The outcome of viral infection is determined by the ability of the virus to redirect cellular systems toward progeny production countered by the ability of the cell to block these viral actions. Thus, an infected culture consists of thousands of cells, each fighting its own individual battle.
KEYWORDS: polyomavirus, BKV, single-cell transcriptomics
ABSTRACT
BK virus (BKV) is a human polyomavirus that is generally harmless but can cause devastating disease in immunosuppressed individuals. BKV infection of renal cells is a common problem for kidney transplant patients undergoing immunosuppressive therapy. In cultured primary human renal proximal tubule epithelial (RPTE) cells, BKV undergoes a productive infection. The BKV-encoded large T antigen (LT) induces cell cycle entry, resulting in the upregulation of numerous genes associated with cell proliferation. Consistently, microarray and transcriptome sequencing (RNA-seq) experiments performed on bulk infected cell populations identified several proliferation-related pathways that are upregulated by BKV. These studies revealed few genes that are downregulated. In this study, we analyzed viral and cellular transcripts in single mock- or BKV-infected cells. We found that the levels of viral mRNAs vary widely among infected cells, resulting in different levels of LT and viral capsid protein expression. Cells expressing the highest levels of viral transcripts account for approximately 20% of the culture and have a gene expression pattern that is distinct from that of cells expressing lower levels of viral mRNAs. Surprisingly, cells expressing low levels of viral mRNA do not progress with time to high expression, suggesting that the two cellular responses are determined prior to or shortly following infection. Finally, comparison of cellular gene expression patterns of cells expressing high levels of viral mRNA with those of mock-infected cells or cells expressing low levels of viral mRNA revealed previously unidentified pathways that are downregulated by BKV. Among these are pathways associated with drug metabolism and detoxification, tumor necrosis factor (TNF) signaling, energy metabolism, and translation.
IMPORTANCE The outcome of viral infection is determined by the ability of the virus to redirect cellular systems toward progeny production countered by the ability of the cell to block these viral actions. Thus, an infected culture consists of thousands of cells, each fighting its own individual battle. Bulk measurements, such as PCR or RNA-seq, measure the average of these individual responses to infection. Single-cell transcriptomics provides a window to the one-on-one battle between BKV and each cell. Our studies reveal that only a minority of infected cells are overwhelmed by the virus and produce large amounts of BKV mRNAs and proteins, while the infection appears to be restricted in the remaining cells. Correlation of viral transcript levels with cellular gene expression patterns reveals pathways manipulated by BKV that may play a role in limiting infection.
INTRODUCTION
BK virus (BKV) (human polyomavirus 1) is a member of the polyomavirus family that establishes lifelong persistent infections in most humans (1, 2). While normally harmless, BKV can cause serious disease in immunosuppressed individuals, especially transplant patients. In fact, BKV is a leading cause of nephropathy and the subsequent loss of kidney transplant. BKV has also been linked to several other pathologies, including hemorrhagic cystitis, interstitial cystitis, and cancer (3–8).
BKV causes a productive infection in cultured primary human epithelial cells derived from the renal proximal tubules (RPTE cells). In this system, BKV infection advances through an ordered series of events. Like other polyomaviruses, following attachment, the virion transits via endosomes and the endoplasmic reticulum (ER), and the viral DNA is delivered to the nucleus (9, 10). The cellular transcription apparatus then engages the early region viral promoter, resulting in the synthesis of large T antigen (LT) and small T antigen (ST). Both LT and ST manipulate key signaling pathways and thereby render the cellular environment more conducive to viral replication. For example, LT binds the retinoblastoma (Rb) tumor suppressor family of proteins, pRb, p130, and p107, and antagonizes their ability to inhibit the E2F family of transcription factors (11–14). Consequently, E2F factors increase the transcription of many genes associated with the cell cycle. Thus, the expression of E2F-regulated genes (ERGs) results in cell cycle entry. Simultaneously, LT binds the tumor suppressor p53 and blocks its ability to induce the transcription of prodeath genes (15–19). As viral protein levels rise, LT then recruits the cellular DNA replication apparatus and positions it on the viral origin of replication, which results in viral DNA replication (20–22). In addition, BKV infection activates the DNA damage response, which in turn facilitates viral DNA replication by keeping infected cells locked in S phase (23–25). Finally, LT activates transcription from the viral late promoter, resulting in the expression of the late proteins agno, VP1, VP2, and VP3 and the subsequent assembly of progeny virions (26, 27).
In RPTE cells, LT mRNA can be detected as early as 12 h postinoculation (hpi) by either transcriptome sequencing (RNA-seq) or reverse transcription-quantitative PCR (RT-qPCR), and its levels continue to rise through the course of the 5- to 6-day infectious cycle (28, 29). Western blot analysis shows that LT protein levels follow a similar pattern. Microarray and RNA-seq experiments show that elevated ERG expression and cell cycle entry occur shortly after LT expression (29, 30). Viral DNA replication and the expression of VP1 mRNA and protein occur at 36 hpi, and the first detectable progeny virions appear by 48 hpi. These bulk molecular approaches are consistent with a view in which infection proceeds through a synchronous series of ordered steps. However, single-cell methods, such as indirect immunofluorescence, suggest a more complicated story. Cells intensively staining for LT are readily detected by 24 hpi. However, the number of these cells is relatively small, and the majority of cells exhibit either very weak staining or no staining at all. As infection proceeds, the number of intensely staining cells eventually increases, approaching 100%. A similar pattern is observed with VP1 expression. These results suggest that BKV infection in RPTE cells proceeds asynchronously, with some cells expressing abundant levels of LT and VP1 by 48 hpi, while the expression of these proteins in other cells is delayed. In order to understand what factors contribute to the asynchronous cellular response to BKV, we examined the infectious cycle using single-cell transcriptomics (SCT).
RESULTS
BKV-infected cells show wide variation in the levels of viral transcripts.
In order to assess gene expression patterns at the single-cell level, we utilized inDrop, a microfluidics-based droplet barcoding platform, in which individual mock- or BKV-inoculated cells were encapsulated at various times postinoculation, followed by library preparation and sequencing (31). Infections of RPTE cells were carried out at a multiplicity of infection (MOI) of 5. Approximately 1,800 to 2,800 cells per sample were analyzed following sequencing and quality control steps. Viral and cellular gene expression was assessed at 2 and 5 days postinoculation (dpi).
Assuming that a typical mammalian cell contains 200,000 mRNA molecules (32), analysis of capture efficiency indicates that mRNAs present at <10 copies/cell are at or below the detection limit (Fig. 1A and B). While the levels of viral mRNAs varied widely among inoculated cells, the levels of cellular transcripts were fairly constant. In contrast, the levels of viral mRNAs varied enormously across the infected cell population at both 2 and 5 dpi (Fig. 1C and D). In some cells, as much as 25% of the total mRNA population was viral, while other cells had no detectable viral transcripts. In total, viral mRNA was detected in about 80% of inoculated cells at 2 dpi and in 65% at 5 dpi.
FIG 1.
