Abstract
Background and Aim:
Respiratory infection due to Mannheimia haemolytica and Pasteurella multocida are responsible for huge economic losses in livestock sector globally and it is poorly understood in ovine population. The study aimed to investigate and characterize M. haemolytica and P. multocida from infected and healthy sheep to rule out the involvement of these bacteria in the disease.
Materials and Methods:
A total of 374 healthy and infected sheep samples were processed for isolation, direct detection by multiplex PCR (mPCR), and antibiotic susceptibility testing by phenotypic and genotypic methods.
Results:
Overall, 55 Pasteurella isolates (27 [7.2%] M. haemolytica and 28 [7.4%] P. multocida) were recovered and identified by bacteriological tests and species-specific PCR assays. Significant correlation between the detection of M. haemolytica (66.6%) with disease condition and P. multocida (19.1%) exclusively from infected sheep was recorded by mPCR. In vitro antibiotic susceptibility testing of 55 isolates revealed higher multidrug resistance in M. haemolytica (25.9%) than P. multocida (7.1%) isolates. Descending resistance towards penicillin (63.6%), oxytetracycline (23.6%), streptomycin (14.5%), and gentamicin (12.7%) and absolute sensitivity towards chloramphenicol were observed in both the pathogens. The antibiotic resistance genes such as strA (32.7%) and sul2 (32.7%) associated with streptomycin and sulfonamide resistance, respectively, were detected in the isolates.
Conclusion:
The study revealed the significant involvement of M. haemolytica together with P. multocida in ovine respiratory infection and is probably responsible for frequent disease outbreaks even after vaccination against hemorrhagic septicemia in sheep population of Karnataka, southern province of India.
Keywords: antimicrobial susceptibility, isolation, Mannheima haemolytica, multiplex PCR, Pasteurella multocida
Introduction
Respiratory disease in ruminant population accounts for substantial economic losses to the livestock sector globally [1]. Pasteurella multocida and Mannheimia haemolytica are the main etiological agents of the disease known to cause 30% of deaths in feedlot cattle and acute outbreaks in sheep population resulting in huge mortality all across the world [2]. The clinical manifestations of the disease in small ruminants include dyspnea, pyrexia, dullness, reduced appetite, anorexia, rapid shallow respiration, profuse mucopurulent nasal, and ocular discharge and death within 12-24 h during the outbreaks [3]. P. multocida and M. haemolytica are Gram-negative, bipolar coccobacillus belonging to the family Pasteurellaceae of gamma proteobacteria [4,5]. Based on the capsular antigen, M. haemolytica has been classified into 12 serotypes (A1, A2, A5-A9, A12- A14, A16, and A17) [5] and based on capsular and somatic antigens, P. multocida is grouped into five serogroups (A, B, D, E, and F) and 16 serotypes [6].
The stress due to unfavorable environmental conditions, animal transportation and bacterial and viral infections are the predisposing factors for respiratory disease in ruminants [7]. The involvement of P. multocida and M. haemolytica in bronchopneumonia and presence of these bacteria as predictors of respiratory disease in ruminants are reported in various studies [8,9]. In ruminants, severity of respiratory infection leading to bronchopneumonia due to treatment failure as result of antimicrobial resistance has also been reported [10]. Various studies have noted the emergence of multidrug resistance to beta-lactams, tetracycline, streptomycin, sulfonamides, macrolides, and sulfamethazine in M. haemolytica and P. multocida [11,12].
Of the 512.05 million livestock population in India, the sheep and goats make up to 65.06 and 135.17 million, respectively [13]. Sheep and goats are extensively distributed all across the agro-ecological terrain of India contributing to the improvement of the socio-economic status of rural population [14]. Respiratory infection in sheep, the actual etiology and epidemiology are scarcely documented in the country. Frequent respiratory infection outbreaks in sheep even after vaccination against hemorrhagic septicemia (HS) in different parts of Karnataka, Southern India prompted us to investigate the etiology of the disease.
The study aimed to rule out the involvement of P. multocida and M. haemolytica in respiratory disease of sheep and to assess the phenotypic and genotypic antibiotic resistance for implementing appropriate therapeutic measures to control the disease.
Materials and Methods
Ethical approval
The study approved by Institutional Animal Ethics Committee, Indian Council of Agricultural Research-National Institute of Veterinary Epidemiology and Disease Informatics (ICAR- NIVEDI), Bengaluru, India and the authors have taken permission from the farm owners to publish the data. All applicable national, and institutional guidelines for the animal’s care were followed during the sample collection.
Sample collection and processing
A total of 374 (nasal-242 and lung-132) samples were collected from five different districts of Karnataka state from April 2015 to December 2016 (Table-1). Among the nasal samples, 59 were collected from apparently healthy sheep from five different flocks with no history of respiratory infection before one month of sample collection. Similarly, 183 nasal samples were collected from sheep exhibiting symptoms of respiratory infection (nasal discharge, lacrimal discharge, fever, weakness, and diarrhea) from 11 different flocks. The deep nasal swabs samples were collected into 2 ml brain heart infusion (BHI) broth. Lung tissue samples (n=132 [healthy-94 and lung with lesions-38]) were collected while slaughtering sheep at Bengaluru Municipal Abattoir, Bengaluru, India, in a tissue collection containers and transported to the laboratory.
Table-1.
