Abstract
Objectives
The main endocrine cell types in pancreatic islets are alpha, beta, and delta cells. Although these cell types have distinct roles in the regulation of glucose homeostasis, inadequate purification methods preclude the study of cell type-specific effects. We developed a reliable approach that enables simultaneous sorting of live alpha, beta, and delta cells from mouse islets for downstream analyses.
Methods
We developed an antibody panel against cell surface antigens to enable isolation of highly purified endocrine subsets from mouse islets based on the specific differential expression of CD71 on beta cells and CD24 on delta cells. We rigorously demonstrated the reliability and validity of our approach using bulk and single cell qPCR, immunocytochemistry, reporter mice, and transcriptomics.
Results
Pancreatic alpha, beta, and delta cells can be separated based on beta cell-specific CD71 surface expression and high expression of CD24 on delta cells. We applied our new sorting strategy to demonstrate that CD71, which is the transferrin receptor mediating the uptake of transferrin-bound iron, is upregulated in beta cells during early postnatal weeks. We found that beta cells express higher levels of several other genes implicated in iron metabolism and iron deprivation significantly impaired beta cell function. In human beta cells, CD71 is similarly required for iron uptake and CD71 surface expression is regulated in a glucose-dependent manner.
Conclusions
This study provides a novel and efficient purification method for murine alpha, beta, and delta cells, identifies for the first time CD71 as a postnatal beta cell-specific marker, and demonstrates a central role of iron metabolism in beta cell function.
Keywords: Islet, Alpha cell, Beta cell, Delta cell, Transferrin receptor, FACS, Iron metabolism
Graphical abstract

Highlights
-
•
CD71 is a marker that is highly expressed in murine pancreatic beta-cells.
-
•
CD71 and CD24 can be used to purify live murine alpha-, beta-, and delta-cells.
-
•
Iron metabolism in murine beta-cells is increased compared to that in alpha-, and delta-cells.
-
•
Human beta-cells regulate CD71 surface expression in a glucose-dependent manner.
1. Introduction
Diabetes is a metabolic disorder that affects more than 400 million people worldwide. The disease is characterized by chronic hyperglycemia resulting from dysfunction or destruction of insulin-secreting beta cells. Together with glucagon-secreting alpha cells and somatostatin-secreting delta cells, beta cells reside in small cell clusters or islets dispersed throughout the pancreas. Paracrine interactions between these endocrine cell types are paramount to a fine-tuned regulation of blood glucose. Phenotyping the different endocrine islet cell types at distinct stages of development or physiology was hitherto not feasible due to a lack of protocols for simultaneous subset purification. Sorting of dispersed islet cells has generally been performed to separate beta from non-beta cells using zinc probes [1,2], cellular granularity [3], autofluorescence [4], or antibodies directed at surface markers [[5], [6], [7]]. Neither of these tools are widely used due to the limited purity of the obtained beta cell-enriched subset, but also the inability to adequately and simultaneously isolate the other endocrine subsets. The best available tools for isolating alpha, beta, and delta cells are fluorescent reporter mice that allow the simultaneous isolation of only two of these cell types (alpha and beta cells, or delta and beta cells) [8]. Unfortunately, these genetic features are difficult to introduce into other mouse models due to the complexity of the crossings and strain-specific susceptibility to environmental factors such as high-fat diet, a model for type 2 diabetes. Single cell technologies may resolve cell specificity in such contexts but still have downsides including lower signal-to-noise ratios compared to bulk analyses [9].
Cell sorting based on antibody panels against surface markers enabled the advanced characterization of subsets in hematopoietic tissues [10]. We recently used a similar approach to reconstruct human pancreatic organogenesis [11,12] and have now applied the same strategy to improve FACS-based purification of endocrine subsets. Using a combination of the surface markers CD24, CD49f, and CD71, we were able to reliably sort live high-purity alpha, beta, and delta cells. The specific surface expression of the transferrin receptor CD71 on beta cells revealed their increased iron metabolism compared to alpha and delta cells. The reported progress is relevant to diabetes pathophysiology because of an elevated risk of diabetes under pathological iron overload conditions [13,14] as well as in those with mutations in iron metabolism genes [15,16] and elevated iron stores within the physiological range [[17], [18], [19]]. We show that both mouse and human beta cells express high CD71 levels for the import of transferrin-bound iron and in a human beta cell line, CD71 surface levels are regulated in a glucose-dependent manner. Furthermore, we provide proof of the detrimental impact of iron deprivation on murine beta cell function.
Our work shows proof of concept for a novel and efficient purification method for mouse alpha, beta, and delta cells, defines CD71 as a beta cell-specific marker, highlights the importance of a fine-tuned iron metabolism for beta cell function, and provides new mechanistic clues for understanding beta cell susceptibility to iron-induced cellular damage.
2. Methods
2.1. Animal procedures
All of the animal studies complied with the ARRIVE guidelines and were conducted in strict accordance with the EU Directive 2010/63/EU for animal experiments and with regard to specific national laws and INSERM guidelines. Mice were maintained on a 12:12 h light–dark cycle at an ambient temperature of 23 °C and provided water and food ad libitum. Mice were killed either by CO2 asphyxiation or cervical dislocation. C57BL/6JRj mice, including pregnant and neonatal mice, were obtained from Janvier Labs (Saint Berthevin, France). All of the adult mice were 12 weeks old and male unless otherwise specified. Fetal pancreatic buds were dissected as previously described [20]. Fetal and neonatal pancreases were procured from mixed litters and pooled independent of sex. Ins1-Cre; Rosa26-LSL-YFP reporter mice (Ins-YFP) were a gift from Harry Heimberg and were obtained by crossing Ins1-Cre mice (#026801, Jackson Laboratory) with Rosa26-LSL-YFP mice (a gift from Patrick Jacquemin, originally #006148, Jackson Laboratory). The mice were genotyped by PCR on tail DNA using the following primers: for Cre (wild-type product 488 bp and mutant product 675 bp), 5′-GGAAGCAGAATTCCAGATACTTG-3’ (common), 5′-GTCAAACAGCATCTTTGTGGTC-3′ (wild-type forward), and 5′-GCTGGAAGATGGCGATTAGC-3' (mutant forward), and for yellow fluorescent protein (YFP) (wild-type product 650 bp, mutant product 250 bp), 5′-AAAGTCGCTCTGAGTTGTTAT-3' (Rosa26 forward), 5′-GGAGCGGGAGAAATGGATATG-3' (Rosa26 reverse), and 5′-CATCAAGGAAACCCTGGACTACTG-3' (SpliAc).
2.2. Isolation of pancreatic islets
To isolate adult islets, 0.5 mg/ml of collagenase P (#C9263, Sigma–Aldrich, St. Louis, MO, USA) diluted in HBSS (#14025, Thermo Fisher Scientific, Waltham, MA, USA) was injected in the main pancreatic duct while the ampulla of Vater was clamped. Next, the pancreas was collected and incubated in 0.5 mg/ml of collagenase P at 37 °C for 20 min. To isolate neonatal islets, the pancreas was cut in ∼5 pieces of equal size prior to incubation in 0.5 mg/ml of collagenase P and incubated for 5 min (P1) or 11 min (P15). Islets were handpicked and cultured overnight in RPMI 1640 (#61870-010, Thermo Fisher Scientific) supplemented with 10% (vol/vol) fetal bovine serum (FBS) (S1810, Biowest, Nuaillé, France).
2.3. Flow cytometry
Islets were dispersed in single cell suspensions by incubation for 3 min at 37 °C in 0.025% trypsin–EDTA (#25300054, Thermo Fisher Scientific). Fetal pancreata were dissociated as previously described [11]. For cell surface staining, cells were incubated with antibodies at 4 °C for 15 min in the dark in FACS medium (HBSS + 2% FBS), rinsed in FACS medium, and resuspended in FACS medium with propidium iodide (1/4000, #P4864, Sigma–Aldrich) before FACS acquisition and sorting. For intracellular staining after cell surface staining, cells were rinsed in 1x PBS, fixed, and permeabilized with a Transcription Factor Staining Buffer Set (#00-5523-00, Thermo Fisher Scientific) according to the manufacturer's instructions. Briefly, cells were incubated with antibodies at room temperature for 30 min in the dark in permeabilization buffer, rinsed twice in permeabilization buffer, and resuspended in FACS medium. A list of primary antibodies is provided in Supplementary Table 1. For each antibody, optimal dilution was determined by titration. Cell sorting was carried out using a FACSAria III (BD Biosciences, San Jose, CA, USA). Data were analyzed using FlowJo 9.9.4 (RRID: SCR_008520). Dead cells were excluded from analyses using propidium iodide, and non-pancreatic lineages were excluded using the pan-hematopoietic marker CD45, the erythroid marker TER119, and the endothelial marker CD31. Cells sorted for qPCR or transcriptomics were collected directly in Buffer RLT for qPCR (Qiagen, Hilden, Germany) or Buffer RL (#51800, Norgen BioTek Corp., Thorold, ON, Canada) for next-generation sequencing.
