The Al-resistance transcription factor STOP1 is mono-SUMOylated at K40, K212, or K395 sites and deSUMOylated by the SUMO protease ESD4, which is involved in the regulation of Al resistance in Arabidopsis.
Abstract
Aluminum (Al) is a primary constraint for crop production on acid soils, which make up more than 30% of the arable land in the world. Al resistance in Arabidopsis (Arabidopsis thaliana) is achieved by malate secretion mediated by the Al-ACTIVATED MALATE TRANSPORTER1 (AtALMT1) transporter. The C2H2-type transcription factor SENSITIVE TO PROTON RHIZOTOXICITY1 (STOP1) is essential and required for Al resistance, where it acts by inducing the expression of Al-resistance genes, including AtALMT1. In this study, we report that STOP1 protein function is modified by SUMOylation. The SMALL UBIQUITIN-LIKE MODIFIER (SUMO) protease ESD4, but not other SUMO proteases, specifically interacts with and deSUMOylates STOP1. Mutation of ESD4 increases the level of STOP1 SUMOylation and the expression of the STOP1-regulated gene AtALMT1, which contributes to the increased Al resistance in esd4. The esd4 mutation does not influence STOP1 protein abundance but increases the association of STOP1 with the AtALMT1 promoter, which might explain the elevated expression of AtALMT1 in esd4. We demonstrate that STOP1 is mono-SUMOylated at K40, K212, or K395 sites, and blocking STOP1 SUMOylation reduces STOP1 stability and the expression of STOP1-regulated genes, leading to the reduced Al resistance. Our results thus reveal the involvement of SUMOylation in the regulation of STOP1 and Al resistance in Arabidopsis.
INTRODUCTION
Aluminum (Al) is the most abundant metal and the third most abundant element after oxygen and silicon in the Earth’s crust. Most Al exists as insoluble aluminosilicates or aluminum oxides, which are nontoxic to plant growth. However, as soils acidify to a pH of 5.5 or lower, Al3+ ions are dissolved and become highly toxic to plant root growth (Kochian, 1995; Kochian et al., 2004; Ma, 2007). Al toxicity is, thus, a main constraint for crop production on acid soils. Since 40 to 50% of the potentially arable lands in the world are acidic, Al toxicity is one of the most serious global problems and the second greatest abiotic restraint to crop production, exceeded only by drought stress (von Uexkull and Mutert, 1995).

To survive in Al-toxic environments, plants have evolved various mechanisms to detoxify Al (Kochian et al., 2004, 2015; Liu et al., 2014). Among them, Al-induced exudation of organic acids, including malate, citrate, and oxalate, is an essential mechanism for Al detoxification in most plant species (Ma et al., 2001; Ryan et al., 2001). Different plant species may secrete distinct organic acids to chelate and detoxify Al. Arabidopsis (Arabidopsis thaliana), for instance, mainly secretes malate to detoxify Al, although it also releases citrate in response to Al stress (Hoekenga et al., 2003; Liu et al., 2009). Transporters required for the malate and citrate secretion were first identified in crops (Sasaki et al., 2004; Furukawa et al., 2007; Magalhaes et al., 2007). Based on the sequence similarity and functional analysis, the malate transporter AtALMT1 (Al-ACTIVATED MALATE TRANSPORTER1) and the citrate transporter AtMATE, for root malate and citrate exudation, respectively, were subsequently identified in Arabidopsis (Hoekenga et al., 2006; Liu et al., 2009). In addition to the organic acid exudation-based Al-resistance mechanism, Arabidopsis plants also utilize the tonoplast-localized half-size ATP binding cassette transporter ALS1 and the bacterial-type ATP binding cassette transporter complex AtSTAR1/ALS3 to sequester Al into the vacuoles or modify the cell wall to detoxify Al (Larsen et al., 2005, 2007; Huang et al., 2010; Dong et al., 2017). The C2H2-type zinc finger transcription factor SENSITIVE TO PROTON RHIZOTOXICITY1 (STOP1) plays an essential role in the regulation of Al resistance by modulating the expression of some Al-resistance genes, including AtALMT1, AtMATE, and ALS3 (Iuchi et al., 2007; Sawaki et al., 2009). Although the transcript levels of the STOP1-downstream genes AtALMT1, AtMATE, and ALS3 are induced by Al stress, the mRNA level of STOP1 itself is not influenced by Al (Iuchi et al., 2007; Sawaki et al., 2009), suggesting that STOP1 might be subjected to posttranscriptional and/or posttranslational regulation. In line with this idea, we recently found that Al stress can stabilize the STOP1 protein and that the F-box protein REGULATION OF ATALMT1 EXPRESSION1 (RAE1) acts as an E3 ubiquitin ligase to promote STOP1 ubiquitination and degradation (Zhang et al., 2019). More recently, the core component of the THO/TREX complex, HPR1, was found to regulate STOP1 at the posttranscriptional level through the modulation of nucleocytoplasmic STOP1 mRNA export (Guo et al., 2020).
The SMALL UBIQUITIN-LIKE MODIFIER (SUMO) is a small polypeptide structurally related to ubiquitin. SUMO can be attached to protein substrates in a process called SUMOylation, which can alter protein function, activity, localization, and/or stability (Wilkinson and Henley, 2010; Jentsch and Psakhye, 2013; Augustine and Vierstra, 2018). Like ubiquitin, SUMO is conjugated to its substrates through an E1-E2-E3 cascade (Saitoh et al., 1997). In Arabidopsis, only one E1 SUMO-activating enzyme heterodimer (SAE1a/b and SAE2), a single E2 SUMO-conjugating enzyme (SCE1), and two E3 ligases (SIZ1 and MMS21/HPY2) have been identified (Miura and Hasegawa, 2010). Recently, two homologous E4-type SUMO ligases, PIAL1 and PIAL2, were identified and demonstrated to be involved in SUMO chain formation (Tomanov et al., 2014). SUMOylation is a reversible process: deSUMOylation is catalyzed by SUMO proteases (Yates et al., 2016; Castro et al., 2018). This reversible conjunction of SUMO to target proteins is particularly important for the regulation of responses to abiotic stresses. For instance, SUMO can target the MYB transcription factor PHR1 through the E3 ligase SIZ1 to regulate phosphate (Pi)-deficiency responses (Miura et al., 2005). SIZ1-mediated SUMOylation plays an important role in the drought stress response through the regulation of drought-responsive genes (Catala et al., 2007). SUMOylation is also involved in the regulation of low-temperature tolerance through targeting the transcription factor ICE1 (Miura et al., 2007).
An important class of plant SUMO proteases is that of the ubiquitin-like proteases (ULPs), which belong to the C48 clade of the Cys protease family (Castro et al., 2018). The Arabidopsis genome is presumed to have eight ULPs (Castro et al., 2018). Since there are substantially more ULPs than SUMO E3 ligases in Arabidopsis, it has been suggested that substrate specificity in the SUMOylation process might be conferred by ULPs and that deSUMOylating might be a major regulatory step to control the effect of SUMOylation (Yates et al., 2016; Verma et al., 2018).
The ULP member ESD4 was the first demonstrated to be crucial for the regulation of SUMOylation in plants, and mutation of ESD4 induces early flowering and severe pleiotropic effects on plant growth (Reeves et al., 2002; Murtas et al., 2003), which are partially caused by salicylic acid overaccumulation (Villajuana-Bonequi et al., 2014). Although ESD4 plays a critical role in the regulation of plant development and stress responses, no direct target substrates of ESD4 have been identified. The two ULP members OTS1/ULP1d and OTS2/ULP1c are the best characterized and have been demonstrated to be redundantly involved in the regulation of various biological processes. For instance, OTS1 and OTS2 modulate salt stress responses partly through deSUMOylation and destabilization of growth-repressing DELLA proteins (Conti et al., 2008, 2014) and regulate light-induced signaling by deSUMOylating the photoreceptor phytochrome B (Sadanandom et al., 2015). OTS1 and OTS2 are also reported to regulate jasmonic acid signaling by modulating JAZ protein SUMOylation and stability (Srivastava et al., 2018). Recently, another ULP member, ULP1a, was demonstrated to mediate the deSUMOylation of BZR1, a key transcription factor of brassinosteroid signaling, to integrate environmental cues into brassinosteroid signaling to shape plant growth (Srivastava et al., 2020). Other ULP members, such as SPF1/ULP2b and/or SPF2/ULP2a, modulate flowering time and fertility partially through regulating FLC and EDA9 stability, respectively (Kong et al., 2017; Liu et al., 2017).
Previously, we generated a transgenic reporter line by fusing the promoter of the STOP1-target gene AtALMT1 to a luciferase (LUC) reporter gene and performed a forward genetic screen to identify genes involved in the regulation of AtALMT1 and/or STOP1 (Zhang et al., 2019). In this study, we characterize two independent mutants of the SUMO protease ESD4 that yield enhanced Al resistance. We conclude that ESD4 directly deSUMOylates STOP1 to regulate the expression of STOP1-downstream genes and Al resistance. We also determined that STOP1 can be mono-SUMOylated at K40, K212, or K395 sites and that the SUMOylation of STOP1 is important for maintaining STOP1 protein levels and Al resistance.
RESULTS
rae5 Mutants Show Increased Expression of the pAtALMT1:LUC Reporter and of the Endogenous AtALMT1
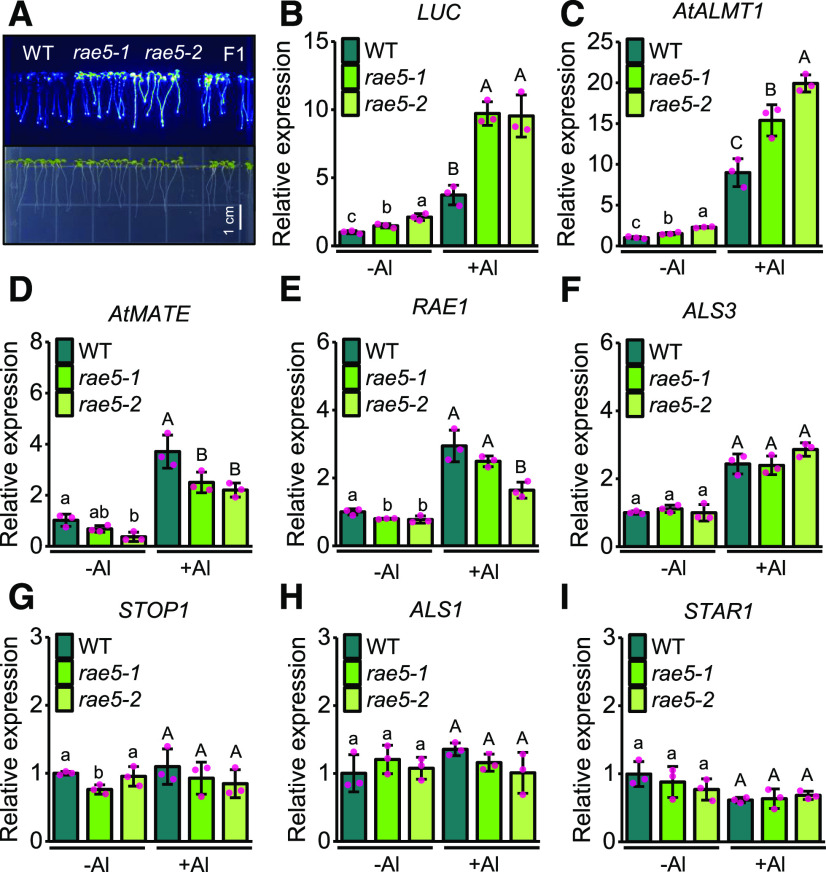
We previously performed a forward genetic screen on an EMS-mutagenized population of the AtALMT1 promoter-driven LUC reporter (pAtALMT1:LUC) line and identified 13 rae mutants with increased LUC signal, which includes eight rae1 mutants (Zhang et al., 2019). In this study, we focused on two other mutants, rae5-1 and rae5-2, which were allelic to each other and showed higher LUC expression than the wild type (Figure 1A).
Figure 1.
