2010; Moorthy et al., 2015; Yang et al., 2019). In present times, we can also add the impact of smoking and environmental combustion pollutants on health outcomes from COVID-19 infection (Li Volti et al., 2020). The metabolism of these chemicals causes tissue injury and carcinogenic mutations (Bolton and Dunlap, 2017; Bostrom et al., 2002; Castano-Vinyals et al., 2004; Moorthy et al., 2015; Yang et al., 2019), and also impacts the immune system (O'Driscoll et al., 2018), notably from effects in the bone marrow (BM) (Larsen et al., 2016; N'Jai A et al., 2011; N'Jai A et al., 2010).
In previous work, we have shown that hematopoietic stem and progenitor cells (HSPC) in mouse BM respond with remarkable speed and selectivity to PAHs (Larsen et al., 2016). These effects have wide systemic consequences, notably in the spleen and thymus (Larsen et al., 2016). This disruption, which is mediated by cytochrome P450 1B1 (CYP1B1), is completed within a few hours. However, PAHs can also generate a rapid protection process. The PAH selectivity of these opposing processes is dependent on specific metabolites. Here, we develop an in vitro model to better understand the molecular processes that contribute to these novel responses.
These PAH responses overlap with physiological roles of CYP1B1 in the BM (Iqbal et al., 2013). Central to these activities are mesenchymal stem cells (MSC), which express CYP1B1 (Lin et al., 2016). MSC provide specific support factors for HSPC, while additionally undergoing self-renewal and differentiation. The alternative mesodermal lineages include osteoblasts (Ichii et al., 2012; Seike et al., 2018), adipocytes and muscle cells (Dorheim et al., 1993).
These MSC functions are modeled by the embryonic OP9 and C3H10T1/2 and the bone marrow stromal, BMS2, cell lines, which derive from, respectively, AGM location of E11.5 embryos, epidermis of E14.5 embryos and BM of 5 week old adult mice (Hanlon et al., 2005b; Kincade et al., 1989; Muller et al., 1994). Each of these lines expresses CYP1B1 (Alexander et al., 1997; Heidel et al., 1998; Rondelli et al., 2016). We have recently established roles for CYP1B1 in neonatal liver development, which depends on partnership with retinol and Srebp transcription factors (Maguire et al., 2020). Stellate cells, which have mesenchymal origins, are early participants (Maguire et al., 2017; Maguire et al., 2020). We examine, here, the capacity of BMS2 cells to model BM MSC, with respect to the effects of PAH on HSPC lineages (Bennett et al., 2018; Kincade et al., 1989; Near et al., 1999; Phinney and Prockop, 2007; Pietrangeli et al., 1988; Ryan et al., 2007; Villa et al., 2017).
CYP1B1 has diverse functions across many cell types, commonly involving local endocrine and immune effects. Protection from oxidative stress is a typical feature that is shown by the effects of CYP1B1 deletion (Gao et al., 2008; Leung et al., 2009; Palenski et al., 2013b). CYP1B1 utilizes multiple physiological substrates, including retinol, estradiol and polyunsaturated fatty acids (Chen et al., 2004; Johansen et al., 2016; Lefevre et al., 2015; Li et al., 2017; Pingili et al., 2016), although typically with only modest activities. CYP1B1 also effectively converts PAHs to dihydrodiol epoxides (PAHDE) (Heidel et al., 2000). These reactive electrophiles produce DNA mutations through adduct formation and double strand breaks (DSB) (Siddens et al., 2015). This work identifies CYP1B1 as the dominant cytochrome P450 in BMS2 cells.
HSPC differentiate into lymphoid, myeloid, and erythroid lineages (Lai and Kondo, 2006) that migrate to sites of injury where they generate inflammatory and repair responses (Li and Ikehara, 2013). MSC provide essential support for HSPC differentiation by releasing specific support cytokines, including Cxcl12, Csf and Ilf7 (Crane et al., 2017). Subsets of MSC, notably leptin receptor positive (Lepr+MSC) cells, undergo self-renewal, directed by Cxcl12 and Kitl/Scf (Galan-Diez and Kousteni, 2018). BMS2 cells lack Lepr expression and the capacity for self-renewal, but effectively support lymphoid progenitors (Rondelli et al., 2016). Here, we show that many of the most abundant genes in Lepr+MSC are also highly expressed in BMS2 cells, including CYP1B1. This led us to hypothesize that PAH metabolism in Lepr+MSC causes the rapid and extensive suppression of HSPC expansion in BM. In this respect, BMS2 cells should provide an informative model for PAH effects in the vascular hematopoietic niche.
BMS2 cells are used in these studies to address the remarkable opposing effects of PAH on BM lymphoid and myeloid progenitor cells (N'Jai A et al., 2011). The adverse effects are realized predominantly through local CYP1B1-mediated bioactivation to PAHDE, rather than by poorly expressed cytochrome P450 1A1 (CYP1A1) (Heidel et al., 1998). 7, 12-dimethylbenz[a]anthracene (DMBA) extensively suppresses specific colony forming activities (CFU) within 6 h, through a process that is CYP1B1-dependent (N'Jai A et al., 2010). The number of colonies quantifies the proportion of initial active progenitors (lymphoid, myeloid or erythroid), while the colony size indicates the rate of expansion. Flow cytometry analyses show matching DMBA effects on HSPC (Larsen et al., 2016). A key feature of the PAH suppression response is a complete insensitivity to induction by PAHs. This matches the novel regulation of CYP1B1 in BMS2 cells, which is likely to extend to Lepr+MSC. Presumably, CYP1B1 has an important, but unrecognized physiological function in these cells. Here, we have extended our previous analyses of CYP1B1 expression to provide insight into this unusual regulation.
Benzo(a)pyrene (BP) is less active in BM metabolism-mediated effects (N'Jai A et al., 2011). However, the protection effects are not produced by MSC, including BMS2 cells (Rondelli et al., 2016). Instead, this protection depends on both Ah receptor (AhR) induction and cytochrome P450 1A1 (CYP1A1) (Galvan et al., 2003), and parallels the rapid appearance of multiple cytokines in BM cells (Larsen et al., 2016; N'Jai A et al., 2011; Rondelli et al., 2016).
Metabolism by CYP1A1 and CYP1B1 is critical to understanding PAH effects mediated by AhR compared to those effected by non-metabolizable, 2,3,7,8-tetrachlorodibenzodioxin (TCDD). CYP1A1 is highly expressed after PAH-mediated activation of AhR in hepatocytes, but not in the BM (Galvan et al., 2003). CYP1B1 is essentially absent from mouse hepatocytes (Bushkofsky et al., 2016; Larsen et al., 2015). AhR-responsive genes demonstrate direct PAH-mediated AhR activation that is complete within 8 h (Hanlon et al., 2005b). Several of the responding genes contain AhR response elements (DRE), while an equal number are novel and cell-type selective. CYP1A1 metabolism in hepatocytes and in the gastro-intestinal tract is the major contributor to PAH clearance (Uno et al., 2004; Uno et al., 2006), but may also contribute circulating BP quinones to effect cytokine stimulation (N'Jai A et al., 2011).
The lack of AhR involvement in the CYP1B1 mediation of PAH toxicity is particularly remarkable because BM CYP1B1 is substantially induced by PAHs and TCDD (Galvan et al., 2003). This finding leads us to hypothesize that the PAH toxicity arises from a sub-population of MSC that do not show this induction but are effective because of their proximity to the HSPC. This hypothesis is fully compatible with recent characterizations of MSC in the BM niche (Galan-Diez and Kousteni, 2018). BMS2 cells, and their primary counterparts, reproduce the substantial CYP1B1 basal expression, without stimulation by AhR activators (Heidel et al., 1998). Importantly, AhR deletion shows that much of the basal expression is lost in AhR-null MEFs (Alexander et al., 1997) and BM MSC (Heidel et al., 1998). This pattern of regulation is also observed in OP9 MSC, while basal expression in MEFs is substantially enhanced by AhR activation (Rondelli et al., 2016).
This basal CYP1B1 expression in BMS2 cells is regulated via dual SP1 complexes in the proximal promoter (Wo et al., 1997), in partnership with complexes in the Ah enhancer region (AhER) and other upstream enhancers (Zhang et al., 1998; Zhang et al., 2003). Constitutive CYP1B1 is also activated by AhR in absence of an exogenous ligand, via disruption of cell adhesion (Cho et al., 2004; Ziegler et al., 2016) and by endogenous ligands formed from tryptophan (Seok et al., 2018; Villa et al., 2017). This work shows that CYP1B1 expression in MSC is highly susceptible to autocrine regulation.
CYP1B1 metabolizes DMBA and BP to PAHDE, which generate DNA adducts and DSB that activate ATM kinase to phosphorylate p53 (Ganesan et al., 2013; Gao et al., 2008). p53 activation plays an essential role in both HSPC suppression (Page et al., 2003; Teague et al., 2010) and stabilization of the MSC-HSPC niche (Phinney and Prockop, 2007). We show, here, that the BMS2 transcriptome exhibits expression asymmetry in the pairing of receptor and their respective activators. We identify several strongly expressed pairs that could potentially affect autocrine regulation, notably Pdgfa and Pdgfrb. By contrast, MSC macrophage cytokines (Ifng, Il1b and Tnf) are absent, but their respective receptors are strongly expressed. This BMS2 model for MSC fits an emerging picture of the hematopoietic niche, driven by signaling from macrophage (Chow et al., 2011), with bidirectional effects on HSPC (Schajnovitz et al., 2011).
To better understand the in vivo MSC responses to DMBA and BP, we examined their effects on gene expression in BMS2 cells. We resolve direct AhR effects produced within 8 h and metabolite-driven responses that only appear after 8 h. This delay accommodates the multistep generation of PAHDE. The participation of the ATM/p53 is evident from the selectivity of gene responses, which are heavily weighted to DMBA. There is only a modest preference seen for general p53 activation.
Single cell sequencing of eluted BM cells shows multiple clusters, based on statistical analyses of mRNA abundance (Lai and Kondo, 2006). Hematopoietic progenitors are retained in the adherent fraction through surface attachment to the far larger, adherent fibroblastic MSC, which contribute only about 5% of the mRNA (Hu et al., 2018). Here, we compare the pattern of highly expressed mRNA in BMS2 cells with the corresponding mRNA in the adherent BM fraction. A set of functional MSC factors are identified in adherent BM cells in the range of (4 ±3%). We also identify factors that are expressed in BMS2 cells but have much lower expression in the MSC of adherent BM cells. We identify a twelve-gene functional core that shares expression in BMS2 cells, the adherent BM fraction and Lepr+MSC. Notably, their expression is highly correlated with CYP1B1, but is not affected by DMBA or BP.
2. Methods
2.1. Cell culture
Primary BM cells were isolated and cultured, as previously described (Larsen et al., 2016). BMS2 cells were a gift from Dr. Paul Kincade (Oklahoma Medical Research Foundation, Oklahoma City, OK) (Kincade et al., 1989; Pietrangeli et al., 1988), while C3H10T1/2 cells were purchased from ATCC (Manassas, VA). Chinese hamster V79 cell lines expressing recombinant human CYP1A1 or CYP1B1 (V79-hCYP1A1 and V79-hCYP1B1, respectively) were provided by Dr. J. Doehmer (Luch et al., 1998; Luch et al., 1999). All cells were cultured under standard conditions (37 °C, 5% CO2 in saturated atmospheric humidity) in FBS-supplemented media (BMS2, RPMI 1640; C3H10T1/2, DMEM; V79, DMEM high glucose supplemented with pyruvate and G418) (Fisher Scientific, Waltham, MA). Physically separated co-cultures, which shared media, were used for the analyses of secreted factor exchange. The cell lines lose CYP1B1 expression when near confluence and, therefore, all cultures were completed at an initial 70% of confluence, unless otherwise stated. Adipogenic differentiation in primary BM cells and the BMS2 cell line was completed as previously described (Jefcoate et al., 2008). Colony forming unit (CFU) assays were completed using kits purchased from Stem Cell Technologies, according to manufacturers' protocol and as previously described (Larsen et al., 2016; Rondelli et al., 2016).
2.2. Microarrays
Microarray analyses were completed in triplicate cultures of BMS2 cells treated with either DMBA, BP, or TCDD. Control cells were treated with vehicle (DMSO). RNA was isolated using Qiagen's RNeasy mini-kit (Hilden, Germany) and quantified using a Nanodrop spectrophotometer (Thermo Fisher Scientific; Waltham, MA). RNA integrity was assessed using denaturing formaldehyde-agarose gel electrophoresis. Cy3/5 labeling was completed using Agilent Technologies' Dual Color Gene Expression kit. Analyses were completed on the Whole Mouse Genome Microarray 4 × 44 slides, using the DNA Microarray Scanner and Feature Extraction Software (Santa Clara, CA). Cy5 values of greater than 50 were considered significantly above background for analysis. Expression is presented as fold change (Cy3/Cy5) from untreated cultures. Analysis was completed using the EDGE3 software package (Vollrath et al., 2009). All p < .01 were considered statistically significant.
2.3. qPCR
Microarray analyses were confirmed via qPCR expression. Total RNA was isolated as described above. Reverse transcription was completed using random oligos and GoTaq DNA polymerase (Promega, Madison, WI), with specific expression determined using Apex qPCR master mix (Genesee Scientific), as per manufacturer's instructions. Signal was detected and integrated using the BioRad CFX Real Time PCR Detection System (Hercules, CA). Primers were obtained from IDT (Coralville, IA): CYP1B1 (F: CCACTATTACGGACATCTTCGG, R: CACAACCTGGTCCAACTCAG); CYP1A1 (F: AGAGCACTACAGGACATTTGAG, R: CCAAAGAGGTCCAAAACAATCG); Aldh3a1 (F: GGCGTGGTCCTTGTCATAG, R: AGGGATAAGTGTTGAAAGCAG); Tiparp (F: CTTTTCCGTTCCTGTTTCATACTG, R: CGTTTCAGGGTAAAAGTTGGC); Ptgs2 (F: CTCACGAAGGAACTCAGCAC, R: GGATTGGAACAGCAAGGATTTG), Cxcl10 (F: TCAGCACCATGAACCCAAG, R: CTATGGCCCTCATTCTCACTG), Cdkn1a (F: CAGATCCACAGCGATATCCAG, R: AGAGACAACGGCACACTTTG), Ccng1 (F: CAGTTCTTTGGCTTTGACACG, R: TTCCTCTTCAGTCGCTTTCAC), Cxcl12 (F: TCCTCAACACTCCAAACTGTG, R: GACTCACACCTCTCACATCTTG), Csf1 (F: ACCCAGGATGAGGACAGAC, R: AGGAAGATGGTAGGAGAGGG). Graphical analysis was completed using GraphPad Prism (La Jolla, CA) software. Ct values were normalized to GAPDH (F: TCAACAGCAACTCCCACTCTTCCA, R: ACCACCCTGTTGCTGTAGCCGTATT) expression.
