Abstract
KDEL receptor cycles between the ER and the Golgi to retrieve ER-resident chaperones that get leaked to the secretory pathway during protein export from the ER. Recent studies have shown that a fraction of KDEL receptor may reside in the plasma membrane and function as a putative cell surface receptor. However, the trafficking itinerary and mechanism of cell surface expressed KDEL receptor remains largely unknown. In this study, we used N-terminally Halo-tagged KDEL receptor to investigate its endocytosis from the plasma membrane and trafficking itinerary of the endocytosed receptor through the endolysosomal compartments. Our results indicate that surface-expressed KDEL receptor undergoes highly complex recycling pathways via the Golgi and peri-nuclear recycling endosomes that are positive for Rab11 and Rab14, respectively. Unexpectedly, KDEL receptor appears to preferentially utilize clathrin-mediated endocytic pathway as well as clathrin-dependent transport carriers for export from the trans-Golgi network. Taken together, we suggest that KDEL receptor may be a bona fide cell surface receptor with a complex, yet well-defined trafficking itinerary through the endolysosomal compartments.
Electronic supplementary material
The online version of this article (10.1007/s00018-020-03570-3) contains supplementary material, which is available to authorized users.
Keywords: KDEL receptor, Golgi, Rab11, Rab14, Clathrin, Membrane trafficking, MANF, Caveolae, EEA1, Lysosome, Late endosomes, Early endosomes, Recycling endosomes
Introduction
KDEL receptor (KDELR) is a seven transmembrane protein and plays an important role in the early secretory pathway by capturing and recycling ER-resident chaperones at the Golgi during protein secretion [1, 2]. Most of the soluble ER-chaperones contain C-terminal tetra-peptide ‘KDEL’ sequence that is recognized by KDELR for the recycling pathway [3]. The receptor also appears to function as a signaling protein that responds to incoming transport carriers from the ER that delivers secretory cargo proteins to the Golgi [4–7]. This role and its seven transmembrane arrangements raised strong arguments that the receptor might be an atypical GPCR at the Golgi, which presumably orchestrates membrane flux and maintain equilibrium in the early secretory pathway by regulating retrograde transport against bulk flow of anterograde transport of lipids and proteins toward the plasma membrane (PM) [6–8]. However, a recently resolved structure of KDEL receptor was found to resemble a SWEET transporter family protein, rather than a GPCR-type family protein [9], adding to the complexity in KDELR function. Although KDELR1 and a eukaryotic SWEET transporter share a poor sequence identity (~24%), their structures are similar. The seven transmembrane (TM) alpha helices of KDELR1 are divided into two internal triple helix bundles (THBs) composed of the first three N- and last three C-terminal helices respectively. Both THBs have helices in a 1–3–2 arrangement and connected invertely by the linker helix TM4, which is a typical structural arrangement of SWEET transporters.
In mammalian cells, three isoforms of KDELR are known to be expressed (KDELR1, KDELR2 and KDELR3). Since the identification of mammalian KDELR in the early 1990s, a vast majority of studies have focused on function and trafficking of KDELR1 mainly using HeLa cells as a model system [10–14]. In HeLa cells, relative mRNA level of KDELR3 was found to be only ~5% of KDELR1, whereas KDELR2 mRNA level was approximately 2-times higher than KDELR1 [15]. The three isoforms appear to bind variants of ‘KDEL’ sequence motif ([KRHQSA]-[DENQ]-E-L) with different specificity that are found in various ER-resident chaperones [15].
Interestingly, recent studies have shown that a small fraction of KDELR1 may also reside in the plasma membrane and function as a putative cell surface receptor for certain growth factors, including mesencephalic astrocyte-derived neurotrophic factor (MANF) [16–18]. MANF is an ER-resident protein that gets secreted during prolonged ER stress and has been reported to promote regeneration of a number of damaged tissues and cells, pointing to a possibility that KDELR/MANF-mediated signaling at the PM may be involved in cellular regeneration during cellular stress or damage [17–21].
However, the trafficking itinerary and mechanism of surface-expressed KDELR1 has remained elusive. As exogenous over-expression of KDELR1 often leads to autoactivation and its retrograde transport to the ER [10, 14], it has been a major challenge to investigate KDELR trafficking at the PM and through the endolysosomal compartments.
To circumvent this issue, we constructed N-terminally Halo-tagged KDELR1 that allowed us to selectively label surface-expressed KDELR1 using membrane impermeable Halo-tag dye on its extracellular side for both fixed cell and live imaging study. Using this strategy, we were able to determine precise trafficking itinerary of KDELR from the plasma membrane to the Golgi via the endolysosomal compartments.
Our results indicate that surface-expressed KDELR1 undergo highly complex recycling pathways through the endolysosomal compartment and the Golgi that involves both fast and slow recycling endosomes. Further, we show that KDELR1 preferentially utilizes clathrin-mediated transport carriers both at the PM for endocytosis and at the TGN for export from the Golgi, respectively. Collectively, these results suggest that KDELR is likely a bona-fide cell surface receptor with a complex, yet well-defined intracellular trafficking itinerary.
Results
To study membrane trafficking of surface-expressed KDELR1, we used human breast carcinoma MCF7 and HeLa cells, both of which appeared to express a high level of KDELR1 on the cell surface, compared to other cell lines examined during our preliminary screening (data not shown).
All three isoforms of KDELR are expressed at the cell surface
Since there are mainly three known isoforms of KDEL-R (KDELR1, KDELR2 and KDELR3) expressed in mammalian cells, we were curious to find out if all the three isoforms can be detected at the cell surface. To this end, we exogenously over-expressed KDELR isoforms tagged with mCherry at their C-terminal tail in MCF7 cells. Briefly, MCF7 cells were transiently transfected with the different KDELR isoforms for 18 h, followed by surface biotinylation protocol to determine the relative amount of KDELR surface expression. The results indicated that all three isoforms were readily detected at the PM (Fig. 1a). Surprisingly, KDELR3 surface expression was 2–3 times higher, compared to KDELR1 and KDELR2. Since KDELR1 is the most representative KDELR that have been studied in the field, we decided to focus on KDELR1 trafficking in this study. Nonetheless, this difference in surface expression pattern for KDELR3 needs to be addressed in a future study, as it might hint at the possibility that KDELR3 may have a distinct role at the PM.
Fig. 1.