Viral gene expression varied greatly among BKV-infected RPTE cells at both 2 and 5 dpi. (A) Using results from the RPTE bulk RNA-seq experiment at 2 dpi, cellular genes in the BKV sample were placed into different groups based on their transcripts per kilobase million (TPM) values. (B) The mean and median percentages of cells expressing genes in each TPM group for the 2-dpi SCT were calculated and plotted (M mean, mock mean percent expression; M median, mock median percent expression; B mean, BKV mean percent expression; B median, BKV median percent expression). Note that the results between mock and BKV are similar, and percent expression remains low (<20%) for genes in <100 TPM groups. (C) Variations in viral early region (ER) and late region (LR) gene expression across 2,199 filtered BKV-inoculated cells at 2 dpi. The numbers of BKV-inoculated cells (y axes) were plotted against the percentages of BKV ER (left) or LR (right) gene counts relative to the total UMI counts in histograms. The areas of the histograms in the red boxes were enlarged to better visualize the distribution of cells with 0 to 0.5% ER gene counts (left, indicated with a block arrow) and cells with 0 to 25% LR gene counts (right, indicated with a block arrow). (D) Variations in viral ER and LR gene expression across 2,205 filtered BKV-inoculated cells at 5 dpi. Histograms of the numbers of cells with specific percentages of BKV ER (left) or LR (right) counts relative to the total UMI counts are shown. The enlarged area of the ER histogram shows the distribution of cells with 0 to 2% ER gene counts (left, indicated with a block arrow), and the enlarged area of the LR histogram shows the number of cells with 0 to 25% LR gene counts (right, indicated with a block arrow). (E) Percentage of total BKV-inoculated cells in each group at 2 and 5 dpi. BKV-inoculated cells were divided into low-, medium-, and high-BKV groups based on the proportions of UMIs that mapped to the viral late transcript relative to those that mapped to cellular transcripts (low-BKV cells, ≤1%; medium-BKV cells, >1% and ≤10%; high-BKV cells, >10%).
For further analysis, the BKV-inoculated cells were divided into low-, medium-, and high-BKV groups based on the proportions of unique molecular identifiers (UMIs) that mapped to the viral late transcript relative to those that mapped to cellular transcripts (low-BKV cells, ≤1%; medium-BKV cells, >1% and ≤10%; high-BKV cells, >10%). At 2 dpi, 78 to 80% of the cells were in the low-expressing group, while 12 to 16% were in the medium group, and 4 to 8% were in the high group. This distribution was consistent in independent experiments using cells derived from a different donor or cells plated at high or medium density. We initially expected that inoculated cells in the low-expression group would progress to the higher-expression groups as infection proceeds. Surprisingly, this distribution did not change at 5 dpi, and the combined medium- and high-expression groups comprised only 17% of the total number of cells (Fig. 1E).
Extensive variation in the levels of viral proteins expressed by BKV-infected cells.
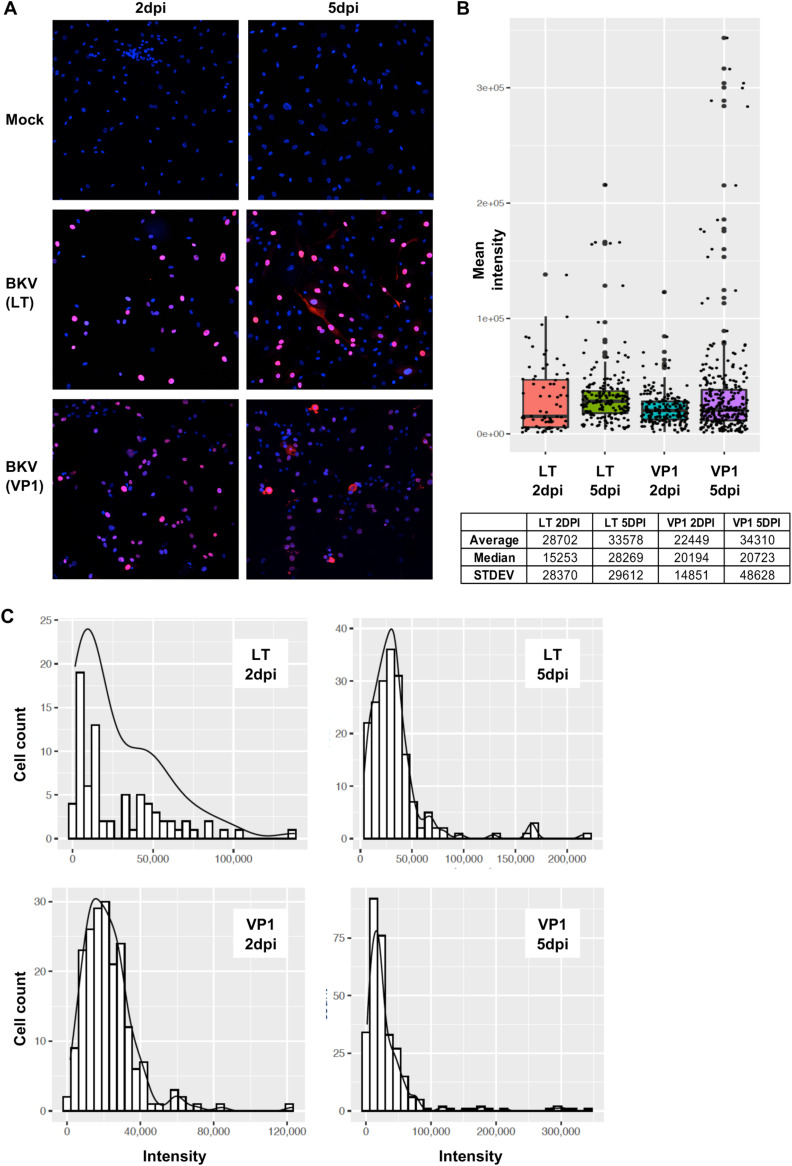
The variation in viral mRNA levels was unexpected, as immunofluorescence experiments from our laboratory and others have suggested that LT, encoded by the viral early region, and VP1, the major late protein, are expressed at high levels in nearly all infected cells. However, in most cases, these studies were optimized for viral protein detection and thus used relatively high levels of antibodies. To determine if the levels of LT and VP1 proteins vary among infected cells, we performed immunofluorescence studies using limiting amounts of LT or VP1 antibodies. Under these conditions, the intensity of both LT and VP1 immunostaining varied considerably among different cells (Fig. 2A).
FIG 2.
BKV-infected RPTE cells differed widely in their expression of viral protein products. RPTE cells were inoculated with BKV at an MOI of 1, and the presence of early (LT) and late (VP1) viral products was monitored by immunofluorescence with specific antibodies. (A) Overlapping images of DAPI (4′,6-diamidino-2-phenylindole) (blue) and viral products (magenta) reveal many different levels of intensity in cells from the same culture and viral inoculation, at 2 and 5 dpi. (B) Box plot of staining intensities of individual cells according to the specific viral product and time postinfection. The table indicates the average, median, and standard deviation values for each of the four experimental conditions shown. (C) Distribution of staining intensity according to the number of cells showing similar protein levels.
We used confocal microscopy to assess these differences. The mean intensity of fluorescence for individual cells expressing LT or VP1 was assessed at 2 and 5 dpi and represented as either histograms or density plots (Fig. 2B and C). The mean levels of both viral proteins were unevenly distributed across the entire range of different intensities, particularly at 5 dpi (Fig. 2), and the vast majority of cells expressed low or medium levels of viral products, while a lower percentage expressed much higher levels (Fig. 2). The LT expression levels varied over 90-fold at 2 dpi and about 60-fold at 5 dpi among BKV-infected cells. Similarly, the VP1 expression levels varied over 70-fold at 2 dpi and more than 200-fold at 5 dpi among BKV-infected cells.