2.4. In cell western
Cells were cultured in 96 well, white-walled plates and treated with the appropriate PAH for 24 h. Cells were fixed in 4% paraformaldehyde and permeabilized with 0.1%Triton X-100. Endogenous peroxides were quenched with 1% H2O2. Primary antibodies against phospho-p53 (Ser15) and phospho-H2AX (Ser139) were obtained from Cell Signaling (Beverly, MA) and used at a 1:500 dilution. Anti-mouse HRP-conjugated secondary antibody was purchased from Promega Corporation. Chemiluminescence was generated using a 1:1 reagent mixture of SuperSignal™ ELISA Femto Substrate (ThermoFisher Scientific) and normalized to DNA content using Hoecht florescence (Sigma-Aldrich, St. Louis, MO). Signal was quantified on a BioTek Synergy 2 plate reader (BioTek, Winooski, VT).
2.5. Western blots
SDS-PAGE (7.5%) immunoblot analyses were completed on total BMS2 and C3H10T1/2 cell lysates, as previously described (Cimafranca et al., 2004). Cells were treated with DMSO, TCDD or 3′-methoxy-4′-nitroflavone (MNF), as indicated, for 24 h prior to isolation and lysate preparation. The CYP1B1 antibody was previously prepared in this laboratory (Savas et al., 1997). A non-specific background band serves as normalization for protein loading.
2.6. Promoter reporters
Reporter constructs were previously prepared, as described (Zhang et al., 2003). These promoter constructs were transfected into BMS2 and C3H10T1/2 cells, at indicated cell densities, using electroporation at 200 V. Promoter activity was measured with the Luciferase Reporter Assay System (Promega Corporation), as per manufacturer's instructions, using a Pharmingen Moonlight 3010 luminometer (BD Biosciences, San Jose, CA). MNF was used as an AhR antagonist, suppressing the AhER promoter activity in response to TCDD (10 nM, 24 h) or DMSO solvent control. Data was expressed as a fold-induction of luminescence relative to untreated cells.
2.7. Co-culture
BMS2 and C3H10T1/2 cells were co-cultured to 80% of confluence in separate nested dishes, which provided by a barrier that allowed effective media exchange. To provide controls, each cell line was individually cultured in both compartments (two controls). The co-culture and control cultures were maintained for 24 h prior to recovery and electroporation at 200 V, as described for the standard promoter analyses. These electroporated cultures were separately stimulated with TCDD and DMSO solvent control for 24 h. In an alternative procedure, either BMS2 cells or C3H10T1/2 cells were similarly transfected with the AhER reporter and co-cultured for 24 h with an equal number of non-transfected cells, either of the same type or opposite type.
2.8. Statistical analyses
Data were graphed using GraphPad Prism software (version 8) and are represented as mean ± SEM, n = 3–6 observations/condition. CFU data from PAH-treated mice are expressed as the percent of the value of vehicle control (olive oil)-treated mice, with the control is set at 100%. Anova statistical analyses, followed by a Tukey post-test was completed unless otherwise stated. Student t-tests were completed using the unpaired, two-tailed constraints. Agilent microarray data was analyzed by the EDGE3 software using the Limma analysis, which assesses significance based on ANOVA statistics (Vollrath et al., 2009)
3. Results
3.1. Partnership of MSC with HSPC in BM. design to model MSC with BMS2 cells
In vivo treatment with DMBA has established that metabolism to reactive PAHDE by CYP1B1 results in 80 and 30% suppression of, respectively, lymphoid (preB) and myeloid (GM) CFU within 6 h in WT mice (Fig. 1A). BP is appreciably less active and all activities are lost in CYP1B1-ko mice (Fig. 1A) (N'Jai A et al., 2011). Intraperitoneal (IP) (WT, Fig. 1A) and oral administration (all WT, Fig. 1B) of DMBA yielded similar 6 h results, but analysis of serum PAH shows that elimination is complete within 24 h after oral administration (N'Jai A et al., 2010), such that substantial restoration of preB and GM CFU occurs within 48 h (Fig. 1B), with near complete reversal after 168 h (Larsen et al., 2016). This recovery is noteworthy because we find that DMBA-induced changes in transcription, mediated by CYP1B1 metabolism, are too slow to account for the acute CFU suppression, but closely parallels the recovery process (Table 1A ). These changes in CFU are paralleled in the T lymphocytes in the thymus, which also derive from BM progenitors, but with a delay of about 40 h (Larsen et al., 2016). Flow cytometry analyses show parallel effects in HSPC progenitor populations (Larsen et al., 2016), which follow the CFU changes, preceding the mature BM populations.
Fig. 1.
DMBA and BP effect changes in adherent BM. BMS2 cells effectively model MSC processes.
A. IP DMBA (left, 1 uM) and BP (right, 1 uM) treatment suppresses CFU activity in C57BL/6 J (WT) lymphoid (pre—B) and myeloid (GM) BM cells within 6 h. Suppression is greater with DMBA treatment. CYP1B1 deletion (KO) attenuates these suppressive effects. Statistical significance: *p < .01, **p < .01.
B. Reversal of lymphoid and myeloid CFU suppression is observable within 48 h post oral DMBA (1 uM) treatment. Statistical significance: ***p < .001.
C. Time course of BP stimulated gene expression. BP (1 uM) maximally induces BM mediators of oxidative stress and inflammatory cytokine expression within 6 h, paralleling CFU activity suppression.
D. The BMS2 cell model effectively models MSC, with respect to adipogenic differentiation. Differentiation (lipid droplet accumulation) was assessed in cultured BMS2 cells 8 days post adipogenic stimulation. The primary BM MSC from C57BL/6 J (BM/6 J) mice were cultured for 7 days prior to stimulation. Differentiation was examined in these primary cultures 8 days post stimulation. 10× magnification.
E. Co-culture of BM MSC with BMS2 cells sustains lymphoid (pre—B) CFU activity for 24 h post BM isolation. Cells were placed in culture with media alone (Media) or in co-culture with BMS2 (BMS2) cells for 24 h prior to methocult culture initiation. Direct cultures (positive control) were placed in methocult media immediately upon isolation from the BM. *Statistical significance, p < .05.
F. BMS2 cells express substantial basal CYP1B1 mRNA (10 μg mRNA/lane, left), relative to C3H10T1/2 embryo fibroblasts, which is modestly induced by 24 h AhR activation (0.1% DMSO control, C, 10 uM DMBA, D; 10 nM TCDD, T). Basal BMS2 microsomal CYP1B1 protein expression parallels primary C57BL/6 J (BM/6 J) BM levels (right), which is sustained with in vivo AhR deletion (BM/6 J AhR−/−). BM cells lack CYP1A1 expression. Cell cultures were treated with 0.1% DMSO (C) or 10 nM TCDD (T) for 24 h prior to microsomal isolation. Immunoblot analyses were completed on two separate blots, visualized with ECL detection. Purified CYP1A1 (2 ng/lane) expression servs as a standard, in common with both membranes, and a normalizing control.
Table 1A.
DMBA and BP effects mediated by direct AhR activation in BMS2 cells.
| Gene | TCDD (8 h) FC |
BP (8 h) FC |
BP (24 h) FC |
DMBA (8 h) FC |
DMBA (24 h) FC |
|---|---|---|---|---|---|
| Stimulation | |||||
| Cyp1a1 | 92.2 | 70.6 | 66.8 | 45.7 | 141.4* |
| Ahrr | 4.7 | 4.6 | 7.9* | 3.8 | 7.8* |
| Aldh3a1 | 4.6 | 2.9 | 1.4ns | 3.5 | 4.5* |
| Tiparp | 6.6 | 4.6 | 4.0 | 3.1 | 3.5 |
| Nqo1 | 2.9 | 2.6 | 2.5 | 1.9 | 2.5* |
| Adh7 | 2.2 | 2.1 | 2.3 | 1.7 | 2.4* |
| Ptx3 | 2.8 | 2.2 | 2.7 | 2.0 | 3.2* |
| Inhbb | 2.7 | 2.4 | 3.0 | 1.9 | 2.7* |
| Arl6ip5 | 2.7 | 2.1 | 1.7 | 1.8 | 1.7 |
| Ccno | 2.0 | 2.1 | 2.4 | 1.8 | 2.5* |
| Gchfr | 2.1 | 2.0 | 1.8 | 1.4 | 1.9* |
| Cxcl5 | 2.3 | 1.8 | 2.9* | 1.2ns | 3.1* |
| Ler3 | 1.8 | 1.6 | 3.1* | 1.6 | 2.3* |
| Cyp1b1c | 1.1ns. | 1.0 ns. | 1.1ns | −1.1ns | 1.3ns. |
CYP1B1 and CYP1A1 convert PAHs, like DMBA and BP, to epoxides and phenols by mono‑oxygenation reactions (Li et al., 2017; Moorthy et al., 2015). Epoxides are unstable but are rapidly converted by microsomal epoxide hydrolase (Ephx1) to trans-dihydrodiols or spontaneously rearrange to phenols (Christou et al., 1990; Gehly et al., 1979; Pottenger and Jefcoate, 1990). In MSC, like BMS2 or C3H10T1/2, Ephx1 is present at about the same levels as CYP1B1 (Christou et al., 1990; Savas et al., 1994; Savas et al., 1993). The metabolism of PAHs to epoxides and then to dihydrodiols appears to be tightly coupled. In liver microsomes, supplemental Ephx1 greatly enhances dihydrodiol formation from CYP1A1, whereas in MSC the proportion of dihydrodiols is high and unaffected (Pottenger and Jefcoate, 1990). Dihydrodiol epoxides (PAHDE) are converted by a further CYP1B1 mono‑oxygenation step that can be delayed until the starting PAH is sufficiently metabolized (Keller et al., 1987).
PAHDE DNA adducts in BM follow similar differences between DMBA and BP and dependence on CYP1B1 (Galvan et al., 2005). Comparison of BP and BP dihydrodiols in blood and BM, measured by HPLC analyses (N'Jai A et al., 2011), are consistent with origin from AhR-induced CYP1A1 in the liver (N'Jai A et al., 2011; Uno et al., 2004). BM levels of BP, BP-dihydrodiol and BPDE adducts in WT and CYP1B1-ko mice establish that they are determined by intra-BM metabolism by CYP1B1 (Heidel et al., 2000; N'Jai A et al., 2011).
In the BM, the functionality of HSPC within their vascular niche is strongly affected by interaction between local MSC and macrophage (Chow et al., 2011). BP stimulates oxidative stress mediators and inflammatory cytokines within 6 h (Fig. 1C), probably from BM macrophage (Chow et al., 2011). Il1b is increased 5-fold within 6 h, before declining to initial levels after 12- and 24 h (Fig. 1C). Tnf and Ccl3 are similarly responsive (Fig. 1C). This cytokine response and the selective BP protection mechanism each depend on AhR and CYP1A1 (Larsen et al., 2016; N'Jai A et al., 2011; Rondelli et al., 2016). DMBA does not produce this cytokine response (Larsen et al., 2016; N'Jai A et al., 2011; Rondelli et al., 2016) .
CYP1A1 additionally converts BP to quinones, as major products (Keller and Jefcoate, 1984), which cause ROS activation of macrophage and cytokine release (Bolton and Dunlap, 2017). DMBA has no equivalent product since the radical-cation pathway that generates BP quinones (Chakravarti et al., 2008) converts DMBA to the 7- and 12-hydroxymethyl derivatives (Gehly et al., 1979). DMBA 8,9-o-quinone forms from secondary dehydrogenation of 8,9-dihydrodiol (Penning, 2014), but too slowly to deliver protective cytokines. The HSPC factor, Cxcl12, increases as the cytokines decline, whereas Csf1 and Pdgfa remain constant (Fig. 1C).
Cells that are eluted from BM include large, extended fibroblastic cells that adhere to plastic and sequester a ten-fold larger number of hematopoietic cells, including the HSPC that deliver colony expansion (N'Jai A et al., 2011). Culture of this mixed population for 3 weeks expands the MSC population, while increasing the basal CYP1B1 content by 100-fold and largely removing induction by PAHs (Heidel et al., 1998; Phinney et al., 1999). The BMS2 cell line, selected from this expansion, provides an informative model of MSC. The enrichment of MSC is readily tracked through the increased proportion of adherent cells that undergo adipogenesis (Hu et al., 2018). The BMS2 cells exhibited cell rounding and lipid droplet formation when treated for 8d with an adipogenic cocktail (Fig. 1D). This response is replicated in adherent BM cells, but only after 7 days cell expansion that enriches MSC (Fig. 1D). This is consistent with the enrichment of MSC over the course of the 15-day culture period (Phinney et al., 2005; Phinney and Prockop, 2007).
The adipose markers are, however, mixed with osteoblast markers in the these cells, much as reported for C3H10T1/2 cells (Kelly et al., 1998). These MSC lines produce adhesion and adipogenic responses that are each blocked by a combination of AhR activation (TCDD) and Mek-Erk stimulation (Egf or Fgf) (Hanlon et al., 2005a; Liu and Jefcoate, 2006). The HSPC CFU activity is sustained in adherent BM cells by co-culture with BMS2 cells (Fig. 1E). This support is further reproduced by media alone that is enriched over 24 h culture with the cells (Fig. 1E). DMBA inhibition is reproduced in vitro from CYP1B1 activation in the MSC cells (Rondelli et al., 2016).
The BMS2 cells express much higher high levels of basal CYP1B1 than C3H10T1/2 cells, as shown by either the 5.2 kb mRNA or the immunoblotted protein (Fig. 1F, left and right, respectively). This basal protein shows only modest induction by AhR activation (TCDD treatment). Basal expression is sustained in enriched AhR-ko primary BM (BM/6 J AhR−/−) cells, but is completely removed in basal mouse embryo fibroblasts (Heidel et al., 1999). BMS2 and primary BM cells are devoid of CYP1A1 protein, even though inducible mRNA is detectable. The CYP1B1 metabolic activity in the MSC lines mediates the in vitro DMBA suppression.
