Abstract
Epimedium tianmenshanensis is a rare perennial herb distributed in China, and it is also an important medicinal plant. Here, we used illumina paired-end sequencing technology to obtain the complete chloroplast genome of E. tianmenshanensis, and compared analysis with related species. The length of the complete chloroplast genome of E. tianmenshanensis is 156,956 bp, which is a relatively conserved quadripartite structure including a large single copy (LSC) region of 88,409 bp, a small single copy (SSC) region of 17,448 bp, and a pair of inverted repeat (IRa/IRb) regions of 25,550 bp. The whole genome contains 132 unique genes, including 85 protein-coding genes, 38 tRNA genes, eight rRNA genes and one pseudogene. 87 simple sequence repeats (SSRs) were identified, and most of them were found to be composed of A/T. In addition, 22,923 codons were detected in 78 protein-coding genes of E. tianmenshanensis, and the overall codon bias pattern in the genome tended to use A/U ending codons. Phylogenetic analysis demonstrated that all the Epimedium species formed a monophyletic clade, and E. tianmenshanensis had the closest relationship to E. dolichostemon. The results of this study provided useful molecular information about the evolution and molecular biology of E. tianmenshanensis.
Electronic supplementary material
The online version of this article (10.1007/s12298-020-00882-3) contains supplementary material, which is available to authorized users.
Keywords: Chloroplast genome, Epimedium, Phylogenetic analysis, RSCU, SSRs
Introduction
Epimedii Folium (EF) is one of the most popular Traditional Chinese Medicine (TCM), with a history of more than 2000 years (Zhang et al. 2016). The dried leaves of Epimedium brevicomu, E. sagittatum, E. pubescens and E. koreanum are recorded as the source plants of EF in the 2020 edition of Chinese Pharmacopoeia (Chinese Pharmacopoeia 2020), however, a total of 15 species of Epimedium are common sources of EF in the market (Ma et al. 2011). The aboveground portion of Epimedium can be used for nourishing kidney, reinforcing yang and regulating bone remodeling (Ma et al. 2011; Jiang et al. 2015). As one of the most important genera in Berberidaceae, the Epimedium comprises more than 50 species (Ying et al. 2011). China is the modern geographical distribution center of Epimedium, in which almost 40 species were found (Li et al. 2018). Furthermore, some reports on new species in China have been published in recent years (Guo et al. 2007; Zhang and Li 2009; He et al. 2010; Sheng and Tian 2011). However, because of high levels of morphological variation, species delimitation in Epimedium has been problematic (Sun et al. 2005). With the development of DNA sequencing technology, more and more researchers have focused on chloroplast genome research (Yang et al. 2019). Compared with nuclear genes, the chloroplast genome is often more conserved (Meng et al. 2018), which has great significance in plant phylogeny and species identification (Zhou et al. 2017; Gu et al. 2018). E. tianmenshanensis is a rare perennial herb, and only occurs in limited areas at Mt. Tianmen, Zhangjiajie City, Hunan Province, China, which was first published in phytotaxa in 2015 (Zhang et al. 2015). To make clear the difference between E. tianmenshanensis and other species of Epimedium, the complete chloroplast genome of E. tianmenshanensis was sequenced and analyzed to provide a basis for further understanding of phylogenetic relationships of Epimedium species and afford some information for further research and application of these medicinal species.
Materials and methods
Plant materials and DNA extraction
Fresh leaves of wild E. tianmenshanensis were collected from Tianmenshan National Geological Park, Zhangjiajie City, Hunan Province, China, on wet slopes under, 29°03′04″ N, and 110°28′34″ E, elevation 1463 m. The voucher specimens were deposited in the Center of Herbarium, institute of Chinese Materia Medica, Hunan Academy of Chinese Medicine, Changsha, China, under accession number HUTM100010. The whole genomic DNA was extracted from fresh leaves using Rapid Plant Genomic DNA Isolation Kit, Sangon Biotech (Shanghai) Co., Ltd. Then, the quality and integrity of DNA were checked by BioPhotometer Plus (Nucleic acid and protein detector, Eppendorf, Germany), and the high- quality DNA was ultimately sent to GENEWIZ (Suzhou, China) for sequencing.