Details of nasal and lung samples collected from different locations of Karnataka.
| No. | Place | Latitude | Longitude | Name of district | Healthy samples | Infected samples | Total samples |
|---|---|---|---|---|---|---|---|
| 1 | Chikkajala | 13.1715° N | 77.6356° E | Bangalore Urban | 13 | 35 | 48 |
| 2 | Channahalli | 13.1790° N | 77.6165° E | Bangalore Rural | 6 | 47 | 53 |
| 3 | Seethakempanahalli | 13.1729° N | 77.5195° E | Bangalore | 0 | 10 | 10 |
| 4 | Chintamani | 13.3862° N | 78.0603° E | Kolar | 9 | 0 | 9 |
| 5 | Kolar | 12.9984° N | 78.0737° E | Kolar | 0 | 42 | 42 |
| 6 | Kalavara | 13.5970° N | 74.7483° E | Chikkaballapur | 7 | 15 | 22 |
| 7 | Gauribidanur | 13.6112° N | 77.5170° E | Chikkaballapur | 24 | 34 | 58 |
| 8. | Municipal Abattoir Shivajinagar, Bangalore | 12.9857° N | 77.6057° E | Bangalore | 94 | 38 | 132 (lung samples) |
| Total number of samples | 153 | 221 | |||||
Bacterial isolation and identification
For isolation of M. haemolytica and P. multocida, nasal samples enriched for 18 h in BHI broth were inoculated onto Tryptic Soya Agar (TSA) supplemented with 5% sheep blood (blood agar) and 15 mg/mL of bacitracin and incubated at 37°C for 24 h [15]. For lung samples, the tissue near to the bronchus was directly inoculated onto blood agar with 15 mg/mL of bacitracin and incubated as mentioned above. The colonies obtained on blood agar were purified once again on blood agar and later on BHI agar. The colonies showing the morphology of M. haemolytica (β-hemolytic, white-grayish, medium-sized round, and non-mucoid) and P. multocida (non-hemolytic, greyish, medium-sized round, and non-mucoid) were processed separately for the identification by bacteriological tests (Gram’s staining, catalase, oxidase, indole, lactose fermentation, and growth on MacConkey lactose agar [MLA]) as per standard protocols [15].
Species confirmation by PCR
The genomic DNA was extracted from pure cultures by DNeasy kit as per the manufacturer’s protocol (Qiagen, Hilden, Germany). The quality and quantity of the extracted DNA was ascertained by NanoDrop2000 (Thermo Scientific, Waltham, USA) and on 0.8% agarose gel electrophoresis. Isolates were confirmed by species-specific multiplex PCR (mPCR) assay targeting lktD, HP; NZ_AASA01000080 and 16S rDNA [16] for M. haemolytica and species-specific PCR using KMT1SP6-KMT1T7 primers for P. multocida [17] (Table-2).
Table-2.
List of the primers used for species identification, capsular, and antibiotics resistance gene typing of M. haemolytica and P. multocida isolates.
| No. | Primer sets | Sequence (5’→3’) | Gene target | Fragment size (bp) | Reference |
|---|---|---|---|---|---|
| M. haemolytica identification | |||||
| 1 | Sod A | (F)AGCAGCGACTACTCGTGTTGGTTCG (R)AAGACTAAAATCGGATAGCCTGAACGCTG |
sodA | 143 | [18] |
| 2 | Lkt | (F)GCAGGAGGTGATTATTAAAGTGG (R)CAGCAGTTATTGTCATACCTGAAC |
lktD | 206 | [16] |
| 3 | Lkt2 | (F)CTCTCTTTAGAAAAGCTGGAAAC (R)TTTTGCCAAGTGGTGTATTGC |
lktD | 179 | |
| 4 | HP | (F)CGAGCAAGCACAATTACATTATGG (R)CACCGTCAAATTCCTGTGGATAAC |
Unknown/NZ_AASA01000080 | 90 | |
| 5 | 16S | (F)GCTAACTCCGTGCCAGCAG (R)CGTGGACTACCAGGGTATCTAAC |
16S rDNA | 304 | |
| P. multocida identification | |||||
| 6 | KMT1T7 KMT1SP6 | (F)ATC CGC TAT TTA CCC AGT GG (R)GCT GTAAAC GAACTC GCC |
KMT1 | 460 | [17] |
| Antibiotics resistance gene typing | |||||
| No. | Antibiotics / gene | Sequence (5’→3’) | Annealing temperature (°C) | Fragment size (bp) | Reference |
| 14 | Tetracyclin/tetH | (F)ATACTGCTGATCACCGT (R)TCCCAATAAGCGACGCT |
60 | 1076 | [20] |
| 15 | Oxytetracycline/ICE tetR | (F)CGGCTTGGGTTAATAATGGCG (R)ATAACGCGAAAAGCTTCCGC |
58 | 425 | |
| 16 | Penicillin/blaOXA-2 | (F)GCAGACGAACGCCAAGCGGA (R)CCCGCACGATTGCCTCCCTC |
64 | 625 | |
| 17 | Ampicillin/blaROB-1 | (F)AATAACCCTTGCCCCAATTC (R)TCGCTTATCAGGTGTGCTTG |
60 | 685 | |
| 18 | Sulfonamide/sul2 | (F)CCAATACCGCCAGCCCGTCG (R)TGCCTTGTCGCGTGGTGTGG |
64 | 489 | |
| 19 | Gentamicin/aadB | (F)TTACGCAGCAGGGCAGTCGC (R)GCGGCACGCAAGACCTCAAC |
66 | 551 | |
| 21 | Streptomycin/strA/strB | (F)AAGGCAAGGCGTTCGCGGTC (R)CCGGCGGCTGATCTGTCTGG |
64 | 506/586 | |