Surface-expressed wild type KDELR1, but not the H12A mutant receptor, binds TAEKDEL peptide. a Comparison of cell surface expression level of three isoforms of KDELR. MCF7 cells were transiently transfected with KDELR1-mCherry, KDELR2-mCherry or KDELR3-mCherry for 18 h. Cell surface expression of KDELR-mCherry was probed by cell surface biotinylation protocol, as described in the methods. b Schematic representation of the integration strategy to generate C-terminally 3xFlag-mCherry/Halo-tagged proteins expressed from the endogenous KDELR1 locus. A double-strand break was created at the last exon of KDELR1 gene by Cas9 RNP. Gene knock-in was mediated by a plasmid DNA donor template that contains 3xFlag-mCherry/Halo sequence, neomycin resistant gene and homology arms. c Cell surface biotinylation of endogenously tagged KDELR1. Cell surface proteins were biotinylated by treatment with (+) or without (−) Sulfo-NHS-LC-Biotin in KDELR1-knock-in (C-terminal 3xFlag-mCherry endogenously tagged) HeLa cells. Biotinylated proteins were isolated by streptavidin-agarose and subjected to western blot analysis using the indicated antibodies. Whole cell lysates (input) served as control to determine the total amount of KDELR1, while GM130 served as a cytosolic marker protein, which showed no biotinylation and EGFR served as a plasma membrane marker protein, respectively. Membrane fraction (surface) illustrates the total fraction of proteins at the cell surface. d Schematic representation of Halo-tagged KDEL receptor. e Expression of Halo-KDELR and Halo-KDELR H12A in MCF7 KDELR KD cells was detected by immunoblotting. f Comparison of the expression level of over-expressed HA-Halo-3xFlag-tagged KDELR1 in HeLa wt, MCF7 wt and MCF KDELR1 KD cells with that of endogenously 3xFlag-Halo-tagged KDELR1 in HeLa cells by immunoblotting. g MCF7 KDELR KD cells were transfected with either Halo-KDELR or Halo-KDELR H12A for 18 h. Living cells were then stained with HaloTag Alexa Fluor 488 and HaloTag TMR ligands at 4 °C for 30 min, followed by fixation and DAPI staining. The subcellular localization of Halo-KDELR or Halo-KDELR H12A was observed using Zeiss LSM880. h–i Halo-KDELR (h) or Halo-KDELR H12A (i) expressed in MCF7 KDELR KD cells was incubated with HaloTag Alexa Fluor 488 ligand and 50 μM TMR-TAEKDEL peptide in DMEM (pH 6.5) supplemented with 10% FBS at 4 °C for 30 min. Cells were then washed 3 times with PBS and incubated at 37 °C for 0, 15, and 30 min, followed by fixation. Confocal images were acquired using Zeiss LSM880. Scale bar: 5 µm
Endogenously tagged KDELR1 is expressed at the cell surface
These results did not prove that endogenous KDELR1 is expressed at the PM. As we could not find a good commercial antibody against KDELR1 that works for either western blot or confocal experiment, we opted to introduce a 3xFlag-mCherry tag at the C-terminal end of endogenous KDELR1 gene using CRISPR-Cas9 technique, as shown in the illustration (Fig. 1b). Note that we used HeLa cells for increased transfection efficiency and successful isolation of monoclonal cell lines, as introducing large tags to endogenous genes is a highly time-consuming and technically challenging feat. After selection with neomycin, we were able to isolate a few monoclonal cell lines and confirmed the specificity of tag insertion position by genome sequencing. We then determined whether endogenously tagged KDELR1 was indeed expressed at the PM using surface biotinylation protocol in this cell line. As shown in Fig. 1c, KDELR1 endogenously tagged with 3xFlag-mCherry was readily detected at the cell surface. As a positive control, we also found epidermal growth factor receptor (EGFR) at the PM of this cell line, whereas a Golgi matrix protein, GM130, was not detected at the PM in the same experiment. Taken together, these results indicated that (1) all isoforms of KDELR can be expressed at the PM; (2) both endogenous and exogenously over-expressed KDELR1 are expressed in sufficient amount at the PM.
Experimental design for N-terminally Halo-tagged KDELR1 at the endogenous expression level
The biggest obstacle to study membrane trafficking of KDELR at the cell surface so far had been the difficulty to selectively label the surface pool of KDELR and follow the receptor endocytosis at the PM and intracellular trafficking through the endolysosomal compartment. To overcome this issue, we devised a N-terminally Halo-tagged KDELR1 construct to minimize the chance of interfering with the “KDEL” motif binding site, which is located in the extracellular (luminal) face of the 7-TM receptor. As only 1–2 amino acids of its N-terminal end were predicted to remain at the extracellular face of KDELR1, we introduced a HA-signal peptide as a membrane insertion signal, followed by introduction of a Halo-3xFlag-tag with a GS-linker to the + 1 position of the first methionine residue of the original human KDELR1 sequence (HA-Halo-3Flag-KDELR1, Fig. 1d). We also generated H12A mutant construct with N-terminal HA-Halo tag as a negative control, which has been reported to abrogate the receptor binding to “KDEL” peptide in a recent KDELR structural study [9].
As an effort to avoid over-expression artifact during the confocal-based study as best as we can, we then stably knock-downed the endogenous KDELR1 in MCF7 cells using shRNA-based lentiviral transduction method, as described in the experimental procedures. Specificity and efficiency of shRNA-dependent knock-down of KDELR1 were confirmed by quantitative RT-PCR (supplementary Fig. 1A). The results showed that KDELR1 knockdown efficiency was ~86%, and that there were a negligible change in mRNA levels of both KDELR2 and KDELR3 in the KDELR1 KD MCF7 cells. Knock-down efficiency of this RNAi targeting sequence was further confirmed in a siRNA form using 3xFlag-mCherry knock-in KDELR1 HeLa cells to show KDELR1 KD at protein level (supplementary Fig. 1B). Then, we rescued the KDELR1 KD MCF7 cells with either wild-type HA-Halo-3Flag-KDELR1 or H12A KDELR1 mutant [9]. Both KDEL receptor constructs showed an expected size of ~52 kDa in western blot analysis (Fig. 1e).
To assess their expression level, we used HeLa cells with endogenously 3xFlag-Halo-tagged KDELR1 at its C-terminus as a standard to compare the expression level of exogenously expressed, N-terminally HA-Halo-3xFlag-tagged KDELR1 in both HeLa and MCF7 cells (Fig. 1f). To this end, we transiently transfected either wt HeLa (Fig. 1f; lane 2) or wt MCF7 (lane 3) or MCF7.KDELR1 KD (lane 4) with HA-Halo-KDELR1 construct for 18 h and assessed their expression levels by western blots using anti-Halo-tag antibody.
The results indicated that both MCF7 wt and MCF KDELR1 KD cells showed similar expression level of HA-Halo-3xFlag-tagged KDELR1 as that of endogenously 3xFlag-Halo-tagged KDELR1 in HeLa cells (Fig. 1f). Overall, these results suggest that N-terminally HA-Halo-tagged KDELR1 expression in MCF7 cells are comparable to that of endogenous KDELR1 in wt HeLa cells.
Both wild type Halo-KDELR1 and the H12A mutant receptors localize to the Golgi and the PM
We then examined their intracellular localization using membrane-permeable ‘HaloTag-TMR’ dye to label both the Golgi-localized and surface-expressed KDELR or membrane impermeable ‘HaloTag-Alexa488′ to selectively label surface-expressed KDELR (Fig. 1g). The results indicated that both the WT and the H12A mutant KDELR1 localized similarly to the Golgi area and the cell surface. Next, we further examined Golgi localization of the WT and the mutant receptor, to confirm that both receptor constructs are normal in their localization within the Golgi apparatus (supplementary Fig. 2). The results clearly showed that there was no significant difference in their co-localization with a cis-Golgi marker, GM130.
During preliminary live imaging experiments, we found that both the WT and the mutant KDELR1 labeled with HaloTag-Alexa488 constantly undergo endocytosis at 37 °C even in the absence of KDEL peptide and found them concentrated in the cytoplasmic punta within ~15 min, which resembled features of early endosomes. To synchronize the ligand binding-induced receptor endocytosis, we decided to incubate the cells with KDEL peptide at 4 °C for 30 min in culture medium adjusted to pH 6.5 [9], followed by temperature shift to 37 °C to allow endocytosis of the receptor as a general protocol during the course of this study.