Illumina sequencing, assembly, and annotation
To ensure accurate and reliable analyses, the raw reads were filtered to obtain high-quality clean reads by Cutadapt v1.9.1 (Martin 2011). The total of 8.76G clean reads were used for the plastome assembly by Velvet v1.2.10 (Zerbino and Birney 2008) and NOVOPlasty v2.7.2 (Dierckxsens et al. 2016), with that of congeneric E. sagittatum (NC_029428.1) as the reference, and the chloroplast genome sequence was annotated using GeSeq tool (Tillich et al. 2017). The annotated result was manually checked, and adjusted by comparison with homologous genes from other sequenced chloroplast genomes. Additionally, the transfer RNA (tRNA) genes were further identified by tRNAscan-SE v2.0 (Lowe and Chan 2016). The circular chloroplast genome map was drawn using the OGDRAW program (Lohse et al. 2007).
Repeat sequences and SSRs analysis
REPuter (Kurtz et al. 2001) was used to analyze repeat sequences, including palindromic and forward repeats within the chloroplast genome, with the following parameters: the minimal repeat size of 30, hamming distance to three and over 90% identity. Tandem repeats were detected using Tandem Repeats Finder with default parameters (Benson 1999). The distributing frequency of chloroplast genome SSRs were analyzed using the microsatellite identification tool MISA (https://webblast.ipk-gatersleben.de/misa/), with the following parameters: 10 for mononucleotide repeats, 5 for dinucleotide repeats, 4 for trinucleotide repeats, and 3 for tetranucleotide repeats, pentanucleotide repeats and hexanucleotide repeats.
Phylogenetic analysis
To construct a phylogenetic tree, nine chloroplast genomes from Berberidaceae were employed (Table 1). Among these nine taxa, Vancouveria hexandra and Sinopodophyllum hexandrum were set as outgroups. The completed chloroplast genome sequences of the selected species were downloaded from the National Center for Biotechnology Information (NCBI). The software MAFFT 7.380 (Katoh and Standley 2013) was used to align the genome sequence, and BioEdit v 7.0.9.0 (Hall 1999) was used to examine and manually adjust the sequence alignment result. Phylogenetic reconstructions were conducted using Maximum Parsimony (MP) and Bayesian Inference (BI) methods. The MP analysis was implemented in PAUP* v 4.0 beta 10 (Swofford 2002), and the BI was implemented in MrBayes 3.2.6 (Ronquist et al. 2012). For the MP analysis, the bootstrap probability of each branch was calculated by 1000 replications. For BI analysis, the best substitution models were tested based on Akaike information criterion (AIC) by MrModeltest v 2.3 (Nylander 2004), and the best-fitting models in the analysis were GTR + I + G. Four Markov Chains Monte Carlo (MCMC) samples were run twice from random starting trees for 1 × 106 generations, and trees were sampled every 100 generations.
Table 1.
