Abstract

Personalized medicine and therapies represent the goal of modern medicine, as drug discovery strives to move away from one-cure-for-all and makes use of the various targets and biomarkers within differing disease areas. This approach, especially in oncology, is often undermined when the cells make use of alternative survival pathways. As such, acquired resistance is unfortunately common. In order to combat this phenomenon, synthetic lethality is being investigated, making use of existing genetic fragilities within the cancer cell. This Perspective highlights exciting targets within synthetic lethality, (PARP, ATR, ATM, DNA-PKcs, WEE1, CDK12, RAD51, RAD52, and PD-1) and discusses the medicinal chemistry programs being used to interrogate them, the challenges these programs face, and what the future holds for this promising field.
1. Introduction
It is commonly known that cancer is a disease of the genome, wherein errors in DNA replication and repair cause failures in cell function, while the mechanisms that would normally deal with these faulty cells are also compromised, resulting in cancer cell survival and ultimately proliferation. Before the advent of the human genome project in 2003, cancer treatments were mainly radiotherapy and nonspecific chemotherapy: two treatment methods that are famed for their lack of specificity and severe side effects. The human genome project has aided the route toward personalized medicine, wherein unique biomarkers in individuals can be detected and treated with tailor-made therapeutics for that specific cancer. There have been numerous success stories, primarily where “oncogene addiction” occurs, in which a particular subtype of cancer is over-reliant for survival on a particular oncogene, and therefore inhibition of this oncogene through small molecules or antibodies causes selected cell death. Examples include imatinib, which targets BCR-ABL that has extended the median survival of patients with chronic myelogenous leukemia to greater than 10 years.1 Another success story can be seen in melanoma, where treatment with the BRAF inhibitor vemurafenib has caused melanoma to change from a mostly untreatable disease to one where over 50% of patients show a meaningful clinical response.2 However, this trend toward precision-based personalized medicine has fallen out of fashion in the cancer drug discovery community. First, not all cancers show a simple oncogene addiction, and their survival mechanisms are far more complex; also, certain oncogenes have proved undruggable by small molecules and antibodies.3 Furthermore, even with initial success, issues of resistance and poor mechanistic understanding have hampered efforts, resulting in oncology still reliant on radiotherapy and nonspecific chemotherapeutics. Attention has turned toward synthetic lethality, exploiting hampered DNA repair mechanisms in cancer and allowing access to previously undruggable targets by indirect inhibition.4
Synthetic lethality (SL) was first discovered in the fruit fly (Drosophila pseudoobscura) in 1946 5 and can be defined as the relationship that can occur between two genes where either one functioning maintains viability of the cell; however upon dysfunction of both genes, the cell becomes unviable.6 In its simplest and most desirable application, this would result in the selective killing of cells that rely on the mechanisms driven by these two genes, i.e., cancer cells, while leaving the healthy cells alive. One synthetically lethal pair, PARP (poly (ADP-ribose) polymerases) and BRCA1/2, are now exploited in standard of care treatments;7 however this was 6 years ago, and still no other SL-based drugs have gained regulatory approval.
Within the field of synthetic lethality there are numerous examples of synthetically lethal gene pairs; therefore in this Perspective we select prominent and interesting examples to use as case studies for the field. Within these sections the targets typical mode of function is described, followed by the proposed mechanisms of synthetic lethality. Subsequently, medicinal chemistry programs that have produced molecules with potential to cause “small molecule-induced synthetic lethality” are discussed, including compounds in early stage, preclinical, and clinical studies. The review ultimately concludes with the challenges facing such small molecules and the drugging of their targets.
2. PARP and BRCA2
PARP is a family of enzymes containing 17 members,8 of which 15 have been shown to catalyze the transfer ADP-ribose to target proteins.9,10 PARP1 and PARP2 play important roles in DNA repair, making them an attractive target for oncology. PARP’s role in DNA repair was first discovered when it was observed that there was a correlation between high amounts of DNA lesions and increased PARP concentration.11 PARP is involved in the repair of single-stranded breaks (SSBs), through base excision repair (BER) (Figure 1), where it forms a part of the BER complex.12 In addition to this, PARP has been observed to play a role in nucleotide excision repair (NER). This and BER are both mechanisms for DNA repair.13 PARP has been observed in cell-free systems to bind tightly to broken DNA, and after auto-poly-ADP-ribosylation, it allows repair enzymes to access the DNA in order to initiate repair.14,15
Figure 1.

Schematic representation of PARP’s role in SSB and DSB repair. Upon formation of a SSB, PARP1 and other acceptor proteins (histone H1, TOP1, and others) are recruited and attached to the lesion site. This recruits other factors involved in DNA repair (Polβ, LigIII, TDP1, and XRCC1) which, through BER, repair the lesion. If PARP is inhibited, the SSB becomes a DSB which causes γH2AX foci formation, which in the presence of BRCA1/2 triggers HR which repairs the DSB. In the absence of BRCA this break becomes synthetically lethal. Adapted from Clinical Cancer Research, Copyright 2010, Vol. 16, Issue (18), , Page 4532, Christophe E. Redon, Asako J. Nakamura, Yong-Wei Zhang, Jiuping (Jay) Ji, William M. Bonner, Robert J. Kinders, Ralph E. Parchment, James H. Doroshow, Yves Pommier, Histone γH2AX and Poly(ADP-Ribose) as Clinical Pharmacodynamic Biomarkers ,22 with permission from AACR.
BRCA2 plays an important role in repairing double-stranded breaks (DSBs), as part of the homologous recombination (HR) pathway, and has been reported to be synthetically lethal when disrupted in combination with PARP,16 which as stated previously is involved in the repairing of SSB. This relationship was first discovered in 2005,17,18 and since this date, numerous papers have been published discussing the mechanism behind this.19−21 In the case of BRCA2 and PARP, in patients with defective BRCA2, PARP inhibitors can be administered to induce synthetic lethality.
Numerous cancers have defects in HR: such tumors include ovarian (50%) breast, prostate, and pancreatic cancers (all 10–20%).23 This is thought to aid cancer initiation and progression, with the hypothesis being that this fault in HR will cause more DNA breaks, resulting in more mutations. Due to cancer cell survival mechanisms, these DNA breaks may not necessarily be fatal to the cell and in fact may allow for more favorable conditions for cell survival.24 Due to the high proportion of cancers that express these HR defects, they are prime candidates for treatment with PARPi. Three main mechanisms have been hypothesized to exploit the synthetically lethal relationship between PARP and BRCA2. The first begins with a PARP inhibitor (PARPi) blocking BER, thus causing the conversion of an SSB to a DSB that BRCA2 would normally fix. If the tumor is genetically deficient in BRCA2, it will not be able to perform HR and therefore cause irreparable DNA damage, leading to cell death.23 In the second mechanism, the PARPi binds to the PARP1 enzyme on the chromatin, thus trapping it in mechanism referred to as PARP trapping; this causes a lesion that must be repaired which, due to compromised HR, is not possible and causes cell death. In the third mechanism, DSBs are resected during the S phase; however, since HR is defective, the alternative microhomology-mediated end joining (MMEJ) pathway attempts to repair the break. Since PARP1 is inhibited by the PARPi, it can no longer recruit POLQ, therefore blocking this pathway and triggering cell death;25 the same effect is thought to be achieved through POLQ dysregulation.26
PARP inhibitors demonstrate the first major success in the field of synthetic lethality and became the first evidence of modulating SL in the clinic when, in 2014, olaparib (1, Figure 2) became the first PARPi approved by the FDA. This was approved for the treatment of ovarian cancer, following a phase II trial in which 34% of patients exhibiting BRCA1/2 mutations showed compelling response rates. The patients had been given at least three rounds of chemotherapy previously and were then treated with 1 as a single agent.27 It was seen in this trial and subsequent ones that platinum-sensitive cancers showed a better overall response rate to PARPi compared to platinum-resistant cancers.28,29 Despite this, 1 has shown some success in platinum-resistant cancers in patients with BRCA1/2 mutations.28,30 This diversifies it from the other approved PARPi’s. 1 was approved in tablet and capsule form for the treatment of advanced ovarian cancer, where the patient had received at least three rounds of prior chemotherapy.
Figure 2.
Approved PARP inhibitors.
The second PARPi to be approved was rucaparib (2, Figure 2) in 2016, which demonstrated similar effects to 1 in phase II trials;31,32 however, the side effects between the two drugs are slightly different, affecting treatment choice. 2 was approved for use in patients with advanced ovarian cancer with deficient BRCA1/2 (either germline or somatic), who had received at least two treatments of chemotherapy in the past. The third FDA approved PARPi for the treatment of advanced ovarian cancer is niraparib (3, Figure 2), and in 2017, it was approved for patients with recurrent epithelial ovarian, fallopian tube, or primary peritoneal cancer in platinum-sensitive tumors.33
PARPi’s have also been approved for the treatment of breast cancer. This was achieved through two phase III trials, studying 1(34) and talazoparib (4, Figure 2)35 in comparison to the clinicians’ choice of chemotherapy in patients with human epidermal growth factor receptor 2 (HER-2) negative breast cancer who had received chemotherapy previously and have seen no progression on the platinum-based therapy. PARP inhibition showed an increase in progress-free survival in comparison to the platinum therapies. The final PARPi to gain regulatory approval was 4 in 2018, which is orally administered to adults with deleterious/suspected deleterious germline BRCA-mutated, (HER2)-negative, locally advanced, or metastatic breast cancer;361 was also approved for the same disease area. Furthermore, 1 was also approved for the treatment of pancreatic cancer as a first line maintenance treatment in cancers with a loss of function of BRCA2, following a double-blind placebo-controlled multicenter trial on 154 patients.37 The median progress-free survival of the participants increased from 3.8 months in the placebo to 7.4 months with 1. Although this is a small increase, any progress in this particularly challenging disease area should be applauded.
Recently (May 2020), 2, followed by 1, gained FDA approval for the treatment of germline and/or somatic BRCA-mutated metastatic castration-resistant prostate cancer. 2 was approved as a result of Triton 2, an ongoing multicenter, single arm clinical trial in 115 patients with BRCA-mutated (germline and/or somatic) mCRPC who had been treated with androgen receptor-directed therapy and taxane-based chemotherapy. 2 showed an objective response rate of 44% in 62 patients with measurable disease; 56% of these showed a duration of response greater than 6 months.381 was approved on the basis of a 2019 phase III trial of 387 men, where 1 increased progression-free survival from 3.6 months to 7.4 months. 1 also decreased the risk of disease progression or death to a median of 5.8 months vs 3.5 months for standard of care treatment.39
The field of PARP inhibition has been covered extensively by the literature; last year, an excellent review summarized the field of PARP inhibitors from a medicinal chemistry perspective, taking advantage of the extensive knowledge of the PARP protein to highlight binding motifs and structural similarities between the various generations of PARP inhibitors.40 The majority of inhibitors discussed in this review could theoretically be applied to inducing the SL phenotype; the field is extensively covered in the review by Jain et al., in which 37 different inhibitors are discussed with the majority of these being discovered after the approval of 1.
PARPi resistance can develop in cancer cells showing the BRCA2 mutation, occurring in one of two ways: the cell either finds a way to re-establish HR repair or repairs the break through alternative means, and both are covered in great detail in the review by D’Andrea.23 These mutations are relevant in a clinical setting, as 46% of platinum-resistant serious grade ovarian cancers showed a mutation that resulted in the restoration of the BRCA2 pathway. This is achieved either through genetic events that cancel the loss of function of the BRCA2 mutation or through genetic inversion of the mutation, resulting in restoration of the wild-type (WT) BRCA2 protein. These resistance mechanisms demonstrate the continuous need for evolution of understanding of PARP inhibition, and the BRCA1/2 pathway. Examples of how increased understanding of the PARP1/BRCA2 pathway could bypass resistance are shown later in this text, as these resistance mechanisms rely on the restoration of BRCA2 function. Through an increased understanding of the relationship between BRCA2 and RAD51, it has been proposed that PARP1 inhibitor efficacy could be restored through interruption of the BRCA2/RAD51 interaction. Therefore, given the crossover between many DNA repair mechanism modulating targets, it is possible that other key interactions for BRCA2 could be elucidated as understanding of these pathways increases, providing new potential SL-tools to restore PARPi activity.
3. ATR, ATM, and DNA-PKcs Inhibitors
ATM and DNA-PKcs exhibit complementary functions in DNA damage repair, notably HR and nonhomologous end joining (NHEJ). The co-dependent nature of these two kinases is evident from the inactivation of these two kinases, causing embryonic lethality.41 A loss of function through either genetic malfunction or chemical modulation can cause synthetic lethality within cells. The inhibition of both targets causes an accumulation of DSB, which in turn causes an increase CTBP interacting protein (CtIP) mediated resection, thus forming large single-stranded DNA (ssDNA) tracts, followed by the triggering of apoptosis through the ATR/CHK1 pathway.42 This has been achieved with inhibition of DNA-PKcs as a monotherapy in ATM-defective lymphomas.43 In addition to ATM, inhibition of DNA-PKcs has been shown to be synthetically lethal in cells in a number of other targets that play a key role in the HR process: BRCA1, BRCA2, CHK2, Rad50, PTIP, and PAXIP.44 The inactivation of ATM and DNA-PKcs has been seen to be more effective in BRCA1/2 deficient cells.45 The link between ATM and BRCA1/2 is not well understood: it has been hypothesized that rather than being dependent on BRCA1/2, ATM is dependent on certain genetic changes this genotype causes.46 Additionally, it has been seen that deficiency of either ATM or DNA-PKcs causes a sensitization to DNA-damaging agents such as topoisomerase I and II poisons or DNA alkylating agents.47 It has been proposed that the observed SL mechanism is not due to a failing of the DNA repair mechanism; this would differentiate this pathway somewhat from other SL pathways. Lastly, ATM has been shown to be synthetically lethal with the kinases MAPK and MEK1/2,48 with ATM playing a role in prosurvival pathways, noticeably the AKT/mTOR pathway: when MAPK or MEK1/2 is inhibited, if ATM is also impaired, this pathway is not open and therefore cell death occurs. This is important, as MEK inhibitors have already gained regulatory approval.49
As shown by its complementary functions with ATM, DNA-PKcs plays a key role in DNA repair in HR and NHEJ; the role of DNA-PKcs in DNA repair has been reviewed comprehensively by Goodwin et al.50 In addition to these well documented DNA damage repair pathways, DNA-PKcs is also involved with various processes that are thought to be important for tumor progression, such as cell cycle progression, transcription, and telomere maintenance,51 making DNA-PKcs an attractive target for cancer therapy.
DNA-PKcs is dysregulated in numerous cancers such as melanoma, where it encourages angiogenesis and tumor migration, and it has been discovered that the DNA-PKcs is associated with the secretion of prometastatic proteins through modification of the tumor microenvironment.52 DNA-PKcs dysregulation has also been observed in hepatocellular carcinoma53 and myeloma54 and is associated with radioresistance in cancers including thyroid,55 oral cavity,56 and cervical cancer.57
ATR is not directly associated with the DNA damage response; however it is involved with protecting cells from replication stress.58 It does this through preventing the collapse of the replication fork and inducing G2/M arrest by activating the checkpoint kinase CHK1. Cancer cells rapidly proliferate and therefore tend to undergo much replication stress and thus are particularly dependent on ATR. ATR has been observed to show SL with ATM/CHK2/p53: due to the amount of DSBs that are formed when ATR is inhibited, inhibiting key players in the DSB pathway is likely to result in an SL relationship. Of these SL relationships, ATR-ATM seems to be the most promising, as ATM is involved in both DNA repair in addition to cell cycle checkpoint activation. Furthermore, a deficiency in ATR has been shown to be synthetically lethal with agents that cause DNA stress, such as nonspecific chemotherapeutics. These SL relationships have been seen in various cancer cells including leukemia,59 pancreatic,60 and gastric cancer;61 in some cases, the SL phenotype was only observed in combination with DNA damaging agents.
ATR has also been shown to be synthetically lethal with CHK1, where small molecule-induced synthetic lethality has been reported, resulting in the selective killing of cancer cells.62 It has been hypothesized that the mechanism for SL of these two targets is that CHK1 inhibition increases replication stress through the deregulation of origin firing.63 The result of this is the stalling of DNA replication forks, increasing the concentration of ssDNA that must be repaired by the ATR-dependent replication protein A; however, due to ATR inhibition, this cannot occur and therefore a large amount of DSB accumulates, thus causing cell death.
ATR, ATM, and DNA-PKcs are promising targets for impeding DNA damage repair, as all are involved in the disruption of the cell cycle and the initiation of the DNA damage repair process (Figure 3). Currently multiple ATR inhibitors are in clinical trials, the data for which can be seen in Table 1.
Figure 3.

Demonstration of ATM, ATR, and DNA-PKcs involvement in DNA repair networks. Adapted from Trends in Cancer, Vol. 4, Issue (11), , Omar L. Kantidze, Artem K. Velichko, Artem V. Luzhin, Nadezhda V. Petrova, Sergey V. Razin, Synthetically Lethal Interactions of ATM, ATR, and DNAPKcs ,42 Page 762, Copyright (2018) with permission from Elsevier.
Table 1. List of Current Clinical Trials Involving Inhibition of ATR.
| trial identifier | drug | phase | summary | status and accession date |
|---|---|---|---|---|
| NCT04266912 | 6 | I/II | A combination study of avelumab with 6/nediseritib in participants with metastatic or inoperable tumors with deficient DNA damage repair. | Recruiting |
| Avelumab (PD-L1 antibody) | February 12, 2020 | |||
| Nedisertib (DNA-PKi) | ||||
| NCT02589522 | 6 | I | Studying the side effects and dose of 6 in combination with whole brain radiation in participants with non-small-cell lung cancer, small cell lung cancer, or neuroendocrine tumors that have spread to the brain. | Recruiting |
| October 28, 2015 | ||||
| NCT02630199 | 11 | I | A combination study assessing the safety and tolerability of 6 and paciltaxel in participating metastatic cancer patients who have failed standard chemotherapy. | Recruiting |
| Paclitaxel (nonspecific chemotherapeutic) | December 15, 2015 | |||
| NCT03896503 | 6 | II | Combination study comparing 6 and topotecan HCl to topotecan HCl as a single agent in participants with relapsed/extrapulmonary small cell lung cancer. | Recruiting |
| Topotecan HCl (nonspecific chemotherapeutic) | April 1, 2019 | |||
| NCT04149145 | 3 | I | A combination study of 3 and 6 in participants with PARP-resistant recurrent ovarian cancer, to assess the safety of the combination, the response rate, and the percentage of participants who proceed to 6 months progression-free survival among patients who have developed PARPi resistance and to assess the indicators of response and progression of this disease following the combination therapy. | Not yet recruiting |
| 6 | November 4, 2019 | |||
| NCT03527147 | AZD9150 | I | A study to investigate various targeted agents’ safety, tolerability, efficacy, and PK/PD profile in participants with relapsed or refractory aggressive non-Hodgkin’s lymphoma. | Recruiting |
| Acalabrutinib | May 17, 2018 | |||
| 11 | ||||
| Hu5F9-G4 | ||||
| Rituximab | ||||
| AZD5153 | ||||
| NCT03462342 | 1 | II | A combination study to assess the safety, tolerability, response rate, and progression-free survival of 1 and 11 in women with recurrent ovarian cancer (platinum-sensitive or platinum-resistant). | Recruiting |
| 11 | March 12, 2018 | |||
| NCT04065269 | 1 | II | A combination trial of 1 and 11 in participants with gynecological cancers with ARId1A loss or no loss, to assess response rates in groups of participants selected based on their cancer cell subtype and the presence of an abnormality in ARID1A gene. | Recruiting |
| 11 | August 22, 2019 | |||
| NCT02723864 | Veliparib | I | A combination study to assess the safety, tolerability, and maximum dose of 6 and veliparib combined with cisplatin in participants with advanced refractory solid tumors. | Recruiting |
| 6 | March 31, 2016 | |||
| NCT04267939 | 3 | I | A combination study to assess the safety, tolerability, and maximum/recommended phase II dose of 8 and 3 in participants with recurrent advanced solid tumors and ovarian cancer. | Recruiting |
| 8 | February 13, 2020 | |||
| NCT03188965 | 8 | I | A study to assess the safety, tolerability, maximum dose, and response rate of 8 in participants with advanced solid tumors and lymphomas. | Recruiting |
| July 16, 2017 | ||||
| NCT04095273 | 8 | I | A combination study to assess the safety/tolerability, PD/PK profile, and optimum dose for 8 and pembrolizumab in participants with advanced solid tumors. | Recruiting |
| Pembrolizumab (PD-1 antibody) | September 19, 2019 | |||
| NCT03641547 | 6 | I | A combination study of 6 with standard of care chemotherapeutics to establish the safety, tolerability, maximum tolerated dose, and efficacy in participants with esophageal and other cancers. | Recruiting |
| Cisplatin | August 22, 2018 | |||
| Capecitabine | ||||
| Radiotherapy (nonspecific chemotherapeutics) | ||||
| NCT02223923 | 11 | I | A study to assess 11 in combination with radiotherapy. The study will investigate 11’s safety, tolerability, dose, and dosing schedule while assessing preliminary drug response rates in participants with solid tumors. | Active, not recruiting |
| Radiotherapy | August 22, 2014 | |||
| NCT03682289 | 1 | II | A study to assess 11 as a monotherapy and in a separate arm of the trial in combination with 1, in participants with metastatic renal cell carcinoma, urothelial carcinoma, all pancreatic cancers, or other solid tumors. This study will assess objective response rate, median duration of response, median progression-free survival, and progression-free survival rate at 6 and 12 months. It will also further assess 11’s safety. | Recruiting, |
| 11 | September 24, 2018 | |||
| NCT03641313 | 6 | II | A combination study to determine the overall response rate of 6 and irinotecan in participants with progressive, metastatic, or unresectable TP53 mutant gastric or gastroesophageal junction cancer. Furthermore, it seeks to assess the duration of response, time to progression, progression-free survival, and OS in participants with the combination of drugs in comparison to irinotecan alone and in participants with other DNA damage repair defects such as mutations in RCA1, BRCA2, MRE11, RAD50, RAD51, RAD52, RAD54L, NBN, ATM, H2AX, PALB2, RPA, BRIP1, BARD1, ATR, ATRX, CHK1, CHK2, MDM2, MDM4, FANCA, FANCC, FANCD2, FANCE, FANCF, FANCG and FANCL. Lastly, it seeks to compare the combination of 6 and irinotecan in participants who are Pt-sensitive and resistant. | Not yet recruiting |
| Irinotecan (topoisomerase inhibitor) | August 22, 2018 | |||
| NCT02627443 | 6 | I/II | A combination study to assess the safety, tolerability, maximum tolerated dose of 6, carboplatin, and gemcitabine HCl in patients with recurrent and metastatic ovarian, primary peritoneal, or fallopian tube cancer. Furthermore, this study seeks to assess the maximum tolerated dose effect on overall survival, duration of response, and progression-free survival. | Recruiting |
| Carboplatin (nonspecific chemotherapeutic) | December 11, 2015 | |||
| Gemcitabine HCl (Cytotoxic chemotherapeutic) | ||||
| NCT02567409 | 6 | II | A combination study to assess whether 6 in combination with standard of care therapeutics improves progression-free survival, in comparison to standard of chemotherapy as a monotherapy in patients with metastatic urothelial cancer. Furthermore, this study seeks to compare overall survival, tumor response rate, and safety between the two study arms. Lastly, to assess the role of p53 in predicting response to 6. | Active, not recruiting |
| Cisplatin (nonspecific chemotherapeutic) | October 5, 2015 | |||
| Gemcitabine HCl (cytotoxic chemotherapeutic) | ||||
| NCT02487095 | 6 | I/II | A study to assess the safety and efficacy of 6 and topotecan in treating small cell lung cancer. | Recruiting |
| Topotecan (nonspecific chemotherapeutic) | July 1, 2015 | |||
| NCT03787680 | 1 | II | A combination study is to test the effectiveness, safety, and tolerability of 1 and 11 for all participants with metastatic castration-resistant prostate cancer. | Recruiting |
| 11 | December 26, 2018 | |||
| NCT02595892 | 6 | II | A combination study to assess the progression-free survival of 6 and gemcitabine in comparison to gemcitabine as a monotherapy in participants with recurrent ovarian, primary peritoneal, or fallopian tube cancer. Across both arms will also be tested overall response rate, safety profiles, progression-free survival at 6 months, clinical benefit rate, duration of response, cancer antigen (CA)125 reduction, and overall survival. | Active, not recruiting |
| Gemcitabine (cytotoxic chemotherapeutic) | November 4, 2015 | |||
| NCT03669601 | 11 | I | A dose escalation trial to assess the safety of 11 in combination with gemcitabine in participants with advanced solid tumors. | Recruiting |
| Gemcitabine (cytotoxic chemotherapeutic) | September 13, 2018 | |||
| NCT03328273 | 11 | I/II | A combination study that evaluates the safety, pharmacokinetics, pharmacodynamics, and efficacy of acalabrutinib and 11. | Recruiting |
| Acalabrutinib (BTK inhibitor) | November 1, 2017 | |||
| NCT04052555 | 6 | I | A combination study to test the recommended dose for phase II trials of 6 in combination with standard of care radiotherapy in participants with triple-negative or estrogen receptor and/or progesterone receptor positive, HER-2 negative breast cancer. | Recruiting |
| Radiotherapy | August 12, 2019 | |||
| NCT02567422 | 6 | I | A combination study to assess the safety, tolerability, and maximum tolerated dose of 6 in combination with standard of care cisplatin and radiotherapy in participants with locally advanced head and neck squamous cell carcinoma. | Recruiting |
| Cisplatin (nonspecific chemotherapeutic) | October 5, 2015 | |||
| Radiotherapy | ||||
| NCT02595931 | 6 | I | A study to assess the safety, tolerability, and maximum tolerated dose of 6 and irinotecan hydrochloride in treating participants with metastatic or unresectable DNA damage repair deficient solid tumors. | Recruiting |
| Irinotecan HCl (topoisomerase inhibitor) | November 4, 2015 | |||
| NCT04266912 | 6 | I/II | A study to assess the safety, tolerability, and maximum tolerated dose of 6 and avelumab in treating participants with metastatic or unresectable DNA damage repair deficient solid tumors. | Recruiting |
| Avelumab (PD-L1 antibody) | February 12, 2020 | |||
| NCT03517969 | 6 | II | A combination study that studies response rate of 20 and carboplatin with and without docetaxal in participants with castration-resistant prostate cancer. | Recruiting |
| Carboplatin (nonspecific chemotherapeutic) | May 8, 2018 | |||
| Docetaxel (microtubules binder) |
VE-821 (5, Figure 4A) (ATR IC50 = 26 nM) is an ATR inhibitor developed by Vertex pharmaceuticals in 2011. This compound is typical of numerous inhibitors from this development program, all of which contain a 2-aminopyrazine binding motif.64 While this compound was a useful tool in understanding the underlying biology associated with ATR inhibition, it did not exhibit good solubility, and its metabolic profile, specifically producing aniline-like metabolites, was undesirable for a clinical candidate; thus Vertex began optimization of 5.65 Initial optimization of 5 involved the replacement of the anilide group with various fused bi-cycles: while these were well-tolerated in terms of ATR inhibition, they also demonstrated a loss in selectivity for ATR against ATM. Through in silico modeling it was theorized that this loss in selectivity was caused by the bulky fused bi-cycles and the gatekeeper residue of the PIKK tyrosine kinase family, causing rotation of the bound inhibitor. This in turn led to a further steric clash between Pro2755 of ATM and the isopropylsulfone group of the inhibitor that caused tighter ATM binding. With this information in hand, Vertex theorized five-membered phenyl substituted heterocycles would avoid these steric clashes and therefore would be able to maintain selectivity, as they more closely resemble the anilide moiety present in 5. Additionally, with substitution upon the phenyl ring, a localized highly negatively charged area of the ATP binding pocket could be exploited to further increase potency, selectivity, and solubility. These SAR studies resulted in the eventual synthesis of VX-970 (6, Figure 4A), which shows picomolar ATR activity (IC50 = 0.17 nM) and >250 degree of selectivity over ATM. In cells, it was able to sensitize colorectal cancer cells to cisplatin, using less than 50 nM 6. Furthermore, it demonstrated a good pharmacokinetic profile (Cl = 26 mL min–1 kg–1; Vss = 21 L/kg; T1/2 = 11.6 h), in addition to good bioavailability, making it ideal for further in vivo studies. 6 is a first-in-class inhibitor and has been tested both as a monotherapy and with nonspecific chemotherapeutics such as topotecan, carboplatin, gemcitabine, and cisplatin.66−696 presented promising results as a monotherapy, showing a complete response in one patient for greater than 19 months, with no associated toxicity. However, in phase I trials with the nonspecific chemotherapeutics, bone marrow toxicity was observed. The toxicity issues observed are somewhat common for nonspecific chemotherapeutics, and in this instance the combinatorial approach yielded no benefit over the monotherapeutic approach. Several trials are still ongoing for 6 in numerous cancer subtypes. 6 has been acquired by Merck KGaA under the new name M6620; additionally Merck has entered a second ATR inhibitor using a different scaffold into clinical trials, M4344 (7, Figure 4B) (ATR Ki ≤ 150 pM).70 The development of 7 has yet to be published, with preclinical data only being presented at a congress.