High levels of viral transcripts correlate with cell cycle entry.
Following infection, one of the actions of LT is to drive quiescent cells into the cell cycle. We used the Seurat package to assess the cell cycle state of mock- and BKV-infected cells expressing different levels of viral mRNAs (Fig. 3). Approximately 25% of mock-infected cells were in the S or G2/M phase of the cell cycle, while the remaining cells were growth arrested at 2 or 5 dpi. In contrast, nearly all cells expressing medium or high levels of viral mRNAs had entered the cell cycle at this time. About 60% of cells in the low-viral-expression group were in the cell cycle, while 40% remained growth arrested, even at 5 dpi. Thus, cell cycle entry correlates with high levels of viral mRNA. Furthermore, a significant number of inoculated cells remained growth arrested at late stages of infection, even though they were producing at least some viral mRNAs and proteins (Fig. 3, G1 phase, low BKV).
FIG 3.
Distribution of cells into specific stages of the cell cycle according to viral load. RPTE1 cells from three different experimental SCT sets (2 dpi high confluence [HC], 2 dpi, and 5 dpi) were grouped according to their viral load (mock [none] or low, medium, or high expression of late viral transcripts). Within each group, the percentages of cells at different stages of the cell cycle (G1, S, or G2/M) are depicted.
BKV infection reprograms the cellular transcriptome.
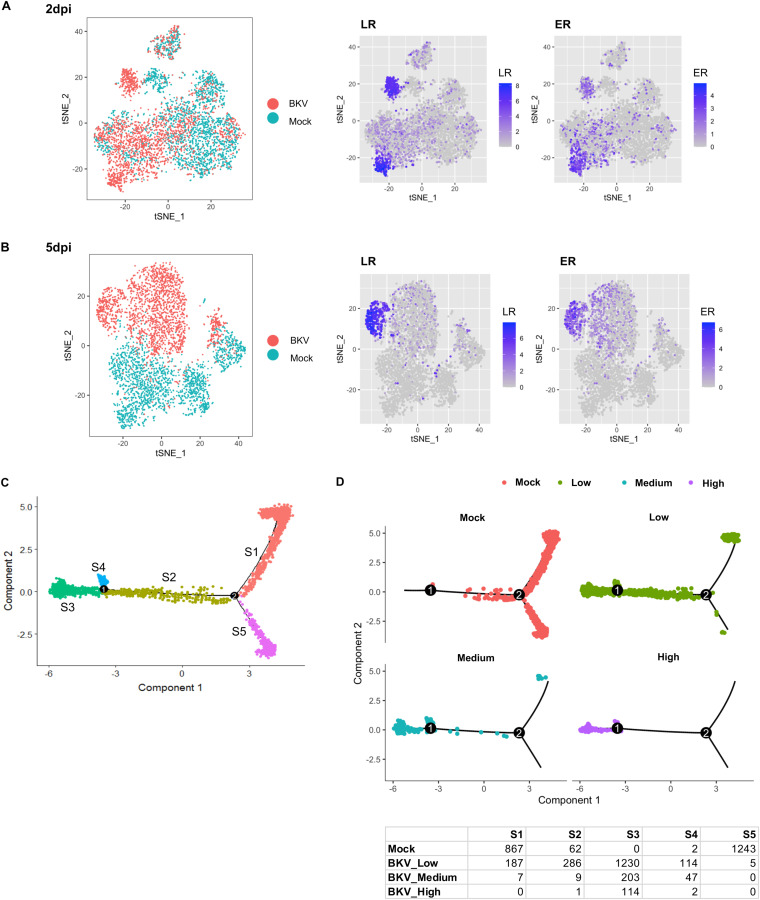
In order to determine the cellular transcriptional responses to BKV, we computationally mixed the sequencing data from mock- and BKV-inoculated cells and analyzed the transcription profiles of cells in the mixture using Seurat software. The gene expression profiles of mock- and BKV-inoculated cells were partially intermixed at 2 dpi (Fig. 4A); however, by 5 dpi, they were completely separated (Fig. 4B). Furthermore, cells expressing the highest levels of viral mRNAs clustered together and were separated from cells expressing lower levels of viral transcripts, suggesting that the cellular gene expression profiles of these two populations are distinct (Fig. 4A and B, right).
FIG 4.
BKV infection reprogramed cellular gene expression in RPTE cells. (A, left) The principal-component analysis of mock- and BKV-inoculated cells at 2 dpi was visualized using a t-distributed stochastic neighbor embedding (tSNE) plot. Each dot represents a cell (BKV-inoculated cells and mock-inoculated). The tSNE plot for mock- and BKV-inoculated cells shows a mixture of cells from the two populations. (Middle and right) Expression levels of the viral early region (ER) (middle) and late region (LR) (right) are also depicted using tSNE plots. Expression levels of ER and LR transcripts in BKV-inoculated cells are colored based on LogFC values. BKV cells with low levels of LR transcripts largely overlapped mock cells. (B) tSNE plots of mock- and BKV-inoculated cells at 5 dpi were also generated and show that even the low-BKV-expressing cells were completely separated from mock cells. (C) Mock- and BKV-inoculated cells at 5 dpi were ordered into a total of 5 distinct states based on their transcriptional profiles (S1 to S5) using the Monocle pseudotime function. (D) The distributions of mock cells and BKV cells in the low, medium, and high groups were visualized separately by their corresponding pseudotime states (top). The cell numbers from the mock and BKV groups in each state are listed in the summary table.
To better understand the relationships between these transcriptional profiles, we performed pseudotime analysis on mock and infected cells at 5 dpi. This analysis assigned the cells to five states (state 1 [S1] to S5) and ordered these states along the pseudotime scale (Fig. 4C). Mock cells were largely limited to S1 and S5, states which also contained a small number of infected cells. However, the majority of the infected cells occupied S2, S3, and S4. Nearly all of the cells expressing the highest levels of viral mRNAs were localized to S3, at the end of pseudotime (Fig. 4D). Cells expressing lower levels of viral transcripts also occupied S3, but a significant number were also present in S2 and S4, indicating that they had only partially transited through pseudotime.
Patterns of cellular gene expression covary with viral transcript levels.
To identify cellular processes regulated by BKV infection, we analyzed the BKV-inoculated cells and identified genes whose mRNA levels covaried with the amount of viral mRNA using the Monocle software package (33–35). Using 100 randomly selected cells from each group of BKV-inoculated cells, we defined high-confidence BKV-regulated genes (q value of <0.01) as those whose mRNA levels covaried with viral transcript levels in four independent SCT experiments (with varying timing and cell confluence). Using these stringent criteria, we found 80 genes whose mRNA levels covaried with VP1 transcript levels (Fig. 5).
FIG 5.
Cellular genes whose levels covaried with BKV late gene expression levels had functions in diverse biological processes. A total of 80 common covarying genes were identified across 4 independent experiments. The changes in RNA levels for these genes are summarized in a heat map generated using LogFC values of these covarying genes as group markers (low-, medium [Med]-, or high-BKV group versus mock) and/or markers by cell cycle (CC) phase. The markers by cell cycle phase were identified by comparing cells from each of the 4 groups of BKV-inoculated cells against mock cells in matching cell cycle phases. Due to the high similarity between the Low* group cells and cells with no detectable viral UMIs, only the Low* group results are presented for markers by phase. Each row corresponds to a gene. The official gene symbols and corresponding cellular processes are listed on the right. Specific BKV groups with or without cell cycle phase specifications and the time points of infection are indicated on the top. The black box highlights the results of bulk RNA-seq (LogFC of BKV/mock) at 2 dpi.