Mutation of RAE5 Alters the Expression of STOP1-Regulated Genes.
(A) Increased LUC signal of pAtALMT1:LUC in rae5 mutants and F1 plants from a cross between rae5-1 and rae5-2. Bar = 1 cm. WT, wild type.
(B) to (I) Effect of rae5 mutations on the mRNA accumulation levels of Al-resistance genes. Seedlings of the wild type (WT), rae5-1, and rae5-2 were exposed to 0 or 30 µM Al for 8 h, and then the roots were excised for mRNA isolation and expression analysis. RT-qPCR analysis was performed to determine the expression of LUC (B), AtALMT1 (C), AtMATE (D), RAE1 (E), ALS3 (F), STOP1 (G), ALS1 (H), and AtSTAR1 (I). Data shown are means ± sd of three biological replicates. Different letters at each treatment indicate values significantly different (P < 0.05, ANOVA followed by Tukey’s test). At least three independent experiments were performed with similar results.
We compared mRNA expression levels of the LUC reporter gene and the endogenous AtALMT1 gene in roots of the wild type and rae5 mutants. In accordance with the increased LUC signal in the mutants, the expression of both genes was significantly higher in the mutants than in the wild type under both –Al and +Al conditions (Figures 1B and 1C). To examine whether the rae5 mutations affect the expression of other Al-resistance genes in the roots, we conducted an RT-qPCR expression analysis on AtMATE, RAE1, ALS3, STOP1, ALS1, and AtSTAR1 in the wild type and the rae5 mutants. AtMATE and RAE1 were expressed at lower levels in the mutants than in the wild type, while ALS3 was expressed at a similar level in the wild type and the mutants (Figures 1D to 1F). The expression levels of STOP1, ALS1, and AtSTAR1, which are not modulated by the transcription factor STOP1 (Iuchi et al., 2007; Sawaki et al., 2009), were similar between the wild type and the mutants (Figures 1G to 1I).
We also compared the expression of STOP1-regulated Al-resistance genes in shoots. In the shoots, mutation of STOP1 nearly abolished the expression of LUC and AtALMT1 (Supplemental Figures 1A and 1B), indicating that STOP1 is also crucial for AtALMT1 expression in the shoots. Nevertheless, the expression of AtMATE, RAE1, and ALS3 was only slightly reduced in the stop1-3 mutant under +Al conditions (Supplemental Figures 1C to 1E), suggesting that STOP1 does not play a critical role in the regulation of the expression of these genes in the shoots. Mutation of RAE5 increased the expression LUC and AtALMT1 but not that of AtMATE, RAE1, and ALS3 (Supplemental Figure 1).
Mutation of RAE5 Increases Malate Secretion and Al Resistance
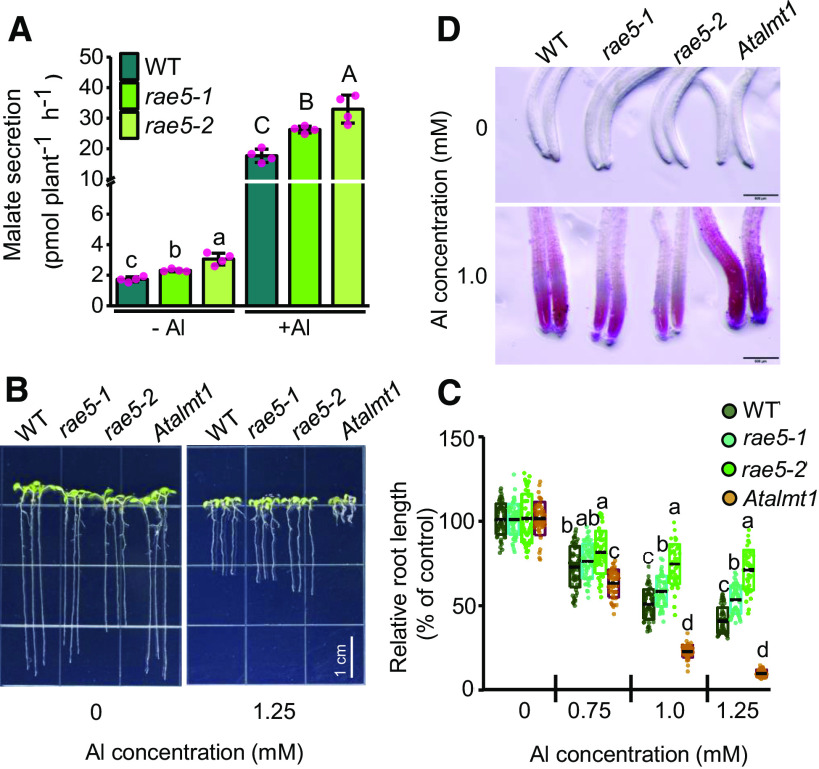
Because of the increased and decreased expression of AtALMT1 and AtMATE in rae5 mutants, respectively, we determined whether malate and citrate release was altered in the mutants. The results showed that the mutants secreted more malate than the wild type under both –Al and +Al conditions, while the release of citrate was lower in the mutants than in wild type under +Al conditions (Figure 2A; Supplemental Figure 2A).
Figure 2.
The rae5 Mutations Increase Malate Secretion and Al Resistance.
(A) Increased malate release in rae5 mutants. Seedlings of the wild type (WT), rae5-1, and rae5-2 were treated with 0 or 10 μM AlCl3 for 12 h, and the root exudates were analyzed for the malate concentrations (pmol plant−1 h−1). Values are means ± sd of four biological replicates.
(B) to (D) Al-resistance ([B] and [C]) and Al-accumulation (D) phenotypes of the wild type (WT), rae5-1, rae5-2, and Atalmt1. Seedlings were grown on a soaked gel medium containing 0, 0.75, 1.0, or 1.25 mM Al for 7 d, and the relative root growth was used to evaluate Al resistance. The Al indicator Eriochrome Cyanine R was used to stain the roots exposed to 0 or 1.0 mM Al (D). Values are means ± sd of relative root length of 48 to 60 seedlings. Bars = 1 cm.
Means with different letters are significantly different (P < 0.05, ANOVA test followed by Tukey’s test). At least three independent experiments were performed with similar results.
To assess the Al-resistance phenotype of rae5 mutants, we grew wild-type, rae5-1, rae5-2, and Atalmt1 plants on agar plates containing different Al concentrations. Under control conditions, the root growth of rae5-1 and rae5-2 was slightly slower than that of the wild type (Figure 2B). rae5-1 showed enhanced resistance to Al than the wild type at 1.25 mM Al, while rae5-2 was more resistant at both 1.0 and 1.25 mM Al, suggesting that rae5-2 is a stronger allele compared with rae5-1 (Figures 2B and 2C). The Atalmt1 control was more sensitive to Al than the wild type at all Al concentrations. Consistent with these results, the rae5 mutants accumulated less Al in root tips than the wild type, based on staining with the Al indicator Eriochrome Cyanine R (Figure 2D), while the Atalmt1 mutant control accumulated more Al in the root tips. These results demonstrate that the rae5 mutations increase malate secretion and Al resistance.
To examine whether rae5 mutants are specifically resistant to Al, we tested the tolerance of rae5 mutants to two other toxic metals, Cd and La. The mutants did not show altered tolerance to Cd or La compared with the wild type (Supplemental Figures 2B and 2C), demonstrating that rae5 mutants are not broadly resistant to all toxic metals.
Mutation of RAE5 also induced morphological defects. The mutants showed reduced plant height and organ size, low fertility, and early flowering (Supplemental Figure 2D). rae5-2 had stronger morphological defects than rae5-1.
RAE5 Encodes the SUMO Protease ESD4
A genetic analysis of the rae5-2 mutation was performed using an F2 population from a cross between rae5-2 and the wild type. Among 356 F2 plants, 75 plants showed increased LUC signal and morphological defects, while the other 281 plants had normal LUC signal and morphology. The ratio of plants with high LUC signal to plants with normal LUC signal fitted to 1:3 (χ2 = 2.93, P > 0.05), suggesting that the increased LUC signal and the morphological defects both were controlled by a single recessive gene.
To clone the RAE5 gene, the same F2 population was used and pooled DNA from 75 F2 plants with increased LUC signal and morphological defects was subjected to whole-genome sequencing. As a control, we used the wild-type DNA sequence, which had been sequenced before (Zhang et al., 2019). By MutMap analysis (Abe et al., 2012), we located the candidate gene to a small region of chromosome 4 (Supplemental Figure 3A). To confirm the results of the MutMap analysis, we developed four derived cleaved amplified polymorphic sequence (dCAPS) markers surrounding the candidate region on the basis of mutations in rae5-2 (Supplemental Table 1) and used 58 F2 plants with increased LUC expression and defective morphologies to conduct a marker linkage analysis. The four markers displayed, to different extents, some linkage to the mutant phenotype (Supplemental Table 1), confirming that the RAE5 gene was indeed located in this region. We mapped the RAE5 gene between D2 and D4 markers with 8 and 12 recombinants, respectively, and the D3 marker, developed on a C-to-T substitution at +1459 bp from the start codon of At4G15880 (ESD4), was completely linked to the mutant phenotype (Supplemental Table 1). This substitution caused an amino acid change from Arg to the stop codon in ESD4 in the mutant. We also sequenced ESD4 in the rae5-1 allele and identified a G-to-A substitution at +1369 bp from the start codon in rae5-1, introducing an amino acid change from Asp to Asn. These results suggest that these mutations in ESD4 are responsible for the increased LUC expression and morphological defects in the rae5 mutants.
To further confirm that RAE5 is ESD4, we performed a complementation test on rae5-1 by transforming a wild-type genomic fragment of ESD4 fused with a 3Flag tag into the mutant. Quantitative expression analysis showed that the increased expression of LUC and AtALMT1 and the reduced expression of AtMATE in rae5-1 were rescued in two complementation lines (Supplemental Figure 3B). The elevated Al resistance and defective plant growth in the mutant were also recovered (Supplemental Figures 3C to 3E). Together, these results demonstrate that RAE5 encodes the SUMO protease ESD4. For clarity, we refer to rae5-1 and rae5-2 as esd4-3 and esd4-4, respectively, in the following sections.
ESD4 Protein Accumulation Is Increased in Response to Al Stress
To investigate whether Al stress affects ESD4 mRNA expression, we exposed wild-type roots to Al stress for different times and then quantified the mRNA levels of ESD4. Although Al stress could highly induce the expression of AtALMT1 control for all time points, the expression of ESD4 was not responsive to the Al stress (Figure 3A). To examine whether ESD4 protein accumulation is influenced by Al stress, we utilized the complementation line (pESD4:ESD4-3Flag esd4-3) for the detection of ESD4 protein using the anti-Flag antibody and exposed the line to –Al or +Al conditions for 0, 2, 4, or 8 h. Al stress was able to increase the protein accumulation of ESD4-3Flag after 4 h of treatment (Figures 3B and 3C).
Figure 3.
Al Stress Promotes ESD4 Protein Accumulation.
(A) Effect of Al on ESD4 mRNA expression. Wild-type seedlings were treated without Al or with 30 μM Al for 2, 4, 6, or 8 h, and roots were sampled for RT-qPCR analysis of AtALMT1 and ESD4.
(B) and (C) Representative gels (B) and relative protein levels (C) of ESD4-3Flag under –Al or +Al conditions. Seedlings of an esd4-3 complementation line (pESD4:ESD4-3Flag) were treated without Al or with 30 μM Al for 2, 4, or 8 h, and roots were sampled for immunoblot analysis of ESD4-3Flag and ACTIN control. ESD4-3Flag protein levels were first normalized to their respective ACTIN controls and then normalized to the protein level under the –Al condition. Data shown are means ± sd of three biological replicates. Asterisks indicate values that are statistically different (Student’s t test, **, P < 0.01). WT, wild type.
Two and three independent experiments were performed with similar results for (A) and ([B] and [C]), respectively.
