Abstract
The species Enterovirus A (EV-A) consists of two conventional clusters and one unconventional cluster. At present, sequence analysis shows no evidence of recombination between conventional and unconventional EV-A types. However, the factors underlying this genetic barrier are unclear. Here, we systematically dissected the genome components linked to these peculiar phenomena, using the viral reverse genetic tools. We reported that viral capsids of the unconventional EV-A types expressed poorly in human cells. The trans-encapsidation outputs across conventional and unconventional EV-A types were also with low efficiency. However, replicons of conventional types bearing exchanged 5′-untranslated region (UTR) or non-structural regions from the unconventional types were replication-competent. Furthermore, we created a viable recombinant EVA71 (conventional type) with its P3 region replaced by that from EVA89 (unconventional type). Thus, our data for the first time reveal the potential for fertile genetic exchanges between conventional and unconventional EV-A types. It also discloses that the mysterious recombination barriers may lie in uncoordinated capsid expression and particle assembly by different EV-A clusters.
Keywords: EVA71, EVA89, recombination, evolution, reverse genetics
Introduction
Human enteroviruses are non-enveloped, positive-sense RNA viruses and members of the genus Enterovirus, and have been classified into seven species, including Enterovirus A-D and Rhinovirus A-C [1]. Enterovirus A-D currently consist of 107 proposed types, and cause various mild to severe symptoms, such as hand, foot and mouth disease (HFMD), gastroenteritis, poliomyelitis and myopericarditis [2]. Species Enterovirus A (EV-A) consists of at least 25 types, of which enterovirus A71 (EVA71) and coxsackievirus A16 (CVA16) are the major pathogens for HFMD [3]. Recently, coxsackievirus A6 (CVA6) and coxsackievirus A10 (CVA10) have emerged as the more popular etiology of HFMD [4]. Notably, a group of EV-A types including enterovirus A76 (EVA76), enterovirus A89 (EVA89), enterovirus A90 (EVA90) and enterovirus A91 (EVA91) previously isolated from acute flaccid paralysis patients, form a unique phylogenetic clade of EV-A, regarded as the ‘unconventional’ cluster, whereas the more commonly seen EV-A types (EVA71, CVA6, CVA10 and CVA16 etc.) form other ‘conventional’ clusters [5].
The enterovirus genome is about 7.5 kb in length, containing 5′-untranslated region (5′ UTR), P1-, P2- and P3-region, 3′UTR, and a poly(A) tail [6]. The P1 region encodes a polyprotein that would be cleaved into four viral structural proteins (VP4, VP2, VP3, VP1). The P2 and P3 regions encode at least seven non-structural proteins (2A, 2B, 2C, 3A, 3B, 3C, 3D) [7], which control the cellular environment, establish the membrane-bound replication complex, and replicate the genome by the formation of a negative-stranded replicative intermediate [8]. Recombination is known to be the critical driver for enterovirus evolution [9]. Previous studies showed that P2 (2AB) or P3 region hosted the most frequent recombination breakpoints for circulating enterovirus strains [10–12]. It is also reported that naturally occurring or artificially constructed 5′-UTR exchanges in enterovirus genomes generated viable recombinants [13, 14]. The relevant recombination mechanism by enteroviruses involves template-switching of the viral RNA-dependent RNA-polymerases (RdRp) during negative-strand RNA synthesis [15]. An alternative process that involves the replication-independent joining of RNA molecules has been proposed, which may be mediated by cellular RNA ligases [16, 17]. Recombination within the P1 region is rare and only occurs between very closely related viruses or involves the extreme termini of VP1, presumably due to restrictions imposed by the correct assembly of icosahedral shell [18].
Currently, the recombination events of EV-A species are incompletely understood. Although circulating EV-A recombinants are mainly formed by intra-species recombination events [12, 19, 20], there is no evidence of any genetic exchange between the conventional and unconventional EV-A types, even though they belong to the same species [5, 21]. The intriguing intraspecific segregation between the two EV-A clusters prompted us to explore the mechanisms of this recombination barrier. Here, using the viral reverse genetic tools, we constructed various vectors for EV-A capsid, subgenomic replicons and viral infectious clones, then found that capsid from unconventional EV-A types expressed poorly without codon-optimization in human cells. The trans-encapsidation system was utilized to study assembly barriers between conventional and unconventional EV-A types. The regions of 5′-UTR, P2 and P3 were exchanged to investigate whether the recombination affected the function of viral replicons. Importantly, viable EV-A recombinant cross the cluster-barriers was successfully rescued. Together, our data for the first time, reveal a potential for fertile genetic exchange between conventional and unconventional EV-A types. It also discloses that the unknown genetic exchange barrier between those two EV-A clusters likely lies in uncoordinated capsid expression and particle assembly.
Methods
Cell culture and virus
Human embryonic kidney 293T (HEK293T) cells, human rhabdomyosarcoma (RD) cells, human neuroblastoma SK-N-SH cells, African green monkey kidney Vero cells and human cervical epithelial Hela cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10 % fetal bovine serum (FBS). All cell lines were from the Cell Bank of the Chinese Academy of Sciences (Shanghai, PR China). SCARB2 knockout RD cells were reported previously [22]. EVA71 (EU703812) and the chimeric vEV71/89P3 virus were propagated in RD cells. Cells and viruses were grown in an incubator with 5 % CO2 at 37 °C. The viral titre was determined by plaque assay.
Antibodies and reagents
Mouse monoclonal antibody against EVA71-VP1 protein was from Abcam (Cambridge, UK). The secondary antibody APC-conjugated rat anti-mouse IgG1 was from BD Biosciences (Franklin Lakes, CA, USA). Fluoroshield Mounting Medium with DAPI was from Abcam. Rabbit anti-GAPDH was from Youke (Shanghai, PR China). Transfection reagents Fugene and Lipo3000 were purchased from Promega (Madison, WI, USA) and ThermoFisher Scientific (Waltham, MA, USA), respectively.