To determine the directionality (up- or downregulation) and the scale of the change for each of the above-mentioned covarying genes, we performed additional and parallel analyses for differentially expressed genes using the Seurat software package. BKV group markers were identified as differentially expressed genes between cells in each BKV group in comparison to mock-inoculated cells (low BKV/mock, medium BKV/mock, and high BKV/mock). The change of each marker gene was specified as average log fold change (LogFC) values, which can be positive (upregulation) or negative (downregulation) values. The processes and pathways associated with these genes were examined further.
BKV induces cell cycle entry, a process that in itself causes profound changes in gene expression profiles. To determine if BKV-induced gene expression changes were the downstream consequence of cell cycle entry, we first scored mock and BKV cells according to their cell cycle phase using Seurat and then compared cells in BKV expression groups with mock cells in the same phase of the cell cycle in order to identify cell cycle phase-matched group markers. Changes in gene expression levels of the common covarying genes were summarized in a heat map using LogFC values associated with general BKV group markers and the cell cycle phase-matched group markers (Fig. 5). As expected, genes related to cell proliferation, including multiple ERGs, were upregulated by BKV infection.
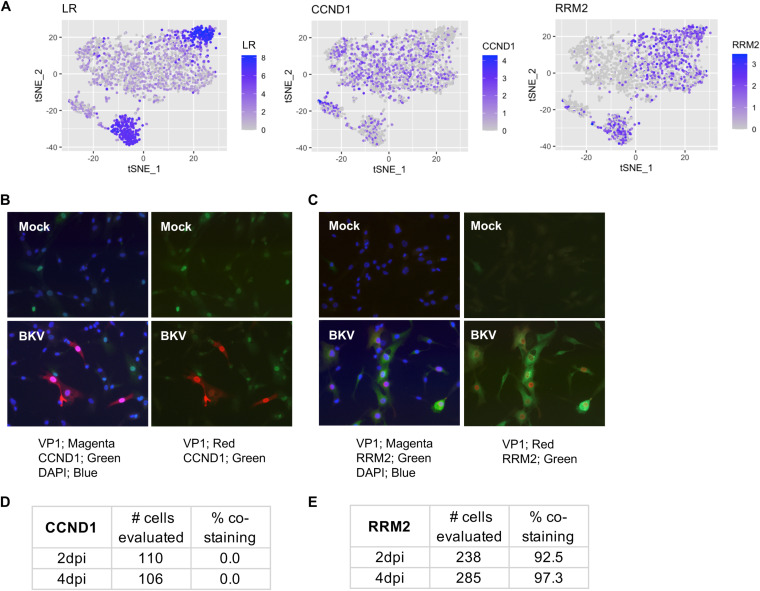
We confirmed the effects of viral expression on two BKV group markers, one upregulated (RRM2) and one downregulated (CCND1), by immunofluorescence analysis of their protein expression in correlation with VP1 expression (Fig. 6). In agreement with our SCT analysis, the expression of CCND1 was reduced in cells showing high levels of viral products, while RRM2 expression directly correlated with the expression of viral products.
FIG 6.
Confirmation of gene expression trends observed by SCT through analysis of specific protein markers. (A) Simplified tSNE plots indicating the expression of late viral products or the indicated markers in RPTE cells 2 dpi after inoculation with BKV. The expression of CCND1 is reduced in cells showing high levels of viral products, while RRM2 expression directly correlates with the expression of viral products. (B) Immunofluorescence analysis of CCND1 protein expression in comparison to VP1 protein in cells inoculated with BKV at 2 dpi. Combined overlapping images of DAPI, CCND1, and VP1 are shown on the left, while overlaps between CCND1 and VP1 are shown on the right. (C) Immunofluorescence analysis of RRM2 protein expression in comparison to VP1 protein in cells inoculated with BKV at 2 dpi. Combined overlapping images of DAPI, RRM2, and VP1 are shown on the left, while overlaps between RRM2 and VP1 are shown on the right. (D) Quantification of CCND1 and VP1 costaining. (E) Quantification of RRM2 and VP1 costaining. Note that CCND1 was a statistically significant covarying gene in all 4 independent SCT experiments, although its adjusted P value was 0.025 in 1 experiment.
We next broadened the analysis to the full set of ERGs (Fig. 7). Strikingly, while most of these genes were also upregulated in the normal cell cycle, the degree of upregulation was much higher in BKV-infected cells, and a more robust upregulation of ERGs was associated with cells expressing high levels of viral products at later time points. Consistent with bulk RNA-seq experiments, there was no evidence for the activation of p53-dependent transcription.
FIG 7.
Broad and strong upregulation of ERGs by BKV infection. (A) More extensive and more robust upregulations of ERGs occurred at the later time points. The overlapped box-and-dot plot shows all ERGs with a LogFC of ≥0.5 in each BKV group (group 1, undetected or 0%, cells from which no viral UMIs were detected; group 2, Low*, cells with viral UMIs that comprised >0% and ≤1% of total cellular UMIs; group 3, Med, cells with viral UMIs comprising >1% and ≤10% of total cellular UMIs; and group 4, High, cells with viral UMIs comprising >10% of total cellular UMIs). (B) Markers by cell cycle phase show greater upregulation of many ERGs in BKV-infected cells than those in the normal cell cycle (columns M_G2M and M_S) at 5 dpi. Due to the high similarity between the Low* group cells and cells with no detectable viral UMIs, only the Low* group results are presented for markers by phase. Note that the markers by phase for the BKV group cells were determined by comparing BKV cells in each group with mock cells in the matching cell cycle. Therefore, the positive LogFC values of BKV markers visualized by the heat map show further upregulations on top of the increased levels of the corresponding genes during the normal cell cycle.