2.4. Conjugated transferrin incorporation
After overnight culture in RPMI 1640 + 10% FBS, islets were serum-starved for 30 min in HBSS supplemented with 1% BSA and 20 mM of glucose. CF-568 conjugated human transferrin (TF-CF568, #00083, Biotium, Fremont, CA, USA) was added to a final concentration of 25 ug/mL and incubated at 37 °C for 30 min in the dark. After transferrin incorporation, islets were rinsed in 1x PBS, dissociated, and stained for FACS analysis as previously described. YFP was excited by a 488 nm laser and analyzed in the FITC channel (515–545 nm). The CF-568 was excited by a 561 nm laser and analyzed in the PE channel (575–589 nm).
2.5. EndoC-βH1 cell culture and treatment
The female human beta cell line EndoC-βH1 (RRID: CVCL_L909) was cultured as previously described and authenticated using short tandem repeats [21]. Twenty hours before FACS staining and conjugated transferrin incorporation, the cells were cultured in DMEM at 0.5 mM or 5.5 mM of glucose and then serum-starved in KRBH buffer (NaCl 115 mM, KCl 5 mM, NaHCO3 24 mM, CaCl2 2.5 mM, MgCl2 1 mM, HEPES 10 mM, and BSA 0.1% w/v) supplemented with 20 mM of glucose for 30 min, followed by TF-CF568 incorporation for 30 min as previously described. The cells were then rinsed in 1x PBS and detached with 0.05% trypsin–EDTA for 3 min at 37 °C, then rinsed in 1x PBS +20% FBS before subsequent FACS staining as previously described. Primary antibodies and their optimal working dilutions are listed in Supplementary Table 1.
2.6. Bulk and single cell quantitative PCR
For bulk qPCR, RNA was isolated with an RNeasy Micro Kit (#74004, Qiagen) and RNA samples were reverse-transcribed using a Maxima First Strand cDNA Synthesis Kit (K1642, Thermo Fisher Scientific) and a Biometra T3 Thermocycler (Analytik Jena, Jena, Germany). All of the kits were used per the manufacturer's instructions. qPCR was conducted with SYBR (for oligonucleotides [Eurofins France, Nantes, France], see Supplementary Table 2) or TaqMan (for assays [Thermo Fisher Scientific], see Supplementary Table 3). The relative expression was calculated using the 2ˆ-ΔCt method, and fold changes were assessed using the 2ˆ-ΔΔCt in the control group using Ppia as a reference gene. qPCR was performed on a QuantStudio 3 (Thermo Fisher Scientific) per the manufacturer's instructions.
For single cell qPCR, single cells were sorted in 96-well PCR plates in 9 μL of RT/pre-amp mix from a CellsDirect One-Step qRT-PCR Kit (#11753500, Thermo Fisher Scientific) and maintained at −20 °C overnight. For each subset analyzed, a control well containing 10 cells was analyzed. Pre-amplified (20 cycles) cDNA was obtained per the manufacturer's instructions and diluted 1:5 in TE buffer and low EDTA (#J75793, Thermo Fisher Scientific) for qPCR reactions. Multiplex qPCR was conducted on a Biomark HD for 40 cycles (Fluidigm, South San Francisco, CA, USA). The same TaqMan assays were used for both RT/pre-amp and qPCR reactions (Supplementary Table 3). Single cell samples where the reference gene was detected before 20 cycles were included in the analysis.
2.7. Next-generation sequencing and bioinformatics
RNA was isolated using a Single cell RNA Purification Kit (#51800, Norgen) per the manufacturer's instructions. RNA quality was verified by electrophoresis using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). For library construction, 20 ng of high-quality (RIN >7) total RNA was processed using an Ovation Solo RNA-seq Kit (#0501-96, NuGEN, Leek, the Netherlands) per the manufacturer's instructions. Briefly, total RNA was treated with DNaseI and reverse-transcribed using random primers. Chemical treatment during second strand synthesis enabled us to achieve strand specificity. After end repair, adaptor ligation, and library amplification, depletion of rRNA was realized using AnyDeplete (NuGEN). Libraries were then quantified with a Qubit HS DNA assay (#Q32855, Thermo Fisher Scientific) and library profiles were assessed using a DNA High Sensitivity LabChip Kit on an Agilent 2100 Bioanalyzer (#5067-4626, Agilent Technologies). Libraries were sequenced on an Illumina Nextseq 500 instrument using 75 base lengths reading V2 chemistry in a paired-end mode. After sequencing, a primary analysis based on AOZAN software (Genomic Paris Centre, Ecole Normale Supérieure, Paris, France) was applied to demultiplex and control the quality of the raw data (based on FastQC modules version 0.11.5). Sequencing reads were mapped to the mouse genome version GenCode M23 (RRID: SCR_014966 and GRCm38.p6 release 93) using STAR v2.5.2b [22] (RRID: SCR_015899). Transcripts were quantified using the RSEM software tool [23] (RRID: SCR_013027). Heatmaps were constructed using the pheatmap R package (RRID: SCR_016418). Gene set enrichment analysis was conducted with GSEA v4.0 software [24,25] (RRID: SCR_003199).
2.8. Immunocytochemistry
Cells were FACS-sorted on precoated Shandon Cytoslides (#5991051, Thermo Fisher Scientific) and allowed to attach for 30 min. The cells were fixed with 4% formaldehyde, washed in 1x PBS, permeabilized with TBS +0.1% Triton, and incubated overnight with primary antibody. Next, the cells were incubated with a peroxidase-labeled secondary antibody from an ImmPRESS HRP Polymer Detection Kit (MP-7401, Vector Laboratories, Burlingame, CA, USA) per the manufacturer's instructions and developed with liquid DAB (SK4100, Vector Laboratories). The cells were counterstained with hematoxylin, rinsed, and mounted. A list of primary and secondary antibodies is supplied in Supplementary Table 1.
2.9. DFO treatment of qPCR and glucose-stimulated insulin secretion
After overnight culture, comparable medium-sized islets were handpicked and treated with or without deferoxamine mesylate (DFO) (#D9533, Sigma–Aldrich, St. Louis, MO, USA) for 24 h. When used for qPCR, the islets were subsequently transferred to RLT buffer. For glucose-stimulated insulin secretion, the islets were preincubated for 1 h in glucose-free Ham's F-10 (Gibco, Thermo Fischer Scientific) followed by incubation for 1 h with 2 and 20 mM of glucose. The DFO treatment continued throughout preincubation and glucose-stimulated insulin secretion. The islets were transferred to RIPA buffer for insulin content analysis. The insulin concentration in the medium and cells was measured by ELISA (#90080, Chrystal Chem Europe, Zaandam, the Netherlands). All of the replicates shown are the mean of duplicates of 20 islets per mouse. Three independent experiments were conducted with n = 3–4 mice per condition and per experiment.
2.10. Statistical analyses
Each n represents an independent biological sample. All of the graphs show means ± SEM.
Statistical analysis was conducted using Prism v8.3 (RRID: SCR_002798, GraphPad Software, San Diego, CA, USA) applying either a Mann–Whitney test, an unpaired t test, or Welch's t test as indicated in the figure legends. A p value < 0.05 was considered statistically significant. Symbols for indicating p values are ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.005; ∗∗∗∗p < 0.001. For gene set enrichment analysis, FDR <25% was considered significantly enriched.
2.11. Deposited data
The datasets generated during this study are available at NCBI Gene Expression Omnibus: GSE149488 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE149488).