Viral sequence analysis
Phylogenetic analysis was carried out in mega5 [23]. Kimura two-parameter distances and maximum-likelihood methods were used to determine the evolutionary relationships on the basis of VP1, P2 and P3 region sequences. Here, prototype sequences of the 23 EV-A types, including 13 conventional (CVA2, CVA3, CVA4, CVA5, CVA6, CVA7, CVA8, CVA10, CVA12, CVA14, CVA16, EVA71, EVA120), 5 unconventional (EVA76, EVA89, EVA90, EVA91, EVA121) EV-A types infecting humans and the other 5 EV-A types (EVA92, EVA122, EVA123, EVA124, EVA125) from non-human hosts were analysed (https://picornaviridae.com/enterovirus/ev-a/ev-a.htm). Strains of CBV3 (M33854)and PV1 (AF111981) were used as the outgroup. Similarity analysis of the complete genome sequence alignments was performed using a sliding window of 250 nucleotides moving in steps of 30 nucleotides in Simplot v3.5.1.
Vectors construction, in vitro transcription and cell transfection
The vectors of EVA71 capsid (pcDNA6-Capsid), subgenomic replicon (PSVA-Luc), and infectious clone (PL451-EVA71) were previously reported [22, 24]. Based on those EVA71 backbone vectors, we constructed the corresponding vectors for other EV-A types through a seamless cloning protocol (Qcbio S and T, Shanghai, PR China) according to the manufacturer’s instructions. Briefly, for constructing the capsid expressing vectors, VP4-VP1 gene segment was seamlessly cloned into the pcDNA6.0-EGFP backbone. Due to low yield in transfected cells, capsid sequences of unconventional EV-A types, including EVA76, EVA89, EVA90 and EVA91, were codon-optimized (Table S1, available in the online version of this article) to increase their translation levels.
The constructions of enterovirus subgenomic replicons were based on the PSVA-Luciferase backbone and consisted of two steps. Firstly, the original EVA71’s 5′-UTR was replaced by 5′-UTRs from other EV-A types, including CVA6, CVA10, CVA16, EVA89, EVA91 and EVA125. Secondly, the non-structural gene segments (from 2A to 3′UTR) were further exchanged. Here, we generated chimeric replicons for CVA6, CVA16, CVA10 and EVA89. Similarly, we generated chimeric infectious clones in which the EVA71 P2 or P3 regions were replaced by those of EVA89, named as vEVA71/89P2 and vEVA71/89P3.
For producing viral transcripts from replicons and infectious clones, plasmids were linearized at the unique SalI restriction site downstream of the 3′ poly(A) tail and purified. Then RNA transcripts were synthesized from those linear templates with a MEGAscript T7 kit (Cat. AM1334, Invitrogen, USA) for 4 h at 37 ℃ and purified using the TIANamp RNA Kit (Cat. SD101, TIANGEN, Beijing, PR China). In vitro-transcribed RNA was quantified and examined by 0.7 % agarose gel analysis. In the current study, the RD cells in 12-well plates were transfected by the in vitro-transcribed RNA (0.5 µg/well), using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA), following the manufacturer’s instruction.
Trans-encapsidation and luciferase assay
Trans-encapsidation was conducted by sequential transfection of HEK293T cells with capsid expressor (2 days) and replicon RNA (1 day). Pseudotyped enteroviruses were harvested at 24 h post-RNA-transfection with two rounds of freeze–thaw cycle. For the luciferase assay, RD cells were transfected with replicon RNA or infected by pseudotyped virus, respectively. Twelve hours post-transfection or infection, the medium was discarded and RD cells were lysed directly on the plates by adding 150 µl cell culture lysis reagent (Promega, Madison, WI, USA). The luciferase activity of the cell lysate was detected in a Lumat LB9507 tube luminescence instrument by the luciferase assay kit (Promega, Madison, WI, USA). All experiments were carried out in duplicates and repeated at least three times.
Monitoring viral infection
Here, we applied various methods to monitor different aspects of the viral activities. Real-time PCR was used to monitor viral genome replication. Immunofluorescence microscopy and flow cytometry were used to monitor viral protein expression levels. Western blot was used to confirm the viral intervening of host cellular machinery. Viral plaque assays and virus growth curve were used to describe the virological characteristics. The experimental procedures were provided as below.
RNA extraction, reverse transcription, real-time PCR
Viral RNA was extracted from infected cell lysates using the Qiagen RNAeasy Mini kit, reverse transcribed by SuperscriptⅣ reverse transcriptase (Invitrogen) and an oligo-dT primer at 50 °C for 40 min and terminated by incubation for 15 min at 85 °C. For quantification, real-time PCR analysis was performed by using the SYBR mix with specific primers (EVA71-VP1-F: 5′- GCAGCCCAAAAGAACTTCAC-3′, EVA71-VP1-R: 5′-ATTTCAGCAGCTTGGAGTGC-3′) of VP1 on the LineGene 9600 Plus (Bori, Hangzhou, PR China). Real-time PCR procedure was conducted as following conditions: 5 mins at 95 °C, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. The quantified pcDNA6.0-EVA71VP1 plasmid was serially diluted for generating a standard curve.
Western blotting
RD cells were harvested and lysed in Radioimmunoprecipitation assay buffer (RIPA buffer) with a mixture of proteinase inhibitors (MedChem Express, Monmouth Junction, NJ, USA). The concentration of proteins was quantified by bicinchoninic acid protein assay (Biosharp, PR China) and equal amounts of proteins were loaded and separated by 10 % SDS polyacrylamide gels, and then transferred to a nitrocellulose membrane. The membrane was blocked with blocking buffer (0.1 % Tween-20 in PBS containing 5 % milk) and then was incubated overnight with mouse anti-EIF4G antibody diluted in blocking buffer at 4 °C. The membrane was then washed three times in 0.1 % Tween-20/TBS and incubated with anti-mouse IgG conjugated to AlexaFluor790 (Jackson Immuno Research, West Grove, PA, USA) for 2 h at room temperature. The immunoblots were visualized using an Odyssey Fc Imager (Lincoln, NE, USA).