Surface-expressed KDELR, but not the H12A mutant receptor, binds TAEKDEL peptide at the PM
When fluorescently labeled KDEL peptide (TAEKDEL-FITC) was added to the medium using this protocol, only the WT receptor was able to bind TAEKDEL peptide (50 μM; pH 6.5; 4 °C), as expected, whereas the H12A mutant receptor failed to bind the ligand under the same condition (Fig. 1h–i), confirming that this single point mutation completely abrogates the receptor-ligand interaction.
Surface-expressed KDELR undergoes clathrin-mediated endocytosis
We then examined whether clathrin may be involved in KDELR endocytosis, as it is currently unknown how surface-expressed KDELR may get endocytosed. For this experiment, we decided to use wt HeLa cells, transiently transfected with HA-Halo-tagged KDELR1, as it was technically challenging to obtain clean live imaging data with MCF7 cells, due to relatively poor cell spreading onto glass surface, which makes imaging of cell periphery more difficult than other cell lines. Thus, HeLa cells were transiently co-transfected with HA-Halo-tagged-KDELR1 and mCherry-clathrin light chain (mCherry-CLC) overnight. We then selectively labeled surface-expressed KDELR1 using membrane-impermeable HaloTag-Alexa488 at 4 °C and allowed the receptor to undergo endocytosis by temperature shift to 37 °C and adding TAEKDEL peptide in pH 6.5 for live imaging study using Zeiss confocal microscope with Airyscan. The results clearly demonstrated that there is a significant overlap between the two proteins during KDELR1 endocytosis from the PM, suggesting that surface-expressed KDELR1 is likely to undergo clathrin-mediated endocytosis (Fig. 2a; white arrowheads in the inset; see line graphs for more detailed analysis). In contrast, we did not observe significant overlap between KDELR1 and mCherry-CLC upon addition of control peptide (TAEAAAA) to the medium.
Fig. 2.
Surface-expressed KDELR undergo clathrin-mediated endocytosis. a Live-cell imaging of clathrin-dependent endocytosis of KDELR1. HeLa cells were co-transfected with Halo-KDELR1 and mCherry-CLC (clathrin light chain) for 18 h. Living cells were preincubated with HaloTag Alexa Fluor 488 ligand at 4 °C for 30 min. Cells were then washed with PBS and observed using Zeiss Airyscan confocal microscope at 37 °C for 30 min in live cell imaging medium (pH 6.5), supplemented with 1% FBS and 50 μM TAEKDEL or TAEAAAA peptides. Arrowheads indicate punctae containing both Halo-KDELR and mCherry-CLC proteins. Scale bars: 10 µm. Insets show a magnified view of the boxed area. Line profiles through regions of interest were analyzed by Fiji. b TIRF imaging showed endocytosis of KDELR1 was dependent of clathrin rather than caveolin. MCF7 cells overexpressing Halo-KDELR were incubated with HaloTag Alexa Fluor 488 ligand and 50 μM TAEKDEL peptide in DEMEM (pH 6.5) supplemented with 10% FBS at 4 °C for 30 min. Cells were then washed 3 times with ice-cold PBS and incubated at 37 °C for 0 and 30 min, followed by fixation and staining by anti-clathrin or anti-caveolin antibodies, respectively. Images were acquired using Nikon TIRF microscope. Box graphs summarize the co-localization of Halo-KDELR1 with clathrin or caveolin, respectively (n = 10 cells). Statistical analysis was performed using two-way ANOVA with a Tukey's post-hoc test for multiple comparisons. Insets show a magnified view of the boxed area. Line profiles through regions of interest were analyzed by Fiji. Scale bars: 10 µm. *** p < 0.001, **** p < 0.0001, ns not significant
We performed additional experiments using fixed cells and total internal reflection fluorescence (TIRF) microscope for more precise quantitative analysis. To this end, MCF7 KDELR1 KD cells were transfected with HA-Halo-KDELR1 or HA-Halo-KDELR1 H12A overnight, followed by labeling with membrane-impermeable HaloTag-Alexa488 and temperature shift protocol for endocytosis. Cells were then fixed after 30 min at 37 °C and stained with either anti-clathrin or anti-caveolin antibodies. The stained cells were examined within ~200 nm from the PM using Nikon TIRF microscope to determine whether surface-expressed KDELR1 may preferentially utilize either of these endocytic routes from the PM.
The results showed that surface-expressed HA-Halo-KDELR1 co-localized with clathrin to a significantly higher extent than caveolin upon addition of KDEL ligand (0.70 ± 0.05 for clathrin vs. 0.23 ± 0.12 for caveolin; see supplementary Table 1), further confirming that KDELR1 may preferentially undergo clathrin-mediated endocytosis from the PM (Fig. 2b). Prior to addition of KDEL peptide to the medium, co-localization between KDELR and clathrin was moderate, but addition of KDEL peptide significantly increased co-localization between the two proteins (Fig. 2b; insets; see line graphs for more detailed analysis). Note that we also routinely observed increased clustering of surface-expressed KDELR as well as clathrin (to a much lesser extent for caveolin) upon addition of KDEL peptide during the experiments, consistent with a previous report by other group [18]. Further statistical analysis by 2-way ANOVA methods found that only KDELR1 and clathrin showed a statistically significant co-localization (Fig. 2b, supplementary Table 1), whereas KDELR1 and caveolin showed a statistically insignificant colocalization (Fig. 2c, supplementary Table 1), confirming that KDELR1 is likely to be endocytosed via clathrin-mediated endocytosis.
Surface-expressed KDELR1 is transported to the early endosomes upon endocytosis
To determine the initial itinerary of surface-expressed KDELR1 upon endocytosis, we used various markers of known endolysosomal compartments, including EEA1 (early endosomes), Mitotracker (mitochondria), Lysotracker (Lysosomes) and Rab7 (late endosomes) for co-localization study. The results showed that EEA1 showed relatively high Pearson coefficient with KDELR1, which further increased upon addition of KDEL ligand, whereas both mitochondrial marker and lysosomal marker showed negligible co-localization index (Fig. 3a–c). Late endosomal marker, Rab7, also showed poor co-localization with endocytosed HA-Halo-KDELR1 (Fig. 3d), suggesting that early endosomes are likely the first major assembly point of endocytosed KDELR1 from the PM.
Fig. 3.
Surface-expressed KDELR is transported to the early endosomes upon endocytosis. Co-localization of endocytosed Halo-KDELR and intracellular organelles was determined using confocal microscopy. a MCF7 KDELR KD cells were transfected with either Halo-KDELR or Halo-KDELR H12A with mCherry-tagged Rab7 (d). After 18 h, cells were treated with DMSO (a and d) or MitoTracker (b) or LysoTracker (c) at 37 °C for 30 min and then incubated with HaloTag Alexa Fluor 488 ligand and TAEKDEL peptide in DMEM (pH 6.5) with 10% FBS at 4 °C for 30 min. Cells were then washed with PBS and transferred to 37 °C for 0, 15, or 30 min, prior to fixation and staining with anti-EEA1 antibody (a). The colocalization between HA-Halo-KDELR (WT or H12A) and various organelles was quantified using Pearson’s coefficient and summarized by line graph (n = 20 cells). Statistical analysis was performed using two-way ANOVA with a Tukey's post-hoc test for multiple comparisons. Scale bars: 5 µm. * p < 0.05, *** p < 0.001, **** p < 0.0001, #p < 0.0001, ns not significant
ER-localization signal peptide “RTDL” of MANF also stimulates trafficking of endocytosed KDELR1 to the early endosomes
Since studies have suggested that surface-expressed KDELR1 may function as one of the MANF receptors, we were curious to know whether “RTDL” peptide could also stimulates delivery of endocytosed KDELR1 to the early endosomes. For this experiment, we used wt MCF7 cells (rather than KDELR1 KD MCF7) to exclude the possibility that KDELR1 knock-down and rescue approach may have introduced undesirable cellular or ER stress in our cell lines, thereby causing artifacts in our observation and analysis. Thus, wt MCF7 cells were transiently transfected with HA-Halo-KDELR1 for 18 h, followed by incubation with either unlabeled KDEL peptide or RTDL peptide or DMSO at 4 °C, pH 6.5 for 30 min. After a temperature shift protocol to 37 °C for 15 min or 30 min, cells were fixed and stained with anti-EEA1 antibody, as described earlier.