3. Results
3.1. Transferrin receptor (CD71) surface expression allows distinction of adult mouse beta cells from alpha and delta cells
To devise a sorting strategy for endocrine islet subsets, we tested combinations of surface markers for which expression in the pancreatic epithelium was previously reported, including CD133 and CD49f [26], CD71 [27], and CD24 [28]. The individual or limited combinatorial use of these surface markers previously did not allow isolation of the different endocrine lineages, but we hypothesized that larger combinations of these surface markers might reveal subsets of interest. We analyzed dispersed islet cells from 12-week-old C57BL/6 mice via flow cytometry using antibodies directed against CD24, CD71 (TFRC), and CD49f (ITGA6) in the EpCam+ epithelial fraction after excluding dead cells and non-pancreatic lineages (Fig S1a; see methods). CD24 defined two subsets in the epithelial fraction: CD24low cells, representing the majority of the EpCam+ compartment (∼85%), and CD24high cells (∼10%) (Figure 1A–B and S1a). CD71 and CD49f further segregated these two fractions into four subsets: CD24lowCD71- and CD24lowCD71+, CD24highCD49f+, and CD24highCD49f− (Figure 1A). Intracellular staining for glucagon and insulin, the hormones produced by alpha and beta cells, respectively, revealed that these endocrine cells were restricted to the CD24low subset (Figure 1B and Fig S1a) and could be discriminated based on their CD71 surface expression (Figure 1C). Of note, fetal liver erythroid cells (FL TER 199+) are known for their high expression of CD71 (the transferrin receptor), which is required for the cellular uptake of iron from transferrin that is used in these cells to produce heme. CD71 surface levels on the beta cell-enriched fraction were at ∼30% of the level on the FL TER 199+ cells, while only at ∼8% on the alpha cell-enriched fraction (Figure 1C).
Figure 1.
Adult pancreatic beta cells are defined by higher expression of the transferrin receptor (CD71) compared to alpha and delta cells. (A) Representative flow cytometry plots of dispersed islets from adult C57BL/6 mice. CD24, CD71, and CD49f expression was analyzed on EpCam+ cells after excluding dead cells (propidium iodide-positive cells) and hematopoietic and endothelial cells (lineage [Lin]: CD45-, TER119-, or CD31-positive cells). The EpCam+ fraction could be divided into CD24high and CD24low cells (middle left panel). The CD24low fraction could be further subdivided into CD71+ and CD71- cells (middle right panel), while the CD24high fraction comprised CD49f+ and CD49f− cells (right panel). Data represent at least 11 independent experiments. (B) Flow cytometry plots of dispersed islets from adult C57BL/6 mice. After surface staining for lineage, EpCam, CD24, CD71, and CD49f, cells were fixed and permeabilized for intracellular staining for insulin and glucagon. The left panel shows the CD24high and CD24low cells within the Lin− Epcam+ fraction. The right panel shows insulin and glucagon staining in Lin−EpCam+CD24lowcells. (C) CD71 surface protein expression levels in insulin+ (blue) and glucagon+ (purple) fractions compared to TER119+ erythrocytes from E14.4 fetal liver (FL14) (green). Plots represent 3 independent experiments. (D) Hormone transcript expression was assessed by qRT-PCR on sorted CD24high (CD49f−) (red), CD24low CD71+ (blue) and CD71- (purple) epithelial fractions. Graphs show Insulin1 (left panel), Glucagon (middle panel), and Somatostatin (right panel) expression from 6 independent sortings. (E) Frequencies of CD24highCD49f−, CD24lowCD71+, and CD24lowCD71- cells among EpCam+ fractions from 10 independent experiments. Each experiment included a pool of islets from 4 mice. (D–E) One-way ANOVA. Full lines indicate mean, and error bars denote s.e.m.
We next analyzed the expression of endocrine and exocrine genes using qRT-PCR in the four FACS-sorted subsets. The CD24lowCD71- and CD24lowCD71+ subsets were consistently enriched in Glucagon (Gcg) and Insulin (Ins1) transcripts, respectively, while the CD24highCD49f− subset was highly enriched in Somatostatin (Sst) transcripts (Figure 1D). Ghrelin (Ghrl) transcripts were expressed at similarly low levels in all of the sorted subsets (data not shown). Low levels of acinar (Cpa1) and ductal (Sox9) transcripts originating from contaminating exocrine cells in the islet preparations were confined to the CD24highCD49f+ subset that was devoid of endocrine transcripts (Fig S1b-c). These results show that alpha, beta, and delta cell-enriched fractions could be sorted based on their surface expression of CD24, CD49f, and CD71. Sorting was highly reproducible between islet preparations. Moreover, the relative abundance of the specific endocrine cell-enriched fractions in the EpCam+ compartment (Figure 1E) was in line with the corresponding abundance of alpha (∼20%), beta (∼75%), and delta cells (∼5%) in adult mouse islets [29].
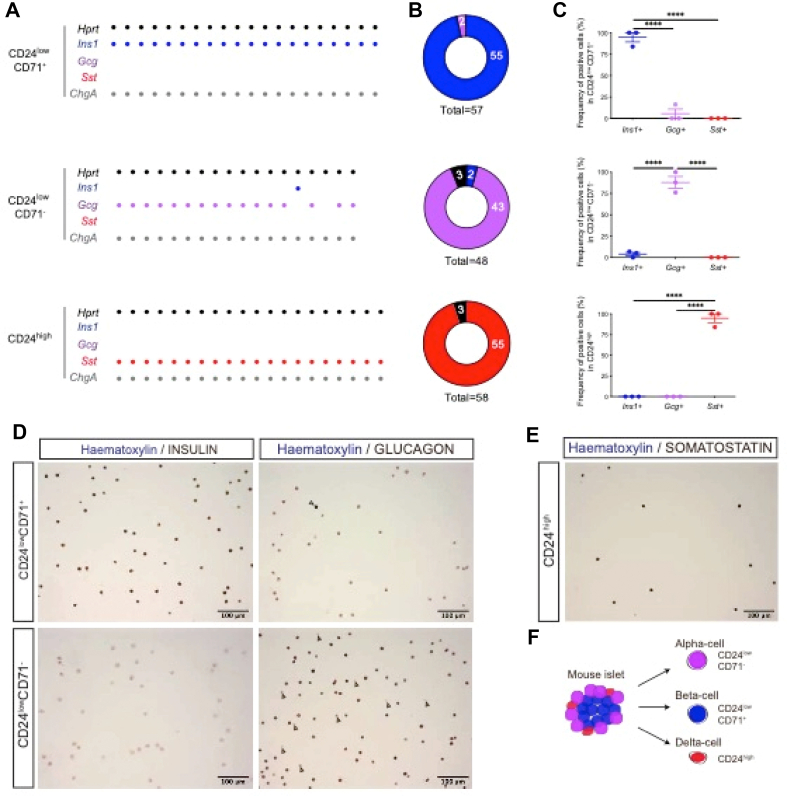
We next assessed the purity of these subsets by analyzing the expression of selected endocrine transcripts using multiplex qRT-PCR on single sorted CD24highCD49f−, CD24lowCD71+, and CD24lowCD71- cells. All of the sorted single cells expressed the pan-endocrine marker Chromogranin A (Chga) and were monohormonal (Figure 2A). The percentage of sorted CD24lowCD71+ cells expressing Ins1 was 95 ± 5%, while Gcg was expressed in only 5 ± 5%. Among the sorted CD24lowCD71- cells, 88 ± 7% expressed Gcg, and 95 ± 5% of the sorted CD24highCD49f− cells expressed Sst (Figure 2B–C). Immunocytochemistry for the endocrine hormones on the FACS-sorted CD24lowCD71-, CD24lowCD71+, and CD24highCD49f− subsets confirmed their high enrichment of glucagon-, insulin-, and somatostatin-positive cells, respectively (Figure 2D–E). These findings demonstrate that the surface markers CD24, CD71, and CD49f enable sorting to high-purity of murine alpha, beta, and delta cells (Figure 2F).
Figure 2.