Immunofluorescence microscopy and flow cytometry
RD cells were seeded in a 12-well plate (3×105 cells/well) with coverslips. The next day, RD cells were incubated with EVA71 or vEVA71/89P3 (m.o.i.=1) for 6 h. Then, RD cells were washed with cold PBS twice, fixed with 4 % paraformaldehyde (Sigma, St Louis, USA) for 15 min, permeabilized with 0.05 % Triton X-100 in 2 % FBS/PBS, and then incubated with mouse anti-VP1 antibody (1 : 1000 dilution) (Cambridge, UK) for 1 h at room temperature. Three washes with 0.01 % Triton X-100 in 2 % FBS/PBS were followed by 30 min incubation with the secondary antibodies conjugated to AlexaFluor 594 (1 : 1000 dilution) at room temperature. After washes with 0.01 % Triton X-100 in 2 % FBS/PBS. The slides were mounted with DAPI and imaged under a fluorescence microscope (Life Technology, Grand Island, NY, USA).
Cells were fixed and permeabilized with fixation/permeabilization solution (BD Biosciences) for 20 min, and then incubated with the mouse anti-VP1 antibody (1 : 1000 dilution, Abcam). After wash, cells were incubated with a rat anti-mouse secondary antibody conjugated to APC (diluted by 1 : 200, BD Biosciences), as previously described [25]. Cells were analysed by BD Accuri C6 (BD Biosciences).
Viral plaque assays and virus growth curve
RD cell monolayers in six or twelve-well culture plates (1.3×106 or 3×105 per well) were washed twice with DMEM containing 2 % FBS, and then infected with 900 or 300 µl per well of 1 : 10 serially diluted viral stocks. The plate was shaken every 15 min for 1 h. Then the inoculums were removed and 2 or 1 ml of DMEM containing 2 % FBS and 1 % low melting point agarose (Promega, Madison, USA) was added to each well, before incubation at 37 °C. Then, 3 to 5 days later, the plates were stained with 0.1 % crystal violet (Sigma, St. Louis, USA) and viral titre were measured by counting the plaques.
RD, Vero, Hela, and SK-N-SH cells were seeded in 12-well plate in triplicate. Next day, cells were infected with EVA71 or vEVA71/89P3 (RD: m.o.i.=0.01, other cells: m.o.i.=5) for 1 h at 4 °C. Then cells were washed twice with PBS before continuous culture in DMEM with 2 % FBS. Cells and supernatants were collected for measuring viral growth every 6 h for a period of total of 48 h.
Statistical analysis
Data were analysed using an unpaired Student’s t-test (*, P<0.05; **, P<0.01; ***, P<0.001) on GraphPad Prism version 8.0 (La Jolla, CA, USA). Values in graphs represent the mean and standard error from experiments performed in triplicate, with three independent experiments.
Results
Phylogenetic analysis reveals distinct clusters of EV-A types
To better understand the recombination rules of the species EV-A, we conducted genome sequence analysis of different EV-A types. VP1 sequence difference determine the enterovirus types, thus we built a phylogenetic tree of the VP1 region (Fig. 1a). As expected, CVB3- and PV1-VP1 formed the outgroup, while all other EV-A types grouped together and formed distinct clusters. Consistent with earlier reports [5, 26], two of those clusters (the conventional I and conventional II cluster) contained commonly identified pathogens for HFMD, while a third cluster (the unconventional cluster) consisted of the uncommon EV-A types, including the EVA76, EVA89, EVA90, EVA91 and EVA121. Intriguingly, among the three different human-infecting EV-A clusters, recombination between the two conventional clusters were frequently observed [19], however, those between conventional and unconventional clusters have never been encountered. To better understand the rules governing genetic exchanges within the species EV-A, we conducted the following analysis.
Fig. 1.
The genomic comparison reveals the clustering structure of the species Enterovirus A. (a) Constructing the evolutionary relationship of 17 human-infecting Enterovirus A types using the complete prototype strain VP1 nucleotide sequences. An unrooted tree was built using the maximum-likelihood algorithm implemented in mega5, as described in Methods. Genetic distance is shown above the branches, and bootstrap values (1000 replicates) are shown below. The branch names indicate Enterovirus types and the prototype GenBank accession number. The rightmost labels indicate distinct clusters of the species Enterovirus A. (b) Similarity plot of the complete genome sequences of the selected human-infecting types from different clusters of Enterovirus A, using a sliding window of 250 nucleotides moving in 30-nucleotide steps. EVA71 BrCr strain was used as the query sequence.
Since larger sequence differences may preclude the recombination, we first conducted similarity analysis of the complete genome sequence of conventional and unconventional EV-A types by plotting the sequence identities (based on the EVA71 query) in a sliding window using SimPlot (Fig. 1b). We observed that the similarity patterns varied at different genomic regions. The capsid gene was the most divergent part, but the three EV-A clusters showed roughly equal divergence. Meanwhile, despite that the 5′-UTR and non-structural regions were more similar, the unconventional cluster did diverge more from the other two conventional clusters (Fig. 1b). These findings were further supported by sequence comparisons between any two viral types from different clusters (Table 1) and phylogenetic analysis of P2 and P3 regions (Fig. S1), which identify conventional and unconventional groups of EV-A types. Thus, our data shows that genetic segregation between the conventional and unconventional EV-A clusters concurs with larger sequence differences in the non-capsid regions between them.
Table 1.
