Abstract
Cutaneous mechanosensory neurons are activated by mechanical loads applied to the skin, and these stimuli are proposed to generate mechanical strain within sensory neurons. Using a microfluidic device to deliver controlled stimuli to intact animals and large, immobile, and fluorescent protein-tagged mitochondria as fiducial markers in the touch receptor neurons (TRNs), we visualized and measured touch-induced mechanical strain in Caenorhabditis elegans worms. At steady state, touch stimuli sufficient to activate TRNs induce an average strain of 3.1% at the center of the actuator and this strain decays to near zero at the edges of the actuator. We also measured strain in animals carrying mutations affecting links between the extracellular matrix (ECM) and the TRNs but could not detect any differences in touch-induced mechanical strain between wild-type and mutant animals. Collectively, these results demonstrate that touching the skin induces local mechanical strain in intact animals and suggest that a fully intact ECM is not essential for transmitting mechanical strain from the skin to cutaneous mechanosensory neurons.
INTRODUCTION
Touch and proprioception are essential to the daily lives of all animals, including humans. Classical examples of mechanotransduction, both of these senses depend on activation of mechano-electrical transduction (MeT) channels arrayed within somatosensory neurons (Katta et al., 2015) and are thought to activate following physical deformation of sensory cells during touch and movement. Consistent with this inference, neurons innervating stretch receptor organs in vertebrates (Bewick and Banks, 2015) and arthropods (Suslak and Jarman, 2015) are activated by experimentally applied mechanical strain (stretch). High-speed volumetric imaging reveals that proprioceptors deform in crawling Drosophila larvae (He et al., 2019; Vaadia et al., 2019) and that physical deformations are correlated with calcium transients. Similarly, movement induces localized calcium transients in the tertiary dendrites of the Caenorhabditis elegans PVD proprioceptors that depend on expression of a DEG/ENaC/ASIC protein (Tao et al., 2019). These studies reinforce the idea that physical deformation of sensory neurons during movement is critical for activation of native MeT channels, but does not address whether or not similar deformations occur in response to touch or measure the extent of mechanical strain that occurs within the sensory neurons themselves.
Using C. elegans touch receptor neurons (TRNs), we investigated the quantitative relationship between touch and physical deformation of sensory neurons in intact living animals. The ready availability of transgenic animals expressing TRN-specific markers and their transparent body make C. elegans an especially useful animal for investigating touch-evoked sensory neuron deformation. Adult animals have six TRNs, consisting of two bilaterally symmetric pairs of TRNs (ALM and PLM) and two neurons that run along the ventral midline (AVM and PVM) (Goodman and Sengupta, 2019). These six neurons extend long, unusually straight sensory neurites (Krieg et al., 2017) that are embedded in epidermal cells and have a distinctive, electron-dense extracellular matrix or ECM as well as hemidesmosome structures attaching the TRNs to the cuticle (Chalfie and Thomson, 1979; Chalfie and Sulston, 1981).
All of the TRNs express the MEC-4 channel, which is required for touch-evoked calcium transients (Suzuki et al., 2003) and touch-evoked MeT currents (O’Hagan et al., 2005). The MEC-4 channels localize to puncta arrayed along the entire length of wild-type TRN sensory neurites (Chelur et al., 2002; Emtage et al., 2004; Cueva et al., 2007; Katta et al., 2019), but are disrupted in ECM mutants (Emtage et al., 2004).
Touch-evoked behavior (Petzold et al., 2013) and MeT channel activation (Eastwood et al., 2015) depend on body indentation rather than the force applied. Slow stimuli fail to activate MeT currents and their size increases with stimulus frequency, indicating that activation of MeT channels in their native environment depends on tissue viscoelasticity (Eastwood et al., 2015; Katta et al., 2019; Sanzeni et al., 2019). Although slow, movement-induced physical deformations of the worm’s body and its neurons are too slow to activate MeT currents in the TRNs (Eastwood et al., 2015; Katta et al., 2019), these undulatory movements produce mechanical strains of up to 40% (Krieg et al., 2014, 2017), indicating that TRNs can withstand significant mesoscale extension and compression in situ. The ability to withstand movement-induced strain is shared by mammalian nerves that experience up to 30% strain during limb movement (Phillips et al., 2004).
Here, we sought to determine whether or not body indentations sufficient to evoke calcium transients in TRNs also generate local strain. To achieve this goal, we visualized steady-state, touch-induced strain in TRNs in living animals restrained within a microfluidic stimulation device equipped with the ability to deliver mechanical stimuli (Nekimken et al., 2017a; Fehlauer et al., 2018). Our approach benefits from large, immobile mitochondria distributed within TRN sensory neurites (Sure et al., 2018), exploits the ability to tag these mitochondria with a red fluorescent protein (Zheng et al., 2014), and borrows analytic principles from traction force microscopy (Ribeiro et al., 2016). Although mitochondria have been used to evaluate neuronal mechanics in culture (O’Toole et al., 2015), we believe this study is the first to make use of mitochondria as mechanical fiducial markers in living animals. We show that body indentation increases local steady-state mechanical strain in the TRNs in a manner that is robust to mutations known to affect attachment of the TRN to the extracellular matrix and to epidermal cells.
RESULTS
Mechanical stimulation in a microfluidic device
To directly observe touch-induced deformation of the TRNs, we sought to develop a method that combines the delivery of controlled mechanical stimuli and three-dimensional optical imaging. To reach this goal, we applied mechanical stimuli to adult worms confined in a microfluidic device (Nekimken et al., 2017a, Figure 1A).Our device has pneumatic actuators that consist of a thin flexible wall that separates a channel filled with air from a worm in the trap channel. When air pressure in the actuator channel is increased using a pressure controller, the thin wall expands like a balloon, deforming the trapped worm and generating indentations sufficient to activate the TRNs (Nekimken et al., 2017a). Other devices that deliver mechanical stimuli to restrained (Cho et al., 2018) or moving (McClanahan et al., 2017) worms in microfluidic chambers have been reported, but they use larger actuator channels and are thus not well matched to our goal of investigating local deformation of the TRNs.