CD71, CD49f, and CD24 allow isolation of high-purity alpha, beta, and delta cell-enriched fractions. (A–B) Single cell multiplex qRT-PCR on sorted CD24lowCD71+, CD24lowCD71-, and CD24highCD49f− pancreatic epithelial cells from adult C57BL/6 mice. Hprt (black), Ins1 (blue), Gcg (purple), Sst (red) and ChgA (gray) expression was analyzed using the Biomark Fluidigm platform. (A) Expression of Hprt, Ins1, Gcg, Sst, and ChgA in single CD24lowCD71+ (top), CD24lowCD71- (middle), and CD24highCD49f− (bottom) cells. Each dot indicates positivity of a single cell for a given gene tested in the horizontal lines. (B) Pie charts show the number of total sorted CD24lowCD71+, CD24lowCD71-, and CD24highCD49f− pancreatic epithelial cells expressing either Ins1 (blue), Gcg (purple), Sst (red), or no hormones (black). (C) Histograms show the frequency of Ins1, Gcg, and Sst expressing single cells in the 3 subsets analyzed. The data shown in (A) represent 3 independent experiments, while the data in (B) and (C) denote the pooled data from these experiments. One-way ANOVA. Full lines indicate mean, and error bars represent s.e.m. (D–E) Immunocytochemical staining for insulin, glucagon, and somatostatin on FACS-sorted CD24lowCD71+, CD24lowCD71-, and CD24highCD49f− pancreatic epithelial cells from adult C57BL/6 mice. (D) CD24lowCD71+ and CD24lowCD71- pancreatic epithelial cells were sorted on precoated slides, fixed, permeabilized, and stained for insulin and glucagon (on two separated slides) (brown) and counterstained with hematoxylin (blue). White arrow indicates a glucagon-positive cell in the CD24lowCD71+ fraction, yellow arrows indicate glucagon-negative cells in the CD24lowCD71- fraction. (E) CD24highCD49f− pancreatic epithelial cells were treated as in (D) and stained for somatostatin (brown) and hematoxylin (blue). The micrographs represent 2 independent experiments. (F) Schematic of alpha, beta, and delta cell surface markers.
3.2. Beta cells import more transferrin than other islet cell types
Given the differential expression of the transferrin receptor (CD71) in beta cells, we examined whether this corresponds to functional differences in transferrin uptake capacity. To test this, we performed transferrin incorporation assays by starving adult mouse islets of iron-containing serum and then incubating them with fluorescently labeled transferrin (TF-CF568). The islets were then dispersed to single cells and analyzed via flow cytometry (Figure 3A). The TF-CF568 signal was strongest in CD24lowCD71+ cells and similarly low in CD24lowCD71- and CD24high cells, implying the highest transferrin uptake in the beta cell-enriched fraction (Figure 3B–C and S1d). Accordingly, beta cells from adult Ins-YFP reporter mice displayed a stronger TF-CF568 signal than YFP− non-beta cells (Figure 3D–E). Taken together, these findings indicate that increased CD71 surface levels on adult beta cells correlates with increased transferrin import compared to other islet cell types.
Figure 3.
Beta cells import more transferrin than other endocrine islet cells. (A) Experimental design for islet transferrin incorporation. After overnight culture in RPMI 10% FBS, islets were serum starved for 30 min at 37 °C. Conjugated human transferrin was then added for incorporation for 30 min at 37 °C and 20 mM of glucose. The islets were rinsed, dispersed, and stained for FACS analysis. (B) Flow cytometry plots of dispersed islets from adult C57BL/6 mice after conjugated transferrin (TF-CF568) incorporation. The left panel shows CD24 and CD71 expression in the live Lin−EpCam+ fraction, middle panel shows TF-CF568 fluorescence and CD71 expression in the live Lin−EpCam+CD24low fraction. The right panel shows TF-CF568 fluorescence on CD71+ (blue) and CD71- (purple) CD24low epithelial cells, and the gray histogram shows the basal fluorescence of unstained islet cells. Data represent 3 independent experiments. (C) Histogram shows the mean fluorescence intensity (MFI) of TF-CF568 in the CD24lowCD71+, CD24lowCD71-, and CD24highCD49f− epithelial fractions from 3 independent experiments. (D) Flow cytometry analysis of dispersed islets from adult (>12 weeks old) Ins-YFP mice after TF-CF568 incorporation. The left panel shows the EpCam expression and TF-CF568 fluorescence in the live Lin−EpCam+ fraction, the middle panel shows the CD24 and YFP expression in live Lin−EpCam+ cells, and the right panel shows TF-CF568 fluorescence in live Lin−EpCam+CD24low fraction. (E) Histogram shows TF-CF568 fluorescence on YFP+ (red) and YFP- (blue) CD24low epithelial cells, and the gray histogram shows basal fluorescence of unstained islet cells. Plots represent 4 independent islet preparations.
3.3. CD71 expression on beta cells is acquired postnatally and increases during maturation
We next assessed whether the same markers could be used to separate the different endocrine subpopulations from fetal mouse pancreas. Endocrine cells are scarce in fetal mouse pancreas and scattered throughout the exocrine compartment. Therefore, endocrine cells were first enriched using a previously described strategy based on the CD133 and CD49f expression [26]. This led us to analyze the CD133 and CD49f expression in the EpCam+ epithelial fraction of E14.5, E16.5, and E18.5 C57BL/6 fetal pancreas, after the exclusion of dead cells and non-pancreatic lineages. The same fractions (Fr.) I, II, and III as described by Suguiyama et al. [26] were identified with Fr. IV being the EpCam− fraction excluded from our protocol (Fig S2a). Within Fr. III, the subset enriched in endocrine hormone-expressing cells, we assessed the CD71 and CD24 expression and cellular granularity (side scatter parameter, SSC). The latter was in line with our previous demonstration of its use to isolate hormone-expressing cells from human fetal pancreas [12]. This allowed us to identify two subsets distinguishable by low or high granularity, Fr. III CD24lowSSClow and Fr. III CD24lowSSChigh, respectively, and a third Fr. III CD24high subset that was present from E16.5 onward. The relative abundance of the Fr. III CD24low SSChigh subset increased from E14.5 (8% of Fr. III) to E18.5 (20% of Fr. III) (Figure 4A). Ins1 and Gcg transcripts were enriched in the CD24+SSChigh subset, while Sst transcripts were enriched in the Fr. III CD24high subset (Figure 4B). The CD71 surface expression in Fr. III CD24+SSChigh that contained fetal Ins1-and Gcg-expressing cells was similar to the adult CD24lowCD71- fraction and considered negative in comparison with the adult CD24lowCD71+ subset (Figure 4C). Taken together, CD71 was absent from immature mouse fetal beta cells but CD24 already allowed the isolation of fetal delta cells.
Figure 4.
CD71 is not differentially expressed on fetal alpha and beta cells. (A) Flow cytometry plots of fetal pancreas from E14.5, E16.5, and E18.5 C57Bl6 embryos showing CD24 expression and granularity (SSC-A) in fraction III (Fr. III, live Lin−EpCam+CD133−CD49flow) containing hormone-expressing cells. Cell preparations for each experiment were pooled from 6 to 18 embryos, and the plots represent 5 independent experiments. (B) Hormone expression was assessed by qRT-PCR on Fr. III CD24high (red), CD24+SSCint (gray), and CD24+SSChigh cells (black) sorted from E16.5 C57BL/6 embryos. Graphs show Ins1 (left), Gcg (middle), and Sst (right) expression from 3 independent experiments. (C) Comparative flow cytometry analysis of fetal pancreas from E16.5 C57BL/6 embryos and dispersed pancreatic islets from adult C57BL/6 mice. Histogram shows CD71 expression levels on Fr. III CD24+SSChigh and CD24low epithelial islet cells.
This led us to determine when differential CD24 and CD71 expression starts defining alpha, beta, and delta cells during postnatal development. We applied our sorting strategy to islets isolated from mice at postnatal days (P)1, P7, and P15. All of the fractions defined in the EpCam+ adult islet cell compartment were detected from P1 onward (Figure 5A). Similar to the adult islets, the Ins1, Gcg, and Sst expression segregated in the CD24lowCD71+, CD24lowCD71-, and CD24highCD49f− subsets, respectively (Figure 5B). At P1, the surface expression of CD71 on the CD24lowCD71+ subset was lower compared to the adults but increased during the first postnatal weeks until P15. In contrast, the CD71 levels on the CD24lowCD71- subset remained low throughout early postnatal life (Figure 5C–D). Although the smaller differential in CD71 surface levels before P15 complicated segregation of the alpha and beta cell-enriched subsets, analysis of CD49f expression distinguished both (Figure 5B). The CD71 expression on beta cells is thus acquired after birth and increases in the first postnatal weeks, an important period for beta cell maturation, to reach adult levels at P15 (Figure 5C–D). These findings suggested that increased iron import could be important for beta cell function.
Figure 5.
