Abstract
Telomerase-free cancer cells employ a recombination-based alternative lengthening of telomeres (ALT) pathway that depends on ALT-associated promyelocytic leukemia nuclear bodies (APBs), whose function is unclear. We find that APBs behave as liquid condensates in response to telomere DNA damage, suggesting two potential functions: condensation to enrich DNA repair factors and coalescence to cluster telomeres. To test these models, we developed a chemically induced dimerization approach to induce de novo APB condensation in live cells without DNA damage. We show that telomere-binding protein sumoylation nucleates APB condensation via interactions between small ubiquitin-like modifier (SUMO) and SUMO interaction motif (SIM), and that APB coalescence drives telomere clustering. The induced APBs lack DNA repair factors, indicating that APB functions in promoting telomere clustering can be uncoupled from enriching DNA repair factors. Indeed, telomere clustering relies only on liquid properties of the condensate, as an alternative condensation chemistry also induces clustering independent of sumoylation. Our findings introduce a chemical dimerization approach to manipulate phase separation and demonstrate how the material properties and chemical composition of APBs independently contribute to ALT, suggesting a general framework for how chromatin condensates promote cellular functions.
INTRODUCTION
Telomeres are repetitive sequences at chromosome ends that shorten with each cell cycle in cells that lack a telomere maintenance mechanism. Critical telomere shortening induces replicative senescence or apoptosis (Harley et al., 1990), whereas cancer cells maintain replicative potential by actively elongating their telomeres. The majority of human cancer cells reactivate the enzyme telomerase, but a significant fraction (10–15%) employ an alternative lengthening of telomeres (ALT) pathway that involves DNA recombination and repair to maintain telomere length (Dilley and Greenberg, 2015; Lazzerini-Denchi and Sfeir, 2016; Sobinoff and Pickett, 2017). The molecular mechanisms underlying ALT are unclear, but one unique characteristic is the presence of ALT telomere-associated promyelocytic leukemia nuclear bodies (APBs), a class of ALT telomere-associated promyelocytic leukemia (PML) nuclear bodies used for ALT diagnosis (Yeager et al., 1999). PML nuclear bodies are dynamic structures in the nucleus that transiently sequester up to 100 different proteins that are implicated in many cellular functions including tumor suppression, DNA replication, gene transcription, DNA repair, viral pathogenicity, cellular senescence, and apoptosis (Lallemand-Breitenbach and de The, 2010). Inhibiting APB formation by knocking down PML protein, an essential component of PML nuclear bodies, leads to telomere shortening (Draskovic et al., 2009; Osterwald et al., 2015; Loe et al., 2020), indicating that APBs contribute to ALT telomere maintenance. In addition to typical PML nuclear body components, APBs contain proteins involved in homologous recombination such as replication protein A (RPA), Rad51, and breast cancer susceptibility protein 1 (BRCA1) (Nabetani and Ishikawa, 2011), which suggests that APBs promote telomere synthesis. Indeed, new telomere DNA synthesis has been detected in APBs (Chung et al., 2011; Cho et al., 2014; O’Sullivan et al., 2014; Sahin et al., 2014; Zhang et al., 2019). While APBs are proposed to be sites of telomere recombination during ALT, the precise functions of these specialized PML nuclear bodies are poorly understood, and whether their formation requires a DNA damage response is unclear.
Telomeres cluster within APBs presumably to provide repair templates for telomere DNA synthesis. Many functionally distinct proteins can initiate APB assembly, leading to the proposal of a multiple-pathway model (Chung et al., 2011). This model is supported by an RNA interference screen that identified close to 30 proteins that affect APB formation, including proteins involved in telomere and chromatin organization, protein sumoylation, and DNA repair (Osterwald et al., 2015). Given such complexity, the mechanisms governing APB assembly and function remain unclear, and limitations include lack of a conceptual model for how they form and tools to manipulate the process for cell biological analyses. We previously showed that introducing DNA damage at telomeres leads to APB formation, telomere clustering within the induced APBs, and telomere elongation (Cho et al., 2014). While DNA damage from either replication stress or telomere DNA double strand breaks can trigger APB formation and telomere clustering (O’Sullivan et al., 2014; Cho et al., 2014), the physical mechanisms underlying telomere clustering within APBs are unknown.
Many nuclear bodies and membrane-free organelles—such as P granules, nucleoli, signaling complexes, and stress granules (Brangwynne et al., 2009, 2011; Altmeyer et al., 2015; Patel et al., 2015; Su et al., 2016)—assemble by liquid–liquid phase separation, in which proteins and/or nucleic acids separate from the surrounding milieu and form a condensed liquid phase (Banani et al., 2017). Components of these condensates are highly concentrated but can dynamically exchange with the diluted phase. Liquid phase separation provides a mechanism for organizing matter in cells, particularly protein interaction networks that do not form stable complexes with fixed stoichiometry. Notably, such stable complexes are relatively rare, and protein–protein interactions are dominated by weak interactions (Hein et al., 2015). In vitro reconstitution has provided valuable insights on how those weak interactions drive the condensation process, but little is known about how liquid phase separation promotes cellular functions.
Tools that can control liquid phase separation in live cells will allow new experiments probing cellular functions. Optogenetic approaches have been developed to control disordered proteins with light to map the phase diagrams and reveal how they restructure the genome (Shin et al., 2017, 2018). However, such tools rely on constant light illumination, limiting their utility for processes on longer time scales and application to biochemical assays that require a population of cells to be treated. Here we develop a chemically induced protein dimerization approach to control APB formation and demonstrate a decoupling of APB functions that rely on liquid material properties and chemical composition.
RESULTS
SUMO–SIM interactions drive APB liquid condensation to cluster telomeres
Previously, we introduced DNA damage on telomeres in ALT cells by fusing the FokI nuclease to the telomere-binding protein TRF1, which induced APB formation, telomere clustering within APBs, and telomere elongation (Cho et al., 2014). With this assay, we observed that APBs exhibit liquid behavior, including coalescence after colliding (Figure 1, A and B) and dynamic exchange of components within APBs and with the surrounding nucleoplasm, as shown by fluorescence recovery after photobleaching (Figure 1C). These phenomena are characteristics of liquid condensates formed by liquid–liquid phase separation, leading us to hypothesize that APBs are liquid droplets condensed on telomeres after DNA damage as a mechanism for telomere clustering and elongation. The liquid nature of APBs would promote telomere clustering via coalescence, and the condensates may serve as platforms to concentrate DNA repair factors to aid telomere synthesis. The switch-like self-assembly and disassembly of liquid droplets would allow APBs to rapidly nucleate as telomeres shorten and subsequently dissolve by reversing the nucleation signal.
