Abstract
Background
Candida species cause a wide spectrum of disease entities. Candida africana and Candida dubliniensis–members of Candida albicans complex-are currently gaining both clinical and epidemiologic significance.
Materials and methods
Totally, 150 pediatric isolates that had previously been identified as C. albicans species complex based on a positive germ tube test were included. The isolates were cultured on CHROMagar Candida medium to ensure their purity and the results of germ tube test. For definitive speciation, PCR amplification and size polymorphism of the hyphal wall protein 1 (HWP1) gene was used. The results of HWP1-PCR were confirmed by sequencing the amplified fragments for randomly selected isolates of C. africana and C. dubliniensis.
Results
All 150 isolates included in this study were reconfirmed as C. albicans complex on chromogenic media. Based on the HWP1 gene size polymorphism, 141 (94%) isolates were identified as C. albicans, 2 (1.33%) as C. africana, and 1 (0.67%) as C. dubliniensis. The remaining 6 (4%) C. albicans complex isolates were a mix of C. albicans and C. africana. All isolates of C. africana and C. dubliniensis have been recovered from cases of candiduria.
Conclusion
C. africana, either alone or mixed with C. albicans, could be a cause of candiduria among pediatric patients and should not be ignored.
Keywords: Candiduria, Candida albicans complex, Candida africana, Candida dubliniensis, pediatrics
1. Introduction
The frequency of fungal infections is increasing worldwide. Species of the genus Candida are opportunist fungi capable of causing mild cutaneous to severe and life-threatening systemic infections in susceptible individuals. Among these species, Candida albicans is the most clinically encountered pathogen and most researched [1],[2]. Other species, such as Candida africana and Candida dubliniensis-closely related to C. albicans-initially referred to as ‘atypical’ Candida species are now fully characterizable using molecular techniques and classified as distinct species in C. albicans species complex [3]–[6]. Of recent, the frequency of ‘atypical’ Candida species among clinical Candida isolates has been rising [7],[8]. C. africana infections have been reported from various countries and genitourinary tract samples, especially vaginal swabs have been found as the most common source of isolation [5],[9]–[12]. Earlier, C. dubliniensis was mainly isolated from oral cavity of HIV/AIDS patients but subsequent epidemiological studies revealed that this species is recoverable from other body sites such as skin, vagina, respiratory tract, and urine of both HIV-infected and non-HIV patients [13]–[16]. Both C. africana and C. dubliniensis are clinically important members of C. albicans complex but their identification using conventional techniques proves to be challenging due to mutual sharing of similar morphological characteristics. Molecular tools suitable for identification of Candida species include the sequence-analysis of the ribosomal DNA (rDNA) ITS region; the use of duplex/multiplex PCR for identification of isolates such as C. albicans and C. dubliniensis; and amplification of the hyphal wall protein 1 (HWP1) gene - the first molecular method used to discriminate C. dubliniensis and C. africana from C. albicans [4]. Molecular studies based on the ITS data sequence revealed that C. africana is a separate species from C. albicans with an interspecies sequence homology of 99.3 to 99.8%; whereas C .dubliniensis differs from C. albicans with an interspecies sequence homology of 91.2–94.4% [17]. Despite their clinical significance, they are rarely sought for in routine laboratory examination-many members of the complex could be misidentified in less suspicious cases-due to their low frequencies in clinical settings [18]. In this study, we aimed to identify the minor species (C. africana and C. dubliniensis) in the C. albicans complex recovered from different clinical specimens using HWP1 gene amplification method among pediatric patients at a tertiary medical center in Tehran, Iran.
2. Material and methods
2.1. Candida albicans complex isolates
One hundred and fifty clinical isolates of C. albicans species complex were included in this study. These isolates had been collected during routine work at the microbiology laboratory of Children's Medical Center, Tehran, Iran. The isolates had previously been identified as C. albicans species complex based on a positive result in germ tube test.
2.2. Re-identification of isolates
To confirm the purity of isolates and the results of germ tube test, the isolates were streaked on CHROMagar Candida medium (CHROMagar, France). The results of germ tube test were confirmed if characteristic green colonies grown on the medium.