3.2. Experimental design for the analysis of BMS2 cells as a model for CYP1B1 participation in BM microvascular niche MSC
Single cell sequencing of adherent BM cells points towards multiple MSC types, including those marked by expression of the leptin receptor (Lepr) (Lepr+MSC) (Severe et al., 2019; Tikhonova et al., 2019). These single cell expression profiles overlap appreciably with BMS2 cells (Tables 2C, S5 and S6). MSC have been estimated to represent about 0.3% of BM cells (Zhou et al., 2014). We confirm this assessment, here, with highly expressed markers, such as Cxcl12 and Csf1. MSC that match BMS2 expression of these markers are present at about 3% of adherent BM cells, which in turn represent about 10% of total eluted BM cells (Boregowda et al., 2016; N'Jai A et al., 2011). Lepr+MSC, which represent a high proportion of these MSC (Severe et al., 2019), have been positioned to distinct BM niche vascular compartments with fluorescent markers and enriched for more in depth sequencing (Tikhonova et al., 2019).
Table 2C.
Overlap of BMS2 cells with BM Lepr+MSC.
| Gene | BMS2 |
Lepr + MSC |
|---|---|---|
| Basal (10−3) | N | |
| Lepr | <0.01 | >300 |
| Cyp1b1 | 6 | >300 |
| Markers | ||
| Cxcl12 | 35 | >300 |
| Csf1 | 35 | 253 |
| Gas6 | 24 | >300 |
| Ccl2 | 25 | 160 |
| Ccl7 | 5 | 117 |
| Ebf3 | 5 | >300 |
| Nnmt | 9.5 | >300 |
| Ptx3 | 5 | 176 |
| Il1rn | 8 | >300 |
| Svep1 | 9 | 110 |
| Fzd1 | 13.5 | 37 |
| Kitl | 5 | >300 |
| Receptors | ||
| Il1r1 | 6 | 61 |
| Tnfrsf1a/Tnfr1 | 18 | 7 |
| Tnfrs11b/Opg | 1.0 | 36 |
| Ifngr1 | 17 | 34 |
| Ifngr2 | 4 | 17 |
| Il10rb | 10 | 18 |
| Fgfr2 | 0.35 | 96 |
| Il3ra | 2 | 66 |
| Tgfbr2 | 4 | 17 |
Lepr+MSC from Severe et al., 2019.
Fig. 2 presents three ways that we have used BMS2 cells to assess how CYP1B1 functions in MSC within the hematopoietic niche: 1) characterize the selectivity of BMS2 gene responses to DMBA and BP. Compare these responses to those derived from equivalent in vivo PAH treatments by using rapidly isolated adherent BM cells; 2) evaluate the anomalous BMS2 constitutive AhR regulation of CYP1B1, notably the high constitutive expression and low PAH induction; 3) examine the overlap of BMS2 mRNA profiles with those of the recently reported single cell clustering of Lepr+MSC populations (Severe et al., 2019; Tikhonova et al., 2019). This overlap tests whether there is a core set of functional markers that are conserved, in contrast to adaptive clusters and shifts in the poise between diverse mesenchymal differentiation fates.
Fig. 2.
Experimental design for the analysis of BMS2 cells as a model for CYP1B1 participation of the BM microvascular niche MSC.
These studies utilize three approaches:
1. Characterization of the selectivity of gene responses to DMBA and BP, in vivo, from rapidly isolated adherent BM cells and in cultured BMS2 cells.
2. Examination of the AhR regulation of CYP1B1 expression in BMS2 cells, with respect to their high basal expression and low PAH-mediated induction.
3. Examination of the overlap of mRNA profiles between BMS2 cells, adherent BM populations and the recently reported, single cell clustering of Lepr+MSC populations.
3.3. Selectivity of acute versus delayed responses to PAHs in BMS2 cells
CYP1B1 induction through AhR activation is a key feature of the PAH-mediated response in most cell types (Li et al., 2017). In BMS2 cells, the appreciable basal CYP1B1 gene expression is unaffected by DMBA over a 24 h period (Figs. 1F, left and 3A). CYP1A1 has insignificant basal expression, but is induced to almost 40% of CYP1B1 levels after a 24 h stimulation (Fig. 3A), while failing to yield detectable protein (Fig. 1F, right) (Heidel et al., 1999). Two other genes with canonical DRE elements, Aldh3a1 and Tiparp, show peak stimulations after 8 h (Fig. 3A). The TCDD-mediated stimulation of AhR-responsive genes in C3H10T1/2 cells also reached maximum induction in 8 h (Hanlon et al., 2005b). Suspected stress response genes (Ptgs2, Cxcl10, Cdkn1a/p21 and Ccng/cyclin G1) failed to respond to DMBA during the first 8 h but showed a secondary response to DMBA between 12- and 24 h (Fig. 3B). Cdkn1a/p21 and Ccng1/Cyclin G1 are both well characterized cell cycle responses to p53 activation (Reinke and Lozano, 1997). These genes do not respond to TCDD or to BP (Fig. 3C).
Fig. 3.
Direct PAH and metabolite-mediated gene expression changes and p53 activation in BMS2 cells.
A. qPCR analysis of gene expression changes induced by DMBA (1 uM, over 24 h) treatment in 4 canonical AhR-responsive genes: CYP1A1, CYP1B1, Aldh3a1, and Tiparp. Basal (0 h) microarray expression levels are normalized to 1, as indicated.
B. qPCR analysis of the expression time course of 4 metabolism-mediated stress response genes with DMBA treatment: Ptgs2, Cxcl10, Cdkn1a/p21 and Ccng1/cyclin G1. Cells were treated with 1 uM DMBA over a 24 h period. Basal (0 h) microarray expression levels are normalized to 1, as indicated.
C. DMBA PAH selectivity in BMS2 cells. Gene expression responses were measured after 24 h treatment by DMBA (1 uM) and BP (1 uM), in comparison to the 8 h TCDD (10 nM) response. Significance (*) was defined as p < .05 relative to DMSO control.
D. Heat maps showing hierarchical cluster analysis of PAH responses in BMS2 cells. Genes are presented relative to their selective response to DMBA (left) and BP treatment (right). Red indicates upregulation (FC>2), green indicates down regulation (FC<-2). Genes are listed to the right, the corresponding relative clustering response hierarchical diagram to the left. Treatment columns are, from left to right: DMSO vehicle control (C), BP (B; 1 M; 24 h), DMBA (D; 1 M; 24 h) and TCDD (T; 10 nM; 8 h). Treatments were completed in triplicate. Only genes with expression differences, p-values<0.05 and Cy5-values >100 were analyzed.
E. In cell western analysis of BMS2 cells show significant p53 phosphorylation in response to 24h PAH treatment. Top: a representative fluorescence image of PAH-mediated p53 and H2AX phosphorylation obtained in the high throughput 96-wellplate assay. BP (left) and DMBA (right) significantly activate p53 phosphorylation in BMS2 cells. Significance (*) was defined as PAH-mediated fold change p<0.05 relative to DMSO vehicle control.
Hierarchical clustering of these gene responses, selected for preferential 24 h responses (FC > 2, p < .05), shows a strong DMBA preference (Figs. 3D). 179 genes showed preferred responses to DMBA (D), divided approximately equally between stimulations and suppressions. 68 genes responded to DMBA, but not to TCDD (T), thereby indicating metabolite mediation. Over half of the DMBA responses failed to show significant BP (B) responses. Only 20 BP stimulations were unmatched by DMBA. 16 genes show direct 8 h stimulations by TCDD that are matched by 24 h stimulations by both PAHs.
Tables 1A–C, S2 and S3 provide a more quantitative perspective of the preferences of BP and DMBA for, respectively, direct AhR activation and metabolite driven responses. DMBA also shows a striking synergy between the two processes. Each of these tables includes the 8 h response to TCDD, which defines the optimal direct AhR stimulation, without effects from metabolites. Table 1A shows direct 8 h gene responses to DMBA and BP that parallel TCDD stimulations, with 12 genes exhibiting direct PAH responses, 5 of which have well established DREs (CYP1A1, Ahrr, Aldh3a1, Tiparp, and Nqo1) (Lee et al., 2015). BP stimulations paralleled TCDD stimulations, while DMBA was less active.
Notably, DMBA compensated with further substantial increases (>30%) between 8- and 24 h (11/12 genes). Table 1B shows the prevalence of this biphasic PAH stimulation, which is also seen for canonical AhR responders, CYP1A1 and Ahrr (Table 1A). This synergy of direct AhR activation, with delayed metabolite stress, is the prevailing mechanism for DMBA. Many AhR-responsive genes carry additional elements for factors that respond to chemical stress (NFκB, p53, and Nrf2) (Kalthoff et al., 2010; Mitchell and Elferink, 2009; Tian et al., 1999; Tijet et al., 2006; Wakabayashi et al., 2010). The consistency of the finding suggests that AhR/ARNT activity is further activated at a single element. This overlap is most extensive for partnership with Nrf2 (Nault et al., 2018).
Table 1B.
Genes in which 24 h DMBA or BP stimulates expression in excess of TCDD in BMS2 cells.
| TCDD (8 h) FC |
BP (8 h) FC |
BP (24 h) FC |
DMBA (8 h) FC |
DMBA (24 h) FC |
|
|---|---|---|---|---|---|
| DMBA select | |||||
| Phlda1 | 1.5 | 1.6 | 1.6 | 1.4 | 2.2* |
| Nfe2l2 | 1.6 | 1.5 | 1.9 | 1.4 | 2.1* |
| Ddit4 | 1.7 | 1.7 | 1.9 | 1.5 | 2.6* |
| Ptgs2 | 1.5 | 1.4 | 1.8 | 1.4 | 3.4* |
| Dual | |||||
| Cxcl5 | 2.3 | 1.8 | 2.9* | 1.2n.s. | 3.1* |
| Ler3 | 1.8 | 1.6 | 3.1* | 1.6 | 2.3* |
| Fosl1 | 1.5 | 1.2 | 2.2* | 1.4 | 2.2* |
| BP-select | |||||
| Gprc5a | 1.6 | 1.5 | 2.8* | 1.4 | 1.6 |
| Slc46a3 | 1.6 | 1.7 | 2.7* | 1.5 | 1.9 |
Comparison of these 12 gene responses to TCDD in C3H10T1/2 cells shows similar shared 8 h stimulations, including CYP1A1, Aldh3a1, Adh7 and Nqo1 (Hanlon et al., 2005a; Hanlon et al., 2005b). There are also AhR targets in C3H10T1/2 cells that are not repeated in BMS2 cells (Ch3l1, Glypican 1 and Sod3).
3.4. Metabolism-mediated stimulations by DMBA far exceed those with BP
DMBA-mediated metabolic activation between 8- and 24 h is much more effective than for BP. Table 1C shows 25 highly expressed genes that were stimulated by DMBA after 24 h, but not after 8 h. Each increase has been previously linked to p53 stimulation (Table S1). An additional 22 DMBA-selective genes are shown in Table S2, several associated with p53 activity. None of these genes were stimulated by TCDD. Many of the genes in Table 1C respond fully to p53 within 8 h, when DSB are produced directly by γ-radiation (Fei and El-Deiry, 2003). The slower DMBA response probably arises from delayed PAHDE generation (Keller et al., 1987). Only 14 genes showed a preferential stimulation by BP (FC > 1.8) (Table S3). A further 7 showed equal responses to BP and DMBA. Suppression was also produced after 8 h, again with preference for DMBA (Table S4).
Table 1C.
Genes selectively stimulated by DMBA in BMS2 cells.
| Gene | DMBA (24 h) FC |
BP (24 h) FC |
TCDD (8 h) FC |
|---|---|---|---|
| p53-associated | |||
| Gdfl5 | 5.3 | 1.7 | ns |
| Gadd45a | 4.0 | ns | ns |
| Trp53inp1 | 3.1 | ns | ns |
| Rnd1 | 2.9 | ns | ns |
| Cdkn1a/p21 | 2.9 | 1.5 | ns |
| Eda2r | 2.7 | ns | ns |
| Ddit4 | 2.6 | 1.9 | 1.7 |
| Ccng1 | 2.5 | ns | ns |
| Slc19a2 | 2.4 | ns | ns |
| Bbc3 | 2.3 | ns | ns |
| Mdm2 | 2.3 | ns | ns |
| Btg2 | 2.1 | 2.1 | ns |
| Klf4 | 2.1 | 1.7 | ns |
| Cgref1/Cgr11 | 2.1 | 1.5 | ns |
| Ckap2 | 2.1 | 1.5 | ns |
| Arhgap11a | 2.1 | ns | ns |
| Sulf2 | 2.0 | ns | ns |
| Ei24 | 1.9 | ns | ns |
| Ctgf | 1.9 | ns | ns |
| Ptp4a1 | 1.9 | ns | ns |
| Psrc1 | 1.8 | 1.5 | ns |
| Fas | 1.8 | ns | ns |
| Plk2 | 1.8 | ns | ns |
| Rad51 | 1.8 | ns | ns |
| Txnip | 2.3 | ns | ns |
* >30% increase at 24 h compared to 8 h.
All fold change values presented are significant (p < .01) compared to control, unless indicated (ns).
3.5. DMBA and BP exhibit similar stimulations of 15Sp53 phosphorylation
There was only a small preference for CYP1B1-mediated DMBA metabolism (24 h), relative to BP metabolism, in BMS2 cells, assessed in the general cell activation of 15Sp53 (Fei and El-Deiry, 2003; Mirzayans et al., 2013) (Figs. 3E and S1A), despite the large DMBA preference for gene effects mediated by p53 (Table 1C). p-H2AX, another more direct marker of ATM kinase activation, showed a similar modest preference for DMBA compared to BP (Fig. 3E). This may result because the distorted DMBA 3,4-dihydrodiol-1,2-epoxide structure is more active than the corresponding more planar BP 7,8-dihydrodiol-9,10-epoxide in the context of gene chromatin structure (Chakravarti et al., 2008; Dreij et al., 2005). This in-cell immunodetection method was validated by the selective inhibition of this p53 activation by specific CYP1B1 inhibitors, TMS and α-NF, in CYP1B1-V79 cells, but not CYP1A1-V79 cells (Figs. S1B and C).
3.6. Anomalous basal CYP1B1 expression in BMS2 cells
Although the basal expression of CYP1B1 in primary MSC from BM or from MEFs depends on AhR (Alexander et al., 1997; Heidel et al., 1998), the low PAH induction in BMS2 cells may derive from unusually high basal AhR activity. AhR induction of CYP1B1 is completely dependent on two highly conserved tandem SP1 sequences and a 265 base Ah-receptor enhancer region (AhER) (Wo et al., 1997; Zheng and Jefcoate, 2005; Zheng et al., 2013). This AhER has three AhR/ARNT complexes working in concert, two of which are substantially suppressed by an overlapping AhR inhibitory complex (AIC) (Zhang et al., 1998; Zhang et al., 2003).