STOP1 Is Mono-SUMOylated at Multiple Sites and DeSUMOylated by ESD4
Because ESD4 is involved in the SUMOylation pathway and the regulation of the expression of STOP1-target genes, we hypothesized that ESD4 might regulate the level of STOP1 SUMOylation. We first determined whether STOP1 can be SUMOylated in Arabidopsis. We coexpressed STOP1-2Flag or GFP-2Flag control with 6Myc-tagged SUMO1 precursor into wild-type Arabidopsis protoplasts and then detected SUMOylated STOP1 using the Myc antibody after Flag immunoprecipitation. We found that STOP1 but not GFP could be SUMOylated, with two major bands (Figure 4A). Nevertheless, when we increased the running time of the protein on the gel, we were able to observe SUMOylated STOP1 of three bands (Supplemental Figure 4A). Since SUMOylation is a reversible modification that normally occurs in less than 5% of the total protein (Creton and Jentsch, 2010), we were unable to detect the SUMOylated forms of STOP1 directly using the anti-Flag antibody. To confirm that STOP1 was covalently modified by mature SUMO, we coexpressed a mature wild-type SUMO1GG or a conjugation-deficient mutant SUMO1AA with STOP1 in protoplasts. STOP1 formed higher order adducts with SUMO1GG but not with SUMO1AA (Figure 4B), confirming that STOP1 is SUMOylated by mature SUMO. To investigate whether STOP1 was modified with a SUMO chain at a single site or mono-SUMOylated at multiple acceptor sites, we generated a mutant SUMO1 (SUMO14KR) in which four Lys residues were changed to Arg (K9, K10, K23, and K42) to abrogate the formation of poly-SUMO chains (Miller et al., 2010). Coexpression of the mutated SUMO14KR with STOP1 did not influence the formation of SUMOylated STOP1 (Figure 4C). These results suggest that STOP1 is subjected to multiple mono-SUMOylation events.
Figure 4.

SUMOylation of STOP1 in the Wild Type and esd4 Mutants.
(A) and (B) Detection of STOP1 SUMOylation with the expression of Myc-tagged SUMO precursor (6Myc-SUMO1; [A]), mature SUMO (6Myc-SUMO1GG), or conjugation-deficient SUMO (6Myc-SUMO1AA; [B]). GFP-2Flag served as a negative control.
(C) STOP1 is mono-SUMOylated. Expression of the SUMO14KR variant, where four different Lys residues are substituted for Arg, which prevents the formation of poly-SUMO chains, does not affect the STOP1 SUMOylation pattern.
(D) SUMOylation levels of STOP1 were increased in the esd4-3 and esd4-4 mutants. For detection of STOP1 SUMOylation in protoplasts, the protoplasts of the wild type (WT) or esd4 mutants were cotransfected with constructs for STOP1-2Flag and different forms of 6Myc-SUMO1. Crude total protein lysates (Input) were used for STOP1 immunoprecipitation (IP) with anti-FLAG magnetic beads, and 6Myc-SUMO1-modified STOP1 was detected using an anti-Myc antibody.
(E) and (F) Al stress reduces the level of STOP1 SUMOylation in Arabidopsis plants. Plants of the wild type (WT) and/or esd4-3 harboring different transgenes were treated with or without 30 μM Al for 8 h. Total root proteins (Input) were immunoprecipitated with anti-HA magnetic beads, and SUMOylated STOP1 was detected using anti-Flag antibody. The total STOP1 proteins were adjusted to a similar level between –Al and +Al treatments for the immunoprecipitation of STOP1-3HA. STOP1 SUMOylation was normalized to the wild-type level under the –Al condition. Data shown in (F) are means ± sd of three biological replicates. Asterisks indicate values that are statistically different (Student’s t test, *, P < 0.05 and **, P < 0.01). Three independent experiments were performed with similar results.
To examine whether the esd4 mutations affected STOP1 SUMOylation, we coexpressed STOP1-2Flag with 6Myc-SUMO1 precursor in wild-type, esd4-3, or esd4-4 protoplasts and then compared the SUMOylation levels of STOP1 between the wild type and the mutants. STOP1 SUMOylation was increased in the two mutants compared with the wild type (Figure 4D). Since ESD4 can be involved in the processing of the SUMO precursor or the deSUMOylation to influence the SUMOylation level of target substrates (Murtas et al., 2003), we cotransformed mature SUMO (6Myc-SUMO1GG) with the STOP1-2Flag into protoplasts to determine whether STOP1 SUMOylation levels are influenced by the esd4 mutations. Like the SUMO precursor, introduction of the mature SUMO1 could also increase the SUMOylation of STOP1 in esd4 protoplasts (Figure 4D), suggesting that ESD4 is involved in the deconjugation of SUMO from STOP1.
To investigate whether STOP1 can be SUMOylated in planta, we transformed SUMO1 promoter-driven 2Flag-SUMO1 into the previously generated pSTOP1:STOP1-3HA transgenic line (Zhang et al., 2019) and selected a transgenic line with single-locus segregation in the 2Flag-SUMO1 transgene for the detection of SUMOylated STOP1 using the anti-Flag antibody after STOP1-3HA immunoprecipitation. We crossed the transgenic line with the esd4-3 mutant to introduce the 2Flag-SUMO1 transgene into the mutant background and also generate the 2Flag-SUMO1 control line without the pSTOP1:STOP1-3HA transgene. Although the level of SUMOylated protein is low and normally occurs in less than 5% of the total protein (Creton and Jentsch, 2010), we were able to detect SUMOylated STOP1 with three observable bands (Figure 4E; Supplemental Figure 4B), demonstrating that STOP1 can be SUMOylated in Arabidopsis plants. Because Al stress promotes STOP1 protein accumulation (Zhang et al., 2019), we normalized the total amount of STOP1 to compare the SUMOylation level of STOP1 under –Al and +Al conditions. Al stress decreased the level of SUMOylated STOP1, although the global levels of protein SUMOylation were not affected by the Al stress (Figures 4E and 4F; Supplemental Figure 4B). In the esd4-3 mutant background, the level of STOP1 SUMOylation was increased compared with that in the wild type (Figures 4E and 4F), consistent with the role of ESD4 in the deSUMOylation of STOP1. The level of SUMOylated STOP1 was also reduced by Al stress in the background of the esd4-3 mutant (Figures 4E and 4F).
Next, we tested whether ESD4 can interact with STOP1 directly. Since the wild-type ESD4 protein is not well expressed in yeast (Saccharomyces cerevisiae) cells (Elrouby and Coupland, 2010), we expressed a catalytically inactive form of ESD4 (C448S) to test the interaction between ESD4 and STOP1 in a yeast two-hybrid assay and found that the inactive variant of ESD4 interacts with STOP1 (Figure 5A). To investigate whether wild-type ESD4 can interact with STOP1 in planta, we performed split-LUC complementation assays (Chen et al., 2008). We used the combinations RAE1ρF-nLUC (F-box domain-deleted RAE1) and cLUC-STOP1, and RAE1-nLUC and cLUC-STOP1, as positive and negative controls, respectively (Zhang et al., 2019). We detected the LUC signal with the combinations ESD4-nLUC and cLUC-STOP1 and with the positive control, but not with other combinations (Figure 5B), and we also detected protein interaction-dependent LUC signal of cLUC-ESD4 and STOP1-nLUC (Figure 5B). These results indicate that ESD4 can interact with STOP1 in planta. To further examine whether ESD4 and STOP1 form protein complexes in Arabidopsis, we conducted coimmunoprecipitation assays in Arabidopsis protoplasts. ESD4-3HA could coimmunoprecipitate with STOP1-2Flag but not with the GFP-2Flag control (Figure 5C). Together, these data indicate that ESD4 can interact with STOP1 in vivo.
Figure 5.
ESD4 Interacts with and deSUMOylates STOP1.
(A) Interaction of STOP1 and ESD4 in a yeast two-hybrid assay. The coding sequences for STOP1 and catalytically inactive (C448S) ESD4 were introduced into pGADT7 and pGBKT7 vectors, respectively. Yeast cells coexpressing STOP1 and ESD4C448S were grown on SD (-Leu/Trp) and SD (-Leu/Trp/His) media.
(B) Interaction of ESD4 with STOP1 as indicated by split-LUC complementation assays. The CaMV 35S promoter-driven construct pairs indicated were coexpressed in N. benthamiana leaves, and then the LUC activity due to LUC reconstitution was measured for the different combinations.
(C) Coimmunoprecipitation of ESD4 with STOP1. ESD4-3HA was coexpressed with STOP1-3Flag or GFP-2Flag in Arabidopsis protoplasts. Crude protein extracts were immunoprecipitated with anti-HA magnetic beads and then detected with anti-Flag antibody.
(D) and (E) ESD4 mediates the deSUMOylation of STOP1. STOP1-2Flag was coexpressed with ESD4-3HA, esd4-3-3HA, esd4-4-3HA, or ESD4C448S-3HA and SUMO precursor (6Myc-SUMO1; [D]) or mature SUMO (6Myc-SUMO1GG; [E]) in Arabidopsis protoplasts. Crude total protein extracts were used to immunoprecipitate STOP1 with anti-FLAG magnetic beads, and the SUMOylated form of STOP1 was detected with anti-Myc antibody.
Two and three independent experiments were performed with similar results for (A), and (B) to (E), respectively.
To investigate whether the interaction of STOP1 with ESD4 is specific, we first used the yeast two-hybrid method to test the interaction of STOP1 with four other ULP SUMO proteases, ELS1, ELS2, OTS1, and OTS2, and observed that STOP1 did not interact with the four SUMO proteases (Supplemental Figure 5A). We then used split-LUC assays to test the interaction of STOP1 with all seven other ULP SUMO proteases. ESD4 but not the other seven ULP SUMO proteases was able to interact with STOP1 (Supplemental Figure 5B). These data indicate that the interaction between STOP1 and ESD4 is specific.
To further test whether ESD4 deSUMOylates STOP1, STOP1-2Flag was coexpressed with wild-type or mutated ESD4-3HA and SUMO precursor (6Myc-SUMO1) or mature SUMO (6Myc-SUMO1GG) in wild-type protoplasts. The results showed that wild-type ESD4-3HA deSUMOylated STOP1-2Flag in both 6Myc-SUMO1- and 6Myc-SUMO1GG-expressing cells (Figures 5D and 5E), whereas mutated versions of ESD4, including esd4-3-3HA, esd4-4-3HA, and catalytically inactive ESD4-3HA (ESD4C448S-3HA), were unable to deSUMOylate STOP1-2Flag. Together, these data indicate that ESD4 interacts with and deSUMOylates STOP1.
Mutation of ESD4 Does Not Affect STOP1 Protein Accumulation but Alters Its Association with the Promoters of Different STOP1-Target Genes
To investigate whether mutation of ESD4 influences STOP1 protein levels, we crossed pSTOP1:STOP1-3HA and pSTOP1:STOP1-GUS transgenic lines, which had been generated previously (Zhang et al., 2019), with esd4-3 to introduce the transgenes into the mutant background. Immunoblot and GUS-staining analyses revealed that the STOP1-3HA/GUS protein levels were not significantly different in the esd4-3 background compared with the wild type under both –Al and +Al conditions (Figures 6A and 6B). To examine whether mutation of ESD4 affects STOP1 stability, we treated plant roots with the protein synthesis inhibitor cycloheximide (CHX) and Al stress for different times. Although CHX treatment was able to inhibit STOP1 protein synthesis in the wild type and esd4-3, the decrease in STOP1 levels did not exhibit a clear difference between the wild type and the mutant (Figure 6C), which suggested that the esd4-3 mutation did not affect STOP1 stability under Al stress conditions. Nevertheless, when the plant roots were first treated with Al to induce STOP1 accumulation and then the plants were transferred to Al-free conditions with the addition of CHX, the STOP1 protein levels decreased more slowly in the mutant than in the wild type (Figure 6D), suggesting that mutation of ESD4 can increase the stability of STOP1 once the Al stress is removed.
Figure 6.
Effects of the esd4-3 Mutation on STOP1 Level and Stability and on the Association of STOP1 with the Promoters of STOP1-Regulated Genes.