Chloroplast genome sequences used for phylogenetic tree construction
| Species name | GenBank ID | Species name | GenBank ID |
|---|---|---|---|
| Epimedium acuminatum | NC_029941.1 | E. sagittatum | NC_029428.1 |
| E. dolichostemon | NC_029942.1 | E. tianmenshanensis | MT028488 |
| E. koreanum | NC_029943.1 | ||
| Outgroups | |||
| E. lishihchenii | NC_029944.1 | Vancouveria hexandra | NC_042222.1 |
| E. pseudowushanense | NC_029945.1 | Sinopodophyllum hexandrum | KT445939.1 |
Results
Chloroplast genome general features
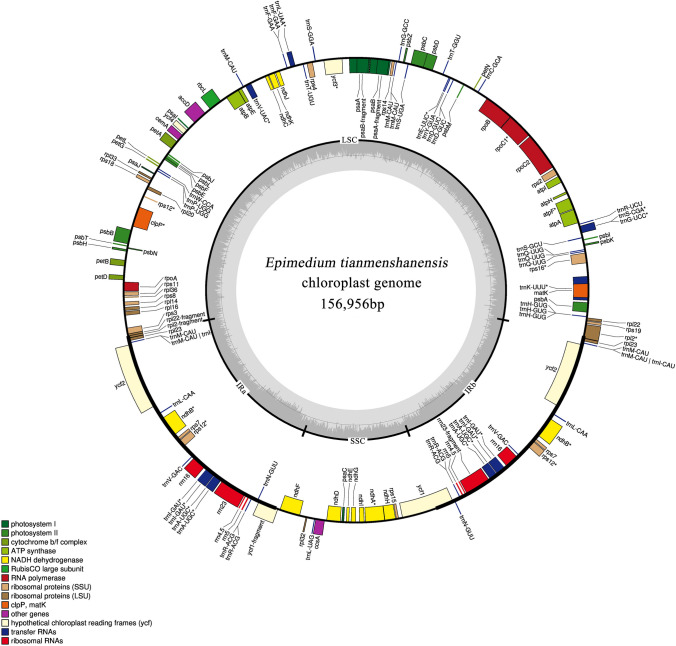
In total, we obtained 8764 Mb short sequence data with a Q20 of 96.75% using the Illumina HiSeq XTen platform. The length of chloroplast genome of E. tianmenshanensis is 156,956 bp with a typical quadripartite structure, including a large single copy region (LSC, 88,409 bp) and a small single copy region (SSC, 17,448 bp), separated by a pair of inverted repeats (IRa/IRb, 25,550 bp) (Fig. 1). The total GC content of chloroplast genome was 38.78%, of which, the IR regions had a higher GC content of 43.23%, while the GC contents of the LSC and SSC were 37.38% and 32.88%, respectively. This genome sequence encoded 132 genes, including 85 protein-coding genes, 38 tRNA genes, eight rRNA genes and one pseudogene (Table 2). The gene ycf1 crossed the SSC/IRb region, and the pseudogene fragment ycf1 was located at the IRa region. Besides, the rpl22 gene located in the IRa/LSC junction region lost their protein-coding ability because of incomplete gene duplication. Sixteen genes had introns, of which 13 contain one intron and three (rps12, ycf3 and clpP) contain two introns. The chloroplast genome of E. tianmenshanensis with gene annotations was submitted to GenBank under the accession number MT028488.
Fig. 1.
Chloroplast genome map of E. tianmenshanensis. Genes drawn inside the circle are transcribed clockwise, genes outside are transcribed counterclockwise. Genes are color coded by their function in the legend. The area in darker gray and lighter gray in the inner circle indicates GC content and AT content, respectively
Table 2.
Functional classification of E. tianmenshanensis chloroplast genes
| Category for genes | Group of genes | Name of genes |
|---|---|---|
| Self-replication | rRNA genes | rrn16 (x2), rrn4.5 (x2), rrn5 (x2), rrn23 (x2) |
| tRNA genes | trnT-CAU (x2), trnL-CAA (x2), trnV-GAC (x2), trnR-ACG (x2), trnN-GUU (x2), trnL-UAG, trnP-UGG, trnW-CCA, trnM-CAU, rnF-GAA, trnT-UGU, trnS-GGA, trnfM-CAU, trnG-GCC, trnS-UGA, trnT-GGU, trnE-UUC, trnY-GUA, trnD-GUC, trnC-GCA, trnR-UCU, trnS-GCU, trnQ-UUG (2), trnG-UCC*, trnH-GUG, trnL-UAA*, trnV-UAC*, trnA-UGC* (2), trnI-GAU* (x2), trnK-UUU* | |
| Small subunit of ribosome | rps19, rps7 (x2), rps18, rps4, rps14, rps2, rps11, rps3, rps16*, rps8, rps15, rps12** (x2) | |
| Large subunit of ribosome | rpl32, rpl20, rpl33, rpl14, rpl2 (x2)*, rpl23 (x2), rpl36, rpl16, rpl22 | |
