Abstract
Data on the draft genome sequence of Erysipelothrix rhusiopathiae strain VR-2 is presented in this report. E. rhusiopathiae strain VR-2 is a commercial attenuated vaccine widely used in Russia and a number European countries for immunization of pigs against swine erysipelas. The draft genome sequence of 1,704,727 bp in length included 1415 protein sequences, 50 tRNA genes and 3 rRNA genes according NCBI Prokaryotic Genomes Automatic Annotation Pipeline results. The draft genome sequence data of E. rhusiopathiae strain VR-2 is available in GenBank under the accession nos. RJTK00000000.1, PRJNA504614 and SAMN10395786 for Genome, Bioproject and Biosample, respectively. The obtained sequence data may be helpful for searching genetic markers of VR-2, aimed to develop assays to discriminate between field isolates and this vaccine strain of E. rhusiopathiae.
Keywords: Erysipelothrix rhusiopathiae, Swine erysipelas, Attenuated vaccine, Draft genome sequence, WGS
Specifications Table
| Subject | Biology |
| Specific subject area | Microbial genomics |
| Type of data | Genomic sequence, predicted genes and annotation |
| How data were acquired | Whole-genome sequencing using 454 pyrosequencing |
| Data format | Raw and analysed |
| Parameters for data collection | The genomic DNA was extracted from E. rhusiopathiae VR-2 vaccine strain with standard phenol-chloroform method and sequenced using 454 next generation sequencing system (Roche, Switzerland). Reads were assembled into 39 contigs using GS de novo Assembler version 2.7. Genome annotation was performed with NCBI PGAP and RAST servers. |
| Description of data collection | Extraction of genomic DNA, fragment library preparation, 454 pyrosequencing, de novo assembly and annotation procedures |
| Data source location | FSBSI «All-Russian Scientific Research and Technological Institute of Biological Industry» (FSBSI VNITIBP RAS, Shchelkovsky district, Moscow Region, Russia). |
| Data accessibility | Raw FastaQ reads and genome assembly data are available in Mendeley Data repository at https://data.mendeley.com/datasets/pvf8cj3zn8/2. The draft genome sequence data of E. rhusiopathiae strain VR-2 is available in GenBank under the accession nos. RJTK00000000.1 (https://www.ncbi.nlm.nih.gov/nuccore/RJTK00000000.1/), PRJNA504614 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA504614) and SAMN10395786 (https://www.ncbi.nlm.nih.gov/biosample/?term=SAMN10395786) for Genome, Bioproject and Biosample, respectively. |
Value of the Data
-
•
Draft genome data may be used for study in depth the genetic features of this E. rhusiopathiae vaccine strain.
-
•
Draft genome data may be helpful for searching genetic markers of E. rhusiopathiae vaccine strain VR-2 aimed to develop genetic assays for differentiation of field isolates and this live vaccine strain.
-
•
This genome sequence data may be used in comparative studies to better understand the genetic diversity and the genomic and molecular attributes underlying the pathogenicity of E. rhusiopathiae strains.
1. Data Description
Erysipelothrix rhusiopathiae is a gram-positive, non-spore-forming, rod-shaped bacterium that causes erysipeloid in humans and erysipelas in animals, including swine erysipelas [1], [2], [3]. Due to economical importance of swine erysipelas in husbandry, an epizootic of E. rhusiopathiae is controlled by administration of commercially available E. rhusiopathiae vaccines in many countries [4], [5], [6], [7], [8]. In Russia and a number of European countries E. rhusiopathiae live attenuated vaccine strain VR-2 is widely used for this purpose [9], [10], [11]. The use of attenuated vaccine strains is associated with risks of their reversion to virulence, caused outbreaks of swine erysipelas, therefore differentiation of E. rhusiopathiae vaccine strain VR-2 and field isolates is especially important for epizootical monitoring of swine erysipelas in countries applying this commercially available live attenuated vaccine for immunization of pigs against swine erysipelas. In this regard whole genome sequencing is a promising approach for searching for genetic markers of E. rhusiopathiae VR-2 to develop the strain-specific genetic assays for differentiation of field isolates and this live vaccine strain.
After assembling the obtained 89,701 sequence reads 39 contigs of 1,704,727 bp total length at 20-fold coverage were generated (Table 1). A 36.5% CG content of VR-2 strain well correlated with CG% of the complete genome sequences of other E. rhusiopathiae strains form GenBank (Table 1). VR-2 genome sequence identity with genome neighbors – E. rhusiopathiae strains WH13013 and Fujisawa – was 94.9695% and 94.7062%, respectively.
Table 1.