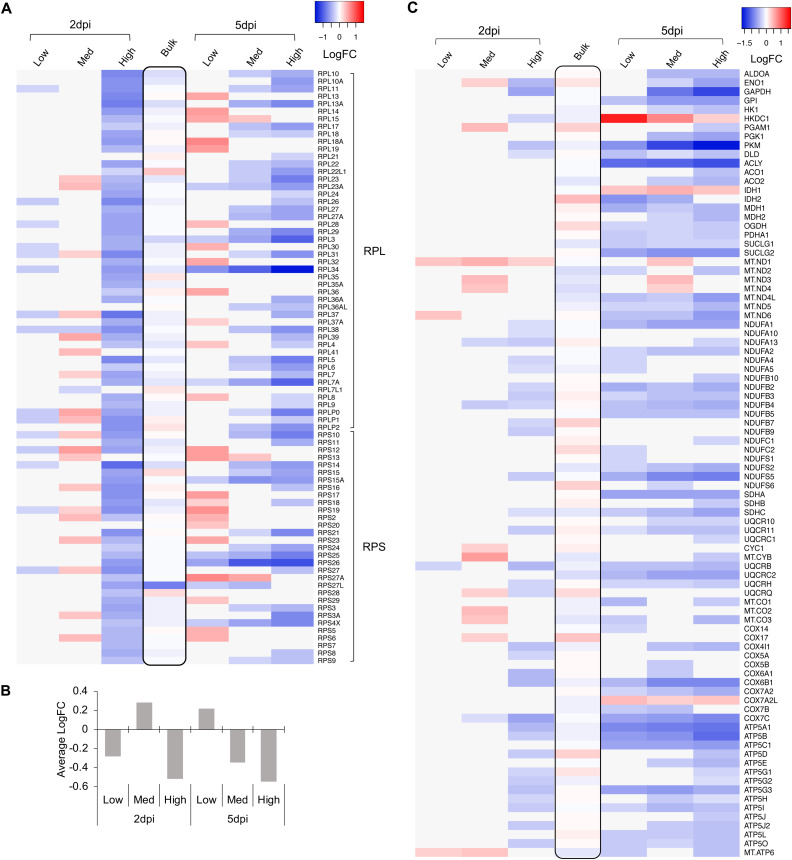
Several of the covarying genes identified in our stringent analysis encoded ribosomal proteins or components of energy metabolism pathways (Fig. 5). Therefore, we expanded the analysis to examine the expression of all components of these pathways. Many of the transcripts from the 85 cytoplasmic ribosomal protein genes were upregulated in the medium-viral-expression group at 2 dpi (Fig. 8A and B). This is consistent with previous reports that simian virus 40 (SV40), a related polyomavirus, upregulates the expression of both rRNAs and ribosomal proteins (36–38). In contrast, nearly all of these genes were coordinately downregulated in cells expressing the highest levels of viral transcripts. Analysis of BKV versus mock cells in matching cell cycle phases confirmed coordinated changes in ribosomal protein genes during BKV infection. Similarly, many genes associated with glycolysis, the tricarboxylic acid (TCA) cycle, and oxidative phosphorylation were downregulated in the high-viral-expression groups at 2 dpi and across all BKV-inoculated cells at 5 dpi (Fig. 8C).
FIG 8.
Dynamic changes of cytoplasmic ribosomal protein (RP) genes were associated with different levels of BKV late gene expression. Genes involved in cellular energy metabolism were extensively regulated during BKV infection. (A) A heat map was generated using LogFC values of group markers at 2 and 5 dpi without scaling. The time points and BKV groups are indicated on the top. RP genes (RPL [ribosomal large subunit] and RPS [ribosomal small subunit]) are labeled on the left. The black box in the middle lane highlights the results of bulk RNA-seq (LogFC of BKV/mock) at 2 dpi. (B) The general trend of changes in cytoplasmic RP genes represented by their average LogFCs is summarized in a chart. (C) Heat maps were generated using LogFC values of group markers involved in energy metabolism at 2 and 5 dpi. The time points and BKV groups are indicated on the top. Gene symbols are listed on the right side of the heat maps.
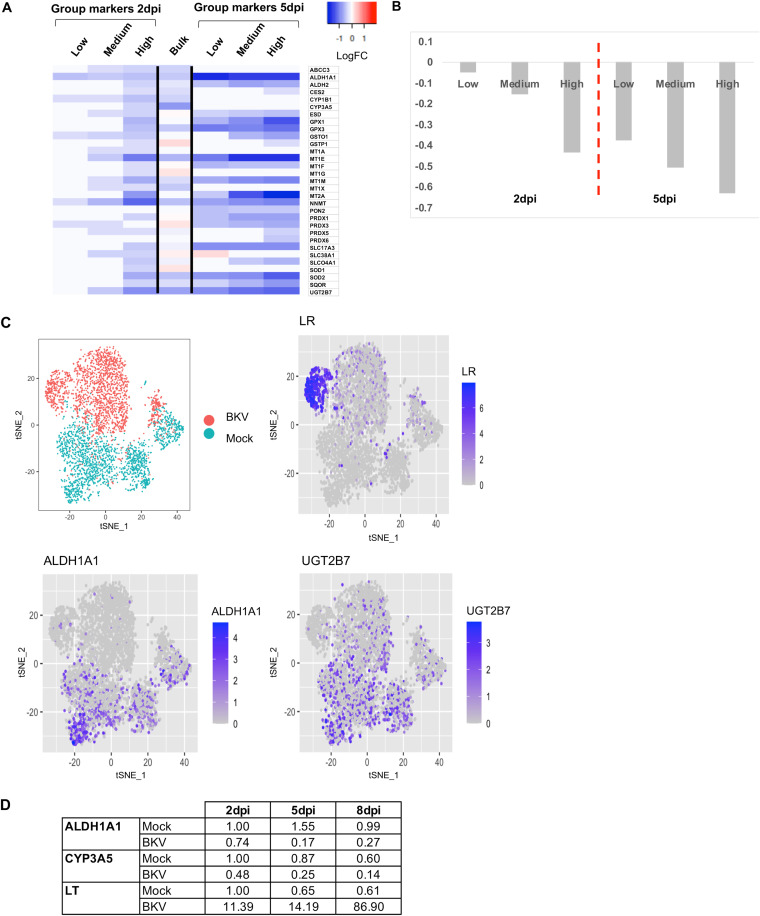
We noticed that several BKV-regulated genes were associated with the modification, secretion, and/or elimination of damaging agents such as xenobiotics and drugs, reactive oxygen species, metals, urea, or alcohol, which we collectively named detox for simplicity. In each of these cases, the mRNA levels of the detox genes showed an inverse correlation with viral transcript levels, especially at 5 dpi (Fig. 9A to C). To confirm this trend, we evaluated the relative levels of two selected detox genes, ALDH1 and CYP3A5, using real-time PCR at 2, 5, and 8 dpi. Consistent with our SCT analysis, we observed that the expression of the tested detox genes decreased considerably upon BKV inoculation, in reverse correlation with the expression of early viral products (Fig. 9D). The downregulation trend was independent of the state of the cell cycle.
FIG 9.
BKV infection resulted in the downregulation of detoxification genes. (A) Expression of detoxification genes in BKV-inoculated RPTE cells according to viral expression levels. A heat map was generated from LogFC values without scaling (red is upregulation, and blue is downregulation). Regulation of detoxification-related transcripts at the bulk RNA level at 2 dpi is included for comparison. (B) Average changes in expression levels (LogFC) of detoxification genes in different cells according to viral load. (C) Simplified tSNE plots indicating cell identity (mock or BKV) and the corresponding expression of late viral products or selected detoxification genes. (D) Relative expression of selected detoxification genes in control and BKV-inoculated cells after 2, 5, and 8 dpi. Values were determined by real-time PCR and normalized against the levels of an endogenous control (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]).
Single-cell analysis reveals potential antagonists of BKV infection.