Comparison of the prototype nucleotide sequences and deduced amino acid sequences among viral types of conventional and unconventional EV-A clusters
|
Region |
Within unconventional cluster (%identity) |
Within conventional cluster (%identity) |
Between different clusters (%identity) |
|||
|---|---|---|---|---|---|---|
|
Nucleotide |
Amino acid |
Nucleotide |
Amino acid |
Nucleotide |
Amino acid |
|
|
5'UTR |
83.88 (77.1–94.2) |
– |
83.95 (80.7–88.5) |
– |
74.61 (69.6–81.3) |
– |
|
VP4 |
80.54 (76.3–85.0) |
95.92 (92.7–100) |
68.47 (61.3–75.8) |
74.84 (62.3–91.3) |
67.45 (62.8–72.4) |
72.73 (63.7–78.2) |
|
VP2 |
70.53 (67.6–72.6) |
81.75 (78.5–86.0) |
68.01 (64.0–100) |
76.35 (69.8–100) |
66.78 (63.6–71.8) |
75.25 (71.8–81.0) |
|
VP3 |
70.74 (69.6–71.8) |
82.86 (79.5–85.6) |
67.72 (63.6–72.5) |
75.22 (68.1–88.3) |
67.19 (64.3–71.1) |
75.42 (69.6–79.7) |
|
VP1 |
67.36 (65.1–70.1) |
72.75 (70.0–80.1) |
59.07 (52.6–73.2) |
61.11 (53.0–83.3) |
59.09 (52.4–63.3) |
60.68 (53.5–65.2) |
|
2A |
71.54 (65.7–91.7) |
77.03 (70.0–98.6) |
79.56 (75.5–86.4) |
96.17 (94.6–98.0) |
67.29 (64.0–71.7) |
72.37 (68.6–80.6) |
|
2B |
80.01 (75.0–87.5) |
95.6 (92.9–98.9) |
80.93 (74.7–91.5) |
96.95 (93.9–100) |
68.48 (62.6–73.4) |
76.1 (73.7–79.7) |
|
2C |
80.73 (74.9–90.7) |
92.4 (87.5–99.0) |
82.43 (77.1–92.7) |
97.4 (96.0–99.0) |
75.16 (72.5–77.7) |
88 (86.3–90.5) |
|
3A |
77.08 (67.4–91.7) |
86 (76.7–100) |
81.63 (76.3–91.8) |
97.03 (93.0–100) |
71.69 (67.8–77.9) |
83.53 (77.9–90.6) |
|
3B |
77.82 (66.6–89.3) |
87.24 (77.2–100) |
78.28 (63.6–93.9) |
92.32 (81.8–100) |
66.28 (56.0–77.2) |
77.45 (63.6–86.3) |
|
3C |
79.52 (72.6–91.6) |
90 (84.1–98.3) |
80.17 (75.0–100) |
94.95 (91.2–100) |
72.89 (69.7–77.2) |
84.99 (83.0–96.3) |
|
3D |
90.24 (89.2–95.5) |
98.44 (97.6–99.3) |
81.23 (76.4–100) |
95.33 (92.4–100) |
72.95 (71.2–74.2) |
83.64 (82.6–86.6) |
|
3'UTR* |
90.53 (88.2–97.8) |
– |
85.8 (75.9–100) |
– |
33.25 (27.3–37.8) |
– |
Conventional EV-A cluster include prototype CVA2, CVA4, CVA5, CVA6, CVA7, CVA8, CVA10, CVA12, CVA14, CVA16 and EVA71.
Unconventional EV-A cluster include prototype EV-A76, EV-A89, EV-A90, EV-A91 and EV-A121.
The average and range of the identities are shown. The 3′-UTR of EVA121 is removed from analysis because of its short length.
Capsid region of unconventional EV-A types expressed poorly in human cell lines
Next, we conducted functional analysis at different genomic segments of the different EV-A clusters. Based on the EVA71-capsid expression vector carrying a fused and cleavable enhanced green fluorescent protein reporter (EGFP), we constructed the capsid expressors for four conventional EV-A types including EVA71, CVA6, CVA16 and CVA10, and four unconventional EV-A types including EVA76, EVA89, EVA90 and EVA91 (Fig. 2a). These plasmids were confirmed by the Sanger sequencing and quality-examined by electrophoresis (Fig. 2b). Then equal quantities of plasmids were introduced into HEK293T cells and the EGFP expression levels were examined by fluorescence microscopy (Fig. 2c, d). To our surprise, although EVA71, CVA6, CVA16 and CVA10 capsid expressors produced brighter EGFP signals, those of EVA76, EVA89, EVA90 and EVA91 were much dimmer, indicating poorer expressions. Interestingly, upon human-codon optimization, EVA76-, EVA89-, EVA90- and EVA91-opti capsid vectors regained strong EGFP signals (Fig. 2e), indicating codon bias as a crucial factor resulting in the lower capsid expression specifically among the unconventional EV-A types. Indeed, the nucleotide composition differed between unconventional and unconventional EV-A types (Table S2). In addition, we tested RD and Hela cells and further confirmed poorer expression by capsid gene from unconventional types (unpublished data).
Fig. 2.
Viruses from the conventional and unconventional Enterovirus A clusters differed in their capsid expression levels in human cells. (a) Schematic representation of the enterovirus capsid expressing vector, which contained a CMV promoter followed by EGFP reporter, VP4, VP2, VP3 and VP1 coding sequences. A viral 2A protease cleavage site (AITTL) was inserted between EGFP and VP4. (b) Image showing the capsid plasmids analysed by agarose gel electrophoresis. ‘Opti-’ stands for the codon-optimized plasmid. (c–e) HEK293T cells were transfected by capsid plasmids and EGFP signals were captured using fluorescence microscopy 2 days later. EGFP expression levels among (c) conventional Enterovirus A types, (d) unconventional Enterovirus A types and (e) those of codon-optimized. (f) Various pseudoviruses were produced by trans-encapsidation using different viral capsids and EVA71 replicon RNA. Pseudoviruses were harvested after two rounds of freeze–thaw cycle, and then added to RD cells. Then, 12 h post-infection, the luciferase activities were recorded. The GnHCl (viral replication inhibitor) treated EVA71-pseudovirus was used as the assay controls. (g) The pseudoviruses of EVA71, CVA10 and EVA91, representative of three Enterovirus A clusters, were generated. These pseudovirus infectivities in SCARB2-expressing (WT) or null (KO) cells were examined by monitoring luciferase activity.