FIGURE 1:
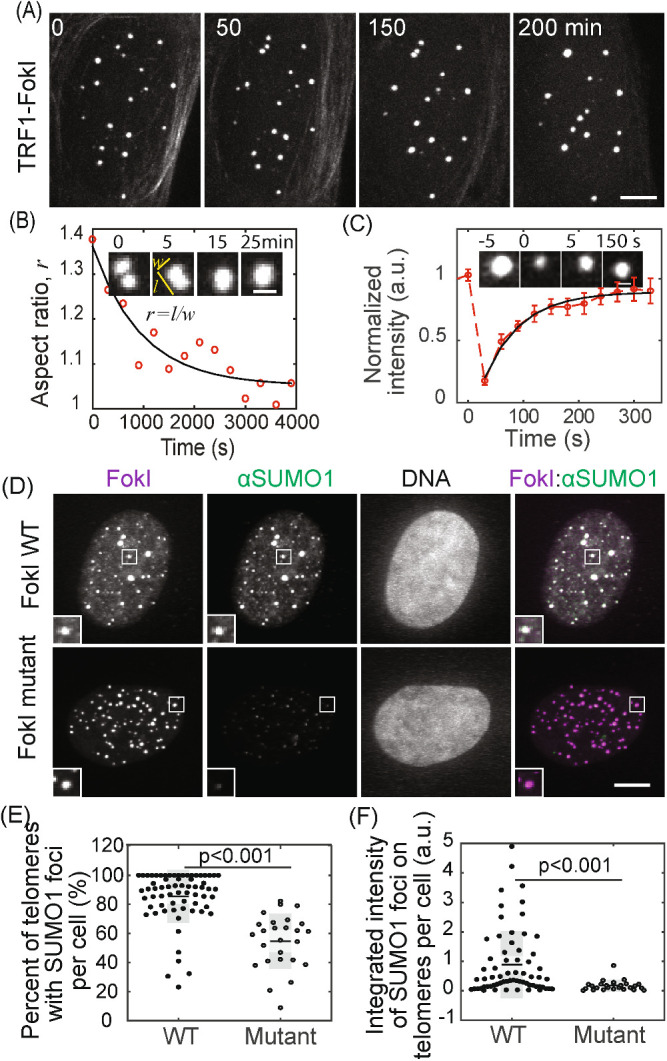
APBs exhibit liquid behavior and concentrate SUMO. APB formation was induced by creating DNA damage on telomeres with TRF1-FokI. (A, B) Cells were imaged live starting 1 h after triggering mCherry-TRF1-FokI import into the nucleus. Images show clustering of TRF1 foci (A) and fusion (B, insets), quantified by change in aspect ratio (defined as length/width) over time (exponential fit: 15 min half time). Time 0 is defined as the time point when two foci touch. (C) Fluorescence recovery after photobleaching (FRAP) of TRF1-FokI-mCherry, representing DNA damage-induced APBs. Insets shows a single TRF1 foci, intensity normalized to the first time point, exponential fit: 44 ± 17 s recovery half time from 14 events. Error bars STD. (D–F) SUMO1 IF for cells expressing TRF1-FokI or a nuclease-dead mutant. The overlay of FokI (purple) and SUMO1 (green) appears white (D, insets two times enlarged). Graphs show the percentage of telomeres with SUMO1 foci and the integrated intensity of SUMO1 foci on telomeres. Each data point represents one cell from two biological replicates, black lines mean, gray bars 95% confidence interval. Scale bars, 5 μm (A, D) or 1 μm (B, C). See also Figure 1–Supplemental Figure S1.
We considered the possibility that sumoylation of telomere-binding proteins (e.g., shelterin complex) triggers APB condensation, driven by multivalent SUMO–SIM interactions. Many APB components are SUMOylated, contain SIM domains, or both (Supplemental Table S1) (Shen et al., 2006; Potts and Yu, 2007; Chung et al., 2011; Shima et al., 2013), and sumoylation of telomere proteins is required for APB formation (Potts and Yu, 2007). Furthermore, synthetic SUMO and SIM peptides can drive liquid droplet formation in vitro (Banani et al., 2016). These findings are consistent with a model in which SUMO–SIM interactions on telomere-binding proteins cooperate during phase separation to drive telomere coalescence into APBs. DNA damage responses triggered by telomere shortening would be a stimulus to induce SUMOylation. Conversely, desumoylation after telomere elongation would lead to APB dissolution. Supporting this idea, we observed enrichment of both SUMO1 and SUMO2/3 after DNA damage induced with FokI, but not with a FokI mutant that lacks nuclease activity (Figure 1, D–F; Figure 1–Supplemental Figure S1).
To test the hypothesis that telomere sumoylation drives APB condensation via SUMO–SIM interactions, we developed a protein dimerization approach to induce de novo APB formation on telomeres without DNA damage. To mimic sumoylation on telomeres and avoid overexpressing SUMO, we recruited SIM to telomeres with a chemical inducer of dimerization. We predicted that SIM recruited to telomeres can bring endogenous SUMO to telomeres to induce APB condensation. The chemical dimerizer consists of two linked ligands: trimethoprim (TMP) and Haloligand, and can dimerize proteins fused to the cognate receptors: Escherichia coli dihydrofolate reductase (eDHFR) and a bacterial alkyldehalogenase enzyme (Haloenzyme), respectively (Figure 2A). An advantage of this system is that it is reversible by adding excess TMP to compete for eDHFR, unlike other chemically induced dimerization systems such as rapamycin (DeRose et al., 2013; Ballister et al., 2014). We fused Haloenzyme to the telomere-binding protein TRF1 to anchor it to telomeres and to GFP for visualization. SIM was fused to eDHFR and to mCherry. After adding the dimerizer to cells expressing Halo-GFP-TRF1 and SIM-mCherry-eDHFR, SIM was recruited to telomeres, which resulted in enrichment of both SUMO1 and SUMO2/3 on telomeres (Figure 2, B–D; Figure 2–Supplemental Figure S1). To confirm that enrichment of SUMO is indeed based on SUMO–SIM interaction, we used a SIM mutant that cannot interact with SUMO (Banani et al., 2016). As predicted, the SIM mutant was recruited to telomeres without SUMO enrichment. To confirm that the sites of SIM recruitment are telomeres, we used fluorescence in situ hybridization (FISH) to visualize telomere DNA directly and observed colocalization of SIM with telomere signal (Figure 2E).
FIGURE 2:
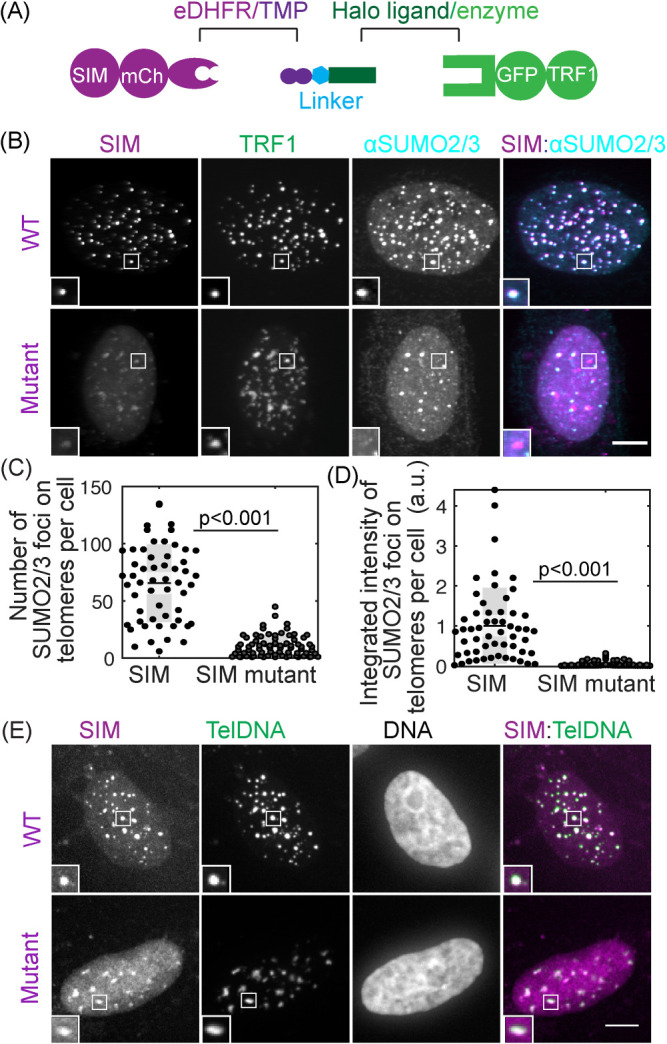
Recruiting SUMO to telomeres through SIM with a chemical dimerizer. (A) Dimerization schematic: SIM is fused to mCherry and eDHFR, and TRF1 is fused to Halo and GFP. The dimerizer is TNH: TMP-NVOC (6-nitroveratryl oxycarbonyl)-Halo (Zhang et al., 2017). (B–D) Cells expressing SIM-mCherry-DHFR (WT) or a SIM mutant that cannot interact with SUMO, together with Halo-GFP-TRF1, were incubated with TNH before fixing and staining for SUMO2/3. The overlay of SIM (purple) and SUMO2/3 (cyan) appears white (B, insets two times enlarged). Graphs show the number of telomeres with SUMO2/3 foci and the integrated intensity of SUMO2/3 foci on telomeres. Note that the integrated intensity of SUMO2/3 foci on telomeres in the SIM mutant is small compared with WT but not zero because of endogenous telomere sumoylation in U2OS cells. Each data represents one cell from two biological replicates, black lines mean, gray bars 95% confidence interval. (E) Telomere FISH images after recruiting SIM or SIM mutant to telomeres. The overlay of SIM (purple) and telomere DNA FISH (green) appears white. The dim SIM mutant foci on telomeres, relatively to the signal in the nucleoplasm, are due to the inability of the SIM mutant to enrich on telomeres through phase separation, combined with reduced protein fluorescent intensity in the FISH experiment. Scale bars, 5 μm. See also Figure 2–Supplemental Figure S1.