2.3. Molecular identification
The isolates were investigated based on the size polymorphism of the HWP1 gene using primers CR-f (GCT ACC ACT TCA GAA TCA TCA TC) and CR-r (GCA CCT TCA GTC GTA GAG ACG). PCR was performed using an initial denaturation at 95 °C or 5 minutes; 35 cycles of denaturation at 94 °C for 45 seconds, annealing at 58 °C for 40 seconds, and extension at 72 °C for 55 seconds; and a final extension at 72 °C for 10 minutes [4]. To fasten the procedure of molecular identification, we used a colony-PCR approach instead of extracting the genomic DNA and using it as template. Briefly, using sterile micropipette tips, an overnight colony of each isolate was transferred into a PCR reaction tube containing 12.5 µL of 2X master mix (Amplicon, Denmark), 1 µL of forward and reverse primers (10 pg/µL), and 10.5 µL of deionized, sterile distilled water. PCR amplicons were visualized using a transilluminator (Syngene, USA) after electrophoresis in 1.5% w/v agarose gel. The PCR product sizes of 941 bp, ∼700 bp, and 569 bp were identical to C. albicans, C. africana, and C. dubliniensis, respectively.
To confirm the results of HWP1 size polymorphism, isolates of C. africana and C. dubliniensis were randomly selected and a fragment of the HWP1 gene was amplified using the same primers and PCR conditions described earlier. The PCR products were subjected to sequencing (Macrogene, South Korea) and the results were analyzed using Basic Local Alignment Search Tool (BLAST) in the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast).
3. Results
The isolation source of the 150 C. albicans species complex isolates is shown in Figure 1. All 150 isolates included in this study were reconfirmed as C. albicans complex on chromogenic media by growing as green colonies. By molecular characterization using HWP1 gene size polymorphism, 141 (94%) isolates were identified as C. albicans, 2 (1.33%) isolates as C. africana, and 1 (0.67%) isolate as C. dubliniensis. The remaining 6 (4%) C. albicans complex isolates were found to be a mix of C. albicans and C. africana. Accordingly, in total, 156 isolates were identified from 150 C. albicans complex isolates, of which 147 (94.23%) were C. albicans, 8 (5.13%) were C. africana, and 1 (0.64%) was C. dubliniensis. Two pure C. africana isolates and the C. dubliniensis isolate were subjected to PCR-sequencing and the obtained results confirmed their identity. The sequences were deposited in GenBank under accession numbers MT829308–MT829310. Regarding the source of isolation, all isolates (pure and mixed) of C. africana and the isolate of C. dubliniensis were recovered from urine specimens. Table 1 shows the distribution of various species based on the isolation source.
Figure 1. Distribution of 150 Candida albicans complex isolates based on the specimen type (BAL: bronchoalveolar lavage; CVC: central venous catheter; CSF: cerebrospinal fluid).
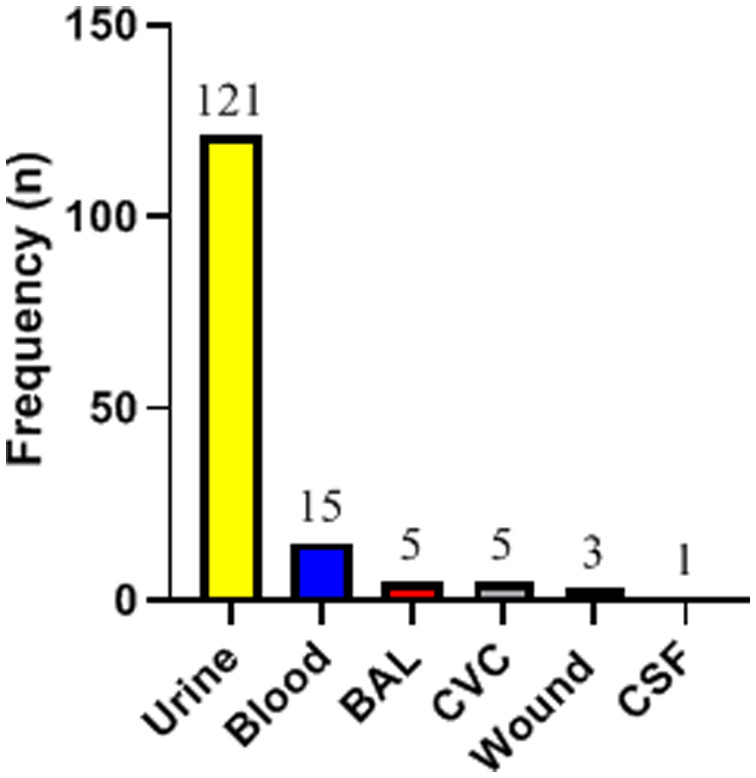
Table 1. Distribution of Candida albicans complex isolates based on the isolation source.
