ABSTRACT
La-related proteins 7 (LARP7) are a class of RNA chaperones that bind the 3′ ends of RNA and are constitutively associated with their specific target RNAs. In metazoa, Larp7 binds to the long non-coding 7SK RNA as a core component of the 7SK RNP, a major regulator of eukaryotic transcription. In the ciliate Tetrahymena the LARP7 protein p65 is a component of telomerase, an essential ribonucleoprotein complex that maintains the telomeric DNA at eukaryotic chromosome ends. p65 is important for the ordered assembly of telomerase RNA (TER) with telomerase reverse transcriptase. Unexpectedly, Schizosaccharomyces pombe Pof8 was recently identified as a LARP7 protein and a core component of fission yeast telomerase essential for biogenesis. LARP7 proteins have a conserved N-terminal La motif and RRM1 (La module) and C-terminal RRM2 with specific RNA substrate recognition attributed to RRM2, first structurally characterized in p65 as an atypical RRM named xRRM. Here we present the X-ray crystal structure and NMR studies of S. pombe Pof8 RRM2. Sequence and structure comparison of Pof8 RRM2 to p65 and human Larp7 xRRMs reveals conserved features for RNA binding with the main variability in the length of the non-canonical helix α3. This study shows that Pof8 has conserved xRRM features, providing insight into TER recognition and the defining characteristics of the xRRM.
KEYWORDS: La protein, LARP, RRM, NMR, X-ray crystallography, telomerase, xRRM, RNA, 7SK
Introduction
The eukaryotic La protein and La-related protein (LARP) superfamily bind to diverse RNA targets and are involved in RNA processing and assembly [1–3]. Genuine La protein recognizes the 3′ UUU-OH terminus of most nascent RNA polymerase III transcripts to protect and stabilize them and in many cases also acts as a chaperone to fold the RNAs into functional complexes [4–10]. LARPs bind to specific RNAs to function in the folding and biogenesis of their ribonucleoproteins (RNPs) [2]. Based on evolutionary and domain organization conservation, they have been classified into four families (LARP1, LARP4, LARP6 and LARP7), and the LARP7 family appears most closely related to genuine La protein [1–3].
All LARP7 proteins identified to date are components of 7SK [11,12] or telomerase RNPs [13–17]. The 7SK RNP sequesters and inactivates the kinase activity of the positive transcription elongation factor b (P-TEFb) to regulate the elongation phase of RNA Polymerase II [18,19] and has been identified in metazoa [20,21]. Larp7 (here lowercase is used to distinguish the protein name in metazoa from the LARP7 family) binds to the long-noncoding 7SK RNA as a core component of the 7SK RNP and is required for 7SK RNP hierarchical assembly with P-TEFb and transcription regulation [11,12,22,23]. Telomerase is an RNP that maintains telomeric DNA repeats at the ends of linear chromosomes [24,25] and has been identified in most eukaryotes [26,27]. Telomerase is comprised of the telomerase RNA (TER) scaffold and template and the telomerase reverse transcriptase (TERT) enzyme, required together for catalytic activity, and additional proteins required for biogenesis and activity that form the holoenzyme in vivo [26,28,29]. In the ciliates Tetrahymena thermophila and Euplotes aediculatus, the respective LARP7 protein p65 and p43 binds to TER and is required for its assembly with TERT [13,14,30–32]. Recently, a LARP7 protein, Pof8 (also called Lar7), was identified in the fission yeast Schizosaccharomyces pombe [15–17]. Pof8 binds to the S. pombe telomerase RNA (TER1) and recruits TER1 processing proteins Lsm2-8 to facilitate assembly of TER1 with TERT, and is essential for telomerase biogenesis and function [15–17].
LARP7s have three structured domains – an N-terminal-winged helix domain (La motif, LaM) followed by an RNA recognition motif (RRM1) that together form a La module, and a C-terminal atypical RRM2 [2] (Fig. 1A). The La module is a conserved feature of genuine La and LARPs that specifically recognizes and binds to the RNA 3′ UUU-OH end [4,5,7], although the genuine La protein and LARP6 La modules sometimes bind alternate sequences [9,33,34]. The La module of ciliate and metazoan LARP7s binds to the 3′ UUU-OH terminus of their TER and 7SK cognate RNAs, respectively [23,35,36]. For both Tetrahymena p65 and human Larp7, the C-terminal RRM2 is essential for specific RNA recognition and RNP assembly [22,31,32,37]. High-resolution structures of these domains in complex with RNA revealed an atypical mode of RNA binding divergent from that of canonical RRMs [38–40], and the domain was named xRRM for extended helical RRM [31,41,42]. Compared to the canonical RRM, the xRRM lacks conserved single-strand RNA recognition sequences RNP1 (K/R-G-F/Y-G/A-F/Y-I/L/V-X-F/Y) and RNP2 (I/L/V-F/Y-I/L/V–X/N/L) on the β-sheet and has an additional C-terminal helix α3 that lies across this surface (Fig. 1B,C) [40,41]. These structures, together with multiple sequence alignment of LARP7s with predicted xRRMs, revealed several conserved features key for RNA recognition: a Y/W-X-D/Q (RNP3) sequence on strand β2 (where underline indicates residues that interact with RNA), a conserved R on strand β3, and charged/aromatic residues on the C-terminal end of helix α3 (Fig. 1B,C) that together form an RNA binding surface on the side of the β-sheet rather than the surface and recognize a combination of base paired and unpaired nucleotides with high affinity and specificity [31,41,42]. Euplotes LARP7 telomerase protein p43, a homolog of p65, binds a similar site in TER as p65 [13,30,43]. Based on sequence and homology modelling it is predicted to contain an xRRM [31,42]. Human genuine La contains an atypical RRM with most of the features of an xRRM (Fig. 1B) and has chaperone activity [44,45], but the RNA-binding mode remains unknown.
Figure 1.