FIGURE 1:
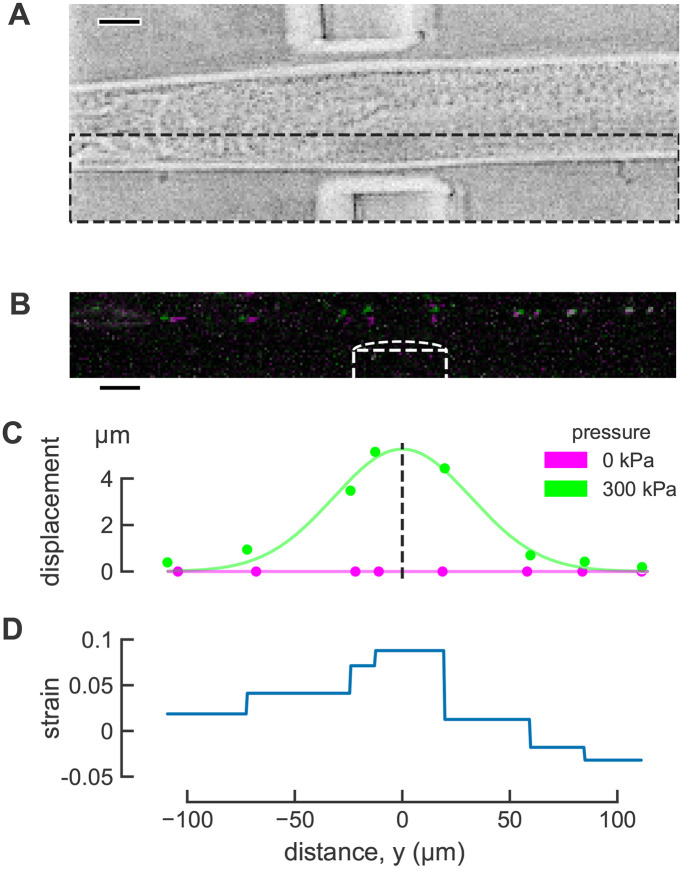
Touch-induced mechanical strain measured using mitochondria as fiducial markers in C. elegans TRNs in vivo. (A) Brightfield image of worm in the microfluidic device. Scale bar 20 μm. (B) Maximum projection image of TRN mitochondria before (magenta) and during (green) mechanical stimulus. Mitochondria that did not move appreciably appear white due to the overlap of magenta and green. Image intensity and contrast was adjusted to improve visualization. Scale bar 20 μm. (C) Displacement in the direction of actuation before (magenta) and during (green) stimulation. The smooth line (green) is a Gaussian fit used to infer the center of actuator. (D) Distribution of touch-induced strain in the TRN for a single actuation trial.
Our device is designed to increase the probability that animals will enter the trap with either their left or their right sides in contact with the bottom of the chamber and their ventral and dorsal sides near the actuator (Nekimken et al., 2017a). Consistent with this expectation and the position of the AVM and ALM neurons within the worm’s body, previous experiments using this device resulted in a higher frequency of AVM activation compared with ALM (Nekimken et al., 2017a). However, the rotational orientation of each worm was variable. We took advantage of this variation to select animals oriented such that ALM (rather than AVM) was near the actuator and used only ALM neurons for this study. In all cases, this variation in worm positioning leads to variation in the size of the effective stimulus. To ensure that all animals received a deforming stimulus, we limited our analysis to animals whose TRNs were deformed enough to observe visually during the experiment.
Mitochondria as fiducial markers for inferring strain
We used fluorescent protein-tagged mitochondria as fiducial markers to measure strain, an approach that uses particle tracking with large (∼1 µm), immobile mitochondria in the TRNs (Sure et al., 2018) serving as natural fiducial markers. We imaged mitochondria position before and after mechanical stimulation (Figure 1, B and C). Next, we used the displacement between adjacent mitochondria to quantify one-dimensional mechanical strain along the long axis of the TRN (Figure 1D) according to: ε = ∆L/L0, where L0 is the resting, undeformed length of an object, and ∆L is the change in length of the object when deformed. For clarity, we refer to this as longitudinal strain. As shown in Figure 1, mitochondria adjacent to the actuator moved more than those anterior and posterior to the actuator (Figure 1, B and C) and the inferred strain was greatest near the center of the actuator (Figure 1D). This method enables direct observation of touch-evoked strain in C. elegans TRNs in a single dimension aligned with the TRN’s longest dimension.
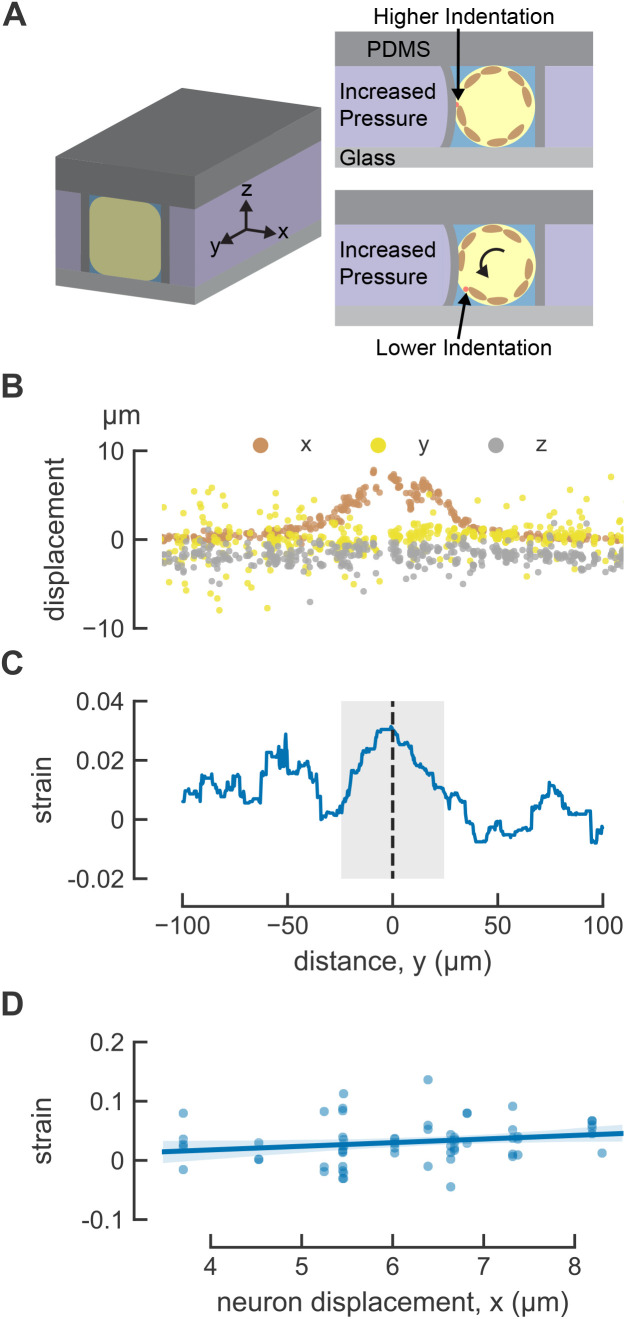
We note that this method is not suitable for detecting shear strain or bending strain. To derive these measurements, we would need to detect angular changes (shear) or relative position (bending) of the mitochondria within the TRNs. Given that the diameter of each mitochondrion is only ∼100 nm and the TRN is diameter is not much larger than this (200–300 nm; see Figure 3C in Cueva et al., 2007), such movements cannot be resolved because these objects are similar in size to the estimated lateral resolution of 254 nm and axial resolution of 632 nm of our imaging system. What about strain in directions orthogonal to the long axis of the TRN dendrite? Although our image stacks contain three-dimensional position data (Figure 2, A and B), the initial distance between adjacent pairs of mitochondria in the x and z directions is small compared with the resulting displacement. Thus, measurement resolutions lead to large uncertainties in the strain calculation for these dimensions. Both limitations can be attributed to the confinement of the mitochondria within the narrow caliber of the TRN and the fact that the TRNs lie primarily within a single focal plane. Thus, the size of mitochondria, the geometry of the TRNs, and the optical resolution of the spinning-disk confocal limit this measurement to one-dimensional strain along the y-dimension that traces the main axis of the TRN.