Figure 4.
(A) SAR study around ATR inhibitor VE-821 (5) to get VX-970 (6). (B) Structure of ATR inhibitor M4344 (7). (C) Structure of BAY1895344 (8) identified by Bayer AG as a selective ATR inhibitor.
BAY1895344 (8, Figure 4C) was developed by Bayer AG as described in a patent (8 is example 111 in patent WO 2016/020320),71 through a high-throughput screen HTS. 8 is a novel potent (7 nM) and selective ATR inhibitor (hitting 6 of 456 kinases tested in the kinome screen). Furthermore, 8 was able to selectively inhibit ATR mediated DNA repair and inhibited proliferation in a range of cancer cell lines, being most active in lymphoma cell lines showing an IC50 of 9 nM.72 It exhibits strong monotherapy efficacy in cancers with impeded DNA damage repair. In addition to this, it synergizes well with nonspecific chemotherapeutics (cisplatin and carboplatin) and other DNA damaging agents (external beam radiotherapy).
It is currently under investigation in clinical trials in patients with advanced solid tumors and lymphomas (Table 1). Lastly, 8 showed synergism with 1 in BRCA1/2 deficient breast cancer cells, suggesting a synthetically lethal interaction; these results were mirrored in vivo in breast cancer models.
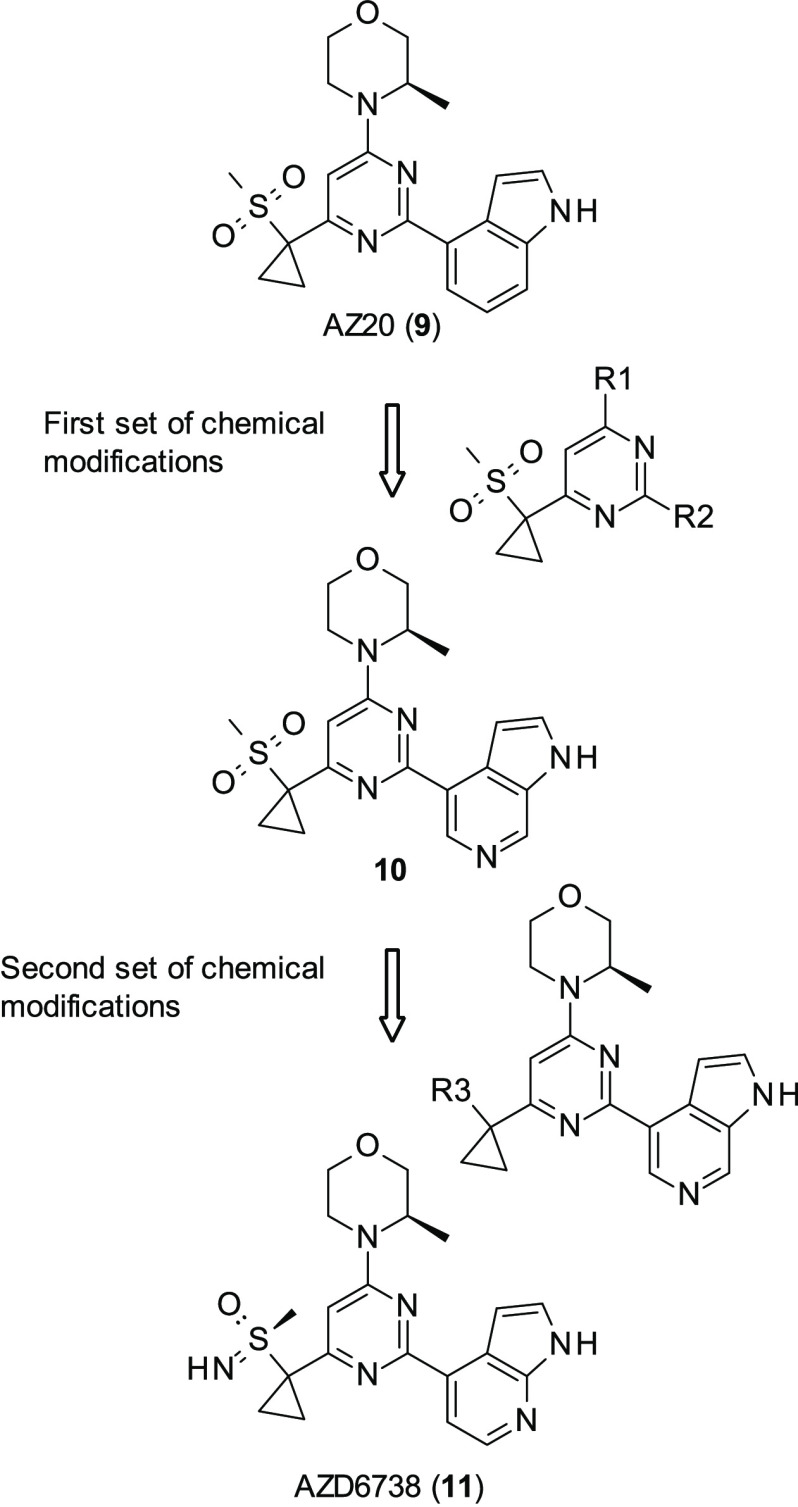
AstraZeneca (AZ) had previously developed a series of potent and selective ATR inhibitors, of which the best performing was AZ20 (9, Figure 5) (ATR IC50 = 5 nM).73 However, while this compound was able to prevent the growth of ATM-deficient xenograph models in tolerated dosages, its aqueous solubility was poor. Furthermore, it was found to hit cytochrome P450 3A4 (CYP3A4), thereby increasing the chance of drug–drug interaction: given the unlikeliness of ATR inhibitors being administered as a monotherapy, this would stop it progressing through further studies. Therefore, a drug-discovery program was initiated, looking to keep 9’s activity but with improved aqueous solubility and lessened CYP3A4 activity.74 Initial SAR looked at making modifications at the R1 and R2 positions (Figure 5); alterations at the R1 position did not reduce CYP3A4 activity and therefore were quickly abandoned. Modifications at the R2 position began with simple ring substitutions; while this did reduce CYP3A4 activity, it also decreased ATR activity, suggesting that the indole group was the reason for CYP3A4 activity. Ring switch to 6- or 7-azaindole and to 2-aminobenzimidazole showed similar ATR activity in cell-based and biochemical assays to AZ20 while reducing CYP3A4 activity. The best of the series was 10 (Figure 5) (ATR IC50 = 5 nM). Exploration of substitution of the benzimidazole was attempted; however, none of the changes at this position proved fruitful as all modifications in this position resulted in either loss in ATR inhibition or physicochemical properties, such as aqueous solubility.
Figure 5.
Chemical modifications introduced in AZ20 (9) to improve solubility and reduce CYP3A4 activity.
Subsequent modifications began at the sulfone group, looking to expand upon 10. While initial replacement of the sulfone with sulfoxide resulted in promising molecules, the metabolic risks associated with sulfoxides were deemed too high, and therefore AZ did not pursue this avenue of research. However, substitution with various sulfoximines resulted in AZD6738 (11, Figure 5) (ATR IC50 = 4 nM), which achieved all goals of the lead-development program, with decreased lipophillicity and improved aqueous solubility, in addition to reduced CYP3A4 activity in comparison to 9.
Compound 11 was taken through to further screening for its biological, physiochemical, and ADME properties. It was found to be the lead compound from this series due to its excellent solubility, permeability, and selectivity (only hitting ∼60% for PIK3C2G and CLK4 out of the 409 kinases tested in the kinome screen) and demonstrated profound antitumor activity. 11 has progressed in multiple phase I/II trials, and while some are in combination with nonspecific chemotherapeutics such as carboplatin (NCT02264678), it has also entered trials with the PARP inhibitor 1, suggesting synthetic lethality (NCT03462342, NCT03330847), with further preclinical studies underway for other combinations with 1.75 Overall, the clinical candidate 11 has a favorable pharmacokinetic profile for once or twice daily dosing, achieving biological efficacy at moderate dosages. The selectivity of this compound should be noted as it makes it an ideal tool for further exploration of ATR’s synthetically lethal partners such as ATM and CHK1.
It has been discovered that homology exists between PI3K and ATR;76 therefore, it was hoped that it could be harnessed for the discovery of novel ATR inhibitors. Ramachandran et al. had previously been working on dual inhibitors of Bruton’s tyrosine kinase (BTK) and PI3K:77 this was used as a starting point for the design of ATR inhibitors.78 From the 299 selected compounds in the BTK series, 11 showed inhibition of >50% at 5 μM while only 1 showed inhibition of >50% at 500 nM. Interestingly, despite the homology between the two kinases, there was poor overlap between the inhibition of both, suggesting that the scaffold utilized in this series is differentiated somewhat from other ATR inhibitors (8, 9–11), which were also developed from PI3K inhibitors. Of the originally tested compounds, 12 (Figure 6A) stood out as the prime target for further optimization not only because of it being the strongest inhibitor of ATR from this series (IC50 = 0.220 μM) but also because it showed relatively low activity against BRK and PI3K (BRK inh = 24%, PI3Kd inh = 52% at 100 nM). 12 was tested as a racemic mixture as there was less than a 2-fold difference in ATR activity between the two enantiomers (IC50 = 0.304 μM vs 0.158 μM). This was tested with three other compounds (13–15, Figure 6) that also showed good activity (in the low micromolar range). All four of these compounds share the same common 4-aminopyrazolopyrimidine scaffold functionalized at N1 with a partially aromatic bicyclic system: it was discovered that removal of the hydrogen bond donors/acceptors present in this core scaffold was not well-tolerated, causing either a partial or complete loss of activity against ATR. In addition to this, replacement of the azaindolyl group with an aminopyridine group which displayed the same hydrogen bond acceptors/donors was not tolerated, suggesting the steric bulk present in the aminopyrazolopyrimidine group is necessary for ATR inhibition. Follow-up SAR was conducted, introducing new bulky groups at the C3 position, and yet none of these were able to improve on the activity of 12. Therefore the focus of the SAR studies turned to alkylation at the N1 position. While the majority of modifications at this position did not yield a significant improvement on the activity of 12, compound 16 (Figure 6B), which takes influence from Vertex’s 5(65) featuring a phenylsulfonyl group, managed to yield the most potent ATR inhibitor in this series to date with a 3-fold improvement in activity (IC50 = 66 nM) over 12.
Figure 6.
(A) Hit compounds from Ramachandran et al. (B) Optimization of 12 to 16.
16 was compared to the clinical candidate ATR inhibitors 6 (IC50 = 15 nM) and 11 (IC50 = 86 nM) and in the same testing conditions showed similar levels of ATR inhibition. The lipophilic efficiency of 16 (5.8) and oxidative stability were in the same region of the two literature compounds. 16 was also tested for selectivity in a kinome screen of 394 kinases: of these, 10 were inhibited at greater than 70% at 1 μM and 76 were inhibited at greater than 70% at the 10 μM, with only breast tumor kinase (BTK) being inhibited to a greater degree than ATR. Lastly, 16 was tested in vivo in mice, where it showed good clearance but poor bioavailability, suggesting further optimization is required. Given ATR’s potential for small molecule-induced synthetic lethality, any optimization of this compound should be monitored with interest.
In 2004, Hickson et al. developed the first selective ATM inhibitors, as up to this date all molecules that had inhibited ATM were nonspecific PIKK and PI3k inhibitors such as caffeine. Through a combinatorial library approach based around the nonspecific PIKK and PI3k inhibitor LY294002 (17, Figure 7A), the group developed the ATP competitive inhibitor KU-55933 (18, Figure 7A), which demonstrated in biochemical assays an IC50 of 12.9 ± 0.1 nM, with at least a 100-fold degree of selectivity over 60 other selected similar kinases.79 In these studies, interesting structural information was gleaned when replacing the oxygen in the morpholine ring (Figure 7). This resulted in a significant drop in potency in KU-58050 (19, Figure 7A) (2.96 ± 0.44 μM), a 200-fold drop in potency in comparison to 18, showing the importance of the oxygen at this position. Furthermore, 18 was able to sensitize cells to ionizing radiation and nonspecific chemotherapeutics, thus increasing the amount of DSB.
Figure 7.
(A) Development of ATM inhibitor LY294002 (17) into KU-60019 (20). (B) Structure of ATM inhibitor CP466722 (21).
Following on from this work, in 2009 Golding et al. developed KU-60019 (20, Figure 7A) by expanding upon the thianthrene group with the aim of improving 18’s bioavailability and pharmacokinetics.8020 was more water-soluble, was able to double the potency with respect to 18 (6.3 nM), and was more selective, showing a 240- and 1600-fold degree of selectivity for DNA-PKcs and ATR, respectively. 20 is better at sensitizing cells to ionizing radiation, showing an enhancement of 4.4 compared to 1.6 from 18, when both compounds were administered to glioma cells at 10 μM. In scratch assays, 20 was able to reduce invasion in vitro in U87 cells and a greater than 70% drop in invasion was observed in a dose-dependent manner. In the more invasive U1242 glioma cells, a greater than 50% decrease in invasion was observed. It is thought that 20 showed this anti-invasive phenotype by acting on AKT and MEK/ERK prosurvival pathways. 20 is currently undergoing a clinical trial (Table 2) in combination studies with CHK2 inhibitors; should this work, it could indicate small molecule-induced synthetic lethality.
Table 2. Current Clinical Trials Involving ATM Inhibitors.
| trial identifer | drug | phase | summary | status and accession date |
|---|---|---|---|---|
| NCT03423628 | 29 | I | A study to assess the safety, tolerability, and PK of increasing doses of 29 in combination with distinct regimens of radiation therapy in participants with brain cancer. | Recruiting |
| Radiotherapy | February 6, 2018 | |||
| NCT03571438 | CX4945 (CK2i) | Not applicable | A combination study to compare CX4945 and 20 to standard of care treatments directly on organotypic cultures of tumors from participants, measuring incidents of cell death. | Recruiting |
| 20 | June 27, 2018 | |||
| Sunitinib (multiple RTKi) | ||||
| Pazopanib (multiple RTKi) | ||||
| Temsirolimus (mTORi) | ||||
| NCT02588105 | 1 | I | A study to test the safety, tolerance PK/PD, and initial efficacy of 28 as a monotherapy and in combination with the other listed drugs. | Active, not recruiting October 27, 2015 |
| 28 | ||||
| Irinotecan (topoisomerase inhibitor) | ||||
| Fluorouracil (thymidylate synthase inhibitor) | ||||
| Folinic acid |
In 2008 Rainey et al., through use of an in vitro kinase screen of approximately 1500 small molecules, identified CP466722 (21, Figure 7B) (ATM IC50 = 12.9 nM).81 This was able to inhibit ATM activity in vitro but did not show activity against the closely related kinases in the PI3k family. 21 showed a lack of toxicity and the ability to inhibit ATM in human and mouse cells, the latter being important as the group wished to test the compounds in vivo in murine models. Furthermore, 21 was able to sensitize cancer cells to ionizing radiation. It was also demonstrated that 21 was a reversible binder, shown to bind quickly and effectively for at least 8 h in tissue culture; however, upon 30 min of wash off, ATM-dependent phosphorylation events were restored. This reversibility is key due to the important role ATM plays in the cell cycle.82 This compound has yet to be used for further studies.
AstraZeneca has acquired KuDOS pharmaceuticals and has continued building upon the progress made by 20. Through optimization of an initial screening hit, 22 (Figure 8) (ATR cell IC50 = 82 nM), AZ was able to develop two further compounds: 23 (Figure 8) (ATR cell IC50 = 46 nM) and 24 (Figure 8) (ATR cell IC50 = 33 nM). 23 and 24 allowed exploration of the ATM inhibitors in vivo.83 At an oral dosage of 100 mg/kg QD, 23 was able to increase sensitivity to ionizing radiation in a HT29 xenograph mouse model.84 Both 23 and 24 were able to enhance the efficacy of irinotecan in a SW620 mouse xenograft models, with 24 able to contribute to tumor regression.83,84 In line with other ATM inhibitors, 23 and 24 were ineffective without a DNA damaging agent to induce DSB. 24 showed a favorable reduction in activity against hERG relative to 23 (IC50 of 22 and 4.5 μM, respectively) and therefore had a lower chance of cardiovascular issues later on in clinical development, suggesting that 24 would be the better clinical candidate going forward.85 Through pharmacological modeling, it was found that 24 would have a low half-life (only 4 h) in humans, and therefore the predicted dose would be 700 mg QD, with a maximum unbound concentration Cmax of 1.3 μM. In addition, other calculated parameters such as predicted clinically efficacious dose and maximum absorbable dose (Dabs) were unfavorable; therefore AZ concluded that the probable chance of attrition was too high. A further development program was initiated with an aim to produce molecules with a reduced predicted clinical dose and an increased Dabs. AZ set about attempting to increase their compounds’ volume of distribution (Vss), as pharmacokinetic half-life is calculated through Vss and clearance (CL). As 24 already showed a low metabolic turnover, further reduction of CL would be challenging; therefore, Vss was chosen as the parameter to optimize. This began with the attempted inclusion of a basic substituent with the molecule, as it has been shown previously that basic compounds increase Vss with respect to acidic or neutral compounds,86 with the idea of keeping other parameters the same. While 25 (ATR cell IC50 = 8.6 nM) showed an increase in ATM inhibition with respect to 24, a drop in permeability to unacceptable levels occurred, meaning that basic substituents would be unlikely to be tolerated going forward. In an effort to increase permeability by reducing the number of hydrogen bond donors, a scaffold hop was attempted. This was done using a pseudo-ring system and a tri-cycle system, imidazo[4,5-c]quinolin-2-one, which was selected from previous AZ in-house data. This scaffold has also seen success in ATM inhibition as an off-target in the dual PI3K/mTOR inhibitor, NVP-BEZ235, which has entered clinical trials.87,88 Of these compounds, 26 and 27 (Figure 8) were the most potent (ATM IC50 = 0.95 and 0.36 μM, respectively), with 27 showing reduced aqueous solubility. Given the promising results with this scaffold, motifs from 23 and 24 were modeled upon the imidazo[4,5-c]quinolin-2-one scaffold to create a second series. Through in-depth SAR of the imidazo[4,5-c]quinolin-2-one scaffold, AZ eventually identified AZD0156 (28, Figure 8) as the lead compound. 28 showed an excellent affinity for ATM (IC50 = 0.04 nM in biochemical assays, 0.57 nM in cells) and an exceptional degree of selectivity, with only 2 of the 397 kinases tested in the kinome screening showing greater than 70% inhibition (mTOR, 93%; LRRK2, 87%). 28 showed desirable levels of unbound drug in human plus two other mammalian species (rat and dog) and acceptable permeability, and it did not target any of the main five isoforms of P450.89 In addition to this, the predicted pharmacokinetics of 28 were favorable, with low to moderate clearance in man (∼8 mL min–1 kg–1), moderate to high Vss (5.8 L/kg), and high oral bioavailability (66%). In docking studies, 28 showed good complementarity to ATM’s ATP binding site, displaying two interactions with the kinase hinge region (Cys2770 and Lys2717) and one in the back binding pocket (Tyr2755). In addition to this, the basic amine, present in 28, is predicted to be surrounded by three acidic residues (Asp2725, Asp2720, and Asp288). 28 demonstrated the ability to increase the effectiveness of DSB-inducing agents such as irinotecan. In combination studies, irinotecan (50 mg/kg ip Q7D) and 28 (dosed orally at 20 mg/kg QD) were tolerated, causing tumor regression in SW620 xenograph models in immunocompromised mice. However, as expected, when 28 was administered as a monotherapy, it showed no effect. All these results indicate that 28 is a first-in-class selective ATM inhibitor with promising potential applications for cancer therapy; it is currently involved in a clinical trial highlighted in Table 2.
Figure 8.
Development by AstraZeneca of ATM inhibitor from hit 22 into candidate AZD1390 (29) through the lead AZD156 (28).
With the 28 series showing success, further screens were conducted on related hits to try and find other clinical candidates. Given rates of attrition in drug development, having a second, backup molecule in the pipeline provides a level of security. AZD1390 (29, Figure 8) was discovered in an in vitro screen, with a similar structure as 28. It was designed to perform well in the following parameters: ATM autophosphorylation activity (IC50 = 0.09 nM); selectivity against closely related PIKK family kinases (ATR, DNA-PK, mTOR (all IC50 ≥ 1 μM)); general selectivity across the kinome; lack of activity in novel dual-transfected human MDR1 and BCRP efflux transporters assays.90 In terms of general selectivity, 29 was tested against a panel of 121 kinases and showed ≥50% inhibition against three targets (CSF1R, NUAK1, and SGK) at 1 μM, with none of the 354 kinases tested showing ≥50% inhibition at 0.1 μM. In a similar fashion to 28, 29 showed minimal hERG activity (>33.3 and 6.55 μM, respectively). 29 demonstrated good blood–brain barrier (BBB) penetration and has favorable physicochemical and PD/PK properties. Furthermore, it was able to interrupt DNA damage repair in glioblastoma and lung cancer cells. In line with previous studies on ATM inhibitors, 29 was more effective in p53-deficient cells.91 It was also demonstrated that treatment of animal orthotropic brain models of glioblastoma and lung cancer brain metastasis (>3 h) with 29 at a concentration above the IC50, in combination with ionizing radiation, blocked tumor growth. It was also observed that a combination of ionizing radiation and 29 caused an increase in apoptosis in comparison to 29 as a monotherapy. In brain cancer models using intracranially implanted xenografts, 29 was tested in combination with ionizing radiation and a triplet combination of 29, ionizing radiation, and the alkylating agent temozolomide.
While the triplet combination showed more success than the doublet, it appeared to be an additive effect, while ionizing radiation and 29’s combination were synergistic. In neither study, any behavioral abnormalities were observed because of the administration of 29, which is key for treatment of the brain. 29 was tested in human-derived glioblastoma models from patients with either temozolomide resistance or sensitivity and either p53 WT or mutant, with the most sensitive to 29 and ionizing radiation as expected being in the p53 mutant models, thus agreeing with their previous data. On the basis of these data, 29 is being considered as a clinical candidate and entered a phase I clinical trial in patients with brain cancer, the information of which are reported in Table 2. These data, in particular the work in glioblastoma, are of great importance to the field. Glioblastoma is the most aggressive cancer of the brain, with poor patient prognosis, a seemingly inevitable rate of recursion, and a median survival rate of 12–14 months with current standard of care therapy.9229 shows the first example of potential SL-related treatment of glioblastoma, one of the most urgent unmet medical needs in oncology.