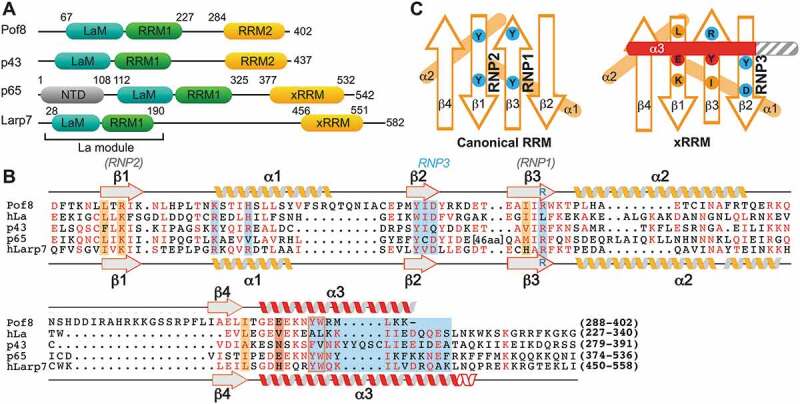
Domains and sequence alignments of LARP7 and La proteins (A) Domain organization of LARP7s from yeast S. pombe (Pof8), ciliate Euplotes aediculatus (p43), ciliate Tetrahymena thermophila (p65), and human (Larp7). Numbers above the sequence schematic indicate known domain boundaries of the La motif (cyan), RRM1 (green) and xRRM (orange). (B) Sequence alignment of LARP7 p65 and Larp7 xRRMs, human La protein (hLa) RRM2, Pof8, and Euplotes p43. Residues with high similarity are coloured red. The secondary structure elements of Pof8 and hLarp7 are shown as helices and sheets above and below the sequence, respectively. Locations of RNP1 and RNP2 in canonical RRMs are indicated above the sequence in brackets for reference. Conserved residues determined by this and previous work to contribute to RNA binding and helix α3–β sheet interaction are sky blue and orange (for β-sheet) and red (for α3), respectively. (C) Cartoon comparing the conserved RNA binding features of a canonical RRM (left) and xRRM (right), based in part on this work. Conserved residues that bind RNA are shown in sky blue and that contribute to the helix α3–β sheet interaction are shown in red and orange, respectively. For the canonical RRM, circled residues are those that most commonly interact with RNA. For the xRRM, the length of helix α3 (shown in grey) varies. Variable regions not involved in RNA binding (β4′ strand and loops) not pictured
The fission yeast LARP7 protein Pof8 was predicted to contain an xRRM at its C-terminus based on sequence similarity to Tetrahymena p65 and human Larp7 [15–17]. Loss of Pof8 severely reduces telomerase activity in vivo and results in critically short telomeres, ultimately leading to uncapped chromosomes and chromosome end fusions [16]. Truncation constructs deleting the putative RRM2 domain reduced TER1–TERT assembly, TER1 levels, and had a similar phenotype to Pof8 knock-down [16]. Previously, association of a LARP7 protein with telomerase RNA was thought to be unique to ciliates, whose TER is an RNA polymerase III transcript with a native 3′ UUU-OH terminus that binds the La module. Although this 3′-end sequence is absent in RNA polymerase II mRNA transcripts, the intron-encoded fission yeast TER1 is spliced resulting in a 3′ UUU-OH terminus [46,47]. However, the Lsm2-8 proteins bind to this region [48], ostensibly preventing La module interaction with the 3′ terminus. The complete secondary structure of the 1213 nt TER1 has not been established and the binding site(s) for Pof8 is unknown. Although less well characterized, there is evidence that hLarp7 plays a role in human telomerase abundance and activity [49], suggesting that LARP7s may be broadly involved in telomerase function.
Here we present a 1.35 Å resolution crystal structure and solution NMR study of the C-terminal domain of Pof8, which reveals features consistent with an xRRM, despite having a shorter helix α3. We compare the structure with p65 and hLarp7 xRRM structures in the absence and presence of their cognate RNAs [31,42,50], as well as with genuine La RRM2 [44], to refine the sequence and structural features of the xRRM class of atypical RRMs, and we propose the RNA binding mode in Pof8.
Results
Crystal structure of the Pof8 C-terminal domain
Based on sequence homology and predicted secondary structure, a La module and RRM2 were predicted at Pof8 N- and C-termini, respectively [15–17] (Fig. 1A). In particular, the C-terminal RRM2 domain has significant sequence homology to the xRRM domains in Tetrahymena p65 [31] and human Larp7 [50] (Fig. 1B). Based on sequence alignment and known RRM topology (Fig. 1C), a Pof8 construct containing residues 282–402 was cloned into a pET vector containing an N-terminal His6-SUMO fusion protein, and recombinant protein was expressed, purified, and screened by solution NMR spectroscopy to identify the presence of a folded domain. The Pof8 construct was crystallized, with crystals diffracting to 1.35 Å in space group P3121 (Table 1). The structure was determined by heavy atom (Hg) phasing using PCMBS-soaked crystals. The electron density of the protein was visible up to 2σ, with weak density for residues 376–378 and no density was observed for N-terminal residues 282–287 or C-terminal residue 402 (Fig. 2A). The crystal structure of Pof8 C-terminal domain revealed an atypical RRM with an overall β1-α1-β2-β3-α2-β4-α3 topology. A four-stranded antiparallel β-sheet consisting of β4 (aa 384–387), β1 (aa 294–298), β3 (aa 339–344) and β2 (aa 329–332) forms the front face with helices α1 (aa 306–320) and α2 (aa 347–359) packed on the back of the β-sheet to form the hydrophobic core and helix α3 (aa 391–401) on top of the β-sheet (Fig. 2B,C). There are four loops: β1-α1 (aa 299–305), α1-β2 (aa 321–328), β2-β3 (aa 333–338), β3-α2 (aa 345–346), α2-β4 (aa 360–383), and β4-α3 (aa 388–390). Helix α3 (not present in canonical RRMs) is positioned orthogonal to the long axis of the β-strands, and lies across where the canonical RNP1 and RNP2 residues are normally found. Canonical RNP1 and RNP2 sequences on β3 and β1, respectively, are absent and there is a Y330–I331–D332 sequence on β2 (RNP3) and R343 on β3, consistent with an xRRM (Figs. 1C, 2D) [41].
Table 1.
Crystallography statistics for S. pombe Pof8 xRRM
| Native | PCMBS soak | ||
|---|---|---|---|
| Data Collection | |||
| Wavelength (Å) | 0.9791 | 0.9791 | |
| Space Group | P31 2 1 | P31 2 1 | |
| Cell dimensions | |||
| a, b, c (Å) | 57.36, 57.36, 70.3 | 56.01, 56.01, 65.29 | |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | |
| Resolution (Å) | 1.35 | 1.2 | |
| Rmerge | 0.044 (0.68) | 0.055 (0.5) | |
| Rmeasure | 0.049 (0.78) | 0.054 (0.58) | |
| Rpim | 0.022 (0.36) | 0.019 (0.17) | |
| I/σ | 16.1 (1.95) | 17.1 (3.92) | |
| Completeness (%) | 99.1 (98.6) | 96.2 (91.0) | |
| No. of total reflections | 136156 | 162043 | |
| No. of unique reflections | 29661 | 34507 | |
| Multiplicity | 4.6 (4.3) | 10.0 (9.3) | |
| Wilson B factor | 19.47 | 12.76 | |
| CC1/2 | 0.999 (0.83) | 0.998 (0.898) | |
| CC* | 1 | 1 | |
| Refinement | |||
| Resolution (Å) | 49.67–1.35 | 48.506–1.2 | |
| No. of reflections | 29647 | 69061 (36157) | |
| Rwork/Rfree | 0.1629/0.1940 | 0.2216/0.2313 | |
| No. of atoms | 1019 | 901 | |
| Protein | 936 | 888 | |
| Nitrate | 4 | 4 | |
| Water | 78 | 13 | |
| Average B factors | 17 | 17 | |
| r.m.s.d | |||
| Bond lengths (Å) | 0.005 | 0.007 | |
| angles (°) | 0.713 | 0.849 | |
| Ramachandran plot | |||
| Favoured (%) | 98.23 | 99.06 | |
| Allowed (%) | 1.77 | 0.94 | |
| Outliers (%) | 0 | 0 |
Figure 2.