To directly test whether SIM recruitment leads to liquid condensation on telomeres, we used live imaging to monitor TRF1 and SIM signals over time (Supplemental Movie S1). We observed that after SIM recruitment, both SIM and TRF1 foci became brighter and bigger (Figure 3, A and B), as predicted for liquid droplet nucleation and growth. In addition, both SIM and TRF1 foci rounded up, indicating formation of liquid condensates. Such liquid behavior is also shown by fusion events and the dynamic exchange of components, similar to DNA damage-induced foci (Figure 3, D and E). Additionally, dimerization-induced condensates can be disrupted by 1,6-hexanediol and NaCl, similar to other membrane-free organelles (Figure 3–Supplemental Figure S1). Droplet fusion also drove telomere clustering, leading to reduced telomere number over time (Figure 3C). Although in previous studies we demonstrated that clustered telomeres were chromosomally attached (Cho et al., 2014), we cannot rule out a contribution from extrachromosomal telomere (ECTR) DNA that exists in ALT cells. In contrast, a SIM mutant that cannot interact with SUMO was recruited to telomeres after dimerization, but did not induce condensation or telomere clustering (Supplemental Movie S2; Figure 3–Supplemental Figure S2). Collectively, these findings support our hypothesis that condensation is driven by SUMO–SIM interactions.
FIGURE 3:
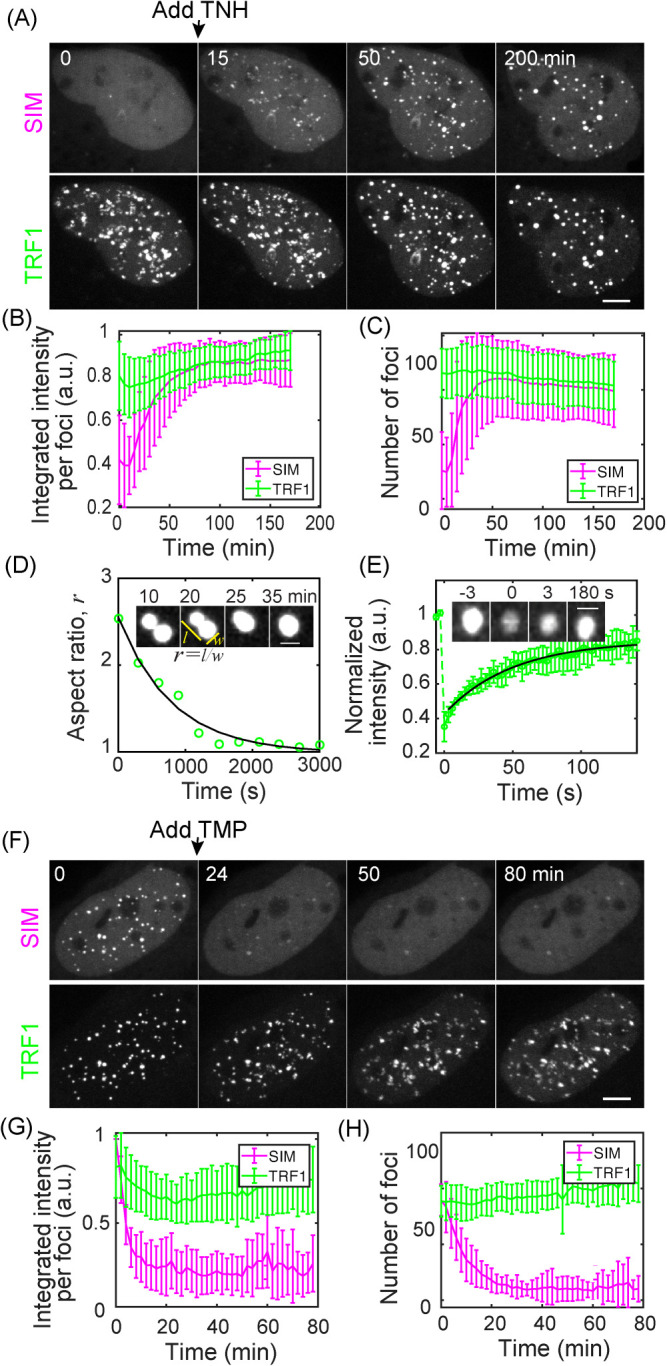
SUMO–SIM interactions drive liquid condensation and telomere clustering. (A–D) TNH was added to U2OS cells expressing SIM-mCherry-DHFR and Halo-GFP-TRF1 after the first time point to induce dimerization. Graphs show mean integrated intensity per TRF1 and SIM foci (B) and number of TRF1 and SIM foci (C) over time; 36 cells from four duplicates; error bars STD. P value between first and last time point for TRF1 foci intensity <0.001, SIM foci intensity <0.001, TRF1 foci number <0.03, SIM foci number <0.001. Insets (D) show an example of a fusion event, with the change in aspect ratio quantified (exponential fit, decay time 13 min). The time when two foci touch is defined as time 0. (E) FRAP of dimerization-induced condensates by bleaching TRF1. Intensity is normalized to the first time point, exponential fit: 35 ± 12 s recovery half time for 12 events. (F–H) After dimerization induced by TNH in U2OS cells expressing SIM-mCherry-DHFR and Halo-GFP-TRF1, TMP was added to release SIM from telomeres; 12 cells from two duplicates, error bars STD. P value between first and last time point for TRF1 foci intensity n.s., SIM foci intensity <0.001, TRF1 foci number <0.02, SIM foci number <0.001. Scale bars, 5 μm. See also Figure 3–Supplemental Figures S1 and S2.
Movie S1.
Recruit SIM to telomeres. Movie for Figure 3A. Left: SIM-mCherry-eDHFR, middle: Halo-GFP-TRF1, right: composite of SIM (magenta) and TRF1 (green). TNH was added to cells after the first time point to induce dimerization. Yellow box highlights a fusion event.
Movie S2.
Recruit SIM mutant to telomeres. Movie for Figure 3-figure supplement 1. Left: SIM mutant-mCherry-eDHFR, middle: TRF1-GFP-Halo, right: composite of SIM mutant (magenta) and TRF1 (green). TNH was added to cells after the first time point to induce dimerization.
Our phase transition model predicts that reversal of the nucleation signal will result in the dissolution of condensates. To test this prediction, we first formed condensates on telomeres by SIM recruitment and then added free TMP to compete with the dimerizer for eDHFR binding to reverse dimerization (Ballister et al., 2014). Condensation and telomere clustering were reversed as the intensity decreased in the foci while increasing in the nucleoplasm (Supplemental Movie S3; Figure 3, F and G) and telomere number increased (Figure 3H), consistent with our model.
Movie S3.
Release SIM from telomeres. Movie for Figure 3F. Left: SIM-mCherry-eDHFR, middle: Halo-GFP-TRF1, right: composite of SIM (magenta) and TRF1 (green). Cells were incubated with TNH to induce dimerization for 2 hours before imaging. TMP was added after the first time point to release SIM from telomeres.
Decoupling of APB functions
APB condensates could promote homology-directed telomere DNA synthesis in ALT by either or both of two mechanisms: 1) concentrating DNA repair factors on telomeres through APB condensation; 2) clustering telomeres for repair templates through APB coalescence. The first mechanism relies on compositional control of phase-separated condensates, while the second mechanism takes advantage of the liquid properties of biomolecular condensates.