The results showed that over-expressed HA-Halo-tagged KDELR1 in KDEL peptide-treated cells behaved similarly with the receptor expressed in KDELR1 KD MCF7 cells (Fig. 4a–c), although average Pearson coefficient was slightly reduced especially at 15 min time-point (0.42 ± 0.07 in wt MCF7 vs. 0.51 ± 0.10 in KD-rescue MCF7). This difference became negligible at 30 min time-point (0.64 ± 0.08 in wt MCF7 vs. 0.61 ± 0.07 in KD-rescue MCF7), suggesting that KDEL peptide-induced delivery of KDELR1 to the early endosome likely reached a plateau at this time-point (Fig. 4d). On the other hand, RTDL peptide further increased colocalization index between endocytosed KDELR1 and EEA1 over KDEL peptide at 15 min time-point (0.42 ± 0.07 in KDEL peptide vs. 0.49 ± 0.05 in RTDL peptide) under same experimental conditions, suggesting that RTDL motif of MANF can be at least equally stimulating as a KDEL motif. Although this result does not confirm KDELR1 as the physiologically relevant MANF receptor at the PM, it does raise a strong possibility that extracellular MANF can be effectively captured and endocytosed by surface-expressed KDELR1.
Fig. 4.
RTDL motif of MANF can also accelerate trafficking of endocytosed KDELR1 to the early endosomes. MCF7 cells were transfected with HA-Halo-KDELR1 for 18 h and incubated with HaloTag Alexa Fluo 488 ligand and 50 μM TAEKDEL (a), 50 μM ASARTDL (b) or DMSO (c) in DMEM (pH 6.5) with 10% FBS at 4 °C for 30 min. Cells were then washed with PBS and transferred to 37 °C for indicated times, prior to fixation and staining with anti-EEA1 antibody. Insets and line graphs show the colocalization details of HA-Halo-KDELR and EEA1. Pearson’s coefficient was used to quantify the colocalization between HA-Halo-KDELR and EEA1. Statistical analysis was performed using two-way ANOVA with a Tukey's post-hoc test for multiple comparisons. Scale bar: 5 µm. ** p < 0.01, **** p < 0.0001, #p < 0.0001, ##p < 0.01, ###p < 0.001
Endocytosed KDELR1 enters the Golgi and is recycled to the endosomal compartments or the PM via clathrin-mediated transport carriers
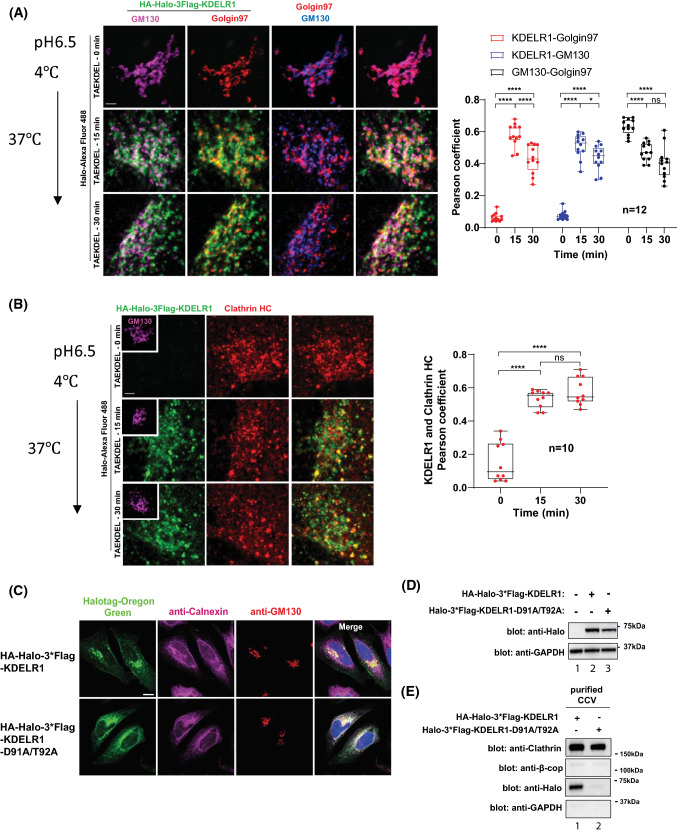
As KDELR1 is normally a Golgi-localized protein, we hypothesized that bulk of endocytosed KDELR1 would be found at the Golgi after ~30 min. However, we found that endocytosed HaloTag-Alexa488 labeled KDELR1 only partially colocalized with both cis-Golgi marker, GM130, and a TGN marker, Golgin97 (Fig. 5a) at that time-point.
Fig. 5.
A fraction of endocytosed KDELR enters the Golgi and is recycled to the endosomal compartments or the PM via clathrin-mediated transport carriers. a Co-localization of endocytosed KDELR with the Golgi apparatus was investigated by Leica confocal microscope in HeLa cells treated with 50 μM TAEKDEL peptide for indicated times and costained with GM130 (cis-Golgi) and Golgin97 (trans-Golgi). n = 12, Statistical analysis was performed using one-way ANOVA with a Tukey's post-hoc test. Scale bars: 2 µm. b Co-localization of endocytosed KDELR with endogenous clathrin in HeLa cells treated with 50 μM TAEKDEL peptide for indicated times was examined by Leica confocal microscopy using antibody against clathrin heavy chain. n = 10, Statistical analysis was performed using one-way ANOVA with a Tukey's post-hoc test. Scale bars: 2 µm. c HeLa cells transfected with HA-Halo-KDELR1 or Halo-KDELR1 D91A/T92A were stained with membrane permeable-Halotag Oregon Green, anti-calnexin antibody, anti-GM130 antibody, and DAPI. Confocal images were obtained by Zeiss LSM880. Scale bar: 5 µm. d The protein expression of KDELR1 WT and D91A/T92A mutant was analyzed by immunoblotting of HeLa cells transfected with HA-Halo-KDELR1 or HA-Halo-KDELR1 D91A/T92A. Endogenous GAPDH was detected as a control. e HeLa cells were transfected with HA-Halo-KDELR1 or HA-Halo-KDELR1 D91A/T92A for 18 h and then collected to purify clathrin-coated vesicles (CCV) according to a published protocol (23). CCVs were further analyzed by SDS-PAGE and western blotting with specific antibodies against Halo, clathrin heavy chain, β-COP and GAPDH. HA-Halo-KDELR1, but not Halo-KDELR1 D91A/T92A, was detected in CCVs. * p < 0.05, **** p < 0.0001, ns not significant
Because it is possible that endocytosed KDELR1 may be recycled back from the Golgi to the endosomal compartments or the PM, we stained the cells with anti-clathrin antibody to see whether clathrin-dependent transport carriers may be involved in rapid recycling of endocytosed KDELR1 from the Golgi. The confocal results clearly indicated that there is a significant overlap between endocytosed KDELR1 from the cell surface and clathrin in peri-nuclear area, which was positive for a cis-Golgi marker, GM130 (Fig. 5b; inset in magenta color). Taken together, these results show that endocytosed KDELR1 may be recycled using clathrin-mediated transport carriers from the TGN en route to the endosomal compartments or the PM.