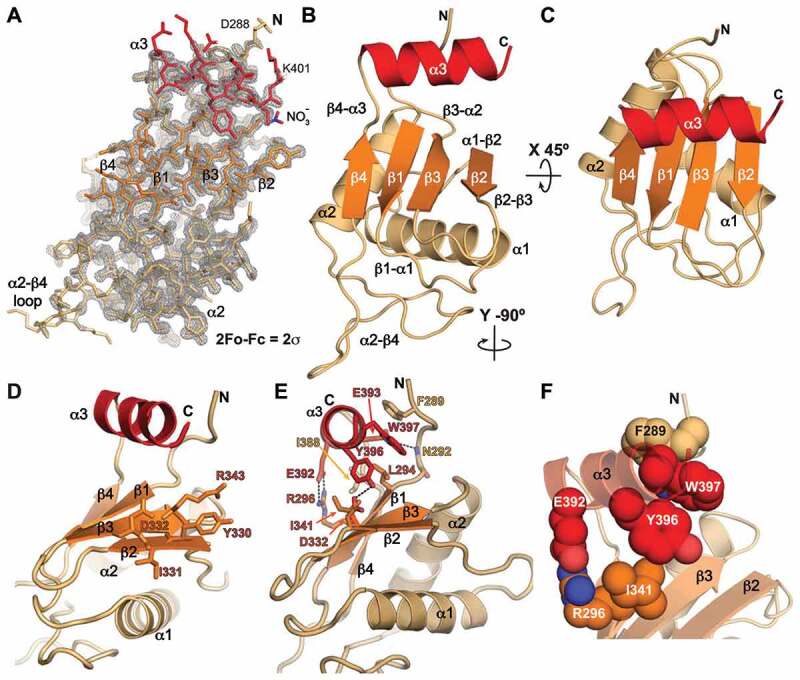
Crystal structure of Pof8 RRM2 at 1.35 Å. (A) 2 Fo-Fc electron density map with crystal structure model shown in stick representation. The map is contoured at 2σ. (B, C) Two views of ribbon representation of the Pof8 xRRM (residues 288–402) crystal structure. The β-sheet is coloured orange, helices α1 and α2 are tan, and helix α3 is red. (D) Ribbon representation with equivalent conserved residues involved in RNA binding in p65 and hLarp7 xRRMs shown as sticks. (E) Ribbon representation with conserved residues involved in stabilizing the α3–β-sheet interactions shown as sticks. (F) Ribbon representation with conserved residues involved in stabilizing the α3–β-sheet interactions shown as space fill to highlight stacking interactions
The Pof8 RRM2 helix α3 has three turns and is positioned through electrostatic and hydrophobic interactions with the β-sheet and the N-terminal residues (aa 289–294) (Fig. 2E,F). There is a salt bridge between R296 (β1) and E392 (α3) side-chains, a hydrogen bond between the N292 backbone amide (N-tail) and the E393 side-chain (α3), and a hydrogen bond between D332 (β2) and Y396 (α3) side-chains. Residues L294 (β1), I341 (β3), I388 (β4-α3 loop), and Y396 (α3) form hydrophobic contacts at the β-sheet – helix α3 interface. The I341 (β3) side-chain stacks below the aromatic ring of Y396 (α3), and the W397 (α3) side-chain has π-π stacking with Y396 (α3) and F289 (N-tail) side-chains (Fig. 2E,F). Overall, Pof8 RRM2 has all of the characteristic features of an xRRM as previously described except for the absence of residues beyond helix α3 that could extend it upon RNA binding.
Conformational flexibility within the Pof8 RRM2
The global and local dynamics of the Pof8 RRM2 were investigated with solution NMR, using the Pof8 construct used for crystal studies. A 2D 1H-15N Heteronuclear Single Quantum Coherence (HSQC) spectrum of the backbone amides shows a well-dispersed set of peaks indicative of a well-folded protein (Fig. 3A). Backbone resonance assignments could be completed for the majority of the Pof8 RRM2, with the exception of α2-β4 loop residues Q350-L383 due to weak peak intensities caused by line broadening, indicative of conformational exchange. Close inspection of the 2D 1H-15N HSQC spectrum of the backbone amides revealed a set of uniformly low-intensity additional peaks that appear to be due to peak doubling, likely caused by a second, lowly populated species in slow exchange with a major species (Fig. 3A,B). 3D resonance assignment confirmed the identities of about half of these peaks. Peak doubling was only observed for the backbone amides. Nearly all of the doubled peaks where the weak peak could be assigned were from residues at the β-sheet – helix α3 interface (T290, N292, K298, E339, I341, E386, E393, W397) or the helix α3 C-terminus (R398, M399, L400, K401) (Fig. 3C). Other weak peaks whose identities could be inferred from proximity to an assigned peak (e.g. L294, T295, R296) also correspond to residues at the β-sheet – helix α3 – N-tail interfaces (Fig. 3A,C).
Figure 3.
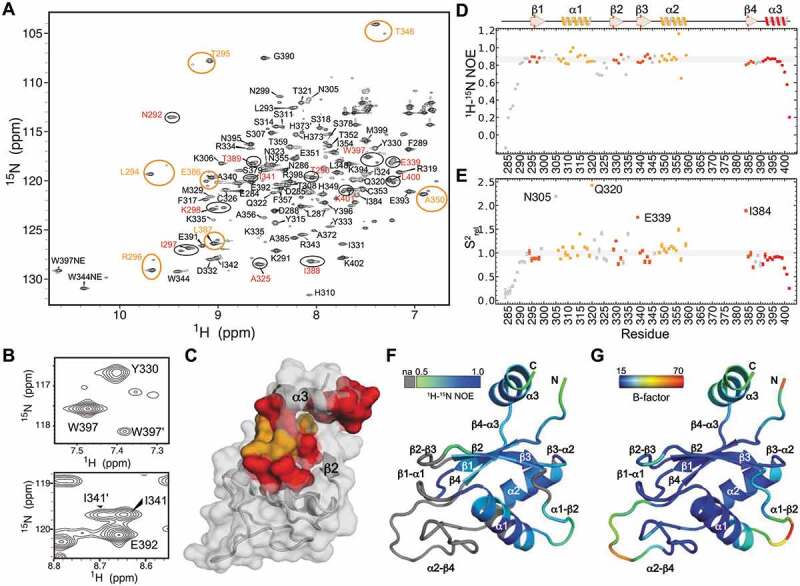
NMR characterization of Pof8 RRM2 (A) 1H-15N HSQC spectrum of Pof8 xRRM. Pairs of amide peaks that are doubled are circled; those for which the minor peak is assigned are labelled in red and inferred by proximity to major peak in orange. (B) Expanded regions from panel A showing peak doubling of amides W397 (helix α3) and I341 (β3). (C) Distribution of residues whose amides show peak doubling, mapped onto the structure. Assigned residues are red, residues with inferred assignments are orange. (D) Plot of heteronuclear NOE values vs residue number. Secondary structure elements are indicated above. (E) Plot of normalized order parameters vs residue number. Residues with values greater than 1.5 are labelled inset. (F) 1H-15N heteronuclear NOE values mapped onto a ribbon structure, with scale ranging from grey (na, not available), 0.5 (green) to 1.0 (blue). (G) Crystal structure B-factor mapped on a ribbon structure, scale ranging from 15.00 (blue) to 70.00 (red)
To determine the conformational flexibility in the major populated conformation of the Pof8 RRM2, we measured 1H-15N heteronuclear nuclear Overhauser effects (NOEs) (Fig. 3D). Significantly reduced values are observed for the backbone amides of N-terminal residues E284-F289 and C-terminal residues L400-K402, indicating that the N- and C-termini are highly flexible relative to the globular RRM fold. This data is consistent with the X-ray crystal electron density map that had missing density for N-terminal residues K282 to L287 and C-terminal residue K402. Reduced 1H-15N heteronuclear NOE values are also observed for residues Q322-C326 (α1-β2 loop) and residues R334-A340 (β2-β3 loop). To further probe these dynamics, we measured 15N R1 and R2 spin relaxation parameters and computed relative order parameters for backbone amide residues [51] (Fig. 3E and Supplemental Table 1). The relative order parameters (S2rel) describe the relative degree of order with values ranging from zero, representing minimum order, and one, representing maximum order within the molecule. Consistent with 1H-15N heteronuclear NOE values, reduced values are observed for N-terminal residues E284-F298 and C-terminal residues L400-K402. However, S2rel values greater than one were observed for several residues including N305 located at the β1-α1 loop; Y315 and Q320 located on the helix α1 surface; I324 located in the α1-β2 loop; K335 and E339, located at the β2-β3 loop and β3 edge; and A356, T359, and I384 adjacent to the α2-β4 loop (Fig. 3E). Inspection of R1 and R2 values indicates that the elevated S2rel values are due to elevated R2 values, likely due to contributions from chemical exchange between two or more states. Consistent with chemical exchange occurring at these residues, 1H-15N heteronuclear NOE, R1, and R2 values could not be measured for the β1-α1 loop, β2-β3 loop, and α2-β4 loop due to extensive line broadening, likely due to chemical exchange (Fig. 3D,E). Consistent with NMR data, the crystallographic B-factors indicate that the hydrophobic core is stable and that the N-terminus, α1-β2 loop, β2-β3 loop, α2-β4 loop, and helix α3 have higher B-factors (Fig. 3F-G). In addition, although the α2-β4 loop has higher B-factors, there is clear albeit weak electron density for it (Fig. 2A), while these residues could not be assigned due to conformational exchange. We attribute these differences to crystal contacts between the α2-β4 loop and another molecule in the crystal lattice. Together, these data indicate that for Pof8 RRM2, the α2-β4 loop is flexible in solution and helix α3 appears to exist in two conformations: a major species where helix α3 is positioned on the β-sheet as in the crystal structure and a second minor species that may have alternate or unstable positioning of helix α3 relative to the β-sheet.