Next, we sought to determine the trans-encapsidation ability for the capsid of each EV-A type. Pseudotyped enteroviruses were produced by sequential transfection of capsid expressor and EVA71 replicon RNAs containing a luciferase reporter. Then the pseudoviruses infectivity was evaluated by luciferase activity. As shown in Fig. 2f, pseudoviruses of the conventional EV-A types manifested much higher activities than those pseudotyped by unconventional types, despite that the codon-optimization improved their yields. Thus, the EVA71-RNA trans-encapsidation by capsids from conventional types was more efficient than those from unconventional types. We further showed that EVA71-typed pseudovirus could infect RD cells but not SCARB2-null cells, while pseudoviruses carrying CVA10 or EVA91 capsid could infect both cell lines (Fig. 2g), confirming their independence of using SCARB2 as receptors.
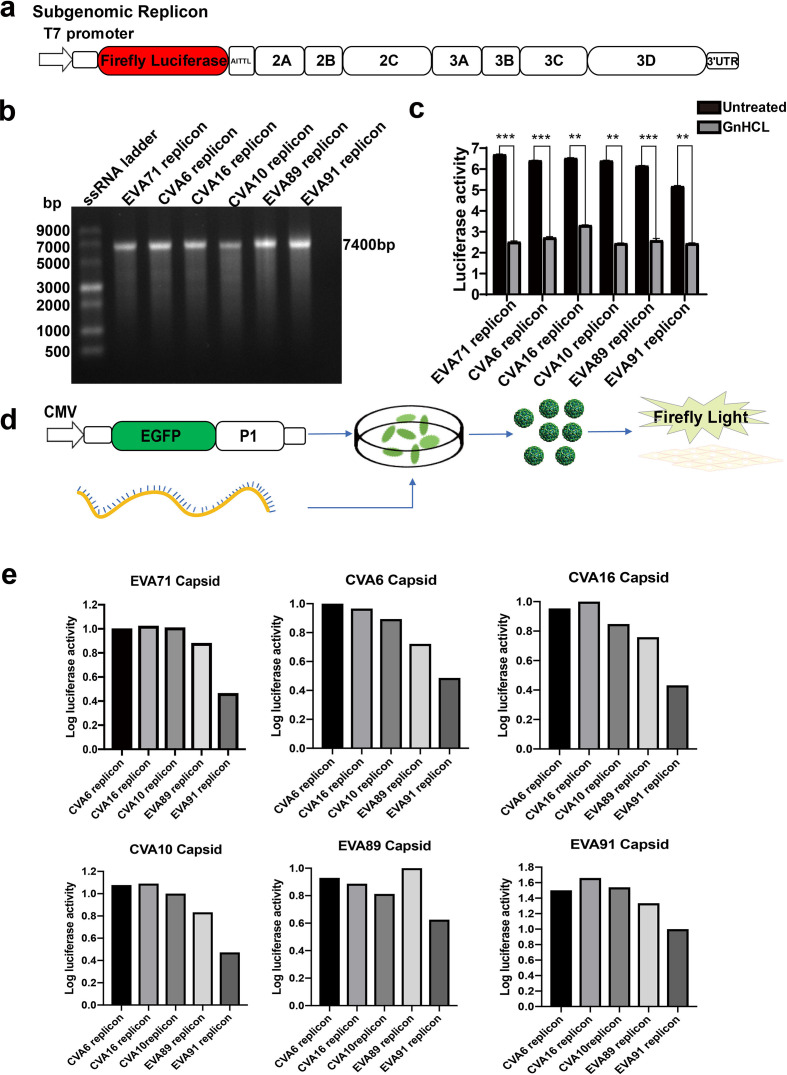
Trans-encapsidation sets' recombination barriers between the conventional and unconventional EV-A types
To further evaluate the trans-encapsidation between conventional and unconventional EV-A types, we systematically constructed viral replicons and capsid expressors of various viral types (capsid expressors for unconventional types were codon-optimized in this experiment) (Fig. 3a, d). The replicon RNAs were generated by in vitro transcription (Fig. 3b) and evaluated after introduction into RD cells. The replicative activities of those enteroviral replicons can be appreciated by its comparison with GnHCl treatment control. Replicons from unconventional EV-A types were capable of replication despite their lower activity than those of conventional types (Fig. 3c). Trans-encapsidation efficiency was then evaluated systematically (Fig. 3d, e). Consistent with their lower levels of replicon activities, EVA89 and EVA91 replicon trans-encapsidation efficiency was generally lower than those from conventional EV-A types, except for EVA89-replicon trans-encapsidated by EVA89-capsid, which led to a comparable pseudovirus activity as other conventional types. When comparing different viral capsids, EVA89 and EVA91 capsids also seemed to show a slightly better trans-encapsidation efficiency towards their own replicons (Fig. 3e). Together, these data did suggest that a weak barrier preventing trans-encapsidation between conventional and unconventional types existed.
Fig. 3.
Systematic testing of trans-encapsidation between conventional and unconventional Enterovirus A clusters. (a) The cartoon showed the construct of the subgenomic enterovirus replicon carrying a luciferase reporter. (b) SalI linearized each replicon vector was in vitro-transcribed by the T7 polymerase in vitro-transcription kit to generate the replicon RNA. The quality of those RNAs was examined by agarose gel electrophoresis. (c) Luciferase activity was monitored at 12 h after introducing replicon RNAs into RD cells. GnHCL treatment was used to determine the baseline luciferase activity. (d) The schematic diagram of generating pseudoviruses by trans-encapsidation. (e) Systematic activity tests of pseudoviruses generated by cross trans-encapsidations between CVA6-, CVA10-, CVA16-, EVA89- and EVA91-replicon RNAs and capsids. The values represent relative luciferase activity normalized to signals from the replicons pseudotyped by its native capsids. Data were summarized as mean±sd (n=3). Unpaired Student’s t-test, **P<0.01; ***P<0.001.