| RNA polymerase subunits | rpoA, rpoC2, rpoB, rpoC1* | |
| Phytosynthesis | NADH dehydrogenase | ndhG, ndhC, ndhJ, ndhB* (x2), ndhD, ndhH, ndhI, ndhF, ndhK, ndhE, ndhA* |
| Photosystem I | psaC, psaJ, psaI, psaA, psaB, ycf4, ycf3** | |
| Photosystem II | psbN, psbT, psbE, psbF, psbL, psbJ, psbZ, psbC, psbD, psbM, psbI, psbA, psbB, psbH, psbK | |
| Cytochrome b/f complex | petG, petN, petA, petB, petL, petD | |
| ATP synthase gene | atpE, atpH, atpF*, atpB, atpI, atpA | |
| Large subunit of rubisco | rbcL | |
| Othergenes | Maturase | matK |
| Protease | clpP** | |
| Membrane protein | cemA | |
| Acetyl-CoA-carboxylase | accD | |
| Cytochrome c biogenesis | ccsA | |
| Translational initiation factor | ΨinfA | |
| Unknown function | Conserved open reading frames | ycf1 (x2), ycf2 (x2) |
*Indicates the genes containing one intron, **indicates the genes containing two introns, (x2) indicates genes duplicated in the IR regions. ΨIndicates the genes was pseudogene
Analysis of codon usage bias
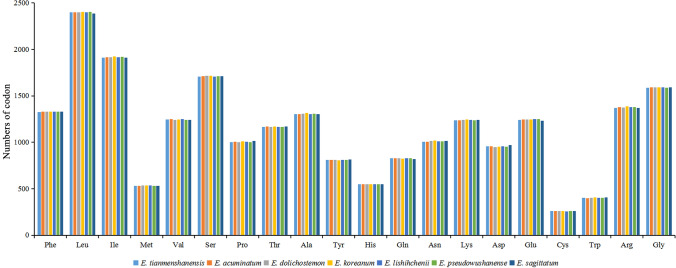
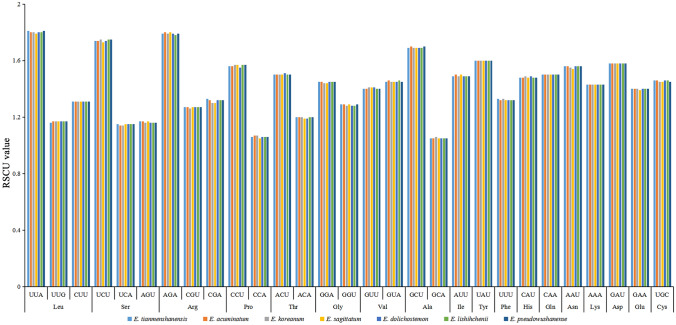
After a long period of development and evolution of organisms, the use of codons will have a certain preference, and the codon preference of different species is also different, that is racial specificity (Wu et al. 2007; Wang et al. 2018). Relative synonymous codon usage (RSCU) is one of the commonly used parameters to measure codon usage bias. Here, RSCU was analyzed using CodonW 1.4.2 (Peden 1999) on the protein-coding genes (remove repeat sequences) in the chloroplast genome of E. tianmenshanensis, and compared with related species. The 78 protein-coding genes (Table S1) of E. tianmenshanensis plastome included 22,923 codons. Among these codons, leucine was the most abundant amino acid, with 2398 (10.46%), while cysteine was the least, with 259 (1.13%) (Table 3). Compared with the other six plants of the same genus, the total number of codons of different amino acids did not differ much (Fig. 2). The RSCU values of 29 codons were greater than 1, and all ended with A/U except UUG and UGC, of which about 45.16% ended in A and about 48.38% ended in U, indicating these codons tend to end in A/U (Table 3). Compared with other related species, the RSCU value of the Epimedium chloroplast genomes exhibit a similar pattern (Fig. 3). This further indicates that the chloroplast genome is more conservative and the difference among related species is small.
Table 3.
Relative synonymous codon usage (RSCU) of the chloroplast genome of E. tianmenshanensis. RSCU values > 1 was shown in bold
| Amino acids | Codon | NO. | RSCU | AA frequency (%) | Amino acids | Codon | NO. | RSCU | AA frequency (%) |
|---|---|---|---|---|---|---|---|---|---|
| Arg | AGA | 408 | 1.79 | 5.98 | Leu | UUA | 722 | 1.81 | 10.46 |
| CGU | 290 | 1.27 | UUG | 465 | 1.16 | ||||