Discussion
Structural comparison of Pof8 with LARP7 xRRMs
The combination of helix α3, that lies across the β-sheet and over where the absent RNP2 and RNP1 would be, and conserved RNP3 sequence on the Pof8 RRM2 is unique to the xRRM class of atypical RRMs, and led us to hypothesize that the Pof8 RRM2 is an xRRM. The human genuine La protein also contains an atypical RRM2 that we previously predicted to be an xRRM [50]. Below we compare the conserved structural and RNA recognition features for the existing examples of defined xRRMs (p65 and hLarp7), Pof8 RRM2, and human La protein (hLa) RRM2.
Pof8, p65, hLarp7, and hLa RRM2 vary in the lengths of the loops between β strands and helices, presence or absence of a β4′, length of helix α3, and number of residues following α3 (Fig. 4). For all, the beginning of helix α3 is enriched in acidic residues that face the β-sheet surface at β4 and β1, followed by aromatic and hydrophobic residues over β3 and β2, respectively (Figs. 1B, 4). Comparison of the three LARP7 RRM2 structures revealed that they have several interactions in common between the β-sheet and helix α3. First, there is a salt bridge between a conserved basic (K/R) residue in the middle of β1 and an acidic residue in the first turn of helix α3, which is conserved in Pof8 (Fig. 4A,E) and p65 (Fig. 4B,F) but not present in hLarp7 (Fig. 4C,G). Second, there is a hydrophobic patch between an I/L residue in β1, an I/L residue in the β4-α3 loop, and conserved Y-W residues in the second turn of helix α3 (Fig. 4A-C,E-G). These hydrophobic contacts may stabilize the β4-α3 loop and help to orient helix α3 on the β-sheet. Third, there is a stacking interaction between a conserved hydrophobic/aromatic residue in β3 with the conserved Y in the second turn of helix α3 (Figs. 2F, 4A-C,E-G). The contacts from β1 and β3 with residues on the first two turns of helix α3 anchor it on the β-sheet. Interestingly, the conserved residues in β1 and β3 that interact with helix α3 are located in the positions of RNP2 and RNP1, respectively, in the canonical RRM (Fig. 1C) [52,53]. It appears that while the xRRM lacks RNP1 and RNP2 sequences that interact with RNA, these sites on the β-sheet have adapted to interact with and stabilize helix α3 on the β-sheet surface.
Figure 4.
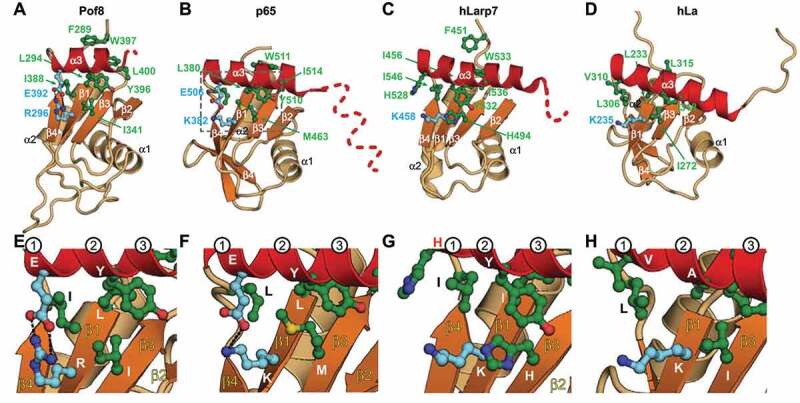
Comparison of structures of LARP7 xRRMs and hLa protein RRM2: helix α3–β sheet interactions. Ribbon representations of RRM2 from (A) S. pombe Pof8, crystal structure (this work), (B) Tetrahymena p65, crystal structure (PDB ID 4EYT) and (C) human Larp7, solution NMR structure (PDB ID 5KNW), (D) human La, solution NMR structure (PDB ID 1OWX). Dashed line indicates disordered residues missing in the density and/or that become helical on binding RNA. (E-H) Zoomed regions of (A-D) highlighting the conserved residues important for α3–β-sheet interactions. The first three turns of helix α3 are numbered. Secondary structure motifs are coloured as in Fig 2. Residue side chains important for α3–β sheet interactions are shown as ball and stick and coloured green (hydrophobic) and cyan (charged)
RNA-binding determinants of the xRRM
Based on the sequence and structural similarity of Pof8 to p65 and hLarp7 xRRMs (Fig. 1B,C), comparison to structures of xRRM-RNA complexes [31,42], and effects of Pof8 amino acid substitutions and deletions on TER1 binding and abundance in vivo and telomere lengths [15,16], we propose a putative RNA-binding interface. Residues involved in RNA binding are shown as sticks on the ribbon model for p65 and hLarp7 (Fig. 5B, C) [31,42]. p65 and hLarp7 xRRMs share a conserved binding pocket formed by residues on the third and fourth turn of helix α3 and the β3-β2 strands (Fig. 5G). The binding pocket recognizes 2 or 3 nts, respectively, including one Gua, that insert into the binding pocket between helix α3 and β3-β2 (Fig. 5E-G). The L on the third turn of helix α3 lies above the Gua, forming the ceiling of the binding pocket (Fig. 5G). Pof8 RRM2 has a near-identical conserved binding pocket, including L on helix α3 turn 3 (Fig. 4A, Fig. 5A). The RNP3 Y-X-D sequence on β2 and R on β3, previously identified as determinants of RNA binding [41], are identical among Pof8, p65, and hLarp7.
Figure 5.