(A) Immunoblot analysis of STOP1-3HA in the wild type (WT) and esd4-3 under –Al or +Al conditions. TUBULIN protein was used as an internal control. Values are means ± sd of three biological replicates.
(B) Quantification of STOP1-GUS expression in the wild type (WT) and esd4-3 under –Al or +Al conditions. 4-MU, 4-methylumbelliferone.
(C) and (D) Effects of esd4-3 mutation on STOP1 stability. WT, wild type.
(C) Roots of the wild type and esd4-3 harboring the pSTOP1:STOP1-3HA transgene were treated without Al or with 30 μM Al and 100 μM CHX for different times as indicated. STOP1-3HA protein levels were detected at each time point with anti-HA antibody. ACTIN protein served as an internal control.
(D) Roots were treated with 30 μM Al for 4 h and then transferred to –Al conditions plus 100 μM CHX for different times as indicated, and then the protein levels of STOP1-3HA at each time point were detected.
(E) to (H) ChIP assay was performed to determine the association of STOP1-3HA with the promoters of STOP1-regulated genes, including AtALMT1 (E), AtMATE (F) and RAE1 (G), and with the promoter of ACTIN7 control (H). Values are means ± sd of three technical replicates. Asterisks indicate values that are statistically different (Student’s t test, *, P < 0.05 and **, P < 0.01). N.S. indicates nonsignificantly different P values. Three independent experiments were performed with similar results. WT, wild type.
ESD4 localizes to the nuclear rim and can interact with the nuclear pore component NUA (Murtas et al., 2003; Xu et al., 2007), which suggests that EDS4 might regulate the nuclear accumulation of STOP1. To test this hypothesis, we generated a single-locus 35S:STOP1-GFP transgenic line in the wild-type Arabidopsis background and then introduced the transgene into the esd4-3 mutant background through crossing. We compared the ratio of GFP fluorescence in the nucleus versus the cytosol to determine whether the esd4-3 mutation alters the subcellular localization of STOP1-GFP. The results revealed that mutation of ESD4 did not affect the subcellular localization of STOP1 under either –Al and +Al conditions (Supplemental Figure 6). Interestingly, Al stress promoted STOP1 accumulation in the nucleus in both the wild type and the mutant, as indicated by the increased nuclear:cytoplasmic ratio of the STOP1 protein (Supplemental Figure 6), although the total STOP1 protein levels were also increased by Al stress (Supplemental Figure 6).
Because mutation of ESD4 increases AtALMT1 expression while concomitantly reducing the expression of AtMATE and RAE1, we performed a chromatin immunoprecipitation (ChIP) assay to determine whether the association of STOP1-3HA to the promoters of these genes was altered in the esd4-3 background. The binding regions of STOP1 in the promoters of AtALMT1 and RAE1 were previously identified (Tokizawa et al., 2015; Zhang et al., 2019), but those in the promoter of AtMATE had not yet been determined. Therefore, we performed ChIP assays using the pSTOP1:STOP1-3HA transgenic line to screen for the STOP1 binding region in the AtMATE promoter. STOP1-3HA preferentially associated with fragments 6 and 7 of the AtMATE promoter (Supplemental Figure 7). We then performed ChIP assays in the wild-type and esd4-3 backgrounds under –Al or +Al conditions. The association of STOP1-3HA with the AtALMT1 promoter was more pronounced in the mutant than in the wild type under both –Al and +Al conditions, whereas the opposite was observed for the AtMATE and RAE1 promoters (Figures 6E to 6H). These results demonstrate that increased SUMOylation had different effects on the association of STOP1 with the promoters of different STOP1-target genes.
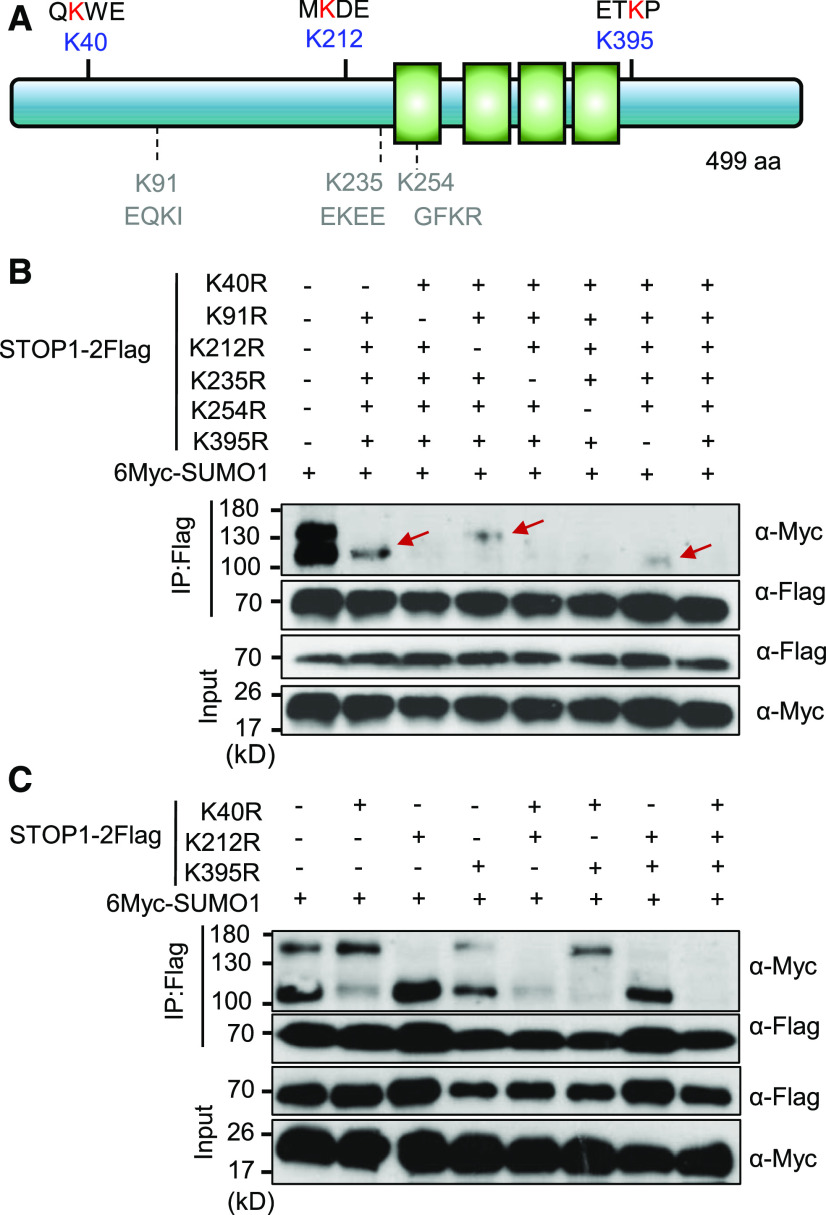
STOP1 Can Be SUMOylated at K40, K212, or K395 Sites
To identify the SUMO acceptor sites in STOP1, we mutated six predicted SUMO sites of STOP1-3HA (Figure 7A) and coexpressed this mutated STOP1-3HA construct with 6Myc-SUMO1 in protoplasts to determine whether mono-SUMOylation still occurred. SUMOylation was fully blocked for the STOP1-3HA mutant, in which these six Lys residues were substituted for Arg (Figure 7B). We then generated six mutant versions of STOP1-3HA in which only five of the six Lys residues were replaced by Arg (leaving each time a different Lys intact) and coexpressed each of them with 6Myc-SUMO1 to examine whether specific Lys residues were required for SUMOylation. We found that three residues (K40, K212, and K395) could be SUMOylated (Figure 7B). To confirm that these three sites are sufficient and required for STOP1 SUMOylation, we constructed a series of mutated versions of STOP1 with single, double, or triple Lys-to-Arg substitutions and then cotransformed each construct with 6Myc-SUMO1 in the protoplasts. Consistently, the combined mutation of all three Lys residues eliminated STOP1 SUMOylation (Figure 7C). Mutating K40 or K395 reduced the STOP1 SUMOylation level at the lower band, while the residue K212 controlled SUMOylation of the upper band of STOP1 (Figures 7B and 7C). These results indicate that STOP1 is mono-SUMOylated at three sites: K40, K212, and K395.
Figure 7.
STOP1 Is SUMOylated at K40, K212, and K395 Sites.
(A) Domain structure of STOP1 with the predicted SUMO acceptor sites. Six predicted SUMO acceptor sites with the cognate motif are shown, including the Lys acceptor. The green boxes indicate the four C2H2 zinc finger domains of STOP1. aa, amino acids.
(B) Effects of the combined replacement of five of the six SUMO acceptor Lys residues in STOP1-2Flag with Arg residues on STOP1 SUMOylation. Red arrows indicate the bands with SUMO-modified STOP1.
(C) Effects of single, double, and triple K-to-R mutations of K40, K212, and K395 on STOP1 SUMOylation. Crude protein extracts of protoplasts coexpressing 6Myc-SUMO1 and mutated STOP1-2Flag were used to immunoprecipitate STOP1 with anti-FLAG antibody, and the SUMOylated forms of STOP1 were detected using anti-Myc antibody. At least three independent experiments were performed with similar results.
Blocking of STOP1 SUMOylation Reduces STOP1 Accumulation and Associated Phenotypes of Al Resistance and Low-Phosphate Response
To determine the biological function of STOP1 SUMOylation in Arabidopsis, we conducted complementation assays on stop1-2 by transforming pSTOP1:STOP1K40R-3HA, pSTOP1:STOP1K212R-3HA, or pSTOP1:STOP1K395R-3HA into their respective mutant backgrounds. We selected single-locus homozygous transgenic lines that showed similar mRNA accumulation levels of STOP1 to a previously generated wild-type STOP1-3HA complementation line (Supplemental Figures 8A to 8C) and then compared the expression of STOP1-regulated genes, STOP1 protein levels, and Al resistance in these lines. Mutation of K40 reduced the expression of AtALMT1 and ALS3 but increased AtMATE expression (Supplemental Figure 8A). Intriguingly, the STOP1 protein level was not significantly affected by this mutation (Figure 8A), although decreased Al resistance was observed in the complementation line pSTOP1:STOP1K40R-3HA (Figures 8D and 8E). The K212R mutation did not affect the expression of STOP1-regulated genes, STOP1 accumulation, and Al resistance (Figures 8B, 8D, and 8E; Supplemental Figure 8B). Unlike the K40R and K212R mutations, the K395R mutation decreased the expression of both AtALMT1 and AtMATE as well as the STOP1 protein level (Figure 8C; Supplemental Figure 8C). Consistent with those results, Al resistance was decreased in the complementation line pSTOP1:STOP1K395R-3HA (Figures 8D and 8E).
Figure 8.
Effects of Mutation of STOP1 SUMOylation Sites on STOP1 Protein Levels and Al Resistance.
Wild-type and mutated versions of STOP1 were transformed into stop1-2 the mutant to generate various complementation lines.
(A) to (C) Immunoblot analysis of STOP1-3HA in STOP1WT (WT), STOP1K40R (K40R; [A]), STOP1K212R (K212R; [B]), and STOP1K395R (K395R; [C]) complementation lines exposed to 0 (–Al) or 30 μM Al (+Al) for 8 h. WT, wild type.
(D) and (E) Al-resistance phenotypes of the wild type (WT) and K40R, K212R, and K395R complementation lines. Seedlings were grown on a soaked gel medium containing 0, 0.75, or 1.0 mM Al for 7 d. Values are means ± sd of root lengths of 21 to 35 seedlings. Means with different letters are significantly different (P < 0.05, ANOVA test followed by Tukey’s test). Three independent experiments were performed with similar results. Bar = 1 cm.
(F) Immunoblot analysis of STOP1-3HA in one STOP1WT (WT) and two STOP1 K40R, K212R, K395R (3KR-1 and 3KR-2) complementation lines under –Al and +Al conditions.
(G) STOP1 stability was increased in the 3KR line. Roots of the wild type (WT) and 3KR lines were treated with 30 μM Al for 4 h and then transferred to –Al conditions with 100 μM CHX for different times as indicated. STOP1-3HA protein levels were detected at each time point with anti-HA antibody.