| CGA | 303 | 1.33 | CUU | 523 | 1.31 | ||||
| CGG | 102 | 0.45 | CUC | 182 | 0.46 | ||||
| AGG | 175 | 0.77 | CUA | 347 | 0.87 | ||||
| CGC | 93 | 0.41 | CUG | 159 | 0.40 | ||||
| Ala | GCU | 551 | 1.69 | 5.68 | Lys | AAA | 886 | 1.43 | 5.39 |
| GCA | 342 | 1.05 | AAG | 350 | 0.57 | ||||
| GCC | 224 | 0.69 | Met | AUG | 533 | 1.00 | 2.33 | ||
| GCG | 186 | 0.57 | Pro | CCU | 392 | 1.56 | 4.37 | ||
| Asn | AAU | 786 | 1.56 | 4.39 | CCA | 264 | 1.05 | ||
| AAC | 221 | 0.44 | CCC | 210 | 0.84 | ||||
| Asp | GAU | 754 | 1.58 | 4.17 | CCG | 136 | 0.54 | ||
| GAC | 203 | 0.42 | Phe | UUU | 878 | 1.32 | 5.78 | ||
| Cys | UGU | 189 | 1.46 | 1.13 | UUC | 448 | 0.68 | ||
| UGC | 70 | 0.54 | Ser | UCU | 496 | 1.74 | 7.44 | ||
| Gly | GGU | 511 | 1.29 | 6.93 | UCA | 326 | 1.15 | ||
| GGA | 577 | 1.45 | AGU | 332 | 1.17 | ||||
| GGC | 191 | 0.48 | UCC | 283 | 1.00 | ||||
| GGG | 310 | 0.78 | UCG | 160 | 0.56 | ||||
| Gln | CAA | 620 | 1.50 | 3.61 | AGC | 109 | 0.38 | ||
| CAG | 207 | 0.50 | Thr | ACU | 438 | 1.50 | 5.09 | ||
| Glu | GAA | 872 | 1.40 | 5.42 | ACA | 349 | 1.20 | ||
| GAG | 370 | 0.60 | ACC | 234 | 0.80 | ||||
| His | CAU | 407 | 1.48 | 2.40 | ACG | 146 | 0.50 | ||
| CAC | 143 | 0.52 | Tyr | UAU | 649 | 1.60 | 3.54 | ||
| Ile | AUU | 951 | 1.49 | 8.34 | UAC | 162 | 0.40 | ||
| AUC | 386 | 0.61 | Trp | UGG | 402 | 1.00 | 1.75 | ||
| AUA | 574 | 0.90 | Val | GUU | 438 | 1.40 | 5.44 | ||
| TERa | UAA | 35 | 1.33 | 3.45 | GUA | 453 | 1.45 | ||
| UAG | 22 | 0.84 | GUC | 154 | 0.49 | ||||
| UGA | 22 | 0.84 | GUG | 202 | 0.65 |
aTermination codon
Fig. 2.
The frequency of codons for protein-coding genes in the chloroplast genomes of seven Epimedium plants
Fig. 3.
Codons with RSCU values greater than 1 based on 78 protein-coding genes in the chloroplast genome of E. tianmenshanensis and six related species
Characterization of SSRs and repeat sequences
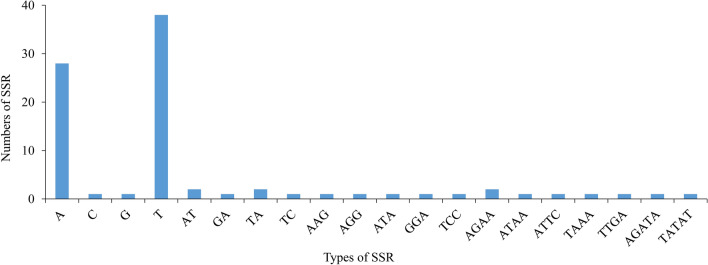
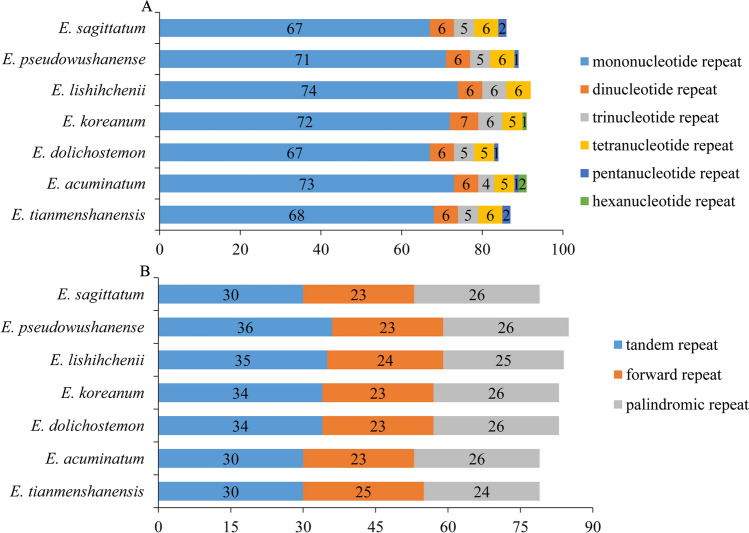
Simple sequence repeats (SSRs), also known as DNA microsatellites, refer to short (1–6 bp) tandemly repeated DNA sequences (Zheng et al. 2019), which are widely used in phylogenetic analysis (Yuan et al. 2017). Here, we identified 87 SSRs loci in the chloroplast genome of E. tianmenshanensis, including 68 mononucleotide SSR loci, six dinucleotide SSR loci, five trinucleotide SSR loci, six tetranucleotide SSR loci and two pentanucleotide SSR loci (Fig. 4). The total numbers of the SSRs were 91, 84, 91, 89, 86 and 92 in E. acuminatum, E. dolichostemon, E. koreanum, E. pseudowushanense, E. sagittatum and E. lishihchenii respectively (Fig. 5a). Similar to E. tianmenshanensis, the majority of the SSRs in these chloroplast genomes were mononucleotides repeats. Additionally, hexanucleotide repeats were present only in E. acuminatum and E. koreanum. Meanwhile, E. koreanum and E. lishihchenii did not have pentanucleotide repeats.