FIGURE 3:
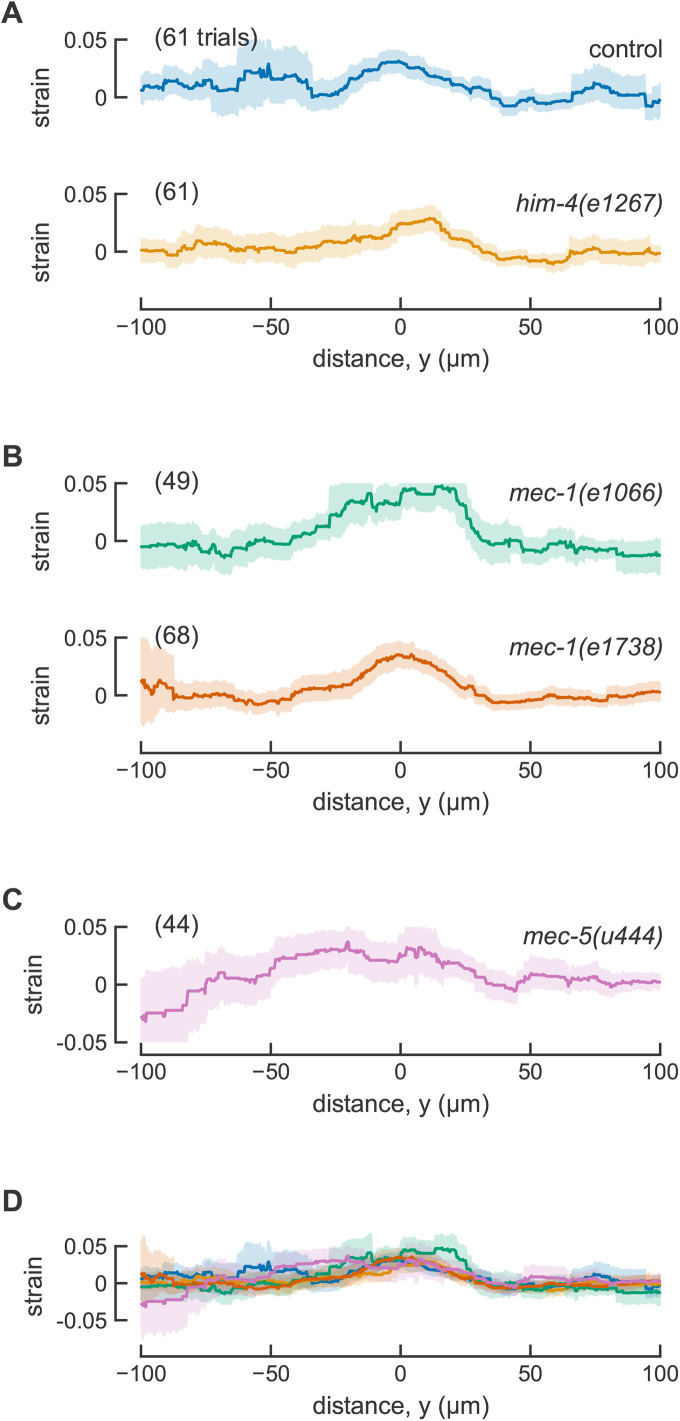
Touch-induced mechanical strain profiles are similar in control animals and ECM mutants. (A) Spatially averaged strain in control ALM neurons with normal TRN attachment (15 animals) and him-4(e1267) with attachment defects (16 animals). The data for control animals are the same data as Figure 2. (B) Spatially averaged strain in mec-1 mutants. 14 independent animals for mec-1(1066) and 16 animals for mec-1(e1738). (C) Spatially averaged strain in mec-5(u444) mutants (17 animals). (D) Overlay of all spatially averaged strain profiles. Smooth lines are the averages across all trials and shaded areas show the error (95% confidence intervals). One ALM neuron was tested in each animal.
FIGURE 2:
Indentation induces local longitudinal strain in C. elegans TRNs. (A) Three-dimensional diagram of the positioning of the worm in the microfluidics trap, the animal-centric coordinate system used to characterize strain, and the consequences of trapping animals in different orientations. TRN is red, and muscles are included in brown as visual aid. (B) Touch-induced displacement in three dimensions. (C) Touch-induced strain, anterior is to the left and posterior is to the right; y = 0 is the center of the actuator; n = 61 trials from N = 15 worms. (D) Mechanical strain at y = 0 as a function of maximum neuron displacement. The line is a linear fit to the data: ε = 0.0061x − 0.0067, where ε is the strain and x is the maximum displacement. The shaded area indicates 95% confidence intervals of the fit. R2 = 0.04.
Local indentation induces local mechanical strain in TRNs
Next, we estimated the spatially averaged distribution of touch-induced strain in the TRN. The mitochondria are sparsely distributed in the TRN and strain is a pairwise measurement between adjacent mitochondria, so our strain measurements result in a series of discontinuous step functions for a single trial rather than a smooth curve (Figure 1D). Each segment of the plot indicates the average strain between two markers, so the maximum local strain on each interval may be larger than our measured strain. The average distance between mitochondria in control jsIs1073 animals was 26 μm (min: 5 µm; max: 64 µm) and we were able to analyze an average of 8.9 intervals per trial (Supplemental Table S1). To obtain averages across trials, we plotted deformation against position in the longitudinal or y direction, fit this profile with a Gaussian function, and defined the position of the maximum as the center of the actuator and y = 0 (see dotted line in Figure 1C). In this coordinate system, positions anterior to y = 0 are negative and positions posterior to y = 0 are positive. Finally, we determined the average strain at each longitudinal position along the neurite across actuation trials (see Figure 2C). Note that the average strain is both larger and noisier for anterior (negative) positions. This is likely to reflect the fact that the head is less constrained in the microfluidic channel than other parts of the body and suggests that displacement and strain on the anterior side of the actuator include both touch- and movement-induced mechanical strain on the neuron.
As expected, strain at the center of the actuator (y = 0) increases with TRN deformation (Figure 2D), but the dependence on TRN deformation was weak. The observed, but modest variation in TRN deformation arises from two aspects of our method. First, because the actuator is fixed to the surrounding material of the device on all sides, its center is the location of maximum deformation. Second, although most animals are trapped with either their left or right side in contact with the coverslip at the bottom of the device, some are rotated along their anterior–posterior axis such that the position of the imaged TRN varies with respect to the actuator (Figure 2A). Thus, the average strain induced between the pair of mitochondria at the center of the actuator was 0.031 ± 0.005 (mean ± SEM, n = 61 trials, N = 15 animals) in transgenic jsIs1073 TRN neurons. Although additional studies exploring a wider range of deformations will be needed to determine the nature relationship between TRN deformation and strain, to our knowledge, these are the first in vivo measurements of touch-induced longitudinal strain in TRNs, and they link local body indentation to local cellular deformation.