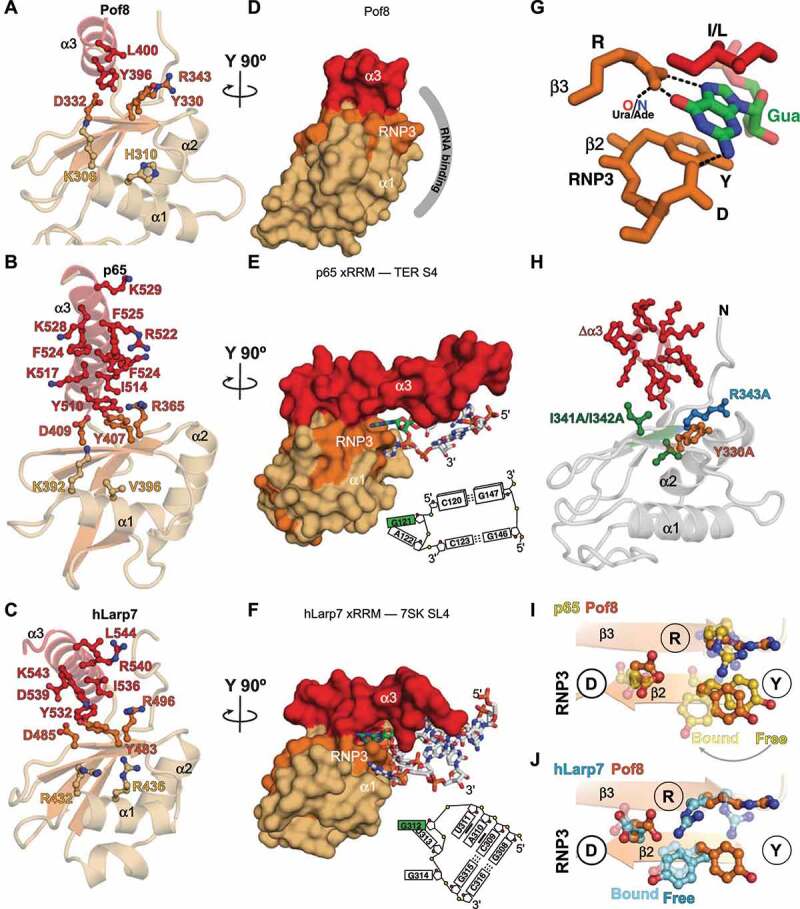
Comparison of structures of LARP7 xRRMs: RNA binding determinants (A-C) Ribbon representations of crystal structures of (A) Pof8 xRRM (this work), (B) p65 xRRM bound to telomerase RNA stem 4 (PDB ID 4ERD), (C) hLarp7 xRRM bound to 7SK stem-loop 4 (PDB ID 6D12). Side chains that interact (B, C) or are predicted to interact (A) with RNA are shown as sticks; (D-F) Surface representations of (D) Pof8 xRRM (this work), (E) p65 xRRM bound to telomerase RNA stem 4 (PDB ID 4ERD), (F) hLarp7 xRRM bound to 7SK stem-loop 4 (PDB ID 6D12), rotated 90° from (A-C). The proposed RNA interacting surface of Pof8 is shown by the grey arc, and for p65 and hLarp7 the interacting RNA nucleotides are shown as sticks, with Gua in green, and an RNA schematic is shown at right; (G) Stick representation of Gua recognition in the xRRM binding pocket. The interaction between β3 R and a Ura O4 (e.g. hLarp7) or Ade N7 (e.g. p65) is also indicated; (H) Ribbon representation of Pof8 with residues whose substitution to alanine or deletion affect RNA binding in vitro or TER abundance and telomere length in vivo [15–17] shown as sticks. Δα3 is deletion of the entire helix. I341A/I342A is a double substitution. (I-J) Ribbon and stick representation illustrating position of RNP3 Y on (I) RNA free and bound p65 xRRM (gold) and RNA free Pof8 xRRM (orange) and (J) RNA free and bound hLarp7 xRRM (argon blue) and RNA free Pof8 xRRM (orange)
For p65 and hLarp7, the RNP3 and R on β3 interact extensively with single-stranded RNA nucleotides, and alanine substitution of either Y (β2, RNP3) or R (β3) significantly impairs binding affinity to substrate RNA [31,42]. The conserved R on β3 has two hydrogen bonds to the Hoogsteen edge of Gua while the conserved D on RNP3 hydrogen bonds to the Watson-Crick face. The Gua is further stabilized in the binding pocket by stacking interactions with helix α3 conserved I (I/L) above and conserved RNP3 Y residue below. The β3 R also hydrogen bonds to AdeN7 (p65) or UraO4 (hLarp7) (Fig. 5G). In p65, the RNP3 Y undergoes a large change in position between the RNA-free and RNA-bound states (Fig. 5I), while in hLarp7 the position of RNP3 Y is already near to the bound state position prior to RNA binding (Fig. 5J). For the RNA-free Pof8 structure reported here, the position of RNP3 Y is close to that of free p65 (Fig. 5I,J).
In p65 and hLarp7, the longer helix α3 also binds to two or one base pairs, respectively. For p65, the long helix inserts between two stems on either side of the Gua-Ade bulge residues recognized in the binding pocket described above, causing a large bend between helices, while for hLarp7 a base pair at the top of the hairpin loop is recognized (Fig. 5E,F). We speculate that in Pof8, the two conserved lysines at the end of helix α3 might hydrogen bond to a base pair. Based on the above analysis, we conclude that Pof8 has a binding pocket for a Gua (or possibly a Ura) and at least one other nucleotide analogous to that for hLarp7 and p65.
Fig. 5H maps mutations and deletions that have been shown to affect Pof8 function in vivo [15–17] onto the crystal structure of Pof8. Deletion of helix α3 (residues 390–402) reduced TER1 levels in a similar manner as Pof8 deletion and caused telomere shortening [16], and I341A-I342A substitution, which would remove the hydrophobic contact between I341 (β3) and Y396 (α3), also resulted in shortened telomeres, reduced TER1 levels, and reduced co-immunoprecipitation of Pof8 with TER1 [15]. These results indicate that helix α3 is essential for RNA binding and stability in vivo and are consistent with a requirement for stable positioning of helix α3 to form the RNA binding pocket. Single residue substitutions Y330A (RNP3) and R343A (β3) had similarly deleterious effects in vivo compared to deletion of helix α3, deletion of the RRM, or full-length knock-down [16], consistent with their predicted importance for nucleotide recognition in the putative Pof8 RNA binding pocket (Fig. 5D).
Finally, we note that in Pof8, p65, hLarp7, and hLa, helix α1 contains a conserved basic (K/R) residue (Fig. 1B) that in hLarp7 interacts with RNA (Fig. 5F) [42]. A similar interaction may be present in the p65–RNA complex, but is not definitive since the sequence in this position in the crystal structure is not native [31]. We propose that this interaction is common to the LARP7 proteins, and may provide further RNA binding affinity and/or specificity. Based on our analysis of the structures, sequence, and mutagenesis data, we conclude that the Pof8 RRM2 is an xRRM, as discussed further below.
Comparison of LARP7 xRRMs to hLa RRM2
La protein generally binds RNA polymerase III transcripts after transcription, but does not remain associated with the RNA, in contrast to LARP7s. The hLa RRM2 has known chaperone activity and has broad RNA substrate recognition [2,10]. Compared to LARP7 RRM2s, hLa RRM2 has an extensive hydrophobic interface between helix α3 and the β-sheet, with the stacking and salt bridge interactions observed for LARP7 xRRMs absent (Fig. 4D,H). The hLa RRM2 helix α3 lies closer to the β-sheet and has an additional contact between β2 and helix α3. hLa has an RNP3 (W261-I262-D263) characteristic of xRRMs, but the R on β3 that is conserved among LARP7 xRRMs is replaced by an L (Figs. 1B, 4D). This R contributes to nucleotide specificity of LARP7 xRRMs through hydrogen bonding to bases, and its replacement in hLa protein with an L might explain the lower binding affinity and lack of specificity of hLa RRM2 vs LARP7 xRRMs [54,55]. As there are no structures to date of hLa RRM2 in complex with RNA, the binding mode is unknown. However, overall hLa RRM2 shares all essential features of an xRRM except for the conserved R (β3). We propose that hLa RRM2 is an xRRM with reduced binding affinity in accordance with its function in binding multiple substrates.