HA-Halo-KDELR1, but not the ER-retained mutant receptor, is found in purified clathrin-coated vesicles
To further corroborate this finding biochemically, we decided to examine purified clathrin-coated vesicles (CCV) from HeLa cells to see whether HA-Halo-KDELR1 is found in the purified CCV-enriched fraction. To this end, we transiently transfected HeLa cells with either wt HA-Halo-KDELR1 or HA-Halo-KDELR1.D91A/T92A mutant constructs, which had been shown to cause strict ER retention of the receptor [11]. This mutant construct was used as a negative control, as inclusion in CCVs requires that KDELR exits the ER and reaches the Golgi. To confirm this previous finding by others, we prepared the D91A/T92A mutant construct and transiently transfected HeLa cells with either wt or the mutant HA-Halo-KDELR1 plasmids for 18 h. After staining the receptors with membrane permeable Halotag-oregon green dye, we examined whether the mutant receptor is indeed retained in the ER by confocal microscope.
The results confirmed that the D91A/T92A mutant receptor was found mostly in the calnexin-positive ER, whereas the wt KDELR1 localized mostly to GM130-positive Golgi and the PM (Fig. 5c). After further checking their expression and size by western blot (Fig. 5d), we carried out CCV purification, according to a published protocol to purify CCVs from HeLa cells [22, 23]. Purified CCVs were then analyzed by western blots for enrichment of clathrin, KDELR1 (Halo-tag) and GAPDH (a negative control). As expected, clathrin (but not β-COP) were highly enriched in CCVs, purified from both the wt and the mutant receptor transfected HeLa cells. Strikingly, only wt HA-Halo-KDELR1 was detected, whereas the D91A/T92A mutant KDELR1 was not found in the purified CCV fraction (Fig. 5e), suggesting that KDELR1 is indeed included in CCVs, isolated from HeLa cells.
Endocytosed KDELR is recycled to the PM via Rab14- and Rab11-positive recycling endosomes
Since only a fraction of endocytosed KDELR1 overlapped with the Golgi and peri-nuclear clathrin, we reasoned that a major portion of endocytosed KDELR1 may go through recycling endosomes, before they are transported back to the PM. To test this hypothesis, we co-transfected HA-Halo-KDELR1 with mCherry-Rab11 or mCherry-Rab4 or mCherry-Rab14, which represent slow (Rab11), intermediate (Rab14) and fast (Rab4) peri-nuclear recycling pathways, respectively [24, 25]. After temperature shift protocol, we examined the cells for the colocalization between endocytosed KDELR1 and the indicated Rab-GTPases by confocal microscopy. The results showed that endocytosed wt KDELR1 showed significantly increased colocalization with mCherry-Rab11 upon addition of KDEL peptide at both 15 and 30 min time-points, compared to the H12A mutant receptor (Fig. 6a; also see supplementary Table 1). Similar increase in colocalization index was observed for Rab14, suggesting that both Rab11 and Rab14-positive recycling endosomes are likely to be involved in recycling of endocytosed KDELR1 upon ligand binding (Fig. 6a, b). Rab4-positive recycling endosomes showed statistically meaningful colocalization index with endocytosed KDELR1 only at 30 min time-points, suggesting that Rab4-positive recycling endosomes are relatively less utilized by endocytosed KDELR1, compared to Rab11 and Rab14-recycling endosomes (0.43 ± 0.12 for wt KDELR1 vs. 0.27 ± 0.05 for H12A KDELR1) (Fig. 6c; supplementary Table 1 ). These results indicate that endocytosed KDELR1 undergoes a highly complex itinerary through the endolysosomal compartments for its recycling back to the PM (Fig. 6a–c).
Fig. 6.
Endocytosed KDELR is recycled via peri-nuclear recycling pathways. Colocalization of endocytosed Halo-KDELR and mCherry-tagged Rab11 (a), Rab14 (b) and Rab4 (c) was determined using confocal microscopy. MCF7 KDELR KD cells transfected with Halo-KDELR or Halo-KDELR H12A and mCherry-tagged Rab 11 (a), Rab14 (b), or Rab4 (c) were incubated with HaloTag Alexa Fluor 488 ligand and TAEKDEL peptide in DMEM (pH 6.5) with 10% FBS at 4 °C for 30 min, followed by washing with PBS and incubated at 37 °C for 0, 15, or 30 min. Cells were then fixed and observed using Zeiss LSM880. Line graphs indicate the colocalization of Halo-KDELR (WT or H12A) and Rab proteins (n = 20 cells). Statistical analysis was performed using two-way ANOVA with a Tukey's post-hoc test for multiple comparisons. Scale bars: 10 µm. * p < 0.05, ** p < 0.01, **** p < 0.0001, #p < 0.0001, ns not significant
Discussion
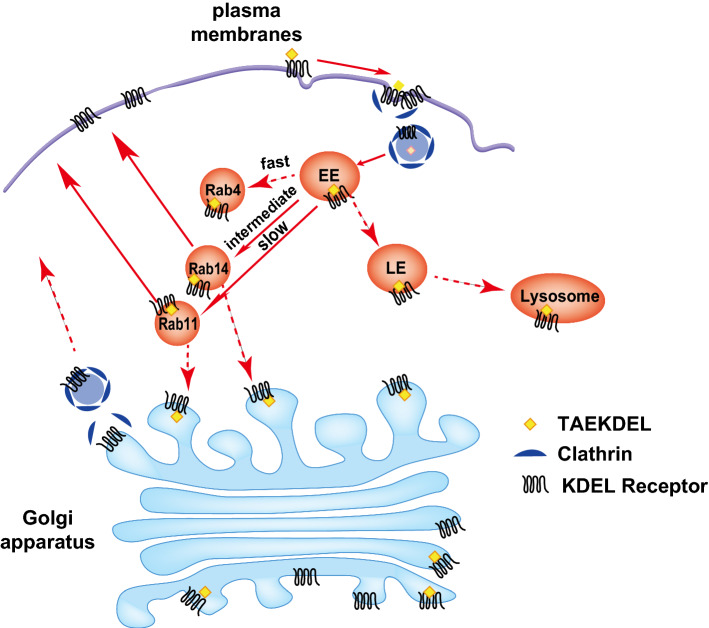
In this study, we demonstrated that surface-expressed KDELR1 undergo accelerated endocytosis upon KDEL ligand binding. Endocytosed KDELR1 is then subjected to recycling through the Golgi apparatus as well as endolysosomal compartments via (1) early endosome (EE); (2) Rab14-positive intermediate recycling pathway; (3) Rab11-positive slow recycling pathway (Fig. 7). We also observed intracellular trafficking of KDELR via clathrin-mediated transport carriers both at the PM and at the Golgi. Interestingly, endocytosed KDELR1 did not significantly co-localized with Rab7-positive late endosomes (LE) and the lysosomes, suggesting that only a negligible amount of endocytosed KDELR1 may be subjected to degradation in the lysosomes, even when the receptor is bound to KDEL ligands. Nonetheless, we cannot exclude a possibility that this negligible amount of lysosome-targeted KDELR fraction may be relevant to regulation of potential KDELR-mediated signaling from the cell surface.