Do tagged mitochondria affect TRN function?
We used classical touch assays to address this question (Chalfie et al., 2014a; Nekimken et al., 2017b) by measuring touch sensitivity in wild-type (N2) and transgenic worms carrying the transgenic mitochondria marker, jsIs1073. In blinded assays of three independent cohorts of 25 animals for each genotype, wild-type and jsIs1073 animals had touch response rates of 0.869 and 0.865, respectively. The difference between the means was 0.00667 (95% CI [–0.0347, 0.048]). Thus, the jsIs1073 transgene does not decrease touch sensitivity.
Selected ECM mutants and their touch sensation and anatomical phenotypes
Having measured touch-induced mechanical strain in control TRNs, next we measured longitudinal strain in four ECM mutants: him-4(e1267), mec-1(e1738), mec-1(e1066), and mec-5(u440). One of these mutants, him-4(e1267), has a partial defect in touch sensitivity, and the others are touch insensitive (Du et al., 1996; Emtage et al., 2004; Vogel and Hedgecock, 2001). We reproduced these previous results using a 10-touch assay (Table 1). Whereas him-4 mutants retain MEC-4 puncta that are grossly wild type, TRN neurites in mec-1 and mec-5 mutants lack prominent MEC-4 puncta (Emtage et al., 2004). In him-4 and in mec-1(e1738) mutants analyzed here, the ALM and PLM neurons are displaced from their normal body position near the lateral midlines and are not properly embedded in the epidermis (Vogel and Hedgecock, 2001; Emtage et al., 2004). This effect is inferred to arise from a defective TRN-ECM attachment (Vogel and Hedgecock, 2001). Consistent with this idea, the electron-dense ECM is not detected in mec-1 mutants (Chalfie and Sulston, 1981). Analyzing these four mutants enables us to evaluate the relationship, if any, between touch-induced longitudinal strain and three other phenotypes: behavioral responses to touch, the distribution of MEC-4 puncta, and TRN-ECM attachment.
TABLE 1:
Touch sensitivity of ECM mutants.
| Genotype | Response probability | Mean difference from control | 95% confidence interval of mean difference |
|---|---|---|---|
| Control | 0.742 | ||
| him-4(e1267) | 0.527 | –0.215 | (–0.275, –0.163) |
| mec-1(e1066) | 0.083 | –0.659 | (–0.698, –0.619) |
| mec-1(e1738) | 0.237 | –0.505 | (–0.551, –0.461) |
| mec-5(u444) | 0.208 | –0.534 | (–0.580, –0.487) |
All mutants we tested were less sensitive to touch than control animals; effect size was smallest for him-4(e1267) and largest for mec-1(e1066). All strains tested contained the transgene jsIs1073 that tags mitochondria in the TRNs with RFP. N = 100 worms for each genotype tested in four independent cohorts of 25 blinded to genotype.
The him-4 gene encodes hemicentin, a conserved ECM protein that is rich in Ig and EGF domains and is expressed by body wall muscles (Vogel and Hedgecock, 2001). The him-4 locus is >30 kb, which complicated determination of the molecular defect encoded by e1267 using classical sequencing methods. We used whole-genome sequencing to circumvent this limitation and found that e1267 encodes a single base indel in the third exon of the him-4 gene, introducing a shift in the reading frame of the encoded protein (Supplemental Figure S1). A second polymorphism was detected in the intron following the 48th exon. Thus, the e1267 allele is a null allele and animals carrying this mutant are likely to lack the HIM-4 protein. The mec-1 gene encodes a large secreted protein rich in kunitz-like domains and is expressed by the TRNs, and many alleles of this gene were found in forward genetic screens for touch-insensitive animals (Chalfie and Sulston, 1981; Chalfie and Au, 1989). The e1066 allele is a null and animals carrying this allele are likely to lack the MEC-1 protein, whereas the e1738 allele encodes a premature stop codon and is proposed to express a truncated MEC-1 protein. Both mec-1 mutants are touch insensitive, but only e1066 lacks proper TRN-ECM attachments (Emtage et al., 2004). The mec-5 encodes an atypical collagen and, unlike mec-1, it is not made by the TRNs (Du et al., 1996). Like other ECM mutants we analyzed here, the u440 allele is null and u440 mutants do not express the MEC-5 protein. The TRN neurites in mec-5 mutants are positioned near the lateral midlines, but are less straight than they are in wild-type animals, meandering such that some segments of the neurite are close to the muscle and other parts closer to the lateral midline (Emtage et al., 2004).
Touch-induced longitudinal strain in TRNs with ECM mutants
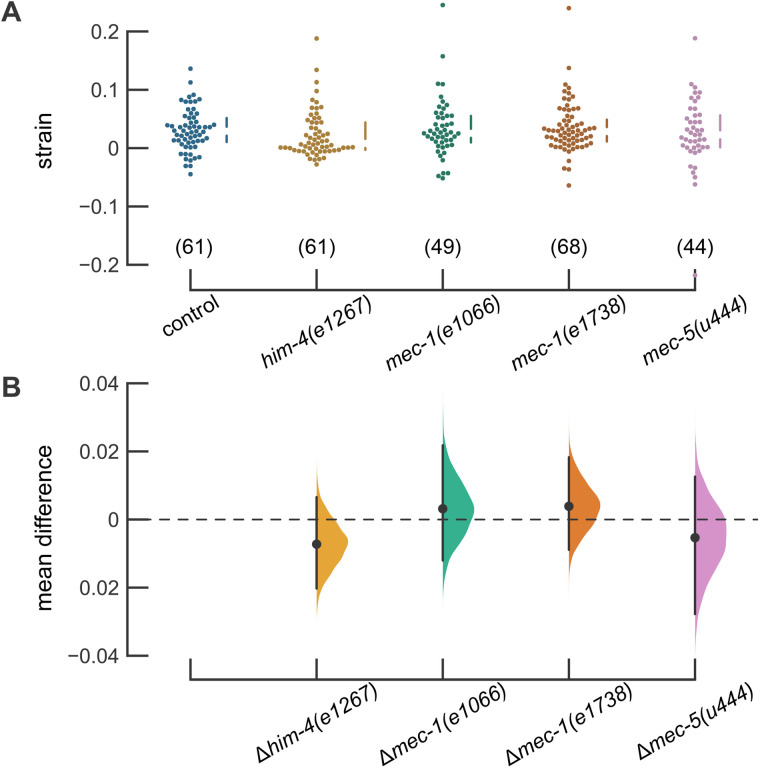
To analyze transmission of mechanical energy from the skin in animals lacking proper TRN-ECM attachment, we introduced the jsIs1073 transgene into him-4 and mec-1 mutants and applied mechanical stimuli to animals trapped in our pneumatic microfluidic device. Touch-induced longitudinal mechanical strain in him-4(e1267) TRNs was indistinguishable from that observed in control animals (Figure 3A; Table 2). Touch-induced longitudinal strain was likewise similar to control in both mec-1 mutants (Figure 3B) and in mec-5 mutants (Figure 3C). Figure 3D shows that the average longitudinal strain profiles for control, him-4, mec-1, and mec-5 mutants are similar across the entire 200-µm segment of the TRN neurite analyzed here. Values for strain measured at the actuator center (y = 0) had similar mean values and distribution for all genotypes tested (Figure 4A) and estimation graphics (Ho et al., 2019) indicate that the mean values for all genotypes are not different than control values (Figure 4B). Except for mec-5 mutants, the number of mitochondria available for tracking and the average distance between mitochondria was similar in control transgenic and mutant animals (Supplemental Table S1). The mec-5 mutants had fewer, more widely spaced mitochondria than control animals, an effect that would be expected to impair the spatial resolution of longitudinal strain measurements. Collectively, these findings suggest that neither severe nor mild defects in TRN-ECM attachment play a significant role in the generation of touch-induced longitudinal mechanical strain at steady state. Future studies and techniques will be needed to resolve dynamic changes in local strain that are expected to occur on the millisecond timescale of MEC-4-dependent MeT channel activation.