Recombinants outside the EV-A capsid regions are replication-competent
Previous studies showed that enterovirus recombination preferentially occurred at 5′-UTR and non-structural regions [9]. Here, we functionally dissected this recombination between different EV-A types. First, we created the 5′-UTR recombinants from the EVA71 replicon backbone (Fig. 4b). Although the 5′-UTR diverged largely between conventional and unconventional EV-A types (Fig. 4a), we found that all 5′-UTR swapping replicons retained comparable replicative capabilities, including the most divergent EVA125-5′-UTR recombinant (Fig. 4c). Furthermore, similar levels of pseudoviruses were generated from those replicons trans-encapsidated with an EVA71 capsid (Fig. 4d), suggesting that the 5′-UTR region was fully exchangeable among different EV-A types.
Fig. 4.
Testing genome recombinations at 5′-UTR and non-structural protein regions. (a) Phylogenetic trees of the 5’UTR sequences from the various Enterovirus A types. (b) Schematic representation of the six chimeric replicons and the parental EVA71. The 5′-UTR of CVA6, CVA16, CVA10, EVA89, EVA91 and EVA125 were differentially coloured. (c) The 5′-UTR chimeric replicon RNAs were obtained by in vitro transcription and transfected into RD cells. Luciferase activity was monitored at 12 h post-transfection. GnHC1 treatment was used as the assay control. (d) The 5′-UTR chimeric replicon RNAs and EVA71-capsid plasmids were sequentially transfected into HEK293T cells to produce trans-encapsidated pseudoviruses. The infectivity of those pseudoviruses was reflected by luciferase activities at 12 h post-infection. (e) P2- or (g) P3-region of EVA71 was replaced by the counterpart of CVA6, CVA16, CVA10 and EVA89 to create the chimeric replicon constructs as indicated. (f, h) Luciferase activities in RNA transfected RD cells reflect the replication activities of those (f) P2- and (h) P3 chimeric replicons. The values represent luciferase activity in three independent experiments (mean±sd). Unpaired Student’s t-test, *P<0.05; **P<0.01; ***P<0.001.
Next, to determine whether the exchange of P2 or P3 region affected the replication activity of EV-A types, we replaced the P2 or P3 in an EVA71 replicon backbone by those from other EV-A types (Fig. 4e, g). Among the recombinants within conventional EV-A types, we found that rEVA71/16P2, rEVA71/10P2 and rEVA71/16P3 were replication-competent, while rEVA71/6P2, rEVA71/6P3 and rEVA71/10P3 were not. However, both rEVA71/89P2 and rEVA71/89P3, to our surprise, were replication-competent (Fig. 4f, g), suggesting that the exchange of the non-structural region across the conventional and unconventional EV-A types could be tolerable.
A viable viral recombinant between conventional and unconventional EV-A types
Based on the aforementioned replicon data, we next sought to determine whether enterovirus recombination across conventional and unconventional types were indeed viable. We constructed infectious clones for viral recombinants, vEVA71/89P2 and vEVA71/89P3, by swapping P2 or P3 in the EVA71 infectious clone with those from EVA89 (Fig. 5a). The viral RNAs were obtained and introduced into RD cells for rescuing the viral recombinants. Cytopathic effect (CPE) post-vEVA71/89P3 transfection was observed after four blind passages, although vEVA71/89P2 was not viable (Fig. 5b). The titre of vEVA71/89P3 was determined to be 1.6 Í107 p.f.u. ml−1 (Fig. 5c). Its infectivity on RD cells could be similarly monitored by immunofluorescence and flow cytometry measuring EVA71-VP1 levels (Fig. 5d, e). Cap-dependent translation factor eIF4G, is cleaved during enterovirus infection [27], here we confirmed that vEVA71/89P3 infection cleaved eIF4G in RD cells by immunoblot, which further verify the viral intervening of host cellular machinery (Fig. 5f). In addition, the growth rates of EVA71 and vEVA71/89P3 in RD, Vero, Hela, and SK-N-SH cells were also similar (Fig. 5g). Thus, the generation of vEVA71/89P3 proved that recombination between conventional and unconventional EV-A types could produce viable EV-A recombinants.
Fig. 5.
Generation of viable EVA71 and EVA89 recombinant by artificial recombination. (a) The cartoon showed the recreation of viral recombinants (vEVA71/89P2 and vEVA71/89P3) through artificially exchanging EVA71’s P2 or P3 by EVA89’s P2 or P3, respectively. (b) RNAs generated from infectious clones were introduced into RD cells. vEVA71/89P3 but not vEVA71/89P2 caused obvious cytopathic effect (CPE) after four blind passages in cell culture. EVA71 was used as a positive control. (c) The litres of rescued vEVA71/89P3 viral stocks were determined by plaque assay. (d) RD cells were infected by vEVA71/89P3 (m.o.i.=1) overnight, and then fixed and stained by an anti-VP1 primary antibody and AlexFlour 647-conjugated secondary antibody for fluorescence microscopy. DAPI was used to visualize the nuclei. (VP1, red; nuclei, blue). (e) RD cells were infected by vEVA71/89P3 (m.o.i.=1) for 6 h. Then the proportion of VP1-expressing cells was measured by flow cytometry. (f) RD cells were infected by EV71 or vEVA71/89P3 (m.o.i.=1) for 6 h. The cleavage of eIF4G was determined by immunoblot. (g) The infectivity of EV71 and vEV71/89P3 was examined in different cell lines. The charts depicted the viral growth curve by monitoring viral RNA levels in infected cultures.
Discussion
Genome recombination is one of the driving forces for enterovirus evolution and emergence of new viral strains [28]. Although inter-species recombination is relatively rare, intra-species recombination among enteroviruses is frequent. Based on VP1 phylogenetics, human-infecting EV-A types consists of three distinct clusters. Two conventional clusters include the commonly identified EV-A types causing HFMD (such as CVA6, CVA10, CVA16 and EVA71), while the unconventional cluster includes those uncommon types (such as EVA76, EVA89, EVA90 and EVA91). Although recombination within the conventional EV-A types was frequently observed [19], those across conventional and unconventional types have never been identified.