| Specimen |
Candida species (n) |
|||
| C. albicans | C. africana | C. dubliniensis | Mix of C. albicans and C. africana | |
| Urine | 112 | 2 | 1 | 6 |
| Blood | 15 | |||
| Central venous catheter | 5 | |||
| Bronchoalveolar lavage | 5 | |||
| Wound discharge | 3 | |||
| Cerebrospinal fluid | 1 | |||
| Total isolates | 141 | 2 | 1 | 12* |
* Including 6 C. africana and 6 C. albicans isolates.
4. Discussion
The ‘C. albicans complex’ is a group of opportunistic fungal pathogens responsible for more than 50% of candidiasis [1],[7]. Species of this group are more often involved in oral and vaginal human infections [9],[10],[12],[19]. However, infections from other body sites such as respiratory tracts, urinary tracts, wounds, and skin, have been reported [12],[13],[20],[21]. Candida infection is a common phenomenon in pediatric subpopulation; frequent cases of childhood candidiasis have been reported [22]–[24].
Previous studies have reported mucous and cutaneous infections as the most common manifestation of pediatric candidiasis. Similarly, a study in Iran recovered Candida isolates predominantly from the oral cavity and stool of neutropenic patients suffering from leukemia [24]. However, in our study, the majority of the isolates were recovered from urine 121/150 (80.67%) and blood 15/150 (10.0%) specimens. Also, this contrasts the report from another medical center in Iran where the majority of Candida isolates among children were recovered from nails (39.5%) and blood (13.9%) infections [25]. Haddadi et al. reported that yeast isolates were the least recovered from urine specimens of neutropenic children [24]. Furthermore, out of the 498 clinical specimens investigated in a multicenter study, the majority of the Candida isolates from adults were recovered from vagina (32.5%) and oral cavity (24.3%) [26]. The observed heterogeneity might have resulted from the peculiarity of the subgroup and the underlying medical conditions of the patients studied.
C. africana and C. dubliniensis are emerging yeasts that attain clinical and epidemiological significance and, together with C. albicans form the C. albicans complex [26]. Discrimination between members of the complex is improbable using the conventional mycological tools. The molecular assay that involves amplification of the HWP1 gene was among the earliest and reliable tools, that stands the test of time, to discriminate between the uncommon species of the complex [4]. Similar to the current study, C. africana, C. dubliniensis, and C. albicans were adequately identified using HWP1 gene amplification [4],[9]–[11],[27]. In our study, C. albicans (94.23%) was the predominant species identified among the C. albicans complex which is in agreement with other studies [9]–[11],[13],[16],[21],[28]. We also identified both C. africana and C. dubliniensis among the urine isolates. Although C. africana initially appeared to be restricted to Africa and Europe and largely isolated from genital samples, it is now clear to have a worldwide distribution and recoverable from various clinical specimens [3],[12],[13],[20]. We found in our investigation that C. africana has a higher isolation rate (5.13%) than the C. dubliniensis (0.64%). This result corroborates the findings reported in Iran-where the frequency C. africana (3%) and C. dubliniensis (0%) was reported among 100 vaginal isolates of C. albicans species complex [10]–and a multicenter study that reported a higher frequency of C. africana (7.2%) than C. dubliniensis (2.9%), among Candida isolates recovered from 498 clinical specimens originated from various patient groups [26]. However, the higher isolation rate of C. dubliniensis over C. africana is being reported from other studies [9],[13],[19],[20]. Whereas C. dubliniensis is being predominantly recovered from oral and oropharyngeal lesions of HIV/AIDS patients, isolates were shown to be recovered from other patients suffering from cancer [14], cystic fibrosis [27], genital and respiratory tracts infections [9]–[13],[19],[20],[28], and from a gastric fluid sample [26]. The reason(s) for the disparate frequencies of isolation might be linked to factors such as the difference in the quality of sampling, the anatomic sites, the nature of the clinical sample, the underlying disease entities, racial or genotypic differences, and the immune status of the individuals involved.