TABLE 2:
Touch-evoked longitudinal strain in the TRNs as a function of genotype.
| Genotype | Mean strain ± SEM | Mean difference from control | 95% confidence interval of mean difference | Stimulation trials | Animals |
|---|---|---|---|---|---|
| Control | 0.031 ± 0.005 | 61 | 15 | ||
| him-4(e1267) | 0.024 ± 0.005 | –0.007 | (–0.020, 0.007) | 61 | 16 |
| mec-1(e1066) | 0.034 ± 0.007 | 0.003 | (–0.012 0.022) | 49 | 14 |
| mec-1(e1738) | 0.035 ± 0.005 | 0.004 | (–0.009 0.018) | 68 | 16 |
| mec-5(u444) | 0.026 ± 0.009 | –0.005 | (–0.028 0.013) | 44 | 17 |
There was no detectable difference in the magnitude of the touch-evoked strain in control and ECM mutant animals.
FIGURE 4:
Strain at the center of the actuator is similar in control animals and ECM mutants. (A) Strain at the center of the actuator. Each point is the result of a single trial. The vertical lines next to the swarm plots indicate the median and quartiles of the data. Data were collected from 15, 16, 14, 16, and 17 animals (left to right). (B) Difference in the mean strain between control and each mutant with a bootstrapped resampled distribution of the data. Estimation plots generated using the DABEST plotting package (Ho et al., 2019).
DISCUSSION
Using mitochondria as natural fiducial markers in C. elegans TRNs, we showed for the first time that body indentation sufficient to activate the TRN causes an increase in longitudinal strain. The magnitude of this strain increases with TRN displacement. In wild-type animals, the touch-induced strain closest to the point of maximum indentation was roughly 3% (Figure 3; Table 2). This change in strain is unlikely to damage these sensory neurons, since they are subjected to mechanical strain on the order of 40% during locomotion (Krieg et al., 2014, 2017). Such global changes in strain are too slow to activate the TRNs (Eastwood et al., 2015; Katta et al., 2019).
The 3% strain we measured may be a lower bound on the true value for longitudinal strain. Simulations predicted an elongated strain field with a peak value of 12% for longitudinal strain (Sanzeni et al., 2019). Unlike our measurements, which used a wide (50 µm), flexible PDMS membrane to indent the worm’s body and have limited spatial resolution, the simulations employed a stiff spherical bead (10 µm diameter) to deliver touch stimuli and a continuous deformation function. The simulations (Sanzeni et al., 2019) predict strains are present in the other two dimensions (along the direction of the stimulus and tangential to the circumference of the worm at the point of the stimulus). Due to the nature of our measurement, however, we were only able to measure one-dimensional strain along the length of the TRN. Nevertheless, these measurements of touch-induced strain can be incorporated into future models of C. elegans touch sensation to improve understanding of the mechanical state of the TRN and mechano-sensitive ion channel complex on touch stimulation.
Origins of mechanical coupling between the skin surface and the somatosensory neurons
We tested the idea that strain transmission would depend on TRN-ECM attachment, be correlated with impaired touch sensation, and be independent of the proper distribution of MEC-4 channels. To our surprise, we found that touch-induced longitudinal mechanical strain was similar in control TRNs and all ECM mutants tested here, including the him-4(e1267) and mec-1(e1066) mutants exhibiting severe defects in TRN-ECM attachments. Thus, the touch-induced longitudinal strain we observed in the ALM neurons is independent of their attachment to other tissues or the expression of ECM proteins MEC-1 and MEC-5, at least at steady state.
This finding implies that mechanical coupling between the skin and the sensory neurons persists in mutants with ECM defects. Friction is one alternative source of mechanical coupling between TRNs and surrounding tissues. In this scenario, all of the worm’s tissues are compacted together by a high hydrostatic pressure and this would elevate friction between tissues. Further compression applied to the outside of the worm during touch stimulation could lead to stiffening of the worm’s body that might be caused by internal structures jamming together (Gilpin et al., 2015). When we and others immobilize worms in microfluidic devices, we fabricate channels small enough to apply gentle compressive forces that the worm cannot overcome. As a result, the worm is compressed on all sides except the nose and tail when in the microfluidic trap, potentially jamming the TRNs against surrounding tissues and further increasing friction. Another possibility is that the generation of touch-induced local mechanical strain is dominated by the cytoskeleton. In support of this idea, mutations that disrupt the expression of MEC-12 α-tubulin and MEC-7 β-tubulin decrease mechanoreceptor currents and increase stimulus amplitude needed to activate these currents (O’Hagan, 2005; Bounoutas et al., 2009). Additional experimental work and methods with improved spatial and temporal resolution will be needed to differentiate among these possibilities.
Spatial and temporal resolution of mechanical strain transmission
The spatial resolution of our strain measurements is limited by the average distance between adjacent, immobile mitochondria, which was 26 µm (Supplemental Table S1). Thus, this method may lack sufficient spatial resolution to detect micro- or nanoscale variations in mechanical strain transmission. The spatial resolution of strain measurements could be improved by using more closely spaced fiducial markers. In principle, MEC-4 channels (with an average spacing of 2–3 µm) tagged with a very bright fluorescent protein would provide a 10-fold increase in resolution. Independent of potential improvements in spatial resolution, the present approach is limited to steady-state measurements because our volume imaging rate is low (15 s per stack). In this regard, it is important to note that wild-type C. elegans TRNs are preferentially activated mostly by high-velocity stimuli (Suzuki et al., 2003; Eastwood et al., 2015; Nekimken et al., 2017a; Katta et al., 2019). Temporal resolution could be improved using other imaging techniques enabling rapid acquisition of imaging volumes. If volumes could be acquired at 20 Hz or faster, it would possible to observe mechanical strain in a 10-Hz buzz stimulus that we previously used to activate the TRNs (Nekimken et al., 2017a).