| (F)TCGCACCTGCTTGATCGCGG (R)GCTCGAATATGCCGGGGAGCG | |||||
| 20 | Chloramphenicol/catAIII | (F)ACCATGTGGTTTTAGCTTAACA (R)GCAATAACAGTCTATCCCCTTC |
64 | 470 | [21] |
M. haemolytica=Mannheimia haemolytica, P. multocida=Pasteurella multocida
Direct detection of P. multocida and M. haemolytica by mPCR
The mPCR was standardized for the rapid detection of P. multocida and M. haemolytica species using set of primers for P. multocida (KMT1SP6-KMT1T7 [460bp]) [17] and M. haemolytica (sodA gene primer for 143bp) [18] (Table-2). The DNA was extracted from samples enriched in BHI broth for 18 h by DNeasy kit as per the manufacturer’s protocol (Qiagen, Hilden, Germany). Extracted DNA (100ng/μl) served as template in 25 μl PCR mixture consisting of 1× hotstart Taq plus master mix (Thermo scientific) and 0.3 μM and 0.5 μM of M. haemolytica and P. multocida primers, respectively. PCR amplification was performed using mastercycler (Eppendorf Ltd., Mississauga, Canada) with the following thermal conditions: Initial denaturing temperature of 95°C for 5 min followed by 30 cycles of 94°C for 30 s, annealing temperature of 56°C for 1 min, extension temperature of 72°C for 1 min, and final extension at 72°C for 8 min. The PCR products were analyzed by electrophoresis on 1.5% agarose gel with 10 μg/ml-1 ethidium bromide and visualized in gel documentation system. M. haemolytica serotype A2 (ATCC No. 29698) and serotype A7, (ATCC No. 33396) and P. multocida type A (ATCC No. 12945) and type B serogroups (ATCC No. 43137) were used as positive controls.
In vitro antimicrobial susceptibility test (ABST)
In vitro ABST was performed for 28 P. multocida and 27 M. haemolytica isolates using Kirby–Bauer disk diffusion method [19] on Muller-Hinton agar as per Clinical and Laboratory Standards Institute (CLSI) with Escherichia coli ATCC 25922 as a quality control organism. The antibiotics commonly used in the treatment of the respiratory ailments were chosen, namely, amoxicillin/clavulanic acid (30 mcg), co-trimoxazole (25 mcg), ampicillin (10 mcg), penicillin (10 U), enrofloxacin (5 mcg), chloramphenicol (30 mcg), gentamicin (10 mcg), streptomycin (10 mcg), oxytetracycline (30 mcg), and tetracycline (30 mcg). The zone of inhibition was interpreted as per the performance standards for ABST specified in 16 informational supplement of CLSI (CLSI, 2006).
Antibiotic resistance marker genotyping
All the isolates irrespective of the resistance pattern in ABST were subjected to antibiotic resistance marker typing for penicillin (blaOXA-2), ampicillin (blaROB-1), sulfonamide (sul2), gentamicin (aadB), chloramphenicol (CatAII), tetracycline (tet H), and streptomycin (strA, strB) by simplex PCR assays [20,21] (Table-2).
Statistical analysis
Statistical analysis was performed with SPSS 16.0 (SPSS Inc., Chicago) and p<0.05 was considered statistically significant.
Results
Isolation and identification of M. haemolytica and P. multocida
Among 374 samples processed, 64 (17.1%) and 35 (9.4%) isolates obtained were identified as M. haemolytica and P. multocida, respectively, by bacteriological tests (Table-3). All of these isolates were Gram-negative, showed bipolar coccobacilli morphology microscopically and positive to oxidase and catalase tests. M. haemolytica isolates showed β-hemolysis on blood agar, variable lactose fermentation reaction on MLA and negative for indole production. Whereas, P. multocida isolates were non-hemolytic, indole positive and failed to grow on MLA. Among these identified isolates, 27 were confirmed as M. haemolytica by amplification of 304, 206, and 90 bps in mPCR (Figure-1a) and 28 isolates as P. multocida by amplification of 460bp (Figure-1b). From 132 lung tissue samples processed, 11 and 14 isolates of M. haemolytica and P. multocida, respectively, were recovered from infected lungs showing a significant correlation between isolation and disease status (p<0.0001). Similarly, 13 P. multocida isolates were recovered solely from the nasal samples of sheep suffering from respiratory infection. Co-isolation of M. haemolytica and P. multocida was observed in 15.7% infected lung samples (Table-4).
Table-3.