As highlighted earlier in the review, DNA-PKcs is thought to be synthetically lethal with a number of DNA repair modulating targets, specifically ATM. In this section we highlight some of the DNA-PKcs inhibitors currently in clinical and preclinical studies.
SU11752 (30, Figure 9) was the first selective DNA-PK inhibitor to be discovered when it was identified from a library of 3-substituted indolin-2-ones. 30 showed a similar in vitro DNA-PK activity to the known PI3K inhibitor wortmanin (31, Figure 9) (30 IC50 = 0.13 μM vs 31 IC50 = 0.1 μM). The binding mode of 30 was then assessed: 31 has been shown previously to irreversibly bind to DNA-PK, a trait that would be undesirable in a cell cycle modulator;93 however 30 was shown to be a reversible ATP competitive inhibitor. Furthermore, 30 was much more selective than 31, as 30 was 500 times less active against 31’s primary target PI3Kγ. Additionally 31 inhibits ATM, while in cells 31 was seen to be a poor inhibitor of ATM. 31 was able to sensitize glioblastoma cells to ionizing radiation and disrupt DNA repair.
Figure 9.
Structure of SU-11752 (30) and wortmannin (31).
17 is a known PI3K inhibitor (PI3Kα, -β, -δ IC50 = 0.5, 0.97, 0.57 μM) that was used as a starting point in the pursuit of novel selective DNA-PK inhibitors in work conducted by Griffin et al. Initial optimization of 17 (Figure 10) resulted in the synthesis of a series of DNA-PK inhibitors, which displayed IC50’s in the low micromolar or nanomolar range. The most potent of this series were pyrimidoisoquinolinone (32, Figure 10) (DNA-PK IC50 = 0.28 μM) and NU7026 (33, Figure 10) (DNA-PK IC50 = 0.23 μM), which were able to sensitize tumor cell lines to ionizing radiation and DNA damaging agents.94
Figure 10.
Optimization of PI3K inhibitor 17 resulting in the discovery of KU-5788 (34).
This work was continued through the use of a pharmacophore mapping approach, replacing or substituting upon the 2-morpholinyl substituent. This approach resulted in the discovery of KU-57788 (34, Figure 10) (formerly NU7441).9534 was shown to be a potent DNA-PK inhibitor (IC50 = 13 nM) with good selectivity against ATM and ATR (no activity at 100 μM); however it also hit mTOR and PI3K (IC50 = 1.7 and 5 nM, respectively). In a similar manner to other DNA-PK inhibitors 34 was able to enhance radiosensitization and the cytotoxicity of etoposide, in Hela cells, while showing no cellular toxicity as a single agent.
AZ set out to discover a selective DNA-PK inhibitor. AZ had observed that many of the current DNA-PK inhibitors showed poor selectivity over related PI3Ks and PIKK family members such as ATM and mTOR. Through use of an in-house screen searching for DNA-PK activity over PI3K, the initial hit (35, Figure 11A) (biochemical data not reported) was discovered. 35 was optimized, improving its potency, physiochemical, and pharmacokinetic properties to give AZD7648 (36, Figure 11A).9636 was a potent inhibitor of DNA-PK (IC50 = 0.6 nM) and showed good selectivity: in a panel of 397 kinases, only 4 showed inhibition of >50% (PK, PI3Kα, PI3Kδ, and PI3Kγ), and of these 4 kinases 36 showed at least a > 60-fold degree of selectivity for DNA-PKcs. In A549 cells 36 inhibits DNA-PKcs autophosphorylation at Ser2056 (IC50 = 90 nM); furthermore its selectively was also repeated in cells, with 36 showing a >90-fold selectivity for DNA-PKcs over ATM, ATR, mTOR, and three PI3K isoforms (α, β, and δ), with a 10-fold selectivity increase over PI3Kγ. The selectivity of 36 was compared to other notable DNA-PKcs inhibitors (including 37) which all had at least one secondary target with <10-fold selectivity. 36 was able to enhance DNA damaging therapies such as radiotherapy, doxorubicin, and 1in vitro and in vivo. Due to its mechanism of acting through inhibition of NHEJ, as opposed to HR, 36 is differentiated from many of the current candidates for SL modulating drugs. This, combined with its potency and selectivity, makes it an interesting compound for further study.
Figure 11.
(A) Optimization of DNA-PK inhibitor 35 to AZD7648 (36) by AZ. (B) Structure of DNA-PK inhibitor M3814 (37) developed by Merck.
M3814 (also known as nedisertib) (37, Figure 11B) is a potent selective DNA-PK inhibitor (DNA-PK IC50 ≤ 3 nM).97 Information on the development of this inhibitor has not been published, and 37 was first described in a patent filed by Merck.9837 has been shown to enhance the antitumor effect of ionizing radiation in solid tumors and leukemia through inhibition of the NHEJ pathway, a common feature of selective potent DNA-PK inhibitors.97,99,100 This effect is much more pronounced in cancer cells that express WT-p53, thought to be due to overactivation of p53/ATM which causes a much higher concentration of p53 in comparison to radiation treatment administered alone, leading to premature cell cycle arrest and senescence. This enhancement of radiotherapy has also been observed in mice, where combination of 37 and radiotherapy caused complete remission.97 Due to these promising results in vitro and in vivo, four clinical trials are currently underway using 37. These are listed in Table 3. To date, 37 is the only selective DNA-PK inhibitor involved in clinical development.
Table 3. List of Current Clinical Trials Involving Inhibition of DNA-PK.
| trial identifier | drug | phase | summary | status and accession date |
|---|---|---|---|---|
| NCT02316197 | 37 | I | A first in human trial to assess the safety, efficacy, PK/PD properties, and tolerability of 37 in patients with advanced solid tumors or chronic lymphocytic leukemia. A secondary aim is to discover the correct dosing regime for 37 for future studies. | Completed (no results posted) |
| December 12, 2014 | ||||
| NCT02516813 | 37 | I | A trial to assess the safety, efficacy, and tolerability of 37 in combination with radiotherapy and chemotherapy in patients with solid tumors. | Recruiting |
| Radiotherapy cisplatin | April 6, 2015 | |||
| NCT03770689 | 37 | I/II | The purpose of this study in phase Ib is to assess the maximum tolerated dose of 37 in combination with capecitabine and radiotherapy in participants with locally advanced rectal cancer. In Phase II the study seeks to evaluate the efficacy of 37 in terms of pathological complete response/clinical complete response in combination with capecitabine and radiotherapy, in comparison with a placebo and capecitabine/radiotherapy administered as monotherapies. | Recruiting |
| Capecitabine radiotherapy | December 10, 2018 | |||
| NCT03724890 | 37 | I | A study to assess the maximum tolerated dose for phase II studies of 37 in combination with avelumab with or without radiotherapy in participants with advanced solid tumors. | Recruiting |
| Avelumab radiotherapy | October 30, 2018 | |||
| NCT01353625 | 39 | I | A first in human trial to assess the safety and efficacy of 39 in participants with advanced solid tumors and hematologic malignancies who are unresponsive to standard of care treatments. The study also seeks to determine optimum dosages for later clinical trials and to assess the bioavailability of tablet and capsule formulations. | Active, not recruiting |
| May 13, 2011 | ||||
| NCT01421524 | 39 | I | A trial to assess the safety and efficacy of 39 in participants with advanced solid tumors, non-Hodgkin’s lymphoma, or multiple myeloma who are unresponsive to standard of care treatments, and to assess optimum dosing regiments for later clinical trials. | Active, not recruiting |
| August 23, 2011 |
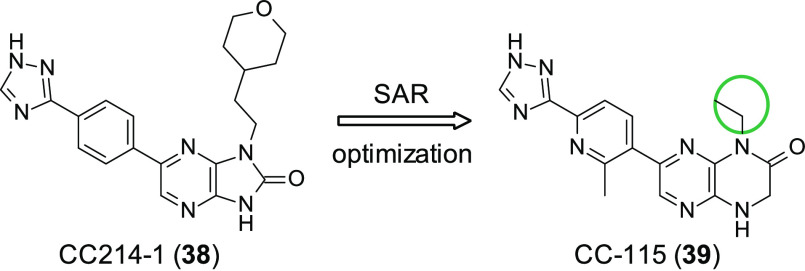
Mortensen et al. attempted to discover novel selective mTOR inhibitors through SAR studies of a series of 4,6- or 1,7-disubstituted-3,4-dihydropyrazino[2,3-b]pyrazine-2(1H)-ones, and in doing so, they discovered the potent dual mTOR/DNA-PK inhibitor CC-115 (39, Figure 12).101 Previous studies had identified CC214-1 (38, Figure 12) which displayed good on-target potency (mTOR IC50 = 2 nM); however it showed poor PK properties, showing negligible oral bioavailability. A focused SAR approach featuring 2-methyl-6-(1H-1,2,4-triazol-3-yl)pyridin-3-yl substituted analogues allowed the identification of small substituents in the N1/N4 position, which kept the excellent potency of the early compounds of the series while improving the PK profile. This culminated in the identification of 39 (mTORIC50 = 21 nM) as a clinical candidate. The DNA-PK activity of 39 was identified in separate work conducted by Tsuji et al., where inhibition of autophosphorylation of DNA-PK led to the blocking of DNA-PK facilitated NHEJ.102 In addition to 39’s activity against mTOR and DNA-PK it was found to inhibit ATM, which as stated earlier is thought to have an SL relationship with DNA-PK; this interaction appears to have been confirmed as 39 is more active in ATM deficient cells, suggesting synthetic lethality between DNA-PK inhibition and ATM deficiency. This in combination with antitumor activity seen in solid tumor and hematopoietic cell lines and apoptosis/antiproliferation in various cell lines has lent support to 39 being explored as a clinical candidate. To date, 39 is featured in two clinical trials, which are reported in Table 3.
Figure 12.
Optimization of mTOR inhibitor CC214-1 (38) leading to the discovery of mTOR/DNA-PK dual inhibitor CC-115 (39). Small substituents in the N1/N4 position resulted in the maintenance of high potency but improved PD/PK properties.
4. WEE1 Inhibitors
WEE1 is a kinase involved in cell cycle progression, where it prevents entry into mitosis in response to DNA damage. Furthermore, it has been implicated in the scheduling of cell division and HR repair.103 WEE1 prevents entry into mitosis through regulation of the G/M and S checkpoints through the phosphorylation of CDK1 and CDK2.104 As stated in this review, in the event of DNA damage, ATR or ATM pathways can be activated: ATR activation (Figure 3) results in the phosphorylation of CDK1, which leads to activation of checkpoint regulators of which WEE1 is one, which phosphorylates and inactivates the CDK1-cylin B complex on Tyr15, causing cell cycle arrest in G2.105 This halting of the cell cycle is preferential for cancer cells, as DNA damage is not repaired before replication in the S phase; therefore mutations can occur that favor proliferation, encouraging cancer growth. As the cancer cells are reliant on disruption of the apoptosis process, it is thought that the G2 checkpoint is vital for their survival, thereby presenting WEE1 as a promising target for cancer therapy. It is thought that healthy cells, containing fewer DNA breaks, will not be as affected by the disruption of the G2 checkpoint, and therefore this treatment would be considered somewhat selective for cancer cells.
WEE1 is highly expressed in numerous cancer types, including glioblastoma,106 breast cancer,107 leukemia,108 and others. Additionally, high levels of WEE1 expression have been observed in the event of severe DNA stress in various cancer types correlating with a poor prognosis.109,110 The cancer types that highly express WEE1 are thought to be extremely reliant on the G2 halting of the cell cycle, making them an attractive target for WEE1 inhibition.
Synthetic lethality has reported between WEE1 and ATR inhibitors. This was achieved through the combination of the WEE1 inhibitor AZD1775 (formally MK-1775) (40, Figure 13A) and two ATR inhibitors 11 and ETP-46464 (41, Figure 13B).111 In this study synergistic killing of cancer cells from various tissues was observed, but untransformed cell remained unaffected. Through mechanistic studies using reversible inhibition of WEE1/ATR it was revealed that inhibition occurred in the G2/M phase. Additionally, through live cell imaging, it was seen that inhibition of WEE1 and ATR caused cells to enter mitosis, and cells with overly damaged DNA underwent mitotic catastrophe.
Figure 13.
(A) SAR and optimization of WEE1 inhibitor 40 to 43. (B) Structure of ATR inhibitor ETP-46464 (41). (C) Structure of first WEE1 inhibitor PD0166285 (42).
SL has been observed between WEE1 and TP53 mutants. TP53 encodes for the tumor suppressor p53 and is the most commonly mutated gene within human cancer and generally is thought to be associated with poor prognosis.112 As the mutations cause a loss of function, TP53 mutations cannot be directly targeted; therefore SL through WEE1 provides an opportunity to drug a previously undruggable target. As cells with TP53 mutations lack an effective G1 checkpoint, they become over-reliant on the G2 checkpoint; therefore inhibition of WEE1, which acts in the G2 checkpoint, is thought to cause SL. This has been demonstrated in TP53 mutant colorectal carcinoma where, after exposure to radiation, inhibition of WEE1 with the preclinical inhibitor PD0166285 (42, Figure 13C) caused the cells to evade G2 arrest and prematurely enter mitosis.113 Furthermore, 40 has shown substantial improvement in carboplatin treatment of advanced solid tumors with p53 mutants in phase I and phase II trials.114,115
Additionally, in a screen for SL partners for CHK1, WEE1 has been identified as a possible SL partner. Combination studies of 40 and the CHK1 inhibitor PF-00477736 in brain, ovarian, colon, and prostate cancer cells confirmed synergism between WEE1 and CHK1, independent of p53 status.116
A number of WEE1 inhibitors have been synthesized to date, with some in clinical studies. Here we will touch upon some of the most prominent inhibitors in the field.
The first WEE1 inhibitor developed was 42 and was identified through use of a HTS looking to find WEE1 inhibitors. 42 is a highly potent ATP competitive WEE1 inhibitor (IC50 = 24 nM) and was able to inhibit CDC2 at Tyr15 (a direct substrate of WEE1) in cells at 0.5 μM.113 Furthermore, it was able to sensitize cells to radiation therapy, with this effect being more pronounced in p53 deficient cells. However, 42 is not a selective inhibitor and shows IC50 < 100 nM for CDK1, MYT1, c-SRC, EGFR, FGFR-1, and PDGFR-β. 42 has been used as a tool compound for examining the effect of WEE1 inhibition in numerous cancers such as colon, lung, and melanoma, among others;105 however, its lack of selectivity has meant its usefulness beyond this is quite limited.
40 was originally identified by Hirai et al. through use of a HTS; initial hits were optimized through SAR to yield the orally available selective potent small molecule 40 (Figure 13A) (WEE1 IC50 = 5.2 nM).117 A linear relationship between IC50 and ATP concentration indicated that 40 acted in an ATP competitive manner. To evaluate selectivity, 40 was tested in a kinase screen, and only 8 of the 223 kinases were inhibited >80% when treated with 1 μM of 41, demonstrating remarkable selectivity for an ATP competitive kinase inhibitor. In cells WEE1 inhibition was observed through 40 inhibiting phosphorylation of CDC12 at Tyr15.
Furthermore, 40 abrogates the G2 DNA damage checkpoint and in combination with the DNA damaging agents gemcitabine, carboplatin, and cisplatin caused apoptosis in p53 deficient cells. 40 has also been observed to induce apoptosis and inhibit proliferation as a monotherapy in numerous cell types (acute lymphoblastic leukemia cells, lung cancer cells, and colorectal cancer cells among others).118−120 Furthermore, in vivo it was seen to inhibit tumor growth without perceived toxicity. As previous stated in the review, 40 has entered numerous phase I and II clinical trials and is the most established WEE1 inhibitor in the field.
As little SAR data exist for 40, Matheson et al. synthesized a series of analogues to evaluate which structural features are necessary for successful WEE1 inhibition.121 The analogues that inhibited WEE1 in the same nM range had lessened cytotoxicity when administered as a single therapy in comparison to 40, and synergism was observed in combination with cisplatin in medulloblastoma cells. CJM061 (43, Figure 13A) was the most active from this analogue series (WEE1 IC50 = 2.8 nM) and its development can be seen in (Figure 13A).
These promising compounds, especially 40, show the potential for WEE1 inhibitors, with at least three confirmed SL relationships. Other than 40 no other WEE1 inhibitor is in clinical development. Should it prove to be unsuccessful in these clinical trials, more work is needed to fully make use of this attractive target for cancer therapy.
5. CDK12 Inhibitors
CDK12 was first identified when investigating the cell cycle regulator CDC2.122 While it has structural similarities to other members of the CDK subfamily, which play a role in cell cycle regulations, CDK12 is a transcription kinase involved in the transcription of genes involved in DNA repair,123 regulating specific genes that respond to stress, heat shock, and DNA damage.124 CDK12 presents a great opportunity to test SL due to the high amount of genomic alterations seen in numerous cancers including high-grade ovarian carcinoma,125,126 HER-2 positive breast cancer,127 and lung adenocarcinoma,128 resulting in loss of function. To date, SL pairs for CDK12 have been identified as PARP,129 MYC,130 and EWS/FLI.131
CDK12 mutations have been shown to sensitize cancers to traditional DNA damaging agents;132 therefore CDK12 inhibitors have gained traction in recent years as a way of implementing small molecule-induced synthetic lethality. A group within Takeda has developed a series of potent and selective CDK12 inhibtors,133 which also hit CDK13, another potential modulator of DNA repair. A modest initial CDK12 inhibitor (44, Figure 14) was identified through a HTS (CDK12 IC50 = 0.36 μM). While it demonstrated good selectivity over other related kinases within the CDK subfamily, it also showed activity against CDK2, which is involved in healthy regulation of the cell cycle; therefore, for 44 to be useful, it would need to be optimized further. Through SAR studies, they discovered that, first, the aminopyridyl moiety was binding in the hinge region of CDK12 and could not be modified. This is a fairly typical moiety for kinase inhibitors. Second, it was discovered through the same SAR studies that at the 5-position of the pyridine ring it is likely that an electron-withdrawing group is required. Through later docking simulations of 44 in both CDK2 and CDK12, it was proposed that the oxygen atom in the sulfonyl moiety interacted with Lys89 in CDK2, which is not conserved in CDK12. This was therefore changed, utilizing experimentally derived conformations from the Cambridge Structural Database, to either a tert-amide or a tert-sulfonamide. Of these, the tert-amide showed better selectivity for CDK12 over CDK2 (compound 45, Figure 14). Some final modifications, such as the introduction of a benzyl group and small modifications to improve physiochemical properties, resulted in the highly selective inhibitor, with it only showing inhibition of over 80% at 1 μM in 3 of the 441 kinases present in the kinome screen. 46 (Figure 14) (CDK12 IC50 = 0.052 μM) was shown to inhibit CDK12 phosphorylation of SER2 in SKBR2 cells (pSer2 IC50 = 0.195 μM). This compound therefore presents a valuable tool in probing SL or could serve as a starting point for further drug discovery programs investigating SL.
Figure 14.
Optimization of CDK12 inhibitor 44 to 46.
Other CDK12 and CDK13 inhibitors have emerged within the same period, in work completed by Zhang et al.134 Previous work conducted by the group yielded the CDK7 inhibitor THZ1 (47, Figure 16A).13547, while being active against CDK7 (IC50 = 3.2 nM), also showed activity against CDK12 (IC50 = 864 nM) and CDK13 (IC50 = 225 nM) and was therefore a starting point for further drug discovery. It was reasoned initially that alteration of the acrylamide moiety would divert the binding toward Cys1039 of CDK12. Through SAR studies that are yet to be reported, the compound THZ531 (48, Figure 16A) was synthesized. 48 has shown IC50’s in biochemical assays of 158 nM and 69 nM for CDK12 and CDK13, respectively, and has a more than 50-fold level of selectivity over the other members of CDK family. Interestingly, when the electrophilic acrylamide was replaced by a moiety incapable of covalently binding, propylamide, the activity of the compound dramatically reduced, suggesting that the ability to covalently bind is necessary for preserving CDK12 and CDK13 activity. Its selectivity over the wider kinome is good, with none of the other 211 kinases tested showing an activity of over 55%.
Figure 16.
(A) Optimization of CDK inhibitor THZ1 (47). (B) Structure of CDK inhibitor dinaciclib (49).
48 has been cocrystallized with CDK12. This has presented some useful binding information, which could serve others in the field, in the pursuit of novel CDK12 inhibitors (Figure 15). Two CDK12-cycklin K complexes were found, each bound to 48 in a different rotamer. These crystallization studies revealed that a labile αK helix can be displaced from CDK12 allowing binding through the linker of 48 with Cys1039. The aminopyrimidine of 48 was found to form two hydrogen bonds to the backbone of M816, with the pendant 3-indolyl and the piperidine groups involved in hydrophobic interactions. The binding of the secondary amide was shown to have two different conformations that caused the solvent exposed groups to pack either against the N-lobe β strand or C-lobe αD helix. The resolution for the exposed groups was too poor to be defined apart from the position of Cys1039. Interestingly Cys312, the equivalent sulfhydryl to Cys1039 in CDK7, was further away from the binding region, suggesting poorer binding for 49 against CDK7 in comparison to CDK12. 49 was able to induce apoptosis in Jurkat cancer cells, in addition to being able to inhibit transcriptional regulation, reducing DNA damage repair and superenhancer gene expression. 49 has been shown to demonstrate a synergistic effect with PARPi’s in Ewing sarcoma in both cells and mouse models, as it was seen that the Ewing sarcoma cell models were particularly sensitive to CHK12 inhibition. Furthermore, the expression of the tumor specific oncogene EWS/FLI shows SL with CHK12 inhibition. The inhibition of CHK12 was also seen to impair DNA damage repair in Ewing sarcoma cells.131
Figure 15.

Cocrystallization of 48 with CDK12 adapted from Zhang et al.134 (a) 48 binds to M816 in the kinase hinge region and connects to Cys1039 in two conformations via the compound’s flexible linker. Solvent-exposed regions of 48 with poor electron density are represented by thin sticks. (b) Omit map contoured at 2.5σ for 48 bound to CDK12 chain C. (c) Omit map contoured at 2.5σ for 48 bound to CDK12 chain D. Reproduced with permission from Springer Nature, Nature Chemical Biology, Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors, Tinghu Zhang, Nicholas Kwiatkowski, Calla M. Olson, Sarah E. Dixon-Clarke, Brian J. Abraham, Ann K. Greifenberg, Scott B. Ficarro, Jonathan M. Elkins, Yanke Liang, Nancy M. Hannett, Theresa Manz, Mingfeng Hao, Bartlomiej Bartkowiak, Arno L. Greenleaf, Jarrod A. Marto, Matthias Geyer, Alex N. Bullock, Richard A. Young, Nathanael S. Gray. ,134 Copyright 2019 Springer Nature.
A known CDK inhibitor, dinaciclib (49, Figure 16B),136 whose primary targets are CDK1, CDK2, CDK5, and CDK9, has shown a documented response in breast cancer137 and in recent years has also been shown to inhibit CDK12. While showing a higher level of promiscuity and also hitting cell cycle kinases within the family, 49 is the most potent CDK12 inhibitor to date, with an IC50 of 40–60 nM in biochemical assays.138 In cell-based assays, 49 showed phenotypic responses typical of CDK12 inhibition, including gene repression of multiple genes involved in HR, whose expression is thought to correlate with CDK12 expression.139 This was achieved with minimal disruption to the cell cycle, indicating CDK12 inhibition as the cause of this observation. Due to the dysregulation of HR influencing genes, it was reasoned that 49 would be able to sensitize triple-negative breast cancer cells to PARPi. This was observed through the combination of 49 with the known PARP inhibitor veliparib, which showed a 2.5- to 12.5-fold increase in its activity as a single-agent treatment, demonstrating a synergistic SL effect. This impairment of HR by 49 was mirrored through knockout of CDK12, showing the same phenotype. Not only was 49 capable of showing a synergistic effect with PARPi’s, it was also able to resensitize triple-negative breast cancer cells that had become resistant to PARP inhibition to 1. These results seem to indicate that the use of 49 can cause an increase in the effectivity of PARP inhibition even if a partial response is seen with the PARPi being administered as a monotherapy.
CDK12’s progress as a target has meant that the idea of utilization of CDK12’s synthetic lethality has moved on to clinical trials. While there is not a great abundance of trials directly using CDK12 inhibitors, other potential SL partners are being modulated in CDK12 deficient cancers, thus giving an SL effect. If these worked, it opens the door for chemically induced small-molecule lethality. The trials are summarized in Table 4. One CDK12 inhibitor has entered clinical trials, and a number of selective CDK12 inhibitors are currently in preclinical studies. CDK12 inhibitors have the potential to be useful in the application of SL to cancer therapy.