To determine how APBs function, we first examined whether dimerization-induced condensates are APBs by observing PML protein, whose localization on telomeres defines APBs. Recruiting SIM to telomeres increased colocalization of PML with telomeres, compared with control cells where SIM was not recruited (Figure 4, A–C). Together with our previous findings that the dimerization-induced condensates contain other known components of APBs—SUMO (Figure 2, B–D; Figure 2–Supplemental Figure S1), telomere DNA (Figure 2E), and TRF1 (Figure 3)—this result indicates that the induced condensates are indeed APBs. Such an increase in PML localization to telomeres was not seen when the SIM mutant was recruited, agreeing with the hypothesis that SUMO–SIM interactions drive APB condensation. We then looked at proteins involved in the DNA damage response and repair pathways: 53BP1, PCNA, and POLD3, which localize to APBs induced by DNA damage (Cho et al., 2014; Dilley et al., 2016; Verma et al., 2018, 2019). None of these factors were recruited after dimerization-induced condensation (Figure 4, D–F; Figure 4–Supplemental Figure S1), indicating that they are recruited to the APB condensates via additional signals emanating from damaged DNA. These are predicted to include recessed three prime ends that arise at recombination intermediates as well as modified chromatin adjacent to the break site. Damage-induced sumoylation of DNA repair factors is also likely to contribute. Indeed, PCNA is enriched in SIM dimerization-induced condensates after fusing it to SUMO1 to mimic sumoylation (Figure 4–Supplemental Figure S2). The lack of DNA repair factors in dimerization-induced condensates suggests that telomere clustering in these condensates is not sufficient to assemble the protein complexes that are responsible for telomere DNA synthesis. Indeed, unlike FokI-induced DNA damage, nascent telomere DNA synthesis in telomere clusters was not observed after SIM dimerization (Figure 4–Supplemental Figure S3).
FIGURE 4:
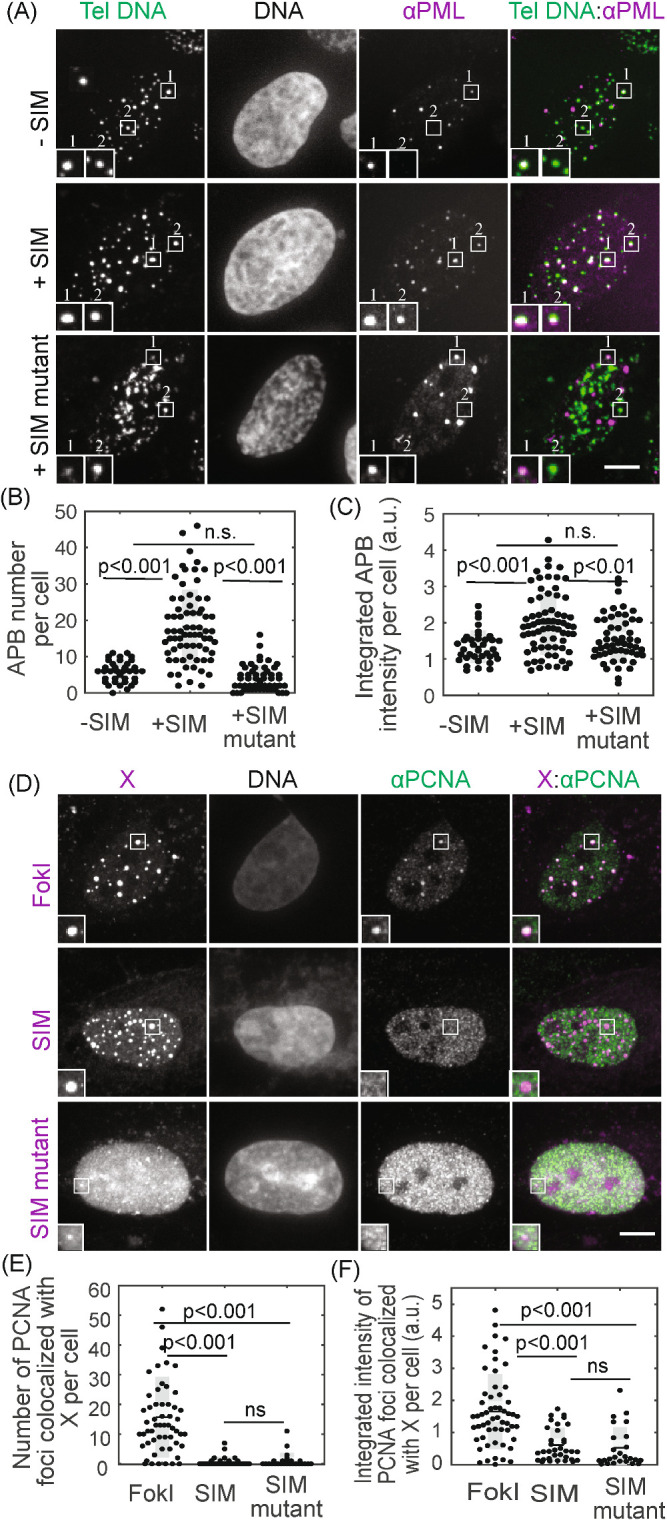
Condensates contain APB scaffold components but not DNA repair factors. (A–C) FISH of telomere DNA and IF of PML for cells with or without SIM recruited to telomeres or with SIM mutant recruited to telomeres. The overlay of PML (purple) and telomere DNA (green) appears white (A, insets two times enlarged), indicating APBs with PML nuclear bodies localized to telomeres. Graphs show APB number and integrated APB intensity per cell. (D–F) Immunofluorescence of PCNA for cells with FokI-induced damage or with SIM or SIM mutant recruited. In representative images (D, insets two times enlarged), X indicates FokI, SIM, or SIM mutant, and colocalization with PCNA appears white in overlay images (right panels). Graphs show the number of PCNA foci colocalized with FokI, SIM, or SIM mutant and integrated intensity. Each data point (B, C, E, F) represents one cell from two biological replicates, black line mean, gray bar 95% confidence interval. Scale bars, 5 μm. See also Figure 4–Supplemental Figures S1–S3.
Our model predicts that the ability to cluster telomeres relies on the liquid material properties of APBs and not the specific chemical composition. To test this prediction, we aimed to induce non-APB liquid droplets with a different chemistry on telomeres and determine whether they can cluster telomeres. Besides multivalent interactions between modular interaction pairs such as SUMO and SIM, another way of driving condensation is through interactions between disordered or low complexity protein domains that behave like flexible polymers (Elbaum-Garfinkle et al., 2015; Lin et al., 2015; Nott et al., 2015; Patel et al., 2015; Zhang et al., 2015). We selected the arginine/glycine-rich (RGG) domain from the P granule component LAF-1, which forms liquid condensates in vitro and in vivo (Elbaum-Garfinkle et al., 2015; Schuster et al., 2018). Recruiting RGG to telomeres resulted in condensation as shown by the increase in telomere foci intensity (Supplemental Movie S4; Figure 5, A and B). The induced condensates exhibited liquid behavior such as the ability to fuse, which led to telomere clustering as shown by the decrease in telomere foci over time (Figure 5, C and D). We also confirmed that the RGG condensates were indeed on telomeres and did not increase PML protein on telomeres compared with cells without RGG recruited (Figure 5, E–G), indicating the induced condensates are not APBs. These results support the model that liquid condensation drives telomere clustering independent of specific protein components of the condensates.