Related to the unclear factors underlying this recombination barrier, we found here that the capsid genes from the unconventional EV-A types expressed poorly in human cells. However, codon-optimization could restore them to comparable levels with those of conventional types. Indeed, our preliminary sequence analysis supports that there is a significant codon-bias by unconventional EV-A types versus conventional ones (Table S2 and unpublished data). These data imply that unconventional and conventional EV-A types likely circulate in different host species. We feel interested to speculate that human is not the natural host for unconventional EV-A types and this could also explain why there is a recombination barrier between conventional and unconventional EV-A types. Actually, there are other EV-A types (such as EVA92, EVA119, EVA122, EVA123, EVA124 and EVA125) isolated from non-human hosts [29]. Including those types into future analysis may reveal more insights about the genomic evolution, codon-bias and host species segregation of the different EV-A types.
Recombination events in enteroviruses more frequently occur at the non-capsid regions [9]. Here, we functionally validated the recombination at non-capsid regions across conventional and unconventional EV-A types. Indeed, the sequences of 5′-UTR and non-structural proteins are more divergent across – than within – the conventional and unconventional clusters, consistent with that they are more genetically segregated. However, two hybrid replicons harboring exchanged 5′-UTR or non-structural regions across conventional and unconventional EV-A clusters (vEVA71/89P2 and vEVA71/89P3) are found to be replication-competent. Thus, our data support that across various EV-A types, the function of non-structural components is likely well preserved and that recombination at the non-capsid regions is unrestricted. More importantly, we were able to rescue vEVA71/89P3 from its infectious clone, the very first viable recombinant enterovirus crossing the conventional and unconventional cluster barrier. The success of generating vEVA71/89P3 further proved the potential for possible fertile genetic exchanges between conventional and unconventional EV-A types. Although unconventional EV-A types are less commonly encountered in human infections, they were previously identified in surveillance program of acute flaccid paralysis [5, 21], suggesting possibility to cause human diseases. Therefore, tracking the emergence and validating potential pathogenesis of the cluster-crossing recombinant EV-A strains should be aimed by future research.
In conclusion, this study deepens our understandings of factors driving enterovirus evolution, showing that viral capsid translation efficiency may set the mysterious recombination barrier segregating conventional and unconventional EV-A clusters, whereas the non-capsid regions remain fully exchangeable.
Supplementary Data
Funding information
This work was supported by the Shanghai Public Health Clinical Center (KY-GW-2018–17)
Acknowledgements
We want to thank Professor Wenhui Li at National Institute of Biomedical Sciences (NIBS) for providing us the capsid, subgenomic replicon and infectious clone of EVA71.
Author contributions
M.W. and L.Z. conducted most of experiments, analysed data and drafted the manuscript; J.F. and J.Y. were involved in experiment design, data analysis and manuscript preparation; Y.D., R.Y. and L.L. participated in some experiments; S.Z. guided the experiment design and data analysis, and finalized the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: CPE, cytopathic effect; CVA2, coxsackievirus A2; CVA3, coxsackievirus A3; CVA4, coxsackievirus A4; CVA5, coxsackievirus A5; CVA6, coxsackievirus A6; CVA7, coxsackievirus A7; CVA8, coxsackievirus A8; CVA10, coxsackievirus A10; CVA12, coxsackievirus A12; CVA14, coxsackievirus A14; CVA16, coxsackievirus A16; DMEM, Dulbecco's modified Eagle's Medium; EGFP, enhanced green fluorescent protein reporter; EV-A, enterovirus A; EV-A71, enterovirus A71; EVA76, enterovirus A76; EVA89, enterovirus A89; EVA90, enterovirus A90; EVA91, enterovirus A91; EVA92, enterovirus A92; EVA120, enterovirus A120; EVA121, enterovirus A121; EVA122, enterovirus A122; EVA123, enterovirus A123; EVA124, enterovirus A124; EVA125, enterovirus A125; FBS, fetal bovine serum; HEK293T, human embryonic kidney 293T; HFMD, hand foot and mouth disease; RD, human rhabdomyosarcoma; RdRp, RNA-dependent RNA-polymerases.
Two supplementary tables and one supplementary figures are available with the online version of this article.
References
- 1.Nikonov OS, Chernykh ES, Garber MB, Nikonova EY. Enteroviruses: classification, diseases they cause, and approaches to development of antiviral drugs. Biochemistry. 2017;82:1615–1631. doi: 10.1134/S0006297917130041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simmonds P, Gorbalenya AE, Harvala H, Hovi T, Knowles NJ, et al. Recommendations for the nomenclature of enteroviruses and rhinoviruses. Arch Virol. 2020;165:793–797. doi: 10.1007/s00705-019-04520-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aswathyraj S, Arunkumar G, Alidjinou EK, Hober D, Hand HD. Hand, foot and mouth disease (HFMD): emerging epidemiology and the need for a vaccine strategy. Med Microbiol Immunol. 2016;205:397–407. doi: 10.1007/s00430-016-0465-y. [DOI] [PubMed] [Google Scholar]
- 4.Fu X, Wan Z, Li Y, Hu Y, Jin X, et al. National epidemiology and evolutionary history of four hand, foot and mouth disease-related enteroviruses in China from 2008 to 2016. Virol Sin. 2020;35:21–33. doi: 10.1007/s12250-019-00169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oberste MS, Maher K, Michele SM, Belliot G, Uddin M, et al. Enteroviruses 76, 89, 90 and 91 represent a novel group within the species human enterovirus a. J Gen Virol. 2005;86:445–451. doi: 10.1099/vir.0.80475-0. [DOI] [PubMed] [Google Scholar]