ChIP analyses showed similar basal AhR binding to the CYP1B1 and CYP1A1 promoters in mammary cells, despite the selective basal expression of CYP1B1 (Yang et al., 2008). Cell-selective basal signaling evidently contributes to the consistent basal CYP1B1 expression. Repression of basal AhR signaling by cell adhesion was shown in C3H10T1/2 cells and b-catenin released from adhesion complexes was shown to be a basal participant (Cho et al., 2004). This participation of WnT/b-catenin signaling in basal CYP1B1 expression has been further established in multiple cell types (Mohamed et al., 2019; Ziegler et al., 2016). A further consideration is that cell selectivity for CYP1B1 and CYP1A1 in C3H10T1/2 and Hepa cells is primarily determined by selective chromatin interactions that are removed by DNA methylation (Beedanagari et al., 2010a; Beedanagari et al., 2010b).
We further examined the basal and induced signaling to CYP1B1 in BMS2 cells with a luciferase reporter comprised of the dual SP1 elements linked to the AhER. Key elements in the AhER are three 12 base sequences (XRE 1, 4 and 5) that each bind the AhR/ARNT complex (Fig. 4A) (Zhang et al., 1998; Zhang et al., 2003).Two matched XREs (1 and 4) are less active due interaction with the AIC (Zhang et al., 2003).
Fig. 4.
Analysis of basal and TCDD-induced expression of the transfected CYP1B1-AhER luciferase reporter in the BMS2 cell line.
A. Diagram of the XRE elements in the CYP1B1-AhER promoter and of the CYP1B1-AhER constructs investigated.
B. Western immunoblot analysis of CYP1B1 protein expression in BMS2 total cell lysates. CYP1B1 expression (arrow) was analyzed in duplicate samples following 24 h TCDD-induction and 3′-methoxy-4′-nitroflavone (MNF) inhibition. A non-specific background band (top) serves as a loading control reference.
C. MNF inhibits TCDD induction (2-fold) of the AhER reporter in BMS2 cells, and further reduces basal expression up to 3-fold in transfected BMS2 cells.
D. Mutations in the AhER construct indicate that both Ebox and XRE4 elements play critical roles in the TCDD (1 nM)-mediated induction of luciferase reporter expression, above vehicle control, suppressing induction in BMS2 transfected cells
An AhR antagonist, 3MNF (Ryan et al., 2007), removed most basal CYP1B1 protein expression (Fig. 4B) and reduced the basal AhER/SP1 reporter activity by three-fold (Fig. 4C). The basal inhibition progressed up to 0.1 uM, but surprisingly reversed at higher concentrations (0.1–10 μM), suggesting alternative binding sites. Low TCDD induction of CYP1B1 in BMS2 cells is matched by the AhER/SP1 reporter. The incomplete suppression of basal expression by MNF is consistent with the appreciable expression in AhR−/− primary BM cells. The AhER/SP1 reporter showed low basal and strong induction in C3H10T1/2 cells, thus paralleling the protein expression (Figs. S2A and B).
We tested the AhER-SP1 interactions in basal and induced cells by introducing mutations that blocked AhR binding to each of the XREs (Fig. S2C). These changes had similar effects on basal and induced activities in BMS2 and C3H10T1/2 cells (Figs. 4D and S2D). The XRE5 is essential for activity (MXRE5). Mutation of the distal XRE1 (MXRE1) and of XRE4 (MXRE4) are ineffective individually, but the double mutation fully removes expression (Zhang et al., 2003). Mutations of the XRE4 (MXRE4), the Ebox (MEBOX) and their double mutation (MEBOX/MXRE4) indicated a partnership between these adjacent complexes (Figs. 4D and S2D).
We also simplified the AhR enhancer by using a tandem quadruple XRE5 repeat (4XRE5), which then removes participation by the AIC (Fig. 4A). The basal and induced activities are appreciably higher (Fig. 4D), consistent with removal of the AIC. The high basal activity in CYP1B1 in BMS2 cells, therefore, derives from effects of cell-selective factors on the SP1/AhR partnership.
3.7. Evidence for paracrine influences on CYP1B1 expression in BMS2 cells
CYP1B1 transcription in C3H10T1/2 cells showed important contributions from cell adhesion, cell density and EGF signaling, each involving AhR participation (Cho et al., 2004; Cho et al., 2005). For AhER and 4XRE5 reporters in BMS2 cells, density increase from 50 to 80% shows large increases in basal activity, but not in TCDD induction (Fig. 5A). However, at 90%, additional cell-cell contacts cause both activities to decline. This increase in basal density-dependent signaling is consistent with increased effects of secreted cell factors. This signaling overlaps the effect of TCDD on the AhR, which is, therefore, not further stimulated. Enhanced cell-cell contacts block both autocrine and TCDD signaling. This suppression by cell-cell adhesion appears as the reverse of the AhR-dependent stimulation when adhesion is disrupted in C3H10T1/2 cells (Cho et al., 2004).
Fig. 5.
Increased cell density stimulates basal CYP1B1 expression in BMS2 cells, in parallel with increased Cxcl12 expression.
A. Effect of cell density on basal and TCDD-induced activities of AhER/0.2 (AhER) and 4XRE5 reporters at different cell densities (50–100% of confluence) in BMS2 cells. The cells were transfected and then plated at the indicated densities. Cells were treated with TCDD (1 nM) or solvent control (DMSO) for 24 h prior analysis of luciferase activity.
B. Expression (relative to β-actin) of basal and TCDD-induced (10 nM) CYP1B1 and CYP1A1 mRNA in BMS2 cells plated at approximately 50 and 80% of confluence and cultured for 6- and 24 h. Statistical significance: **p < .01, ***p < .001 for high relative to low density culture.
C. Effects of co-culture of BMS2 and C3H10T1/2 cells on their respective basal and TCDD-induced activities. Each cell type was transfected with AhER/0.2 (AhER-SP1) and plated at 80% of confluence in separate compartments that allowed effective media exchange. Each line was also plated in both compartments (two controls). During the 24 h co-culture, the cells were separately stimulated with either TCDD (10 nM) or DMSO (solvent control) prior to analysis of luciferase activities. Significance (*) was defined as p < .05 for TCDD treatment as compared to the respective DMSO control.
D. Expression of basal Cxcl12 and Csf1 (relative to β-actin) at 50 and 80% of confluent cell density. Statistical significance: ** p < .01 for high density relative to low density culture.
For CYP1B1 and CYP1A1, mRNA expression induction, but not basal expression, was elevated by an increase in cell density (Fig. 5B). The natural genes are more responsive to ligand-free AhR than the reporters, but less responsive to TCDD-activated AhR, particularly at the low density. At 80% of confluence, the autocrine boost enhances the TCDD-activated AhR more than the basal expression. At this density, the reporters and genes then respond similarly.
CYP1A1 also responds similarly, suggesting that AhR is directly affected. The opposing effects of cell density and cell contacts suggests that the balance of effects may differ between chromatin-free reporters and the histone sequestered gene. C3H10T1/2 cells showed similar trends of cell density for reporters and natural CYP1B1 (Fig. S2E). Low activities in confluent C3H10T1/2 cells are activated by removal of cell-cell contact, in parallel with nuclear translocation of the AhR (Cho et al., 2004).
We also showed that co-culture with BMS2 cells elevated basal reporter activity in C3H10T1/2 cells (Fig. 5C). BMS2 basal activity declined. Thus, BMS2 cells release net activators, while the C3H10T1/2 cells release net suppressor factors. Again, the maximum TCDD-induced activities are unaffected. Thus, autocrine/paracrine factors activate the AhR partnership with SP1 to maximum levels produced by TCDD complex formation.
Cxcl12, a MSC participant in HSPC support that is also regulated by SP1 and adhesion (Schajnovitz et al., 2011), showed an even larger cell density stimulation effect in BMS2 cells (Fig. 5D). Csf1, another HSPC stimulant, was unaffected (Fig. 5D). This density increase in Cxcl12 was also shown in C3H10T1/2 cells (Fig. S2E).
3.8. Expression of autocrine and asymmetric paracrine pairing of receptors in BMS2 cells
A search of the BMS2 transcriptome for secreted activator proteins and paired receptors identified eight pairs with potential for autocrine regulation (Table 2A ). The asymmetry of gene expression between secreted factors and their cognate receptor is repeated across six distinct Lepr+MSC clusters (Tikhonova et al., 2019). Thus, the paracrine pairings Pdgfa/Pdgfrb, Fgf7/Fgfr2 are retained as potential autocrine stimulants of Mek/Erk (Andrae et al., 2008; Chen et al., 2013; Noriega-Guerra and Freitas, 2018). In C3H10T1/2 cells, EGF activates Mek-Erk and stimulates AhR induction of CYP1B1 by TCDD (Hanlon et al., 2005b). TCDD also functions in concert with Mek-Erk to enhance focal adhesion signaling,thereby to suppress MSC adipogenic differentiation (Hanlon et al., 2005b). SP1 is phosphorylated through this pathway suggesting a likely activation mechanism (Karkhanis and Park, 2015).
Table 2A.
Autocrine and paracrine regulation of BMS2 and BM MSC.
| Secreted protein (SP) | BMS2 expression Cy3 × 10−3 |
BM stimulation FC-BP (6 h) |
Receptor | BMS2 Expression Cy3 × 10−3 |
|---|---|---|---|---|
| A) Autocrine | ||||
| Pdgfa | 19 | ns | Pdgfrb | 30 |
| Tgfb3 | 6 | ns | Tgfbr2 | 4 |
| Wnt2 | 1.2 | ns | Fzd1 | 13.5 |
| Cxcl12 | 35 | ns | Cxcr7 | 2.5 |
| Hgf | 1.2 | 2 | Met | 3 |
| Fgf7 | 25 | ns | Fgfr2 | 1 |
| Cxcl10 | 1.5 | 10 | Cxcr6 | 0.5 |
| Il6 | 0.6 | 5 | Il6ra | 0.1 |
| B) Receptor | ||||
| Tnfa | 0 | 6 | Tnfrsf1a | 18 |
| Tnfsf11/Rankl | 0 | 3 | Tnfrsf11b/Opg | 1 |
| Il1b | 0 | 5 | Il1r1 | 6 |
| Il4 | 0 | ns | Il4ra | 2 |
| Il3 | 0 | ns | Il3ra | 2 |
| Il10 | 0 | 5 | IL10rb | 10 |
| Il17d | 0.5 | ns | 1l17ra | 14 |
| Ifng | 0 | 3.5 | Ifngr1 | 17 |
| C) Donor | ||||
| Ccl2 | 25 | 2 | Ccr2 | 0 |
| Ccl7 | 5 | 4 | ||
| Il7 | 1 | ns | Il7r | 0 |
| Kitl | 5 | ns | c-kit | 0 |
Macrophage cytokines show striking asymmetry in MSC expression. They are largely absent in the BMS2 cells, while their cognate receptors are highly expressed (Tnfrsf1a, Ifngr1, Il1r1) (Table 2A). Ccl2 and Ccl7 are retained as the only C—C chemokines but, again, with absence of their receptors (Ccr2). Many of these paracrine receptors correspond to cytokines that are highly stimulated by BP, in vivo, within 6 h (Fig. 1C). This asymmetry of expression supports a model in which BP-stimulation of cytokines in niche macrophage produce changes in MSC signaling that may contribute to stress protection. Thus, macrophage stimulation can potentially contribute to a selective redistribution of the clusters.
The third asymmetric display comprises paracrine donors that target other cells, as the BMS2 cells lack endogenous expression of the paired receptor (Table 2A). For example, Kitl, which controls HSPC, is expressed without the corresponding c-Kit receptor. Likewise, Ccl2 functions as an angiogenic and immune stimulant (Lim et al., 2016). These pairings are highly selective among the clusters.
3.9. Functional characterization of BMS2 mRNA in relation to adherent BM mRNA
In Table 2B , we compare expression of abundant functional genes from BMS2 cells to their expression in adherent BM cells. mRNA from MSC represents less than 5% of the total in this fraction, which largely derives from hematopoietic cells attached to the MSC (Phinney and Prockop, 2007). These highly expressed BMS2 genes (Cy3 > 3500) and adherent BM mRNA are ranked by proportion of the BMS2 content (Supplement Excel file). Approximately 100 genes appear in the 1–6% range, including Csf1 (4%), Gas6 (2%), Ccl2 (6%), Cxcl12 (3%), Kitl and Ebf3 (1–2%) (Table 2B). Each has been linked to MSC activity, thus suggesting that they are markers for BMS-like cells within the adherent BM-cell fraction (at 3% of the content).
Table 2B.
BMS2 and adherent BM expression of key functional genes.
| Gene |
Basal (Cy3x 10−3) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Cytokine/Stress | BMS2 | BM | Matrix Regulators | BMS2 | BM | Matrix Proteins | BMS2 | BM |
| Cxcl1 | 0.4 | 0.2 | Ctgf/Ccn2 | 76 | 0.3 | Col1a1, | 61 | 1.4 |
| Cxcl9 | 1.9 | 0.07 | Wisp1/Ccn4 | 6.9 | 0.01 | Col1a2 | 94 | 1.2 |
| Cxcl10 | 1.5 | 1.0 | Wisp2/Ccn5 | 8 | 0.04 | Col5a1 | 56 | 1.0 |
| Gas6 | 24 | 0.5 | Periostin/postn | 11 | 0.04 | Col8a1 | 27 | 0.1 |
| Ccl2 | 25 | 1.5 | Thbs1 | 1.8 | 1.0 | Col3a1 | 40 | 0.07 |
| Ccl7 | 5 | 0.1 | Thbs2 | 13 | 0.09 | Col2a1, | 7 | 0.08 |
| Gdf15 | 0.5 | 0.6 | Sparc | 60 | 0.2 | Fn1/Fibronectin | 36 | 1.0 |
| Spp1 | 100 | 9.0 | Sparcl | 7.5 | 0.07 | Fbln2/Fibulin | 53 | 0.03 |
| IL6 | 0.4 | 0.3 | *Bmp1 | 7.9 | 0.1 | Exosome Marker | ||
| Ptgs2 | 1.8 | 0.2 | Timp1 | 94 | 0.6 | Cd63 | 73 | 17 |
| Timp2 | 37 | 2.1 | Cd81 | 40 | 15 | |||
| Timp3 | 36 | 0.3 | Cd9 | 30 | 12 | |||
| Mmp14 | 57 | 1.2 | Other Markers | |||||
| Mgp | 72 | 0.09 | Cyp1b1 | 6 | 0.03 | |||
| Lpl | 4 | 0.6 | ||||||
| Development | BMS2 | BM | BMS2 | BM | |
|---|---|---|---|---|---|
| Cxcl12 | 35 | 0.9 | Pdgfa | 19 | 0.7 |
| IL7 | 1.0 | 0.01 | Tgfb3 | 6 | 0.3 |
| Csf1 | 35 | 1.4 | Hgf | 1.2 | 0.2 |
| Fgf7 | 25 | 0.05 | Angiotensin/Agt | 11 | 0.1 |
| Cgref1 | 4.4 | 0.05 | Osbpl3 | 3.3 | 0.2 |
| Wnt2 | 1.2 | 0.04 | Inhba | 11 | 1.8 |
| Igfbp6 | 34 | 0.2 | Kitl | 5 | 0.03 |
| Igfbp7 | 52 | 0.5 | Ebf3 | 5 | 0.08 |
Several matrix proteins, including Col1a1, Col1a2, Col2a1 and Fn1 are also expressed in this range, but CYP1B1 and a substantial proportion of matrix proteins and regulators (Fbln2, Ctgf, Wisp2) have far higher expression in BMS2 cells (Table 2B).