(H) and (I) Al-resistance phenotypes of the wild type (WT) and 3KR complementation lines. Values are means ± sd of relative root lengths of 32 to 43 seedlings. Means with different letters are significantly different (P < 0.05, ANOVA test followed by Tukey’s test). Three independent experiments were performed with similar results. Bar = 1 cm.
We also mutated all three sites of STOP1 to conduct a complementation test of stop1. We selected two complementation lines with the triple mutation 3KR (K40R, K212R, and K395R), which showed similar mRNA levels of STOP1 to the wild-type complementation line (Supplemental Figure 8D). The results revealed that mutating all three SUMO acceptor sites reduced the expression of STOP1-target genes, AtALMT1, AtMATE, and ALS3, in the presence of Al (Supplemental Figure 8D). Consistent with the reduced expression of its target genes, STOP1 protein levels were decreased in both 3KR complementation lines (Figure 8F). To examine the stability of STOP13KR in vivo, we first exposed the plants with Al stress to increased STOP1 levels and then transferred the plants to –Al conditions with the addition of CHX to inhibit protein synthesis. STOP13KR levels decreased more quickly than the STOP1WT protein levels (Figure 8G), which suggests that SUMOylation of STOP1 stabilizes the STOP1 protein. We then evaluated Al resistance in these lines, and the results showed that the Al resistance was reduced in the STOP13KR complementation lines compared with the STOP1WT line (Figures 8H and 8I). Together, these results indicate that a block of STOP1 SUMOylation destabilizes STOP1 and reduces Al resistance.
Since STOP1 is required for both Al resistance and proton tolerance (Iuchi et al., 2007), we also compared the proton tolerance in these complementation lines. The results showed that K40R, K212R, or K395R single mutations did not affect the tolerance to proton toxicity (Supplemental Figures 9A and 9B). Nevertheless, the triple mutation 3KR reduced the proton tolerance (Supplemental Figures 9C and 9D). Recently, AtALMT1-mediated malate secretion was reported to promote apoplastic Fe toxicity and consequently inhibit root growth under low-Pi conditions, and STOP1 is involved in the low-Pi-induced root growth inhibition through the modulation of AtALMT1 expression (Balzergue et al., 2017; Mora-Macías et al., 2017). In accordance with the reduced AtALMT1 expression, the K40R and K395R complementation lines were less sensitive to low-Pi-induced inhibition of root growth compared with the wild-type STOP1 complementation lines (Supplemental Figures 10A and 10B), while the K212 mutation did not affect the low-Pi response. Similarly, the 3KR complementation lines also showed reduced sensitivity to the low-Pi-induced inhibition of root growth (Supplemental Figures 10C and 10D). These results demonstrate that a block of STOP1 SUMOylation also decreases proton tolerance and the low-Pi response.
DISCUSSION
The transcription factor STOP1 is crucial for Al resistance in Arabidopsis (Iuchi et al., 2007). Previous work has demonstrated that STOP1 is regulated by Al stress at posttranslational levels and that the F-box protein RAE1 is involved in the regulation of STOP1 ubiquitination and degradation (Zhang et al., 2019). In this study, we unravel another layer of posttranslational regulation of STOP1 by SUMOylation. We show that Al resistance can be modulated via a SUMO conjugation switch on the STOP1 transcription factor (Figure 9). STOP1 is mono-SUMOylated at the sites K40, K212, and K395 and deSUMOylated by the SUMO protease ESD4. This dynamic SUMOylation of STOP1 is involved in the control of the expression of downstream genes and Al resistance. Prevention of the ESD4-mediated deSUMOylation of STOP1 elevates STOP1 SUMOylation levels, which leads to altered association to the promoters of downstream genes, increasing binding to the promoter of AtALMT1 and decreasing binding to the promoters of AtMATE. These differences in physical association to the promoters in turn lead to the increased expression of AtALMT1 and the reduced expression of AtMATE, which contributes to the enhanced Al resistance. Blocking of STOP1 SUMOylation at K395 or all three sites (K40, K212, and K395), on the other hand, destabilizes STOP1 and causes reduced expression of STOP1-regulated genes and decreased Al resistance (Figure 9). Interestingly, contrary to the effect of increased STOP1 SUMOylation on the expression of STOP1-regulated genes, blocking of STOP1 SUMOylation at K40 decreases the expression of AtALMT1 and ALS3 but increases AtMATE expression. SUMO modification has been suggested to affect the assembly of transcription factors and the recruitment of chromatin-modifying proteins (Hay, 2005; Wilkinson and Henley, 2010), which can cause transcriptional activation as well as transcriptional repression. For instance, SUMOylation of heat shock transcription factors HSF1 and HSF2 increases DNA binding activity (Goodson et al., 2001; Hong et al., 2001), while SUMO modification on Elk-1 transcription factor can cause the recruitment of histone deacetylases to repress transcription (Yang and Sharrocks, 2004). Likewise, SUMOylation of some transcription factors such as Smad4 can activate transcription on some promoters and repress transcription on other promoters (Long et al., 2004), which is dependent on a complex interplay between promoter context and the intrinsic capability of SUMO to regulate transcription (Hay, 2005). Therefore, the different effects of SUMOylated and unSUMOylated STOP1 on the expression of AtALMT1 and AtMATE might also be attributed to different assemblies of STOP1 and its associated proteins and/or the recruitment of different chromatin-modifying proteins on the promoters of AtALMT1 and AtMATE, although the exact underlying mechanism remains to be demonstrated in the future.
Figure 9.
Model for the Regulation of STOP1 Function and Stability by SUMOylation.
STOP1 is mono-SUMOylated at three sites, K40, K212, and K395, presumably through the SUMOylation pathway. The STOP1 SUMOylation is reversible, and it can be deSUMOylated by the SUMO protease ESD4, which is increased in response to Al stress. Mutation of ESD4 elevates STOP1 SUMOylation levels and consequently increases and decreases the association of STOP1 with the promoters of AtALMT1 and AtMATE, respectively, which causes the increased AtALMT1 expression and the reduced expression of AtMATE and RAE1, and finally contributes to the increased Al resistance. Mutation of STOP1 SUMOylation at K40 increases AtALMT1 and decreases AtMATE expression, which leads to reduced Al resistance. Mutation at K212 does not affect STOP1 function and Al resistance. Blocking of STOP1 SUMOylation at K395 or all three (K40, K212, and K395) sites reduces STOP1 protein level and the expression of AtALMT1 and AtMATE, leading to the decreased Al resistance.
Mutation of ESD4 causes reduced plant growth and increased accumulation of the stress hormone salicylic acid (Reeves et al., 2002; Villajuana-Bonequi et al., 2014). Here, we found that dysfunction of ESD4 could enhance Al resistance (Figure 2). These observations suggest that ESD4 might be involved in balancing plant growth and stress tolerance. Intriguingly, Al stress increases instead of decreases ESD4 protein accumulation (Figure 3), although ESD4 plays a negative role in Al resistance. The increased ESD4 accumulation under Al stress conditions did not affect global protein SUMOylation (Figure 4E), suggesting that, under Al stress conditions, the elevated ESD4 protein might be inefficient in the deSUMOylation of ESD4-target proteins. Thus, we speculate that the increased ESD4 accumulation might help plants to rapidly deSUMOylate target proteins to attenuate Al resistance responses and ensure growth robustness once the Al stress is removed.
This work establishes STOP1 as a bona fide ESD4 target substrate. We demonstrate that ESD4 directly interacts with and deSUMOylates STOP1. Mutation of RAE5/ESD4 increases STOP1 SUMOylation (Figure 4), which causes the exudation of more malate but less citrate in the mutants than in the wild type (Figure 2; Supplemental Figure 2A). Since AtALMT1-mediated malate secretion plays a predominant role in the detoxification of Al compared with AtMATE-mediated citrate exudation (Hoekenga et al., 2003, 2006; Liu et al., 2009), the increased malate secretion might contribute significantly to the enhanced Al resistance in the esd4 mutants, while the negative effect of reduced citrate secretion on the Al resistance is negligible. As mutation of ESD4 also causes pleiotropic development phenotypes under control conditions (Figure 2C; Supplemental Figure 2D), we cannot exclude the possibility that the esd4 mutations affect the SUMOylation and function of other unidentified proteins, which in turn affects plant development and also contributes to the increased Al resistance in the mutants.
Since two major bands were observed for SUMOylated STOP1, STOP1 might be modified with a poly-SUMO chain at a single acceptor site or mono-SUMOylated at multiple sites. Given that mutation of four Lys residues in SUMO1 (SUMO14KR), which are required for the formation of poly-SUMO chains (Miller et al., 2010), does not affect the two major bands of SUMOylated STOP1 (Figure 4C), STOP1 is likely to be mono-SUMOylated at multiple sites. Further site-directed mutagenesis revealed that SUMOylation at the K212 site of STOP1 is responsible for the upper band, while SUMOylation at the K40 or K395 site is required for the lower band (Figures 7B and 7C). Although the topology of SUMOylated STOP1 is still unknown, the apparent larger size of SUMOylated STOP1 than predicted could be attributed to the anomalous running of branched SUMO-modified proteins in SDS-PAGE, which has been documented before (Chupreta et al., 2007; Park-Sarge and Sarge, 2009). The SUMOylation at the K212 site, which is in the middle of STOP1, may be particularly inclined to such anomalous behavior, since the SUMOylated STOP1 at this site migrates substantially slower than would correspond to its predicted size (Figures 7B and 7C). Despite the unknown reason underpinning the extremely slow migration in SDS-PAGE, the SUMOylated STOP1 at K212 does not influence STOP1 function and Al resistance (Figures 8B and 8E).
Previous work has demonstrated that Al and Fe can trigger STOP1 accumulation in the nucleus under Pi-deficient conditions (Balzergue et al., 2017; Godon et al., 2019). Nevertheless, it is not clear whether the increased nuclear STOP1 accumulation is solely caused by the increase in total STOP1 levels. We found that Al stress increases the nuclear:cytoplasmic ratio of STOP1 as well as the total STOP1 levels (Supplemental Figure 6), which suggests that Al stress might promote STOP1 movement from the cytoplasm to the nucleus or preferentially inhibit the STOP1 degradation in the nucleus. Intriguingly, although elimination of SUMOylation destabilizes STOP1, Al stress reduces rather than increases this posttranslational modification in STOP1 (Figures 4E and 4F; Supplemental Figure 4B). While Al-induced ESD4 accumulation might contribute to the decreased STOP1 SUMOylation (Figure 2), Al stress likely also influences other components to reduce SUMO modification on STOP1, since the SUMOylation levels of STOP1 were also decreased in the esd4-3 mutant under Al stress conditions (Figures 4E and 4F). In addition, although loss of STOP1 SUMOylation reduces STOP1 protein levels (Figure 8), the increased SUMOylation in esd4 did not significantly influence STOP1 accumulation and stability under Al stress conditions (Figures 6A to 6C). Nevertheless, when Al-stressed roots were transferred to Al-free solution, STOP1 protein stability was higher in esd4 than in the wild type (Figure 6D). Together, these results suggest that Al might affect other posttranslational modifications of STOP1, which can interfere with and override the STOP1 SUMOylation to trigger the increased nuclear accumulation of this transcription factor at a similar level in the wild type and esd4, and that once Al stress is removed, the SUMO modification on STOP1 can exert an effect on stabilizing STOP1 protein. Our results also imply that the Al-induced reduction of STOP1 SUMOylation might enable plants to accelerate the degradation of overaccumulated STOP1 to alleviate Al-resistance responses when the Al stress is absent. Uncovering additional layers of posttranslational regulation and dissecting the interactions among these potential regulatory mechanisms will be a stepping stone to engineer Al resistance in crops in the future.