Fig. 4.
Numbers and types of SSR in the chloroplast genomes of E. tianmenshanensis
Fig. 5.
Types and numbers of repeat in the chloroplast genomes of seven plastomes of Epimedium. a Numbers and types of SSR. b total of the three repeat types
In addition, a total of 79 repeats including tandem, palindrome and forward repeats were found in the E. tianmenshanensis chloroplast genome. Similarly, 79, 83, 83, 85, 79 and 84 repeats were detected in E. acuminatum, E. dolichostemon, E. koreanum, E. pseudowushanense, E. sagittatum and E. lishihchenii respectively (Fig. 5b). Among these repeat sequences, tandem repeats had larger numbers than others. The numbers of palindromic repeats were more than forward repeats in E. tianmenshanensis, and less than forward repeats in other species.
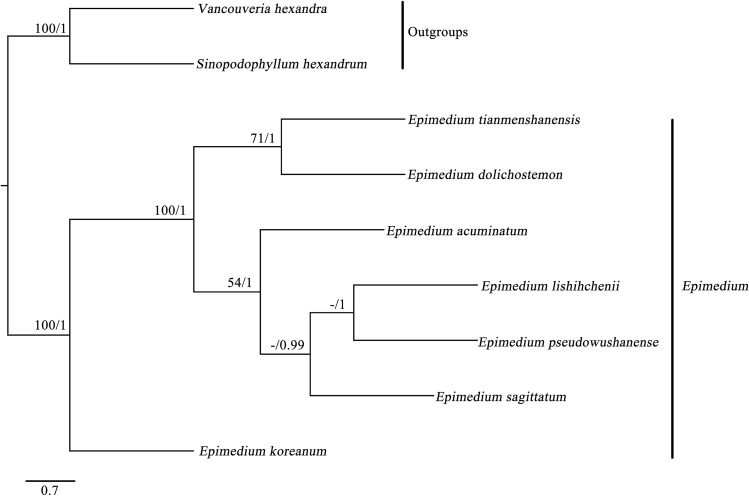
Phylogenetic analysis
To analyze the phylogenetic position of E. tianmenshanensis in Epimedium, A phylogenetic analysis was performed based on nine complete chloroplast genomes of species from Berberidaceae (including two outgroups). The result of molecular analysis based on BI and MP methods revealed the same topology, and the Bayesian tree was presented (Fig. 6). All species of Epimedium were grouped together with higher support (PP = 1, BS = 100), in which E. tianmenshanensis and E. dolichostemon were reciprocally monophyletic, and the two species formed a clade (PP = 1, BS = 71). Our study will provide valuable genetic information for phylogenetic studies of Epimedium.
Fig. 6.
The partitioned Bayesian phylogenetic tree based on nine complete chloroplast genome sequences. Parsimony bootstrap values (before the slash markers) higher than 50% and Bayesian posterior probabilities (after the slash markers) more than 0.95 were indicated along branches
Discussion
Chloroplast genome has been widely used in species identification, some scholars have proposed the concept of ‘super bar code’,that is the whole chloroplast genome was used as the standard sequence for species identification (Boynton et al. 1988). In this study, we obtained the complete chloroplast sequence of E. tianmenshanensis and compared to the chloroplast genomes from different species of the same genus. The length of chloroplast genome of E. tianmenshanensis is 156,956 bp with a typical quadripartite structure, including 132 unique genes encoding 85 proteins, eight rRNAs, 38 tRNAs and one pseudogenes (infA). Based on comparison of the complete chloroplasts genomes of Epimedium plants, we found that the genome length, gene number, gene order, and chloroplast genome structure were similar. This further indicated that the chloroplast genome is often conserved.
Codon usage bias analysis showed that the RSCU values of 29 codons were greater than 1, and tended to end in A/U, which is similar to the codon use bias in plants of the same genus and also similar to other plants, such as Cinnamomum camphora (Chen et al. 2017), Wurfbainia villosa (An et al. 2020) and Aconitum hemsleyanum (Meng et al. 2018). Those further indicate that dicotyledons’s codon tend to end in A/T (Kawabe and Miyashita 2003).