Expression of CD71 by beta cells is acquired postnatally and increases as beta cells mature. (A) Flow cytometry plots of dispersed islet cells from 1-day-old (P1), 7-day-old (P7), and 15-day-old (P15) newborn C57BL/6 male and female pups and 12-week-old C57BL/6 male mice. The top panels show the CD24 and CD71 expression among the live Lin−EpCam+ fraction, and the bottom panels show the CD71 and CD49f expression among the Live Lin−EpCam+CD24low fraction. Plots represent 3 independent experiments; each experiment was a pool of 4–12 mice. (B) Hormone expression was assessed by qRT-PCR on CD24high ((CD49f−) (red), CD24lowCD71+ (blue), and CD71- (purple) epithelial islet fractions (Live Lin−EpCam+) sorted from P1 and P15 newborn C57BL/6 males and females. Graphs show Ins1 (left panel), Gcg (middle panel), and Sst (right panel) expression from 3 independent experiments. (C) Flow cytometry analysis of fetal pancreas from E16.5 C57BL/6 embryos and pancreatic islets from P1, P7, and 12-week-old C57BL/6 mice. Histograms show CD71 surface expression levels on the epithelial Fraction (Fr.) III CD24+SSChigh (blue) and CD24lowCD71+CD49f+ beta cell-enriched islet fraction at P1 (green), P7 (orange), and 12 weeks (red). (D) MFI of CD71 staining on CD24lowCD71+ (blue) and CD71- (purple) epithelial islet fractions (Live Lin−EpCam+) from P1, P7, and P15 C57BL/6 male and female pups and adult C57BL/6 male mice from 3 independent experiments; each experiment was a pool of 4–12 mice. Two-way ANOVA. Full lines indicate mean, error bars for s.e.m.
3.4. Transcriptomic profiling of alpha, beta, and delta cell-enriched subsets reveals a signature of increased iron metabolism in beta cells
We generated transcriptomes of the CD24lowCD71-, CD24lowCD71+, and CD24highCD49f− subsets from adult mouse islets. The transcriptomes of all of the subsets were devoid of hematopoietic and endothelial transcripts underpinning the robust lineage negative exclusion by FACS (Fig S3a). All of the subsets of interest contained abundant epithelial transcripts (Epcam) and were enriched in the genes encoding the surface markers used for the sorts: that is, Cd24a (CD24), Itga6 (CD49f), and Tfrc (CD71) (Fig S3b). Interestingly, Tfrc encoding the transferrin receptor CD71 was differentially expressed between the beta cell-enriched subset and the non-beta cells, but below the log2 fold change of two. The discrepancy of the small differences in Tfrc transcript levels with the markedly different CD71 surface levels suggested a differential post-transcriptional regulation of the transferrin receptor in the CD71+ and CD71- subsets. We next analyzed a set of known alpha, beta, and delta cell markers within the sorted subsets. The alpha cell gene set, including Gcg, Mafb, Irx1, Irx2, Arx, Ttr, Slc7a2, Pou3f4, Pou6f2, and Sstr2, was enriched in the CD24lowCD71- fraction. The beta cell gene set, including Ins1, Ins2, Mafa, Ucn3, Nkx6-1, Syt4, Prlr, Gad1, G6pc2, and Glp1r, was enriched in the CD24lowCD71+ fraction. The delta cell gene set, including Sst, Hhex, Rbp4, Crhr2, Ghsr, and Neurog3, was enriched in the CD24lowCD49f− fraction (Figure 6A). Note that while Neurog3 is not a classical delta cell marker, others have also found it to be more abundant in delta cells compared to alpha and beta cells [8]. Next, we compared the dataset generated by DiGruccio et al. (2016) that used hormone reporter mice with ours and found both to be highly comparable in terms of the relative expression of the main hormone transcripts in the different endocrine subsets, indicating similar purity (Fig S3c). Collectively, the expression patterns of these gene sets confirmed the enrichment of bona fide beta cells in the CD24lowCD71+ fraction, alpha cells in the CD24lowCD71- fraction, and delta cells in the CD24lowCD49f− fraction.
Figure 6.
Iron metabolism-related genes are enriched in beta cells. (A) Heatmap showing the relative mRNA levels of selected alpha, beta, and delta cell genes in the CD24lowCD71+ (blue), CD24lowCD71- (purple), and CD24high (red) fractions. Each column represents a different biological replicate (n = 3 per fraction from 3 independent experiments). The Z score normalized mRNA expression is color coded: blue (−2) over white (0) to red (+2). (B–D) Enrichment plots for 3 iron transport gene sets comparing the CD24lowCD71+ (beta cell-enriched) with the CD24lowCD71- (alpha cell-enriched) fraction. (E) Heatmap showing the relative mRNA levels of selected iron metabolism genes in the CD24lowCD71+, CD24lowCD71-, and CD24high fractions. Color code as in (A). (F) Schematic of the major proteins for which the iron metabolism genes analyzed in (E) encode colored red when more expressed in the beta cell-enriched fraction and blue when less expressed compared to the alpha and delta cell-enriched fraction. (G–H) The Tfrc and Ftl1 expression was assessed by RT-qPCR in (G) adult CD24lowCD71+ (blue), CD24lowCD71- (purple), and CD24high (red) fractions (G) P1 and adult CD24lowCD71+ fractions. Data are a pool of 3 independent experiments. One-way ANOVA for (G) and unpaired t-test for (H). (i)Tfr1, Ins1, Iapp, Pdx1, Nkx6-1, Mafa, and Gcg transcript expression in control and DFO-treated islets. Mann–Whitney test for Tfrc and unpaired t-test for all of the other genes. (J) Glucose-stimulated insulin secretion (left) and insulin content (right) from control and DFO-treated islets. Two-way ANOVA. Full lines indicate mean, error bars for s.e.m, and n = 10–11 from 3 independent experiments. Circles, control; black circles, DFO-treated.
To investigate whether iron metabolism was higher in the beta cell-enriched CD24lowCD71+ fraction, we performed an enrichment analysis for iron-related gene sets. We found that three such gene sets were enriched in the beta cell-enriched fraction compared to the alpha cell-enriched fraction (Figure 6B–D, Supplementary Tables 4-6). Common genes from these sets that were implicated in cellular iron import (Tfrc), regulation (Steap2 and Ireb2), and storage (Fth1 and Ftl1) were expressed at higher levels in the beta cell-enriched fraction, while Slc40a1 that encodes ferroportin, an iron exporter, was more abundant in the alpha cell-enriched fraction (Figure 6E). Steap2 encodes a metalloreductase that reduces iron from its ferric (Fe3+) to its ferrous (Fe2+) state to support the uptake of ferric iron substrates. Ireb2 encoded iron-responsive element-binding protein 2, an RNA-binding protein that regulates intracellular iron levels by regulating the translation and stability of mRNAs implicated in iron homeostasis. Fth1 and Ftl1 encoded the ferritin heavy and light chain protein subunits of the main intracellular iron storage protein, ferritin (Figure 6F). We validated via qRT-PCR that Ftl1 transcripts were enriched in the CD24lowCD71+ fraction compared to the other islet subsets (Figure 6G). However, we found no significant enrichment of Tfrc transcripts by qRT-PCR in CD24lowCD71+ vs CD71- subsets, suggesting again that post-transcriptional regulation or endosomal recycling kinetics could differentially regulate CD71 surface levels in the alpha and beta cell fractions (Figure 6G).
Based on the progressive increase in the CD71 surface levels on beta cells during the first two weeks after birth, we next hypothesized that iron import and storage increase in maturing beta cells and compared the Tfrc and Flt1 expression in P1 vs adult (12 weeks) CD24lowCD71+ fractions via qRT-PCR. The expression of Tfrc was not significantly different although it tended to increase from P1 to adult; Flt1, however, was significantly higher expressed in the adult CD71+ fraction compared to P1 (Figure 6H). Collectively, these data suggest that adult beta cells display a higher iron storage capacity compared to early neonatal beta cells (Figure 6H).
3.5. Depriving beta cells of iron impairs their function
To study the role of CD71 in beta cell function, we deprived mouse islets of iron by culturing them in the presence of an iron scavenger, deferoxamine mesylate (DFO). As expected, iron-deprived islets cells upregulated Tfrc expression. Interestingly, they also downregulated Ins1 and Iapp expression. Comparable expression of Pdx1, Gcg, Nkx6-1, Mafa, and Slc2a2 was maintained excluding the hypothesis that DFO treatment decreased the relative proportion of beta cells in islets (Figure 6I). In addition, iron-deprived islets secreted 45% less insulin in response to high glucose stimulation, while the total insulin content did not vary, highlighting the functional importance of iron metabolism for insulin secretion in beta cells (Figure 6J).