A refined definition of xRRM
Pof8 and p65 are constitutive components of S. pombe and Tetrahymena telomerase, required for biogenesis and assembly of TER with TERT. The structure of Pof8 RRM2 reported here is the third example from the LARP7 family, and provides new insights into RNA binding by these domains and those of the related La proteins. The xRRM was first structurally characterized in p65 in the absence and presence of its cognate RNA [31], and subsequently in hLarp7 [42,50]. We note that the RRM2 of Larp7 and La protein have recently been alternatively designated as RRM2α [2]; based on the work reported here we conclude that the xRRM and RRMα are the same RRM variant. Comparing the three LARP7 RRM2 structures shows that they share all the features of an xRRM first defined for p65 except for the strikingly variable length and sequence of helix α3 residues extending past the β-sheet (Fig. 4). Although in the absence of RNA the p65 helix α3 is a similar length to that of Pof8, upon RNA binding the helix is extended from four to eight turns (Fig. 4B). In free hLarp7 α3 has 5 turns and is extended by an additional turn on binding RNA. For both p65 and hLarp7 xRRMs, truncation of these extra turns of helix α3 resulted in significantly reduced binding affinity to substrate RNA [31,50], indicating that the region of helix α3 that extends beyond the β-sheet contributes to high affinity binding.
The sequence differences in helix α3 between p65 and hLarp7 appear to aid in substrate discrimination; the aromatic residues in p65 helix α3 insert orthogonal to the helical axis in the TER SL4 major groove to induce a sharp bend between the helices bracketing the GA bulge, while the basic residues in the hLarp7 helix α3 interact with and insert parallel to the major groove at the RNA apical loop [31,42]. The Pof8 helix α3 is three turns long and ends at the protein C-terminus with two consecutive lysines, the last of which is disordered. It seems likely that the short Pof8 helix α3 will result in a weaker binding affinity compared to the xRRMs of p65 (30 nM) [31] and hLarp7 (100 nM) [42,50] to their cognate substrates. Additional affinity may be provided by interactions between conserved residues on helix α1 and RNA. In summary, this work has defined the structure of the recently discovered yeast telomerase holoenzyme protein Pof8 C-terminal domain and provided a more definitive description of the xRRM and its conserved versus variable determinants of RNA specificity and affinity.
Methods
Protein expression and purification
The gene encoding Pof8 RRM2 (residues 282–402) was codon optimized for E. coli and synthesized into a gBlock (Integrated DNA Technologies) and subsequently cloned into a pET-His6-SUMO vector using Gibson ligation (New England Biolabs) [56]. The recombinant plasmid was subsequently transformed into Escherichia coli BL21 (DE3) cells (Agilent Technologies) for protein expression. Bacterial cultures were grown in LB media with 50 μg/ml kanamycin at 37°C until OD600 reached 0.6, then induced with a final concentration of 0.5 mM IPTG for 18–24 h at 18°C. Cells were harvested, sonicated in resuspension buffer (10% glycerol, 50 mM Tris pH 7.5, 750 mM NaCl, 30 mM imidazole, 3 mM NaN3, 0.1% Triton-X 100, 1 mM 2-carboxyethyl phosphine (TCEP), sonicated and centrifuged at 17,000 rpm for 45 mins. Clarified cell lysate containing His6-tagged protein was loaded onto a Ni–Sepharose affinity column (HisTrap HP; GE Healthcare). The column was washed with wash buffer (5% glycerol, 50 mM Tris pH 7.5, 750 mM NaCl, 30 mM imidazole, 3 mM NaN3, 1 mM TCEP), then eluted with elution buffer (5% glycerol, 50 mM Tris pH 7.5, 750 mM NaCl, 300 mM imidazole, 3 mM NaN3, 1 mM TCEP) to release bound His6-SUMO-tagged Pof8 RRM2. The eluate was incubated with SUMO protease (expressed and purified in-house from a pET28a vector with an N-terminal His6 tag) (approximately 0.5 mg protease to 5–10 mg Pof8 RRM2) for 3–4 hours at ambient temperature while dialysing against a buffer containing 20 mM Tris pH 7.5, 250 mM NaCl, 5 mM β-mercaptoethanol. The cleaved Pof8 RRM2 was purified by loading the cleavage reaction onto the Ni–Sepharose affinity column and collecting the flow-through. After further purification by size-exclusion chromatography (HiLoad 26/600 Superdex 75; GE Healthcare) in crystallization buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 1 mM TCEP) or NMR buffer (20 mM NaPO4 pH 6.1, 50 mM KCl, 1 mM TCEP), protein peak fractions were measured, pooled and concentrated using 3 KDa cut-off Amicon filters (Millipore Sigma). Unlabelled protein was purified from cells grown in LB media (Fisher Scientific) and uniformly 15N- or 15N,13C- labelled proteins were purified from cells grown in M9 minimal media with 15N ammonium chloride and/or 13C D-glucose (Cambridge Isotope Labs) as the sole nitrogen and/or carbon source, respectively.
NMR spectroscopy
NMR samples were concentrated to 0.1–0.8 mM in NMR buffer plus 5% D2O. NMR experiments were performed at 298 K on AVANCE 800 MHz Bruker spectrometer equipped with HCN cryoprobe. Nearly complete backbone (N, H, C, Cα, Cβ, Hα, Hβ) assignments were obtained for all residues except residues in the α2-β4 loop (E361-L383), using standard triple resonance assignment experiments [57,58]. Briefly, 3D CBCACONH, HNCACB, HNCA, HBHACONH, HNCO and HNCACO experiments from the Bruker experimental suite were acquired on Topspin 4.0.7 (Bruker) using non-uniform sampling with 25% sparsity and a poisson-gap sampling schedule [59]. The experiments were processed with Topspin 4.0.7 and analysed using NMRFAM-SPARKY 1.414 [60] to assign backbone resonances [61,62]. After partial manual assignment, the I-PINE web server was used to validate assignments and obtain additional assignments for residues in the β1-α1 loop L300 and T304, α1-β2 loop E327, β2-β3 loop K335-T338, β3-α2 loop K345, and α2 residue R358 [63]. The 15N- 3D NOESY experiment (noesyhsqcetf3gp3d) from the Bruker experimental suite was acquired on Topspin 4.0.7 (Bruker) with a mixing time of 120 ms to obtain NOE crosspeaks to further validate assignments. For I-PINE, atomic coordinates from the crystal structure, pre-assignments from manually assigned resonances, and peak lists from 1H-15N HSQC, 1H-13C HSQC, HNCA, HNCACB, CBCACONH, CCONH, HBHACONH, HNCO, HNCACO, and 15N-NOESY spectra were included as input. The 1H-15N heteronuclear NOE experiment (hsqcnoef3gpsi) from the Bruker experimental suite was acquired on Topspin 4.0.7 (Bruker), processed with NMRPipe [64], analysed with NMRFAM-SPARKY 1.414 [60] and Office Excel (Microsoft), and plotted using matplotlib. The 15N T1 (hsqct1etf3gpsi3d) and T2 spin relaxation experiments (hsqct2etf3gpsitc3d) from the Bruker experimental suite were acquired on Topspin 4.0.7 (Bruker), with interscan delays of 2.5 s and 3 s for T1 and T2 experiments, respectively. For T1, the relaxation delays used were: 20 ms (in duplicate), 60 ms, 200 ms, 400 ms (in duplicate), 600 ms, 800 ms, and 1 s (in duplicate). For T2, the relaxation delays were: 17 ms (in duplicate), 33.9 ms, 67.8 ms, 199 ms (in duplicate), 204 ms, 237 ms, and 271 ms (in duplicate). Data was processed with Topspin 4.0.7 and analysed with NMRFAM-SPARKY 1.414 and Office Excel (Microsoft). The measured T1 and T2 values were converted to R1 and R2 values and used to compute relative order parameters [51] using the equation S2 = (2R2 – R1). Relative order parameters (S2rel) were normalized to residue A340, located on β3 in the hydrophobic core.