Due to high variability, the SSRs have been used for the study of population genetics (Pauwels et al. 2012; Asaf et al. 2016). There were 87 SSRs in chloroplast genome of E. tianmenshanensis, and detected five types of SSR. By comparing SSRs of the other six species of the same genus, the number of SSR ranged from 84 to 92. The SSRs identified in the chloroplast genome of Epimedium can be used for developing lineage-specific cpSSR markers.
There are many plants in the genus of Epimedium, which contains nearly 50 species in Flora of China (FOC), and some unknown species. Hence, species identification and phylogenetic studies of this genus are particularly important. In this study, the phylogenetic position of E. tianmenshanensis was performed by analyzing the complete chloroplast genome among nine species. The results shown that E. tianmenshanensis showed the closest relationship to E. dolichostemon. In addition, the chloroplast genome contains a large amount of information, which deserves further study. The chloroplast genomes of this study are helpful for the identification and phylogenetic analysis of Epimedium species.
Conclusion
Overall, we successfully assembled, annotated and analyzed the complete chloroplast sequence of E. tianmenshanensis for the purpose of making clear its difference from other Epimedium plants and analyzing its phylogenetic position in Epimedium. The complete chloroplast genome is 156,956 bp in length, containing 132 genes, and the codons tend to end in A/U, which is very similar to other Epimedium species. These indicate that the chloroplast genome of Epimedium are highly conserved in the evolutionary process. We also identified 87 SSRs loci in the chloroplast genome, which can be used to develop lineage-specific markers. The phylogenetic analyses showed that E. tianmenshanensis has the closest relationship to E. dolichostemo among the studied taxa. The complete chloroplast genome of E. tianmenshanensis will be conducive to population genomic studies of Epimedium and provide a valuable insight for species identification and conservation of E. tianmenshanensis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by Special project of “National Chinese materia medica (CMM) resources survey” ([2017] 66), Key project at central government level: The ability establishment of sustainable use for valuable Chinese medicine resources (2060302), “Double First-Class” University project (CPU2018GY11) and National Natural Science Foundation of China (81660639).
Author’s contribution
HL conceived and designed the study; HH reviewed draft of the manuscript; JJ and SZ analyzed the data; BH and XL performed the experiments; LW performed the experiments, analyzed the data and wrote the manuscript.
Availability of data
The DNA sequences was submitted to GenBank under the accession number MT028488.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- An WL, Li J, Yang ZR, Huang YY, Huang S, Zheng XS. Characteristics analysis of the complete Wurfbainia villosa chloroplast genome. Physiol Mol Biol Plants. 2020;26:747–758. doi: 10.1007/s12298-019-00748-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaf S, Khan AL, Khan AR, Muhammad W, Kang SM, Khan MA, et al. Complete chloroplast genome of Nicotiana otophora and its comparison with related species. Front Plant Sci. 2016;7:843. doi: 10.3389/fpls.2016.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton JE, Gillham NW, Harris EH, et al. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988;240:1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- Chen CH, Zheng YJ, Liu SA, Zhong YD, Wu YF, Li J, et al. The complete chloroplast genome of Cinnamomum camphora and its comparison with related Lauraceae species. PeerJ. 2017;5:e3820. doi: 10.7717/peerj.3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese Pharmacopoeia Editorial Committee . Chinese Pharmacopoeia (part I) Bejing: China Medical Science and Technology Press; 2020. [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2016;45:1–9. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu CH, Tembrock LR, Zheng SY, Wu ZQ. The complete chloroplast genome of Catha edulis: a comparative analysis of genome features with related species. Int J Mol Sci. 2018;19:525. doi: 10.3390/ijms19020525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo BL, Fe SZ, Zhong GY, Xiao PG. Two new species of Epimedium (Berberidaceae) from China. Acta Phytotaxon Sin. 2007;45:813–821. doi: 10.1360/aps06138. [DOI] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- He SZ, Wang YY, Guo BL, Xu WF. Epimedium pudingense (Berberidaceae), a new species from Guizhou, China. Ann Bot Fenn. 2010;47:226–228. doi: 10.5735/085.047.0308. [DOI] [Google Scholar]