The continuous enrichment of primary MSC, which precedes the generation of BMS2, is enabled by matrix processes that may facilitate this enrichment. CYP1B1 protects vascular cells from oxygen-induced stress, which is generated by culture under ambient oxygen (20%) conditions (Palenski et al., 2013b; Rondelli et al., 2016). Select genes, including exosome markers, Lpl and Thbs1 (thrombospondin 1), show high expression in both adherent BM and BMS2 cells (Table 2B), likely reflecting alternative sites of expression in adherent BM cells.
3.10. Identification of core Lepr + MSC markers shared with BMS2 cells
The genes with BMS2/adherent BM overlap appropriate to MSC expression were further compared to the single cell profile of Lepr+MSC and other vascular niche cells (Severe et al., 2019) (Table 2C ). This cluster of cells express CYP1B1 as one of the 30 most abundant mRNA (N > 300). Lepr is absent from BMS2 cells (Table 2C), but is also low in other MSC clusters (Severe et al., 2019). Twelve highly expressed BMS2 genes and CYP1B1 show specificity for the Lepr+MSC (Table 2C). Each of the 12 markers has a functional basis for a presence as a core MSC gene (Figs. 6A and S3). The expression of these BMS2 mRNA markers correlated with their expression in BM adherent cells (r2 = 0.86) (Fig. 6B).
Fig. 6.
Matrix proteins and secreted regulatory proteins expressed in BMS2 cells correlate with adherent BM cell expression.
A. Expression of key MSC regulatory factors in BMS2 cells (Cy3) and adherent BM, (percent of BMS2; Supplement Excel file).
B. Correlation of mRNA expression for key MSC regulatory factors in BMS2 cells and in adherent BM cells, at equivalent mitochondrial mRNA (mrpl10) and GAPDH expression.
C. Effects of in vitro DMBA (1 uM) treatment (12h) on adherent select core factors (left) and the three principle DMBA targets (middle) with reference standards, GAPDH and Mrpl10 (right). Statistical significance: *p < .05, **p < .01, ***p < .001 for DMBA treatment relative to basal expression.
The expression levels for 28 genes from BMS2 cells have matches among the most highly expressed genes identified by single cell sequencing of Lepr+MSC in BM (Table S5) (Severe et al., 2019). BMS2 cells, like Lepr+MSC, also express markers for differentiated mesenchymal cell types in the BM niche (Table S6), including osteoblast–like cells (OLC), a chondrocyte (Chond) and a fibroblast-like cluster (Fib) as well as representative endothelia (EC) and pericytes (Per). Several of the MSC expressed genes are highly expressed in OLC, but less than in Lepr+MSC. High expression of Vcam1 and Pdgfrb is shared by MSC and, respectively, EC and Per (Table S5). We also show extensive overlap of cytokine receptors between Lepr+MSC and the other cell types from the BM vascular niche. The distribution of these receptors among different cell types matches the broad diversity of cytokines that are generate by BP (Fig. 1C).
The improved availability was provided by Lepr+ enrichment, which delivered six distinct populations (P1–5 and C) of Lepr+MSC (Table S7) (Tikhonova et al., 2019). Some of the Lepr+MSC clusters show expression biases to either adipoblast or osteoblast differentiation. We used the supplementary single sequencing data of these Lepr-GFP select clusters to show that CYP1B1 expression was highly correlated not only with Lepr, but also with Cxcl12, Kitl, Csf1 and Gas6 (Fig. 6B, Table S7). Ebf3, Ptx3 and Il1rn additionally have high expression in the C cluster (proliferative cluster) and, thus, have weaker correlation with the other core genes. IL7, Fzd1, Svep1 and Nnmt have relatively low expression in the P1 cluster, which is specifically marked by Ccl2 and Ccl7. IL7, the major lymphoid stimulant, demonstrates low expression in the adherent BM fraction and BMS2 cells and only appears as a core gene after Lepr+ enrichment. Spp1 marks P4 and osteoblast clusters. Lpl, an adipocyte marker, is selectively expressed in P2 and P5.
3.11. DMBA, in vivo, does not affect core Lepr + MSC expression markers
Even though DMBA mediates extensive suppression of HSPC expansion (Fig. 1A), scarcely any gene expression changes were seen in cells eluted from treated mice, despite the extensive parallel effects on cytokines produced from macrophage (N'Jai A et al., 2011). We examined 12 h DMBA-induced changes previously observed in adherent BM (N'Jai A et al., 2011) for parallels to BMS2 responses (Fig. 6C). This time point demonstrated full suppression of lymphoid and myeloid CFU. Surprisingly, there were fewer gene responses to DMBA than for BP (N'Jai A et al., 2011), which we previously identified as dominated by cytokines (Fig. 1C).
This absence of stress changes in the cultured BM cells that are seen in BMS2 cells is consistent with stress responses that are not broadly distributed among the total BM population. They are local to the niche presence of MSC and CYP1B1, which our data indicates to be only about 3% of adherent cells. Thus, only highly expressed stress response markers are to be expected. Ccng1, Sulf2, Phlda3 (each p53-induced, Tables 1C and S1) are retained the DMBA stimulation seen in BM cells (Fig. 6C). Other genes that showed strong p53 stress responses to DMBA in BMS2 cells (Gdf15, Gadd45a and Txnip) (Table 1C) did not respond but were predominantly expressed in other adherent cells types (data not shown).
Highly expressed markers shared by BMS2 cells and Lepr+MSC (Cxcl12, Csf1, Gas6) (Table 2C) were scarcely affected by DMBA in the BM cells. Ccl2, Spp1 and Lpl, which each mark more polarized MSC clusters (respectively, P1, P4 and P5; Table S7), show significant increases, as if stress is redistributing the Lepr+MSC (Fig. 6C).
4. Discussion
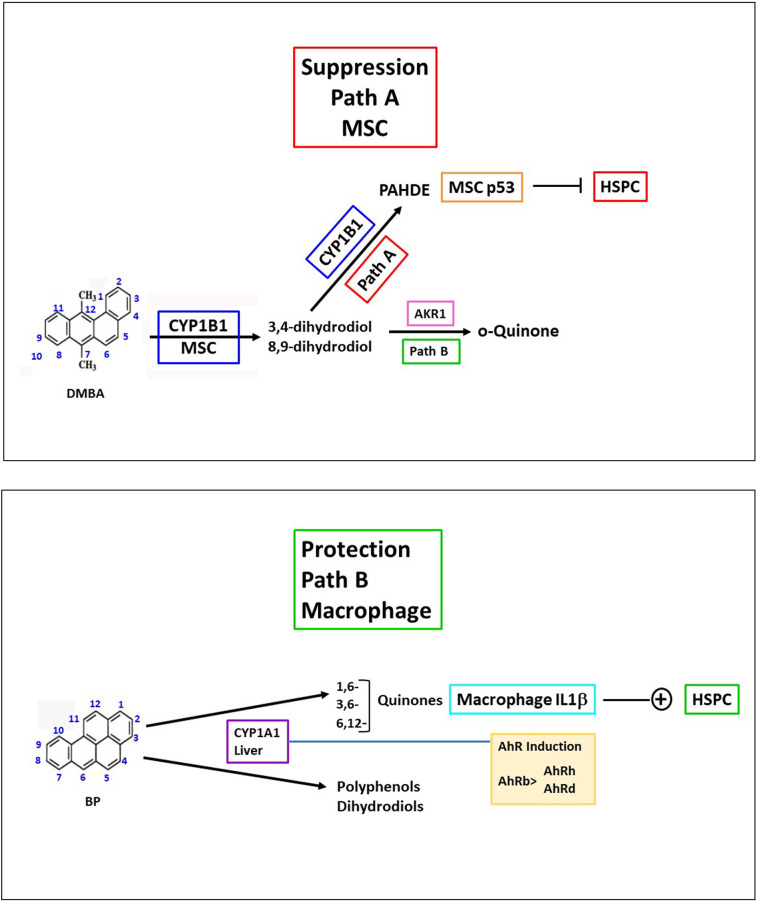
PAHs produce diverse effects on the immune system, particularly on T-lymphocytes and dendritic cells (O'Driscoll et al., 2018; O'Driscoll and Mezrich, 2018). PAHs activate the AhR, which regulates the balance between effector and regulatory T cells (O'Driscoll and Mezrich, 2018) and induces CYP1A1 and CYP1B1. Opposing effects of PAHs on HSPC, typified by DMBA and BP and mediated by, respectively, CYP1B1 and CYP1A1, occur in BM within 6 h (Fig. 7 : Paths A and B). Both focus on the support of HSPC by Lepr+MSC within the BM vascular niche. The BMS2 cells line effectively models critical roles by CYP1B1 and other regulatory proteins in the Lepr+ MSC.
Fig. 7.
Two Reaction Paths for PAHs lead to opposing effects on HSPC in the BM vascular niche. Metabolites produced by CYP1B1 and CYP1A1 direct these effects.
Path A, typified by DMBA, HSPC suppression. CYP1B1 and Ephx1 (microsomal epoxide hydrolase) provide a concerted three step metabolism to generate 3,4-dihydrodiol-1,2-epoxide isomers (PAHDE), marked by DNA adducts in MSC and adjacent pre-B cells (Heidel et al., 2000). These DNA reactions cause DNA DSB, recognized by rapid activation of ATM kinase and p53 phosphorylation. Hypothesis: p53 activation in MSC and attached HSPC in the vascular niche directly effects apoptosis lineage suppression. PAH dihydrodiols are also converted to PAH ortho-quinones via AKR dehydrogenase that also cause DSB. Evidence for their participation in MSC remains to be shown. Environmental PAH share, with synthetic DMBA, structurally restrained, but more reactive PAHDE, including dibenzo(a,l)pyrene (Fjord ring configuration).
Path B, Typified by BP, HSPC protection. CYP1A1, which is induced rapidly to high levels in liver, generates 10% of a mix of BP quinones that are highly active in redox cycling to ROS. BP, in combination with AhR induction, generates very high levels of Il1b and other macrophage inflammatory cytokines (Tnf) that activate their specific MSC receptors. The activation is marked by parallel responses induced by Il1b in MSC (Cxcl1, Il6). Il1b stimulates MSC proliferation responses that protect these cells from stress damage. Hypothesis:Il1b mediates early changes that diminish apoptosis in HSPC. Many PAH undergo 1e- oxidation at CYP1A1 (for example, benz(a)anthracene), but have low AhR inducing activity. Mice have two types of AhR. Inducible b-type (C57Bl/6 mice) and resistant d–type (DBA and 129 strains) in which liver CYP1A1 is very low. BP then functions like DMBA with exclusive CYP1B1-mediated suppression (N'Jai A et al., 2011).
Path A requires effective activation of the PAH, typified by CYP1B1-mediated metabolism of DMBA to PAHDE in MSC, which extensively suppresses HSPC and, within 48 h, mature lymphocytes in BM, thymus and spleen (Fig. 1) (Larsen et al., 2016). In the BM, HSPC, including lymphoid progenitors, are controlled by factors released from Lepr+MSC within the vascular niche (Agarwala and Tamplin, 2018; Seike et al., 2018; Severe et al., 2019; Tikhonova et al., 2019). Factors from BMS2 cells sustain HSPC freshly isolated from the BM (Fig. 1). Metabolism of DMBA by CYP1B1 in these MSC removes lymphoid progenitors (Rondelli et al., 2016). DMBA metabolism in BMS2 cells does not remove support factors, but instead generates PAHDE, which produce DNA adducts, DSB and associated p53 activation in MSC and adjacent HSPC (Heidel et al., 2000). Primary BM MSC and BMS2 cells express high basal levels of CYP1B1, with unusually weak AhR activation (Fig. 1, Fig. 3), even though other AhR marker genes are extensively induced by TCDD, BP and DMBA (Fig. 3, Fig. 6, Tables 1A-C and 2C).
In vivo, a second type of PAH activity, typified by BP, completely prevents this suppression of HSPC (Fig. 7: Path B). This protection arises from systemic metabolism by CYP1A1 from outside the vascular niche, probably the liver (Larsen et al., 2016; N'Jai A et al., 2011; N'Jai A et al., 2010). The BP protection is completely dependent on AhR (N'Jai A et al., 2011). BP protection matches an extensive AhR- and CYP1A1-dependent stimulation of cytokines (IL1b, TNF and IFNg) from macrophage in the BM within 6 h (Fig. 1). CYP1A1 is scarcely present in BM or BMS2 cells (Fig. 1), but AhR induction by PAHs yields high levels in the liver (Galvan et al., 2005). Thus, in vivo BP metabolism and circulating metabolites, including quinones, peak within 6 h, just as BM cytokines appear (Fig. 1) (Larsen et al., 2016; N'Jai A et al., 2011). Equivalent quinones are not matched for DMBA (Gehly et al., 1979). Radiation-induced ROS in macrophage initiates NFkB activation that increases similarly high levels of IL1b (Bigildeev et al., 2013). PCB quinone stimulation of RAW 264.7 macrophage generates p65 NFkB phosphorylation that activates the same set of cytokines (Yang et al., 2019). Il1b stimulates proliferation of MSC via the Il1 receptors and activates over 400 genes (Amann et al., 2019). BP effects on BM produce other increases (Il6, Ccl7, Cxcl1) (Larsen et al., 2016; N'Jai A et al., 2011) that match reported direct stimulation of MSC by Il1b (Hengartner et al., 2015).