3.6. Glucose levels regulate CD71 expression and transferrin import in a human beta cell line
We next evaluated the CD71 surface expression on human beta cells using the EndoC-βH1 cell line and found that it was expressed, albeit at lower levels compared to the mouse beta cell-enriched fraction (Figure 7A–C). Accordingly, the EndoC-βH1 cells readily incorporated conjugated transferrin and, similar to our findings in mouse islets, the incorporation correlated with CD71 surface levels (Figure 7A–C). We hypothesized that the iron requirements of beta cells vary with the extracellular glucose levels and that beta cells regulate intracellular iron levels through transferrin import and CD71 expression. To test this, we compared the CD71 surface expression on EndoC-βH1 cultured for 20 h at low (0.5 mM) or normal (5.5 mM) glucose and found higher expression in the cells cultured in low glucose (Figure 7D, E). Glucose-starved EndoC-βH1, which expressed higher surface levels of CD71, also incorporated more transferrin compared to EndoC-βH1 kept at 5.5 mM of glucose (Figure 7F, G). Taken together, these findings show that CD71 is expressed in EndoC-βH1 cells and that CD71 surface expression is regulated by extracellular glucose levels, modulating transferrin import into human beta cells.
Figure 7.
Glucose levels regulate CD71 expression in a human beta cell line. (A–C) Flow cytometry analysis of the surface CD71 expression and conjugated transferrin incorporation in the EndoC-βH1 human beta cell line. Conjugated transferrin (TF-CF568) incorporation and CD71 staining were conducted as described in Figure 3A. (A) The left panel shows basal fluorescence of CD71 and TF-CF568 levels on unstained EndoC-βH1 and the right panel shows their staining patterns. (B) CD71 surface expression and (C) TF-CF568 fluorescence on unstained (gray) or stained (red) EndoC-βH1. Data represent 5 experiments. (D–E) CD71 surface expression (D) and CD71 MFI (E) in EndoC-βH1 cells starved at 0.5 mM (blue) or maintained at regular 5.5 mM of glucose (red) for 20 h before staining. (F–G) EndoC-βH1 was starved at 0.5 mM or maintained at 5.5 mM of glucose for 20 h before conjugated transferrin (TF-CF568) incorporation at 20 mM of glucose. After incorporation, the cells were stained for surface CD71 and analyzed via flow cytometry. (F) Histogram shows TF-CF568 fluorescence after incorporation in EndoC-βH1 cells that were previously starved at 0.5 mM (blue) or maintained at 5.5 mM (red). Gray curve shows the basal fluorescence of unstained EndoC-βH1 cells. (G) TF-CF568 MFI after incorporation at 20 mM of glucose on previously starved at 0.5 mM (blue) or maintained at 5.5 mM (red) EndoC-βH1. Data are a pool of 3 independent experiments. (E and G) Unpaired t-test. Full lines indicate mean, and the error bars denote s.e.m.
4. Discussion
In this study, we developed an innovative approach to isolate highly purified subsets of alpha, beta, and delta cells from dispersed mouse islets using cell surface markers. Our results demonstrate that: (i) the expression of CD24, CD49f, and CD71 can be used to separate high-purity alpha (CD24lowCD71-), beta (CD24lowCD71+), and delta cells (CD24highCD49f−); (ii) murine beta cells import more transferrin-bound iron and express higher levels of iron import and storage genes compared to alpha and delta cells; (iii) depriving mouse islets of iron hampers normal glucose-stimulated insulin secretion; and (iv) human beta cells in the EndoC-βH1 cell line increase the expression of the transferrin receptor CD71 when deprived of glucose, resulting in increased transferrin uptake.
The cell surface markers described in this study (CD24, CD49f, and CD71) were reported previously but not combined as in the current study. They were employed individually in either fetal or adult mouse pancreas or adult human pancreas. CD133 and CD49f were reported as markers of ductal epithelial cells, including Neurog3+ progenitors and nascent acinar cells in fetal mouse pancreas [26]. We reproduced these findings in fetal pancreas but found that in adult pancreas, CD49f was additionally expressed on an exocrine-enriched subset. CD133 and CD71 were previously used as markers for a mixed subset in adult pancreas enriched in ductal, alpha, and beta cell transcripts, whereas CD71 alone defined an acinar-enriched fraction [27]. Contrary to these data, we found CD71 to be a beta cell-specific surface marker. This discrepancy could be related to the use of total digested pancreas as starting material by Jin et al. whereas we employed purified endocrine islets [27]. Furthermore, the contaminating exocrine fraction that we defined as CD24highCD49f+ expresses CD71, suggesting that without islet purification and combining antibodies, the CD71+ fraction would indeed contain a mix of beta cells and acinar or ductal cells. Notably, we also found CD71 expression on human beta cells in the EndoC-βH1 line. CD133 did not improve subset purification in our sorting strategy. The higher expression of CD24 on exocrine cells was used to sort, by negative selection, an endocrine-enriched fraction from adult human pancreas [28]. In line with these data, we found high CD24 expression in exocrine cells, but using CD49f we could split the CD24high fraction into an exocrine subset that was CD24highCD49f+ and an endocrine delta cell-enriched subset that was CD24highCD49f−. We found that delta cells specifically expressed high CD24 levels as early as E16. This marker thus allows the isolation of this understudied yet functionally important cell type from fetal, neonatal, and adult mouse pancreas for future studies of their development and maturation [30]. Of note, although available transcriptomes of delta cells confirm enrichment in Cd24a transcripts compared to alpha and beta cells, the marker was not previously exploited for delta cell purification [8,28].
Immunostaining for CD71 has previously shown specific expression in the endocrine compartment of the human pancreas [31], but its intracellular abundance makes it difficult to assess intra-islet cellular specificity of CD71 surface levels. A previous transcriptomic approach identified but did not highlight Tfrc as being differentially expressed in beta cells compared to other islet cell types [8]. Based on the differential in transcript abundance (log2 fold change < 2), Tfrc would not be selected as a candidate surface marker for isolating beta cells. Yet the regulation of CD71 surface expression is well characterized and involves extensive endosomal recycling, which may explain how small differences in transcript abundance are amplified at the protein level by differential post-transcriptional regulation [32].
Distinct requirements for iron in beta cells have not been demonstrated unequivocally but could already be inferred from their high ferritin expression in both mouse [33] and human [28]. Additionally, mice lacking IREB2, an RNA-binding protein that regulates intracellular iron levels by modulating the translation and stability of Tfrc, Ftl1, and Fth1 (the latter two encoding the light and heavy chains that comprise ferritin), were recently found to display a specific functional iron deficiency in beta cells resulting in impaired proinsulin processing and hence in reduced insulin content and secretion [34]. This study revealed a crucial role of iron for beta cell function that is further supported by our findings. Indeed, beta cells not only expressed higher CD71 surface levels and took up more transferrin in comparison to alpha and delta cells, but also expressed higher levels of key iron import and storage genes, which suggests distinct cellular iron needs. We found that depriving mouse islets of iron decreased glucose-stimulated insulin secretion. This functional impairment was unrelated to changes in insulin content but was rather accompanied by a decreased abundance of insulin transcripts, suggesting that iron is important for insulin secretion and regulates insulin transcription or mRNA stability. Note that these functional studies were conducted using the iron chelator DFO, which among commercially available iron chelators has a high affinity for iron and a low affinity for zinc. Nevertheless, deleterious effects due to zinc chelation by DFO are not fully excluded.
Iron deficiency is a common micronutrient deficiency that is well known to cause anemia, but in clinical studies, deleterious effects on beta cell function are not obvious. Yet one case report linked both iron deficiency and diabetes with autoreactive antibodies against CD71 [35,36]. In contrast, there is ample clinical evidence demonstrating that excess iron confers an increased risk of developing diabetes and that in patients with type 2 diabetes, it worsens glucometabolic control [14]. Of note, therapeutic phlebotomy, which is used to reduce iron levels, was found to improve beta cell function in patients with pathological iron overload [37,38] but intriguingly also improves glucose control and insulin secretion in patients with high-ferritin type 2 diabetes [39], implying a detrimental effect of high iron levels within the normal range on beta cell function. Upregulation of the divalent metal transporter Slc11a2 (DMT1) in islet inflammation has been suggested to render beta cells prone to ROS-mediated inflammatory damage [40]. Alpha and beta cells, however, do not differ significantly in their expression of Slc11a2 [8,41,42] but do differ in their expression of CD71 as we have shown herein. Our findings could therefore explain the particular vulnerability of beta cells to iron overload. Indeed, we showed that beta cells not only express high levels of transferrin receptor related to their elevated iron needs for oxidative respiration, but also display low expression of the cellular iron exporter Slc40a1 (ferroportin), altogether favoring iron accumulation.