Isolation and mPCR detection of M. haemolytica and P. multocida from various types of sheep samples.
| Type of samples | Number of samples | M. haemolytica | P. multocida | ||||||
|---|---|---|---|---|---|---|---|---|---|
| mPCR positives | χ2/p-value | Number of isolates | χ2/p-value | mPCR positives | χ2/p-value | Number of isolates | χ2/p-value | ||
| Nasal samples | 242 | 143 (59) | 0/1 | 14 (5.8) | 1.80/ 0.179 | 35 (14.5) | 0.91/ 0.340 | 13 (5.4) | 3.75/ 0.052 |
| Lung samples | 132 | 78 (59) | 13 (9.8) | 25 (18.9) | 15 (11.3) | ||||
| Total samples | 374 | 221 (59) | 27 (7.2) | 60(16.0) | 28 (7.5) | ||||
| Nasal samples | |||||||||
| Nasal samples from healthy animals | 59 | 21 (35.6) | 5.13/0.023* | 2 (3.4) | 0.74/ 0.389 | 0 | 0 | ||
| Nasal samples from respiratory infection | 183 | 122 (66.7) | 12 (6.6) | 35 (19.1) | 13(7.1) | ||||
| Total samples | 242 | 143 (59) | 14 (5.8) | 35 (14.5) | 13(5.4) | ||||
| Lung samples | |||||||||
| Healthy lungs | 94 | 57 (60.6) | 0.08/0.777 | 2 (2.1) | 16.48/<0.0001* | 22 (23.4) | 3.07/0.079 | 1 (1.1) | 24.55/<0.0001* |
| Infected lungs | 38 | 21 (55.3) | 11 (28.9) | 3 (7.9) | 14 (36.8) | ||||
| Total samples | 132 | 78 (59) | 13 (9.8) | 25 (18.9) | 15 (11.4) | ||||
The values within the parentheses indicates percentage;
Significance at p<0.05
Figure-1.
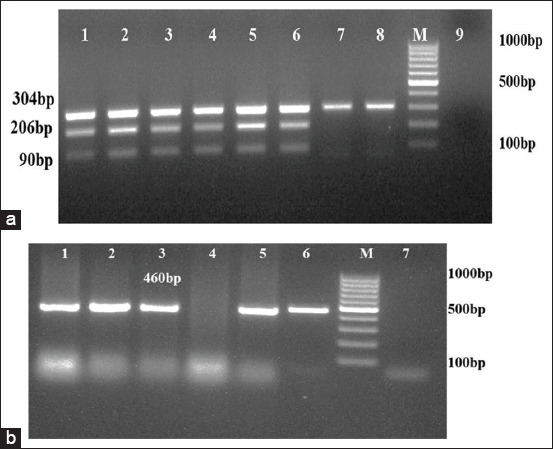
(a) Multiplex PCR amplification for Mannheimia haemolytica. Lane 3-8: DNA extracted from pure cultures; Lane 1-2: Positive controls; Lane 9: No template control; Lane M: 100bp molecular marker; (b) PCR amplification for Pasteurella multocida.Lane1-5: DNA extracted from field isolates; Lane 6: Positive control; Lane 7: No template control; Lane M: 100bp molecular marker.
Table-4.
Concurrence of M. haemolytica and P. multocida by isolation and mPCR in nasal and lung sheep samples.
| Samples | Total samples | mPCR co-positives | Co-isolations |
|---|---|---|---|
| Total nasal samples | 242 | 29 (12) | 0 |
| Total lung samples | 132 | 25 (18.9) | 6 (4.5) |
| Cumulative total samples | 374 | 54 (14.4) | 6 (1.6) |
| Nasal samples | |||
| Nasal samples from healthy sheep | 59 | 0 | 0 |
| Nasal samples from sheep with respiratory infection | 183 | 29 (15.8) | 0 |
| Total samples | 242 | 29 (12) | |
| Lung samples | |||
| Healthy lung samples | 94 | 22 (23.4) | 0 |
| Infected lung samples | 38 | 3 (7.9) | 6 (15.8) |
| Total samples | 132 | 25 (18.9) | 6 (4.5) |
The values within the parentheses indicate percentage. M. haemolytica=Mannheimia haemolytica, P. multocida=Pasteurella multocida, mPCR=Multiplex PCR
Direct detection of M. haemolytica and P. multocida by mPCR
Out of 374 samples processed, 221 (59%) and 60 (16.04%) samples were positive for M. haemolytica and P. multocida, respectively, by mPCR. Of which 66.6% M. haemolytica and 19.1% P. multocida were from nasal samples collected from sheep suffering from respiratory infection. Among 94 and 38 healthy and infected lungs samples processed respectively, higher percentage of both M. haemolytica (60.6%) and P. multocida (23.4%) were detected in healthy lungs (Table-3 and Figure-2) and co-detection of both M. haemolytica and P. multocida was observed in 18.9% of lung samples (Table-4).
Figure-2.
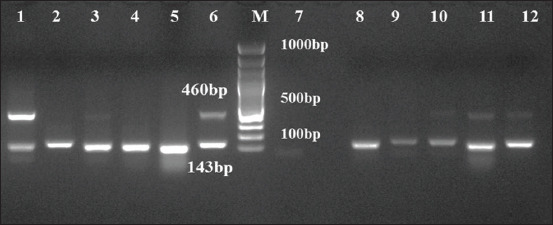
Multiplex PCR amplification for Mannheimia haemolytica and Pasteurella multocida. Lane1-5 and 8-12: DNA extracted from 18 h BHI broth enriched samples; Lane 6: Positive control; Lane 7: No template control; Lane M: 100bp molecular marker.
In vitro antimicrobial susceptibility testing
Among ten different antibiotics tested, chloramphenicol (100%), ampicillin (98.9%), and amoxicillin/clavulanic acid (96.4%) were found most effective drugs against M. haemolytica and P. multocida (Table-5). M. haemolytica isolates showed 81.5%, 40.7%, and 22.2% resistance towards penicillin, oxytetracycline, and streptomycin, respectively, whereas, P. multocida isolates showed resistance only towards penicillin (46.4%) and 7.1% each for gentamicin, oxytetracycline, and streptomycin. Among 55 isolates tested, 22 (81.5%) and 13 (46.4%) isolates of M. haemolytica and P. multocida, respectively, were resistant to at least one antibiotic. Multiple drug resistance against three or more than three antibiotics was noticed in 7.1% (2/28) of P. multocida and 25.9% (7/27) of M. haemolytica isolates (Figure-3).