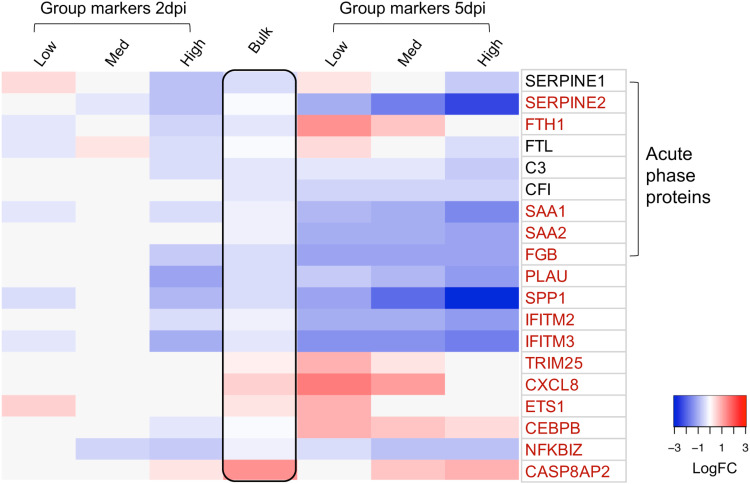
Next, we examined components of innate immune and cell death pathways that are known to antagonize viral infections. BKV infection did not result in significant changes in the expression of interferon (IFN) genes, nor was there a global upregulation of interferon-stimulated genes (ISGs). Thus, consistent with previous studies, BKV does not activate an interferon response in RPTE cells (29, 30, 39). However, we identified several genes involved in immunity whose expression correlated with the levels of viral transcripts (Fig. 10). A subset of these genes was upregulated in cells with low levels of viral transcripts but downregulated in cells with high levels of viral mRNA. These included two transcription factors, ETS1 and CEBPB, involved in the expression of cytokines. Interestingly, the expression of the cytokine CXCL8 followed the same pattern. Other immune genes were downregulated in all BKV-infected cells, with the degree of downregulation being more severe in the high-BKV-expression groups. These include IFITM2 and IFITM3 as well as a number of components of the acute-phase response. All of these genes were regulated by BKV independently of the cell cycle state.
FIG 10.
BKV infection orchestrated the regulation of multiple immune genes. The heat map depicts variations in immune genes, generated using LogFC values of group markers at 2 and 5 dpi. The time points and BKV groups are indicated on the top. Gene symbols corresponding to individual rows are listed on the right. All genes encoding acute-phase proteins are enclosed in the left bracket. Genes highlighted in red are the top regulated genes (genes with LogFC values greater than or equal to 0.75 or less than or equal to −0.75 in at least one group). The black box in the middle lane highlights the results of bulk RNA-seq (LogFC of BKV/mock) at 2 dpi.
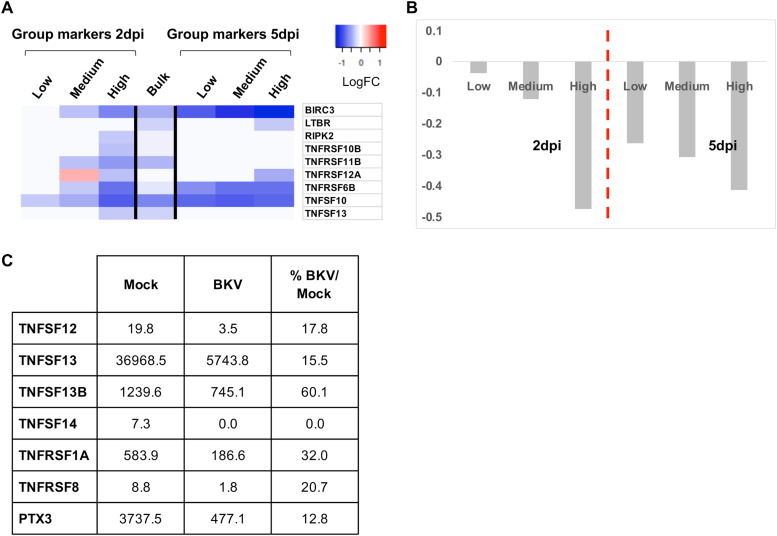
In addition, we observed a significant decrease in transcripts of several members of the tumor necrosis factor (TNF) pathway, including ligands and receptors, upon BKV infection (Fig. 11A). The reduction in the levels of TNF-associated mRNAs was significant at 2 dpi and was more severe in cells expressing higher levels of viral genes (Fig. 11A and B). At 5 dpi, the downregulation was further expanded to many or most BKV-inoculated cells (Fig. 11A and B) and was independent of the cell cycle stage. To confirm these observations, we examined the expression of several TNF-pathway-associated genes using a Luminex multiplex immunoassay. Again, we observed a significant protein reduction of members of the pathway, both ligands and receptors, in the supernatants of RPTE cells inoculated with BKV in comparison to the levels observed in mock supernatants (Fig. 11C). We conclude that BKV infection results in the coordinated downregulation of multiple components of the TNF signaling pathway and that this regulation correlates with the levels of viral mRNA present in the cell.
FIG 11.
Downregulation of members of the TNF pathway in BKV-inoculated RPTE cells. (A) Regulation of TNF-related transcript levels according to viral expression in two SCT experimental sets. The heat map was generated from LogFC values. Red indicates upregulation, and blue reflects downregulation. Regulation of TNF-related transcripts at the bulk RNA level at 2 dpi is included for comparison. (B) Average expression levels (LogFC) of TNF-related genes in different cells according to viral load. (C) Expression of different TNF-related factors in supernatants from mock- or BKV-inoculated cells at 2 dpi, as determined by a multiplex enzyme-linked immunosorbent assay (ELISA). For each factor, values are expressed as picograms per milliliter of culture supernatant.
DISCUSSION
We used inDrop, a microfluidics-based single-cell transcriptomics platform, to examine the response of individual permissive cells to BKV infection. Primary human RPTE cultures infected with BKV (MOI = 5) were analyzed at 2 and 5 dpi. These experiments revealed that the level of viral mRNAs present in infected cells varied over 1,000-fold. While in most cells, viral transcripts accounted for <1% of the total mRNAs present, other cells expressed high levels of viral genes, with viral transcripts accounting for 10 to 25% of the total mRNA. This distribution did not change between 2 and 5 dpi, indicating that cells expressing low levels of viral mRNA do not progress to high levels of expression. Rather, it appears that the high- and low-expressing populations of cells are present by 2 dpi and remain in those states until cell death. To determine if the observed differences in viral mRNA levels were reflected in the amount of viral proteins present, we performed quantitative immunofluorescence using limiting amounts of LT or VP1 antibodies. While most cells expressed viral proteins, different cells in the infected population varied over 90-fold at 2 dpi and 60-fold at 5 dpi in LT expression and over 70-fold at 2 dpi and 200-fold at 5 dpi in VP1 expression. We conclude that BKV-infected RPTE cells vary widely in the amounts of viral proteins expressed and that this variation is likely due to different levels of viral mRNAs.
Examination of cellular gene expression patterns revealed heterogeneous responses to infection that correlated with viral transcript levels. As evidenced by t-distributed stochastic neighbor embedding (tSNE) and pseudotime analyses, all BKV-inoculated cells had gene expression patterns distinct from those of mock cells by 5 dpi. In addition, infected cells expressing high levels of viral transcripts displayed gene expression profiles distinct from those of cells containing low levels of viral mRNA. Some of these differences could be attributed to the cell cycle state, and to control for this, we compared gene expression levels between mock and infected cells that were in the same stage of the cell cycle. We found that cell proliferation-associated genes, including those regulated by E2F, were upregulated in cells expressing moderate or high levels of viral mRNAs, even compared to mock cells, in the S or G2/M stage of the cell cycle. In contrast, the transcript levels of these genes were not drastically altered in cells expressing low levels of viral mRNA. This is consistent with the observation that many BKV-infected cells expressing low levels of viral mRNA remained in G1 arrest, even at 5 dpi.