Conclusion
We performed the first in vivo measurements of touch-induced mechanical strain in C. elegans TRNs. We used mechanically stable mitochondria in the TRNs as natural fiducial markers to observe deformation of the TRN and found that local touch stimuli applied in a microfluidic device induces local strain in the TRN. Defects in the ECM surrounding the TRN did not alter the steady-state mechanical strain in the TRN, suggesting that explicit attachments are not necessary for deformation of the TRN and that the bulk properties of tissues are sufficient to sustain significant mechanical energy transfer from the skin surface to the embedded neurons. In light of the temporal limitations of our measurements, however, we cannot exclude the possibility that TRN-ECM attachments contribute to dynamic aspects of mechanical strain transmission. Collectively, these findings provide an empirical basis for the idea that mechanical stimuli applied to the skin stretch embedded sensory neurons that may be shared by the sensory neurons that innervate the skin and other tissue in mammals.
METHODS AND MATERIALS
Nematode strains
For all experiments measuring mechanical strain, we used animals carrying jsIs1073 (mec-7p::TagRFP-mito::CBunc-119), a transgene that drives expression of TagRFP in the mitochondria in the TRNs (Zheng et al., 2014), alone or together with ECM mutants (Table 3). We relied on visualization of the jsIs1073 transgene and behavioral phenotypes (for mec genes) or anatomical defects (for him-4) to perform genetic crosses. We used gene sequencing to verify the presence of ECM mutants in all strains created for this study. The primer pairs we used to amplify the relevant segments of each gene are:
mec-1(e1066): Forward–catcttccacgccgcaaagtc, Reverse–aatcctctctgccctcatgttcc
mec-1(e1738): Forward–tcacagtcagacgtgcctcg, Reverse–cattgcctcacaccaacttccac
mec-5(u444): Forward–cagaatactatgtacgtaacttgggatc; Reverse–ctcatgggtacgcaaatgatactc
him-4(e1267): Forward–tttcgtgatgactggtgactgtgg; Reverse–ttaaagtcaacagcaccgtgacc
TABLE 3:
C. elegans strains.
| Strain name | Genotype | Source |
|---|---|---|
| N2 (Bristol) | wild-type | CGC |
| NM3573 | jsIs1073 | Zheng et al. (2014) |
| GN885 | jsIs1073;him-4(e1267) | This study |
| GN886 | jsIs1073;mec-1(e1066) | This study |
| GN887 | jsIs1073;mec-1(e1738) | This study |
| GN906 | jsIs1073;mec-5(u444) | This study |
Immobilization and mechanical stimulation with a pneumatic microfluidics device
To provide a repeatable mechanical stimulus that is compatible with imaging the mitochondria of the TRN, we used a microfluidic device made of the transparent elastomer PDMS for simultaneous mechanical stimulation and imaging of C. elegans (Nekimken et al., 2017a). Using a pressure controller (Elveflow OB1), we applied 300 kPa of pressure to one of the device’s actuators to create a mechanical stimulus. We fabricated and operated devices designed for use with young adult worms as described in our previous work (Nekimken et al., 2017a; Fehlauer et al., 2018). Because worms immobilized in this trap are uniformly and partially deformed by the channel, we refer to this as the rest configuration.
Sample preparation and inclusion criteria
We performed all experiments using young adult animals that were synchronized by hypochlorite treatment (Stiernagle, 2006) and cultured for 3 days on NGM agar plates seeded with OP50 bacteria for food. We transferred animals from the agar plates with a platinum wire pick to a drop of imaging medium (see below) in a small Petri dish. After using the pick to push away large particles that might clog the microfluidic device, we aspirated worms into polyethylene tubing (PE50, Intramedic brand, Becton-Dickson) with a syringe and connected the tubing to the device’s inlet. For each trial, we pushed a worm into the trap channel using the syringe and then evaluated whether to use this worm for an experiment based on our inclusion criteria. We included animals that fit in the microfluidic trap with minimal movement and could be ejected through the narrow opening at the head of the trap. When performing experiments with him-4 mutants, we chose animals whose gross morphology was wild type and avoided animals whose intestines were everted. In all cases, we only acquired images from animals whose ALM neurons were oriented adjacent to the mechanical actuation channels.
To improve image quality by reducing reflections generated by the walls of the microfluidic trap, we designed a nontoxic imaging medium with a refractive index similar to PDMS (1.4). The imaging medium was a 70%:30% (vol/vol) mixture of physiological saline and iodixanol (Optiprep, Sigma-Aldrich), a nontoxic density-gradient medium (Boothe et al., 2017). We used the same physiological saline as that used for electrophysiological recordings from C. elegans neurons (O’Hagan et al., 2005), which contains (in mM): NaCl (145), KCl (5), MgCl2 (5), CaCl2 (1), and Na-HEPES (10), adjusted to a pH of 7.2 with NaOH. The imaging medium has an osmolality of 300–325 mOsm (Fiske Micro-Osmometer Model 210), so it does not cause large osmotic shocks to C. elegans. By contrast with other fluids we tested (e.g., glycerol, halocarbon oil), this medium has a viscosity that appeared to be similar to that of physiological saline.
Image acquisition
Although the mechanical stimulus is mostly in the horizontal direction, there is enough deformation in the z-direction to move the neuron out of plane during stimulation in some cases. In initial experiments with a traditional epifluorescence microscope, the mitochondria often moved out of focus during stimulation due to movement in the z-direction. To account for this problem, we used a spinning disk confocal microscope (Nikon TiE, Yokogawa CSU-X1, 40×/NA 1.4 oil objective, and Photometrics Prime95B sCMOS camera) to acquire z-stacks, which provided adequate resolution in the z-direction.
For each trial, we acquired 11 z-stacks containing the neuron of interest, with 300 kPa of pressure applied during the even-numbered stacks, and 0 kPa applied during the odd-numbered stacks. We started acquisition of a stack every 15 s, although the time to acquire each given stack was ∼12 s. The exact time varied depending on the height of the z-stack, which was manually set to accommodate observed motion in the z direction during the test actuation. During the short delay between acquisition of stacks, we toggled the applied pressure.
Image analysis
To detect mitochondria, we used a particle-tracking algorithm implemented in Python (Allan et al., 2018), based on work by Crocker and Grier (Crocker and Grier, 1996). Briefly, the algorithm involves applying a spatial band pass filter, finding peaks, refining the position of peaks by finding their center of mass, and linking particles across timepoints into trajectories. In some stacks, the algorithm failed to detect the mitochondria because they were too close to the top or bottom of the stack, were not bright enough, overlapped with the mitochondria of another TRN, or were blurred due to the motion of the worm. We discarded all stacks subsequent to a stack where the image processing failed, because the strain measurement requires comparison to a previous stack. Additionally, not all of the stacks from each trial were usable, since the fluorophores bleached over time. As a result, not all actuation events yielded strain measurements (see Supplemental Figure S2).