We found that CD71 surface levels and Ftl1 transcripts are both upregulated in the beta cell-enriched subset during the early postnatal weeks that are important for the functional maturation of beta cells, suggesting increasing iron requirements during this period. We hypothesize that iron metabolism plays a role in beta cell maturation. Indeed, functional maturation of beta cells is accompanied by a firm decrease in proliferation and a metabolic switch from aerobic glycolysis to oxidative phosphorylation [43,44]. The required increase in mitochondrial oxidative phosphorylation to generate sufficient ATP in turn most likely depends on an increase in iron supplies. If this hypothesis is true, modulating iron metabolism in differentiation protocols of stem cell-derived beta cells is an attractive strategy to improve the maturation of these therapeutically interesting surrogate beta cells. Intriguingly, in yeast, the switch between fermentative and oxidative metabolism, called the diauxic shift, is associated with increased expression of genes involved in iron uptake. The driving factor for this phenomenon is not a change in extracellular iron levels, but glucose exhaustion itself [45]. It is in this line that we interpret the upregulation of CD71 surface levels in the human beta cell line following culture in low glucose. We propose that beta cells require fine-tuned intracellular levels of iron to support and maintain their function and maturation and that iron import is regulated through changes in the surface expression of the transferrin receptor, which in turn depends on extracellular glucose levels.
Our demonstration that CD24, CD49f, and CD71 expression can be used to sort purified live high-purity alpha, beta, and delta cells provides a novel improved and powerful tool to study these different cell types in the various mouse models used in diabetes research. In particular, the ability to purify alpha and delta cells will allow a more detailed study of these understudied cell types and how they are affected in models of type 1 and type 2 diabetes. Of note, while we strongly encourage adopting our experimental protocol for diverse mouse lines and disease models, we also call for rigor by stressing that careful validation of the identity and purity of the sorted subsets remains crucial. Finally, the iron metabolism of islet cells and beta cells, in particular, should be investigated further as it may provide a molecular explanation for the susceptibility of beta cells to impaired iron metabolism and for the causative link between iron and diabetes risk. As such, it may pave the way for new diabetes therapies.
Contribution statement
Conceptualization: C.B., W.S., and R.S. Methodology: C.B., W.S., and R.S. Investigation: C.B. and W.S. Validation: C.B. and W.S. Formal analysis: C.B. and W.S. Writing the original draft: C.B. and W.S. Writing, review, and editing: C.B, W.S, and R.S. Visualization: C.B. and W.S. Funding acquisition: R.S.
Funding
W.S. is supported by a postdoctoral grant from Agence Nationale de la Recherche (Laboratoire d’Excellence Revive, Investissement d’Avenir; ANR-10-LABX-73). The RS laboratory received funding from Agence Nationale de la Recherche (ANR BromoBeta), the Innovative Medicines Initiative 2 Joint Undertaking Rhapsody under grant agreement no. 115881 supported by the European Union’s Horizon 2020 research and innovation program EFPIA and the Swiss State Secretariat for Education‘ Research, and Innovation (SERI) under contract number 16.0097, resources of which are composed of a financial contribution from the European Union's Seventh Framework Program (FP7/2007–2013); and the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement no. 115797 (INNODIA) and no. 945268 (INNODIA HARVEST). This joint undertaking receives support from the Union's Horizon 2020 research and innovation program EFPIA, JDRF, and the Leona M. and Harry B. Helmsley Charitable Trust, the Fondation Bettencourt Schueller, the Laboratoire d’Excellence consortium Revive, Fondation pour la Recherche Médicale (EQU201903007793), the Dutch Diabetes Research Foundation, the DON Foundation, and Fondation Francophone pour la Recherche sur le Diabetes (FFRD).
Acknowledgments
We thank L. Rachdi, P. Launay, C. Peyssonnaux, H. Heimberg, N. De Leu, Y. Heremans, and J. van den Ameele for discussion and comments on the manuscript; S. Pechberty and C. Lourenço for assistance with some experiments; H. Heimberg and G. Leuckx (Vrije University Brussels, Belgium) for providing the Ins-YFP mice; the Genomic Platform at Cochin Institute for the transcriptome analyses; and G. Gu (Vanderbilt University Medical School, USA) for sharing their protocol for isolating neonatal pancreatic islets. We also thank the CRT platform at the Pasteur Institute (Paris, France) for access to their single cell qPCR design.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.101060.
Contributor Information
C. Berthault, Email: Claire.berthault@inserm.fr.
R. Scharfmann, Email: raphael.scharfmann@inserm.fr.
Conflict of interest
The authors declare no competing interests.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Jindal R.M., Gray D.W., McShane P., Morris P.J. Zinc-specific N-(6-methoxy-8-quinolyl)-para-toluenesulfonamide as a selective nontoxic fluorescence stain for pancreatic islets. Biotechnic & Histochemistry. 1993;68(4):196–205. doi: 10.3109/10520299309104698. [DOI] [PubMed] [Google Scholar]
- 2.Lukowiak B., Vandewalle B., Riachy R., Kerr-Conte J., Gmyr V., Belaich S. Identification and purification of functional human beta-cells by a new specific zinc-fluorescent probe. Journal of Histochemistry and Cytochemistry. 2001;49(4):519–528. doi: 10.1177/002215540104900412. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen D.A., Lernmark A., Berelowitz M., Bloom G.D., Steiner D.F. Sorting of pancreatic islet cell subpopulations by light scattering using a fluorescence-activated cell sorter. Diabetes. 1982;31(4 Pt 1):299–306. doi: 10.2337/diab.31.4.299. [DOI] [PubMed] [Google Scholar]
- 4.Pipeleers D.G., in't Veld P.A., Van de Winkel M., Maes E., Schuit F.C., Gepts W. A new in vitro model for the study of pancreatic A and B cells. Endocrinology. 1985;117(3):806–816. doi: 10.1210/endo-117-3-806. [DOI] [PubMed] [Google Scholar]
- 5.Dorrell C., Grompe M.T., Pan F.C., Zhong Y., Canaday P.S., Shultz L.D. Isolation of mouse pancreatic alpha, beta, duct and acinar populations with cell surface markers. Molecular and Cellular Endocrinology. 2011;339(1–2):144–150. doi: 10.1016/j.mce.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorrell C., Abraham S.L., Lanxon-Cookson K.M., Canaday P.S., Streeter P.R., Grompe M. Isolation of major pancreatic cell types and long-term culture-initiating cells using novel human surface markers. Stem Cell Research. 2008;1(3):183–194. doi: 10.1016/j.scr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Saunders D.C., Brissova M., Phillips N., Shrestha S., Walker J.T., Aramandla R. Ectonucleoside triphosphate diphosphohydrolase-3 antibody targets adult human pancreatic beta cells for in vitro and in vivo analysis. Cell Metabolism. 2019;29(3):745–754. doi: 10.1016/j.cmet.2018.10.007. e744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiGruccio M.R., Mawla A.M., Donaldson C.J., Noguchi G.M., Vaughan J., Cowing-Zitron C. Comprehensive alpha, beta and delta cell transcriptomes reveal that ghrelin selectively activates delta cells and promotes somatostatin release from pancreatic islets. Molecular Metabolism. 2016;5(7):449–458. doi: 10.1016/j.molmet.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mawla A.M., Huising M.O. Navigating the depths and avoiding the shallows of pancreatic islet cell transcriptomes. Diabetes. 2019;68(7):1380–1393. doi: 10.2337/dbi18-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eaves C.J. Hematopoietic stem cells: concepts, definitions, and the new reality. Blood. 2015;125(17):2605–2613. doi: 10.1182/blood-2014-12-570200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramond C., Glaser N., Berthault C., Ameri J., Kirkegaard J.S., Hansson M. Reconstructing human pancreatic differentiation by mapping specific cell populations during development. Elife. 2017;6 doi: 10.7554/eLife.27564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramond C., Beydag-Tasoz B.S., Azad A., van de Bunt M., Petersen M.B.K., Beer N.L. Understanding human fetal pancreas development using subpopulation sorting, RNA sequencing and single-cell profiling. Development. 2018;145(16) doi: 10.1242/dev.165480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swaminathan S., Fonseca V.A., Alam M.G., Shah S.V. The role of iron in diabetes and its complications. Diabetes Care. 2007;30(7):1926–1933. doi: 10.2337/dc06-2625. [DOI] [PubMed] [Google Scholar]