Table-5.
In vitro antibiotic susceptibility of P. multocida and M. haemolytica isolates from sheep.
| No. | Antibiotics (Conc./disc) | P. multocida (n=28) | M. haemolytica (n=27) | Total (n=55) | |||
|---|---|---|---|---|---|---|---|
| Resistant | Intermediate resistant | Resistant | Intermediate resistant | Resistant | Intermediate resistant | ||
| 1 | Amoxicillin/clavulanic acid (30 mcg) | 1 (3.6) | 0 | 1 (3.7) | 0 | 2 (3.6) | 0 |
| 2 | Co-trimoxazole (25 mcg) | 1 (3.6) | 0 | 4 (14.8) | 0 | 5 (9.0) | 0 |
| 3 | Ampicillin (10 mcg) | 0 | 0 | 0 | 1 (3.7) | 0 | 1 (1.8) |
| 4 | Penicillin (10 U) | 13 (46.4) | 0 | 22 (81.5) | 0 | 35 (63.6) | 0 |
| 5 | Enrofloxacin (5 mcg) | 0 | 2 (7.1) | 0 | 13 (48.1) | 0 | 15 (27.2) |
| 6 | Chloramphenicol (30 mcg) | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | Gentamicin (10 mcg) | 2 (7.1) | 1(3.5) | 5 (18.5) | 4 (14.8) | 7 (12.7) | 5 (9.0) |
| 8 | Streptomycin (10 mcg) | 2 (7.1) | 1(3.5) | 6 (22.2) | 11 (40.7) | 8 (14.5) | 12 (21.8) |
| 9 | Oxytetracycline (30 mcg) | 2 (7.1) | 0 | 11 (40.7) | 0 | 13 (23.6) | 0 |
| 10 | Tetracycline (30 mcg) | 0 | 0 | 3 (11.1) | 0 | 3 (5.5) | 0 |
The values within the parentheses indicate percentage. M. haemolytica=Mannheimia haemolytica, P. multocida=Pasteurella multocida
Figure-3.
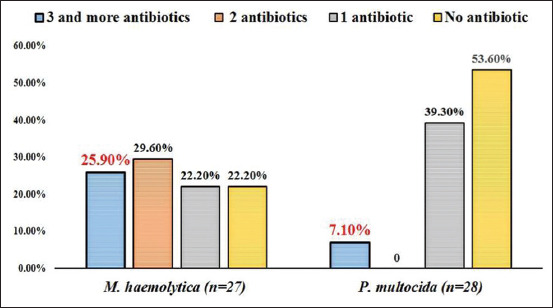
Multi drug resistance profile in Mannheimia haemolytica and Pasteurella multocida isolates.
Antibiotic resistance marker typing
Antibiotic resistance genes by PCR screening did not show any agreement with phenotypic in vitro antibiotic resistance profiles. However, the resistant genes for different antibiotics were higher in M. haemolytica compared to P. multocida isolates similar to phenotypic in vitro method. Out of 28 P. multocida isolates, 14 and 12 isolates showed the presence of strA and sul2 genes. Among 27 M. haemolytica isolates, one, three, four, and six isolates showed the presence of blaROB-1, blaOXA-2, strA, and sul2 genes, respectively (Table-6).
Table-6.
Detection of antibiotic resistance gene markers in P. multocida and M. haemolytica isolates.
| M. haemolytica isolates | ||
|---|---|---|
| Antibiotic resistance markers | No. of positive isolates in nasal samples (n=14) | No. of positive isolates in lung samples (n=13) |
| Penicillin (blaOXA-2) | 3 (21.4) | 0 |
| Ampicillin (blaROB-1) | 1 (7.1) | 0 |
| Streptomycin (strA, strB) | 1 (7.1) (strA) | 3 (23) |
| Chloramphenicol (catAII) | 0 | 0 |
| Tetracycline (tetH) | 0 | 0 |
| Sulfamethoxazole (sul2) | 2 (14.3) | 4 (30.8) |
| Gentamicin (aadB) | 0 | 0 |
| P. multocida isolates | ||
| Antibiotic resistance markers | No. of positive isolates in nasal samples (n=13) | No. of positive isolates in lung samples (n=15) |
| Penicillin (blaOXA-2) | 0 | 0 |
| Ampicillin (blaROB-1) | 0 | 0 |
| Streptomycin (strA, strB) | 5 (38.5) (strA) | 9 (60) (strA) |
| Chloramphenicol (catAII) | 0 | 0 |
| Tetracycline (tetH) | 0 | 0 |
| Sulfamethoxazole (sul2) | 3 (23) | 9 (60) |
| Gentamicin (aadB) | 0 | 0 |
The values within the parentheses indicate percentage. M. haemolytica=Mannheimia haemolytica, P. multocida=Pasteurella multocida
Discussion
Small ruminants are the continuous source of income for the rural populace in India. Increasing prevalence of respiratory infection in small ruminants even after HS and PPR vaccination was a serious concern. Recently respiratory infection outbreaks due to M. haemolytica and P. multocida have been reported in various states of India [22,23]. In the present study, we investigated the involvement of M. haemolytica and P. multocida with the respiratory infection of sheep.