Table 4. Clinical Trials Involving Loss of CDK12 Function.
| trial identifier | drug | phase | summary | status and accession date |
|---|---|---|---|---|
| NCT01434316 | Veliparib (PARPi) and 49 | I | Studying the safety profile of veliparib and 49 in participants with advanced solid tumors. | Recruiting |
| September 14, 2011 | ||||
| NCT04272645 | Abemaciclib and atezolizumab (both PD-L1 immunotheraputics) | II | Studying the effectivity and safety of abemaciclib and atezolizumab in participants with metastatic castration-resistant prostate cancer with and without “CDK12 loss” mutation. | Withdrawn (coordinating site change) |
| February 17, 2020 | ||||
| NCT03570619 | Nivolumab and ipilimumab (both PD-L1 immunotheraptics) | II | Studying the efficacy of nivolumab/ipilimumab in combination and nivolumab as a monotherapy in participants with castration-resistant metastatic prostate carcinoma or other solid tumor histologies, with CDK12 loss of function. | Recruiting |
| June 27, 2018 | ||||
| NCT04104893 | Pembrolizumab (PD-L1 immunotheraptic) | II | A study to assess the activity and efficacy of pembrolizumab in participants with progressive metastatic castration-resistant prostate cancer, characterized by a mismatch repair deficiency or biallelic CDK12 inactivation. | Recruiting |
| September 26, 2019 |
6. PARP and RAD51
RAD51 is a member of the RAD52 epistasis group, which is made up of RAD50, RAD51, RAD52, RAD54, RAD55, RAD57, RAD59, MRE11, and XRS2.140In vitro studies have shown that RAD51 promotes homologous pairing and strand transfer reactions. Following treatment by DNA-damaging agents, RAD51 was seen to be upregulated, indicating its role in HR. Early on during HR, DSB or SSB results in the formation of a length of ssDNA. This strand is then paired with recombinases such as RAD51 and afterward is paired with a homologous duplex to form a DNA joint which is termed the D loop.141 RAD51 then goes on to promote an ATP-mediated strand exchange reaction by polymerizing on DNA, resulting in a helical filament. The formation of this is completed in two steps, nucleation and extension, promoting HR. RAD51 has also been shown to cause polymerization of dsDNA; however the reason for this has yet to be elucidated.142
It has been hypothesized that interrupting BRCA2 and RAD51’s interaction would mirror the synthetically lethal effect seen in utilizing PARPi in BRCA2 deficient tumors143 (Figure 17). This would provide a shortcut to modulating the activity of BRCA2 which, to date, has been thought to be undruggable. While RAD51 inhibitors do exist,144−146 they have not been seen to interfere with the BRCA2–RAD51 interaction. For instance, structural data have shown that BRC4 (the fourth BRC repeat of BRCA2) binds to RAD51 in two hydrophobic binding pockets: one of these is critical for RAD51 multimerization (FxxA). Utilizing high-throughput docking, our group began to work developing modulators for this interaction. From the initial hit compounds identified from the HTS, 50 (Figure 18) was the best performing (EC50 = 53 ± 3 μM), and as such, an SAR program was initiated, at first investigating the optimal length of the alkyl chain between the triazole and the phenyl ring. From these studies, 51 (Figure 18) (EC50 = 25 ± 2 μM) which features a propyl phenyl ring was selected for further biological evaluation. 51 was tested in combination with 1 in two pancreatic cancer cell lines: Capan-1 which lacks functional BRCA2; BxPC-3 which is BRCA2-positive. Unsurprisingly as a monotherapy, 1 was more effective in the Capan-1 cells, due to the lack of BRCA2. Upon combination with 51, no effect was seen in the Capan-1 cells; however, in the BxPC-3 cells, a synergistic effect was seen between 51 and 1, suggesting small molecule-induced synthetic lethality. 51 was tested on the same cell lines that had undergone prior treatment by cisplatin: it demonstrated no effect in the BRCA2-negative cells, whereas in the BRCA2-positive cell line, a small increase in a phenotypic indicator for DNA damage was observed, confirmed by silencing of RAD51 which demonstrated a similar effect to 51. This work could potentially provide a way of inducing SL in patients who lack the BRCA1/2 mutation, increasing the scope of PARPi’s.
Figure 17.

(A) PARP inhibitors triggering synthetic lethality in BRCA-deficient cells. (B) Proposed triggering of small molecule-induced synthetic lethality using PARP inhibitors in combination with RAD51-BRCA2 disruptors. Adapted from European Journal of Medicinal Chemistry, Vol. 165, Marinella Roberti, Fabrizio Schipani, Greta Bagnolini, Domenico Milano, Elisa Giacomini, Federico Falchi, Andrea Balboni, Marcella Manerba, Fulvia Farabegoli, Francesca De Franco, Janet Robertson, Saverio Minucci, Isabella Pallavicini, Giuseppina Di Stefano, S. Girotto, R. Pellicciari, A. Cavalli, Rad51/BRCA2 disruptors inhibit homologous recombination and synergize with olaparib in pancreatic cancer cells ,143 Page 81, Copyright (2019), with permission from Elsevier.
Figure 18.
Depiction of the SAR strategy for the further development of 52. This primarily focused on three areas within the molecule highlighted in green, red, and blue. The proposed moieties in these regions are shown in their accompanying text.
Further SAR optimization of 51 was undertaken.143 These focused around adjusting the chain length of the N4 nitrogen and introducing substitutes on the phenyl ring. Other modifications included changes at the C3 position of the triazole, introducing a series of heterocycles, saturated and unsaturated rings with differing links. Lastly, changes occurred at the C5 position of the triazole, changing the heterocycle present at the position or removing this substituent entirely, replacing it with either a methyl group or a proton; these alterations are summarized in Figure 18. Modifications at the C5 and N4 positions were not well-tolerated, with a drop in activity usually observed. A minor tweak, changing the cyclopentyl group for a cyclohexyl group, resulted in the best compound from the series, compound 52 (Figure 18) (EC50 = 8 ± 2 μM).
52 inhibited the RAD51–BRCA2 interaction, preventing RAD51 function in cells. Furthermore, it significantly improved the function of 1 in BRCA2-positive cells and increased the amount of DSBs when administered in combination with 1. However, due to its low potency or the presence of a mutant from p53 that prevents apoptosis, true synthetic lethality could not be observed, and showing further optimization was needed.143 In order to increase diversity within the field of Rad51–BRCA2 inhibitors, our group ran a second binding screening at the other binding site (LFDE).147 Mutations in this binding pocket are associated with cellular lethality, in addition to the failure of RAD51 assembly in nuclear foci at the point of DNA breaks in vivo, suggesting a key role in RAD51’s function.148 Of the compounds tested, the dihydroquinolone pyrazoline derivative 53 (Figure 19) (EC50 = 16 ± 4 μM) was the most active, and an SAR program was initiated around this compound (this work will be published in a subsequent paper), eventually yielding compound 54 (Figure 19) (EC50 = 19 ± 1 μM). Like 52 from the previous work, 54 was able to bind to RAD51 inhibiting its function, but unlike 52, 54 was able to trigger SL in a dose response manner by impeding HR in pancreatic cancer cells that expressed BRCA2. However, due to 54’s poor solubility, it is unlikely to be able to be tested it in vivo, and therefore further optimization is required before these class of compounds can realize their potential. This work is ongoing and demonstrates one of the first programs targeting small molecule-induced synthetic lethality and a way to expand the use of PARPi’s beyond BRCA1/2 deficient cancers, increasing their usefulness.
Figure 19.
Optimization of 53 to 54. All compounds from this series were tested as racemic mixtures after both enantiomers of 53 showed the same biochemical activity and binding mode.147
A number of other RAD51 inhibitors have been reported in the literature,144−146,149−156 the most noteworthy of which are shown in Figure 20. Only IBR2 (55, Figure 20) (RAD51 IC50 not reported) and BO2 (56, Figure 20) (RAD51 IC50= 27.4 μM) have been directly linked to small molecule-induced synthetic lethality.155,15755 was observed to synergize with multiple drugs (notably imatinib, regorafenib, EGFR inhibitors (including erlotinib, gefitinib, afatinib, and osimertinib), and vincristine) with differing molecular targets, which were not involved in the RAD51 mediated HR pathway. However, the combination of 55 with DNA-damaging agents such as cis-platin was not synergistic.155 In the same study 55 was compared to 56, which was able to synergize with imatinib and vincristine, thereby opening the door for these two compounds to be explored for SL. At a recent conference, Maclay et al. reported a novel RAD51 inhibitor: CYT01B.158 Neither the structure of this compound nor the data associated with it have been published to date; however a patent has been released by the parent company Cytier Therapeutics, which appears to be indicative of the structure.159 CYT01B has been shown to cause DNA stress through DNA replication fork damage. Therefore, it was tested to see if CYT01B could sensitize cells to current therapeutics for the treatment of solid tumors. This was done through combination assays with CYT01B (concentration range of 20 nm to 5 μM) in three cell lines with six targeted agents, the results of which are summarized in Table 5. These studies, in particular the success seen with the platinum-based chemotherapy carboplatin, in addition to PARPi’s, seem to indicate a degree of synthetic lethality. Therefore, this compound should be monitored as it progresses through in vivo models and beyond. As little is known about the compound’s selectivity panel, it is impossible to say whether the observed SL phenotype is due to RAD51 inhibition; however, should this information come to light, it would no doubt be of interest to the field of not only RAD51 inhibitors but also small molecule-induced synthetically lethality.
Figure 20.
Examples of Rad51 inhibitors reported in the literature.
Table 5. Summary of Cell-Based Combination Studies Featuring CYT01B.
| cell line | drug combination | response observed |
|---|---|---|
| ARPE19/HPV16 (HPV immortalized normal epithelial cell line) | V822 (ATRi) (concentration range of 39 nM to 2.5 μM) and CYT01B (20 nM to 5 μM) | Synergism observed at all concentrations. |
| TDRL-505 (RPAi) (concentration range of 39 nM to 5 μM) and CYT01B (20 nM to 5 μM) | Antagonism observed at all concentrations. | |
| Bortezomib (proteasome inhibitor) (concentration range of 39 nM to 2.5 μM) and CYT01B (20 nM to 5 μM) | Synergism observed at all concentrations. | |
| Carboplatin (nonspecific chemotherapeutic) (concentration range of 156 nM to 10 μM) and CYT01B (20 nM to 5 μM) | Synergism observed at all concentrations. | |
| 1 (concentration range of 78 nM to 5 μM) and CYT01B (20 nM to 5 μM) | Synergism observed at all concentrations but stronger synergy than that of niraparib. | |
| Niraparib (3) (PARPi) (concentration range of 78 nM to 5 μM) and CYT01B (20 nM to 5 μM) | Synergism observed at all concentrations. | |
| KYSE-70 (head and neck cancer cell line) | V822 (ATRi) (concentration range of 39 nM to 2.5 μM) and CYT01B (20 nM to 5 μM) | Synergy was observed with CYT01B at 39 nM, but an antagonistic relationship between V822 and CYT01B was seen at low and high concentrations. |
| TDRL-505 (RPAi) (concentration range of 39 nM to 5 μM) and CYT01B (20 nM to 5 μM) | Weak synergy observed at 156 and 312 nM of CYT01B. | |
| Bortezomib (proteasome inhibitor) (concentration range of 39 nM to 2.5 μM) and CYT01B (20 nM to 5 μM) | Antagonism observed at all concentrations. | |
| Carboplatin (nonspecific chemotherapeutic) (concentration range of 156 nM to 10 μM) and CYT01B (20 nM to 5 μM) | Synergism observed at all concentrations. | |
| 1 (concentration range of 78 nM to 5 μM) and CYT01B (20 nM to 5 μM) | Synergism observed at all concentrations but stronger synergy than that of niraparib. | |
| Niraparib (3) (PARPi) (concentration range of 78 nM to 5 μM) and CYT01B (20 nM to 5 μM) | Synergism observed at all concentrations. | |
| Daudi (Burkitt’s lymphoma cell line) | V822 (ATRi) (concentration range of 39 nM to 2.5 μM) and CYT01B (20 nM to 5 μM) | Antagonistic effect at high concentrations but additive at low. |
| TDRL-505 (RPAi) (concentration range of 39 nM to 5 μM) and CYT01B (20 nM to 5 μM) | Antagonism observed at all concentrations. | |
| Bortezomib (proteasome inhibitor) (concentration range of 39 nM to 2.5 μM) and CYT01B (20 nM to 5 μM) | Antagonism observed at all concentrations. | |
| Carboplatin (nonspecific chemotherapeutic) (concentration range of 156 nM to 10 μM) and CYT01B (20 nM to 5 μM) | Synergism observed at all concentrations. | |
| 1 (concentration range of 78 nM to 5 μM) and CYT01B (20 nM to 5 μM) | Synergism observed at all concentrations but stronger synergy than that of niraparib. | |
| Niraparib (3) (PARPi) (concentration range of 78 nM to 5 μM) and CYT01B (20 nM to 5 μM) | Synergism observed at all concentrations. |
7. RAD52 Inhibitors
The RAD52 protein is able to bind to ssDNA and plays a key role in repairing of single-strand and double-strand breaks.160 Within the HR response, it is essential in more simple organisms such as bacteria or yeast, where it acts in aiding RAD51 attachment to ssDNA. However, in more complex organisms such as animals this is largely completed by other proteins such as BRCA1/2. Indeed, knockout of RAD52 in mice does not cause a drop in viability or fertility of the mouse. However, upregulation of RAD52 can have a protecting effect on DNA from ionizing radiation, suggesting it is important to the HR process. Common synthetic lethality partners are BRCA1/2, which play a vital role in annealing RAD51 to ssDNA; however mutations in these proteins are quite common,161 and when this occurs, the cancer cell can compensate by utilizing RAD52 to perform the task instead. This is represented in Figure 21. This makes RAD52 an appealing target for cancer therapy, as it will only have key function in HR when BRCA1/2 are inactive: this will therefore have a selective effect on cancer cells, without influencing health cells.
Figure 21.

RAD52, SL interactions with PARP inhibitors. This figure shows classic SL with PARP inhibition in BRCA2 deficient-cancer cells. These are prone to acquired mutation. The implementation of RAD52 in combination with PARPi has no effect in BRCA2 proficient healthy cells; however, in BRCA2-deficient cancer cells, small-molecule-induced synthetic lethality occurs. Adapted from Cancers, 2019, Vol. 11, Issue (10), , Monika Toma, Katherine Sullivan-Reed, Tomasz Śliwiński, Tomasz Skorski, RAD52 as a Potential Target for Synthetic Lethality-Based Anticancer Therapies ,162 Page 1569, with use of the Attribution 4.0 International (CC BY 4.0) open access license.
One of the mechanisms of synthetic lethality proposed for both RAD52 and BRCA1/2 inhibition is that endonuclease/exonuclease/phosphatase family domains containing protein 1 (EEPD1) mediate DNA cleavage in the absence of BRCA1/2 and RAD52 and cause the production of toxic intermediates that trigger cell death.163 This theory is supported by the fact that suppression of EEPD1 in BRCA1/2- and RAD52-compromised cells causes a drop in the level of synthetic lethality.162 With RAD52 appearing to be a promising target for SL-based drugs, a few examples of small molecule inhibitor discovery programs are discussed below.
The first attempt to modulate RAD52 activity was not done through use of small molecules but rather use of an aptamer.164 This was achieved through use of the F79 aptamer that demonstrated activity in BRCA1/2 deficient cells but had no effect on HR in normal cells. F79 demonstrated SL in leukemia but also breast, pancreatic, and ovarian cells. In vivo tests in mice showed a lengthening of lifespan in mice with BCR-ABL1-positive leukemia. Lastly, RAD52 showed a synergistic effect with nonspecific chemotherapeutics, suggesting that, utilized together, the dose of the chemotherapeutic could be lowered, thereby reducing the risk of side effects. While not being investigated further, aptamers could play a key role in SL targets, specifically those in HR, as elsewhere aptamers have been reported to inhibit the DNA strand exchange of RAD51.165 Again, this work has not been expanded upon and could provide an untapped resource for the synthetic lethality.
6-OH-dopa (57, Figure 22) is a RAD52 inhibitor that was identified from a HTS of 18 304 drug-like compounds combined with 1280 from the Sigma Lopac collection. These compounds were screened through a high-throughput fluorescence polarization assay, and 10 compounds were identified that prevented RAD52 binding to ssDNA at a greater degree than 60%, at IC50 < 5 μM.166 Of these 10 compounds, only one was able to inhibit single-strand annealing in cells: 57. It was later tested to see if it could inhibit HR. Predictably, it did not, due to RAD52’s weak role in healthy cells, therefore indicating some degree of selectivity for RAD52 over other key HR proteins. Furthermore, in biochemical assays, 57 was shown to have a significant degree of selectivity for RAD52 (1.1 μM) over RAD51 (not determined). 57 was also observed to halt proliferation in two cell lines deficient in BRCA1/2 (a Chinese hamster cell line and a pancreatic cancer cell line). In addition, separate studies also showed an increased level of apoptosis and DNA damage in BRCA1/2 cells. Despite being a promising compound, 57 is a dopaminergic derivative, which has been reported to heighten the chance of Parkinson’s disease by degenerating mitral neurons.167 Therefore, within cancer therapy, this is highly unlikely to be explored further. Huang et al. set out to develop small molecule inhibitors of RAD52 through use of a HTS.168 The hit compounds from this screen were tested for RAD52 selectivity, especially over RAD51, and in BRCA1/2 positive and deficient pancreas, ovarian, and triple negative breast cancer cells. Two hit compounds, D-G09 (58) and D-103 (59) (Figure 22), showed the expected phenotype for RAD52 inhibition in these cell lines, while another three compounds D-105 (60), D-K17 (61), and D-G23 (62) (Figure 22) showed activity against two of the cells lines but were of a structurally diverse scaffold, giving more opportunity for chemical space exploration. The best-performing compound of these, 59 (inhibits RAD52 ssDNA annealing at IC50 = 5 μM), was also tested in BRCA1/2-positive and -negative chronic myeloid leukemia cells, showing preferential inhibition of growth of the BRCA1/2-negative cells. Through SPR these compounds were confirmed to inhibit DNA, annealing through direct binding with RAD52 as opposed to DNA substrates. It was also demonstrated that 59 shows an absence of a nonspecific effect on RAD-51 foci production in response to cisplatin, reinforcing the selectivity of the compound.
Figure 22.

Structures of different RAD52 inhibitors.
Hengel et al. embarked on a discovery program for small molecule inhibitors of RAD52, through use of a HTS of the MicroSource SPECTRUM collection.169 Initial hits were tested for RAD52 binding in FRET-based assays, giving five hits that showed an IC50 in or near the nanomolar range. Of these hits, two compounds, (−)-epigallocatechin (63, Figure 22) (RAD52 DNA binding IC50 = 1.8 ± 0.1 μM) and epigallocatechin-3-monogallate (64, Figure 22) (RAD52 DNA binding IC50 = 277 ± 22 nM), were shown to physically bind to RAD52 by NMR and were shown to prevent binding and annealing to RPA-coated ssDNA. Virtual screens of 63 and 64 within the RAD52 ssDNA binding groove seem to indicate that these compounds show binding in this region. Notable interactions include Arg55, Lys65, Arg153, and Arg156 in the vicinity of 63 and 64, which when docked have previously been shown to impact ssDNA binding. Additionally Lys141 and Lys144 have been shown to be important previously in RAD52 function in yeast and are thought to be somewhat evolutionarily conserved; some of these binding interactions are schematically reported in Figure 23. 63 and 64 were shown to dysregulate DSB repair in BRCA1/2-depleted and MUS81-depleted cells. Furthermore, both compounds showed activity in killing BRCA1/2-depleted cells in addition to MUS81-depleted cells. This phenomenon of lower cell viability in MUS81-deficient cells has been reported previously.170 By use of 63 and 64’s predicted docking models, an in silico screen was embarked upon, using the AnalytiCon Discovery MEGx Natural Products Screen Library, which is made up of natural products from plants, fungal, and antimicrobial sources. From this library, NP-004255 (65, Figure 22) (RAD52 DNA binding IC50 = 1.5 ± 0.2 μM) was identified, which had the same binding mode as 63 and 64. 65 underwent the same binding analysis as 63 and 64, where it was shown to bind to RAD52 and RPA and to inhibit the binding of RAD52 to ssDNA in addition to the ssDNA–RPA complex. However, 65 did not affect RAD52’s binding to dsDNA or affect DNA binding by RPA.
Figure 23.

Proposed binding regions of the RAD52 inhibitors: (A) 63 and (B) 64. Adapted from eLIFE, 2016, Vol. 5, e14740, Sarah R. Hengel, Eva Malacaria, Laura Folly da Silva Constantino, Fletcher E. Bain, Andrea Diaz, Brandon G. Koch, Liping Yu, Meng Wu, Pietro Pichierri, M. Ashley Spies, Maria Spies, Small-molecule inhibitors identify the RAD52-ssDNA interaction as critical for recovery from replication stress and for survival of BRCA2 deficient cells ,169 with use of the Attribution 4.0 International (CC BY 4.0) open access license.
Li et al. investigated new RAD52 inhibitors, beginning with use of a virtual screen, by docking the compounds inside the RAD52 monomer.171 This HTS consisted of 47 737 compounds, with the 30 top-performing compounds being sorted into groups of 5 in relation to their chemotype. These 30 compounds were tested for their ADMET properties, and 4 were selected due to their druglikeness for further testing: F779-0434 (66), F848-0436 (67), G640-1014 (68), and D207-0130 (69) (Figure 22), with 66 appearing to be the most promising RAD52 inhibitor. Similar to the work conducted by Spies et al., all 4 compounds tested were seen to bind in the ssDNA binding pocket. Key binding interactions were observed between the 4 compounds and Arg 55, Lys 152, Arg 165, and Tyr 65. Notably the interaction with Arg 55 is shared between 61 and 66, suggesting this is a key RAD52 interaction. These binding interactions can be seen in more detail in Figure 24. 66 was shown to be able to selectivity inhibit growth in a BRCA1/2-dependent pancreas cancer cell to 50% at 10 μM, whereas in the same concentration over 90% of cells from a healthy cell line survived. Furthermore, it was shown to inhibit RAD52 ssDNA annulation beginning at 5 μM, and at 20 μM complete inhibition of the process could be seen. However, the other three compounds selected by the virtual screening did not show as promising an effect in these assays.
Figure 24.

Proposed binding regions of 66. Reproduced with permission of The Royal Society of Chemistry, from Li J.; Yang Q.; Zhang Y.; Huang K.; Sun R.; Zhao Q.. Compound F779-0434 causes synthetic lethality in BRCA2-deficient cancer cells by disrupting RAD52–ssDNA association. RSC Advances, Vol. 8, Issue (34), , pp 18859–18869,171 Copyright 2018, permission conveyed through Copyright Clearance Center, Inc.
To date, no RAD52 inhibitors have entered clinical trials. However, the numerous successes seen in this section, particularly in vitro, and the greater wealth of information known about the binding pocket and binding modes suggest that the potential for RAD52 inhibitors to work as SL agents is vast.
8. PD-1 and Synthetic Lethality
Since the late 19th century, the application of the immune response in cancer therapy has been theorized;172 while it was quite controversial through much of this time, it is now seen as a promising approach to anticancer therapy. One of the targets that has gained interest is programmed cell death protein 1 (PD-1), which is responsible for fine-tuning T-cell function, the maintenance of homeostasis within the immune response, acting as a natural break, and initiating the checkpoint response usually associated with periphery tolerance.173 However, in cancer, tumor cells take advantage of this and use the mechanism to suppress and evade the immune response. Therefore, the concept of a checkpoint blockade has been proposed, where inhibition of PD-1 causes reactivation of the immune response toward the cancerous cell, utilizing the body’s defenses to fight cancer. Several PD-1 monoclonal antibodies have been successful in clinical trials, leading to FDA approval for several monoclonal antibodies, namely, nivolumab, pembrolizumab, atezolizumab, avelumab, and durvalumab. In various cancers, anti PD-1 agents are standard of care therapies for many malignancies including melanoma, small-cell lung cancer, non-small-cell lung cancer, colorectal cancer, and many more.
It is known that blocking DNA repair mechanisms increases genetic instability, which in turn increases epitope expression on the cancer cell surface, which is usually immunodominant.174 It is therefore possible that this increased genetic instability could make the cancer cells more susceptible to the immune response if heightened, making PD-1’s combination with targets involved in the DNA repair appealing as a potential source of synergism, perhaps even to the extent of synthetic lethality. In fact, some DNA-damage regulators such as PARP have been to seen to cause an increase in PD-1 expression, suggesting a likelihood of synthetic lethality. Due to this, PARP and PD-1 joint inhibition has been explored, with synergism seen in preclinical trials in addition to early stage clinical trials, of which there are more than 30 currently in progress in a wide of range of malignancies.174 The potential for PD-1 modulation in combination with other targets is evident throughout this review, with numerous ATR inhibitor clinical trials and trials featuring CDK12 loss of function mutants utilizing PD-1 antibodies. While this approach is still new, with the first work being conducted in 2017,175 it could be expanded to other targets mentioned within this review such as RAD51. The theory is that any target which, when modulated, causes a reduction in DNA repair could potentially be more susceptible to T-cells involved in the immune response, once the checkpoint response has been modulated.
In this section, numerous SL targets and strategies have been highlighted. With the rapidly expanding number of known SL partners, it would be impossible to cover them all in one review. The majority of the highlighted discovery programs focus on developing a selective potent modulator of a specific target and work to optimize through a classical medicinal chemistry approach. It is usually at a later stage of the screening cascade where SL is investigated, initially though combination studies in cells; thus, fewer compounds are screened for SL, which renders it difficult to comment on whether or not the SAR approach is in fact increasing SL. This is understandable as, despite being known for many years, SL approaches are still in their infancy; however, the number of promising compounds in late-stage clinical and preclinical studies gives hope that one of these targets will eventually be employed in an SL approach within the clinic.