FIGURE 5:
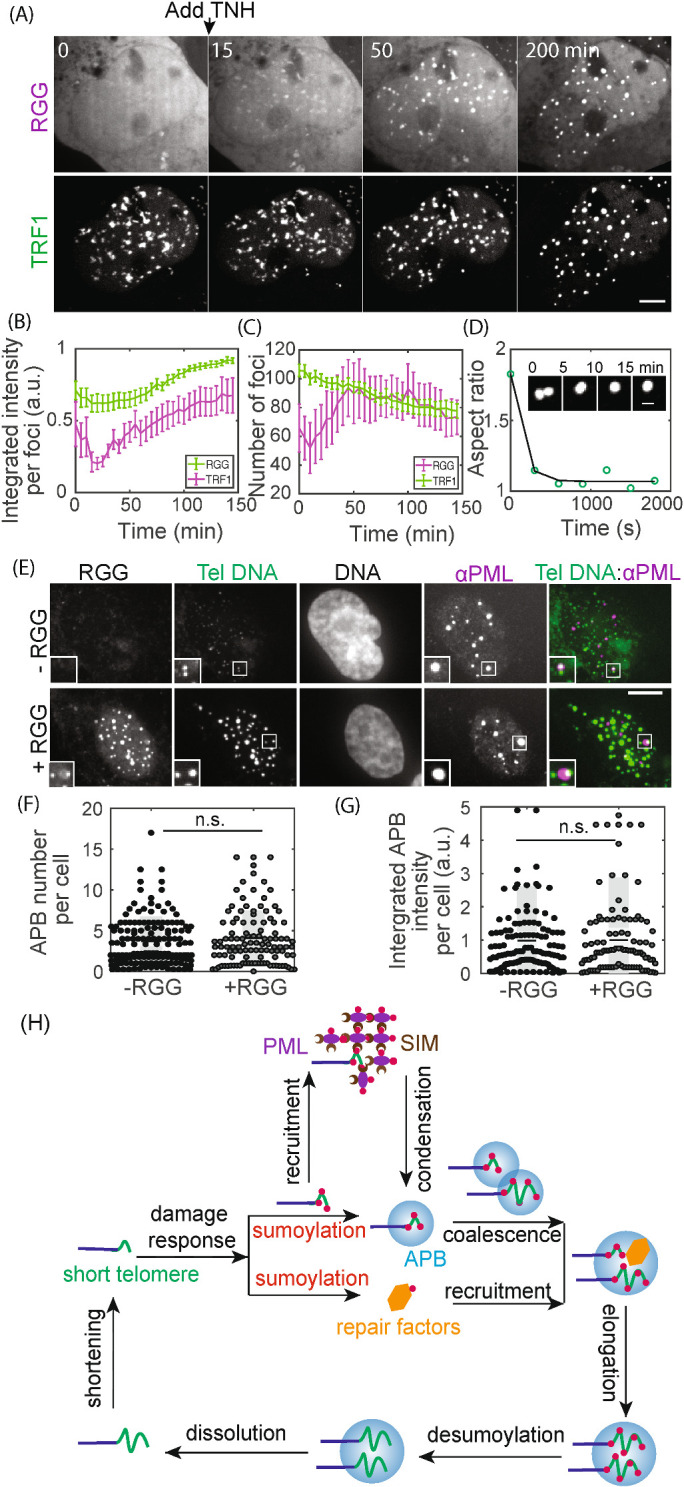
Non-APB condensation on telomeres drives telomere clustering. (A–D) TNH was added to cells expressing RGG-mCherry-RGG-eDHFR and Halo-GFP-TRF1 to induce dimerization and condensation. Graphs show integrated intensity per TRF1 and RGG foci (B) and the number of TRF1 and RGG foci (C) over time; 15 cells from two duplicates, error bars SEM. P value between first and last time point for TRF1 foci intensity <0.001, RGG foci intensity n.s., TRF1 foci number <0.001, RGG foci number n.s. Insets (D) show an example of a fusion event, with the change in aspect ratio quantified (exponential fit, decay time 6 min). (E–G) FISH of telomere DNA and IF of PML for cells with or without RGG recruitment. In representative images (E) the overlay of PML (purple) and telomere DNA (green) appears white, indicating APBs with PML nuclear bodies localized to telomeres. Insets (two times enlarged) show two telomere foci, one with an APB and one without, indicating the basal level of APBs in these cells. Graphs show APB number per cell and integrated APB intensity per cell. Each data point (F, G) represents one cell from two biological replicates, black line mean, gray bar 95% confidence interval. (H) Model for APB condensation and function. Telomere shortening (or replication stress) triggers a DNA damage response, where telomere sumoylation nucleates APB condensation and drives telomere clustering while another aspect of the damage response pathway recruits DNA repair factors to APB condensates. Together the clustered telomeres and enriched DNA repair factors within APBs lead to homology-directed telomere synthesis in ALT. Scale bars, 5 μm.
Movie S4.
Recruit RGG to telomeres. Movie for Figure 5A. Left: RGG-mCherry-RGG-eDHFR, middle: Halo-GFP-TRF1, right: composite of RGG (magenta) and TRF1 (green). TNH was added to cells after the first time point to induce dimerization. Yellow box highlights a fusion event.
DISCUSSION
We established a chemical inducer system to control liquid–liquid phase separation in live cells. With this assay we induced de novo APB formation and provide direct evidence in live cells that APBs, like many membrane-less organelles, are molecular condensates formed following liquid–liquid phase separation. A previous study proposed a multi-pathway model for APB formation because APBs can be induced by tethering many proteins to telomeres (Chung et al., 2011). We propose a unified model for APB formation: a liquid–liquid phase separation triggered by telomere sumoylation via SUMO–SIM interactions as part of a DNA damage response at telomeres (Figure 5H). Tethering different proteins to induce APB formation represents multiple ways to cross the phase boundary, through contributing to sumoylation or directly enriching SUMO and SIM on telomeres. We also find that releasing SIM from telomeres reverses APB condensation. These findings indicate that APB condensates are nucleated on telomeres via sumoylation and can be dissolved via desumoylation. Other posttranslational modifications known to regulate phase separation, such as phosphorylation (Snead and Gladfelter, 2019), may also play a role in APB condensation or dissolution, either by directly controlling de/sumoylation or by modulating SUMO–SIM interaction strength (Chang et al., 2011; Hendriks et al., 2017).
Sumoylation has long been observed as part of the DNA damage response (Hendriks and Vertegaal, 2015). Our observation that sumoylation nucleates APB condensates as a mechanism for ALT telomere clustering may lead to future insights on the roles of sumoylation in DNA repair in other contexts (Xu et al., 2003; Sarangi and Zhao, 2015). Indeed, sumoylation is proposed to generate a glue that holds DNA repair factors together (Psakhye and Jentsch, 2012), which may form through SUMO–SIM driven phase separation as observed here. In addition, PARylation and transcription can drive phase separation of DNA repair factors at damage sites (Altmeyer et al., 2015; Kilic et al., 2019; Pessina et al., 2019; Singatulina et al., 2019). It remains to be determined how sumoylation coordinates with PARylation, transcription, and other DNA damage signaling to facilitate DNA repair through phase separation. As PARylation is one of the earliest events during DNA damage recognition, it is possible that a temporal order of signals beginning with PARP activity and culminating in SUMO–SIM interactions is responsible for phase separation of DNA damage foci. Furthermore, PML bodies associate with genomic loci other than telomeres in non-ALT cells to regulate multiple functions including DNA repair, transcription, viral genome replication, and heterochromatin domain formation (Dellaire and Bazett-Jones, 2004; Eskiw et al., 2004; Ching et al., 2005; Luciani et al., 2006; Shastrula et al., 2019). Our work demonstrates local sumoylation as a mechanism for generating telomere association of PML bodies by either directly nucleating PML bodies or enabling sumoylated telomeres to fuse with existing PML bodies to form APBs. Similarly, protein sumoylation at other genomic loci may trigger PML association. Supporting this notion, a recent study finds that viral protein sumoylation is required for association of PML bodies with viral replication centers (Stubbe et al., 2020).
Our findings that disruption of SUMO–SIM interactions produced a disassembly of telomere condensates suggests approaches to target pathological processes that arise from this type of phase transition. Since sumoylation is involved in many cellular functions, globally targeting sumoylation to prevent APB condensation would have many side effects. Instead, approaches to disrupt APB liquid properties or recruitment of important factors to APBs would be more attractive. For example, pushing APB condensates into gel or solid phase (Shin et al., 2017) by increasing molecule density or interaction strength within APBs would prevent reversible telomere clustering, inhibit dynamic retention of DNA repair factors within APBs and thus prevent telomere elongation.