Fig. 7.
Schematic illustration that depicts intracellular itinerary of surface-expressed KDELR. Our results indicate that surface-expressed KDELR undergoes clathrin-mediated endocytosis, which could be further accelerated upon binding of KDEL ligand to the receptor. Endocytosed KDELR then travels to early endosomes (EE), where initial sorting of the endocytosed receptor seems to occur. From the EE, bulk of endocytosed KDELR seems to undergo recycling through peri-nuclear recycling pathways via Rab14- and Rab11-positive recycling endosomes. A fraction of endocytosed KDELR travels to the Golgi and is rapidly recycled to the endosomal compartments or the PM via clathrin-mediated transport carriers at the TGN. It is currently unknown what fraction of endocytosed KDELR in the Rab14- and Rab11-positive endosomes may transit through the Golgi, prior to recycling to the cell surface
Binding of KDEL ligand or RTDL peptide from MANF to surface-expressed KDELR1 significantly enhanced the receptor endocytosis and its transport to the early endosomes, suggesting that ligand binding (i.e., secreted ER-chaperones or MANF, etc.) may play a pivotal role in signaling and/or sorting of endocytosed KDELR. Because cytoplasmic domains of KDELR lack any clathrin adaptor binding motifs, it is currently unclear how KDELR utilize clathrin-mediated endocytosis and transport carriers at the PM and the Golgi, respectively. It is possible that KDELR may utilize ubiquitination of its loops or tail for endocytosis from the PM, as it is now known that certain GPCRs may use ubiquitination of their intracellular loops or the C-terminal tail for clathrin-mediated endocytosis [26, 27]. Further studies are required to shed light on this subject.
Although not quite identical in its trafficking itinerary, surface-expressed KDELR is reminiscent of cation-independent mannose-6-phosphate receptor (CI-MPR), which mediates delivery of lysosomal hydrolases to the lysosome, but was later found to be a cell surface receptor for insulin-like growth factor-2 (IGF-2) [28–30]. Both KDELR and CI-MPR typically reside at the Golgi as a cargo receptor and had been shown later to function as signaling proteins. As KDELR had been shown to be involved in activation of autophagy in neuronal cell lines and ER stress response [31, 32], it is plausible to posit that signaling by surface-expressed KDELR may also be a part of inter-organelle communication, which maintains cellular homeostasis.
Interestingly, a recent study has reported that activation of KDELR signaling at the Golgi in response to cargo secretion is closely linked to lysosome repositioning toward the Golgi and autophagy-dependent modulation of lipid droplet turnover [33], raising a possibility that more broad inter-organelle communication encompassing most major organelles (including the PM) may exist in mammalian cells.
In summary, in contradiction to the prevalent view in the field, we propose that KDELR is likely a bona fide cell surface receptor with a well-defined trafficking itinerary, which contributes to the maintenance of cellular homeostasis using inputs from the extracellular environment.
Experimental procedures
Reagents and antibodies
HaloTag Alexa Fluor 488, Oregon green and HaloTag TMR ligands were purchased from Promega. LysoTracker and MitoTracker were purchased from Thermo Fisher Scientific. All other reagents were purchased from Merck unless otherwise indicated.
The following antibodies were used: antibody against Halo was obtained from Promega; antibodies against β-actin, mCherry, clathrin, β-COP and caveolin were obtained from Abcam; antibody against GM130 was obtained from BD bioscience; antibodies against EGFR, Golgin-97 and EEA1 were obtained from Cell Signaling Technology. Anti-Rabbit Alexa Fluor 488 (A21441), Alexa Fluor 568 (A10042), Alexa Fluor 647 (A21245) and anti-Mouse Alexa Fluor 488 (A21200), Alexa Fluor 568 (A10037), Alexa Fluor 647 (A21236) for IF were obtained from ThermoFisher.
Cell culture and transfection
HeLa and MCF7 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Corning) supplemented with 10% fetal calf serum (FBS, Gibco) at 37 °C in 5% CO2. DMEM (pH 6.5) was prepared from DMEM powder (high glucose, pyruvate, Thermo Fisher Scientific) in 3.7 g/L sodium bicarbonate and adjusted its pH to 6.5 with hydrochloric acid.
For gene overexpression experiments, HeLa and MCF7 cells were transfected with Halo-tagged KDELR and mCherry-tagged clathrin light chain, Rab4, Rab7, Rab11, or Rab14 using Lipofectamine 3000 (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions. The H12A mutation in KDELR was generated by PCR with Phusion High-Fidelity DNA Polymerase (New England Biolabs).
Generating KDELR1 stable knockdown MCF7 cell lines
The stable knockdown of KDELR1 was achieved by infecting MCF7 cells using lentivirus expression of KDELR1-shRNA (GGTTGCCAAACACTAAATCTG, targeting 3′-UTR). The lentivirus was packaged and commercially provided by Shanghai GenePharma, China. Cells were infected with the lentivirus expressing KDELR1-shRNA using Polybrene (Sigma) overnight. Two days after infection, the cells were cultured in puromycin (0.3–1 µg/ml, ThermoFisher) for two weeks.
Cell surface biotinylation
Cells grown in 6-well plates to 80% confluency were transfected using 1 μg plasmid DNA and 2 μl Lipofectamine 2000 for 18 h. During the biotinylation procedure, all reagents and cell cultures were kept on ice. Cells were washed twice in ice-cold PBS and subsequently incubated in 1 ml/well of a 1 mM Sulfo-NHS-LC-Biotin (APExBIO) in PBS solution for 30 min on ice. The cells were then washed in quenching buffer (100 mM glycine in PBS), and incubated in 1 ml/well of quenching buffer for 15 min on ice. The cells were washed twice with PBS and then lysed in 300 μl 95 °C 1% SDS and sonicated for 20 s. Finally, the lysate was centrifuged for 10 min at 15,000×g. An equal volume of PBS was added in the supernatants and then were incubated with 30 μl of Streptavidin Agarose beads (S1638, Sigma Aldrich) with constant rocking for 1 h at RT. The samples were washed three times with PBS, then eluted with 2× SDS-sample buffer for 10 min at 95 °C and used for Western blot.
CRISPR/Cas9-mediated knock-in of 3×Flag-mCherry and 3×Flag-Halo fusion proteins at KDELR1 loci
We used the CRISPR/Cas9 system to create knock-in cell lines stably expressing C-terminal 3×Flag-mCherry or 3×Flag-Halo fusion proteins at KDELR1 locus. The pX330 plasmid with the KDELR1 single guide RNA (sgRNA) was a kind gift from Francesca Botanelli. Design of the guide RNAs was carried out using the CRISPR Design Tool from the Zhang lab website (https://crispr.mit.edu) to minimize potential off-target effects. The KDELR1 genomic locus (Gene ID 10945) was targeted with the following guide RNA: 5′-GAGAGAGATGGAGAGGACCG-3′ located just after the stop codon in the KDELR1 coding sequence. The guide RNA was encoded in bicistronic expression plasmids pX330 (addgene plasmid #42230). The homologous repair plasmid for genome editing was generated using these four PCR products: the pEGFP-N1 plasmid backbone, the left and right homology arms (~1000 bp), and the reporter/selection cassette. The left homology arm fusing with 3×Flag-mCherry or 3×Flag-Hao were synthesized (Genscript) and subcloned into pEGFP-N1 using AseI and NotI. Then the right homology arm was synthesized and subcloned after the Neor/Kanr resistance cassette to integrate the whole cassette and allow selection of positive recombinants with the drug G418/Geneticin (Thermo Fisher Scientific). The PAM site was mutagenized to avoid re-cutting by the Cas9. A glycin/serin rich linker was added (GSSGRDPGSGSG) before 3×Flag-mCherry or 3×Flag-Halo.