Overall, Pasteurella species recovered from samples were 55 (14.7%), of which, 51% and 49% were identified as P. multocida and M. haemolytica, respectively, and majority of the isolates were from sheep suffering from respiratory infection. Similarly, Miller et al. [9] noted 80% biovariants of M. haemolytica, P. multocida and P. trehalosi species from the diseased sheep indicating the importance of these pathogens in the respiratory infections. In mPCR, M. haemolytica was detected in 59% of the samples and significant correlation was observed between M. haemolytica detection (66.7%) with the diseased condition (p=0.023) which substantiates its role in the disease. Whereas, 35.6% of M. haemolytica PCR positives were also detected in nasal samples collected from apparently healthy sheep. This may be because of the fact that M. haemolytica is generally present in the upper respiratory tract of ruminants which multiplies rapidly along with P. multocida upon exposure to the stress thereby resulting in respiratory illness [7]. Compared to M. haemolytica, the detection of P. multocida by mPCR was less (16.04%) but was detected solely from the infected sheep (14.4%) which acknowledges the primary role of P. multocida in respiratory infection. The pathogenic role of P. multocida in ovine pasteurellosis causing serious outbreaks was reported earlier [2]. Usually, in stressed and immunocompromised host, these secondary bacterial pathogens proliferate and increase in number in the upper respiratory tract and by gravitational drainage they reach to the ventral bronchi, bronchioles, and alveoli to cause bronchopneumonia [7]. Therefore we tried to detect these pathogens in lungs samples by mPCR. M. haemolytica was detected in both infected and healthy lung samples, whereas the higher number of P. multocida were recorded in healthy lung samples. The correlation of the disease status of the slaughtered animals to the detection of the pathogens was difficult as the animals were under transportation stress and their clinical symptoms were unknown.
As a result of ineffective immunoprophylactic measure for respiratory infection, antimicrobial treatment is considered important prophylactic agent for the control of the disease. Due to extensive use of antibiotics as supplements in animal feed both for prophylaxis and growth promotion, antimicrobial resistance among these pathogens was observed [11]. Among ten different antibiotics tested, chloramphenicol (100%), ampicillin (98.9%), and amoxicillin/clavulanic acid (96.4%) were found the most effective drugs against M. haemolytica and P. multocida isolates. In the present work, M. haemolytica and P. multocida isolates showed only 18.5% and 7.1% resistance to gentamicin, respectively. Whereas, the study from Ethiopia reported gentamycin as totally inactive against M. haemolytica and P. multocida isolates for the treatment of ovine pasteurellosis [24]. Lamm et al. [10] reported high resistance to tetracycline in M. haemolytica and P. multocida isolates from bronchopneumonic cattle, whereas, only 11.1% of ovine M. haemolytica isolates were resistant to tetracycline in the present study. Klima et al. [15] noted high resistance towards oxytetracycline, ampicillin, and amoxicillin/clavulanic acid among bovine M. haemolytica isolates. Similarly in our study, ovine M. haemolytica isolates showed 40.7% resistance to oxytetracycline with least resistance to ampicillin and amoxicillin/clavulanic acid. Absolute sensitivity towards chloramphenicol and absolute resistance towards sulfa drug in avian P. multocida stains was reported from India [25]. However, in the present study, absolute sensitivity towards both chloramphenicol and sulfa drugs was observed. Multiple drug resistance (three or more than three antibiotics) was detected higher in M. haemolytica than P. multocida isolates. Andrés-Lasheras et al. [26] and Sarangi et al. [27] have also noted multiple drug resistance in M. haemolytica isolates from BRD infected European cattle and Indian P. multocida isolates from small ruminants. So, periodical antibiotic susceptibility testing is essential to identify the drug/s of choice for the treatment in different host/s and to set guidelines for the prudent use of antibiotic/s in the disease endemic regions.
Along with the external factors such as geographical location, antibiotic pre-treatment, and dosages, antibiotic resistance among Pasteurella species also depends on accessibility of the isolates to the resistance genes in the gene pool. Genes such as blaROB-1, tetH, tetO, tetB, and strA associated with β-lactam, tetracycline, and streptomycin resistance, respectively, were detected in P. multocida pig isolates from Spain [28]. The tetH, blaROB-1 genes were also identified in M. haemolytica isolates from cattle treated for BRD [15]. In the present study, blaOXA-2, blaROB-1, strA, and sul 2 genes in M. haemolytica isolates and only strA and sul2 resistance genes in P. multocida isolates were observed. Plasmids [28], chromosome [11], and integrative conjugative elements [29,30] are the associated factors for the interspecies and inter-genic antibiotic resistance genes transmission in Pasteurellaceae family. Presence of different antibiotic resistance genes in M. haemolytica is as serious concern as these genes can be transferred to P. multocida and other respiratory pathogens by horizontal gene transfer which may cause severe infections.
Conclusion
The study emphasized the significance of M. haemolytica together with P. multocida in causing respiratory infection in sheep. The study suggests to include M. haemolytica while diagnosing respiratory disease which has been ignored or overlooked. Higher drug resistance in M. haemolytica than P. multocida together with the presence of strA and sul2 like antibiotic resistance genes was observed in the isolates. Further studies involving samples from multiple hosts from different geographical and anatomic locations are needed to understand the serotypic, pathogenic, and genotypic variants causing the disease outbreaks in the country.