- 6.Baggen J, Thibaut HJ, Strating JRPM, van Kuppeveld FJM. The life cycle of non-polio enteroviruses and how to target it. Nat Rev Microbiol. 2018;16:368–381. doi: 10.1038/s41579-018-0005-4. [DOI] [PubMed] [Google Scholar]
- 7.Gao Y, Sun S-Q, Guo H-C. Biological function of foot-and-mouth disease virus non-structural proteins and non-coding elements. Virol J. 2016;13:107. doi: 10.1186/s12985-016-0561-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin J-Y, Chen T-C, Weng K-F, Chang S-C, Chen L-L, et al. Viral and host proteins involved in picornavirus life cycle. J Biomed Sci. 2009;16:103. doi: 10.1186/1423-0127-16-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikolaidis M, Mimouli K, Kyriakopoulou Z, Tsimpidis M, Tsakogiannis D, et al. Large-Scale genomic analysis reveals recurrent patterns of intertypic recombination in human enteroviruses. Virology. 2019;526:72–80. doi: 10.1016/j.virol.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Lukashev AN, Lashkevich VA, Ivanova OE, Koroleva GA, Hinkkanen AE, et al. Recombination in circulating enteroviruses. J Virol. 2003;77:10423–10431. doi: 10.1128/JVI.77.19.10423-10431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bessaud M, Joffret M-L, Holmblat B, Razafindratsimandresy R, Delpeyroux F, et al. Genetic relationship between cocirculating human enteroviruses species C. PLoS One. 2011;6:e24823. doi: 10.1371/journal.pone.0024823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kyriakopoulou Z, Pliaka V, Amoutzias GD, Markoulatos P. Recombination among human non-polio enteroviruses: implications for epidemiology and evolution. Virus Genes. 2015;50:177–188. doi: 10.1007/s11262-014-1152-y. [DOI] [PubMed] [Google Scholar]
- 13.Schibler M, Gerlach D, Martinez Y, Van Belle S, Turin L, et al. Experimental human rhinovirus and enterovirus interspecies recombination. J Gen Virol. 2012;93:93–101. doi: 10.1099/vir.0.035808-0. [DOI] [PubMed] [Google Scholar]
- 14.Muslin C, Joffret M-L, Pelletier I, Blondel B, Delpeyroux F. Evolution and emergence of enteroviruses through intra- and inter-species recombination: plasticity and phenotypic impact of modular genetic exchanges in the 5' untranslated region. PLoS Pathog. 2015;11:e1005266. doi: 10.1371/journal.ppat.1005266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowry K, Woodman A, Cook J, Evans DJ. Recombination in enteroviruses is a biphasic replicative process involving the generation of greater-than genome length 'imprecise' intermediates. PLoS Pathog. 2014;10:e1004191. doi: 10.1371/journal.ppat.1004191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodman A, Lee K-M, Janissen R, Gong Y-N, Dekker NH, et al. Predicting Intraserotypic recombination in enterovirus 71. J Virol. 2019;93 doi: 10.1128/JVI.02057-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallei A, Pankraz A, Thiel H-J, Becher P. Rna recombination in vivo in the absence of viral replication. J Virol. 2004;78:6271–6281. doi: 10.1128/JVI.78.12.6271-6281.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martín J, Samoilovich E, Dunn G, Lackenby A, Feldman E, et al. Isolation of an intertypic poliovirus capsid recombinant from a child with vaccine-associated paralytic poliomyelitis. J Virol. 2002;76:10921–10928. doi: 10.1128/JVI.76.21.10921-10928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukashev AN, Shumilina EY, Belalov IS, Ivanova OE, Eremeeva TP, et al. Recombination strategies and evolutionary dynamics of the human enterovirus a global gene pool. J Gen Virol. 2014;95:868–873. doi: 10.1099/vir.0.060004-0. [DOI] [PubMed] [Google Scholar]
- 20.Schibler M, Piuz I, Hao W, Tapparel C, López S. Chimeric rhinoviruses obtained via genetic engineering or artificially induced recombination are viable only if the polyprotein coding sequence derives from the same species. J Virol. 2015;89:4470–4480. doi: 10.1128/JVI.03668-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang K, Zhang Y, Song Y, Cui H, Yan D, et al. Antigenic characteristics and genomic analysis of novel EV-A90 enteroviruses isolated in Xinjiang, China. Sci Rep. 2018;8:10247. doi: 10.1038/s41598-018-28469-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang M, Yan J, Zhu L, Wang M, Liu L, et al. The establishment of infectious clone and single round infectious particles for coxsackievirus A10. Virol Sin. 2020;205 doi: 10.1007/s12250-020-00198-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen P, Song Z, Qi Y, Feng X, Xu N, et al. Molecular determinants of enterovirus 71 viral entry: cleft around GLN-172 on VP1 protein interacts with variable region on scavenge receptor B 2. J Biol Chem. 2012;287:6406–6420. doi: 10.1074/jbc.M111.301622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan M, Yan J, Xun J, Chen C, Zhang Y, et al. Enhanced human enterovirus 71 infection by endocytosis inhibitors reveals multiple entry pathways by enterovirus causing hand-foot-and-mouth diseases. Virol J. 2018;15:1. doi: 10.1186/s12985-017-0913-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deshpande JM, Sharma DK, Saxena VK, Shetty SA, Qureshi T, et al. Genomic characterization of two new enterovirus types, EV-A114 and EV-A121. J Med Microbiol. 2016;65:1465–1471. doi: 10.1099/jmm.0.000380. [DOI] [PubMed] [Google Scholar]
- 27.Hanson PJ, Ye X, Qiu Y, Zhang HM, Hemida MG, et al. Cleavage of DAP5 by coxsackievirus B3 2A protease facilitates viral replication and enhances apoptosis by altering translation of IRES-containing genes. Cell Death Differ. 2016;23:828–840. doi: 10.1038/cdd.2015.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muslin K. Bessaud, Blondel, Delpeyroux. recombination in enteroviruses, a multi-step modular evolutionary process. Viruses. 2019;11 doi: 10.3390/v11090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fieldhouse JK, Wang X, Mallinson KA, Tsao RW, Gray GC. A systematic review of evidence that enteroviruses may be zoonotic. Emerg Microbes Infect. 2018;7:1–9. doi: 10.1038/s41426-018-0159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.