An advantage of our chemical dimerization system is that it allows for sustained recruitment after dimerizer addition. This makes it suitable for single cell live imaging for a prolonged time as well as treatment of a population of cells for fixed cell or biochemical analyses, both difficult to achieve with the currently available optogenetic systems that require constant illumination for phase separation (Shin et al., 2017, 2018). We find that the induced condensates contain the APB signature component PML but not DNA repair factors such as 53BP1, PCNA, and POLD3 (Figure 4; Figure 4–Supplemental Figure S1), indicating that the repair factors are recruited to the APB condensates by DNA damage response signaling other than the telomere sumoylation that nucleates APBs (Figure 5H). Many DNA repair factors such as 53BP1 and PCNA undergo posttranslational modifications including ubiquitylation, phosphorylation, and sumoylation (Dantuma and van Attikum, 2016; Garvin and Morris, 2017). Posttranslational modifications of those DNA repair factors, not captured in our dimerization approach, may be the endogenous stimuli that promote their recruitment to APBs. Supporting this notion, PCNA fused with SUMO1 is enriched in RGG dimerization-induced APB condensates (Figure 4–Supplemental Figure S2). In addition, it has been shown that client recruitment in phase-separated condensate scaffolds is affected by scaffold stoichiometries (Banani et al., 2016). Therefore, the chemistry of APB scaffolds could also be important for repair factor enrichment. It is reported that BLM helicase was recruited to synthetic condensates formed by polySUMO and polySIM only when the condensate was SUMO rich (Min et al., 2019). A recent study showed that the presence of PML protein is required for the recruitment of BTR complex to telomeres for ALT telomere maintenance (Loe et al., 2020). It is possible that the chemistry of APB condensates is actively regulated during the course of telomere elongation to selectively recruit different factors based on direct SUMO–SIM interactions or indirect interactions with existing APB components.
Coalescence of APB liquid droplets that drives telomere clustering (Figure 3, A–E) may provide repair templates for homology-directed telomere DNA synthesis in ALT. ALT cells contain ECTR DNAs that may be either linear or circular, but their functional contribution to ALT is unknown (Cesare and Griffith, 2004). They share sequence identity with telomeres and cannot be differentiated with our TRF1 probe or other labeling techniques targeting telomere DNA sequence. Therefore, the clustering we observe may involve APBs nucleated on both telomeres and ECTRs. Since ECTRs are more mobile, they may be more efficient in clustering with telomeres to provide homology directed-repair templates. Damaged telomeres generate more ECTRs than SIM dimerization, strongly arguing that a majority of events we observe are due to chromosomally attached telomere coalescence in response to SUMO–SIM interactions. We previously showed that DNA damage increases telomere mobility of chromosomally attached telomeres (Cho et al., 2014), indicating that DNA damage not only nucleates APB condensates to enable telomere clustering through droplet coalescence but also actively modulates clustering efficiency by increasing the chance of APB collision. Nuclear actin polymerization increases the mobility of DNA damage sites to cluster DNA damage foci for homology-directed DNA repair (Schrank et al., 2018). It remains to be determined whether actin polymerization increases telomere mobility in response to DNA damage in ALT cells and whether and how it depends on telomere protein sumoylation or APB condensation. In addition, due to the attachment of telomeres to the rest of the chromatin fiber, other mechanisms such as a block copolymer microphase separation (Leibler et al., 1983) may contribute to telomere clustering after initiation by APB phase separation. Further studies dissecting the physical mechanisms underlying telomere clustering and the role of ECTRs will increase our understanding of templating in ALT. We also demonstrated that the ability to cluster telomeres depends only on the liquid properties of APB condensates, not their chemical composition (Figure 5). This finding provides an opportunity to target the physical–chemical properties of APBs for cancer therapy in ALT without affecting the function of their DNA repair components that also contribute to genome integrity in normal cells.
Liquid–liquid phase separation can contribute to cellular functions by multiple mechanisms. For example, the high sensitivity of the phase separation process to environmental factors makes it ideal for sensing stress (Munder et al., 2016; Riback et al., 2017), and concentrating and confining molecules into one compartment can increase the kinetics of biochemistry (Case et al., 2019). The hallmark of such phase separation is the liquid properties of the resulting condensates, which have been carefully characterized in reconstituted systems. The functional significance of these in vitro findings in cells has been widely implied but not yet demonstrated (Shin and Brangwynne, 2017). With an optogenetic system to induce synthetic condensates with disordered proteins, it was shown that condensates can pull targeted chromatin loci together through coalescence (Shin et al., 2018). With chemical dimerization-induced condensation, we show that this general mechanism is applicable to a biologically relevant condensate, namely APBs, to cluster telomeres for homology-directed telomere synthesis in ALT cancer cells, independent of condensate chemistry. In addition, DNA repair factors required for telomere DNA synthesis may be selectively retained in APBs by regulating chemical properties of APB condensate scaffold and client molecules. Our findings may represent a general strategy for reversible genome organization, such as clustering of gene loci for transcription and DNA repair, and suggest a dual function model for chromatin condensates: concentrating factors for biochemistry through composition control while clustering distinct chromatin domains via coalescence. This chemical approach can be used to study how material properties and chemical composition of other condensates contribute to cellular functions.
MATERIALS AND METHODS
Plasmids
The plasmids for inducing DNA damage at telomeres (mCherry-ER-DD-TRF1-FokI or Fok1 mutant) were previously published (Cho et al., 2014). For recruiting SIM to telomeres, TRF1 was substituted for SPC25 in the published 3xHalo-GFP-SPC25 plasmid (Zhang et al., 2017). SIM (or SIM mutant) for SIM-mCherry-eDHFR is from plasmids gifted by Michael Rosen (Banani et al., 2016). The RGG insert for RGG-mCherry-RGG-eDHFR is from a plasmid gifted by Benjamin Schuster (Schuster et al., 2018). The vector containing mCherry-eDHFR is from our published plasmid Mad1-mCherry-eDHFR (Zhang et al., 2017). All other plasmids in this study are derived from a plasmid that contains a CAG promoter for constitutive expression, obtained from E. V. Makeyev (Khandelia et al., 2011).
Cell culture
All experiments were performed with U2OS acceptor cells, originally obtained from E.V. Makayev (Nanyang Technological University, Singapore; Khandelia et al., 2011). Cells were cultured in growth medium (DMEM with 10% fetal bovine serum [FBS] and 1% penicillin/streptomycin) at 37°C in a humidified atmosphere with 5% CO2. The TRF1 constructs (3xHalo-GFP-TRF1, 3xHalo-TRF1, or mCherry-ER-DD-TRF1-FokI) and the eDHFR constructs (SIM, SIM mutant, or RGG) were transiently expressed by transfection with Lipofectamine 2000 (Invitrogen) 24 h prior to imaging, following the manufacturer’s protocol.
Dimerization and damage on telomeres
To recruit proteins to telomeres, cells transfected with 3xHalo-GFP-TRF1 or 3xHalo-TRF1 and one of the mCherry-eDHFR plasmids (SIM, SIM mutant, or RGG) were treated with the dimerizer TNH: TMP-NVOC (6-nitroveratryl oxycarbonyl)-Halo (Zhang et al., 2017). For live imaging, 100 nM TNH was added directly to cells on the microscope stage. For immunofluorescence (IF) or FISH, 100 nM TNH was added to cells and incubated for 2 h before fixing. To induce damage on telomere in cells transfected with mCherry-ER-DD-TRF1-FokI, Shield-1 (Cheminpharma LLC), and 4-hydroxytamoxifen (4-OHT) (Sigma-Aldrich) at 1 μM were added for 1 h to allow TRF1 to enter the nucleus prior to live imaging or 2 h prior to fixing, as previously described (Cho et al., 2014).