9. Challenges and Opportunity in Developing New SL Drugs
To date, numerous potential synthetic lethal genes have been identified through statistical screening,176 genetic screening,177 high-throughput phenotypic screening,178 and combinational approaches. However, this has yet to correlate with improved medicines in the clinic, with only one SL pair being exploited to date and four drugs acting through the same PARP1 SL mechanism (1, 2, 3, and 4). One of the largest barriers to the development of more drugs acting on SL is the heterogeneity of cancer,179 with many SL targets only existing in a small number of cancer subtypes. This therefore means sample sizes are dramatically lowered, be they statistical data points or patient samples for in vitro/in vivo experiments crucial for validating the SL phenotype. Second, more mechanistic detail must be obtained to evaluate how best to drug SL targets. As shown in this review, the mechanistic understanding of many SL relationships is poor, with PARP/BRCA2 being one of the only synthetically lethal gene pairs to have received extensive study. With mechanistic understanding of what drives SL still not fully understood, creative approaches have been employed to investigate this vital piece of the puzzle. Li et al. employed a machine learning approach to discover what “characteristic functional features” are required for a protein-coding gene to have the potential for synthetic lethality.180 The methodologies involved are outside the scope of this review; however through this study Li et al. discovered 15 key characteristic functional features, 14 of which had previously been reported to play a role in SL to some degree in the literature (these were given a descriptor and can be seen in Table 6). These descriptors are generated using machine learning algorithms, which through an enrichment system derived from gene ontology terms and the Kyoto encyclopedia of genes and genome pathways represent functional features. It is hoped that this study can be utilized in both computational and experimental methods for selecting potential genes to investigate for SL. Other big data approaches181 have been employed to try to aid the “hypothesis driven” methods that have been used to discover SL pairs previously, with a collated and curated list of SL interactions available for free online via BioGrid.182
Table 6. Key Characteristics Discovered through a Machine Learning Approach Conducted by Li et al.180.
| maximum relevance and minimum redundancy (mRMR) feature name | description of function |
|---|---|
| GO: 0097505 | This term represents the cellular component named RAD6–18 complex: a previous study in Saccharomyces cerevisiae confirmed RAD6 and RAD18 exhibit SL. The function in humans is similar to that in yeast, and therefore it is likely that these two are SL in humans.190 |
| GO: 0070449 | This term represents a cellular component named the elongin complex which consists of the elongin transcription factors elongin A, B, and C. These have been seen previously to be SL with the von Hippel–Lindau protein complex.191 |
| GO: 0033503 | This term represents the histone H2B ubiquitination complex. It generally modulates heterochromatin-independent histone methylation. The individual components of this complex perform similar and redundant tasks involved in the regulation of cell survival.192 |
| GO: 1901136 | A catabolic process associated with carbohydrate derivation, this consists of chemical reactions that contribute to the digestion and breakdown of carbohydrates. The relationship between catabolic processes for carbohydrates and SL has been previously reported, with genes like BCL-2 being involved both in cancer and in carbohydrate digestion.193 |
| GO: 0008234 | A molecular function involving cytosine protease and thiol protease activity that plays a role in oncogene addiction and is involved in the maintenance of cell viability suggesting a potential for SL.194 |
| GO: 1903772 | A complex involved in the regulation of viral budding. Genes involved in the formation of this complex such as SKD1, LIP5, and IST1-LIKE1 are involved in specific SL mechanisms.195 |
| GO: 0003383 | The biological process of the actin-mediated contraction of the apical end of a polarized epithelial cell. Recently it was discovered that Lin-44 and Wnt participate in SL processes in addition to regulating apical constriction; therefore it is theorized that dysfunctional apical constriction could lead to SL.196 |
| GO: 1903333 | This describes the negative regulation of protein folding: it is theorized that this may cause toxic intermediates under the control of chaperones that may cause further cell death.197 |
| GO: 0070202 | This describes the regulation of protein localization in chromosomes. Two groups of genes, which encode for either the structural maintenance of chromosome proteins or chromosome stability proteins including BRCA, show the SL phenotype.198 |
| GO: 0070087 | A biological process of chromoshadow domain binding is predicted to be involved with SL, as the specific heterodomain protein HP1γ, which is involved in SL, is predicted to bind at the chromoshadow domain, thus connecting this domain to cell viability.199 |
| GO: 0004883 | This feature describes the activity of a glucocorticoid receptor or the participation in transmission and reaction of a glucocorticoid. Recent research shows p53 and GRα/ß are involved in these biological functions and are SL in non-small-cell lung cancer.200 |
| GO: 2001034 | The biological process of DNA repair through HR or n-HEJ, such as PARP.19 |
| GO: 0000209 | The process of protein polyubiquitination; two examples of genes involved in this process are Slx5 and Slx8, which have also been reported to show synthetic lethality with each other. This suggests a connection between protein polyubiquitination and SL.201 |
| Hsa04612 | This represents antigen processing and presentation in the human immune response. Two genes indicated in this response, BRAF and NRAS, have been seen to be synthetically lethal with each other in melanoma.202 |
The ability to generate compounds capable of induced small-molecule synthetic lethality can be hampered by the complexity and size of the binding regions of the targets associated with SL. One such example noted earlier in the text is the attempted induction of SL through the blocking of RAD51–BRCA2 interaction; this is a protein–protein interaction that has in the past been difficult to modulate with small molecules, due to the size of the compound in comparison to the large binding region between the proteins. Work performed earlier in the decade by Hyvönen et al. attempted to modulate the RAD51–BRCA2 interaction through a fragment-based approach. Through protein engineering of RADA, the RAD51 analogue present in archaea, a monomeric form of RAD51 was able to be synthesized.183 A fragment-based approach in 2013 demonstrated, through use of isothermal titration calorimetry (ITC), NMR, and X-ray crystallography, the first small molecule binding at the FxxA site of BRC4, a key binding region for the protein–protein interaction between RAD51 and BRCA2. However, a low success rate of only 0.2% was seen in the ITC assay in comparison to previous enzymatic assays. Two fragments were identified from this screen with low mM affinity; while low, it was a good starting point for further evaluation. This work was expanded upon in 2015, resulting in a 500-fold increase in affinity from the initial hits. Although the most promising hits from this series were unable to be crystallized, they performed well in the ITC assay, which seemed to indicate binding at the same site.184 Taken together, this fragment-based approach, utilizing biophysical assays such as ITC, NMR, SPR, and X-ray crystallography, demonstrates another possible tool in the pursuit of challenging SL targets. The genetic screening approach to evaluate possible SL pairs has gained new tools since the turn of the century. Previously, this was achieved through use of RNA interference (RNAi), where small-interfering RNAs (siRNAs) and/or short hairpin RNAs (shRNA) were employed to knockout genes to investigate whether the simultaneous knockout of two genes caused SL. This has been achieved in a high-throughput manner. An example of such is Novartis’s DRIVE program,177 which screened 7837 genes in 398 cell types. Numerous potential SL relationships were discovered in these studies, and these data could serve as a valuable tool for future drug-discovery programs, as all Novartis’s data on the DRIVE project is freely available through an online portal (https://oncologynibr.shinyapps.io/drive/). However, RNAi-based studies tend to suffer from poor reproducibility and comparison between studies, which can result in false positives and an inability to distinguish between on-target and off-target effects. Recent years have also seen the development of CRISPR-CAS9, which is functionally similar to RNAi but offers a higher-degree of reproducibility. This method involves a strand of single guided RNA (sgRNA) that allows the CAS9 endonuclease to bind with a sequence specific to that strand of sgRNA, thus enabling further modulation or regulation.185 Its advantages over RNAi are the following: it depletes genes in a more consistent manner; it has a higher degree of sensitivity with genes that have low levels of mRNA or short mRNA half-lives; it reduced RNA interference from heterogeneity among different cell lines.186 This technique has been employed in studies investigating synthetic lethality. As an example, Wang et al.187 performed CRISPR analysis in 3 different cell lines that had been treated by the ATR inhibitor, 24, in order to discover more SL pairs for ATR. From these studies, it was discovered that RNASEH2 was SL with ATR inhibition in vitro and in vivo. RNASEH2 is rarely upregulated in cancer subtypes; however this was in fact observed in prostate adenocarcinoma, suggesting a potential new biomarker that could be explored in this disease area. The same CRISPR-CAS9 studies reported that ATR inhibitors were SL with numerous other genes involved in the ATR pathway, suggesting that the whole ATR pathway is critical for cell survival; furthermore, members from the BRCA family were observed to be synthetic lethal with ATR, showing potential SL between ATR and HR-mediating targets. Overall, this demonstrates the potential of the CRISPR-CAS9 technique to be employed in the discovery of previously unknown targets for SL and guides further research. Furthermore, CRISPR-CAS9 in theory could be employed for treatment of cancers. This field is quite controversial with many ethical dilemmas that need to be addressed; however to date, a number of CRISPR-CAS9 based therapies have entered clinical trials.188 Should these be successful, it is possible in the future that CRISPR-CAS9 could be used in a therapeutic way to trigger SL.
Testing SL using in vitro models can be difficult. The biological models that are used for the development of such molecules usually include pairs of isogenic cell-based models. These are not an entirely accurate prediction of potential SL phenotypes, as it has been shown that SL pairs can be dependent on context specificity, with cancer presenting a vast array of genotypes. Small differences in the genotype can mean that a pair of small molecules modulating an SL interaction may be effective in one circumstance but not another. Examples of this phenomenon can be seen in colorectal cancer, where the presence of KRAS and NRAS mutations is a predictor for resistance to anti-EGFR antibodies.189 This is already being used in the clinic and demonstrates the importance of understanding the impact of context specificity on the application of SL-dependent treatments.
As it has been shown in the regulatory body approval of the PARP inhibitors, these drugs can be very effective in treating tumors with “BRCAness”, i.e., a tumor where the patient shows impeded function of one of the BRCA family genes involved in HR.203 Unfortunately, while a number of cancers (e.g., ovarian cancer)204 feature the BRCAness trait, the ability to administer a single inhibitor to induce SL is diminished in cancers that do not express BRCAness. To have these compounds truly be effective and initiate small-molecule-induced synthetic lethality, modulators must be administered in combination with one another in order to see a synergistic effect.147 This has been demonstrated previously with use of the PARP1 inhibitor 2 and temozolomide, an orally bioavailable monofunctional DNA-alkylating agent.205 The use of combinational therapy can be challenging, as the number of therapeutics in combination increases the likelihood of unwanted side effects, in addition to the potential for unknown drug–drug interactions.206 In order to minimize these issues, it is vitally important that the mechanism of action of the compounds and their safety profiles be well understood. If this is the case, the potential for combinational treatment is vast. Work has been conducted into evaluating more potential drug combinations that currently exist by Heinzel et al.207 Through a bioinformatics approach, they investigated 358 ovarian cancer trials utilizing the clinicaltrials.gov Web site. The search was refined further by use of keywords, eventually resulting in 68 trials that showed 61 unique drug interactions. These data allowed them to identify numerous combinations that are not currently being investigated in clinical trials but have the potential for SL and merit further investigation. These data illustrate that while combination treatments present challenges, the wealth of data on some of these well-known compounds in several clinical trials can be used as an asset, and there are great opportunities that can still be explored.
It is hypothesized by the authors that the use of small molecule-induced synthetic lethality should be approached with caution. In traditional SL approaches, where one of the genes lack function, i.e., through mutation in certain cancers, use of an inhibitor for the other SL gene targets the cancer cell and leaves the healthy cell employing a somewhat selective therapy. However, if this approach is replicated with small molecules, it is possible that the mechanisms these genes perform in DNA repair are utilized throughout healthy cells in addition to cancerous cells. Therefore, while cancer cells are more rapidly dividing and therefore will be reliant on HR, noncancerous cells will also use these mechanisms to some degree, resulting in unwanted side effects. Indeed, cancer cells are less genetically stable than healthy cells and therefore feature more DNA breaks, thus being more reliant on HR. This likely confers to small molecule-induced synthetic lethality a degree of selectivity for cancerous cells over healthy ones. The implications for this approach to be employed safely are that the underlying biology of the targets in healthy and cancer cells should be fully understood so as to apply this innovative approach to suitable target pairs.
Synthetic lethality, as shown throughout this review, has been primarily employed in oncology, attempting to disrupt the survival pathways of cancerous cells; however, this is not the only potential application of synthetically lethal therapies. The discovery of new antibiotics is an urgent need, as strains of resistant bacteria such as methicillin-resistant Staphylococcus aureus (MRSA) continue to be a major risk of infection in hospitals throughout the world.208 Due to bacteria being relatively simple organisms, they are reliant on HR for DNA damage repair;209 therefore SL has been looked at as a possible way of developing antibacterials. Charusanti et al. utilized a systems biology-guided approach to discover SL pairs for bacteria.210 The SL pairs identified from the virtual screens employed in these studies were confirmed experimentally with a precision rate of 25–43%. A virtual screen of compounds that were able to dock with 4 pairs of targets was performed subsequently. Two pairs of targets were found through their studies to be synthetically lethal (hemF/hemN, lpdA/sucC), one pair where both genes were found to be singularly essential (glyA/serA), and one pair where only one of the genes was known to be singularly essential (mdh/ppc). These studies did not yield compounds capable of inducing synthetic lethality; however the ability to identify SL pairs in bacteria opens the door to focused drug-discovery programs that may have more success.
Furthermore, additional potential applications can be seen in antiparasitics to treat diseases such as drug-resistant malaria. Due to the high level of conservation of genome integrity and cell cycle genes between species, it has been possible to conduct large screens in yeast and then use the SL pairs discovered to apply to other organisms, using the analogue genes in for example plasmodium.211 Using this approach, Lee et al. were able to compare yeast SL genes to their analogues present in the malaria parasites Plasmodium falciparum and Plasmodium vivax for the potential SL treatment of malaria in humans. Genes were selected that do not have an analogue in mammals, so as to minimize the chance of cross-species promiscuity. Through these studies five SL partners in the plasmodium were discovered. These findings warranted further investigation. To aid this, the group identified a number of commercially available compounds that already act upon these targets, showing a potential application of SL in antiparasitics. Many of the SL relationships discussed in this text have analogues in bacteria and parasites that play key roles in HR due to a high degree of evolutionary conservation. These examples, such as RAD51’s analogue in bacteria RecA or TbRAD51 in T. brucei, perform similar roles in their associated organisms and are heavily involved in HR and DNA repair.212,213 Of course, selectivity between species would be vitally important should any of these targets be explored, which could be challenging due to the large degree of homology between them; however, if this selectivity issue could be resolved, it could expand the usefulness of drugs that function through an SL mechanism greatly and warrants further exploration within the field.
10. Future Prospects and Final Thoughts
It has clearly emerged that the field of SL has much potential for oncology in a need that the personalized medicine from the previous 20 years has struggled to fill. SL offers a way to treat cancer that, in theory, leaves healthy cells unaffected and therefore has reduced side effects compared to nonspecific chemotherapy. However, the application of SL modulators still has a long way to go, as not all cancer subtypes have clearly defined deficiency, such as has been seen with the application of PARP1 and BRCA1/2 inhibitors or the 29 clinical trials currently involving ATR inhibitors. As the fields’ understanding of the underlying cancer biology increases, in addition to the growing network of tools to discover more SL pairs and, importantly, the mechanisms involved in SL, the viability of SL related drugs will increase. For this to occur, cross-disciplinary communication is vital, from target selection to compound design and optimization and beyond into in vitro/in vivo and into the clinic, with a willingness to risk and explore previously uninvestigated targets. Furthermore, the way in which SL drugs are implemented will be key to their success. For example, if full small molecule-induced synthetic lethality is to be used, i.e., inhibiting two SL pairs selectively at once, one must consider the greater potential for resistance as the cancer cell survival pathways adapt. Either it must be accepted that these compounds have a short window of effectiveness, or these mechanisms of resistance must be studied and anticipated so that further treatment can be administered to restore sensitivity, as is attempting to be achieved with PARP1 resistance through the restoration of function of BRCA1/2 using RAD51-BRCA2 inhibitors.143 Alternatively, if SL compounds can be implemented to impede HR in cancers that have undergone DNA stress, such as alkylation agents, nonspecific chemotherapeutics, or ionization, it is possible that through this method SL could be employed to lessen the dosage and therefore side effects of these treatments, with a reduced risk of acquired resistance from the SL pathway. Additionally, it has been theorized that SL drugs would be most effective in early stage cancers, as it is hypothesized that premalignant cancers are less heterogenic, and therefore this is where SL is likely to have the greatest impact.214 Obviously, for this to be viable, early detection of cancers in the clinic has to be achievable, and this chemopreventive approach is very much still in its infancy, existing primarily in animal models.215 With these considerations in place, it can be seen that the future for SL modulations could be bright, and in years to come the number of projects shining light on this interesting method of combating cancer will surely grow, and hopefully this will be reflected in the clinic. We hope this work inspires the medicinal chemistry community to develop novel compounds that can be exploited to discover innovative SL pathways in a chemical biology framework and, through utilization in drug discovery programs, identify SL-based anticancer compounds.
Glossary
Abbreviations Used
- ATRi
ATR inhibitor
- AZ
AstraZeneca
- BER
base excision repair
- BTK
Bruton’s tyrosine kinase
- CL
clearance
- CtIP
CTBP interacting protein
- CYP3A4
cytochrome P450 3A4
- Dabs
maximum absorbable dose
- DNA-PKi
DNA-PK inhibitor
- DSB
double-stranded break
- HER2
human epidermal growth factor 2
- HR
homologous recombination
- HTS
high-throughput screen
- ITC
isothermal titration calorimetry
- MMEJ
microhomology-mediated end joining
- MRSA
methicillin-resistant Staphylococcus aureus
- NER
nucleotide excision repair
- NHEJ
nonhomologous end joining
- PARP
poly (ADP-ribose) polymerase
- PARPi
PARP inhibitor
- RAD52i
RAD52 inhibitor
- RNAi
RNA interference
- RPAi
RPA inhibitor
- sgRNA
single-guided RNA
- shRNA
short hairpin RNA
- siRNA
small-interfering RNA
- SL
synthetic lethality
- SSB
single-stranded break
- ssDNA
single-stranded DNA
- Vss
volume of distribution
- WT
wild-type
Biographies
Samuel H. Myers received his Ph.D. in Medicinal Chemistry from the University of Edinburgh (U.K.) in 2016. After postdoctoral work in medicinal chemistry and chemical biology at University College London (UCL) (U.K.), in 2018 he joined the Italian Institute of Technology, Genova (Italy), where he works as a postdoc in medicinal chemistry.
Jose Antonio Ortega got his Ph.D. degree in Chemistry from the University of Barcelona (UB) in 2006 in the field of nucleic acids research. After working as a chemistry researcher in industrial medicinal chemistry projects at the Scientific Park of Barcelona (PCB), in 2012 he joined the Italian Institute of Technology (IIT) to participate in drug discovery projects mainly focused on the design and synthesis of small molecules for cancer treatment. Within the different strategies applied for the design of new molecular entities, the development of protein–protein interaction disruptors to chemically induce synthetic lethality is the center of his most recent research activities in the computational and chemical biology group at IIT.
Andrea Cavalli is Professor of Medicinal Chemistry at the University of Bologna and Director of Computational and Chemical Biology at the Italian Institute of Technology, Genova, where he is also Deputy Director for Computational Sciences. Prof. Cavalli’s research has combined computational chemistry with drug discovery, focusing on neurodegenerative diseases, cancer, and neglected tropical diseases. In particular, he has been a pioneer in the use of molecular dynamics approaches to drug discovery. These new methods have led to the identification and characterization of lead candidates within the framework of multitarget drug discovery and polypharmacology. He is an author of more than 230 scientific papers and inventor in several international patents. He has delivered more than 120 invited lectures and seminars.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.0c00766.
Molecular formula string (CSV)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
This work was supported in part by the Italian Association for Cancer Research (AIRC) through Grant IG 2018 Id.21386 and the Istituto Italiano di Tecnologia (IIT).
The authors declare no competing financial interest.
Supplementary Material
References
- Hochhaus A.; Larson R. A.; Guilhot F.; Radich J. P.; Branford S.; Hughes T. P.; Baccarani M.; Deininger M. W.; Cervantes F.; Fujihara S.; Ortmann C. E.; Menssen H. D.; Kantarjian H.; O’Brien S. G.; Druker B. J. Long-Term outcomes of Imatinib treatment for chronic myeloid leukemia. N. Engl. J. Med. 2017, 376 (10), 917–927. 10.1056/NEJMoa1609324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman P. B.; Hauschild A.; Robert C.; Haanen J. B.; Ascierto P.; Larkin J.; Dummer R.; Garbe C.; Testori A.; Maio M.; Hogg D.; Lorigan P.; Lebbe C.; Jouary T.; Schadendorf D.; Ribas A.; O’Day S. J.; Sosman J. A.; Kirkwood J. M.; Eggermont A. M.; Dreno B.; Nolop K.; Li J.; Nelson B.; Hou J.; Lee R. J.; Flaherty K. T.; McArthur G. A. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011, 364 (26), 2507–2516. 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini R.; Shao W.; Sellers W. R. Oncogene addiction: pathways of therapeutic response, resistance, and road maps toward a cure. EMBO Rep. 2015, 16 (3), 280–296. 10.15252/embr.201439949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A.; Garraway L. A.; Ashworth A.; Weber B. Synthetic lethality as an engine for cancer drug target discovery. Nat. Rev. Drug Discovery 2020, 19 (1), 23–38. 10.1038/s41573-019-0046-z. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics of natural populations. Xiii. Recombination and variability in populations of Drosophila pseudoobscura. Genetics 1946, 31, 269–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parameswaran S.; Kundapur D.; Vizeacoumar F. S.; Freywald A.; Uppalapati M.; Vizeacoumar F. J. A road map to personalizing targeted cancer therapies using synthetic lethality. Trends Cancer 2019, 5 (1), 11–29. 10.1016/j.trecan.2018.11.001. [DOI] [PubMed] [Google Scholar]
- Deeks E. D. Olaparib: first global approval. Drugs 2015, 75 (2), 231–240. 10.1007/s40265-015-0345-6. [DOI] [PubMed] [Google Scholar]
- Ame J. C.; Spenlehauer C.; de Murcia G. The PARP superfamily. BioEssays 2004, 26 (8), 882–893. 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- D’Amours D.; Desnoyers S.; D’Silva I.; Poirier G. G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999, 342 (2), 249–268. 10.1042/bj3420249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupte R.; Liu Z.; Kraus W. L. PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. 2017, 31 (2), 101–126. 10.1101/gad.291518.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skidmore C. J.; Davies M. I.; Goodwin P. M.; Halldorsson H.; Lewis P. J.; Shall S.; Zia’ee A. A. The involvement of poly(ADP-ribose) polymerase in the degradation of NAD caused by gamma-radiation and N-methyl-N-nitrosourea. Eur. J. Biochem. 1979, 101 (1), 135–142. 10.1111/j.1432-1033.1979.tb04225.x. [DOI] [PubMed] [Google Scholar]
- Caldecott K. W.; Aoufouchi S.; Johnson P.; Shall S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res. 1996, 24 (22), 4387–4394. 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales J.; Li L.; Fattah F. J.; Dong Y.; Bey E. A.; Patel M.; Gao J.; Boothman D. A. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit. Rev. Eukaryotic Gene Expression 2014, 24 (1), 15–28. 10.1615/CritRevEukaryotGeneExpr.2013006875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M. S.; Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature 1992, 356 (6367), 356–358. 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- Langelier M.-F.; Zandarashvili L.; Aguiar P. M.; Black B. E.; Pascal J. M. NAD+ analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains. Nat. Commun. 2018, 9 (1), 844. 10.1038/s41467-018-03234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C. J.; Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science 2017, 355 (6330), 1152–1158. 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant H. E.; Schultz N.; Thomas H. D.; Parker K. M.; Flower D.; Lopez E.; Kyle S.; Meuth M.; Curtin N. J.; Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005, 434 (7035), 913–917. 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Farmer H.; McCabe N.; Lord C. J.; Tutt A. N.; Johnson D. A.; Richardson T. B.; Santarosa M.; Dillon K. J.; Hickson I.; Knights C.; Martin N. M.; Jackson S. P.; Smith G. C.; Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434 (7035), 917–921. 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol. Oncol. 2011, 5 (4), 387–393. 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer S. M.; van Attikum H. PARP inhibitor resistance: a tug-of-war in BRCA-mutated cells. Trends Cell Biol. 2019, 29 (10), 820–834. 10.1016/j.tcb.2019.07.008. [DOI] [PubMed] [Google Scholar]
- Giovannini S.; Weller M. C.; Repmann S.; Moch H.; Jiricny J. Synthetic lethality between BRCA1 deficiency and poly(ADP-ribose) polymerase inhibition is modulated by processing of endogenous oxidative DNA damage. Nucleic Acids Res. 2019, 47 (17), 9132–9143. 10.1093/nar/gkz624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon C. E.; Nakamura A. J.; Zhang Y. W.; Ji J. J.; Bonner W. M.; Kinders R. J.; Parchment R. E.; Doroshow J. H.; Pommier Y. Histone gammaH2AX and poly(ADP-ribose) as clinical pharmacodynamic biomarkers. Clin. Cancer Res. 2010, 16 (18), 4532–4542. 10.1158/1078-0432.CCR-10-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea A. D. Mechanisms of PARP inhibitor sensitivity and resistance. DNA Repair 2018, 71, 172–176. 10.1016/j.dnarep.2018.08.021. [DOI] [PubMed] [Google Scholar]
- Cannan W. J.; Pederson D. S. Mechanisms and consequences of double-strand DNA break formation in Chromatin. J. Cell. Physiol. 2016, 231 (1), 3–14. 10.1002/jcp.25048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R.; Liu J. C.; Amunugama R.; Hajdu I.; Primack B.; Petalcorin M. I. R.; O’Connor K. W.; Konstantinopoulos P. A.; Elledge S. J.; Boulton S. J.; Yusufzai T.; D’Andrea A. D. Homologous-recombination-deficient tumours are dependent on Pol theta-mediated repair. Nature 2015, 518 (7538), 258–U306. 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Song Y.; Li S.; Kurian S.; Xiang R.; Chiba T.; Wu X. DNA polymerase theta (POLQ) is important for repair of DNA double-strand breaks caused by fork collapse. J. Biol. Chem. 2019, 294 (11), 3909–3919. 10.1074/jbc.RA118.005188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domchek S. M.; Aghajanian C.; Shapira-Frommer R.; Schmutzler R. K.; Audeh M. W.; Friedlander M.; Balmana J.; Mitchell G.; Fried G.; Stemmer S. M.; Hubert A.; Rosengarten O.; Loman N.; Robertson J. D.; Mann H.; Kaufman B. Efficacy and safety of olaparib monotherapy in germline BRCA1/2 mutation carriers with advanced ovarian cancer and three or more lines of prior therapy. Gynecol. Oncol. 2016, 140 (2), 199–203. 10.1016/j.ygyno.2015.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong P. C.; Yap T. A.; Boss D. S.; Carden C. P.; Mergui-Roelvink M.; Gourley C.; De Greve J.; Lubinski J.; Shanley S.; Messiou C.; A’Hern R.; Tutt A.; Ashworth A.; Stone J.; Carmichael J.; Schellens J. H.; de Bono J. S.; Kaye S. B. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J. Clin. Oncol. 2010, 28 (15), 2512–2519. 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- Audeh M. W.; Carmichael J.; Penson R. T.; Friedlander M.; Powell B.; Bell-McGuinn K. M.; Scott C.; Weitzel J. N.; Oaknin A.; Loman N.; Lu K.; Schmutzler R. K.; Matulonis U.; Wickens M.; Tutt A. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet 2010, 376 (9737), 245–251. 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- Kaufman B.; Shapira-Frommer R.; Schmutzler R. K.; Audeh M. W.; Friedlander M.; Balmana J.; Mitchell G.; Fried G.; Stemmer S. M.; Hubert A.; Rosengarten O.; Steiner M.; Loman N.; Bowen K.; Fielding A.; Domchek S. M. Olaparib monotherapy in patients with advanced cancer and a Germline BRCA1/2 mutation. J. Clin. Oncol. 2015, 33 (3), 244–250. 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristeleit R.; Shapiro G. I.; Burris H. A.; Oza A. M.; LoRusso P.; Patel M. R.; Domchek S. M.; Balmana J.; Drew Y.; Chen L. M.; Safra T.; Montes A.; Giordano H.; Maloney L.; Goble S.; Isaacson J.; Xiao J.; Borrow J.; Rolfe L.; Shapira-Frommer R. A Phase I-II study of the oral PARP inhibitor Rucaparib in patients with Germline BRCA1/2-mutated ovarian carcinoma or other solid tumors. Clin. Cancer Res. 2017, 23 (15), 4095–4106. 10.1158/1078-0432.CCR-16-2796. [DOI] [PubMed] [Google Scholar]
- Swisher E. M.; Lin K. K.; Oza A. M.; Scott C. L.; Giordano H.; Sun J.; Konecny G. E.; Coleman R. L.; Tinker A. V.; O’Malley D. M.; Kristeleit R. S.; Ma L.; Bell-McGuinn K. M.; Brenton J. D.; Cragun J. M.; Oaknin A.; Ray-Coquard I.; Harrell M. I.; Mann E.; Kaufmann S. H.; Floquet A.; Leary A.; Harding T. C.; Goble S.; Maloney L.; Isaacson J.; Allen A. R.; Rolfe L.; Yelensky R.; Raponi M.; McNeish I. A. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017, 18 (1), 75–87. 10.1016/S1470-2045(16)30559-9. [DOI] [PubMed] [Google Scholar]
- Mirza M. R.; Monk B. J.; Herrstedt J.; Oza A. M.; Mahner S.; Redondo A.; Fabbro M.; Ledermann J. A.; Lorusso D.; Vergote I.; Ben-Baruch N. E.; Marth C.; Madry R.; Christensen R. D.; Berek J. S.; Dorum A.; Tinker A. V.; du Bois A.; Gonzalez-Martin A.; Follana P.; Benigno B.; Rosenberg P.; Gilbert L.; Rimel B. J.; Buscema J.; Balser J. P.; Agarwal S.; Matulonis U. A. Niraparib maintenance therapy in Platinum-sensitive, recurrent ovarian cancer. N. Engl. J. Med. 2016, 375 (22), 2154–2164. 10.1056/NEJMoa1611310. [DOI] [PubMed] [Google Scholar]
- Robson M.; Im S.-A.; Senkus E.; Xu B.; Domchek S. M.; Masuda N.; Delaloge S.; Li W.; Tung N.; Armstrong A.; Wu W.; Goessl C.; Runswick S.; Conte P. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N. Engl. J. Med. 2017, 377, 523–533. 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- Litton J. K.; Rugo H. S.; Ettl J.; Hurvitz S. A.; Goncalves A.; Lee K. H.; Fehrenbacher L.; Yerushalmi R.; Mina L. A.; Martin M.; Roche H.; Im Y. H.; Quek R. G. W.; Markova D.; Tudor I. C.; Hannah A. L.; Eiermann W.; Blum J. L. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N. Engl. J. Med. 2018, 379 (8), 753–763. 10.1056/NEJMoa1802905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy S. M. Talazoparib: first global approval. Drugs 2018, 78 (18), 1939–1946. 10.1007/s40265-018-1026-z. [DOI] [PubMed] [Google Scholar]
- Golan T.; Hammel P.; Reni M.; Van Cutsem E.; Macarulla T.; Hall M. J.; Park J. O.; Hochhauser D.; Arnold D.; Oh D. Y.; Reinacher-Schick A.; Tortora G.; Algul H.; O’Reilly E. M.; McGuinness D.; Cui K. Y.; Schlienger K.; Locker G. Y.; Kindler H. L. Maintenance Olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl. J. Med. 2019, 381 (4), 317–327. 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration. https://www.fda.gov/drugs/fda-grants-accelerated-approval-rucaparib-brca-mutated-metastatic-castration-resistant-prostate (accessed Jul 7, 2020).