Contrary to our findings, other studies that attempted to identify the cryptic species of C. albicans complex were able to identify, in addition to C. albicans, only C. africana [10],[16],[28] or C. dubliniensis [11],[27]. Interestingly, Gumral et al, evaluated vaginal Candida isolates using HWP1 gene polymorphism and found neither C. dubliniensis nor C. africana among the C. albicans complex they investigated [29]. While any member of the complex can independently colonize its preferential anatomic sites, isolation of dual or multiple species have been reported from a single clinical sample [11],[24]. Similarly, we identified C. albicans in combination with C. africana in six of our urine isolates. An effort to discriminate species in mixed infection/colonization, especially in children, is very important for clinicians because each species tends to show a variable level of virulence and different susceptibility profile to antifungal agents.
5. Conclusions
Because they are not routinely sought for, the prevalence of C. dubliniensis and/or C. africana might be underestimated. However, it is evident that they are currently gaining both clinical and epidemiologic significance. Clinicians and scientists shall pay attention to and explore the significance of the cryptic species of C. albicans complex especially in pediatric settings. To our knowledge, we report the first case of C. africana candiduria among pediatrics in Tehran. Further studies are needed to support its clinical and pathologic significance in causing urinary tract infection among children.
Acknowledgments
This study was financially supported by Tehran University of Medical Sciences, Tehran, Iran (grant No. 96-03-27-37410).
Footnotes
Conflict of interest: All authors declare no conflicts of interest in this paper.
References
- 1.Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charsizadeh A, Mirhendi H, Nikmanesh B, et al. Microbial epidemiology of candidaemia in neonatal and paediatric intensive care units at the Children's Medical Center, Tehran. Mycoses. 2018;61:22–29. doi: 10.1111/myc.12698. [DOI] [PubMed] [Google Scholar]
- 3.Odds FC, Bougnoux ME, Shaw DJ, et al. Molecular phylogenetics of Candida albicans. Eukaryotic cell. 2007;6:1041–1052. doi: 10.1128/EC.00041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romeo O, Criseo G. First molecular method for discriminating between Candida africana, Candida albicans, and Candida dubliniensis by using hwp1 gene. Diagn Microbiol Infect Dis. 2008;62:230–233. doi: 10.1016/j.diagmicrobio.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Zhu Y, Shi Y, Fan S, et al. Multilocus sequence typing analysis of Candida africana from vulvovaginal candidiasis. BMC Infect Dis. 2019;19:461. doi: 10.1186/s12879-019-4071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scordino F, Giuffrè L, Felice MR, et al. Genetic diversity of Candida albicans isolates recovered from hospital environments and patients with severe acquired brain injuries. Infect Genet Evol. 2019;76:104068. doi: 10.1016/j.meegid.2019.104068. [DOI] [PubMed] [Google Scholar]
- 7.Sardi J, Scorzoni L, Bernardi T, et al. Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J Med Microbiol. 2013;62:10–24. doi: 10.1099/jmm.0.045054-0. [DOI] [PubMed] [Google Scholar]
- 8.Das KH, Getso MI, Azeez-Ak, et al. Distribution of Candida albicans and non-albicans Candida in clinical samples and their intrinsic biofilm production status. Int J Med Sci Public Health. 2016;5:2443–2447. [Google Scholar]
- 9.Hana S, Latifa M, Camilia C, et al. Characterization of the ‘Candida albicans Complex’: First Report of Candida africana in Tunisia. J Med Microb Diagn. 2020;9:307. [Google Scholar]
- 10.Farahyar S, Izadi S, Razmjou E, et al. Low prevalence of antifungal resistant Candida africana, in the C. albicans complex causing vulvovaginal candidiasis. Heliyon. 2020;6:e03619. doi: 10.1016/j.heliyon.2020.e03619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mucci MJ, Cuestas ML, Landanburu MF, et al. Prevalence of Candida albicans, Candida dubliniensis and Candida africana in pregnant women suffering from vulvovaginal candidiasis in Argentina. Rev Iberoam Micol. 2017;34:72–76. doi: 10.1016/j.riam.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Aghaei Gharehbolagh S, Fallah B, Izadi A, et al. Distribution, antifungal susceptibility pattern and intra-Candida albicans species complex prevalence of Candida africana: A systematic review and meta-analysis. PLoS ONE. 2020;15:e0237046. doi: 10.1371/journal.pone.0237046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharifynia S, Badali H, Sharifi Sorkherizi M, et al. In vitro antifungal susceptibility profiles of Candida albicans complex isolated from patients with respiratory infections. Acta Med Iran. 2016;54:376–381. [PubMed] [Google Scholar]