BMS2 cells effectively model Lepr+MSC, which also express substantial CYP1B1 (Fig. 6 and Tables 2C, S5 and S6). Thus, Lepr+ MSC and BMS2 cells share expression of a further 12 marker genes that have probable hematopoietic functions (Fig. 6 and Tables 2C, S5 and S6). Inflammatory cytokines (Il1b, Tnf, Ifng) are not expressed in either BMS2 cells or Lepr+MSC, while there are high levels of the corresponding receptors (Table 2A). This cluster of 12 key genes link to BMS2 cells in three ways: the expression overlap between BMS2 and adherent BM expression (Fig. 6); their shared prevalence in BMS2 cells and in a Lepr+MSC cluster derived from a total BM single cell transcription (Tables 2C, S5 and S6) (Severe et al., 2019); and their close correlation across Lepr+ − enriched MSC clusters in BM (Table S7) (Tikhonova et al., 2019).
In both Lepr+ selections, CYP1B1 emerges as an equal member with the other 12 genes (Tables 2C, S5, S6, S7). The lymphoid progenitor stimulant, Il7, has low expression, but emerges as a cluster member after the Lepr+ enrichment (Tikhonova et al., 2019). These relationships point to a cluster of BMS2-like cells that are present at 3% of adherent BM cells and 0.3% of total BM content. The low content of CYP1B1 in adherent BM cells compared to BMS2 cells probably derived from selective enrichment driven by the protection from oxygen stress that is provided by CYP1B1 (Palenski et al., 2013a). Equivalent adaptive enrichments in BMS2 expression likely apply to several ECM-associated genes.
In BMS2 cells, CYP1B1 metabolism of DMBA that produces PAHDE adducts (Heidel et al., 2000) also stimulates numerous genes, but only after 8 h (Tables 1A-C and S2). About half have previous reported evidence for p53 activation (Tables 1C and S1), which is a necessary part of the HSPC suppression process (Page et al., 2003). BP is much less effective but is more active in direct 8 h AhR activations (Table 1A). For DMBA, the smaller direct AhR activation is almost invariably boosted by a second phase of metabolite activation, even for CYP1A1 (Table 1A, Fig. 3). This second phase is likely to derive from supplementary Nrf2 activity on the same AhR/ARNT elements (Nault et al., 2018).
This selectivity by DMBA is enhanced by the chromatin environment of the gene since DMBA and BP were similarly effective in the total cell activations of H2AX and p53 (Fig. 3). Gene selectivity for individual PAHDE is structure dependent (Chakravarti et al., 2008; Dreij et al., 2005). The 8 h delay almost certainly arises from the time to generate the PAHDE (Keller et al., 1987), since the same genes are maximally activated by gamma-radiation in 8 h (Fei and El-Deiry, 2003; Mirzayans et al., 2013).
DMBA responses in BMS2 cells are rarely seen in BM (N'Jai A et al., 2011; N'Jai A et al., 2010), because they are so selective to small numbers of specific BM cells. DMBA-induced transcriptional increases in BMS2 cells are strongly indicative of DSB, p-53-mediated interventions (Table 1C) but are far too slow to mediate the 6 h progenitor suppression times that are produced in the Lepr+MSC the BM niche (Fig. 1). Evidently, more direct effects of DMBA metabolites produce faster, non-transcriptional steps (Pietrocola et al., 2013). For example, apoptosis initiated through Bcl2 in the mitochondrial outer membrane (Vaseva and Moll, 2009) is a favored p53 intervention in stem cells (TeSlaa et al., 2016). The observed 48 h recovery from DMBA toxicity (Fig. 1) may, however, derive from this p53-mediated transcription. Most of the BMS2 genes that respond to DMBA (Tables 1A-C, S1 and S2) are insufficiently expressed to be detectable in MSC at 3% of the adherent BM cells. Only three increases (Ccng1, Sulf2 and Phlda3) parallel activations seen in BMS2 cells (Fig. 6, Tables 1B and C). Ccng1 is a potential marker for p53 activation in BM MSC (Reinke and Lozano, 1997). Ccng1 is highly selective to DMBA in both BMS2 cells (Fig. 3, Table 1C) and adherent BM (N'Jai A et al., 2011), with expression at a level consistent with about half expressed in the BM MSC population.
The novel, high constitutive CYP1B1 expression in BMS2 cells (Fig. 1) is predominantly controlled by a strong coupling between the three AhR/ARNT complexes on the DRE of the AhER and the dual SP1 repeats close to the transcription start site (Fig. 4, Fig. 8 ). In the basal state, an autocrine boost by growth factor signaling to SP1 is sufficient to avoid external AhR activation. This cooperation may be enhanced by release of β-catenin from cell junction sites (Cho et al., 2005; Ziegler et al., 2016). Cell-cell adhesion may suppress CYP1B1 expression by withdrawing β-catenin from the nucleus to the junction sites. SP1 is an important regulator of MSC, including for Cxcl12, which parallels CYP1B1 in the cell density stimulation (Fig. 5). SP1 is commonly activated by Erk phosphorylation, including through activation by Pdgfa (Gong et al., 2017; Schajnovitz et al., 2011). This Pdgfa/Pdgfrb pairing is highly expressed, but Hgf/Met and Fgf7/Fgfr2 pairing also activate Erk (Fig. 8). Cxcl12, Tgfb3 and Wnt2 provide additional pairings. Wnt2/Fzd1 activates β-catenin, possibly participating in the adhesion associated AhR changes.
Fig. 8.
Differential AhR Regulation of CYP1B1 and CYP1A1 in BMS2 cells directs selective activation of p53 by DMBA.
BMS2 cells show high AhR-mediated basal CYP1B1 expression and low induction by TCDD and PAHs. Basal CYP1A1 is undetectable, but highly inducible (40% of CYP1B1). Co-culture of BMS2 with C3H10T1/2 cells cause crossover stimulation of basal CYP1B1 in C3H10T1/2 cells, indicative of BMS2 release of secreted paracrine factors (SF) that stimulate C3H10T1/2 cells. Cell density effects suggest that SF contribute to the high basal CYP1B1 in BMS2 cells. Table 5 lists 40 candidate SFs (examples in yellow box). SF bind receptors (SFR) to transmit intracellular signaling. SF/SFR pairs include PDGFa/Pdgfrb, Fg7/Fgfr2 and Hgf/Met, which activate MEK-Erk kinases, that in turn phosphorylate and thereby activate SP1. Wnt2/Frz1 activates β-catenin, which can partner AhR. This endogenous SF/SFR mechanism is selective for CYP1B1, in part because of weaker SP1 promoter site in CYP1A1. Exogenous AhR/ARNT canonical activation by exogenous agonists (purple box) supplements constitutive activation. Paracrine signaling to CYP1B1 differs appreciably in C3H10T1/2 cells, consistent with changes in CYP1B1 that directly activates DMBA to 3,4 dihydrodiol, and subsequently to the PAHDE that forms DNA adducts. PAHDE transfer rapidly to HSPC to directly suppress their replication within 6 h. Ensuing DSBs activate ATM-kinase, which phosphorylates p53, producing slower gene transcription changes (>8 h). CYP1A1 induction by BP additionally generates BP quinones (BP-Q) that do not form directly from DMBA. BP-Q inhibit both CYP1B1 and CYP1A1, thereby slowing PAHDE synthesis.
The strong correlations of CYP1B1 with the other 12 MSC markers (Fig. 6) suggest a physiological role in the MSC regulation of the vascular niche. Other vascular roles include estradiol hydroxylation (Dempsie et al., 2013; Pingili et al., 2017), epoxidation of polyunsaturated fatty acids (Lefevre et al., 2015; Li et al., 2015) and suppression of vascular oxidative stress (Palenski et al., 2013a).
In summary, two distinct but opposing effects of PAHs have been identified that impact MSC control of HSPC within the BM vascular niche (Graphic Abstract, Fig. 7). Path A suppression is mediated within 6 h by high levels of CYP1B1 and Ephx1 in Lepr+MSC that generate PAHDE. DMBA and environmental PAHs that each generate sterically hindered PAHDE are most active (Chakravarti et al., 2008). The high basal CYP1B1 in BM MSC result from partnership between AhR and SP1, boosted by autocrine signaling (Fig. 8). Path B protection derives from PAH quinones, delivered to BM by hepatic CYP1A1 metabolism, which in turn depends on PAH induction via AhR. These products depend on radical cation generation, which is typical for other multi-ring PAHs (Benz(a)anthracene, Chrysene). AhR activation is, however, higher for BP than most other PAH, including DMBA. The mouse AhR genotype is a major factor (N'Jai A et al., 2011), with strains divided between PAH-responsive AhRb and resistant AhRd alleles, due to binding site sequence variation (Seok et al., 2018) that, however, does not affect constitutive activity. In AhRd strains, BP provides no protection and indeed delivers enough PAHDE to match DMBA as a suppressor (N'Jai A et al., 2011).
The mode of protection by PAH quinones probably derives from Il1b, which is stimulated in BM macrophage. Stimulation of MSC through their IL1R receptors (Bigildeev et al., 2013) could, for example, attenuate the ATM/p53 response to PAHDE. We have identified BM increases that match with IL1b/Il1R activation of human MSC (Hengartner et al., 2015). Thus, BM increases in IL6 and Ccl7 track with IL1b increases (Fig. 1, Table 2A). We can now test effects of IL1b on the DMBA p53–mediated responses in BMS2 cells or the HSPC/BMS2 co-culture model.
To assess environmental exposures on BM vascular niches, the effects of CYP1B1/Ephx1 generation of PAHDE in BMS2 can now be compared to PAH quinone stimulation of IL1b and other cytokines in BM macrophages or Raw 264.7 cells (Bolton and Dunlap, 2017; Yang et al., 2019). V79 cells that express human CYP1B1 and CYP1A1 provide a means to probe species shifts in metabolic selectivity (Luch et al., 1998; Luch et al., 1999; Schmalix et al., 1993). Environmental exposure impacts AhR induction of CYP1A1 and, thereby, metabolic increases in the flux of PAH quinones to the BM macrophage that then stimulate cytokine production. Combustion pollution mixtures also include diverse PAH quinones and AhR activators (Bostrom et al., 2002; Layshock et al., 2010).
BMS2 cells and Lepr+MSC share CYP1B1 and a very limited core of Lepr+MSC functional markers as well as a set of cytokine receptors that complement the macrophage cytokine production (Fig. 6, Tables 2A and C). CYP1B1 expression in Lepr+MSC is highly correlated with four secreted HSPC regulatory factors (Cxcl12, Csf1, Kitl, Svep1 and Il7), two secreted immune modulators (Gas6, Ptx3) and Fzd1, which directs Wnt2 activity (Fig. 6). These genes are notably very resistant to Path A and Path B signaling. BMS2 cells express many additional genes that appear to be gained from in vitro selection. The effectiveness of OP9/BMS2 co-culture with BM HSPC (Rondelli et al., 2016) provides confidence that they indeed model PAH effects on Lepr+MSC.
Credit author statement
MCL contributed to experimental design, completion of bone marrow isolation and microarray, adipogenic and CFU experimental analyses and writing of the manuscript.
AA completed bone marrow isolation and cell density experimental analyses.
TT completed bone marrow microarray analyses.
CMR contributed to writing and completed experimental analyses on control of HSPC activity by MSCs.
RJS completed the CYP1B1 promoter construct experiments.
CRJ completed the conceptual design and writing of the manuscript.
Declaration of Competing Interest
The authors acknowledge no conflicts of interest with these studies.
Acknowledgements
We thank Dr. Owen Tamplin for his insightful discussions relating to the BM Lepr+MSC, which facilitated the identification of the 12 core Lepr+MSC genes in the BMS2 cell model.
This publication was made possible by grants from the National Institute of Diabetes and Digestive and Kidney Disease (NIDDK) grant numbers DK072749 and DK090249, and the National Institute of Environmental Health Sciences (NIEHS) grant number ES007015. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. Ahmed Almeldin was partially supported by the Egyptian Cultural & Educational Bureau.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.taap.2020.115111.
Appendix A. Supplementary data
Comparison of BMS2 and Adherent BM gene expression.