Crystallization and data processing
The 2D 1H-15N HSQC of the Pof8 backbone amide resonances showed well-dispersed peaks, indicating a folded protein. The protein was concentrated to 25 mg/ml and crystallized in the hanging drop vapour diffusion method with reservoir solution 14% PEG 3350, 0.1 M NaNO3, 0.1 M Tris pH 8.5 and drops containing 1 μl of protein and 1 μl reservoir. Rod shaped crystals appeared in one day, were cryoprotected in reservoir solutions containing 35% PEG 3350 and flash frozen in liquid nitrogen. The native crystals diffracted to 1.35 Å. Crystals were soaked in 0.1 mM 4-chloro-mercuric-benzene-sulphonate (PCMBS) for 3–5 hours to obtain heavy atom (Hg) based experimental phases. All datasets were collected remotely at the Advanced Photon Source at Argonne National Labs, beamline 24-ID-C on a DECTRIS PILATUS 6 M detector. The Matthews coefficient suggested that there was one molecule of Pof8 RRM2 in an asymmetric unit. XDS/XSCALE [65,66] was used to index, integrate and scale the data. Conservative resolution limits were applied based on I/σ, CC1/2 and Rsym values at the highest resolution shell. SAD phasing was performed by SHELXC/D/E [67] and HKL2MAP [68]. Four Hg sites were identified by SHELXD. SHELXE was used to assign the handedness of the model, produce initial phases and solvent flattening. Phases were further improved with SHARP [69], and an initial model was generated with AUTOSHARP [70] and BUCCANEER [71]. Native crystal structure was solved by Molecular Replacement using the Hg-model. The model was initially refined using Phenix version 1.13.2998 [72] and Coot [73], with final refinement performed using PHENIX with TLS refinement [74].
Multiple sequence alignment
The protein sequence alignment was performed with MUSCLE v 3.8 (EMBL-EBI, https://www.ebi.ac.uk/Tools/msa/muscle/). Alignments are displayed and annotated using ESPript 3 (http://espript.ibcp.fr/) with residues coloured by residue similarity. p65 residues 418–460 (β2-β3 loop), which are not involved in RNA recognition, were omitted from the sequence alignment due to lack of sequence homology to other LARP7 xRRMs.
Supplementary Material
Acknowledgments
This work was supported by NSF grant MCB1517625 and NIH grant R35GM131901 to J.F. The authors acknowledge NMR equipment grants NIH S10OD016336 and S10OD025073 and DOE grant DE-FC0202ER63421 for partial support of NMR and X-ray core facilities. The authors thank M. Capel, K. Rajashankar, N. Sukumar, F. Murphy, I. Kourinov and J. Schuermann of Northeastern Collaborative Access Team (NE-CAT) beamline ID-24 at the Advanced Photon Source (APS) of Argonne National Laboratory, which are supported by NIH grants P41 RR015301 and P41 GM103403. Use of the APS is supported by DOE under Contract DE-AC02-06CH11357. The authors thank Dr. Lukas Sušac for help in the early stages of this work.
Funding Statement
This work was supported by the National Institutes of Health [R35GM131901]; National Science Foundation [MCB1517625]; National Institutes of Health [S10OD016336], [S10OD025073], [P41 RR015301], [P41 GM103403]; Department of Energy [DE-FC0202ER63421], [DE-AC02-06CH11357].
Disclosure of potential conflicts of interest
No potential conflict of interest was reported by the authors.
Highlights
The structure of the S. pombe LARP7 Pof8 C-terminal domain is an xRRM.
Ciliates, human, and fission yeast contain LARP7 proteins with xRRMs involved in telomerase biogenesis.
With three examples of xRRM structures, we refine the definition of xRRM.
Data availability
Atomic coordinates and structure factors have been deposited in the Protein Data Bank with the accession code 6TZN. NMR chemical shift assignments have been deposited in the BioMagnetic Resonance Bank with the accession code 50002. All other data generated or analyzed in this study are included in the published article or are available from the corresponding author upon reasonable request.
Author Contributions
R.B. purified, crystallized, solved and analyzed the crystal structure of Pof8 xRRM; C.D.E. collected and analyzed NMR data and the structure; R.C. cloned, expressed, and purified Pof8 xRRM and collected NMR data; R.P. collected and analyzed NMR data; J.F. analyzed the structure and supervised the project; R.B., C.D.E., and J.F. wrote the manuscript; all authors edited the manuscript..
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Bousquet-Antonelli C, Deragon JM.. A comprehensive analysis of the La-motif protein superfamily. RNA. 2009;15:750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Maraia RJ, Mattijssen S, Cruz-Gallardo I, et al. The La and related RNA-binding proteins (LARPs): structures, functions, and evolving perspectives. Wiley Interdiscip Rev RNA. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bayfield MA, Yang R, Maraia RJ. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs). Biochim Biophys Acta. 2010;1799:365–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Stefano JE. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3ʹ termini of RNA polymerase III transcripts. Cell. 1984;36:145–154. [DOI] [PubMed] [Google Scholar]
- [5].Pruijn GJ, Slobbe RL, van Venrooij WJ. Analysis of protein–RNA interactions within Ro ribonucleoprotein complexes. Nucleic Acids Res. 1991;19:5173–5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Belisova A, Semrad K, Mayer O, et al. RNA chaperone activity of protein components of human Ro RNPs. RNA. 2005;11:1084–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Teplova M, Yuan YR, Phan AT, et al. Structural basis for recognition and sequestration of UUU(OH) 3ʹ temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol Cell. 2006;21:75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Vakiloroayaei A, Shah NS, Oeffinger M, et al. The RNA chaperone La promotes pre-tRNA maturation via indiscriminate binding of both native and misfolded targets. Nucleic Acids Res. 2017;45:11341–11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bayfield MA, Vinayak J, Kerkhofs K, et al. La proteins couple use of sequence-specific and non-specific binding modes to engage RNA substrates. RNA Biol. 2019;1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Blewett NH, Maraia RJ. La involvement in tRNA and other RNA processing events including differences among yeast and other eukaryotes. Biochim Biophys Acta Gene Regul Mech. 2018;1861:361–372. [DOI] [PubMed] [Google Scholar]
- [11].Krueger BJ, Jeronimo C, Roy BB, et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36:2219–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Markert A, Grimm M, Martinez J, et al. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 2008;9:569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Aigner S, Postberg J, Lipps HJ, et al. The Euplotes La motif protein p43 has properties of a telomerase-specific subunit. Biochemistry. 2003;42:5736–5747. [DOI] [PubMed] [Google Scholar]
- [14].Witkin KL, Collins K. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev. 2004;18:1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Collopy LC, Ware TL, Goncalves T, et al. LARP7 family proteins have conserved function in telomerase assembly. Nat Commun. 2018;9:557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mennie AK, Moser BA, Nakamura TM. LARP7-like protein Pof8 regulates telomerase assembly and poly(A)+TERRA expression in fission yeast. Nat Commun. 2018;9:586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Paez-Moscoso DJ, Pan L, Sigauke RF, et al. Pof8 is a La-related protein and a constitutive component of telomerase in fission yeast. Nat Commun. 2018;9:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nguyen VT, Kiss T, Michels AA, et al. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. [DOI] [PubMed] [Google Scholar]
- [19].Yang Z, Zhu Q, Luo K, et al. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. [DOI] [PubMed] [Google Scholar]
- [20].Yazbeck AM, Tout KR, Stadler PF. Detailed secondary structure models of invertebrate 7SK RNAs. RNA Biol. 2018;15:158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Marz M, Donath A, Verstraete N, et al. Evolution of 7SK RNA and its protein partners in metazoa. Mol Biol Evol. 2009;26:2821–2830. [DOI] [PubMed] [Google Scholar]