Authors’ Contribution
RS and SS were involved in planning, execution of the work, and writing of the manuscript. SS, KN, AP, and TK performed disease investigation, sample collection, bacterial isolation, and characterization. BRS and HR provided the conceptual and technical support to formulate the work. All authors read and approved the final manuscript.
Acknowledgments
The authors acknowledge the Director and Scientists, ICAR-National Institute of Veterinary Epidemiology and Disease Informatics, Bengaluru, for providing the facilities and support during this work. The authors are thankful to the Department of Animal Husbandry and Veterinary Services, Government of Karnataka for providing permission for the sample collection. This work was funded by the Department of Biotechnology, Government of India, New Delhi through the twinning project entitled “Aetio-Pathology and molecular epidemiology of bacterial and viral diseases associated with the respiratory problems of yak in the North Eastern Regions of India” (Grant No: BT/391/NE/TBP/2012).
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published institutional affiliation.
References
- 1.Jesse F.F.A, Amira N.A, Isa K.M, Maqbool A, Ali N.M, Chung E.L.T, Lila M.A.M. Association between Mannheimia haemolytica infection with reproductive physiology and performance in small ruminants:A review. Vet. World. 2019;12(7):978. doi: 10.14202/vetworld.2019.978-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohamed R.A, Addelsalam E.B. A review on pneumonic pasteurellosis (Respiratory mannheimiosis) with emphasis on pathogenesis, virulence mechanisms and predisposing factors. Bulg. J. Vet. Med. 2008;11(3):139–160. [Google Scholar]
- 3.Lacasta D, Ferrer L.M, Ramos J.J, Gonzalez J.M, Heras D.L.M. Influence of climatic factors on the development of pneumonia in lambs. Small Rumin. Res. 2008;80(1-3):28–32. [Google Scholar]
- 4.Dabo S.M, Taylor J.D, Confer A.W. Pasteurella multocida and bovine respiratory disease. Anim. Health. Res. Rev. 2007;8(2):129–150. doi: 10.1017/S1466252307001399. [DOI] [PubMed] [Google Scholar]
- 5.Sahay S, Shome R, Sankarasubramanian J, Vishnu U.S, Prajapati A, Natesan K, Shome B.R, Rahman H, Rajendhran J. Genome sequence analysis of the Indian strain Mannheimia haemolytica serotype A2 from ovine pneumonic pasteurellosis. Ann. Microbiol. 2019;69(2):151–160. [Google Scholar]
- 6.Sahay S, Shome R, Sankarasubramanian J, Vishnu U.S, Prajapati A, Natesan K, Shome B.R, Rahman H, Rajendhran J. Insights into the genome sequence of ovine Pasteurella multocida type a strain associated with pneumonic pasteurellosis. Small Rumin. Res. 2018;169:167–175. [Google Scholar]
- 7.García-Alvarez A, Fernández-Garayzábal J.F, Chaves F, Pinto C, Cid D. Ovine Mannheimia haemolytica isolates from lungs with and without pneumonic lesions belong to similar genotypes. Vet. Microbiol. 2018;219:80–86. doi: 10.1016/j.vetmic.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Taylor J.D, Holland B.P, Step D.L, Payton M.E, Confer A.W. Nasal isolation of Mannheimia haemolytica and Pasteurella multocida as predictors of respiratory disease in shipped calves. Res. Vet. Sci. 2015;99:41–45. doi: 10.1016/j.rvsc.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 9.Miller D.S, Weiser G.C, Ward A.C.S, Drew M.L, Chapman P.L. Domestic sheep (Ovis aries) Pasteurellaceae isolates from diagnostic submissions to the caine veterinary teaching center (1990-2004) Vet. Microbiol. 2011;150(3-4):284–288. doi: 10.1016/j.vetmic.2011.01.024. [DOI] [PubMed] [Google Scholar]
- 10.Lamm C.G, Love B.C, Krehbiel C.R, Johnson N.J, Step D.L. Comparison of antemortem antimicrobial treatment regimens to antimicrobial susceptibility patterns of post-mortem lung isolates from feedlot cattle with bronchopneumonia. J. Vet. Diagn. Invest. 2012;24(2):277–282. doi: 10.1177/1040638711428149. [DOI] [PubMed] [Google Scholar]
- 11.Kehrenberg C, Tham N.T.T, Schwarz S. New plasmid-borne antibiotic resistance gene cluster in Pasteurella multocida. Antimicrob. Agents Chemother. 2003;47(9):2978–2980. doi: 10.1128/AAC.47.9.2978-2980.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anholt R.M, Klima C, Allan N, Matheson-Bird H, Schatz C, Ajitkumar P, Otto S.J, Peters D, Schmid K, Olson M, McAllister T. Antimicrobial susceptibility of bacteria that cause bovine respiratory disease complex in Alberta, Canada. Front. Vet. Sci. 2017;4:207. doi: 10.3389/fvets.2017.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DADH. All India Report of 19th Livestock Census-2012, Ministry of Agriculture, Department of Animal Husbandry, Dairying and Fisheries (DADH), Government of India. 2012. Available from: http://www.dahd.nic.in/sites/default/filess/livestock%20%205_0.pdf?. Retrieved on 10-08-2018.