Supplementary material 2
References
- Agarwala S., Tamplin O.J. Neural crossroads in the hematopoietic stem cell niche. Trends Cell Biol. 2018;28(12):987–998. doi: 10.1016/j.tcb.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D.L., Eltom S.E., Jefcoate C.R. Ah receptor regulation of CYP1B1 expression in primary mouse embryo-derived cells. Cancer Res. 1997;57(20):4498–4506. [PubMed] [Google Scholar]
- Amann E.M. Inflammatory response of mesenchymal stromal cells after in vivo exposure with selected trauma-related factors and polytrauma serum. PLoS One. 2019;14(5) doi: 10.1371/journal.pone.0216862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrae J., Gallini R., Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22(10):1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beedanagari S.R. Role of epigenetic mechanisms in differential regulation of the dioxin-inducible human CYP1A1 and CYP1B1 genes. Mol. Pharmacol. 2010;78(4):608–616. doi: 10.1124/mol.110.064899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beedanagari S.R., Taylor R.T., Hankinson O. Differential regulation of the dioxin-induced Cyp1a1 and Cyp1b1 genes in mouse hepatoma and fibroblast cell lines. Toxicol. Lett. 2010;194(1–2):26–33. doi: 10.1016/j.toxlet.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett J.A. Conditional deletion of Ahr alters gene expression profiles in hematopoietic stem cells. PLoS One. 2018;13(11) doi: 10.1371/journal.pone.0206407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigildeev A.E. Interleukin-1 beta is an irradiation-induced stromal growth factor. Cytokine. 2013;64(1):131–137. doi: 10.1016/j.cyto.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Bolton J.L., Dunlap T. Formation and biological targets of Quinones: cytotoxic versus Cytoprotective effects. Chem. Res. Toxicol. 2017;30(1):13–37. doi: 10.1021/acs.chemrestox.6b00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boregowda S.V., Krishnappa V., Phinney D.G. Isolation of mouse bone marrow mesenchymal stem cells. Methods Mol. Biol. 2016;1416:205–223. doi: 10.1007/978-1-4939-3584-0_11. [DOI] [PubMed] [Google Scholar]
- Bostrom C.E. Cancer risk assessment, indicators, and guidelines for polycyclic aromatic hydrocarbons in the ambient air. Environ. Health Perspect. 2002;110(Suppl. 3):451–488. doi: 10.1289/ehp.110-1241197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushkofsky J.R. Cyp1b1 affects external control of mouse hepatocytes, fatty acid homeostasis and signaling involving HNF4alpha and PPARalpha. Arch. Biochem. Biophys. 2016;597:30–47. doi: 10.1016/j.abb.2016.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano-Vinyals G. Biomarkers of exposure to polycyclic aromatic hydrocarbons from environmental air pollution. Occup. Environ. Med. 2004;61(4) doi: 10.1136/oem.2003.008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti D. The role of polycyclic aromatic hydrocarbon-DNA adducts in inducing mutations in mouse skin. Mutat. Res. 2008;649(1–2):161–178. doi: 10.1016/j.mrgentox.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.H. Resveratrol inhibits TCDD-induced expression of CYP1A1 and CYP1B1 and catechol estrogen-mediated oxidative DNA damage in cultured human mammary epithelial cells. Carcinogenesis. 2004;25(10):2005–2013. doi: 10.1093/carcin/bgh183. [DOI] [PubMed] [Google Scholar]
- Chen G. The fibroblast growth factor receptor 2-mediated extracellular signal-regulated kinase 1/2 signaling pathway plays is important in regulating excision repair cross-complementary gene 1 expression in hepatocellular carcinoma. Biomed Rep. 2013;1(4):604–608. doi: 10.3892/br.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.C., Zheng W., Jefcoate C.R. Disruption of cell-cell contact maximally but transiently activates AhR-mediated transcription in 10T1/2 fibroblasts. Toxicol. Appl. Pharmacol. 2004;199(3):220–238. doi: 10.1016/j.taap.2003.12.025. [DOI] [PubMed] [Google Scholar]
- Cho Y.C. Differentiation of pluripotent C3H10T1/2 cells rapidly elevates CYP1B1 through a novel process that overcomes a loss of ah receptor. Arch. Biochem. Biophys. 2005;439(2):139–153. doi: 10.1016/j.abb.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Chow A. Bone marrow CD169+ macrophages promote the retention of hematopoietic stem and progenitor cells in the mesenchymal stem cell niche. J. Exp. Med. 2011;208(2):261–271. doi: 10.1084/jem.20101688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christou M. Differences in the modulation of P450IA1 and epoxide hydratase expression by benz[a]anthracene and 2,3,7,8-tetrachlorodibenzo-p-dioxin in mouse embryo versus mouse hepatoma-derived cell lines. Carcinogenesis. 1990;11(10):1691–1698. doi: 10.1093/carcin/11.10.1691. [DOI] [PubMed] [Google Scholar]
- Cimafranca M.A., Hanlon P.R., Jefcoate C.R. TCDD administration after the pro-adipogenic differentiation stimulus inhibits PPARgamma through a MEK-dependent process but less effectively suppresses adipogenesis. Toxicol. Appl. Pharmacol. 2004;196(1):156–168. doi: 10.1016/j.taap.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Crane G.M., Jeffery E., Morrison S.J. Adult haematopoietic stem cell niches. Nat. Rev. Immunol. 2017;17(9):573–590. doi: 10.1038/nri.2017.53. [DOI] [PubMed] [Google Scholar]
- Dempsie Y. Dexfenfluramine and the oestrogen-metabolizing enzyme CYP1B1 in the development of pulmonary arterial hypertension. Cardiovasc. Res. 2013;99(1):24–34. doi: 10.1093/cvr/cvt064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorheim M.A. Osteoblastic gene expression during adipogenesis in hematopoietic supporting murine bone marrow stromal cells. J. Cell. Physiol. 1993;154(2):317–328. doi: 10.1002/jcp.1041540215. [DOI] [PubMed] [Google Scholar]
- Dreij K., Seidel A., Jernstrom B. Differential removal of DNA adducts derived from anti-diol epoxides of dibenzo[a,l]pyrene and benzo[a]pyrene in human cells. Chem. Res. Toxicol. 2005;18(4):655–664. doi: 10.1021/tx0497090. [DOI] [PubMed] [Google Scholar]
- Fei P., El-Deiry W.S. P53 and radiation responses. Oncogene. 2003;22(37):5774–5783. doi: 10.1038/sj.onc.1206677. [DOI] [PubMed] [Google Scholar]
- Galan-Diez M., Kousteni S. A bone marrow niche-derived molecular switch between osteogenesis and hematopoiesis. Genes Dev. 2018;32(5–6):324–326. doi: 10.1101/gad.314013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan N. Bone marrow cytotoxicity of benzo[a]pyrene is dependent on CYP1B1 but is diminished by ah receptor-mediated induction of CYP1A1 in liver. Toxicol. Appl. Pharmacol. 2003;193(1):84–96. doi: 10.1016/s0041-008x(03)00338-7. [DOI] [PubMed] [Google Scholar]
- Galvan N. Induction of CYP1A1 and CYP1B1 in liver and lung by benzo(a)pyrene and 7,12-d imethylbenz(a)anthracene do not affect distribution of polycyclic hydrocarbons to target tissue: role of AhR and CYP1B1 in bone marrow cytotoxicity. Toxicol. Appl. Pharmacol. 2005;202(3):244–257. doi: 10.1016/j.taap.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Ganesan S., Bhattacharya P., Keating A.F. 7,12-Dimethylbenz[a]anthracene exposure induces the DNA repair response in neonatal rat ovaries. Toxicol. Appl. Pharmacol. 2013;272(3):690–696. doi: 10.1016/j.taap.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J. p53 and ATM/ATR regulate 7,12-dimethylbenz[a]anthracene-induced immunosuppression. Mol. Pharmacol. 2008;73(1):137–146. doi: 10.1124/mol.107.039230. [DOI] [PubMed] [Google Scholar]
- Gehly E.B. The metabolism of benzo(alpha)pyrene by cytochrome P-450 in transformable and nontransformable C3H mouse fibroblasts. J. Biol. Chem. 1979;254(12):5041–5048. [PubMed] [Google Scholar]
- Gong J. Paired related homeobox protein 1 regulates PDGF-induced chemotaxis of hepatic stellate cells in liver fibrosis. Lab. Investig. 2017;97(9):1020–1032. doi: 10.1038/labinvest.2017.65. [DOI] [PubMed] [Google Scholar]
- Hanlon P.R. Microarray analysis of early adipogenesis in C3H10T1/2 cells: cooperative inhibitory effects of growth factors and 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Appl. Pharmacol. 2005;207(1):39–58. doi: 10.1016/j.taap.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Hanlon P.R. Identification of novel TCDD-regulated genes by microarray analysis. Toxicol. Appl. Pharmacol. 2005;202(3):215–228. doi: 10.1016/j.taap.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Heidel S.M., Czuprynski C.J., Jefcoate C.R. Bone marrow stromal cells constitutively express high levels of cytochrome P4501B1 that metabolize 7,12-dimethylbenz[a]anthracene. Mol. Pharmacol. 1998;54(6):1000–1006. doi: 10.1124/mol.54.6.1000. [DOI] [PubMed] [Google Scholar]
- Heidel S.M. Bone marrow stromal cell cytochrome P4501B1 is required for pre-B cell apoptosis induced by 7,12-dimethylbenz[a]anthracene. Mol. Pharmacol. 1999;56(6):1317–1323. doi: 10.1124/mol.56.6.1317. [DOI] [PubMed] [Google Scholar]
- Heidel S.M. Cytochrome P4501B1 mediates induction of bone marrow cytotoxicity and preleukemia cells in mice treated with 7,12-dimethylbenz[a]anthracene. Cancer Res. 2000;60(13):3454–3460. [PubMed] [Google Scholar]
- Hengartner N.E. Crucial role of IL1beta and C3a in the in vitro-response of multipotent mesenchymal stromal cells to inflammatory mediators of polytrauma. PLoS One. 2015;10(1) doi: 10.1371/journal.pone.0116772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y. Comparative study on in vitro culture of mouse bone marrow mesenchymal stem cells. Stem Cells Int. 2018;2018:6704583. doi: 10.1155/2018/6704583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichii M. The canonical Wnt pathway shapes niches supportive of hematopoietic stem/progenitor cells. Blood. 2012;119(7):1683–1692. doi: 10.1182/blood-2011-07-369199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J. Smoke carcinogens cause bone loss through the aryl hydrocarbon receptor and induction of Cyp1 enzymes. Proc. Natl. Acad. Sci. U. S. A. 2013;110(27):11115–11120. doi: 10.1073/pnas.1220919110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefcoate Colin R., Wang Suqing, Liu Xueqing. Methods that resolve different contributions of clonal expansion to Adipogenesis in 3T3-L1 and C3H10T1/2 cells. In: Yang K., editor. Adipose Tissue Protocols. Humana Press; Totowa, NJ: 2008. pp. 173–193. [DOI] [PubMed] [Google Scholar]
- Johansen A.K. The serotonin transporter promotes a pathological estrogen metabolic pathway in pulmonary hypertension via cytochrome P450 1B1. Pulm Circ. 2016;6(1):82–92. doi: 10.1086/685023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalthoff S. Interaction between oxidative stress sensor Nrf2 and xenobiotic-activated aryl hydrocarbon receptor in the regulation of the human phase II detoxifying UDP-glucuronosyltransferase 1A10. J. Biol. Chem. 2010;285(9):5993–6002. doi: 10.1074/jbc.M109.075770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkhanis M., Park J.I. Sp1 regulates Raf/MEK/ERK-induced p21(CIP1) transcription in TP53-mutated cancer cells. Cell. Signal. 2015;27(3):479–486. doi: 10.1016/j.cellsig.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G.M., Jefcoate C.R. Benzo(a)pyrene activation to 7,8-dihydrodiol 9,10-oxide by rat liver microsomes. Control by selective product inhibition. J. Biol. Chem. 1984;259(22):13770–13776. [PubMed] [Google Scholar]
- Keller G.M. Product inhibition of benzo[a]pyrene metabolism in uninduced rat liver microsomes: effect of diol epoxide formation. Chem. Biol. Interact. 1987;61(2):159–175. doi: 10.1016/0009-2797(87)90037-8. [DOI] [PubMed] [Google Scholar]
- Kelly K.A. Murine bone marrow stromally derived BMS2 adipocytes support differentiation and function of osteoclast-like cells in vitro. Endocrinology. 1998;139(4):2092–2101. doi: 10.1210/endo.139.4.5915. [DOI] [PubMed] [Google Scholar]
- Kincade P.W. Cells and molecules that regulate B lymphopoiesis in bone marrow. Annu. Rev. Immunol. 1989;7:111–143. doi: 10.1146/annurev.iy.07.040189.000551. [DOI] [PubMed] [Google Scholar]
- Lai A.Y., Kondo M. Asymmetrical lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. J. Exp. Med. 2006;203(8):1867–1873. doi: 10.1084/jem.20060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M.C. Cytochrome P450 1B1: an unexpected modulator of liver fatty acid homeostasis. Arch. Biochem. Biophys. 2015;571:21–39. doi: 10.1016/j.abb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M.C. Cyp1b1-mediated suppression of lymphoid progenitors in bone marrow by polycyclic aromatic hydrocarbons coordinately impacts spleen and thymus: a selective role for the ah receptor. Pharmacol. Res. Perspect. 2016;4(4) doi: 10.1002/prp2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layshock J.A., Wilson G., Anderson K.A. Ketone and quinone-substituted polycyclic aromatic hydrocarbons in mussel tissue, sediment, urban dust, and diesel particulate matrices. Environ. Toxicol. Chem. 2010;29(11):2450–2460. doi: 10.1002/etc.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Male and female mice show significant differences in hepatic transcriptomic response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. BMC Genomics. 2015;16:625. doi: 10.1186/s12864-015-1840-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre L. LRH-1 mediates anti-inflammatory and antifungal phenotype of IL-13-activated macrophages through the PPARgamma ligand synthesis. Nat. Commun. 2015;6:6801. doi: 10.1038/ncomms7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung G.S. Developmental expression and endocrine regulation of CYP1B1 in rat testis. Drug Metab. Dispos. 2009;37(3):523–528. doi: 10.1124/dmd.108.025635. [DOI] [PubMed] [Google Scholar]
- Li M., Ikehara S. Bone-marrow-derived mesenchymal stem cells for organ repair. Stem Cells Int. 2013;2013:132642. doi: 10.1155/2013/132642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Volti G., Caruso M., Polosa R. Smoking and SARS-CoV-2 disease (COVID-19): dangerous liaisons or confusing relationships? J. Clin. Med. 2020;9(5) doi: 10.3390/jcm9051321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P. Epoxyeicosatrienoic acids enhance embryonic haematopoiesis and adult marrow engraftment. Nature. 2015;523(7561):468–471. doi: 10.1038/nature14569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Zhu W., Gonzalez F.J. Potential role of CYP1B1 in the development and treatment of metabolic diseases. Pharmacol. Ther. 2017;178:18–30. doi: 10.1016/j.pharmthera.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.Y. Targeting the CCL2-CCR2 signaling axis in cancer metastasis. Oncotarget. 2016;7(19):28697–28710. doi: 10.18632/oncotarget.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.M. Target disruption of ribosomal protein pNO40 accelerates aging and impairs osteogenic differentiation of mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2016;469(4):903–910. doi: 10.1016/j.bbrc.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Liu X., Jefcoate C. 2,3,7,8-tetrachlorodibenzo-p-dioxin and epidermal growth factor cooperatively suppress peroxisome proliferator-activated receptor-gamma1 stimulation and restore focal adhesion complexes during adipogenesis: selective contributions of Src, rho, and Erk distinguish these overlapping processes in C3H10T1/2 cells. Mol. Pharmacol. 2006;70(6):1902–1915. doi: 10.1124/mol.106.026534. [DOI] [PubMed] [Google Scholar]