IF, FISH, and EdU labeling
Cells were fixed in 4% formaldehyde for 10 min at room temperature, followed by permeabilization in 0.5% Triton X-100 for 10 min. Cells were incubated with primary antibody at 4°C in a humidified chamber overnight and then with secondary antibody for 1 h at room temperature before washing and mounting. Primary antibodies were anti-SUMO1 (Ab32058, Abcam,1:200 dilution), anti-SUMO2/3 (Asm23, Cytoskleton, 1:200 dilution), anti-PCNA (P10, Cell Signaling, 1:1000 dilution), anti-53BP1(NB100-904, Novus Biologicals, 1:1000 dilution), anti-PML (sc966, Santa Cruz, 1:50 dilution), and anti-POLD3 (H00010714-M01, Abnova, 1:100 dilution). For IF-FISH, coverslips were first stained with primary and secondary antibody, then fixed again in 4% formaldehyde for 10 min at room temperature. Coverslips were then dehydrated in an ethanol series (70%, 80%, 90%, 2 min each) and incubated with 488-telG PNA probe (Panagene) at 75°C for 5 min and then overnight in a humidified chamber at room temperature. Coverslips were then washed and mounted for imaging. For EdU assay, cells were first incubated with 10 µM EdU and TNH for SIM or SIM mutant transfected cells or EdU and Shield1 and 4-OHT for FokI transfected cells, fixed, then labeled with Click-iT EdU Alexa Fluor 647 Imaging Kit (Thermal Fisher).
Image acquisition
For live imaging, cells were seeded on 22 × 22-mm glass coverslips (no. 1.5; Fisher Scientific) coated with poly-d-lysine (Sigma-Aldrich) in single wells of a 6-well plate. When ready for imaging, coverslips were mounted in magnetic chambers (Chamlide CM-S22-1, LCI) with cells maintained in L-15 medium without phenol red (Invitrogen) supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C on a heated stage in an environmental chamber (Incubator BL; PeCon GmbH). Images were acquired with a spinning disk confocal microscope (DM4000; Leica) with a 100 × 1.4 NA objective, an XY Piezo-Z stage (Applied Scientific Instrumentation), a spinning disk (Yokogawa), an electron multiplier charge-coupled device camera (ImageEM; Hamamatsu Photonics), and a laser merge module equipped with 488 and 593 nm lasers (LMM5; Spectral Applied Research) controlled by MetaMorph software (Molecular Devices). Images were taken with 0.5 µm spacing for a total of 6 µm and a 5-min time interval for 2–4 h for both GFP and mCherry channels. Fixed cells were imaged using a 100 × 1.4 NA objective on an inverted fluorescence microscope (DM6000, Leica Micro- systems) equipped with an automated XYZ stage (Ludl Electronic Products), a charge-coupled device camera (QuantEM 512SC, Photometrics), an X-LIGHT Confocal Imager (Crisel Electrooptical Systems), and an IDI high performance fluorescence illuminator equipped with 405, 445, 470, 520, 528, 555, and 640 nm lasers (89 North and Cairn Research), controlled by Metamorph Software (MDS Analytical Technologies). Images were taken with 0.3 µm spacing for a total of 8 µm.
Image processing
All images shown are maximum-intensity projections from all slices in z-stacks generated in ImageJ (Schneider et al., 2012). Quantifications of images and plotting of figures were done in MATLAB (MathWorks). For live imaging, TRF1 foci in the GFP channel were identified with a 3D bandpass filter with custom MATLAB code modified based on gift code from Stephanie Weber (Berry et al., 2015). The number of segmented TRF1 foci and integrated fluorescence intensity per foci were calculated at each time point. The integrated fluorescence intensity per foci was calculated by first summing up the total intensity over all Z slices in the foci and then calculating the average value over all foci in the cell. For colocalization analysis of fixed images, both channels were segmented with a 3D bandpass filter. The number of colocalized foci and the total fluorescence intensity summed over all Z slices and over all colocalized foci in one cell were plotted.
Statistical analyses
All p values were generated with two-sample t test in MATLAB with function ttest2.
Supplementary Material
Acknowledgments
We thank Stephanie Weber for sharing MATLAB code for analyzing foci intensity in 3D and Michael Rosen and Benjamin Schuster for sharing plasmids. We thank members of the Greenberg lab and Lampson lab for helpful discussions. This work was supported by the National Institutes of Health (GM122475 to M.A.L., GM118510 to D.M.C., U54-CA193417 to Physical Sciences Oncology Center at Penn, 1K22CA237632-01 to H.Z., and GM101149 and CA17494 to R.A.G.) and Emerson Collective Cancer Research Fund for M.A.L.
Abbreviations used:
- ALT
alternative lengthening of telomeres
- APB
ALT telomere-associated promyelocytic leukemia nuclear body
- ECTR
extrachromosomal telomere DNA
- eDHFR
Escherichia coli dihydrofolate reductase
- FBS
fetal bovine serum
- FISH
fluorescence in situ hybridization
- IF
immunofluorescence
- PML
promyelocytic leukemia
- RGG
arginine/glycine-rich
- SIM
sumo interaction motif
- SUMO
small ubiquitin-like modifier
- TMP
trimethoprim.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-10-0589) on June 24, 2020.
REFERENCES
- Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grøfte M, Druedahl Rask M-B, Streicher W, Jungmichel S, Nielsen ML, et al (2015). Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat Commun , 8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballister ER, Aonbangkhen C, Mayo AM, Lampson MA, Chenoweth DM. (2014). Localized light-induced protein dimerization in living cells using a photocaged dimerizer. Nat Commun , 5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK. (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol , 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, Rosen MK. (2016). Compositional control of phase-separated cellular bodies. Cell , 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP. (2015). RNA transcription modulates phase transition-driven nuclear body assembly. Proc Natl Acad Sci USA , E5237–E5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Jülicher F, Hyman AA. (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science , 1729–1732. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, Hyman AA. (2011). Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci USA , 4334–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Zhang X, Ditlev JA, Rosen MK. (2019). Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science (80-) , 1093–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Griffith JD. (2004). Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol , 9948–9957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-C, Naik MT, Huang Y-S, Jeng J-C, Liao P-H, Kuo H-Y, Ho C-C, Hsieh Y-L, Lin C-H, Huang N-J, et al (2011). Structural and Functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol Cell , 62–74. [DOI] [PubMed] [Google Scholar]
- Ching RW, Dellaire G, Eskiw CH, Bazett-Jones DP. (2005). PML bodies: A meeting place for genomic loci? J Cell Sci , 847–854. [DOI] [PubMed] [Google Scholar]
- Cho NW, Dilley RL, Lampson MA, Greenberg RA. (2014). Interchromosomal homology searches drive directional ALT telomere movement and synapsis. Cell , 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung I, Leonhardt H, Rippe K. (2011). De novo assembly of a PML nuclear subcompartment occurs through multiple pathways and induces telomere elongation. J Cell Sci , 3603–3618. [DOI] [PubMed] [Google Scholar]
- Dantuma NP, van Attikum H. (2016). Spatiotemporal regulation of posttranslational modifications in the DNA damage response. EMBO J , 6–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaire G, Bazett-Jones DP. (2004). PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. BioEssays , 963–977. [DOI] [PubMed] [Google Scholar]
- DeRose R, Miyamoto T, Inoue T. (2013). Manipulating signaling at will: chemically-inducible dimerization (CID) techniques resolve problems in cell biology. Pflugers Arch , 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilley RL, Greenberg RA. (2015). ALTernative Telomere Maintenance and Cancer. Trends in Cancer , 145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilley RL, Verma P, Cho NW, Winters HD, Wondisford AR, Greenberg RA. (2016). Break-induced telomere synthesis underlies alternative telomere maintenance. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draskovic I, Arnoult N, Steiner V, Bacchetti S, Lomonte P, Londoño-Vallejo A. (2009). Probing PML body function in ALT cells reveals spatiotemporal requirements for telomere recombination. Proc Natl Acad Sci USA , 15726–15731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC-H, Eckmann CR, Myong S, Brangwynne CP. (2015). The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci USA , 7189–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskiw CH, Dellaire G, Bazett-Jones DP. (2004). Chromatin contributes to structural integrity of promyelocytic leukemia bodies through a SUMO-1-independent mechanism. J Biol Chem , 9577–9585. [DOI] [PubMed] [Google Scholar]
- Garvin AJ, Morris JR. (2017). Sumo, a small, but powerful, regulator of double-strand break repair. Philos Trans R Soc B Biol Sci , 20160281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW. (1990). Telomeres shorten during ageing of human fibroblasts. Nature , 458–460. [DOI] [PubMed] [Google Scholar]
- Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, et al (2015). A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell , 712–723. [DOI] [PubMed] [Google Scholar]
- Hendriks IA, Lyon D, Young C, Jensen LJ, Vertegaal ACO, Nielsen ML. (2017). Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat Struct Mol Biol , 325–336. [DOI] [PubMed] [Google Scholar]
- Hendriks IA, Vertegaal ACO. (2015). SUMO in the DNA damage response. Oncotarget , 15734–15735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelia P, Yap K, Makeyev EV. (2011). Streamlined platform for short hairpin RNA interference and transgenesis in cultured mammalian cells. Proc Natl Acad Sci USA , 12799–12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic S, Lezaja A, Gatti M, Bianco E, Michelena J, Imhof R, Altmeyer M. (2019). Phase separation of 53 BP 1 determines liquid-like behavior of DNA repair compartments. EMBO J , e101379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, de The H. (2010). PML nuclear bodies. Cold Spring Harb Perspect Biol , a000661–a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzerini-Denchi E, Sfeir A. (2016). Stop pulling my strings-what telomeres taught us about the DNA damage response. Nat Rev Mol Cell Biol , 364–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibler L, Orland H, Wheeler JC. (1983). Theory of critical micelle concentration for solutions of block copolymers. J Chem Phys , 3550–3557. [Google Scholar]
- Lin Y, Protter DSW, Rosen MK, Parker R. (2015). Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell , 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loe T, Li JSZ, Zhang Y, Azeroglu B, Boddy MN, Denchi EL. (2020). Telomere length heterogeneity in ALT cells is maintained by PML-dependent localization of the BTR complex to telomeres. Genes Dev , 650–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciani JJ, Depetris D, Usson Y, Metzler-Guillemain C, Mignon-Ravix C, Mitchell MJ, Megarbane A, Sarda P, Sirma H, Moncla A, et al (2006). PML nuclear bodies are highly organised DNA-protein structures with a function in heterochromatin remodelling at the G2 phase. J Cell Sci , 2518–2531. [DOI] [PubMed] [Google Scholar]
- Min J, Wright WE, Shay JW. (2019). Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev , 814–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munder MC, Midtvedt D, Franzmann T, Ulbricht E, Muller P, Nuske E, Otto O, Herbig M, Taubenberger A, Maharana S, et al (2016). A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. Elife , e09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabetani A, Ishikawa F. (2011). Alternative lengthening of telomeres pathway: recombination-mediated telomere maintenance mechanism in human cells. J Biochem , 5–14. [DOI] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al (2015). Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell , 936–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwald S, Deeg KI, Chung I, Parisotto D, Wörz S, Rohr K, Erfle H, Rippe K. (2015). PML induces compaction, TRF2 depletion and DNA damage signaling at telomeres and promotes their alternative lengthening. J Cell Sci , 1887–1900. [DOI] [PubMed] [Google Scholar]
- O’Sullivan RJ, Arnoult N, Lackner DH, Oganesian L, Haggblom C, Corpet A, Almouzni G, Karlseder J. (2014). Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat Publ Gr , 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee H, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al (2015). A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell , 1066–1077. [DOI] [PubMed] [Google Scholar]
- Pessina F, Giavazzi F, Yin Y, Gioia U, Vitelli V, Galbiati A, Barozzi S, Garre M, Oldani A, Flaus A, et al (2019). Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat Cell Biol , 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR, Yu H. (2007). The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol , 581–590. [DOI] [PubMed] [Google Scholar]
- Psakhye I, Jentsch S. (2012). Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell , 807–820. [DOI] [PubMed] [Google Scholar]
- Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, Drummond DA. (2017). Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell , 1028–1040.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U, Ferhi O, Jeanne M, Benhenda S, Berthier C, Jollivet F, Niwa-Kawakita M, Faklaris O, Setterblad N, de Thé H, et al (2014). Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins. J Cell Biol , 931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangi P, Zhao X. (2015). SUMO-mediated regulation of DNA damage repair and responses. Trends Biochem Sci , 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods , 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrank BR, Aparicio T, Li Y, Chang W, Chait BT, Gundersen GG, Gottesman ME, Gautier J. (2018). Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature , 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster BS, Reed EH, Parthasarathy R, Jahnke CN, Caldwell RM, Bermudez JG, Ramage H, Good MC, Hammer DA. (2018). Controllable protein phase separation and modular recruitment to form responsive membraneless organelles. Nat Commun , 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastrula PK, Sierra I, Deng Z, Keeney F, Hayden JE, Lieberman PM, Janicki SM. (2019). PML is recruited to heterochromatin during S phase and represses DAXX-mediated histone H3.3 chromatin assembly. J Cell Sci , jcs220970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen TH, Lin H-K, Scaglioni PP, Yung TM, Pandolfi PP. (2006). The Mechanisms of PML-Nuclear Body Formation. Mol Cell , 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima H, Suzuki H, Sun J, Kono K, Shi L, Kinomura A, Horikoshi Y, Ikura T, Ikura M, Kanaar R, et al (2013). Activation of the SUMO modification system is required for the accumulation of RAD51 at sites of DNA damage. J Cell Sci , 5284–5292. [DOI] [PubMed] [Google Scholar]
- Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, Brangwynne CP. (2017). Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell , 159–171.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP. (2017). Liquid phase condensation in cell physiology and disease. Science , eaaf4382. [DOI] [PubMed] [Google Scholar]
- Shin Y, Chang YC, Lee DSW, Berry J, Sanders DW, Ronceray P, Wingreen NS, Haataja M, Brangwynne CP. (2018). Liquid nuclear condensates mechanically sense and restructure the genome. Cell , 1481–1491.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singatulina AS, Hamon L, Sukhanova MV, Bouhss A, Lavrik OI, Pastré D. (2019). PARP-1 activation directs FUS to DNA damage sites to form PARG-reversible compartments enriched in damaged DNA. Cell Rep . [DOI] [PubMed] [Google Scholar]
- Snead WT, Gladfelter AS. (2019). The control centers of biomolecular phase separation: how membrane surfaces, PTMs, and active processes regulate condensation. Mol Cell , 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobinoff AP, Pickett HA. (2017). Alternative lengthening of telomeres: DNA repair pathways converge. Trends Genet , 921–932. [DOI] [PubMed] [Google Scholar]
- Stubbe M, Mai J, Paulus C, Christian Stubbe H, Berscheminski J, Karimi M, Hofmann S, Weber E, Hadian K, Hay R, et al (2020). Viral DNA binding protein SUMOylation promotes PML nuclear body localization next to viral replication centers. MBio , e00049-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, King DS, Taunton J, Rosen MK, Vale RD. (2016). Phase separation of signaling molecules promotes T cell receptor signal transduction. Science , 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P, Dilley RL, Gyparaki MT, Greenberg RA. (2018). Direct quantitative monitoring of homology-directed DNA repair of damaged telomeres. In: Methods in Enzymology, ed. Spies M., Malkova A., New York: Elsevier, 107–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P, Dilley RL, Zhang T, Gyparaki MT, Li Y, Greenberg RA. (2019). RAD52 and SLX4 act nonepistatically to ensure telomere stability during alternative telomere lengthening. Genes Dev , 221–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z-X, Timanova-Atanasova A, Zhao R-X, Chang K-S. (2003). PML colocalizes with and stabilizes the DNA damage response protein TopBP1. Mol Cell Biol , 4247–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. (1999). Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res , 4175–4179. [PubMed] [Google Scholar]
- Zhang H, Aonbangkhen C, Tarasovetc EV, Ballister ER, Chenoweth DM, Lampson MA. (2017). Optogenetic control of kinetochore function. Nat Chem Biol , 1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS. (2015). RNA controls PolyQ protein phase transitions. Mol Cell , 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JM, Yadav T, Ouyang J, Lan L, Zou L. (2019). Alternative lengthening of telomeres through two distinct break-induced replication pathways. Cell Rep , 955–968.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