The pX330 plasmid with the KDELR1 guide and the the corresponding homologous recombinationplasmid were co-transfected in HeLa cells using Lipofectamine 2000. G418 was added to the cells a week after transfection. After two weeks of selection, cells were subjected to single cell sorting into 96-well plates by dilution. Clones were genotyped via western blot and PCR. Two oligos (KDELR1-forward 5′-CATTTCGAGGGCTTCTTCGACCTCATC-3′ and KDELR1-reverse 5′-GTCACCCCTGGATGGGAAAGCTCTTCA-3′) were used to amplify the genomic region around the cut and then to genotype the single clones.
Immunoblotting
Cells were lysed in lysis buffer (25 mM HEPES, pH 7.5, 150 mM NaCl, 10 mM MgCl2·6H2O, 0.1 mM EDTA, 1% NP-40, 1× protease inhibitor cocktail (Roche)) at 4 °C. Crude lysates were subjected to centrifugation at 15,000×g for 15 min at 4 °C. Supernatants were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto a nitrocellulose membrane (Merck). Proteins on the membrane were probed with specific primary antibodies and then with peroxidase-conjugated secondary antibodies. The protein bands were visualized with chemiluminescence (ECL, Bio-Rad) on a ChemiDoc Imagin System (Bio-Rad). Representative blots are shown from several experiments.
RNA isolation and Realtime-PCR
Total RNA from cultured cells was extracted with RNA Isolation kit (Beyotime), cDNA synthesis was carried out using HiScript II Q RT SuperMix (Vazyme). Realtime-PCR was performed using ChamQTM Universal SYBR qPCR Master Mix (Vazyme) and a QuantStudio 3 real-time PCR system (ThermoFisher). GAPDH mRNA was used for normalization. The relative expression of each examined gene was determined with triplicate independent experiments. Primers are listed as follows: KDELR1 forward: AGCCACTACTTGTTTGCGCTA, KDELR1 reverse: CCTGCCACAATGGCGATGA; KDELR2 forward: GCACTGGTCTTCACAACTCGT, KDELR2 reverse: AGATCAGGTACACTGTGGCATA; KDELR3 forward: TCCCAGTCATTGGCCTTTCC, KDELR3 reverse: CCAGTTAGCCAGGTAGAGTGC; GAPDH forward: ACCACAGTCCATGCCATCAC, GAPDH reverse: TCCACCACCCTGTTGCTGTA.
Immunofluorescence staining and confocal microscopy
HeLa or MCF7 cells grown to on glass coverslips were fixed in 4% paraformaldehyde for 10 min, permeabilized with PBS containing 0.5% Triton X-100 for 10 min, blocked in blocking buffer (PBS containing 0.05% Triton X-100 and 2% BSA) for 30 min. Then cells were incubated with primary and secondary antibodies and examined using Zeiss LSM880 with a 63× oil immersion objective or Leica TCS SP8 confocal or Nikon TIRF. For staining of Halo-tagged proteins, HaloTag Alexa Fluor 488 and TMR ligands were directly added to the growth medium of the living cells before fixation in accordance with the manufacturer’s instructions.
Live cell imaging
HeLa cells were seeded on a glass-bottom dish (35-mm diameter, In Vitro Scientific) coated with fibronectin (Millipore). After 18-h co-transfection with HA-Halo-KDELR1 and mCherry-Clathrin light chain, cells were first incubated with a cell-impermeant HaloTag® Alexa Fluor® 488 ligand at 4 °C for 30 min, then after three washes with PBS, the cells were incubated with 50 μM TAEAAAA or TAEKDEL peptide in pH 6.5 live-cell imaging solution (Thermo). Images were acquired immediately with a 63× objective on a Zeiss LSM 880 Airyscan confocal microscope in an atmosphere of 5% CO2 at 37 °C. Images were acquired every 5 s for 30 min.
Image processing and statistical analysis
Pearson coefficient was analyzed by Fiji software. Results are displayed as mean ± SD (standard deviation) of results from each experiment or dataset, as indicated in figure legends. Analyses were performed with GraphPad Prism 8.0 software. Statistical analysis was performed using one-way or two-way ANOVA with a Tukey's post-hoc test for multiple comparisons. N (number of individual experiments) is noted in the figure legends.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary file1. Supplementary Fig. 1(A) KDELR1 knockdown efficiency in MCF7 KDELR KD cells was determined by quantitative real-time PCR in three independent experiments and summarized by histogram. KDELR2 and KDELR3 mRNA levels were also determined as controls. Y axis indicates the relative mRNA level of KDEL receptor in MCF7 KDELR KD cells compared to MCF7 control cells. **: P < 0.01 (B) KDELR1 protein knockdown efficiency by siRNA was determined. KDELR1-3Flag-mCherry knock-in cells were transfected with scramble or KDELR1 siRNA for 72 h before lysed and analyzed by western blotting with anti-mCherry and anti-GAPDH antibodies. Supplementary Fig. 2(H) Confocal images of overexpressed Halo-KDELR or Halo-KDELR H12A in the Golgi region of HeLa cells. Endogenous GM130 was stained as a Golgi marker. Scale bar: 1 µm. Bar graph summarize the co-localization of Halo-KDELRs with GM130 (n = 15 cells). ns not significant. Statistical analysis was performed using Students’ t test. Supplementary Table 1. Summary of the Pearson coefficient values (mean ± SD) in the graphs (PDF 134 kb)
Author contributions
IL, XY, YQ designed the study. JJ, XY, YQ, LZ, BG, SJ, YW performed the experiments. JJ, XY, YQ and IL wrote the manuscript.
Funding
This project was sponsored by Shanghai Pujiang Program (16PJ1407 to YQ). Financial support of this study was provided exclusively by the ShanghaiTech University.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jie Jia and Xihua Yue contributed equally to this work.
Contributor Information
Yi Qian, Email: Qianyi@shanghaitech.edu.cn.
Intaek Lee, Email: Leeintaek@shanghaitech.edu.cn.