- Luch A. Stable expression of human cytochrome P450 1B1 in V79 Chinese hamster cells and metabolically catalyzed DNA adduct formation of dibenzo[a,l]pyrene. Chem. Res. Toxicol. 1998;11(6):686–695. doi: 10.1021/tx970236p. [DOI] [PubMed] [Google Scholar]
- Luch A. Metabolic activation of dibenzo[a,l]pyrene by human cytochrome P450 1A1 and P450 1B1 expressed in V79 Chinese hamster cells. Chem. Res. Toxicol. 1999;12(4):353–364. doi: 10.1021/tx980240g. [DOI] [PubMed] [Google Scholar]
- Maguire M. Cyp1b1 deletion and retinol deficiency coordinately suppress mouse liver lipogenic genes and hepcidin expression during post-natal development. Mol. Cell. Endocrinol. 2017;454:50–68. doi: 10.1016/j.mce.2017.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire M. Cyp1b1 directs Srebp-mediated cholesterol and retinoid synthesis in perinatal liver; association with retinoic acid activity during fetal development. PLoS One. 2020;15(2) doi: 10.1371/journal.pone.0228436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzayans R. Ionizing radiation-induced responses in human cells with differing TP53 status. Int. J. Mol. Sci. 2013;14(11):22409–22435. doi: 10.3390/ijms141122409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell K.A., Elferink C.J. Timing is everything: consequences of transient and sustained AhR activity. Biochem. Pharmacol. 2009;77(6):947–956. doi: 10.1016/j.bcp.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed H.T. Inflammatory breast cancer: activation of the aryl hydrocarbon receptor and its target CYP1B1 correlates closely with Wnt5a/b-beta-catenin signalling, the stem cell phenotype and disease progression. J. Adv. Res. 2019;16:75–86. doi: 10.1016/j.jare.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorthy B., Chu C., Carlin D.J. Polycyclic aromatic hydrocarbons: from metabolism to lung cancer. Toxicol. Sci. 2015;145(1):5–15. doi: 10.1093/toxsci/kfv040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A.M. Development of hematopoietic stem cell activity in the mouse embryo. Immunity. 1994;1(4):291–301. doi: 10.1016/1074-7613(94)90081-7. [DOI] [PubMed] [Google Scholar]
- Nault R. Comparison of hepatic NRF2 and aryl hydrocarbon receptor binding in 2,3,7,8-Tetrachlorodibenzo-p-dioxin-treated mice demonstrates NRF2-independent PKM2 induction. Mol. Pharmacol. 2018;94(2):876–884. doi: 10.1124/mol.118.112144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near R.I. Regulation of preB cell apoptosis by aryl hydrocarbon receptor/transcription factor-expressing stromal/adherent cells. Proc. Soc. Exp. Biol. Med. 1999;221(3):242–252. doi: 10.3181/00379727-221-44410. [DOI] [PubMed] [Google Scholar]
- N’Jai A U. Bone marrow lymphoid and myeloid progenitor cells are suppressed in 7,12-dimethylbenz(a)anthracene (DMBA) treated mice. Toxicology. 2010;271(1–2):27–35. doi: 10.1016/j.tox.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- N’Jai A U. Acute disruption of bone marrow hematopoiesis by benzo(a)pyrene is selectively reversed by aryl hydrocarbon receptor-mediated processes. Mol. Pharmacol. 2011;79(4):724–734. doi: 10.1124/mol.110.070631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noriega-Guerra H., Freitas V.M. Extracellular matrix influencing HGF/c-MET signaling pathway: impact on Cancer progression. Int. J. Mol. Sci. 2018;19(11) doi: 10.3390/ijms19113300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll C.A., Mezrich J.D. The aryl hydrocarbon receptor as an immune-modulator of atmospheric particulate matter-mediated autoimmunity. Front. Immunol. 2018;9:2833. doi: 10.3389/fimmu.2018.02833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll C.A. Polycyclic aromatic hydrocarbons (PAHs) present in ambient urban dust drive proinflammatory T cell and dendritic cell responses via the aryl hydrocarbon receptor (AHR) in vitro. PLoS One. 2018;13(12) doi: 10.1371/journal.pone.0209690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page T.J. 7,12-Dimethylbenz[a]anthracene-induced bone marrow toxicity is p53-dependent. Toxicol. Sci. 2003;74(1):85–92. doi: 10.1093/toxsci/kfg115. [DOI] [PubMed] [Google Scholar]
- Palenski T.L. Cyp1B1 expression promotes angiogenesis by suppressing NF-kappaB activity. Am. J. Physiol. Cell Physiol. 2013;305(11):C1170–C1184. doi: 10.1152/ajpcell.00139.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenski T.L. Lack of Cyp1b1 promotes the proliferative and migratory phenotype of perivascular supporting cells. Lab. Investig. 2013;93(6):646–662. doi: 10.1038/labinvest.2013.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penning T.M. Human aldo-keto reductases and the metabolic activation of polycyclic aromatic hydrocarbons. Chem. Res. Toxicol. 2014;27(11):1901–1917. doi: 10.1021/tx500298n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phinney D.G., Prockop D.J. Concise review: mesenchymal stem/multipotent stromal cells: the state of transdifferentiation and modes of tissue repair--current views. Stem Cells. 2007;25(11):2896–2902. doi: 10.1634/stemcells.2007-0637. [DOI] [PubMed] [Google Scholar]
- Phinney D.G. Plastic adherent stromal cells from the bone marrow of commonly used strains of inbred mice: variations in yield, growth, and differentiation. J. Cell. Biochem. 1999;72(4):570–585. [PubMed] [Google Scholar]
- Phinney D.G. Murine mesenchymal and embryonic stem cells express a similar Hox gene profile. Biochem. Biophys. Res. Commun. 2005;338(4):1759–1765. doi: 10.1016/j.bbrc.2005.10.140. [DOI] [PubMed] [Google Scholar]
- Pietrangeli C.E., Hayashi S., Kincade P.W. Stromal cell lines which support lymphocyte growth: characterization, sensitivity to radiation and responsiveness to growth factors. Eur. J. Immunol. 1988;18(6):863–872. doi: 10.1002/eji.1830180606. [DOI] [PubMed] [Google Scholar]
- Pietrocola F. Regulation of autophagy by stress-responsive transcription factors. Semin. Cancer Biol. 2013;23(5):310–322. doi: 10.1016/j.semcancer.2013.05.008. [DOI] [PubMed] [Google Scholar]
- Pingili A.K. 6beta-Hydroxytestosterone, a cytochrome P450 1B1-testosterone-metabolite, mediates angiotensin II-induced renal dysfunction in male mice. Hypertension. 2016;67(5):916–926. doi: 10.1161/HYPERTENSIONAHA.115.06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingili A.K. 2-Methoxyestradiol reduces angiotensin II-induced hypertension and renal dysfunction in Ovariectomized female and intact male mice. Hypertension. 2017;69(6):1104–1112. doi: 10.1161/HYPERTENSIONAHA.117.09175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottenger L.H., Jefcoate C.R. Characterization of a novel cytochrome P450 from the transformable cell line, C3H/10T1/2. Carcinogenesis. 1990;11(2):321–327. doi: 10.1093/carcin/11.2.321. [DOI] [PubMed] [Google Scholar]
- Reinke V., Lozano G. Differential activation of p53 targets in cells treated with ultraviolet radiation that undergo both apoptosis and growth arrest. Radiat. Res. 1997;148(2):115–122. [PubMed] [Google Scholar]
- Rondelli C.M. PAHs target hematopoietic linages in bone marrow through Cyp1b1 primarily in mesenchymal stromal cells but not AhR: a reconstituted in vitro model. Stem Cells Int. 2016;2016:1753491. doi: 10.1155/2016/1753491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan E.P. Environmental toxicants may modulate osteoblast differentiation by a mechanism involving the aryl hydrocarbon receptor. J. Bone Miner. Res. 2007;22(10):1571–1580. doi: 10.1359/jbmr.070615. [DOI] [PubMed] [Google Scholar]
- Savas U., Christou M., Jefcoate C.R. Mouse endometrium stromal cells express a polycyclic aromatic hydrocarbon-inducible cytochrome P450 that closely resembles the novel P450 in mouse embryo fibroblasts (P450EF) Carcinogenesis. 1993;14(10):2013–2018. doi: 10.1093/carcin/14.10.2013. [DOI] [PubMed] [Google Scholar]
- Savas U. Mouse cytochrome P-450EF, representative of a new 1B subfamily of cytochrome P-450s. Cloning, sequence determination, and tissue expression. J. Biol. Chem. 1994;269(21):14905–14911. [PubMed] [Google Scholar]
- Savas U., Carstens C.P., Jefcoate C.R. Biological oxidations and P450 reactions. Recombinant mouse CYP1B1 expressed in Escherichia coli exhibits selective binding by polycyclic hydrocarbons and metabolism which parallels C3H10T1/2 cell microsomes, but differs from human recombinant CYP1B1. Arch. Biochem. Biophys. 1997;347(2):181–192. doi: 10.1006/abbi.1997.0339. [DOI] [PubMed] [Google Scholar]
- Schajnovitz A. CXCL12 secretion by bone marrow stromal cells is dependent on cell contact and mediated by connexin-43 and connexin-45 gap junctions. Nat. Immunol. 2011;12(5):391–398. doi: 10.1038/ni.2017. [DOI] [PubMed] [Google Scholar]
- Schmalix W.A. Stable expression of human cytochrome P450 1A1 cDNA in V79 Chinese hamster cells and metabolic activation of benzo[a]pyrene. Eur. J. Pharmacol. 1993;248(3):251–261. doi: 10.1016/0926-6917(93)90052-r. [DOI] [PubMed] [Google Scholar]
- Seike M. Stem cell niche-specific Ebf3 maintains the bone marrow cavity. Genes Dev. 2018;32(5–6):359–372. doi: 10.1101/gad.311068.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok S.H. Trace derivatives of kynurenine potently activate the aryl hydrocarbon receptor (AHR) J. Biol. Chem. 2018;293(6):1994–2005. doi: 10.1074/jbc.RA117.000631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severe N. Stress-induced changes in bone marrow stromal cell populations revealed through single-cell protein expression mapping. Cell Stem Cell. 2019;25(4):570–583. doi: 10.1016/j.stem.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddens L.K. Cytochrome P450 1b1 in polycyclic aromatic hydrocarbon (PAH)-induced skin carcinogenesis: Tumorigenicity of individual PAHs and coal-tar extract, DNA adduction and expression of select genes in the Cyp1b1 knockout mouse. Toxicol. Appl. Pharmacol. 2015;287(2):149–160. doi: 10.1016/j.taap.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teague J.E. Proximal events in 7,12-dimethylbenz[a]anthracene-induced, stromal cell-dependent bone marrow B cell apoptosis: stromal cell-B cell communication and apoptosis signaling. J. Immunol. 2010;185(6):3369–3378. doi: 10.4049/jimmunol.0902541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TeSlaa T., Setoguchi K., Teitell M.A. Mitochondria in human pluripotent stem cell apoptosis. Semin. Cell Dev. Biol. 2016;52:76–83. doi: 10.1016/j.semcdb.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y. Ah receptor and NF-kappaB interactions, a potential mechanism for dioxin toxicity. J. Biol. Chem. 1999;274(1):510–515. doi: 10.1074/jbc.274.1.510. [DOI] [PubMed] [Google Scholar]
- Tijet N. Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol. Pharmacol. 2006;69(1):140–153. doi: 10.1124/mol.105.018705. [DOI] [PubMed] [Google Scholar]
- Tikhonova A.N. The bone marrow microenvironment at single-cell resolution. Nature. 2019;569(7755):222–228. doi: 10.1038/s41586-019-1104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno S. Oral exposure to benzo[a]pyrene in the mouse: detoxication by inducible cytochrome P450 is more important than metabolic activation. Mol. Pharmacol. 2004;65(5):1225–1237. doi: 10.1124/mol.65.5.1225. [DOI] [PubMed] [Google Scholar]
- Uno S. Oral benzo[a]pyrene in Cyp1 knockout mouse lines: CYP1A1 important in detoxication, CYP1B1 metabolism required for immune damage independent of total-body burden and clearance rate. Mol. Pharmacol. 2006;69(4):1103–1114. doi: 10.1124/mol.105.021501. [DOI] [PubMed] [Google Scholar]
- Vaseva A.V., Moll U.M. The mitochondrial p53 pathway. Biochim. Biophys. Acta. 2009;1787(5):414–420. doi: 10.1016/j.bbabio.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa M. Aryl hydrocarbon receptor is required for optimal B-cell proliferation. EMBO J. 2017;36(1):116–128. doi: 10.15252/embj.201695027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollrath A.L. EDGE(3): a web-based solution for management and analysis of Agilent two color microarray experiments. BMC Bioinformatics. 2009;10:280. doi: 10.1186/1471-2105-10-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi N. When NRF2 talks, who’s listening? Antioxid. Redox Signal. 2010;13(11):1649–1663. doi: 10.1089/ars.2010.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wo Y.Y., Stewart J., Greenlee W.F. Functional analysis of the promoter for the human CYP1B1 gene. J. Biol. Chem. 1997;272(42):26702–26707. doi: 10.1074/jbc.272.42.26702. [DOI] [PubMed] [Google Scholar]
- Yang X. Constitutive regulation of CYP1B1 by the aryl hydrocarbon receptor (AhR) in pre-malignant and malignant mammary tissue. J. Cell. Biochem. 2008;104(2):402–417. doi: 10.1002/jcb.21630. [DOI] [PubMed] [Google Scholar]
- Yang B. Polychlorinated biphenyl Quinone promotes macrophage-derived foam cell formation. Chem. Res. Toxicol. 2019;32(12):2422–2432. doi: 10.1021/acs.chemrestox.9b00184. [DOI] [PubMed] [Google Scholar]
- Zhang L. Characterization of the mouse Cyp1B1 gene. Identification of an enhancer region that directs aryl hydrocarbon receptor-mediated constitutive and induced expression. J. Biol. Chem. 1998;273(9):5174–5183. doi: 10.1074/jbc.273.9.5174. [DOI] [PubMed] [Google Scholar]
- Zhang L., Zheng W., Jefcoate C.R. Ah receptor regulation of mouse Cyp1B1 is additionally modulated by a second novel complex that forms at two AhR response elements. Toxicol. Appl. Pharmacol. 2003;192(2):174–190. doi: 10.1016/s0041-008x(03)00276-x. [DOI] [PubMed] [Google Scholar]
- Zheng W., Jefcoate C.R. Steroidogenic factor-1 interacts with cAMP response element-binding protein to mediate cAMP stimulation of CYP1B1 via a far upstream enhancer. Mol. Pharmacol. 2005;67(2):499–512. doi: 10.1124/mol.104.005504. [DOI] [PubMed] [Google Scholar]
- Zheng W. Stimulation of mouse Cyp1b1 during adipogenesis: characterization of promoter activation by the transcription factor Pax6. Arch. Biochem. Biophys. 2013;532(1):1–14. doi: 10.1016/j.abb.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.O. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15(2):154–168. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler N. Beta-catenin is required for endothelial Cyp1b1 regulation influencing metabolic barrier function. J. Neurosci. 2016;36(34):8921–8935. doi: 10.1523/JNEUROSCI.0148-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of BMS2 and Adherent BM gene expression.
Supplementary material 2