- 14.Simcox J.A., McClain D.A. Iron and diabetes risk. Cell Metabolism. 2013;17(3):329–341. doi: 10.1016/j.cmet.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raffield L.M., Louie T., Sofer T., Jain D., Ipp E., Taylor K.D. Genome-wide association study of iron traits and relation to diabetes in the Hispanic Community Health Study/Study of Latinos (HCHS/SOL): potential genomic intersection of iron and glucose regulation? Human Molecular Genetics. 2017;26(10):1966–1978. doi: 10.1093/hmg/ddx082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meidtner K., Podmore C., Kroger J., van der Schouw Y.T., Bendinelli B., Agnoli C. Interaction of dietary and genetic factors influencing body iron status and risk of type 2 diabetes within the EPIC-InterAct study. Diabetes Care. 2018;41(2):277–285. doi: 10.2337/dc17-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huth C., Beuerle S., Zierer A., Heier M., Herder C., Kaiser T. Biomarkers of iron metabolism are independently associated with impaired glucose metabolism and type 2 diabetes: the KORA F4 study. European Journal of Endocrinology. 2015;173(5):643–653. doi: 10.1530/EJE-15-0631. [DOI] [PubMed] [Google Scholar]
- 18.Podmore C., Meidtner K., Schulze M.B., Scott R.A., Ramond A., Butterworth A.S. Association of multiple biomarkers of iron metabolism and type 2 diabetes: the EPIC-InterAct study. Diabetes Care. 2016;39(4):572–581. doi: 10.2337/dc15-0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang R., Manson J.E., Meigs J.B., Ma J., Rifai N., Hu F.B. Body iron stores in relation to risk of type 2 diabetes in apparently healthy women. Journal of the American Medical Association. 2004;291(6):711–717. doi: 10.1001/jama.291.6.711. [DOI] [PubMed] [Google Scholar]
- 20.Attali M., Stetsyuk V., Basmaciogullari A., Aiello V., Zanta-Boussif M.A., Duvillie B. Control of beta-cell differentiation by the pancreatic mesenchyme. Diabetes. 2007;56(5):1248–1258. doi: 10.2337/db06-1307. [DOI] [PubMed] [Google Scholar]
- 21.Ravassard P., Hazhouz Y., Pechberty S., Bricout-Neveu E., Armanet M., Czernichow P. A genetically engineered human pancreatic beta cell line exhibiting glucose-inducible insulin secretion. Journal of Clinical Investigation. 2011;121(9):3589–3597. doi: 10.1172/JCI58447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genetics. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 26.Sugiyama T., Rodriguez R.T., McLean G.W., Kim S.K. Conserved markers of fetal pancreatic epithelium permit prospective isolation of islet progenitor cells by FACS. Proceedings of the National Academy of Sciences of the U S A. 2007;104(1):175–180. doi: 10.1073/pnas.0609490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin L., Gao D., Feng T., Tremblay J.R., Ghazalli N., Luo A. Cells with surface expression of CD133highCD71low are enriched for tripotent colony-forming progenitor cells in the adult murine pancreas. Stem Cell Research. 2016;16(1):40–53. doi: 10.1016/j.scr.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muraro M.J., Dharmadhikari G., Grun D., Groen N., Dielen T., Jansen E. A single-cell transcriptome atlas of the human pancreas. Cell Systems. 2016;3(4):385–394. doi: 10.1016/j.cels.2016.09.002. e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cabrera O., Berman D.M., Kenyon N.S., Ricordi C., Berggren P.O., Caicedo A. The unique cytoarchitecture of human pancreatic islets has implications for islet cell function. Proceedings of the National Academy of Sciences of the U S A. 2006;103(7):2334–2339. doi: 10.1073/pnas.0510790103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rorsman P., Huising M.O. The somatostatin-secreting pancreatic delta-cell in health and disease. Nature Reviews Endocrinology. 2018;14(7):404–414. doi: 10.1038/s41574-018-0020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gatter K.C., Brown G., Trowbridge I.S., Woolston R.E., Mason D.Y. Transferrin receptors in human tissues: their distribution and possible clinical relevance. Journal of Clinical Pathology. 1983;36(5):539–545. doi: 10.1136/jcp.36.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Domenico I., McVey Ward D., Kaplan J. Regulation of iron acquisition and storage: consequences for iron-linked disorders. Nature Reviews Molecular Cell Biology. 2008;9(1):72–81. doi: 10.1038/nrm2295. [DOI] [PubMed] [Google Scholar]
- 33.MacDonald M.J., Cook J.D., Epstein M.L., Flowers C.H. Large amount of (apo)ferritin in the pancreatic insulin cell and its stimulation by glucose. The FASEB Journal. 1994;8(10):777–781. doi: 10.1096/fasebj.8.10.8050678. [DOI] [PubMed] [Google Scholar]
- 34.Santos M., Anderson C.P., Neschen S., Zumbrennen-Bullough K.B., Romney S.J., KahleStephan M. Irp2 regulates insulin production through iron-mediated Cdkal1-catalyzed tRNA modification. Nature Communications. 2020;11(1):296. doi: 10.1038/s41467-019-14004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hyman E.S. Acquired iron-deficiency anaemia due to impaired iron transport. Lancet. 1983;1(8316):91–95. doi: 10.1016/s0140-6736(83)91741-5. [DOI] [PubMed] [Google Scholar]
- 36.Larrick J.W., Hyman E.S. Acquired iron-deficiency anemia caused by an antibody against the transferrin receptor. New England Journal of Medicine. 1984;311(4):214–218. doi: 10.1056/NEJM198407263110402. [DOI] [PubMed] [Google Scholar]
- 37.Abraham D., Rogers J., Gault P., Kushner J.P., McClain D.A. Increased insulin secretory capacity but decreased insulin sensitivity after correction of iron overload by phlebotomy in hereditary haemochromatosis. Diabetologia. 2006;49(11):2546–2551. doi: 10.1007/s00125-006-0445-7. [DOI] [PubMed] [Google Scholar]
- 38.Hatunic M., Finucane F.M., Norris S., Pacini G., Nolan J.J. Glucose metabolism after normalization of markers of iron overload by venesection in subjects with hereditary hemochromatosis. Metabolism. 2010;59(12):1811–1815. doi: 10.1016/j.metabol.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 39.Fernandez-Real J.M., Penarroja G., Castro A., Garcia-Bragado F., Hernandez-Aguado I., Ricart W. Blood letting in high-ferritin type 2 diabetes: effects on insulin sensitivity and beta-cell function. Diabetes. 2002;51(4):1000–1004. doi: 10.2337/diabetes.51.4.1000. [DOI] [PubMed] [Google Scholar]
- 40.Hansen J.B., Tonnesen M.F., Madsen A.N., Hagedorn P.H., Friberg J., Grunnet L.G. Divalent metal transporter 1 regulates iron-mediated ROS and pancreatic beta cell fate in response to cytokines. Cell Metabolism. 2012;16(4):449–461. doi: 10.1016/j.cmet.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 41.Koch R.O., Zoller H., Theuri I., Obrist P., Egg G., Strohmayer W. Distribution of DMT 1 within the human glandular system. Histology & Histopathology. 2003;18(4):1095–1101. doi: 10.14670/HH-18.1095. [DOI] [PubMed] [Google Scholar]
- 42.Segerstolpe A., Palasantza A., Eliasson P., Andersson E.M., Andreasson A.C., Sun X. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metabolism. 2016;24(4):593–607. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshihara E., Wei Z., Lin C.S., Fang S., Ahmadian M., Kida Y. ERRgamma is required for the metabolic maturation of therapeutically functional glucose-responsive beta cells. Cell Metabolism. 2016;23(4):622–634. doi: 10.1016/j.cmet.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stolovich-Rain M., Enk J., Vikesa J., Nielsen F.C., Saada A., Glaser B. Weaning triggers a maturation step of pancreatic beta cells. Developmental Cell. 2015;32(5):535–545. doi: 10.1016/j.devcel.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 45.Haurie V., Boucherie H., Sagliocco F. The Snf1 protein kinase controls the induction of genes of the iron uptake pathway at the diauxic shift in Saccharomyces cerevisiae. Journal of Biological Chemistry. 2003;278(46):45391–45396. doi: 10.1074/jbc.M307447200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.