- Astra Zeneca. https://www.astrazeneca.com/media-centre/press-releases/2019/lynparza-more-than-doubled-the-time-without-disease-progression-in-patients-with-brca1-2-atm-mutated-metastatic-castration-resistant-prostate-cancer-30092019.html (accessed Jul 7, 2020).
- Jain P. G.; Patel B. D. Medicinal chemistry approaches of poly ADP-Ribose polymerase 1 (PARP1) inhibitors as anticancer agents - A recent update. Eur. J. Med. Chem. 2019, 165, 198–215. 10.1016/j.ejmech.2019.01.024. [DOI] [PubMed] [Google Scholar]
- Gurley K. E.; Kemp C. J. Synthetic lethality between mutation in Atm and DNA-PK(cs) during murine embryogenesis. Curr. Biol. 2001, 11 (3), 191–194. 10.1016/S0960-9822(01)00048-3. [DOI] [PubMed] [Google Scholar]
- Kantidze O. L.; Velichko A. K.; Luzhin A. V.; Petrova N. V.; Razin S. V. Synthetically lethal interactions of ATM, ATR, and DNA-PKcs. Trends Cancer 2018, 4 (11), 755–768. 10.1016/j.trecan.2018.09.007. [DOI] [PubMed] [Google Scholar]
- Riabinska A.; Daheim M.; Herter-Sprie G. S.; Winkler J.; Fritz C.; Hallek M.; Thomas R. K.; Kreuzer K.-A.; Frenzel L. P.; Monfared P.; Martins-Boucas J.; Chen S.; Reinhardt H. C. Therapeutic targeting of a robust non-oncogene addiction to PRKDC in ATM-defective tumors. Sci. Transl. Med. 2013, 5 (189), 189ra78–189ra78. 10.1126/scitranslmed.3005814. [DOI] [PubMed] [Google Scholar]
- Dietlein F.; Thelen L.; Jokic M.; Jachimowicz R. D.; Ivan L.; Knittel G.; Leeser U.; van Oers J.; Edelmann W.; Heukamp L. C.; Reinhardt H. C. A functional cancer genomics screen identifies a druggable synthetic lethal interaction between MSH3 and PRKDC. Cancer Discovery 2014, 4 (5), 592–605. 10.1158/2159-8290.CD-13-0907. [DOI] [PubMed] [Google Scholar]
- Chen C.-C.; Kass E. M.; Yen W.-F.; Ludwig T.; Moynahan M. E.; Chaudhuri J.; Jasin M. ATM loss leads to synthetic lethality in BRCA1 BRCT mutant mice associated with exacerbated defects in homology-directed repair. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (29), 7665–7670. 10.1073/pnas.1706392114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albarakati N.; Abdel-Fatah T. M.; Doherty R.; Russell R.; Agarwal D.; Moseley P.; Perry C.; Arora A.; Alsubhi N.; Seedhouse C.; Rakha E. A.; Green A.; Ball G.; Chan S.; Caldas C.; Ellis I. O.; Madhusudan S. Targeting BRCA1-BER deficient breast cancer by ATM or DNA-PKcs blockade either alone or in combination with cisplatin for personalized therapy. Mol. Oncol. 2015, 9 (1), 204–217. 10.1016/j.molonc.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R.; Miller K. M.; Forment J. V.; Bradshaw C. R.; Nikan M.; Britton S.; Oelschlaegel T.; Xhemalce B.; Balasubramanian S.; Jackson S. P. Small-molecule–induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012, 8 (3), 301–310. 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smida M.; Fece de la Cruz F.; Kerzendorfer C.; Uras I. Z.; Mair B.; Mazouzi A.; Suchankova T.; Konopka T.; Katz A. M.; Paz K.; Nagy-Bojarszky K.; Muellner M. K.; Bago-Horvath Z.; Haura E. B.; Loizou J. I.; Nijman S. M. B. MEK inhibitors block growth of lung tumours with mutations in ataxia–telangiectasia mutated. Nat. Commun. 2016, 7 (1), 13701. 10.1038/ncomms13701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.; Tian H. Current development status of MEK inhibitors. Molecules 2017, 22 (10), 1551. 10.3390/molecules22101551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin J. F.; Knudsen K. E. Beyond DNA repair: DNA-PK function in cancer. Cancer Discovery 2014, 4 (10), 1126–1139. 10.1158/2159-8290.CD-14-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohiuddin I. S.; Kang M. H. DNA-PK as an emerging therapeutic target in Cancer. Front. Oncol. 2019, 9, 635. 10.3389/fonc.2019.00635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotula E.; Berthault N.; Agrario C.; Lienafa M. C.; Simon A.; Dingli F.; Loew D.; Sibut V.; Saule S.; Dutreix M. DNA-PKcs plays role in cancer metastasis through regulation of secreted proteins involved in migration and invasion. Cell Cycle 2015, 14 (12), 1961–1972. 10.1080/15384101.2015.1026522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell L.; Munck J. M.; Alsinet C.; Villanueva A.; Ogle L.; Willoughby C. E.; Televantou D.; Thomas H. D.; Jackson J.; Burt A. D.; Newell D.; Rose J.; Manas D. M.; Shapiro G. I.; Curtin N. J.; Reeves H. L. DNA-PK-A candidate driver of hepatocarcinogenesis and tissue biomarker that predicts response to treatment and survival. Clin. Cancer Res. 2015, 21 (4), 925–933. 10.1158/1078-0432.CCR-14-0842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero A. B.; San Miguel J.; Gutierrez N. C. Deregulation of DNA double-strand break repair in multiple myeloma: implications for genome stability. PLoS One 2015, 10 (3), e0121581 10.1371/journal.pone.0121581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara M.; Ashizawa K.; Shichijo K.; Kudo T. Expression of the DNA-dependent protein kinase catalytic subunit is associated with the radiosensitivity of human thyroid cancer cell lines. J. Radiat. Res. 2019, 60 (2), 171–177. 10.1093/jrr/rry097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani S.; Mihara M.; Li C.; Nakahara Y.; Hino S.; Nakashiro K.; Hamakawa H. Up-regulation of DNA-dependent protein kinase correlates with radiation resistance in oral squamous cell carcinoma. Cancer Sci. 2003, 94 (10), 894–900. 10.1111/j.1349-7006.2003.tb01372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beskow C.; Skikuniene J.; Holgersson A.; Nilsson B.; Lewensohn R.; Kanter L.; Viktorsson K. Radioresistant cervical cancer shows upregulation of the NHEJ proteins DNA-PKcs, Ku70 and Ku86. Br. J. Cancer 2009, 101 (5), 816–821. 10.1038/sj.bjc.6605201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldivar J. C.; Cortez D.; Cimprich K. A. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 2017, 18 (10), 622–636. 10.1038/nrm.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok M.; Davies N.; Agathanggelou A.; Smith E.; Oldreive C.; Petermann E.; Stewart G.; Brown J.; Lau A.; Pratt G.; Parry H.; Taylor M.; Moss P.; Hillmen P.; Stankovic T. ATR inhibition induces synthetic lethality and overcomes chemoresistance in TP53- or ATM-defective chronic lymphocytic leukemia cells. Blood 2016, 127 (5), 582–595. 10.1182/blood-2015-05-644872. [DOI] [PubMed] [Google Scholar]
- Fokas E.; Prevo R.; Pollard J. R.; Reaper P. M.; Charlton P. A.; Cornelissen B.; Vallis K. A.; Hammond E. M.; Olcina M. M.; Gillies McKenna W.; Muschel R. J.; Brunner T. B. Targeting ATR in vivo using the novel inhibitor VE-822 results in selective sensitization of pancreatic tumors to radiation. Cell Death Dis. 2012, 3 (12), e441 10.1038/cddis.2012.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min A.; Im S.-A.; Jang H.; Kim S.; Lee M.; Kim D. K.; Yang Y.; Kim H.-J.; Lee K.-H.; Kim J. W.; Kim T.-Y.; Oh D.-Y.; Brown J.; Lau A.; O’Connor M. J.; Bang Y.-J. AZD6738, A novel oral inhibitor of ATR, induces synthetic lethality with ATM deficiency in gastric cancer cells. Mol. Cancer Ther. 2017, 16 (4), 566–577. 10.1158/1535-7163.MCT-16-0378. [DOI] [PubMed] [Google Scholar]
- Sanjiv K.; Hagenkort A.; Calderon-Montano J. M.; Koolmeister T.; Reaper P. M.; Mortusewicz O.; Jacques S. A.; Kuiper R. V.; Schultz N.; Scobie M.; Charlton P. A.; Pollard J. R.; Berglund U. W.; Altun M.; Helleday T. Cancer-specific synthetic lethality between ATR and CHK1 kinase activities. Cell Rep. 2016, 17 (12), 3407–3416. 10.1016/j.celrep.2016.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo L. I.; Altmeyer M.; Rask M. B.; Lukas C.; Larsen D. H.; Povlsen L. K.; Bekker-Jensen S.; Mailand N.; Bartek J.; Lukas J. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 2013, 155 (5), 1088–1103. 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- Charrier J. D.; Durrant S. J.; Golec J. M.; Kay D. P.; Knegtel R. M.; MacCormick S.; Mortimore M.; O’Donnell M. E.; Pinder J. L.; Reaper P. M.; Rutherford A. P.; Wang P. S.; Young S. C.; Pollard J. R. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J. Med. Chem. 2011, 54 (7), 2320–2330. 10.1021/jm101488z. [DOI] [PubMed] [Google Scholar]
- Knegtel R.; Charrier J. D.; Durrant S.; Davis C.; O’Donnell M.; Storck P.; MacCormick S.; Kay D.; Pinder J.; Virani A.; Twin H.; Griffiths M.; Reaper P.; Littlewood P.; Young S.; Golec J.; Pollard J. Rational design of 5-(4-(Isopropylsulfonyl)phenyl)-3-(3-(4-((methylamino)methyl)phenyl)isoxazol-5-yl)pyrazin-2-amine (VX-970, M6620): optimization of intra- and intermolecular polar interactions of a new ataxia telangiectasia mutated and Rad3-related (ATR) kinase inhibitor. J. Med. Chem. 2019, 62 (11), 5547–5561. 10.1021/acs.jmedchem.9b00426. [DOI] [PubMed] [Google Scholar]
- O’Carrigan B.; de Miguel Luken M. J.; Papadatos-Pastos D.; Brown J.; Tunariu N.; Perez Lopez R.; Ganegoda M.; Riisnaes R.; Figueiredo I.; Carreira S.; Hare B.; Yang F.; McDermott K.; Penney M. S.; Pollard J.; Lopez J. S.; Banerji U.; De Bono J. S.; Fields S. Z.; Yap T. A. Phase I trial of a first-in-class ATR inhibitor VX-970 as monotherapy (mono) or in combination (combo) with carboplatin (CP) incorporating pharmacodynamics (PD) studies. J. Clin. Oncol. 2016, 34 (15_suppl), 2504. 10.1200/JCO.2016.34.15_suppl.2504. [DOI] [Google Scholar]
- Yap T. A.; de Miguel Luken M. J.; O’Carrigan B.; Roda D.; Papadatos-Pastos D.; Lorente D.; Tunariu N.; Lopez R. P.; Gayle S.; Riisnaes R.; Figueiredo I.; Miranda S.; Carreira S.; Yang F.; Karan S.; Penney M.; Pollard J.; Molife L. R.; Banerji U.; Asmal M.; Fields S. Z.; de Bono J. S. Abstract PR14: Phase I trial of first-in-class ataxia telangiectasia-mutated and Rad3-related (ATR) inhibitor VX-970 as monotherapy (mono) or in combination with carboplatin (CP) in advanced cancer patients (pts) with preliminary evidence of target modulation and antitumor activity. Mol. Cancer. Ther. 2015, 14 (12, Suppl. 2), PR14 10.1158/1535-7163.TARG-15-PR14. [DOI] [Google Scholar]
- Shapiro G.; Wesolowski R.; Middleton M.; Devoe C.; Constantinidou A.; Papadatos-Pastos D.; Fricano M.; Zhang Y.; Karan S.; Pollard J.; Penney M.; Asmal M.; Renshaw F. G.; Fields S. Z.; Yap T. A. Abstract CT012: phase 1 trial of first-in-class ATR inhibitor VX-970 in combination with cisplatin (Cis) in patients (pts) with advanced solid tumors (NCT02157792). Cancer Res. 2016, 76 (14, Suppl.), CT012 10.1158/1538-7445.AM2016-CT012. [DOI] [Google Scholar]
- Plummer E. R.; Dean E. J.; Evans T. R. J.; Greystoke A.; Herbschleb K.; Ranson M.; Brown J.; Zhang Y.; Karan S.; Pollard J.; Penney M. S.; Asmal M.; Fields S. Z.; Middleton M. R. Phase I trial of first-in-class ATR inhibitor VX-970 in combination with gemcitabine (Gem) in advanced solid tumors (NCT02157792). J. Clin. Oncol. 2016, 34 (15_suppl), 2513. 10.1200/JCO.2016.34.15_suppl.2513. [DOI] [Google Scholar]
- Zenke F. T.; Zimmermann A.; Dahmen H.; Elenbaas B.; Pollard J.; Reaper P.; Bagrodia S.; Spilker M. E.; Amendt C.; Blaukat A. Abstract 369: Antitumor activity of M4344, a potent and selective ATR inhibitor, in monotherapy and combination therapy. Cancer Res. 2019, 79 (13, Suppl.), 369. 10.1158/1538-7445.AM2019-369. [DOI] [Google Scholar]
- Wortmann L.; Lucking U.; Lefranc J.; Briem H.; Koppitz M.; Eis K.; Von Nussbaum F.; Bader B.; Wegner A. M.; Siemeister G.; Bone W.; Lienau P.; Grudzinska-Goebel J.; Mossmayer D.; Eberspächer U.; Schick H.. 2-Morpholin-yl)-1,7-naphythridines. WO/2016/020320, 2016.