- Jiang J, Song J, Jia XB. Phytochemistry and ethnopharmacology of Epimedium L. species. CHM. 2015;7:204–222. [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe A, Miyashita NT. Patterns of codon usage bias in three dicot and four monocot plant species. Genes Genet Syst. 2003;78:343–352. doi: 10.1266/ggs.78.343. [DOI] [PubMed] [Google Scholar]
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Guo MY, Pang XH. Identification and classification of medicinal plants in Epimedium. CHM. 2018;10:249–254. [Google Scholar]
- Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, He X, Yang Y, Li M, Hao D, Jia Z. The genus Epimedium: an ethnopharmacological and phytochemical review. J Ethnopharmacol. 2011;134:519–541. doi: 10.1016/j.jep.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Meng J, Li XP, Li HT, Yang JB, Wang H, He J. Comparative analysis of the complete chloroplast genomes of four Aconitum medicinal species. Molecules. 2018;23:1015–1017. doi: 10.3390/molecules23051015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA (2004) MRMODELTEST version 2.1. Computer program distributed by the author. Uppsala University, Uppsala
- Pauwels M, Vekemans X, Godé C, Frérot H, Castric V, Saumitoulaprade P. Nuclear and chloroplast DNA phylogeography reveals vicariance among European populations of the model species for the study of metal tolerance, Arabidopsis halleri (Brassicaceae) New Phytol. 2012;193:916–928. doi: 10.1111/j.1469-8137.2011.04003.x. [DOI] [PubMed] [Google Scholar]
- Peden JF (1999) CodonW. PhD Dissertation, University of Nottingham, Nottinghamshire, UK
- Ronquist F, Teslenko M, Van DMP, Ayres DL, Darling A, Sebastian H, et al. Mrbayes3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng MY, Tian XJ. A new species of Epimedium (Berberidaceae) with 24 chromosomes from Guizhou, China. Novon. 2011;21:262–265. doi: 10.3417/2009081. [DOI] [Google Scholar]
- Sun Y, Fung KP, Leung PC, Shaw PC. A phylogenetic analysis of Epimedium (Berberidaceae) based on nuclear ribosomal DNA sequences. Mol Phylogenet Evol. 2005;35:287–291. doi: 10.1016/j.ympev.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Swofford DL (2002) PAUP*: Phylogenetic Analysis using Parsimony (* and Other Methods), version 4.0b 10. Sinauer, Sunderland
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, et al. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WB, Yu H, Qiu XP. Analysis of repeat sequence and codon bias of chloroplast genome in Scutellaria baicalensis. Mol Plant Breed. 2018;16:2445–2452. [Google Scholar]
- Wu XM, Wu SF, Ren DM, Zhu YP, He FC. The analysis method and progress in the study of codon bias. Hereditas. 2007;29:420–426. doi: 10.1360/yc-007-0420. [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhang Y, Chen YK, Gul JM, Zhang JW, Liu Q, et al. Complete chloroplast genome sequence of the mangrove species Kandelia obovata and comparative analyses with related species. PeerJ. 2019;7:e7713. doi: 10.7717/peerj.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying TS, Boufford DE, Brach AR. Berberidaceae. In: Wu ZY, Raven PH, editors. Flora of China. Beijing and St. Louis: Science Press and Missouri Botanical Garden Press; 2011. pp. 714–800. [Google Scholar]
- Yuan C, Zhong WJ, Mou FS, Gong YY, Pu DQ, Ji PC, Huang HY, Yang ZH, Zhang C. The complete chloroplast genome sequence and phylogenetic analysis of Chuanminshen (Chuanminshenviolaceum Sheh et Shan) Physiol Mol Biol Plants. 2017;23:35–41. doi: 10.1007/s12298-016-0395-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Li JQ. A new species of Epimedium (Berberidaceae) from Hubei, China. Novon. 2009;19:567–569. doi: 10.3417/2007161. [DOI] [Google Scholar]
- Zhang DG, Deng T, Changkyun K, Zhang JW, Nie ZL, Sun H. Epimedium tianmenshanensis (Berberidaceae), a new species from Hunan, China. Phytotaxa. 2015;222:33–43. doi: 10.11646/phytotaxa.222.1.3. [DOI] [Google Scholar]
- Zhang YJ, Du LW, Liu A, Chen JJ, Wu L, Hu WM, et al. The complete chloroplast genome sequences of five Epimedium species: lights into phylogenetic and taxonomic analyses. Front Plant Sci. 2016;7:306. doi: 10.3389/fpls.2016.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XH, Ren CW, Huang S, Li J, Zhao Y. Structure and features of the complete chloroplast genome of Melastoma dodecandrum. Phys Mol Biol Plants. 2019;25:1043–1054. doi: 10.1007/s12298-019-00651-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JG, Chen XL, Cui YX, Sun W, Li YH, Wang Y, et al. Molecular structure and phylogenetic analyses of genomes of two Aristolochia medicinal species. Int J Mol Sci. 2017;18:1839. doi: 10.3390/ijms18091839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DNA sequences was submitted to GenBank under the accession number MT028488.