Single-cell analysis also identified key pathways that were downregulated in cells expressing moderate or high levels of viral mRNAs relative to mock-infected cells or cells with low levels of viral gene expression. Numerous genes associated with energy metabolism and translation, including key components of oxidative phosphorylation, and almost all genes encoding cytoplasmic ribosomal proteins were downregulated by infection. It is not clear if this downregulation is due to the specific action of BKV or is the indirect consequence of cells overwhelmed with large amounts of viral mRNA. Multiple genes involved in drug metabolism/detoxification were also downregulated by BKV infection. In particular, members of all three phases of metabolic detoxification (functionalization, conjugation, and elimination) were coordinately downregulated as viral transcript levels increased. This is consistent with previous studies demonstrating that the overexpression of SV40 LT downregulates the xenobiotic detoxification pathway in cells (40, 41). It also raises the intriguing possibility that BKV-infected cells respond differently to drugs and/or toxins, including antivirals, than uninfected cells. In addition, multiple components of the TNF cell death signaling pathway, including both ligands and receptors, were downregulated in cells expressing moderate or high levels of viral mRNAs. Consistently, we observed that the levels of these proteins were reduced in the media of BKV-infected cells relative to mock. The mRNA levels of the TNF pathway genes were mostly unchanged in BKV-infected cells expressing low levels of viral transcripts. In addition, specific immune-related genes were downregulated in cells containing high levels of viral mRNA, including several key components involved in the acute-phase response.
Most infected cells expressed detectable levels of viral transcripts and proteins, and nearly all of these cells died. However, the levels of viral gene expression, and presumably the amount of progeny virions produced, varied widely within the infected cell population. Similar large variations in viral gene expression have been observed across multiple SCT studies of cells infected by several viruses, including herpes simplex virus 1, influenza virus, cytomegalovirus, and dengue and Zika viruses (42–45). These results suggest that heterogeneous cellular responses to viral infection may likely reflect the dynamic battles between individual infected cells and the virus. We considered two hypotheses to explain the heterogeneous response of primary human RPTE cells to BKV infection. In the first model, the wide differences in viral transcript levels and cellular responses to BKV are due to an asynchronous infection. Thus, cells in the infected population are at different stages of the infectious cycle. In this model, cells expressing low levels of viral transcripts should eventually move to high expressors as infection proceeds. However, this model is not consistent with the observation that the distribution of cells between low and high expressors is largely unchanged, even at late stages of infection. Rather, the data are consistent with an alternative model in which commitment to high or low expression occurs prior to or early after infection. In this scenario, the high- and low-expressing cell populations remain relatively stable until cell death, and the majority of progeny virions are produced from a minority of the infected cells. In this view, BKV inoculation of a permissive culture results in thousands of one-on-one battles, with some cells limiting infection and others succumbing to the virus. If this is the case, identifying and understanding the factors that limit viral infection are paramount.
MATERIALS AND METHODS
BKV infection.
Primary human RPTE cells were purchased from Lonza and maintained according to the manufacturer’s instructions using growth media provided by Lonza. For infection, cells were plated in 6-well plates at a density of 150,000 cells/well the day before inoculation. For infection of cells at high confluence, BKV inoculation was done at 4 days postplating. All BKV infections for SCT were performed according to the synchronized entry protocol at an MOI of 5 (9). Briefly, cells were precooled at 4°C for 15 min before the growth medium was removed. The inoculum containing virus in complete growth medium was then added. The inoculate and the cells were incubated at 4°C for 1 h with occasional gentle rocking. After incubation, the inoculate was removed, and fresh growth media were added to the wells. The plates were then placed into cell culture incubators at 37°C until the cells were ready to be harvested for microfluidics sorting and subsequent scRNA-seq experiments. Mock infections were carried out along with inoculation except that the mock-inoculated cells were not exposed to any virus.
For immunofluorescence, RPTE cells were plated on poly-l-lysine-coated coverslips in 12-well plates at a density of 70,000 cells/well the day before inoculation. Infections were carried out according to the same protocol as the one described above. The coverslips were fixed with 4% formaldehyde and stained using specific antibodies (BKV LT, Pab416 [46]; BKV VP1, MAB3204 [Abnova]; cyclin D, EBP5S [Cell Signaling]; RRM2, E7Y9J [Cell Signaling]), followed by staining with the corresponding Alexa Fluor 488-, Alexa Fluor 568-, or fluorescein isothiocyanate (FITC)-labeled secondary antibodies. Detailed protocols for immunostaining were previously described (29).
Quantification of LT and VP1 staining.
After antibody staining, microscopic images were taken using a Leica TCS SP5 confocal/multiphoton or Leica TCS/SP8 Clarity imaging system. The fluorescence intensity corresponding to each specific antibody was measured for at least 100 individual cells.
Quantification of inflammation markers in cell culture supernatants.
The levels of specific biomarkers of inflammation in the media from cells either noninoculated or inoculated with BKV were monitored with Bio-Plex Pro human inflammation panel 1 (Bio-Rad), which detects proteins of the TNF and IFN families, regulatory T cell (Treg) cytokines, and matrix metalloproteinases.
Library preparation and sequencing.
Mock- and BKV-inoculated cells were collected after trypsinization, and individual cells and hydrogel beads were encapsulated into droplets. Barcoding of single cells and preparation of sequencing libraries were performed according to previously reported inDrop protocols (31, 47). In short, single-cell transcriptomes were first barcoded by reverse transcription in droplets. Next, emulsions were split into fractions containing <4,000 cells for inDrop library preparation using the inDrop single-cell RNA-seq kit from 1Cell-Bio. This library preparation process includes the following six steps: (i) enzymatic cleanup of excess primers, primer-dimers, and hairpins; (ii) second-strand synthesis; (iii) amplification by in vitro transcription; (iv) fragmentation of amplified RNA; (v) reverse transcription; and (vi) PCR amplification of libraries. At the last PCR step, each library was indexed with a unique primer. Libraries were then pooled and sequenced on a NextSeq Illumina platform with a configuration of 36 bp on read 1, 6 bp on the index read, and 50 bp on read 2.
Processing and analysis of scRNA-seq data.
Indrops software (https://github.com/indrops/indrops) was used to process the raw reads. The ENSEMBL hg38 release 90 version of the human genome, together with the BKV genome (GenBank accession number NC_001538.1), was used for alignment. The –min-reads argument for the “quantify” step of Indrops was set independently for each library to obtain a number of cells similar to the number of cells sequenced.
All analyses in R were conducted with R version 3.5 or 3.6. The resulting Indrops UMI count tables were processed with Seurat (version 2.3.4) (48) in the following manner: a minimum expression value of 2 counts was used, and we removed poor-quality cells as those that had >2,500 unique genes expressed or that had too few or too many UMIs per cell (≥2 standard deviations). Additionally, we applied a minimum number of unique genes expressed filter, which ranged from 50 to 200 depending on the sample, in order to obtain at least 1,800 cells. This resulted in 1,800 to 2,800 mock cells and 1,600 to 2,300 BKV cells (depending on the sample). The data were normalized by a scale factor of 10,000, and the result was log transformed. Next, noise from UMI variation was regressed out of the data, and principal-component analysis (PCA) was performed using the set of highly variable genes as determined by Seurat. Next, t-distributed stochastic neighbor embedding (tSNE) was calculated using the number of PCA dimensions as determined by manual inspection of Seurat’s elbow plot. Finally, cell populations were visualized with the TSNEPlot function in Seurat.