- [22].He N, Jahchan NS, Hong E, et al. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol Cell. 2008;29:588–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Muniz L, Egloff S, Kiss T. RNA elements directing in vivo assembly of the 7SK/MePCE/Larp7 transcriptional regulatory snRNP. Nucleic Acids Res. 2013;41:4686–4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. [DOI] [PubMed] [Google Scholar]
- [25].Blackburn EH, Collins K. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Linger BR, Price CM. Conservation of telomere protein complexes: shuffling through evolution. Crit Rev Biochem Mol. 2009;44:434–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lai AG, Pouchkina-Stantcheva N, Di Donfrancesco A, et al. The protein subunit of telomerase displays patterns of dynamic evolution and conservation across different metazoan taxa. BMC Evol Biol. 2017;17:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chan H, Wang Y, Feigon J. Progress in human and tetrahymena telomerase structure determination. Annu Rev Biophys. 2017;46:199–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang Y, Feigon J. Structural biology of telomerase and its interaction at telomeres. Curr Opin Struct Biol. 2017;47:77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Aigner S, Lingner J, Goodrich KJ, et al. Euplotes telomerase contains an La motif protein produced by apparent translational frameshifting. Embo J. 2000;19:6230–6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Singh M, Wang Z, Koo BK, et al. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol Cell. 2012;47:16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Akiyama BM, Loper J, Najarro K, et al. The C-terminal domain of Tetrahymena thermophila telomerase holoenzyme protein p65 induces multiple structural changes in telomerase RNA. RNA. 2012;18:653–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Martino L, Pennell S, Kelly G, et al. Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module. Nucleic Acids Res. 2015;43:645–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Vinayak J, Marrella SA, Hussain RH, et al. Human La binds mRNAs through contacts to the poly(A) tail. Nucleic Acids Res. 2018;46:4228–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Uchikawa E, Natchiar KS, Han X, et al. Structural insight into the mechanism of stabilization of the 7SK small nuclear RNA by LARP7. Nucleic Acids Res. 2015;43:3373–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jiang J, Miracco EJ, Hong K, et al. The architecture of Tetrahymena telomerase holoenzyme. Nature. 2013;496:187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Egloff S, Van Herreweghe E, Kiss T. Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol Cell Biol. 2006;26:630–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nagai K, Oubridge C, Jessen TH, et al. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990;348:515–520. [DOI] [PubMed] [Google Scholar]
- [39].Oubridge C, Ito N, Evans PR, et al. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. [DOI] [PubMed] [Google Scholar]
- [40].Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. Febs J. 2005;272:2118–2131. [DOI] [PubMed] [Google Scholar]
- [41].Singh M, Choi CP, Feigon J. xRRM: a new class of RRM found in the telomerase La family protein p65. RNA Biol. 2013;10:353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Eichhorn CD, Yang Y, Repeta L, et al. Structural basis for recognition of human 7SK long noncoding RNA by the La-related protein Larp7. Proc Natl Acad Sci U S A. 2018;115:E6457–E6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Aigner S, Cech TR. The Euplotes telomerase subunit p43 stimulates enzymatic activity and processivity in vitro. RNA. 2004;10:1108–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jacks A, Babon J, Kelly G, et al. Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure. 2003;11:833–843. [DOI] [PubMed] [Google Scholar]
- [45].Ali N, Pruijn GJ, Kenan DJ, et al. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J Biol Chem. 2000;275:27531–27540. [DOI] [PubMed] [Google Scholar]
- [46].Box JA, Bunch JT, Tang W, et al. Spliceosomal cleavage generates the 3ʹ end of telomerase RNA. Nature. 2008;456:910–914. [DOI] [PubMed] [Google Scholar]
- [47].Rubtsova MP, Vasilkova DP, Naraykina YV, et al. Peculiarities of Yeasts and Human Telomerase RNAs Processing. Acta Naturae. 2016;8:14–22. [PMC free article] [PubMed] [Google Scholar]
- [48].Tang W, Kannan R, Blanchette M, et al. Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature. 2012;484:260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Holohan B, Kim W, Lai TP, et al. Impaired telomere maintenance in Alazami syndrome patients with LARP7 deficiency. BMC Genomics. 2016;17:749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Eichhorn CD, Chug R, Feigon J. hLARP7 C-terminal domain contains an xRRM that binds the 3ʹ hairpin of 7SK RNA. Nucleic Acids Res. 2016;44:9977–9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- [52].Martin-Tumasz S, Richie AC, Clos LJ 2nd, et al. A novel occluded RNA recognition motif in Prp24 unwinds the U6 RNA internal stem loop. Nucleic Acids Res. 2011;39:7837–7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Li H, Tong S, Li X, et al. Structural basis of pre-mRNA recognition by the human cleavage factor Im complex. Cell Res. 2011;21:1039–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Martino L, Pennell S, Kelly G, et al. Analysis of the interaction with the hepatitis C virus mRNA reveals an alternative mode of RNA recognition by the human La protein. Nucleic Acids Res. 2012;40:1381–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Brown KA, Sharifi S, Hussain R, et al. Distinct dynamic modes enable the engagement of dissimilar ligands in a promiscuous atypical RNA recognition motif. Biochemistry. 2016;55:7141–7150. [DOI] [PubMed] [Google Scholar]
- [56].Gibson DG, Young L, Chuang RY, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–U341. [DOI] [PubMed] [Google Scholar]
- [57].Reid DG, MacLachlan LK, Edwards AJ, et al. Introduction to the NMR of proteins. Methods Mol Biol. 1997;60:1–28. [DOI] [PubMed] [Google Scholar]
- [58].Cavanagh J. Protein NMR spectroscopy: principles and practice. 2nd ed. Amsterdam; Boston: Academic Press; 2007. [Google Scholar]
- [59].Hyberts SG, Takeuchi K, Wagner G. Poisson-gap sampling and forward maximum entropy reconstruction for enhancing the resolution and sensitivity of protein NMR data. J Am Chem Soc. 2010;132:2145–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lee W, Tonelli M, Markley JL. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Cavanagh J, Fairbrother WJ, Palmer AGI, et al. Protein NMR Spectroscopy. San Diego, CA: Academic Press; 1996. [Google Scholar]
- [62].Marion D. An introduction to biological NMR spectroscopy. Mol Cell Proteomics. 2013;12:3006–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lee W, Bahrami A, Dashti HT, et al. I-PINE web server: an integrative probabilistic NMR assignment system for proteins. J Biomol NMR. 2019;73:213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Delaglio F, Grzesiek S, Vuister GW, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. [DOI] [PubMed] [Google Scholar]
- [65].Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66:133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Pape T, Schneider TR. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J Appl Crystallogr. 2004;37:843–844. [Google Scholar]
- [69].Bricogne G, Vonrhein C, Flensburg C, et al. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr. 2003;59:2023–2030. [DOI] [PubMed] [Google Scholar]
- [70].Vonrhein C, Blanc E, Roversi P, et al. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. [DOI] [PubMed] [Google Scholar]
- [71].Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. [DOI] [PubMed] [Google Scholar]
- [72].Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. [DOI] [PubMed] [Google Scholar]
- [74].Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the Protein Data Bank with the accession code 6TZN. NMR chemical shift assignments have been deposited in the BioMagnetic Resonance Bank with the accession code 50002. All other data generated or analyzed in this study are included in the published article or are available from the corresponding author upon reasonable request.