METHODS
Plant Materials and Growth Conditions
Wild-type Arabidopsis (Arabidopsis thaliana) plants (Columbia ecotype genetic background; Col-0) carrying a homozygous AtALMT1 promoter-driven LUC transgene (pAtALMT1:LUC; Zhang et al., 2019) were treated with EMS to generate a mutation M2 population. The rae5-1 (esd4-3), rae5-2 (esd4-4), and stop1-3 mutants with increased LUC signal were previously identified from this mutagenized seed population by Zhang et al. (2019). The T-DNA insertion line Atalmt1 (SALK_00962) and stop1-2 (SALK_114108) was obtained from the Arabidopsis Biological Resource Center.
Seeds were grown on a half-strength Murashige and Skoog (MS) medium with 1.2% (w/v) agar and 1% (w/v) Suc for 7 d, and then the seedlings were observed for luminescence signal by a CCD imaging apparatus (Lumazone P1300B, Roper Scientific). Plants were grown on one-tenth-strength Hoagland hydroponic culture or soil culture conditions in a growth chamber (CU-36L4, Perivical) or growth room at 22°C with 14 h of light (100 μmol m−2 s−1; Philips TLD26W865 cool daylight tubes) and 10 h of darkness.
Measurement of Malate and Citrate Secretion in Roots
Ten-day-old seedlings of the wild type, rae5-1, and rae5-2 grown on 1.2% agar plates with half-strength MS nutrient medium and 1% (w/v) Suc were pretreated with a 2% (w/w) MGRL solution (Fujiwara et al., 1992) containing 1% (w/v) Suc for 2 h at pH 4.8 and then treated with the same solution containing 0 or 10 μM AlCl3 at pH 4.8 for 12 h on a 12-well plate. The root exudates from 15 seedlings in each of four biological replicates in the treated solution were collected and then concentrated by lyophilizing (CHRIST ALPHA 1-2 LDplus). The NAD/NADH enzymatic cycling method (Hampp et al., 1984) was used to determine the malate and citrate concentrations in the root exudates.
Evaluation of Al Resistance, Low-Pi Response, and Low pH Tolerance
A modified soaked gel medium method was adopted to assess Al resistance according to Larsen et al. (2005). A gel medium consisting of 50 mL (pH 5.0) of 0.25 mM (NH4)2SO4, 1 mM KNO3, 0.2 mM KH2PO4, 2 mM MgSO4, 1 mM Ca(NO3)2, 1 mM CaSO4, 1 mM K2SO4, 1 µM MnSO4, 5 µM H3BO3, 0.05 µM CuSO4, 0.2 µM ZnSO4, 0.02 µM NaMoO4, 0.1 µM CaCl2, 0.001 µM CoCl2, 1% (w/v) Suc, and 0.4% (w/v) Gellan gum (G1910, Sigma-Aldrich) was prepared. The solidified gel medium was then soaked with 25 mL of the same nutrient solution without Gellan gum containing 0, 0.5, 0.75, 1.0, or 1.25 mM AlCl3 at pH 3.6. After 2 d of soaking, the solution was discarded, and the soaked gel medium was dried and used for seedling growth. After 7 d, the seedlings grown on the soaked gel medium were imaged and root lengths were measured by ImageJ software. Relative root length was used to assess Al resistance, which was determined as a percentage (root length with Al treatment/root length without Al × 100). For Eriochrome Cyanine R staining of Al accumulation, 7-d-old seedlings of the wild type, rae5-1, rae5-2, and Atalmt1 grown on the soaked gel medium containing 1.0 mM Al were stained with 0.1% (w/v) Eriochrome Cyanine R for 30 s. Roots were then washed with distilled water for 1 min three times and photographed using a stereomicroscope.
For the evaluation of low-Pi response, seeds were grown on 0.35% (w/v) Gellan gum medium (pH 5.7) containing 1% (w/v) Suc and 1× Hoagland nutrient solution with 40 µM Fe(III)-EDTA, 0.5 mM (NH4)2SO4, and 0, 15 μM, or 1 mM NH4H2PO4 (Pi). After growth for 7 d, the seedlings were subjected to photography and root length measurement. Relative root length normalized to the 1 mM Pi condition was used to evaluate the response to low-Pi-induced root growth inhibition.
To evaluate the tolerance to low pH, a nutrient medium consisting of full-strength Hoagland nutrient solution, 1% (w/v) Suc, and 0.4% (w/v) Gellan gum was prepared with pH at 5.6 or 4.2 after autoclaving. Seedlings were grown on the nutrient medium plate for 7 d, and then root length was measured. Relative root length normalized to pH 5.6 was used for the evaluation of low-pH tolerance.
Evaluation of the Tolerance to Cd and La Stresses
To compare the Cd and La tolerance in the wild type, rae5-1, and rae5-2, seeds were grown on a plastic mesh floating on one-thirtieth-strength Hoagland nutrient solution with 1 mM CaCl2, 0 or 2 µM CdCl2, or 0.6 µM LaCl3 at pH 5.0 in a growth chamber at 22°C. After growth for 7 d, root lengths of 25 to 33 seedlings in each treatment were measured by ImageJ, and relative root growth as described above was used to assess their tolerance to Cd or La.
Cloning of RAE5
The rae5-2 mutant was crossed with the wild-type background to construct an F2 population for genetic analysis and mapping-by-sequencing. A total of 356 F2 plants were used, and 75 plants showing both increased LUC expression and morphological defects were pooled for high-throughput DNA sequencing using the Illumina HiSeq4000 system that produces 150-bp paired-end reads, which was performed by a commercial company (Shanghai Hanyu Biotech; accession number in the National Center for Biotechnology Information: SRR9290163). The sequencing data had a depth of around 41-fold coverage of the Arabidopsis genome. Clean reads were mapped to the Arabidopsis Col-0 reference genome (The Arabidopsis Information Resource) by using bwa software (version 0.7.10) with default parameters. The aligned reads were processed by GATK (version 3.5; Broad Institute) for calling single-nucleotide polymorphisms. The MutMap method was adopted to search the candidate region of rae5-2 (Abe et al., 2012). To confirm the candidate region of RAE5, four dCAPS polymorphic markers were designed based on single-nucleotide polymorphisms in the RAE5 candidate region of chromosome 4 (Supplemental Table 1). Fifty-eight F2 plants with increased LUC signal and morphological defects were subjected to linkage analysis by using the four dCAPS markers. The dCAPS maker D3 based on the C-to-T substitution in At4g15880 (ESD4) was completely linked to the F2 mutant phenotype (Supplemental Table 1), suggesting that RAE5 encodes the SUMO protease ESD4.
For the complementation test of rae5-1, a DNA fragment consisting of a 1.7-kb promoter and the gene of RAE5/ESD4 (At4g15880) without stop codon was amplified by a primer pair (Supplemental Table 2) and cloned in frame (HindIII and DraIII restriction sites) with a 3Flag tag into the pCAMBIA3301 vector by using the ClonExpress II One Step Cloning Kit (C112-02, Vazyme Biotech). The construct was then introduced into the rae5-1 mutant by Agrobacterium tumefaciens (strain GV3101)-mediated transformation, and T3 plants harboring the homozygous transgene were subjected to expression analysis and phenotypic analysis for the complementation test.
RNA Isolation and RT-qPCR Analysis
To conduct expression analysis of Al-resistance genes, seeds were sown on agar plates containing half-strength MS medium and 1% (w/v) Suc and allowed to grow for 10 d. The seedlings were then exposed to a 0.5 mM CaCl2 solution with 0 or 30 µM AlCl3 at pH 4.7. After 8 h of treatment, roots and/or shoots of around 24 seedlings were excised and pooled for RNA extraction in each of three sets of seedlings. Total RNA of each root sample was extracted using the TaKaRa M iniBEST plant RNA Extraction Kit (catalog number 9769) and then digested with DNase I to remove contaminating DNA. Approximately 1 µg of total RNA was used for first-strand cDNA synthesis by the HiScript 1st Strand cDNA Synthesis Kit (Vazyme Biotech). One-twentieth of the cDNA products were used for real-time PCR analysis by the SYBR Green Master Mix kit (Vazyme Biotech) and gene-specific primers (Supplemental Table 2). UBQ10 was used as an internal control for sample normalization (reference gene) in the real-time RT-PCR analysis (2ΔΔCT method). Real-time RT-PCR data were recorded and analyzed using the CFX96 Touch real-time PCR detection system (Bio-Rad).
Yeast Two-Hybrid Assay and Split-LUC Complementation Analysis
For yeast two-hybrid experiments, the coding sequence (CDS) of STOP1 was introduced into pGADT7 vector, while the CDSs of catalytically inactive SUMO proteases ESD4C448S, ELS1C461S, ELS2C300S, OTS2C512S, and OTS1C525S were cloned into pGBKT7 vector. STOP1 and each SUMO protease were coexpressed into yeast strain AH109, and then the yeast cells were grown on SD (-Leu/Trp) and SD (-Leu/Trp/His) media to determine the protein-protein interaction.
For split-LUC complementation analysis, the CDS of STOP1 was cloned into pCAMBIA-nLUC or pCAMBIA-cLUC vectors, and the CDSs of ESD4, ELS1, ELS2, OTS2, OTS1, FUG1, SPF1, and SPF2 were also introduced into pCAMBIA-nLUC or pCAMBIA-cLUC (Chen et al., 2008). STOP1-nLUC and cLUC-STOP1 were respectively cotransformed with cLUC- and nLUC-fused ESD4, ELS1, ELS2, OTS1, OTS2, FUG1, SPF1, or SPF2 into fully expanded Nicotiana benthamiana leaves by A. tumefaciens-mediated transformation, respectively. The transformed plants were grown in the dark for 1 d and in the long-day conditions for 2 d and then subjected for LUC signal detection to determine whether the two proteins interacted with each other. The primers used for vector construction are listed in Supplemental Table 2.
Observation of STOP1-GFP Subcellular Localization in Wild-Type and esd4-3 Roots
The coding sequence of STOP1 was amplified and fused in-frame with the GFP gene and then inserted into pCAMBIA1301 vector (SpeI and PstI restriction sites) harboring 35S promoter and hygromycin resistance marker using the ClonExpress II One Step Cloning Kit (C112-02, Vazyme Biotech). The resultant vector was transformed into Arabidopsis wild-type plants. A T2 line showing single-locus segregation on hygromycin resistance was screened out and crossed with esd4-3 to introduce the 35S:STOP1-GFP transgene into the mutant background. To compare the subcellular localization of STOP1-GFP in the wild type and esd4-3 under different Al conditions, 35S:STOP1-GFP seedlings of the wild type and esd4-3 were grown on half-strength MS medium plates for 5 d and then transferred to the soaked gel medium described above with or without 1 mM AlCl3 overnight. Roots were first stained with 10 μM propidium iodide solution for 10 s and then subjected to GFP and propidium iodide fluorescence observation by using a Leica confocal microscope (Leica SP8). ImageJ software was used to quantify GFP fluorescence intensity, which was integrated from all pixels in selected areas. To measure the ratio between the nuclear and cytoplasmic signals for each cell, the entire cellular and nuclear area was selected to quantify fluorescence intensity. The cytoplasmic intensity was calculated by subtracting the value for the nuclear area from the whole cell. Thirteen root tip cells of each of five roots were used for the calculation of the ratio of the GFP signal in nucleus to cytosol.
Transient Expression in Protoplasts
Arabidopsis protoplasts were isolated from 14-d-old seedlings according to the method described by Zhai et al. (2009). To determine the SUMOylation status of STOP1, 2-mL protoplasts (∼2 × 106 cells/mL) of the wild type or esd4 mutants was cotransfected with 50 μg of 35S:6Myc-SUMO1 (CDS of the SUMO1 precursor, mature [GG], or mutated SUMO1 [4KR, AA] constructed into pHBT vector) and with 50 μg of 35S:STOP1-2Flag (wild-type or mutated STOP1) and with or without wild-type or mutated ESD4-3HA under the control of the 35S promoter control. The total protein fraction was extracted by macerating frozen tissue in protein extraction buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM MgCl2, 0.25% [v/v] Nonidet P-40, 0.25% [v/v] Triton X-100, 0.05% [w/v] SDS, 1 mM DTT, 50 μM MG132 [A2585, ApexBio], 20 mM N-ethylmaleimide, 50 μM PR-619, and 1× complete protease inhibitor mixture [4693132001, Sigma-Aldrich]) and were immunoprecipitated with 20 μL of anti-FLAG M2 magnetic beads (M8823, Sigma-Aldrich). Next, the beads were washed with 1 mL of protein extraction buffer three times, and the immunoprecipitated proteins were eluted with 1× SDS loading buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% [v/v] glycerol, 0.1% [w/v] bromophenol blue, and 1% [v/v] β-mercaptoethanol) for immunoblot analysis. The SUMOylated form of STOP1 was detected with anti-Myc-HRP antibody (A00704-100, Genscript Biotech).