References
- 1.Munro S, Pelham HR. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 2.Lewis MJ, Pelham HR. A human homologue of the yeast HDEL receptor. Nature. 1990;348:162–163. doi: 10.1038/348162a0. [DOI] [PubMed] [Google Scholar]
- 3.Lewis MJ, Sweet DJ, Pelham HR. The ERD2 gene determines the specificity of the luminal ER protein retention system. Cell. 1990;61:1359–1363. doi: 10.1016/0092-8674(90)90699-F. [DOI] [PubMed] [Google Scholar]
- 4.Pulvirenti T, et al. A traffic-activated Golgi-based signalling circuit coordinates the secretory pathway. Nat Cell Biol. 2008;10:912–922. doi: 10.1038/ncb1751. [DOI] [PubMed] [Google Scholar]
- 5.Capitani M, Sallese M. The KDEL receptor: new functions for an old protein. FEBS Lett. 2009;583:3863–3871. doi: 10.1016/j.febslet.2009.10.053. [DOI] [PubMed] [Google Scholar]
- 6.Giannotta M, et al. The KDEL receptor couples to Galphaq/11 to activate Src kinases and regulate transport through the Golgi. EMBO J. 2012;31:2869–2881. doi: 10.1038/emboj.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solis GP, et al. Golgi-resident Galphao promotes protrusive membrane dynamics. Cell. 2017;170:939–955. doi: 10.1016/j.cell.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 8.Cancino J, et al. Control systems of membrane transport at the interface between the endoplasmic reticulum and the Golgi. Dev Cell. 2014;30:280–294. doi: 10.1016/j.devcel.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 9.Brauer P, et al. Structural basis for pH-dependent retrieval of ER proteins from the Golgi by the KDEL receptor. Science. 2019;363:1103–1107. doi: 10.1126/science.aaw2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis MJ, Pelham HR. Ligand-induced redistribution of a human KDEL receptor from the Golgi complex to the endoplasmic reticulum. Cell. 1992;68:353–364. doi: 10.1016/0092-8674(92)90476-S. [DOI] [PubMed] [Google Scholar]
- 11.Townsley FM, Wilson DW, Pelham HR. Mutational analysis of the human KDEL receptor: distinct structural requirements for Golgi retention, ligand binding and retrograde transport. EMBO J. 1993;12:2821–2829. doi: 10.1002/j.1460-2075.1993.tb05943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson DW, Lewis MJ, Pelham HR. pH-dependent binding of KDEL to its receptor in vitro. J Biol Chem. 1993;268:7465–7468. doi: 10.1016/S0021-9258(18)53197-5. [DOI] [PubMed] [Google Scholar]
- 13.Aoe T, et al. The KDEL receptor, ERD2, regulates intracellular traffic by recruiting a GTPase-activating protein for ARF1. EMBO J. 1997;16:7305–7316. doi: 10.1093/emboj/16.24.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths G, et al. Localization of the Lys, Asp, Glu, Leu tetrapeptide receptor to the Golgi complex and the intermediate compartment in mammalian cells. J Cell Biol. 1994;127:1557–1574. doi: 10.1083/jcb.127.6.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raykhel I, et al. A molecular specificity code for the three mammalian KDEL receptors. J Cell Biol. 2007;179:1193–1204. doi: 10.1083/jcb.200705180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson MJ, Richie CT, Airavaara M, Wang Y, Harvey BK. Mesencephalic astrocyte-derived neurotrophic factor (MANF) secretion and cell surface binding are modulated by KDEL receptors. J Biol Chem. 2013;288:4209–4225. doi: 10.1074/jbc.M112.400648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindahl M, et al. MANF is indispensable for the proliferation and survival of pancreatic beta cells. Cell Rep. 2014;7:366–375. doi: 10.1016/j.celrep.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker B, et al. Cargo binding promotes KDEL receptor clustering at the mammalian cell surface. Sci Rep. 2016;6:28940. doi: 10.1038/srep28940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neves J, et al. Immune modulation by MANF promotes tissue repair and regenerative success in the retina. Science. 2016;353:aaf3646. doi: 10.1126/science.aaf3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norisada J, Hirata Y, Amaya F, Kiuchi K, Oh-hashi K. A comparative analysis of the molecular features of MANF and CDNF. PLoS ONE. 2016;11:e0146923. doi: 10.1371/journal.pone.0146923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindahl M, Saarma M, Lindholm P. Unconventional neurotrophic factors CDNF and MANF: structure, physiological functions and therapeutic potential. Neurobiol Dis. 2017;97:90–102. doi: 10.1016/j.nbd.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Borner GH, et al. Multivariate proteomic profiling identifies novel accessory proteins of coated vesicles. J Cell Biol. 2012;197:141–160. doi: 10.1083/jcb.201111049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borner GH, Fielding AB. Isolating HeLa cell fractions enriched for clathrin-coated vesicles. Cold Spring Harb Protoc. 2014;2014:1184–1187. doi: 10.1101/pdb.prot083147. [DOI] [PubMed] [Google Scholar]
- 24.Linford A, et al. Rab14 and its exchange factor FAM116 link endocytic recycling and adherens junction stability in migrating cells. Dev Cell. 2012;22:952–966. doi: 10.1016/j.devcel.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koike S, Jahn R. SNAREs define targeting specificity of trafficking vesicles by combinatorial interaction with tethering factors. Nat Commun. 2019;10:1608. doi: 10.1038/s41467-019-09617-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dores MR, Schnell JD, Maldonado-Baez L, Wendland B, Hicke L. The function of yeast epsin and Ede1 ubiquitin-binding domains during receptor internalization. Traffic. 2010;11:151–160. doi: 10.1111/j.1600-0854.2009.01003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dores MR, Trejo J. Endo-lysosomal sorting of G-protein-coupled receptors by ubiquitin: diverse pathways for G-protein-coupled receptor destruction and beyond. Traffic. 2019;20:101–109. doi: 10.1111/tra.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hari J, et al. The receptor for insulin-like growth factor II mediates an insulin-like response. EMBO J. 1987;6:3367–3371. doi: 10.1002/j.1460-2075.1987.tb02658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgan DO, et al. Insulin-like growth factor II receptor as a multifunctional binding protein. Nature. 1987;329:301–307. doi: 10.1038/329301a0. [DOI] [PubMed] [Google Scholar]
- 30.Roth RA, et al. Interactions of the receptor for insulin-like growth factor II with mannose-6-phosphate and antibodies to the mannose-6-phosphate receptor. Biochem Biophys Res Commun. 1987;149:600–606. doi: 10.1016/0006-291X(87)90410-4. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto K, et al. The KDEL receptor modulates the endoplasmic reticulum stress response through mitogen-activated protein kinase signaling cascades. J Biol Chem. 2003;278:34525–34532. doi: 10.1074/jbc.M304188200. [DOI] [PubMed] [Google Scholar]
- 32.Wang P, Li B, Zhou L, Fei E, Wang G. The KDEL receptor induces autophagy to promote the clearance of neurodegenerative disease-related proteins. Neuroscience. 2011;190:43–55. doi: 10.1016/j.neuroscience.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 33.Tapia D, et al. KDEL receptor regulates secretion by lysosome relocation- and autophagy-dependent modulation of lipid-droplet turnover. Nat Commun. 2019;10:735. doi: 10.1038/s41467-019-08501-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1. Supplementary Fig. 1(A) KDELR1 knockdown efficiency in MCF7 KDELR KD cells was determined by quantitative real-time PCR in three independent experiments and summarized by histogram. KDELR2 and KDELR3 mRNA levels were also determined as controls. Y axis indicates the relative mRNA level of KDEL receptor in MCF7 KDELR KD cells compared to MCF7 control cells. **: P < 0.01 (B) KDELR1 protein knockdown efficiency by siRNA was determined. KDELR1-3Flag-mCherry knock-in cells were transfected with scramble or KDELR1 siRNA for 72 h before lysed and analyzed by western blotting with anti-mCherry and anti-GAPDH antibodies. Supplementary Fig. 2(H) Confocal images of overexpressed Halo-KDELR or Halo-KDELR H12A in the Golgi region of HeLa cells. Endogenous GM130 was stained as a Golgi marker. Scale bar: 1 µm. Bar graph summarize the co-localization of Halo-KDELRs with GM130 (n = 15 cells). ns not significant. Statistical analysis was performed using Students’ t test. Supplementary Table 1. Summary of the Pearson coefficient values (mean ± SD) in the graphs (PDF 134 kb)