- 14.Mokaddas E, Khan ZU, Ahmad S. Prevalence of Candida dubliniensis among cancer patients in Kuwait: a 5-year retrospective study. Mycoses. 2011;54:e29–e34. doi: 10.1111/j.1439-0507.2009.01822.x. [DOI] [PubMed] [Google Scholar]
- 15.Loreto ÉS, Scheid LA, Nogueira CW, et al. Candida dubliniensis: epidemiology and phenotypic methods for identification. Mycopathologia. 2010;169:431–443. doi: 10.1007/s11046-010-9286-5. [DOI] [PubMed] [Google Scholar]
- 16.Ngouana TK, Krasteva D, Drakulovski P, et al. Investigation of minor species Candida africana, Candida stellatoidea and Candida dubliniensis in the Candida albicans complex among Yaoundé (Cameroon) HIV-infected patients. Mycoses. 2015;58:33–39. doi: 10.1111/myc.12266. [DOI] [PubMed] [Google Scholar]
- 17.Ciardo D, Schär G, Böttger E, et al. Internal transcribed spacer sequencing versus biochemical profiling for identification of medically important yeasts. J Clin Microbiol. 2006;44:77–84. doi: 10.1128/JCM.44.1.77-84.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romeo O, Criseo G. Candida africana and its closest relatives. Mycoses. 2011;54:475–486. doi: 10.1111/j.1439-0507.2010.01939.x. [DOI] [PubMed] [Google Scholar]
- 19.Theill L, Dudiuk C, Morano S, et al. Prevalence and antifungal susceptibility of Candida albicans and its related species Candida dubliniensis and Candida africana isolated from vulvovaginal samples in a hospital of Argentina. Rev Argent Microbiol. 2016;48:43–49. doi: 10.1016/j.ram.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Rezazadeh E, Moazeni M, Sabokbar A. Use of cost effective and rapid molecular tools for identification of Candida species, opportunistic pathogens. Curr Med Mycol. 2016;2:1–4. doi: 10.18869/acadpub.cmm.2.3.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gajdács M, Dóczi I, Ábrók M, et al. Epidemiology of candiduria and Candida urinary tract infections in inpatients and outpatients: Results from a 10-year retrospective survey. Cent European J Urol. 2019;72:209–214. doi: 10.5173/ceju.2019.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu JF, Lai MY, Lee CW, et al. Comparison of the incidence, clinical features and outcomes of invasive candidiasis in children and neonates. BMC Infect Dis. 2018;18:194. doi: 10.1186/s12879-018-3100-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noni M, Stathi A, Vaki I, et al. Changing epidemiology of invasive candidiasis in children during a 10-year period. J Fungi (Basel) 2019;5:19. doi: 10.3390/jof5010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haddadi P, Zareifar S, Badiee P, et al. Yeast colonization and drug susceptibility pattern in the pediatric patients with neutropenia. Jundishapur J Microbiol. 2014;7:e11858. doi: 10.5812/jjm.11858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohammadi R, Ataei B. Candidiasis in pediatrics; identification and in vitro antifungal susceptibility of the clinical isolates. Iran J Ped Hematol Oncol. 2016;6:43–51. [PMC free article] [PubMed] [Google Scholar]
- 26.Romeo O, Criseo G. Molecular epidemiology of Candida albicans and its closely related yeasts Candida dubliniensis and Candida africana. J Clin Microbiol. 2009;47:212–214. doi: 10.1128/JCM.01540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solimani P, Salari S, Hassanzad M, et al. Use of PCR-RFLP and PCR-HWP1 for identification of Candida species isolated from cystic fibrosis patients. Res Mol Med. 2014;2:23–27. [Google Scholar]
- 28.Yazdanparast SA, Khodavaisy S, Fakhim H, et al. Molecular characterization of highly susceptible Candidaafricana from vulvovaginal candidiasis. Mycopathologia. 2015;180:317–323. doi: 10.1007/s11046-015-9924-z. [DOI] [PubMed] [Google Scholar]
- 29.Gumral R, Sancak B, Guzel AB, et al. Lack of Candida africana and Candida dubliniensis in vaginal Candida albicans isolates in Turkey using HWP1 gene polymorphisms. Mycopathologia. 2011;172:73–76. doi: 10.1007/s11046-011-9401-2. [DOI] [PubMed] [Google Scholar]