General features of the E. rhusiopathiae strain VR-2 genome sequence.
| Feature | Value |
|---|---|
| Size | 1,704,727 |
| Coverage | 20x |
| GC Content | 36.5% |
| N50 | 83,301 |
| L50 | 7 |
| Number of contigs | 39 |
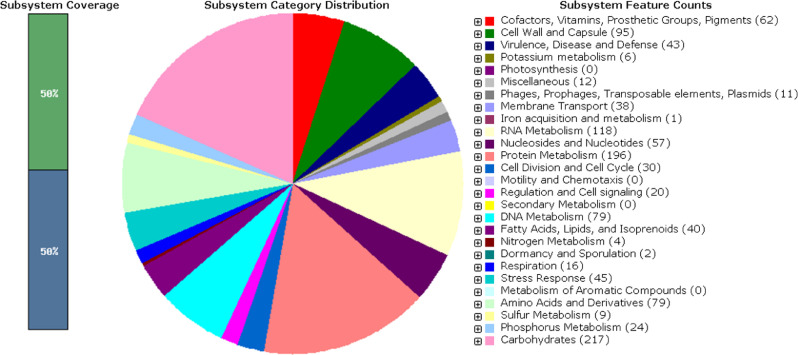
Annotation of the assembled data with NCBI Prokaryotic Genomes Annotation Pipeline [12,13] revealed 1415 protein sequences, 50 tRNA genes and 3 rRNA genes. Annotation with the RAST server [14,15] revealed 264 subsystems. An overview of the subsystem statistics for the draft genome sequence of E. rhusiopathiae strain VR-2 generated by Rapid Annotation System Technology (RAST) is shown in Fig. 1.
Fig. 1.
An overview of the subsystem statistics for draft genome sequence of E. rhusiopathiae strain VR-2 generated by Rapid Annotation System Technology (RAST).
Raw FastaQ reads, the genome assembly data and results of the draft genome sequence annotation with RAST server are available in Mendeley Data repository at https://data.mendeley.com/datasets/pvf8cj3zn8/2.
2. Experimental Design, Materials and Methods
DNA sample of E. rhusiopathiae VR-2 vaccine strain were kindly provided by FSBSI «All-Russian Scientific Research and Technological Institute of Biological Industry» (FSBSI VNITIBP RAS, Shchelkovsky district, Moscow Region, Russia). The genomic DNA was extracted and purified with standard phenol-chloroform method.
The genomic DNA was sequenced using 454 next generation sequencing system (Roche, Switzerland). In brief, 500 mg of the genomic DNA was fragmented by nebulization using Rapid Library Nebulizers (Roche, Cat. No. 05233780001) and Rapid Library Buffers (Roche, Cat. No. 05619181001). The fragmented DNA was purified on the columns from the Qiagen MiniElutePCR Purification kit (Cat. No. 28004) and ligated with DNA adaptor using Rgt/Adaptors kit (Roche Cat. No. 05619203,001) according to the manufacturer's instructions. Small DNA fragments and unligated DNA adaptor molecules were removed using Agencount AMPure XP Beads (Beckman Coulter, Cat. No. A63880). The obtained DNA library was qualified using Lonza FlashDoc (Cat. No. 57025) and FlashGel DNA Cassette (Lonza, Cat. No. 57023). The DNA fraction with predominant sizes ranging from 500 to 800 bp was quantified with Quant-it Picogreen dsDNA Assay Kit (Invitrogen, Cat. No. P7589) and QuantiFluor-ST Fluorometer (Promega, Model E6090) and then amplified by an emulsion PCR with the use of emPCR Reagents Lib-L Kit (Cat.#05996503001). The prepared DNA library was added to the emulsion PCR at a ratio of two DNA molecules per bead. After emPCR completion the emulsion was breaking with the use of Oil and Breaking Kit (Roche Cat. No. 05996511001). The beads with the amplified DNA was isolated with Bead Recovery Reagents (Roche, Cat. No. 05996490001). Sequencing of the DNA library was performed using Sequencing Buffers (Roche, Cat. No. 05996589001), Reagents and Enzymes (Roche, Cat. No. 05996562001), Packing Beads & Supplement CB (Roche, Cat. No. 05996597001) and PicoTiterPlate Kit (Roche, Cat. No. 05996619001). The beads with the amplified DNA as well as layers of packing, enzyme and PPiase beads were loaded into the wells of PicoTiterPlate according to the manufacturer's instructions and sequenced using GS Junior instrument (Roche, Switzerland). Reads were assembled into contigs using the GS de novo Assembler (version 2.7) with default parameters.
Genome annotation was performed using the NCBI Prokaryotic Genomes Automatic Annotation Pipeline [12,13] and Rapid Annotation System Technology (RAST) server [14,15]
Author Statement
The authors contributed equally to this work.