For the prediction of SUMOylation sites in STOP1, Joined Advanced SUMOylation site and SIM Analyzer and GPS-SUMO online services were both used, and the predicted sites were combined. As a result, six potential SUMOylation sites of STOP1 were predicted. We then generated mutated versions of STOP1 with different numbers of mutation sites on the six potential SUMOylation sites by site-directed mutagenesis using wild-type 35S:STOP1-2Flag as a template and oligonucleotide primers listed in Supplemental Table 2. The resultant different versions of 35S:STOP1-2Flag vectors were individually coexpressed with 35S:6Myc-SUMO1 in the protoplasts and subjected to the detection of STOP1 SUMOylation sites as described above.
For coimmunoprecipitation assays, 2 mL of protoplasts (∼2 × 106 cells/mL) were cotransfected with 50 μg of 35S:ESD4-2HA and with 50 μg of 35S:STOP1-2Flag or 35S:GFP-2FLAG. The total protein fraction was extracted using the protein extraction buffer (100 μL), and 20 μL of the protein extract solution was used as the input control. The remaining cell extracts were diluted to 1 mL and incubated with 20 μL of anti-FLAG M2 magnetic beads for 3 h at 4°C with gentle rotation. The extracts were then washed three times with 1 mL of protein extraction buffer, and the bound proteins were eluted with 1× SDS loading buffer for immunoblot analysis using anti-Flag-HRP (A8592, Sigma-Aldrich) or HA-HRP (12013819001, lot 44323100, Roche) antibodies. The primers used for the vector construction are listed in Supplemental Table 2.
Determination of STOP1 SUMOylation and Protein Levels in Roots
To detect STOP1 SUMOylation in planta, 10-d-old transgenic seedlings harboring pSUMO1:2Flag-SUMO1 and/or pSTOP1:STOP1-3HA transgenes were exposed to a 0.5 mM CaCl2 solution containing 0 or 30 μM AlCl3 at pH 4.7 for 8 h. The roots (∼600 mg) were excised and frozen for protein extraction using the extraction buffer composed of 50 mM HEPES (pH 7.5), 100 mM NaCl, 5 mM EDTA, 10% (v/v) glycerol, 0.25% (v/v) Nonidet P-40, 0.5% (v/v) Triton X-100, 0.05% (w/v) SDS, 50 μM MG132, 20 mM N-ethylmaleimide, 50 μM PR-619, and 1× complete protease inhibitor mixture. The extracted proteins were immunoprecipitated with anti-HA magnetic beads (B26202, Biomake), and the immunoprecipitates were then washed with 1 mL of protein extraction buffer five times and subsequently eluted with 1× SDS loading buffer for immunoblot analysis. The SUMOylated form of STOP1 was detected with anti-Flag-HRP antibody. The ACTIN protein detected by anti-ACTIN antibody (CW0264M, CoWin Biosciences) was used as the loading control.
For the determination of ESD4 and STOP1 protein levels in roots, 10-d-old seedlings were exposed to a 0.5 mM CaCl2 solution containing 0 or 30 μM AlCl3 at pH 4.7 for 0, 2, 4, or 8 h. To examine the effect of esd4-3 mutation or STOP13KR on the STOP1 stability, seedlings were treated with 100 μM CHX (A8244, ApexBio) and with 0 or 30 μM AlCl3 for different times. Total proteins were extracted from the roots (∼100 mg) using the extraction buffer (150 μL per 100 mg of tissue) composed of 20 mM Tris-HCl (pH 7.4), 300 mM NaCl, 5 mM MgCl2 (pH 8.0), 5 mM DTT, 0.5% (v/v) Nonidet P-40, 50 μM MG132, and 1× Complete Protease inhibitor tablets EDTA-free (5892791001, Roche). The total proteins were separated by 8% (w/v) SDS-PAGE, and the resolved ESD4-3Flag and STOP1-3HA proteins were analyzed by standard immunoblot using anti-Flag-HRP (A8592, Sigma-Aldrich) and HA-HRP (12013819001, lot 44323100, Roche) antibodies, respectively. Protein bands on the membranes were captured by the ChemiDoc Touch Imaging System (1708370, Bio-Rad). ACTIN or TUBULIN proteins were used as the loading control, which were detected by anti-ACTIN or anti-TUBULIN (M0267-1-HRP, Abiocode) antibodies.
To examine the effect of mutation of STOP1 SUMOylation sites on STOP1 accumulation, constructs of pSTOP1:STOP1K40R-3HA, pSTOP1:STOP1K212R-3HA, pSTOP1:STOP1K395R-3HA, and pSTOP1:STOP1K40R, K212R, K395R-3HA were made by site-directed mutagenesis using wild-type pSTOP1:STOP1-3HA as a template and oligonucleotide primers listed in Supplemental Table 2, and the constructs were then transformed in the stop1-2 mutant background. Single-locus homozygous transgenic lines showing similar expression levels of STOP1 were screened. Ten-day-old seedlings of these transgenic lines were exposed to a 0.5 mM CaCl2 solution containing 0 or 30 μM AlCl3 at pH 4.7 for 8 h, and then STOP1 protein levels in roots were determined by using anti-HA-HRP antibody.
For quantification of STOP1-GUS levels, 7-d-old seedlings of the wild type and esd4-3 harboring the pSTOP1:STOP1-GUS transgene were treated with 0.5 mM CaCl2 solution containing 0 or 30 μM AlCl3 at pH 4.7. After 8 h of treatment, the roots were collected, ground to a fine powder, and then mixed with GUS extraction buffer (50 mM sodium phosphate at pH 7.0, 10 mM EDTA, 0.1% [w/v] SDS, 0.1% [v/v] Triton X-100, 10 mM 2-mercaptoethanol, and 25 μg/mL PMSF). The mixed solution was centrifuged at 15,000 rpm for 10 min at 4°C, and subsequently, a portion of the supernatant was used for the reaction with the GUS extraction buffer containing 1 mM 4-methylumbelliferyl-β-d-glucuronide (A602251, Sangon Biotech) at 37°C. After 1 h, Na2CO3 solution (0.2 M) was added to stop the reaction. The fluorochrome 4-methylumbelliferone reaction product was detected for its fluorescence, with excitation and emission wavelengths of 350 and 455 nm, respectively. GUS activity in each sample was normalized to the total protein.
ChIP Assay
Eight-day-old plants of the wild type and esd4-3 harboring pSTOP1:STOP1-3HA were exposed to a 0.5 mM CaCl2 solution with 0 or 30 µM AlCl3 at pH 4.7 for 8 h. The roots (∼1.2 g) were excised for the ChIP analysis. The ChIP method was according to Saleh et al. (2008) with slight modifications for the sonication step, in which isolated nuclei were sonicated for 55 cycles by a Bioruptor (Diagenode) with 15 s on and 15 s off on high-power set. Protein immunoprecipitation was performed by using the anti-HA antibody (H3663, Sigma-Aldrich) or IgG control (B3001S, Abmart) followed by adding 100 μL of salmon DNA preequilibrium beads (SM00405, Smart Life Science). The protein-bound DNA was quantified by real-time PCR using gene-specific primers (Supplemental Table 2).
Statistical Analysis
Two-tailed Student’s t test was used to do statistical analysis on two lines, while for more than two lines, one-way ANOVA followed by Tukey’s test was used for statistical analysis. The results of statistical analyses and the list of individual data for the statistical analyses are shown in Supplemental Data Sets 1 and 2, respectively.
Accession Numbers
Gene sequence data for this article can be found in The Arabidopsis Information Resource (TAIR database under the following accession numbers: At1g34370 for STOP1 and At4g15880 for ESD4. Whole-genome sequence data of pooled F2 rae5-2 mutant plants can be found in the National Center for Biotechnology Information under accession number SRR9290163.
Supplemental Data
Supplemental Figure 1. Effect of rae5 mutations on the expression of STOP1-regulated genes in shoots (supports Figure 1).
Supplemental Figure 2. Effect of rae5 mutations on citrate exudation, Cd and La tolerance, and plant morphology (supports Figure 2).
Supplemental Figure 3. MutMap analysis and complementation test of rae5 mutants (supports Figures 1 and 2).
Supplemental Figure 4. STOP1 is SUMOylated with three bands and Al stress reduces STOP1 SUMOylation (supports Figure 4).
Supplemental Figure 5. Interaction analysis of STOP1 with SUMO proteases (supports Figure 5).
Supplemental Figure 6. Effect of the esd4-3 mutation and Al treatment, alone and in combination, on the subcellular localization of STOP1 (supports Figure 6).
Supplemental Figure 7. ChIP analysis of the association of STOP1 with different regions of AtMATE promoter (supports Figure 6E).
Supplemental Figure 8. Effect of mutation of STOP1 SUMOylation sites on the expression of STOP1-regulated genes (supports Figure 8).
Supplemental Figure 9. Effect of mutation of STOP1 SUMOylation sites on low-pH tolerance (supports Figure 8).
Supplemental Figure 10. Effect of mutation of STOP1 SUMOylation sites on low-phosphate (Pi) response (supports Figure 8).
Supplemental Table 1. List of mutations in rae5-2 and linkage analysis.
Supplemental Table 2. Primers used in this study.
Supplemental Data Set 1. Statistics summary of quantitative data in all the figures.
Supplemental Data Set 2. List of individual data for the statistical analyses.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
AUTHOR CONTRIBUTIONS
Q.F., J.Z., and C.-F.H. designed the experiments; Q.F., J.Z., N.F., and Y.Z. performed the experiments; H.A.B. commented on the research and the article; C.-F.H. wrote the article.
Acknowledgments
We thank Rosa Lozano-Duran for commenting on the article and Yan Guo for providing us the 6Myc-SUMO1 plasmid. This work was supported by Shanghai Natural Science Foundation (grant 20ZR1466500), the National Natural Science Foundation of China (grants 31570253 and 31870223 to C.-F.H.), the Shanghai Center for Plant Stress Biology, the Chinese Academy of Sciences, and the National Key Laboratory of Plant Molecular Genetics.