- Wengner A. M.; Siemeister G.; Lucking U.; Lefranc J.; Wortmann L.; Lienau P.; Bader B.; Bomer U.; Moosmayer D.; Eberspacher U.; Golfier S.; Schatz C. A.; Baumgart S. J.; Haendler B.; Lejeune P.; Schlicker A.; von Nussbaum F.; Brands M.; Ziegelbauer K.; Mumberg D. The novel ATR inhibitor BAY 1895344 is efficacious as monotherapy and combined with DNA damage-inducing or repair-compromising rherapies in preclinical cancer models. Mol. Cancer Ther. 2020, 19 (1), 26–38. 10.1158/1535-7163.MCT-19-0019. [DOI] [PubMed] [Google Scholar]
- Foote K. M.; Blades K.; Cronin A.; Fillery S.; Guichard S. S.; Hassall L.; Hickson I.; Jacq X.; Jewsbury P. J.; McGuire T. M.; Nissink J. W.; Odedra R.; Page K.; Perkins P.; Suleman A.; Tam K.; Thommes P.; Broadhurst R.; Wood C. Discovery of 4-{4-[(3R)-3-Methylmorpholin-4-yl]-6-[1-(methylsulfonyl)cyclopropyl]pyrimidin-2-yl}-1H-indole (AZ20): a potent and selective inhibitor of ATR protein kinase with monotherapy in vivo antitumor activity. J. Med. Chem. 2013, 56 (5), 2125–2138. 10.1021/jm301859s. [DOI] [PubMed] [Google Scholar]
- Foote K. M.; Nissink J. W. M.; McGuire T.; Turner P.; Guichard S.; Yates J. W. T.; Lau A.; Blades K.; Heathcote D.; Odedra R.; Wilkinson G.; Wilson Z.; Wood C. M.; Jewsbury P. J. Discovery and characterization of AZD6738, a potent inhibitor of ataxia telangiectasia mutated and Rad3 related (ATR) kinase with application as an anticancer agent. J. Med. Chem. 2018, 61 (22), 9889–9907. 10.1021/acs.jmedchem.8b01187. [DOI] [PubMed] [Google Scholar]
- Lloyd R.; Falenta K.; Wijnhoven P. W.; Chabbert C.; Stott J.; Yates J.; Lau A. Y.; Young L. A.; Hollingsworth S. J. Abstract 337: The PARP inhibitor olaparib is synergistic with the ATR inhibitor AZD6738 in ATM deficient cancer cells. Cancer Res. 2018, 78 (13, Suppl.), 337. 10.1158/1538-7445.AM2018-337. [DOI] [Google Scholar]
- Barsanti P. A.; Aversa R. J.; Jin X.; Pan Y.; Lu Y.; Elling R.; Jain R.; Knapp M.; Lan J.; Lin X.; Rudewicz P.; Sim J.; Taricani L.; Thomas G.; Xiao L.; Yue Q. Structure-Based drug design of novel potent and selective tetrahydropyrazolo[1,5-a]pyrazines as ATR inhibitors. ACS Med. Chem. Lett. 2015, 6 (1), 37–41. 10.1021/ml500353p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujala B.; Agarwal A. K.; Middya S.; Banerjee M.; Surya A.; Nayak A. K.; Gupta A.; Khare S.; Guguloth R.; Randive N. A.; Shinde B. U.; Thakur A.; Patel D. I.; Raja M.; Green M. J.; Alfaro J.; Avila P.; Pérez de Arce F.; Almirez R. G.; Kanno S.; Bernales S.; Hung D. T.; Chakravarty S.; McCullagh E.; Quinn K. P.; Rai R.; Pham S. M. Discovery of pyrazolopyrimidine derivatives as novel dual inhibitors of BTK and PI3Kδ. ACS Med. Chem. Lett. 2016, 7 (12), 1161–1166. 10.1021/acsmedchemlett.6b00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S. A.; Jadhavar P. S.; Singh M. P.; Sharma A.; Bagle G. N.; Quinn K. P.; Wong P. Y.; Protter A. A.; Rai R.; Pham S. M.; Lindquist J. N. Discovery of pyrazolopyrimidine derivatives as novel inhibitors of ataxia telangiectasia and rad3 related protein (ATR). Bioorg. Med. Chem. Lett. 2017, 27 (4), 750–754. 10.1016/j.bmcl.2017.01.045. [DOI] [PubMed] [Google Scholar]
- Hickson I.; Zhao Y.; Richardson C. J.; Green S. J.; Martin N. M.; Orr A. I.; Reaper P. M.; Jackson S. P.; Curtin N. J.; Smith G. C. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004, 64 (24), 9152–9159. 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- Golding S. E.; Rosenberg E.; Valerie N.; Hussaini I.; Frigerio M.; Cockcroft X. F.; Chong W. Y.; Hummersone M.; Rigoreau L.; Menear K. A.; O’Connor M. J.; Povirk L. F.; van Meter T.; Valerie K. Improved ATM kinase inhibitor KU-60019 radiosensitizes glioma cells, compromises insulin, AKT and ERK prosurvival signaling, and inhibits migration and invasion. Mol. Cancer Ther. 2009, 8 (10), 2894–2902. 10.1158/1535-7163.MCT-09-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey M. D.; Charlton M. E.; Stanton R. V.; Kastan M. B. Transient inhibition of ATM kinase is sufficient to enhance cellular sensitivity to ionizing radiation. Cancer Res. 2008, 68 (18), 7466–7474. 10.1158/0008-5472.CAN-08-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi P.; Foiani M.; Kumar A. ATM and ATR signaling at a glance. J. Cell Sci. 2015, 128 (23), 4255–4262. 10.1242/jcs.169730. [DOI] [PubMed] [Google Scholar]
- Degorce S. L.; Barlaam B.; Cadogan E.; Dishington A.; Ducray R.; Glossop S. C.; Hassall L. A.; Lach F.; Lau A.; McGuire T. M.; Nowak T.; Ouvry G.; Pike K. G.; Thomason A. G. Discovery of novel 3-quinoline carboxamides as potent, selective, and orally bioavailable inhibitors of ataxia telangiectasia mutated (ATM) kinase. J. Med. Chem. 2016, 59 (13), 6281–6292. 10.1021/acs.jmedchem.6b00519. [DOI] [PubMed] [Google Scholar]
- Karlin J.; Allen J.; Ahmad S. F.; Hughes G.; Sheridan V.; Odedra R.; Farrington P.; Cadogan E. B.; Riches L. C.; Garcia-Trinidad A.; Thomason A. G.; Patel B.; Vincent J.; Lau A.; Pike K. G.; Hunt T. A.; Sule A.; Valerie N. C. K.; Biddlestone-Thorpe L.; Kahn J.; Beckta J. M.; Mukhopadhyay N.; Barlaam B.; Degorce S. L.; Kettle J.; Colclough N.; Wilson J.; Smith A.; Barrett I. P.; Zheng L.; Zhang T.; Wang Y.; Chen K.; Pass M.; Durant S. T.; Valerie K. Orally bioavailable and blood-brain barrier-penetrating ATM inhibitor (AZ32) radiosensitizes intracranial gliomas in mice. Mol. Cancer Ther. 2018, 17 (8), 1637–1647. 10.1158/1535-7163.MCT-17-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike K. G.; Barlaam B.; Cadogan E.; Campbell A.; Chen Y.; Colclough N.; Davies N. L.; de-Almeida C.; Degorce S. L.; Didelot M.; Dishington A.; Ducray R.; Durant S. T.; Hassall L. A.; Holmes J.; Hughes G. D.; MacFaul P. A.; Mulholland K. R.; McGuire T. M.; Ouvry G.; Pass M.; Robb G.; Stratton N.; Wang Z.; Wilson J.; Zhai B.; Zhao K.; Al-Huniti N. The identification of potent, selective, and orally available inhibitors of ataxia telangiectasia mutated (ATM) kinase: the discovery of AZD0156 (8-{6-[3-(Dimethylamino)propoxy]pyridin-3-yl}-3-methyl-1-(tetrahydro-2 H-pyran-4-yl)-1,3-dihydro-2 H-imidazo[4,5- c]quinolin-2-one). J. Med. Chem. 2018, 61 (9), 3823–3841. 10.1021/acs.jmedchem.7b01896. [DOI] [PubMed] [Google Scholar]
- Smith D. A.; Beaumont K.; Maurer T. S.; Di L. Volume of distribution in drug design. J. Med. Chem. 2015, 58 (15), 5691–5698. 10.1021/acs.jmedchem.5b00201. [DOI] [PubMed] [Google Scholar]
- Maira S. M.; Stauffer F.; Brueggen J.; Furet P.; Schnell C.; Fritsch C.; Brachmann S.; Chene P.; De Pover A.; Schoemaker K.; Fabbro D.; Gabriel D.; Simonen M.; Murphy L.; Finan P.; Sellers W.; Garcia-Echeverria C. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol. Cancer Ther. 2008, 7 (7), 1851–1863. 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- Mukherjee B.; Tomimatsu N.; Amancherla K.; Camacho C. V.; Pichamoorthy N.; Burma S. The dual PI3K/mTOR inhibitor NVP-BEZ235 is a potent inhibitor of ATM- and DNA-PKCs-mediated DNA damage responses. Neoplasia 2012, 14 (1), 34–43. 10.1593/neo.111512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riches L. C.; Trinidad A. G.; Hughes G.; Jones G. N.; Hughes A. M.; Thomason A. G.; Gavine P.; Cui A.; Ling S.; Stott J.; Clark R.; Peel S.; Gill P.; Goodwin L. M.; Smith A.; Pike K. G.; Barlaam B.; Pass M.; O’Connor M. J.; Smith G.; Cadogan E. B. Pharmacology of the ATM Inhibitor AZD0156: Potentiation of irradiation and olaparib responses preclinically. Mol. Cancer Ther. 2020, 19 (1), 13–25. 10.1158/1535-7163.MCT-18-1394. [DOI] [PubMed] [Google Scholar]
- Durant S. T.; Zheng L.; Wang Y.; Chen K.; Zhang L.; Zhang T.; Yang Z.; Riches L.; Trinidad A. G.; Fok J. H. L.; Hunt T.; Pike K. G.; Wilson J.; Smith A.; Colclough N.; Reddy V. P.; Sykes A.; Janefeldt A.; Johnstrom P.; Varnas K.; Takano A.; Ling S.; Orme J.; Stott J.; Roberts C.; Barrett I.; Jones G.; Roudier M.; Pierce A.; Allen J.; Kahn J.; Sule A.; Karlin J.; Cronin A.; Chapman M.; Valerie K.; Illingworth R.; Pass M. The brain-penetrant clinical ATM inhibitor AZD1390 radiosensitizes and improves survival of preclinical brain tumor models. Sci. Adv. 2018, 4 (6), eaat1719 10.1126/sciadv.aat1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddlestone-Thorpe L.; Sajjad M.; Rosenberg E.; Beckta J. M.; Valerie N. C.; Tokarz M.; Adams B. R.; Wagner A. F.; Khalil A.; Gilfor D.; Golding S. E.; Deb S.; Temesi D. G.; Lau A.; O’Connor M. J.; Choe K. S.; Parada L. F.; Lim S. K.; Mukhopadhyay N. D.; Valerie K. ATM kinase inhibition preferentially sensitizes p53-mutant glioma to ionizing radiation. Clin. Cancer Res. 2013, 19 (12), 3189–3200. 10.1158/1078-0432.CCR-12-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor O. G.; Brzozowski J. S.; Skelding K. A. Glioblastoma multiforme: an overview of emerging therapeutic targets. Front. Oncol. 2019, 9, 963. 10.3389/fonc.2019.00963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoyanova S.; Bulgarelli-Leva G.; Kirsch C.; Hanck T.; Klinger R.; Wetzker R.; Wymann M. P. Lipid kinase and protein kinase activities of G-protein-coupled phosphoinositide 3-kinase gamma: structure-activity analysis and interactions with wortmannin. Biochem. J. 1997, 324 (2), 489–495. 10.1042/bj3240489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin R. J.; Fontana G.; Golding B. T.; Guiard S.; Hardcastle I. R.; Leahy J. J.; Martin N.; Richardson C.; Rigoreau L.; Stockley M.; Smith G. C. Selective benzopyranone and pyrimido[2,1-a]isoquinolin-4-one inhibitors of DNA-dependent protein kinase: synthesis, structure-activity studies, and radiosensitization of a human tumor cell line in vitro. J. Med. Chem. 2005, 48 (2), 569–585. 10.1021/jm049526a. [DOI] [PubMed] [Google Scholar]
- Hardcastle I. R.; Cockcroft X.; Curtin N. J.; El-Murr M. D.; Leahy J. J. J.; Stockley M.; Golding B. T.; Rigoreau L.; Richardson C.; Smith G. C. M.; Griffin R. J. Discovery of potent chromen-4-one inhibitors of the DNA-dependent protein kinase (DNA-PK) using a small-molecule library approach. J. Med. Chem. 2005, 48 (24), 7829–7846. 10.1021/jm050444b. [DOI] [PubMed] [Google Scholar]
- Fok J. H. L.; Ramos-Montoya A.; Vazquez-Chantada M.; Wijnhoven P. W. G.; Follia V.; James N.; Farrington P. M.; Karmokar A.; Willis S. E.; Cairns J.; Nikkilä J.; Beattie D.; Lamont G. M.; Finlay M. R. V.; Wilson J.; Smith A.; O’Connor L. O.; Ling S.; Fawell S. E.; O’Connor M. J.; Hollingsworth S. J.; Dean E.; Goldberg F. W.; Davies B. R.; Cadogan E. B. AZD7648 is a potent and selective DNA-PK inhibitor that enhances radiation, chemotherapy and olaparib activity. Nat. Commun. 2019, 10 (1), 5065. 10.1038/s41467-019-12836-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke F. T.; Zimmermann A.; Sirrenberg C.; Dahmen H.; Vassilev L.; Pehl U.; Fuchss T.; Blaukat A. Abstract 1658: M3814, a novel investigational DNA-PK inhibitor: enhancing the effect of fractionated radiotherapy leading to complete regression of tumors in mice. Cancer Res. 2016, 76 (14, Suppl.), 1658. 10.1158/1538-7445.AM2016-1658. [DOI] [Google Scholar]
- Fuchss T.; Emde U.; Buchstaller H.-P.; Merski W.. Arylquinazolines. WO2014183850, 2014.
- Sun Q.; Guo Y.; Liu X.; Czauderna F.; Carr M. I.; Zenke F. T.; Blaukat A.; Vassilev L. T. Therapeutic implications of p53 status on cancer cell fate following exposure to ionizing radiation and the DNA-PK inhibitor M3814. Mol. Cancer Res. 2019, 17 (12), 2457–2468. 10.1158/1541-7786.MCR-19-0362. [DOI] [PubMed] [Google Scholar]
- Carr M. I.; Zimmermann A.; Chiu L.-Y.; Zenke F. T.; Blaukat A.; Vassilev L. T. DNA-PK inhibitor, M3814, as a new combination partner of Mylotarg in the treatment of acute myeloid leukemia. Front. Oncol. 2020, 10, 127. 10.3389/fonc.2020.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen D. S.; Perrin-Ninkovic S. M.; Shevlin G.; Elsner J.; Zhao J.; Whitefield B.; Tehrani L.; Sapienza J.; Riggs J. R.; Parnes J. S.; Papa P.; Packard G.; Lee B. G. S.; Harris R.; Correa M.; Bahmanyar S.; Richardson S. J.; Peng S. X.; Leisten J.; Khambatta G.; Hickman M.; Gamez J. C.; Bisonette R. R.; Apuy J.; Cathers B. E.; Canan S. S.; Moghaddam M. F.; Raymon H. K.; Worland P.; Narla R. K.; Fultz K. E.; Sankar S. Optimization of a series of triazole containing mammalian target of rapamycin (mTOR) kinase Inhibitors and the discovery of CC-115. J. Med. Chem. 2015, 58 (14), 5599–5608. 10.1021/acs.jmedchem.5b00627. [DOI] [PubMed] [Google Scholar]
- Tsuji T.; Sapinoso L. M.; Tran T.; Gaffney B.; Wong L.; Sankar S.; Raymon H. K.; Mortensen D. S.; Xu S. CC-115, a dual inhibitor of mTOR kinase and DNA-PK, blocks DNA damage repair pathways and selectively inhibits ATM-deficient cell growth in vitro. Oncotarget 2017, 8 (43), 74688–74702. 10.18632/oncotarget.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Kelly R.; Martín Y.; Koundrioukoff S.; Tanenbaum M. E.; Smits V. A. J.; Medema R. H.; Debatisse M.; Freire R. Wee1 controls genomic stability during replication by regulating the Mus81-Eme1 endonuclease. J. Cell Biol. 2011, 194 (4), 567–579. 10.1083/jcb.201101047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do K.; Doroshow J. H.; Kummar S. Wee1 kinase as a target for cancer therapy. Cell Cycle 2013, 12 (19), 3159–3164. 10.4161/cc.26062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson C. J.; Backos D. S.; Reigan P. Targeting WEE1 kinase in cancer. Trends Pharmacol. Sci. 2016, 37 (10), 872–881. 10.1016/j.tips.2016.06.006. [DOI] [PubMed] [Google Scholar]
- Music D.; Dahlrot R. H.; Hermansen S. K.; Hjelmborg J.; de Stricker K.; Hansen S.; Kristensen B. W. Expression and prognostic value of the WEE1 kinase in gliomas. J. Neuro-Oncol. 2016, 127 (2), 381–389. 10.1007/s11060-015-2050-4. [DOI] [PubMed] [Google Scholar]
- Iorns E.; Lord C. J.; Grigoriadis A.; McDonald S.; Fenwick K.; Mackay A.; Mein C. A.; Natrajan R.; Savage K.; Tamber N.; Reis-Filho J. S.; Turner N. C.; Ashworth A. Integrated functional, gene expression and genomic analysis for the identification of cancer targets. PLoS One 2009, 4 (4), e5120 10.1371/journal.pone.0005120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter C. C.; Kim J.; Fosmire S.; Gearheart C. M.; van Linden A.; Baturin D.; Zaberezhnyy V.; Patel P. R.; Gao D.; Tan A. C.; DeGregori J. Integrated genomic analyses identify WEE1 as a critical mediator of cell fate and a novel therapeutic target in acute myeloid leukemia. Leukemia 2012, 26 (6), 1266–1276. 10.1038/leu.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnussen G. I.; Holm R.; Emilsen E.; Rosnes A. K.; Slipicevic A.; Florenes V. A. High expression of Wee1 is associated with poor disease-free survival in malignant melanoma: potential for targeted therapy. PLoS One 2012, 7 (6), e38254 10.1371/journal.pone.0038254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slipicevic A.; Holth A.; Hellesylt E.; Trope C. G.; Davidson B.; Florenes V. A. Wee1 is a novel independent prognostic marker of poor survival in post-chemotherapy ovarian carcinoma effusions. Gynecol. Oncol. 2014, 135 (1), 118–124. 10.1016/j.ygyno.2014.07.102. [DOI] [PubMed] [Google Scholar]
- Bukhari A. B.; Lewis C. W.; Pearce J. J.; Luong D.; Chan G. K.; Gamper A. M. Inhibiting Wee1 and ATR kinases produces tumor-selective synthetic lethality and suppresses metastasis. J. Clin. Invest. 2019, 129 (3), 1329–1344. 10.1172/JCI122622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman J. E.; Chung C. H. CHK it out! Blocking WEE kinase routs TP53 mutant cancer. Clin. Cancer Res. 2014, 20 (16), 4173–4175. 10.1158/1078-0432.CCR-14-0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Li J.; Booher R. N.; Kraker A.; Lawrence T.; Leopold W. R.; Sun Y. Radiosensitization of p53 mutant cells by PD0166285, a novel G(2) checkpoint abrogator. Cancer Res. 2001, 61 (22), 8211–8217. [PubMed] [Google Scholar]
- Leijen S.; van Geel R. M. J. M.; Pavlick A. C.; Tibes R.; Rosen L.; Razak A. R. A.; Lam R.; Demuth T.; Rose S.; Lee M. A.; Freshwater T.; Shumway S.; Liang L. W.; Oza A. M.; Schellens J. H. M.; Shapiro G. I. Phase I study evaluating WEE1 inhibitor AZD1775 as monotherapy and in combination with gemcitabine, cisplatin, or carboplatin in patients with advanced solid tumors. J. Clin. Oncol. 2016, 34 (36), 4371–4380. 10.1200/JCO.2016.67.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leijen S.; van Geel R. M.; Sonke G. S.; de Jong D.; Rosenberg E. H.; Marchetti S.; Pluim D.; van Werkhoven E.; Rose S.; Lee M. A.; Freshwater T.; Beijnen J. H.; Schellens J. H. Phase II study of WEE1 inhibitor AZD1775 plus carboplatin in patients with TP53-mutated ovarian cancer refractory or resistant to first-line therapy within 3 months. J. Clin. Oncol. 2016, 34 (36), 4354–4361. 10.1200/JCO.2016.67.5942. [DOI] [PubMed] [Google Scholar]
- Carrassa L.; Chilà R.; Lupi M.; Ricci F.; Celenza C.; Mazzoletti M.; Broggini M.; Damia G. Combined inhibition of Chk1 and Wee1: in vitro synergistic effect translates to tumor growth inhibition in vivo. Cell Cycle 2012, 11 (13), 2507–2517. 10.4161/cc.20899. [DOI] [PubMed] [Google Scholar]
- Hirai H.; Iwasawa Y.; Okada M.; Arai T.; Nishibata T.; Kobayashi M.; Kimura T.; Kaneko N.; Ohtani J.; Yamanaka K.; Itadani H.; Takahashi-Suzuki I.; Fukasawa K.; Oki H.; Nambu T.; Jiang J.; Sakai T.; Arakawa H.; Sakamoto T.; Sagara T.; Yoshizumi T.; Mizuarai S.; Kotani H. Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol. Cancer Ther. 2009, 8 (11), 2992–3000. 10.1158/1535-7163.MCT-09-0463. [DOI] [PubMed] [Google Scholar]
- Duan Y.; Dong X.; Nie J.; Li P.; Lu F.; Ma D.; Ji C. Wee1 kinase inhibitor MK-1775 induces apoptosis of acute lymphoblastic leukemia cells and enhances the efficacy of doxorubicin involving downregulation of Notch pathway. Oncol. Lett. 2018, 16 (4), 5473–5481. 10.3892/ol.2018.9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richer A. L.; Cala J. M.; O’Brien K.; Carson V. M.; Inge L. J.; Whitsett T. G. WEE1 kinase inhibitor AZD1775 has preclinical efficacy in LKB1-deficient non-small cell lung cancer. Cancer Res. 2017, 77 (17), 4663–4672. 10.1158/0008-5472.CAN-16-3565. [DOI] [PubMed] [Google Scholar]
- Webster P. J.; Littlejohns A. T.; Gaunt H. J.; Prasad K. R.; Beech D. J.; Burke D. A. AZD1775 induces toxicity through double-stranded DNA breaks independently of chemotherapeutic agents in p53-mutated colorectal cancer cells. Cell Cycle 2017, 16 (22), 2176–2182. 10.1080/15384101.2017.1301329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson C. J.; Venkataraman S.; Amani V.; Harris P. S.; Backos D. S.; Donson A. M.; Wempe M. F.; Foreman N. K.; Vibhakar R.; Reigan P. A WEE1 inhibitor analog of AZD1775 maintains synergy with cisplatin and demonstrates reduced single-agent cytotoxicity in medulloblastoma cells. ACS Chem. Biol. 2016, 11 (7), 2066–2067. 10.1021/acschembio.6b00466. [DOI] [PubMed] [Google Scholar]
- Marques F.; Moreau J. L.; Peaucellier G.; Lozano J. C.; Schatt P.; Picard A.; Callebaut I.; Perret E.; Geneviere A. M. A new subfamily of high molecular mass CDC2-related kinases with PITAI/VRE motifs. Biochem. Biophys. Res. Commun. 2000, 279 (3), 832–837. 10.1006/bbrc.2000.4042. [DOI] [PubMed] [Google Scholar]
- Li X.; Chatterjee N.; Spirohn K.; Boutros M.; Bohmann D. Cdk12 is a gene-selective RNA polymerase II kinase that regulates a subset of the transcriptome, including Nrf2 target genes. Sci. Rep. 2016, 6 (1), 21455. 10.1038/srep21455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui G. Y. L.; Grandori C.; Kemp C. J. CDK12: an emerging therapeutic target for cancer. J. Clin. Pathol. 2018, 71 (11), 957–962. 10.1136/jclinpath-2018-205356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi P. M.; Sutor S. L.; Huntoon C. J.; Karnitz L. M. Ovarian cancer-associated mutations disable catalytic activity of CDK12, a kinase that promotes homologous recombination repair and resistance to cisplatin and poly(ADP-ribose) polymerase inhibitors. J. Biol. Chem. 2014, 289 (13), 9247–9253. 10.1074/jbc.M114.551143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekumi K. M.; Paculova H.; Lenasi T.; Pospichalova V.; Bosken C. A.; Rybarikova J.; Bryja V.; Geyer M.; Blazek D.; Barboric M. Ovarian carcinoma CDK12 mutations misregulate expression of DNA repair genes via deficient formation and function of the Cdk12/CycK complex. Nucleic Acids Res. 2015, 43 (5), 2575–2589. 10.1093/nar/gkv101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo K.; Wai P. T.; Maguire S. L.; Daley F.; Haider S.; Kriplani D.; Campbell J.; Mirza H.; Grigoriadis A.; Tutt A.; Moseley P. M.; Abdel-Fatah T. M. A.; Chan S. Y. T.; Madhusudan S.; Rhaka E. A.; Ellis I. O.; Lord C. J.; Yuan Y.; Green A. R.; Natrajan R. Evaluation of CDK12 protein expression as a potential novel biomarker for DNA damage response-targeted therapies in breast cancer. Mol. Cancer Ther. 2018, 17 (1), 306–315. 10.1158/1535-7163.MCT-17-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas R.; Gao S.; Cultraro C. M.; Maity T. K.; Venugopalan A.; Abdullaev Z.; Shaytan A. K.; Carter C. A.; Thomas A.; Rajan A.; Song Y.; Pitts S.; Chen K.; Bass S.; Boland J.; Hanada K. I.; Chen J.; Meltzer P. S.; Panchenko A. R.; Yang J. C.; Pack S.; Giaccone G.; Schrump D. S.; Khan J.; Guha U. Genomic profiling of multiple sequentially acquired tumor metastatic sites from an “exceptional responder” lung adenocarcinoma patient reveals extensive genomic heterogeneity and novel somatic variants driving treatment response. Cold Spring Harb. Mol. Case Stud. 2016, 2 (6), a001263 10.1101/mcs.a001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajrami I.; Frankum J. R.; Konde A.; Miller R. E.; Rehman F. L.; Brough R.; Campbell J.; Sims D.; Rafiq R.; Hooper S.; Chen L.; Kozarewa I.; Assiotis I.; Fenwick K.; Natrajan R.; Lord C. J.; Ashworth A. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res. 2014, 74 (1), 287–297. 10.1158/0008-5472.CAN-13-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson L.; Muniz L.; West S. 3′ end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes Dev. 2014, 28 (4), 342–356. 10.1101/gad.231274.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniguez A. B.; Stolte B.; Wang E. J.; Conway A. S.; Alexe G.; Dharia N. V.; Kwiatkowski N.; Zhang T.; Abraham B. J.; Mora J.; Kalev P.; Leggett A.; Chowdhury D.; Benes C. H.; Young R. A.; Gray N. S.; Stegmaier K. EWS/FLI confers tumor cell synthetic lethality to CDK12 inhibition in Ewing sarcoma. Cancer Cell 2018, 33 (2), 202–216. 10.1016/j.ccell.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamman K.; Bradley M.; Marineau J.; Choi Y.; Malojcic G.; Orlando D.; Ren Y.; Ke N.; Hu S.; Olson E.; Fritz C.; Roberts C. Targeting the transcriptional kinases CDK12 and CDK13 in breast and ovarian cancer. FASEB J. 2017, 31 (1_supplement), 938.9. 10.1096/fasebj.31.1_supplement.938.9. [DOI] [Google Scholar]
- Ito M.; Tanaka T.; Toita A.; Uchiyama N.; Kokubo H.; Morishita N.; Klein M. G.; Zou H.; Murakami M.; Kondo M.; Sameshima T.; Araki S.; Endo S.; Kawamoto T.; Morin G. B.; Aparicio S. A.; Nakanishi A.; Maezaki H.; Imaeda Y. Discovery of 3-benzyl-1-(trans-4-((5-cyanopyridin-2-yl)amino)cyclohexyl)-1-arylurea derivatives as novel and selective cyclin-dependent kinase 12 (CDK12) Inhibitors. J. Med. Chem. 2018, 61 (17), 7710–7728. 10.1021/acs.jmedchem.8b00683. [DOI] [PubMed] [Google Scholar]
- Zhang T.; Kwiatkowski N.; Olson C. M.; Dixon-Clarke S. E.; Abraham B. J.; Greifenberg A. K.; Ficarro S. B.; Elkins J. M.; Liang Y.; Hannett N. M.; Manz T.; Hao M.; Bartkowiak B.; Greenleaf A. L.; Marto J. A.; Geyer M.; Bullock A. N.; Young R. A.; Gray N. S. Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat. Chem. Biol. 2016, 12 (10), 876–884. 10.1038/nchembio.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski N.; Zhang T.; Rahl P. B.; Abraham B. J.; Reddy J.; Ficarro S. B.; Dastur A.; Amzallag A.; Ramaswamy S.; Tesar B.; Jenkins C. E.; Hannett N. M.; McMillin D.; Sanda T.; Sim T.; Kim N. D.; Look T.; Mitsiades C. S.; Weng A. P.; Brown J. R.; Benes C. H.; Marto J. A.; Young R. A.; Gray N. S. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 2014, 511 (7511), 616–620. 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry D.; Guzi T.; Shanahan F.; Davis N.; Prabhavalkar D.; Wiswell D.; Seghezzi W.; Paruch K.; Dwyer M. P.; Doll R.; Nomeir A.; Windsor W.; Fischmann T.; Wang Y.; Oft M.; Chen T.; Kirschmeier P.; Lees E. M. Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Mol. Cancer Ther. 2010, 9 (8), 2344–2353. 10.1158/1535-7163.MCT-10-0324. [DOI] [PubMed] [Google Scholar]
- Mita M. M.; Joy A. A.; Mita A.; Sankhala K.; Jou Y. M.; Zhang D.; Statkevich P.; Zhu Y.; Yao S. L.; Small K.; Bannerji R.; Shapiro C. L. Randomized phase II trial of the cyclin-dependent kinase inhibitor dinaciclib (MK-7965) versus capecitabine in patients with advanced breast cancer. Clin. Breast Cancer 2014, 14 (3), 169–176. 10.1016/j.clbc.2013.10.016. [DOI] [PubMed] [Google Scholar]
- Johnson S. F.; Cruz C.; Greifenberg A. K.; Dust S.; Stover D. G.; Chi D.; Primack B.; Cao S.; Bernhardy A. J.; Coulson R.; Lazaro J. B.; Kochupurakkal B.; Sun H.; Unitt C.; Moreau L. A.; Sarosiek K. A.; Scaltriti M.; Juric D.; Baselga J.; Richardson A. L.; Rodig S. J.; D’Andrea A. D.; Balmana J.; Johnson N.; Geyer M.; Serra V.; Lim E.; Shapiro G. I. CDK12 inhibition reverses de novo and acquired PARPinhibitor resistance in BRCA wild-type and mutated models of triple-negative breast cancer. Cell Rep. 2016, 17 (9), 2367–2381. 10.1016/j.celrep.2016.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek D.; Kohoutek J.; Bartholomeeusen K.; Johansen E.; Hulinkova P.; Luo Z.; Cimermancic P.; Ule J.; Peterlin B. M. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011, 25 (20), 2158–2172. 10.1101/gad.16962311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P.; West S. C. The human Rad51 protein: polarity of strand transfer and stimulation by hRP-A. EMBO J. 1997, 16 (17), 5198–5206. 10.1093/emboj/16.17.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 1994, 265 (5176), 1241–1243. 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- Zaitseva E. M.; Zaitsev E. N.; Kowalczykowski S. C. The DNA binding properties of saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 1999, 274 (5), 2907–2915. 10.1074/jbc.274.5.2907. [DOI] [PubMed] [Google Scholar]
- Roberti M.; Schipani F.; Bagnolini G.; Milano D.; Giacomini E.; Falchi F.; Balboni A.; Manerba M.; Farabegoli F.; De Franco F.; Robertson J.; Minucci S.; Pallavicini I.; Di Stefano G.; Girotto S.; Pellicciari R.; Cavalli A. Rad51/BRCA2 disruptors inhibit homologous recombination and synergize with olaparib in pancreatic cancer cells. Eur. J. Med. Chem. 2019, 165, 80–92. 10.1016/j.ejmech.2019.01.008. [DOI] [PubMed] [Google Scholar]
- Budke B.; Tueckmantel W.; Miles K.; Kozikowski A. P.; Connell P. P. Optimization of drug candidates that inhibit the D-loop activity of RAD51. ChemMedChem 2019, 14 (10), 1031–1040. 10.1002/cmdc.201900075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand A.; Riviere E.; Renodon-Corniere A. Identification and characterization of human Rad51 inhibitors by screening of an existing drug library. Biochem. Pharmacol. 2014, 91 (3), 293–300. 10.1016/j.bcp.2014.07.033. [DOI] [PubMed] [Google Scholar]
- Zhu J.; Chen H.; Guo X. E.; Qiu X. L.; Hu C. M.; Chamberlin A. R.; Lee W. H. Synthesis, molecular modeling, and biological evaluation of novel RAD51 inhibitors. Eur. J. Med. Chem. 2015, 96, 196–208. 10.1016/j.ejmech.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnolini G.; Milano D.; Manerba M.; Schipani F.; Ortega J. A.; Gioia D.; Falchi F.; Balboni A.; Farabegoli F.; De Franco F.; Robertson J.; Pellicciari R.; Pallavicini I.; Peri S.; Minucci S.; Girotto S.; Di Stefano G.; Roberti M.; Cavalli A. Synthetic lethality in pancreatic cancer: discovery of a new RAD51-BRCA2 small molecule disruptor that inhibits homologous recombination and synergizes with Olaparib. J. Med. Chem. 2020, 63 (5), 2588–2619. 10.1021/acs.jmedchem.9b01526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendra E.; Venkitaraman A. R. Two modules in the BRC repeats of BRCA2 mediate structural and functional interactions with the RAD51 recombinase. Nucleic Acids Res. 2010, 38 (1), 82–96. 10.1093/nar/gkp873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv W.; Budke B.; Pawlowski M.; Connell P. P.; Kozikowski A. P. Development of small molecules that specifically inhibit the D-loop activity of RAD51. J. Med. Chem. 2016, 59 (10), 4511–4525. 10.1021/acs.jmedchem.5b01762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budke B.; Logan H. L.; Kalin J. H.; Zelivianskaia A. S.; Cameron McGuire W.; Miller L. L.; Stark J. M.; Kozikowski A. P.; Bishop D. K.; Connell P. P. RI-1: a chemical inhibitor of RAD51 that disrupts homologous recombination in human cells. Nucleic Acids Res. 2012, 40 (15), 7347–7357. 10.1093/nar/gks353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budke B.; Kalin J. H.; Pawlowski M.; Zelivianskaia A. S.; Wu M.; Kozikowski A. P.; Connell P. P. An optimized RAD51 inhibitor that disrupts homologous recombination without requiring Michael acceptor reactivity. J. Med. Chem. 2013, 56 (1), 254–263. 10.1021/jm301565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanvatirth P.; Jeeves R. E.; Bacon J.; Besra G. S.; Alderwick L. J. Utilisation of the Prestwick chemical library to identify drugs that inhibit the growth of mycobacteria. PLoS One 2019, 14 (3), e0213713 10.1371/journal.pone.0213713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaku M.; Kainuma T.; Ishida-Takaku T.; Ishigami S.; Suzuki H.; Tashiro S.; van Soest R. W.; Nakao Y.; Kurumizaka H. Halenaquinone, a chemical compound that specifically inhibits the secondary DNA binding of RAD51. Genes Cells 2011, 16 (4), 427–436. 10.1111/j.1365-2443.2011.01494.x. [DOI] [PubMed] [Google Scholar]
- Zhu J.; Zhou L.; Wu G.; Konig H.; Lin X.; Li G.; Qiu X. L.; Chen C. F.; Hu C. M.; Goldblatt E.; Bhatia R.; Chamberlin A. R.; Chen P. L.; Lee W. H. A novel small molecule RAD51 inactivator overcomes imatinib-resistance in chronic myeloid leukaemia. EMBO Mol. Med. 2013, 5 (3), 353–365. 10.1002/emmm.201201760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson P. J.; Vincent M. D.; Koropatnick J. Synergistic antiproliferative activity of the RAD51 inhibitor IBR2 with inhibitors of receptor tyrosine kinases and microtubule protein. J. Pharmacol. Exp. Ther. 2018, 364 (1), 46–54. 10.1124/jpet.117.241661. [DOI] [PubMed] [Google Scholar]
- Huang F.; Motlekar N. A.; Burgwin C. M.; Napper A. D.; Diamond S. L.; Mazin A. V. Identification of specific inhibitors of human RAD51 recombinase using high-throughput screening. ACS Chem. Biol. 2011, 6 (6), 628–635. 10.1021/cb100428c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wéra A. C.; Lobbens A.; Stoyanov M.; Lucas S.; Michiels C. Radiation-induced synthetic lethality: combination of poly(ADP-ribose) polymerase and RAD51 inhibitors to sensitize cells to proton irradiation. Cell Cycle 2019, 18 (15), 1770–1783. 10.1080/15384101.2019.1632640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclay T.; Vacca J.; McComas C.; Castro A.; Day M.; Mills K. CYT01B, a novel RAD51 inhibitor, act synergistically with both targeted and chemotherapeutic anti-cancer agents. Blood 2018, 132 (Suppl. 1), 3963. 10.1182/blood-2018-99-119373. [DOI] [Google Scholar]
- Castro A.; McComas C.; Vacca J.; Maclay T.. RAD51 Inhibitors. US20190077799 A1, 2019.