CRediT authorship contribution statement
Svetlana Kovalchuk: Conceptualization, Methodology, Formal analysis, Writing - original draft. Anna Babii: Investigation, Visualization, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Acknowledgments
We would like to thank Dr. Anatoly Ya. Samuylenko from FSBSI «All-Russian Scientific Research and Technological Institute of Biological Industry» (FSBSI VNITIBP RAS, Shchelkovsky district, Moscow Region, Russia) for kindly providing us with the genomic DNA sample of E. rhusiopathiae VR-2.
The study was funded by Ministry of Science and Higher Education of the Russian Federation (No. 075-01250-20-01).
References
- 1.Wood L.R. Erysipelas. In: Straw B.E., Zimmermann J.J., D'Allaire S., Taylor D.J., editors. Diseases of Swine. 9th ed. Blackwell Publishing Professional; 2006. pp. 629–638. [Google Scholar]
- 2.Shimoji Y. Pathogenicity of Erysipelothrix rhusiopathiae: virulence factors and protective immunity. Microb. Infect. 2000;2:965–972. doi: 10.1016/S1286-4579(00)00397-X. [DOI] [PubMed] [Google Scholar]
- 3.Wang Q., Chang B.J., Riley T.V. Erysipelothrix rhusiopathiae. Vet. Microbiol. 2010;140:405–417. doi: 10.1016/j.vetmic.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Seto K., Nishimura Y., Fujiki M., Azechi H., Suzuki K. Studies on acriflavine-fast attenuated Erysipelothrix insidiosa. Comparison on pathogenicity and immunogenicitybetween mice and pigs. Jpn. J. Vet. Sci. 1971;33:161–171. doi: 10.1292/jvms1939.33.161. [DOI] [PubMed] [Google Scholar]
- 5.Mansfield K.G., Fox J.G. Bacterial diseases. In: Marini R., Wachtman L., Tardif S., Mansfield K., Fox J., editors. Common Marmoset in Captivity and Biomedical Research. Elsevier; 2019. pp. 265–287. [DOI] [Google Scholar]
- 6.Brack M., Rensing S., Gatesman T.J. Erysipelothrix insidiosa infection in callitrichids kept behind a barrier system. Infect. Dis. Rev. 1999;1 15e9. [Google Scholar]
- 7.Zou Y., Zhu X., Muhammad H.M., Jiang P., Li Y. Characterization of Erysipelothrix rhusiopathiae strains isolated from acute swine erysipelas outbreaks in Eastern China. J. Vet. Med. Sci. 2015;77:653–660. doi: 10.1292/jvms.14-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi T., Takagi M., Sawada T., Seto K. Cross protection in mice and swine immunized with live erysipelas vaccine to challenge exposure with strains of Erysipelothrix rhusiopathiae of various serotypes. Am. J. Vet. Res. 1984;45:2115–2118. [PubMed] [Google Scholar]
- 9.Majdan S., Krzyszowski M., Kocik T. Anti-erysipelas vaccine from the VR2 strain. Veterinariia. 1972;9:116–118. [PubMed] [Google Scholar]
- 10.Bodurova T. Properties of the lyophilized anti-swine-erysipelas vaccine (VR2) Vet. Med. Nauk. 1973;10:65–69. [PubMed] [Google Scholar]
- 11.Astashova E.A. Morphocytochemical study of Erysipelothrix rhusiopathiae of the vaccine strain VR-2. Veterinariia. 1975;8:29–30. [PubMed] [Google Scholar]
- 12.Haft D.H., DiCuccio M., Badretdin A., Brover V., Chetvernin V., O'Neill K., Li W., Chitsaz F., Derbyshire M.K., Gonzales N.R., Gwadz M., Lu F., Marchler G.H., Song J.S., Thanki N., Yamashita R.A., Zheng C., Thibaud-Nissen F., Geer L.Y., Marchler-Bauer A., Pruitt K.D. RefSeq: an update on prokaryotic genome annotation and curation. Nucl. Acids Res. 2017;46:D851–D860. doi: 10.1093/nar/gkx1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L., Lomsadze A., Pruitt K.D., Borodovsky M., Ostell J. NCBI prokaryotic genome annotation pipeline. Nucl. Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aziz R.K., Bartels D., Best A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., Meyer F., Olsen G.J., Olson R., Osterman A.L., Overbeek R.A., McNeil L.K., Paarmann D., Paczian T., Parrello B., Pusch G.D., Reich C., Stevens R., Vassieva O., Vonstein V., Wilke A., Zagnitko O. The RAST Server: rapid annotations using subsystems technology. BMC Genom. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Overbeek R., Olson R., Pusch G.D., Olsen G.J., Davis J.J., Disz T., Edwards R.A., Gerdes S., Parrello B., Shukla M., Vonstein V., Wattam A.R., Xia F., Stevens R. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST) Nucl. Acids Res. 2014;42 doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]