Due to variability across trials, we manually selected a region of interest around the TRN and tuned parameters in the particle tracking algorithm. Primarily, we changed the minmass threshold, which filters out particles where the sum of pixel values within the boundaries of the particle is below the chosen threshold, and the search radius for the linking step, which specifies how far a particle can travel between timepoints and still be identified as the same particle. Less frequently, we changed the cutoff size for the bandpass filter or the brightness percentile threshold, which sets a minimum value for the brightest pixel in a particle as a percentile of the brightness of pixels in the image. We tuned these parameters until the particles found and linked included only particles along the location of the neuron and not autofluorescent spots. Supplemental Figure S3 shows the range of parameters we used.
Touch assays
To test the touch sensitivity of the mutants used in our strain transmission experiments, we performed touch assays by lightly stroking an eyebrow hair across the body of a worm and scoring its behavioral response (Chalfie et al., 2014b). For each session of touch assays, we tested 25 worms from a plate, performing 10 touches per worm. We performed the touch assays blinded with respect to genotype. For each touch event, we counted a response consisting of reversing direction or speeding up to move away from the stimulus as a positive response.
Whole-genome sequencing of him-4(e1267)
Our whole-genome sequencing protocol involved four subprotocols: 1) DNA extraction, 2) sequencing library preparation, 3) sequencing, and 4) analysis. We isolated DNA from CB1267 him-4(e1267) X animals with a phenol chloroform isoamyl alcohol (PCI) extraction. First, we washed worms off a mostly starved plate using M9 buffer, rinsed the animals twice in M9 buffer, resuspended them in EN buffer (0.1 M NaCl and 20 mM EDTA), removed the supernatant, and flash-froze the sample in liquid nitrogen. Next, we added 450 μl of worm lysis buffer (0.1 M Tris, pH 8.5, 0.1 M NaCl, 50 mM EDTA, and 1% SDS) and 40 μl of proteinase K (10 mg/ml) to 50 μl of frozen worms and incubated at 62°C for 45 min, vortexing occasionally. Then, we performed the PCI extraction in a phase lock gel tube (VWR) by adding 500 μl of PCI to the sample, vortexing, spinning for 5 min at 10,000 rpm, and then collecting the upper phase. We repeated the PCI extraction step and then extracted twice using chloroform. We precipitated the DNA by adding 40 μl of 5 M sodium acetate and 1 ml of ethanol, spinning for 5 min at 10,000 rpm, removing the supernatant, washing in 70% ethanol, and resuspending in 50 μl of TE buffer at pH 7.4.
We created a sequencing library according to manufacturer instructions (Nextera DNA Library Prep Kit, Illumina). Briefly, this includes tagmentation of the DNA using the Illumina Tagment DNA Buffer and Enzyme, cleanup of the tagmented DNA using a Zymo DNA Clean and Concentrator Kit, PCR to add the library indices to the tagmented DNA, and PCR cleanup by gel extraction using a Qiagen MinElute Kit. We did a quality control step to confirm that the average length of DNA fragments was in the expected range of 300–500 base pairs using the Agilent Bioanalyzer at the Stanford Protein and Nucleic Acid Biotechnology Facility. Sequencing was completed on an Illumina NextSeq sequencer in the Stanford Functional Genomics Facility.
We analyzed the sequencing data using the computing cluster of the Stanford Center for Genomics and Personalized Medicine. Briefly, we mapped reads using Bowtie2; used Picard to sort reads, mark duplicates, and prepare read groups; then used GATK to select high quality SNPs and INDELs; and SnpEff to annotate the results.
Code availability
Two kinds of data were analyzed using custom code: image analysis (Python, https://github.com/anekimken/SSN_ImageAnalysis) and whole-genome sequence analysis available (scripts, https://github.com/wormsenseLab/whole_genome_sequencing).
Supplementary Material
Acknowledgments
We thank Chloe Girard, Chantal Akerib, Melissa Pickett, and Anne Villenueve for the gift of their training and assistance in whole genome sequencing; Michael Nonet for NM3573, the strain with labeled mitochondria in the TRNs; Ehsan Rezeai for help with software testing; Michael Lin for the loan of a spinning disk confocal for pilot experiments. Some of the experiments were performed at Stanford core facilities, including the Cell Sciences Imaging Facility, Functional Genomics Facility (funded in part by a Shared Instrumentation Grant, National Institutes of Health [NIH] S10OD018220), the Genetics Bioinformatics Service Center, and the nano@Stanford labs (supported by the National Science Foundation as part of the National Nanotechnology Coordinated Infrastructure under award ECCS-1542152). This work was funded by an NIH fellowship to A.L.N. (F31NS100318) and grant to M.B.G. (R35NS105092).
Abbreviations used:
- ECM
extracellular matrix
- MeT
mechano-electrical transduction
- PCI
phenol chloroform isoamyl alcohol
- TRN
touch receptor neuron.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-01-0049) on June 24, 2020.
REFERENCES
- Allan DB, Caswell T, Keim NC, van der Wel CM. (2018). trackpy: Trackpy v0.4.1. 10.5281/ZENODO.1226458. [DOI]
- Bewick GS, Banks RW. (2015). Mechanotransduction in the muscle spindle. Pflügers Arch Eur J Physiol , 175–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothe T, Hilbert L, Heide M, Berninger L, Huttner WB, Zaburdaev V, Vastenhouw NL, Myers EW, Drechsel DN, Rink JC. (2017). A tunable refractive index matching medium for live imaging cells, tissues and model organisms. Elife . 10.7554/eLife.27240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounoutas A, O’Hagan R, Chalfie M. (2009). The multipurpose 15-protofilament microtubules in C. elegans have specific roles in mechanosensation. Curr Biol , 10.1016/j.cub.2009.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Au M. (1989). Genetic control of differentiation of the Caenorhabditis elegans touch receptor neurons. Science , 1027–1033. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Hart AC, Rankin CH, Goodman MB. (2014a). Assaying Mechanosensation. WormBook, 10.1895/wormbook.1.62.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Hart AC, Rankin CH, Goodman MB. (2014b). Assaying Mechanosensation. WormBook, 10.1895/wormbook.1.172.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Sulston J. (1981). Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev Biol , 358–370. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Thomson JN. (1979). Organization of neuronal microtubules in the nematode Caenorhabditis elegans. J Cell Biol , 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelur DS, Ernstrom GG, Goodman MB, Yao CA, Chen L, O’Hagan R., Chalfie M, O’Hagan R, Chalfie M. (2002). The mechanosensory protein MEC-6 is a subunit of the C. elegans touch-cell degenerin channel. Nature , 669–673. [DOI] [PubMed] [Google Scholar]
- Cho Y, Oakland DN, Lee SA, Schafer WR, Lu H. (2018). On-chip functional neuroimaging with mechanical stimulation in: Caenorhabditis elegans larvae for studying development and neural circuits. Lab Chip , 601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker JC, Grier DG. (1996). Methods of digital video microscopy for colloidal studies. J Colloid Interface Sci , 298–310. [Google Scholar]