- Xue C.; Greene E. C. New roles for RAD52 in DNA repair. Cell Res. 2018, 28 (12), 1127–1128. 10.1038/s41422-018-0105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norquist B.; Wurz K. A.; Pennil C. C.; Garcia R.; Gross J.; Sakai W.; Karlan B. Y.; Taniguchi T.; Swisher E. M. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J. Clin. Oncol. 2011, 29 (22), 3008–3015. 10.1200/JCO.2010.34.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma M.; Sullivan-Reed K.; Sliwinski T.; Skorski T. RAD52 as a potential target for synthetic lethality-based anticancer therapies. Cancers 2019, 11 (10), 1561. 10.3390/cancers11101561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hromas R.; Kim H. S.; Sidhu G.; Williamson E.; Jaiswal A.; Totterdale T. A.; Nole J.; Lee S. H.; Nickoloff J. A.; Kong K. Y. The endonuclease EEPD1 mediates synthetic lethality in RAD52-depleted BRCA1 mutant breast cancer cells. Breast Cancer Res. 2017, 19, 122. 10.1186/s13058-017-0912-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer-Morales K.; Nieborowska-Skorska M.; Scheibner K.; Padget M.; Irvine D. A.; Sliwinski T.; Haas K.; Lee J.; Geng H.; Roy D.; Slupianek A.; Rassool F. V.; Wasik M. A.; Childers W.; Copland M.; Muschen M.; Civin C. I.; Skorski T. Personalized synthetic lethality induced by targeting RAD52 in leukemias identified by gene mutation and expression profile. Blood 2013, 122 (7), 1293–1304. 10.1182/blood-2013-05-501072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez S. F.; Renodon-Corniere A.; Nomme J.; Eveillard D.; Fleury F.; Takahashi M.; Weigel P. Targeting human Rad51 by specific DNA aptamers induces inhibition of homologous recombination. Biochimie 2010, 92 (12), 1832–1838. 10.1016/j.biochi.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Chandramouly G.; McDevitt S.; Sullivan K.; Kent T.; Luz A.; Glickman J. F.; Andrake M.; Skorski T.; Pomerantz R. T. Small-molecule disruption of RAD52 rings as a mechanism for precision medicine in BRCA-deficient cancers. Chem. Biol. 2015, 22 (11), 1491–1504. 10.1016/j.chembiol.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olney J. W.; Zorumski C. F.; Stewart G. R.; Price M. T.; Wang G. J.; Labruyere J. Excitotoxicity of L-dopa and 6-OH-dopa: implications for Parkinson’s and Huntington’s diseases. Exp. Neurol. 1990, 108 (3), 269–272. 10.1016/0014-4886(90)90134-E. [DOI] [PubMed] [Google Scholar]
- Huang F.; Goyal N.; Sullivan K.; Hanamshet K.; Patel M.; Mazina O. M.; Wang C. X.; An W. F.; Spoonamore J.; Metkar S.; Emmitte K. A.; Cocklin S.; Skorski T.; Mazin A. V. Targeting BRCA1- and BRCA2-deficient cells with RAD52 small molecule inhibitors. Nucleic Acids Res. 2016, 44 (9), 4189–4199. 10.1093/nar/gkw087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengel S. R.; Malacaria E.; Folly da Silva Constantino L.; Bain F. E.; Diaz A.; Koch B. G.; Yu L.; Wu M.; Pichierri P.; Spies M. A.; Spies M. Small-molecule inhibitors identify the RAD52-ssDNA interaction as critical for recovery from replication stress and for survival of BRCA2 deficient cells. eLife 2016, 5, e14740 10.7554/eLife.14740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfuni I.; Basile G.; Subramanyam S.; Malacaria E.; Bignami M.; Spies M.; Franchitto A.; Pichierri P. Survival of the replication checkpoint deficient cells requires MUS81-RAD52 function. PLoS Genet. 2013, 9 (10), e1003910 10.1371/journal.pgen.1003910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Yang Q.; Zhang Y.; Huang K.; Sun R.; Zhao Q. Compound F779-0434 causes synthetic lethality in BRCA2-deficient cancer cells by disrupting RAD52–ssDNA association. RSC Adv. 2018, 8 (34), 18859–18869. 10.1039/C8RA01919C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coley W. B. II. Contribution to the knowledge of sarcoma. Ann. Surg. 1891, 14 (3), 199–220. 10.1097/00000658-189112000-00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.; Gu Z.; Chen Y.; Chen B.; Chen W.; Weng L.; Liu X. Application of PD-1 blockade in cancer immunotherapy. Comput. Struct. Biotechnol. J. 2019, 17, 661–674. 10.1016/j.csbj.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A.; Yi M.; Qin S.; Chu Q.; Luo S.; Wu K. Prospects for combining immune checkpoint blockade with PARP inhibition. J. Hematol. Oncol. 2019, 12 (1), 98. 10.1186/s13045-019-0784-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao S.; Xia W.; Yamaguchi H.; Wei Y.; Chen M. K.; Hsu J. M.; Hsu J. L.; Yu W. H.; Du Y.; Lee H. H.; Li C. W.; Chou C. K.; Lim S. O.; Chang S. S.; Litton J.; Arun B.; Hortobagyi G. N.; Hung M. C. PARP inhibitor upregulates PD-L1 expression and enhances cancer-associated immunosuppression. Clin. Cancer Res. 2017, 23 (14), 3711–3720. 10.1158/1078-0432.CCR-16-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Meur N.; Gentleman R. Modeling synthetic lethality. Genome Biol. 2008, 9 (9), R135 10.1186/gb-2008-9-9-r135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald E. R. 3rd; de Weck A.; Schlabach M. R.; Billy E.; Mavrakis K. J.; Hoffman G. R.; Belur D.; Castelletti D.; Frias E.; Gampa K.; Golji J.; Kao I.; Li L.; Megel P.; Perkins T. A.; Ramadan N.; Ruddy D. A.; Silver S. J.; Sovath S.; Stump M.; Weber O.; Widmer R.; Yu J.; Yu K.; Yue Y.; Abramowski D.; Ackley E.; Barrett R.; Berger J.; Bernard J. L.; Billig R.; Brachmann S. M.; Buxton F.; Caothien R.; Caushi J. X.; Chung F. S.; Cortes-Cros M.; deBeaumont R. S.; Delaunay C.; Desplat A.; Duong W.; Dwoske D. A.; Eldridge R. S.; Farsidjani A.; Feng F.; Feng J.; Flemming D.; Forrester W.; Galli G. G.; Gao Z.; Gauter F.; Gibaja V.; Haas K.; Hattenberger M.; Hood T.; Hurov K. E.; Jagani Z.; Jenal M.; Johnson J. A.; Jones M. D.; Kapoor A.; Korn J.; Liu J.; Liu Q.; Liu S.; Liu Y.; Loo A. T.; Macchi K. J.; Martin T.; McAllister G.; Meyer A.; Molle S.; Pagliarini R. A.; Phadke T.; Repko B.; Schouwey T.; Shanahan F.; Shen Q.; Stamm C.; Stephan C.; Stucke V. M.; Tiedt R.; Varadarajan M.; Venkatesan K.; Vitari A. C.; Wallroth M.; Weiler J.; Zhang J.; Mickanin C.; Myer V. E.; Porter J. A.; Lai A.; Bitter H.; Lees E.; Keen N.; Kauffmann A.; Stegmeier F.; Hofmann F.; Schmelzle T.; Sellers W. R. Project DRIVE: A compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell 2017, 170 (3), 577–592. 10.1016/j.cell.2017.07.005. [DOI] [PubMed] [Google Scholar]
- Beetham H.; Chen A.; Telford B. J.; Single A.; Jarman K. E.; Lackovic K.; Luxenburger A.; Guilford P. A high-throughput screen to identify novel synthetic lethal compounds for the treatment of E-cadherin-deficient cells. Sci. Rep. 2019, 9 (1), 12511. 10.1038/s41598-019-48929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofree M.; Shen J. P.; Carter H.; Gross A.; Ideker T. Network-based stratification of tumor mutations. Nat. Methods 2013, 10 (11), 1108–1115. 10.1038/nmeth.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Lu L.; Zhang Y. H.; Liu M.; Chen L.; Huang T.; Cai Y. D. Identification of synthetic lethality based on a functional network by using machine learning algorithms. J. Cell. Biochem. 2019, 120 (1), 405–416. 10.1002/jcb.27395. [DOI] [PubMed] [Google Scholar]
- Benstead-Hume G.; Wooller S. K.; Pearl F. M. G. ‘Big data’ approaches for novel anti-cancer drug discovery. Expert Opin. Drug Discovery 2017, 12 (6), 599–609. 10.1080/17460441.2017.1319356. [DOI] [PubMed] [Google Scholar]
- Oughtred R.; Stark C.; Breitkreutz B. J.; Rust J.; Boucher L.; Chang C.; Kolas N.; O’Donnell L.; Leung G.; McAdam R.; Zhang F.; Dolma S.; Willems A.; Coulombe-Huntington J.; Chatr-Aryamontri A.; Dolinski K.; Tyers M. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019, 47 (D1), D529–D541. 10.1093/nar/gky1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. E.; Ehebauer M. T.; Pukala T.; Marsh M.; Blundell T. L.; Venkitaraman A. R.; Abell C.; Hyvonen M. Using a fragment-based approach to target protein-protein interactions. ChemBioChem 2013, 14 (3), 332–342. 10.1002/cbic.201200521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. E.; Coyne A. G.; Venkitaraman A.; Blundell T. L.; Abell C.; Hyvonen M. Small-molecule inhibitors that target protein-protein interactions in the RAD51 family of recombinases. ChemMedChem 2015, 10 (2), 296–303. 10.1002/cmdc.201402428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S.; Lai L. Synthetic lethality in drug development: the dawn is coming. Future Med. Chem. 2018, 10 (18), 2129–2132. 10.4155/fmc-2018-0227. [DOI] [PubMed] [Google Scholar]
- Zhu S.; Zhou Y.; Wei W. Genome-wide CRISPR/Cas9 screening for high-throughput functional genomics in human cells. Methods Mol. Biol. 2017, 1656, 175–181. 10.1007/978-1-4939-7237-1_11. [DOI] [PubMed] [Google Scholar]
- Wang C.; Wang G.; Feng X.; Shepherd P.; Zhang J.; Tang M.; Chen Z.; Srivastava M.; McLaughlin M. E.; Navone N. M.; Hart G. T.; Chen J. Genome-wide CRISPR screens reveal synthetic lethality of RNASEH2 deficiency and ATR inhibition. Oncogene 2019, 38 (14), 2451–2463. 10.1038/s41388-018-0606-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtmauer E. A.; Fraietta J. A.; Davis M. M.; Cohen A. D.; Weber K. L.; Lancaster E.; Mangan P. A.; Kulikovskaya I.; Gupta M.; Chen F.; Tian L.; Gonzalez V. E.; Xu J.; Jung I.-y.; Melenhorst J. J.; Plesa G.; Shea J.; Matlawski T.; Cervini A.; Gaymon A. L.; Desjardins S.; Lamontagne A.; Salas-Mckee J.; Fesnak A.; Siegel D. L.; Levine B. L.; Jadlowsky J. K.; Young R. M.; Chew A.; Hwang W.-T.; Hexner E. O.; Carreno B. M.; Nobles C. L.; Bushman F. D.; Parker K. R.; Qi Y.; Satpathy A. T.; Chang H. Y.; Zhao Y.; Lacey S. F.; June C. H. CRISPR-engineered T cells in patients with refractory cancer. Science 2020, 367 (6481), eaba7365 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lièvre A.; Bachet J.-B.; Le Corre D.; Boige V.; Landi B.; Emile J.-F.; Côté J.-F.; Tomasic G.; Penna C.; Ducreux M.; Rougier P.; Penault-Llorca F.; Laurent-Puig P. KRAS mutation status is predictive of response to Cetuximab therapy in colorectal cancer. Cancer Res. 2006, 66 (8), 3992–3995. 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- Shiomi N.; Mori M.; Tsuji H.; Imai T.; Inoue H.; Tateishi S.; Yamaizumi M.; Shiomi T. Human RAD18 is involved in S phase-specific single-strand break repair without PCNA monoubiquitination. Nucleic Acids Res. 2006, 35 (2), e9 10.1093/nar/gkl979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi J.; Inoue M.; Fukuda M.; Uchida Y.; Yasukawa T.; Conaway R. C.; Conaway J. W.; Aso T.; Kitajima S. Transcriptional properties of mammalian elongin A and its role in stress response. J. Biol. Chem. 2013, 288 (34), 24302–24315. 10.1074/jbc.M113.496703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M.; Grewal S. I. HULC, a histone H2B ubiquitinating complex, modulates heterochromatin independent of histone methylation in fission yeast. J. Biol. Chem. 2007, 282 (19), 14065–14072. 10.1074/jbc.M700292200. [DOI] [PubMed] [Google Scholar]
- Nguyen M.; Cencic R.; Ertel F.; Bernier C.; Pelletier J.; Roulston A.; Silvius J. R.; Shore G. C. Obatoclax is a direct and potent antagonist of membrane-restricted Mcl-1 and is synthetic lethal with treatment that induces Bim. BMC Cancer 2015, 15, 568. 10.1186/s12885-015-1582-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freije J. M.; Fraile J. M.; Lopez-Otin C. Protease addiction and synthetic lethality in cancer. Front. Oncol. 2011, 1, 25. 10.3389/fonc.2011.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buono R. A.; Paez-Valencia J.; Miller N. D.; Goodman K.; Spitzer C.; Spalding E. P.; Otegui M. S. Role of SKD1 regulators LIP5 and IST1-LIKE1 in endosomal sorting and plant development. Plant Physiol. 2016, 171 (1), 251–264. 10.1104/pp.16.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin J.; King R. S. The long and the short of Wnt signaling in C. elegans. Curr. Opin. Genet. Dev. 2008, 18 (4), 362–367. 10.1016/j.gde.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinovyev A.; Kuperstein I.; Barillot E.; Heyer W. D. Synthetic lethality between gene defects affecting a single non-essential molecular pathway with reversible steps. PLoS Comput. Biol. 2013, 9 (4), e1003016 10.1371/journal.pcbi.1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson L. R.; Chen H.; Collins A. R.; Connell M.; Damia G.; Dasgupta S.; Malhotra M.; Meeker A. K.; Amedei A.; Amin A.; Ashraf S. S.; Aquilano K.; Azmi A. S.; Bhakta D.; Bilsland A.; Boosani C. S.; Chen S.; Ciriolo M. R.; Fujii H.; Guha G.; Halicka D.; Helferich W. G.; Keith W. N.; Mohammed S. I.; Niccolai E.; Yang X.; Honoki K.; Parslow V. R.; Prakash S.; Rezazadeh S.; Shackelford R. E.; Sidransky D.; Tran P. T.; Yang E. S.; Maxwell C. A. Genomic instability in human cancer: Molecular insights and opportunities for therapeutic attack and prevention through diet and nutrition. Semin. Cancer Biol. 2015, 35 (Suppl), S5–S24. 10.1016/j.semcancer.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott S.; Coustham V.; Simonet T.; Bedet C.; Palladino F. Unique and redundant functions of C. elegans HP1 proteins in post-embryonic development. Dev. Biol. 2006, 298 (1), 176–187. 10.1016/j.ydbio.2006.06.039. [DOI] [PubMed] [Google Scholar]
- Patki M.; Gadgeel S.; Huang Y.; McFall T.; Shields A. F.; Matherly L. H.; Bepler G.; Ratnam M. Glucocorticoid receptor status is a principal determinant of variability in the sensitivity of non-small-cell lung cancer cells to pemetrexed. J. Thorac. Oncol. 2014, 9 (4), 519–526. 10.1097/JTO.0000000000000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J. R.; Brill S. J. Activation of the Slx5-Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates. J. Biol. Chem. 2008, 283 (29), 19912–19921. 10.1074/jbc.M802690200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petti C.; Molla A.; Vegetti C.; Ferrone S.; Anichini A.; Sensi M. Coexpression of NRASQ61R and BRAFV600E in human melanoma cells activates senescence and increases susceptibility to cell-mediated cytotoxicity. Cancer Res. 2006, 66 (13), 6503–6511. 10.1158/0008-5472.CAN-05-4671. [DOI] [PubMed] [Google Scholar]
- Byrum A. K.; Vindigni A.; Mosammaparast N. Defining and modulating ‘BRCAness’. Trends Cell Biol. 2019, 29 (9), 740–751. 10.1016/j.tcb.2019.06.005. [DOI] [PubMed] [Google Scholar]
- Muggia F.; Safra T. ‘BRCAness’ and its implications for platinum action in gynecologic cancer. Anticancer Res. 2014, 34 (2), 551–556. [PMC free article] [PubMed] [Google Scholar]
- Plummer R.; Jones C.; Middleton M.; Wilson R.; Evans J.; Olsen A.; Curtin N.; Boddy A.; McHugh P.; Newell D.; Harris A.; Johnson P.; Steinfeldt H.; Dewji R.; Wang D.; Robson L.; Calvert H. Phase I study of the poly(ADP-ribose) polymerase inhibitor, AG014699, in combination with temozolomide in patients with advanced solid tumors. Clin. Cancer Res. 2008, 14 (23), 7917–7923. 10.1158/1078-0432.CCR-08-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poleksic A.; Xie L. Database of adverse events associated with drugs and drug combinations. Sci. Rep. 2019, 9 (1), 20025. 10.1038/s41598-019-56525-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel A.; Marhold M.; Mayer P.; Schwarz M.; Tomasich E.; Lukas A.; Krainer M.; Perco P. Synthetic lethality guiding selection of drug combinations in ovarian cancer. PLoS One 2019, 14 (1), e0210859 10.1371/journal.pone.0210859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassmannshausen R.; Deurenberg R. H.; Kock R.; Hendrix R.; Jurke A.; Rossen J. W.; Friedrich A. W. MRSA prevalence and associated risk factors among health-care workers in non-outbreak situations in the dutch-german EUREGIO. Front. Microbiol. 2016, 7, 1273. 10.3389/fmicb.2016.01273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos M. Why do bacteria engage in homologous recombination?. Trends Microbiol. 2009, 17 (6), 226–232. 10.1016/j.tim.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Aziz R. K.; Monk J. M.; Lewis R. M.; In Loh S.; Mishra A.; Abhay Nagle A.; Satyanarayana C.; Dhakshinamoorthy S.; Luche M.; Kitchen D. B.; Andrews K. A.; Fong N. L.; Li H. J.; Palsson B. O.; Charusanti P. Systems biology-guided identification of synthetic lethal gene pairs and its potential use to discover antibiotic combinations. Sci. Rep. 2015, 5, 16025. 10.1038/srep16025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. J.; Seo E.; Cho Y. Proposal for a new therapy for drug-resistant malaria using plasmodium synthetic lethality inference. Int. J. Parasitol.: Drugs Drug Resist. 2013, 3, 119–128. 10.1016/j.ijpddr.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002, 66 (4), 630–670. 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelso A. A.; Waldvogel S. M.; Luthman A. J.; Sehorn M. G. Homologous recombination in protozoan parasites and recombinase inhibitors. Front. Microbiol. 2017, 8, 1716. 10.3389/fmicb.2017.01716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil N. J.; Bailey M. L.; Hieter P. Synthetic lethality and cancer. Nat. Rev. Genet. 2017, 18 (10), 613–623. 10.1038/nrg.2017.47. [DOI] [PubMed] [Google Scholar]
- Walcott F. L.; Patel J.; Lubet R.; Rodriguez L.; Calzone K. A. Hereditary cancer syndromes as model systems for chemopreventive agent development. Semin. Oncol. 2016, 43 (1), 134–145. 10.1053/j.seminoncol.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.