References
- Abe A., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30: 174–178. [DOI] [PubMed] [Google Scholar]
- Augustine R.C., Vierstra R.D.(2018). SUMOylation: Re-wiring the plant nucleus during stress and development. Curr. Opin. Plant Biol. 45: 143–154. [DOI] [PubMed] [Google Scholar]
- Balzergue C., et al. (2017). Low phosphate activates STOP1-ALMT1 to rapidly inhibit root cell elongation. Nat. Commun. 8: 15300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro P.H., Bachmair A., Bejarano E.R., Coupland G., Lois L.M., Sadanandom A., van den Burg H.A., Vierstra R.D., Azevedo H.(2018). Revised nomenclature and functional overview of the ULP gene family of plant deSUMOylating proteases. J. Exp. Bot. 69: 4505–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala R., Ouyang J., Abreu I.A., Hu Y., Seo H., Zhang X., Chua N.H.(2007). The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19: 2952–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M.(2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chupreta S., Brevig H., Bai L., Merchant J.L., Iñiguez-Lluhí J.A.(2007). Sumoylation-dependent control of homotypic and heterotypic synergy by the Kruppel-type zinc finger protein ZBP-89. J. Biol. Chem. 282: 36155–36166. [DOI] [PubMed] [Google Scholar]
- Conti L., Nelis S., Zhang C., Woodcock A., Swarup R., Galbiati M., Tonelli C., Napier R., Hedden P., Bennett M., Sadanandom A.(2014). Small Ubiquitin-like Modifier protein SUMO enables plants to control growth independently of the phytohormone gibberellin. Dev. Cell 28: 102–110. [DOI] [PubMed] [Google Scholar]
- Conti L., Price G., O’Donnell E., Schwessinger B., Dominy P., Sadanandom A.(2008). Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20: 2894–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creton S., Jentsch S.(2010). SnapShot: The SUMO system. Cell 143: 848–848.e1. [DOI] [PubMed] [Google Scholar]
- Dong J., Piñeros M.A., Li X., Yang H., Liu Y., Murphy A.S., Kochian L.V., Liu D.(2017). An Arabidopsis ABC transporter mediates phosphate deficiency-induced remodeling of root architecture by modulating iron homeostasis in roots. Mol. Plant 10: 244–259. [DOI] [PubMed] [Google Scholar]
- Elrouby N., Coupland G.(2010). Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc. Natl. Acad. Sci. USA 107: 17415–17420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Hirai M.Y., Chino M., Komeda Y., Naito S.(1992). Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiol. 99: 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa J., Yamaji N., Wang H., Mitani N., Murata Y., Sato K., Katsuhara M., Takeda K., Ma J.F.(2007). An aluminum-activated citrate transporter in barley. Plant Cell Physiol. 48: 1081–1091. [DOI] [PubMed] [Google Scholar]
- Godon C., Mercier C., Wang X., David P., Richaud P., Nussaume L., Liu D., Desnos T.(2019). Under phosphate starvation conditions, Fe and Al trigger accumulation of the transcription factor STOP1 in the nucleus of Arabidopsis root cells. Plant J. 99: 937–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson M.L., Hong Y., Rogers R., Matunis M.J., Park-Sarge O.K., Sarge K.D.(2001). Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J. Biol. Chem. 276: 18513–18518. [DOI] [PubMed] [Google Scholar]
- Guo J., Zhang Y., Gao H., Li S., Wang Z.Y., Huang C.F.(2020). Mutation of HPR1 encoding a component of the THO/TREX complex reduces STOP1 accumulation and aluminium resistance in Arabidopsis thaliana. New Phytol. 228: 179–193. [DOI] [PubMed] [Google Scholar]
- Hampp R., Goller M., Füllgraf H.(1984). Determination of compartmented metabolite pools by a combination of rapid fractionation of oat mesophyll protoplasts and enzymic cycling. Plant Physiol. 75: 1017–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay R.T.(2005). SUMO: A history of modification. Mol. Cell 18: 1–12. [DOI] [PubMed] [Google Scholar]
- Hoekenga O.A., et al. (2006). AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 9738–9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekenga O.A., Vision T.J., Shaff J.E., Monforte A.J., Lee G.P., Howell S.H., Kochian L.V.(2003). Identification and characterization of aluminum tolerance loci in Arabidopsis (Landsberg erecta × Columbia) by quantitative trait locus mapping: A physiologically simple but genetically complex trait. Plant Physiol. 132: 936–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Rogers R., Matunis M.J., Mayhew C.N., Goodson M.L., Park-Sarge O.K., Sarge K.D.(2001). Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. 276: 40263–40267. [DOI] [PubMed] [Google Scholar]
- Huang C.F., Yamaji N., Ma J.F.(2010). Knockout of a bacterial-type ATP-binding cassette transporter gene, AtSTAR1, results in increased aluminum sensitivity in Arabidopsis. Plant Physiol. 153: 1669–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Koyama H., Iuchi A., Kobayashi Y., Kitabayashi S., Kobayashi Y., Ikka T., Hirayama T., Shinozaki K., Kobayashi M.(2007). Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. USA 104: 9900–9905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S., Psakhye I.(2013). Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu. Rev. Genet. 47: 167–186. [DOI] [PubMed] [Google Scholar]
- Kochian L.V.(1995). Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46: 237–260. [Google Scholar]
- Kochian L.V., Hoekenga O.A., Pineros M.A.(2004). How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 55: 459–493. [DOI] [PubMed] [Google Scholar]
- Kochian L.V., Piñeros M.A., Liu J., Magalhaes J.V.(2015). Plant adaptation to acid soils: The molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 66: 571–598. [DOI] [PubMed] [Google Scholar]
- Kong X., Luo X., Qu G.P., Liu P., Jin J.B.(2017). Arabidopsis SUMO protease ASP1 positively regulates flowering time partially through regulating FLC stability. J. Integr. Plant Biol. 59: 15–29. [DOI] [PubMed] [Google Scholar]
- Larsen P.B., Cancel J., Rounds M., Ochoa V.(2007). Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 225: 1447–1458. [DOI] [PubMed] [Google Scholar]
- Larsen P.B., Geisler M.J.B., Jones C.A., Williams K.M., Cancel J.D.(2005). ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J. 41: 353–363. [DOI] [PubMed] [Google Scholar]
- Liu J., Magalhaes J.V., Shaff J., Kochian L.V.(2009). Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 57: 389–399. [DOI] [PubMed] [Google Scholar]
- Liu J., Piñeros M.A., Kochian L.V.(2014). The role of aluminum sensing and signaling in plant aluminum resistance. J. Integr. Plant Biol. 56: 221–230. [DOI] [PubMed] [Google Scholar]
- Liu L., Jiang Y., Zhang X., Wang X., Wang Y., Han Y., Coupland G., Jin J.B., Searle I., Fu Y.F., Chen F.(2017). Two SUMO proteases SUMO PROTEASE RELATED TO FERTILITY1 and 2 are required for fertility in Arabidopsis. Plant Physiol. 175: 1703–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J., Wang G., He D., Liu F.(2004). Repression of Smad4 transcriptional activity by SUMO modification. Biochem. J. 379: 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.F.(2007). Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int. Rev. Cytol. 264: 225–252. [DOI] [PubMed] [Google Scholar]
- Ma J.F., Ryan P.R., Delhaize E.(2001). Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 6: 273–278. [DOI] [PubMed] [Google Scholar]
- Magalhaes J.V., et al. (2007). A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat. Genet. 39: 1156–1161. [DOI] [PubMed] [Google Scholar]
- Miller M.J., Barrett-Wilt G.A., Hua Z., Vierstra R.D.(2010). Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 16512–16517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Hasegawa P.M.(2010). Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends Cell Biol. 20: 223–232. [DOI] [PubMed] [Google Scholar]
- Miura K., Jin J.B., Lee J., Yoo C.Y., Stirm V., Miura T., Ashworth E.N., Bressan R.A., Yun D.J., Hasegawa P.M.(2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Rus A., Sharkhuu A., Yokoi S., Karthikeyan A.S., Raghothama K.G., Baek D., Koo Y.D., Jin J.B., Bressan R.A., Yun D.J., Hasegawa P.M.(2005). The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. USA 102: 7760–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Macías J., Ojeda-Rivera J.O., Gutiérrez-Alanís D., Yong-Villalobos L., Oropeza-Aburto A., Raya-González J., Jiménez-Domínguez G., Chávez-Calvillo G., Rellán-Álvarez R., Herrera-Estrella L.(2017). Malate-dependent Fe accumulation is a critical checkpoint in the root developmental response to low phosphate. Proc. Natl. Acad. Sci. USA 114: E3563–E3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtas G., Reeves P.H., Fu Y.F., Bancroft I., Dean C., Coupland G.(2003). A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell 15: 2308–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park-Sarge O.K., Sarge K.D.(2009). Detection of sumoylated proteins. Methods Mol. Biol. 464: 255–265. [DOI] [PubMed] [Google Scholar]
- Reeves P.H., Murtas G., Dash S., Coupland G.(2002). early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development 129: 5349–5361. [DOI] [PubMed] [Google Scholar]
- Ryan P., Delhaize E., Jones D.(2001). Function and mechanism of organic anion exudation from plant roots. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 527–560. [DOI] [PubMed] [Google Scholar]
- Sadanandom A., Ádám É., Orosa B., Viczián A., Klose C., Zhang C., Josse E.M., Kozma-Bognár L., Nagy F.(2015). SUMOylation of phytochrome-B negatively regulates light-induced signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 112: 11108–11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H., Pu R.T., Dasso M.(1997). SUMO-1: Wrestling with a new ubiquitin-related modifier. Trends Biochem. Sci. 22: 374–376. [DOI] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z.(2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Sasaki T., Yamamoto Y., Ezaki B., Katsuhara M., Ahn S.J., Ryan P.R., Delhaize E., Matsumoto H.(2004). A wheat gene encoding an aluminum-activated malate transporter. Plant J. 37: 645–653. [DOI] [PubMed] [Google Scholar]
- Sawaki Y., Iuchi S., Kobayashi Y., Kobayashi Y., Ikka T., Sakurai N., Fujita M., Shinozaki K., Shibata D., Kobayashi M., Koyama H.(2009). STOP1 regulates multiple genes that protect Arabidopsis from proton and aluminum toxicities. Plant Physiol. 150: 281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A.K., Orosa B., Singh P., Cummins I., Walsh C., Zhang C., Grant M., Roberts M.R., Anand G.S., Fitches E., Sadanandom A.(2018). SUMO suppresses the activity of the jasmonic acid receptor CORONATINE INSENSITIVE1. Plant Cell 30: 2099–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M., Srivastava A.K., Orosa-Puente B., Campanaro A., Zhang C., Sadanandom A.(2020). SUMO conjugation to BZR1 enables brassinosteroid signaling to integrate environmental cues to shape plant growth. Curr. Biol. 30: 1410–1423.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokizawa M., Kobayashi Y., Saito T., Kobayashi M., Iuchi S., Nomoto M., Tada Y., Yamamoto Y.Y., Koyama H.(2015). SENSITIVE TO PROTON RHIZOTOXICITY1, CALMODULIN BINDING TRANSCRIPTION ACTIVATOR2, and other transcription factors are involved in ALUMINUM-ACTIVATED MALATE TRANSPORTER1 expression. Plant Physiol. 167: 991–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomanov K., Zeschmann A., Hermkes R., Eifler K., Ziba I., Grieco M., Novatchkova M., Hofmann K., Hesse H., Bachmair A.(2014). Arabidopsis PIAL1 and 2 promote SUMO chain formation as E4-type SUMO ligases and are involved in stress responses and sulfur metabolism. Plant Cell 26: 4547–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma V., Croley F., Sadanandom A.(2018). Fifty shades of SUMO: Its role in immunity and at the fulcrum of the growth-defence balance. Mol. Plant Pathol. 19: 1537–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villajuana-Bonequi M., Elrouby N., Nordström K., Griebel T., Bachmair A., Coupland G.(2014). Elevated salicylic acid levels conferred by increased expression of ISOCHORISMATE SYNTHASE 1 contribute to hyperaccumulation of SUMO1 conjugates in the Arabidopsis mutant early in short days 4. Plant J. 79: 206–219. [DOI] [PubMed] [Google Scholar]
- von Uexkull H.R., Mutert E.(1995). Global extent, development and economic-impact of acid soils. Plant Soil 171: 1–15. [Google Scholar]
- Wilkinson K.A., Henley J.M.(2010). Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 428: 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.M., Rose A., Muthuswamy S., Jeong S.Y., Venkatakrishnan S., Zhao Q., Meier I.(2007). NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell 19: 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.H., Sharrocks A.D.(2004). SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell 13: 611–617. [DOI] [PubMed] [Google Scholar]
- Yates G., Srivastava A.K., Sadanandom A.(2016). SUMO proteases: Uncovering the roles of deSUMOylation in plants. J. Exp. Bot. 67: 2541–2548. [DOI] [PubMed] [Google Scholar]
- Zhai Z., Jung H.I., Vatamaniuk O.K.(2009). Isolation of protoplasts from tissues of 14-day-old seedlings of Arabidopsis thaliana. J. Vis. Exp. 1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang J., Guo J., Zhou F., Singh S., Xu X., Xie Q., Yang Z., Huang C.F.(2019). F-box protein RAE1 regulates the stability of the aluminum-resistance transcription factor STOP1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 116: 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]