- Cueva JG, Mulholland A, Goodman MB. (2007). Nanoscale organization of the MEC-4 DEG/ENaC sensory mechanotransduction channel in Caenorhabditis elegans touch receptor neurons. J Neurosci , 14089–14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Gu G, William CM, Chalfie M. (1996). Extracellular proteins needed for C. elegans mechanosensation. Neuron , 183–194. [DOI] [PubMed] [Google Scholar]
- Eastwood AL, Sanzeni A, Petzold BC, Park S-J, Vergassola M, Pruitt BL, Goodman MB. (2015). Tissue mechanics govern the rapidly adapting and symmetrical response to touch. Proc Natl Acad Sci , E6955–E6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emtage L, Gu G, Hartwieg E, Chalfie M. (2004). Extracellular proteins organize the mechanosensory channel complex in C. elegans touch receptor neurons. Neuron , 795–807. [DOI] [PubMed] [Google Scholar]
- Fehlauer H, Nekimken AL, Kim AA, Pruitt BL, Goodman MB, Krieg M. (2018). Using a microfluidics device for mechanical stimulation and high resolution imaging of C. Elegans. J Vis Exp . 10.3791/56530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpin W, Uppaluri S, Brangwynne CP. (2015). Worms under pressure: bulk mechanical properties of C. elegans are independent of the cuticle. Biophys J , 1887–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M, Sengupta P. (2006). How C. elegans senses mechanical stress, temperature, and other physical stimuli. Genetics, 10.1534/genetics.118.300241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Gulyanon S, Mihovilovic Skanata M, Karagyozov D, Heckscher ES, Krieg M, Tsechpenakis G, Gershow M, Tracey WD. (2019). Direction Selectivity in Drosophila proprioceptors requires the mechanosensory channel Tmc. Curr Biol , 945–956.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J, Tumkaya T, Aryal S, Choi H, Claridge-Chang A. (2019). Moving beyond P values: data analysis with estimation graphics. Nat Methods 10.1038/s41592-019-0470-3 [DOI] [PubMed] [Google Scholar]
- Katta S, Krieg M, Goodman MB. (2015). Feeling force: physical and physiological principles enabling sensory mechanotransduction. Annu Rev Cell Dev Biol. 10.1146/annurev-cellbio-100913-013426. [DOI] [PubMed] [Google Scholar]
- Katta S, Sanzeni A, Das A, Vergassola M, Goodman MB. (2019). Progressive recruitment of distal MEC-4 channels determines touch response strength in C. elegans. J Gen Physiol 10.1085/jgp.201912374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg M, Dunn AR, Goodman MB. (2014). Mechanical control of the sense of touch by β-spectrin. Nat Cell Biol , 224–233. 10.1038/ncb2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg M, Stühmer J, Cueva JG, Fetter R, Spilker K, Cremers D, Shen K, Dunn AR, Goodman MB. (2017). Genetic defects in β-spectrin and tau sensitize C. Elegans axons to movement-induced damage via torque-tension coupling. Elife . 10.7554/eLife.20172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClanahan PD, Xu JH, Fang-Yen C. (2017). Comparing Caenorhabditis elegans gentle and harsh touch response behavior using a multiplexed hydraulic microfluidic device. Integr Biol 10.1039/C7IB00120G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekimken AL, Fehlauer H, Kim AA, Manosalvas-Kjono SN, Ladpli P, Memon F, Gopisetty D, Sanchez V, Goodman MB, Pruitt BL, Krieg M. (2017a). Pneumatic stimulation of C. elegans mechanoreceptor neurons in a microfluidic trap , 1116–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekimken AL, Mazzochette EA, Goodman MB, Pruitt BL. (2017b). Forces applied during classical touch assays for Caenorhabditis elegans. PLoS One . 10.1371/journal.pone.0178080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hagan R. (2005). Components of a mechanotransduction complex in C. elegans touch receptor neurons: An in vivo electrophysiology study. PhD thesis, Columbia University. [Google Scholar]
- O’Hagan R, Chalfie M, Goodman MB. (2005). The MEC-4 DEG/ENaC channel of Caenorhabditis elegans touch receptor neurons transduces mechanical signals. Nat Neurosci , 43–50. [DOI] [PubMed] [Google Scholar]
- O’Toole M, Lamoureux P, Miller KE. (2015). Measurement of subcellular force generation in neurons. Biophys J , 1027–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold BC, Park S-J, Mazzochette EA, Goodman MB, Pruitt BL. (2013). MEMS-based force-clamp analysis of the role of body stiffness in C. elegans touch sensation. Integr Biol (Camb) , 853–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JB, Smit X, Zoysa NDe, Afoke A, Brown RA. (2004). Peripheral nerves in the rat exhibit localized heterogeneity of tensile properties during limb movement. J Physiol , 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro AJS, Denisin AK, Wilson RE, Pruitt BL. (2016). For whom the cells pull: Hydrogel and micropost devices for measuring traction forces. Methods , 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanzeni A, Katta S, Petzold B, Pruitt BL, Goodman MB, Vergassola M. (2019). Somatosensory neurons integrate the geometry of skin deformation and mechanotransduction channels to shape touch sensing. Elife , 1–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T. (2006). Maintenance of C. elegans. WormBook. 10.1895/wormbook.1.101.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sure GR, Chatterjee A, Mishra N, Sabharwal V, Devireddy S, Awasthi A, Mohan S, Koushika SP. (2018). UNC-16/JIP3 and UNC-76/FEZ1 limit the density of mitochondria in C. elegans neurons by maintaining the balance of anterograde and retrograde mitochondrial transport. Sci Rep , 8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suslak TJ, Jarman AP. (2015). Stretching the imagination beyond muscle spindles - stretch-sensitive mechanisms in arthropods. J Anat , 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Kerr R, Bianchi L, Frøkjær-Jensen C, Slone D, Xue J, Gerstbrein B, Driscoll M, Schafer WR. (2003). In vivo imaging of C. elegans mechanosensory neurons demonstrates a specific role for the MEC-4 channel in the process of gentle touch sensation. Neuron , 1005–1017. [DOI] [PubMed] [Google Scholar]
- Tao L, Porto D, Li Z, Fechner S, Lee SA, Goodman MB, Xu XZS, Lu H, Shen K. (2019). Parallel Processing of Two Mechanosensory Modalities by a Single Neuron in C. elegans. Dev Cell , 617–631.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaadia RD, Li W, Voleti V, Singhania A, Hillman EMC, Grueber WB. (2019). Characterization of proprioceptive system dynamics in behaving Drosophila larvae using high-speed volumetric microscopy. Curr Biol , 935–944.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel BE, Hedgecock EM. (2001). Hemicentin, a conserved extracellular member of the immunoglobulin superfamily, organizes epithelial and other cell attachments into oriented line-shaped junctions. Development , 883–894. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Ahlawat S, Schaefer A, Mahoney T, Koushika SP, Nonet ML. (2014). The vesicle protein SAM-4 regulates the processivity of synaptic vesicle transport. PLoS Genet , e1004644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.