- 14.Mohini M, Malla B.A, Mondal G. Small ruminant sector in India:Present status, feeding systems and greenhouse gas emissions. EC Vet. Sci. 2018;3(1):281–289. [Google Scholar]
- 15.Klima C.L, Alexander T.W, Read R.R, Gow S.P, Booker C.W, Hannon S, Sheedy C, McAllister A.T, Selinger L.B. Genetic characterization and antimicrobial susceptibility of Mannheimia haemolytica isolated from the nasopharynx of feedlot cattle. Vet. Microbiol. 2011;149(3-4):390–398. doi: 10.1016/j.vetmic.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 16.Alexander T.W, Cook S.R, Yanke L.J, Booker C.W, Morley P.S, Read R.R, Gow S.P, McAllister T.A. A multiplex polymerase chain reaction assay for the identification of Mannheimia haemolytica, Mannheimia glucosidal and Mannheimia ruminalis. Vet. Microbiol. 2008;130(1-2):165–175. doi: 10.1016/j.vetmic.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Townsend K.M, Frost A.J, Lee C.W, Papadimitriou J.M, Dawkins H.J. Development of PCR assays for species-and type-specific identification of Pasteurella multocida isolates. J. Clin. Microbiol. 1998;36(4):1096–1100. doi: 10.1128/jcm.36.4.1096-1100.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guenther S, Schierack P, Grobbel M, Becker A.L. Real-time PCR assay for the detection of species of the genus Mannheimia. J. Microbiol. Methods. 2008;75(1):75–80. doi: 10.1016/j.mimet.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Bauer A.W, Kirby W.M.M, Sherhis J.C, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966;45(4):493–496. [PubMed] [Google Scholar]
- 20.Klima C.L, Zaheer R, Cook S.R, Booker C.W, Hendrick S, Alexander T.W, McAllister T.A. Pathogens of bovine respiratory disease in North American feedlots conferring multidrug resistance via integrative conjugative elements. J. Clin. Microbiol. 2014;52(2):438–448. doi: 10.1128/JCM.02485-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kehrenberg C, Schwarz S. Occurrence and linkage of genes coding for resistance to sulfonamides, streptomycin and chloramphenicol in bacteria of the genera Pasteurella and Mannheimia. FEMS Microbiol. Lett. 2001;205(2):283–290. doi: 10.1111/j.1574-6968.2001.tb10962.x. [DOI] [PubMed] [Google Scholar]
- 22.Kumar J, Dixit S.K, Kumar R. Rapid detection of Mannheimia haemolytica in lung tissues of sheep and from bacterial culture. Vet. World. 2015;8(9):1073. doi: 10.14202/vetworld.2015.1073-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rawat N, Gilhare V.R, Kushwaha K.K, Hattimare D.D, Khan F.F, Shende R.K, Jolhe D.K. Isolation and molecular characterization of Mannheimia haemolytica and Pasteurella multocida associated with pneumonia of goats in Chhattisgarh. Vet. World. 2019;12(2):331–336. doi: 10.14202/vetworld.2019.331-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marru H.D, Anijajo T.T, Hassen A.A. A study on ovine pneumonic pasteurellosis:Isolation and identification of Pasteurellae and their antibiogram susceptibility pattern in Haramaya district, Eastern Hararghe, Ethiopia. BMC Vet. Res. 2013;9(1):1–8. doi: 10.1186/1746-6148-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shivachandra S.B, Kumar A.A, Biswas A, Ramakrishnan M.A, Singh V.P, Srivastava S.K. Antibiotic sensitivity patterns among Indian strains of avian Pasteurella multocida . Trop. Anim. Health Prod. 2004;36(8):743–750. doi: 10.1023/b:trop.0000045950.35070.7f. [DOI] [PubMed] [Google Scholar]
- 26.Andrés-Lasheras S, Zaheer R, Klima C, Sanderson H, Polo R.O, Milani M.R.M, Vertenten G, Mc Allister T.A. Serotyping and antimicrobial resistance of Mannheimia haemolytica strains from European cattle with bovine respiratory disease. Res. Vet. Sci. 2019;124:10–12. doi: 10.1016/j.rvsc.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 27.Sarangi L.N, Thomas P, Gupta S.K, Priyadarshini A, Kumar S, Nagaleekar V.K, Kumar A, Singh V.P. Virulence gene profiling and antibiotic resistance pattern of Indian isolates of Pasteurella multocida of small ruminant origin. Comp. Immunol. Microbiol. Infect. Dis. 2015;38:33–39. doi: 10.1016/j.cimid.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 28.San Millan A, Escudero J.A, Gutierrez B, Hidalgo L, Garcia N, Llagostera M, Dominguez L, Gonzalez-Zorn B. Multi-resistance in Pasteurella multocida is mediated by coexistence of small plasmids. Antimicrob. Agents Chemother. 2009;53(8):3399–3404. doi: 10.1128/AAC.01522-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eidam C, Poehlein A, Leimbach A, Michael G.B, Kadlec K, Liesegang H, Daniel R, Sweeney M.T, Murray R.W, Watts J.L, Schwarz S. Analysis and comparative genomics of ICEMh1, a novel integrative and conjugative element (ICE) of Mannheimia haemolytica. J. Antimicrob. Chemother. 2015;70(1):93–97. doi: 10.1093/jac/dku361. [DOI] [PubMed] [Google Scholar]
- 30.Peng Z, Liang W, Liu W, Chen H, Wu B. Genome characterization of Pasteurella multocida subspecies septica and comparison with Pasteurella multocida subspecies multocida and gallicida. Arch. Microbiol. 2017;199(4):635–640. doi: 10.1007/s00203-017-1341-x. [DOI] [PubMed] [Google Scholar]