(i) Pseudotime analysis.
The mock and BKV Seurat objects were merged using the Seurat MergeSeurat function and imported into Monocle 2 using the importCDS function (33–35). The union of the Seurat highly variable genes from mock and BKV was used in the setOrderingFilter function. Next, a cell trajectory was determined by using the orderCells function in Monocle.
(ii) Alignment procedure.
We used the Seurat alignment procedure to combine mock and BKV cells together. Rare nonoverlapping cells were removed from further analysis as described previously (48). All analyses described below used the cells remaining after alignment.
(iii) BKV group analysis.
Grouping of cells by BKV late region expression is used to identify genes regulated by viral infection. BKV-inoculated cells are placed into 3 groups based on the proportion of UMIs that mapped to viral late transcripts relative to the proportion that mapped to cellular mRNA: low BKV, cells containing ≤1% of BKV late transcripts; medium BKV, cells with >1% and ≤10% of BKV late transcripts; and high BKV, cells with >10% of BKV late transcripts. Group markers were then determined by comparing cells in each BKV group with the mock cells using the FindMarkers function in Seurat (version 2.3.4).
(iv) Cell cycle phase analysis.
For cell cycle phase analysis, Seurat version 3.1.2 was used. To determine cell cycle phases for individual cells, quantitative scores are calculated based on their expression of S and G2/M marker genes. The scores are assigned using the Seurat CellCycleScoring function, and individual cells are then classified as G1-, S-, or G2/M-phase cells. Markers per BKV group by cell cycle phase were determined by comparing each BKV group (group 1, Undetected or 0%, cells from which no viral UMIs were detected; group 2, Low*, cells with viral UMIs comprising >0% and ≤1% of total cellular UMIs; group 3, Med, cells with viral UMIs comprising >1% and ≤10% of total cellular UMIs; and group 4, High, cells with viral UMIs comprising >10% of total cellular UMIs) with mock in matching cell cycle phases.
(v) Covarying gene analysis.
To determine covarying genes, i.e., cellular genes that change in expression due to BKV late region expression, we randomly selected 100 cells from four BKV groups (0%, >0% and ≤1%, >1% and ≤0%, and >10%) and used differentialGeneTest in Monocle 2. We set the fullModelFormulaStr parameter to the fraction of late region expression in each cell. Next, the union of significant covarying genes (q value of ≤0.01) across the four independent experiments with varying time points and confluence was determined, and this resulted in a total of 80 genes.
ACKNOWLEDGMENTS
This research was supported in part by the University of Pittsburgh Center for Research Computing through the resources provided. This work was supported by the NIH (RO1 AI153156). Sequencing and RNA analysis services were provided by the Genomics Research Core at the University of Pittsburgh.
We thank the Genomics Analysis Core at the University of Pittsburgh for providing bioinformatics support.
REFERENCES
- 1.Egli A, Infanti L, Dumoulin A, Buser A, Samaridis J, Stebler C, Gosert R, Hirsch HH. 2009. Prevalence of polyomavirus BK and JC infection and replication in 400 healthy blood donors. J Infect Dis 199:837–846. doi: 10.1086/597126. [DOI] [PubMed] [Google Scholar]
- 2.Kean JM, Rao S, Wang M, Garcea RL. 2009. Seroepidemiology of human polyomaviruses. PLoS Pathog 5:e1000363. doi: 10.1371/journal.ppat.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonsson A, Pawlita M, Feltkamp MCW, Bouwes Bavinck JN, Euvrard S, Harwood CA, Naldi L, Nindl I, Proby CM, Neale RE, Waterboer T. 2013. Longitudinal study of seroprevalence and serostability of the human polyomaviruses JCV and BKV in organ transplant recipients. J Med Virol 85:327–335. doi: 10.1002/jmv.23472. [DOI] [PubMed] [Google Scholar]
- 4.Nickeleit V, Mihatsch MJ. 2006. Polyomavirus nephropathy in native kidneys and renal allografts: an update on an escalating threat. Transpl Int 19:960–973. doi: 10.1111/j.1432-2277.2006.00360.x. [DOI] [PubMed] [Google Scholar]
- 5.Koskenvuo M, Dumoulin A, Lautenschlager I, Auvinen E, Mannonen L, Anttila VJ, Jahnukainen K, Saarinen-Pihkala UM, Hirsch HH. 2013. BK polyomavirus-associated hemorrhagic cystitis among pediatric allogeneic bone marrow transplant recipients: treatment response and evidence for nosocomial transmission. J Clin Virol 56:77–81. doi: 10.1016/j.jcv.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Kenan DJ, Mieczkowski PA, Latulippe E, Cote I, Singh HK, Nickeleit V. 2017. BK polyomavirus genomic integration and large T antigen expression: evolving paradigms in human oncogenesis. Am J Transplant 17:1674–1680. doi: 10.1111/ajt.14191. [DOI] [PubMed] [Google Scholar]
- 7.Nickeleit V, Singh HK, Goldsmith CS, Miller SE, Kenan DJ. 2013. BK virus-associated urinary bladder carcinoma in transplant recipients: productive or nonproductive polyomavirus infections in tumor cells? Hum Pathol 44:2870–2871. doi: 10.1016/j.humpath.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winter BJ, O’Connell HE, Bowden S, Carey M, Eisen DP. 2015. A case control study reveals that polyomaviruria is significantly associated with interstitial cystitis and vesical ulceration. PLoS One 10:e0137310. doi: 10.1371/journal.pone.0137310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang M, Abend JR, Tsai B, Imperiale MJ. 2009. Early events during BK virus entry and disassembly. J Virol 83:1350–1358. doi: 10.1128/JVI.02169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moriyama T, Sorokin A. 2008. Intracellular trafficking pathway of BK virus in human renal proximal tubular epithelial cells. Virology 371:336–349. doi: 10.1016/j.virol.2007.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyson N, Buchkovich K, Whyte P, Harlow E. 1989. The cellular 107K protein that binds to adenovirus E1A also associates with the large T antigens of SV40 and JC virus. Cell 58:249–255. doi: 10.1016/0092-8674(89)90839-8. [DOI] [PubMed] [Google Scholar]
- 12.Harris KF, Christensen JB, Imperiale MJ. 1996. BK virus large T antigen: interactions with the retinoblastoma family of tumor suppressor proteins and effects on cellular growth control. J Virol 70:2378–2386. doi: 10.1128/JVI.70.4.2378-2386.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sullivan CS, Baker AE, Pipas JM. 2004. Simian virus 40 infection disrupts p130-E2F and p107-E2F complexes but does not perturb pRb-E2F complexes. Virology 320:218–228. doi: 10.1016/j.virol.2003.10.035. [DOI] [PubMed] [Google Scholar]
- 14.Srinivasan A, McClellan AJ, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky JL, Pipas JM. 1997. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol Cell Biol 17:4761–4773. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linzer DI, Levine AJ. 1979. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 16.Lane DP, Crawford LV. 1979. T antigen is bound to a host protein in SV40-transformed cells. Nature 278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 17.Bargonetti J, Reynisdottir I, Friedman PN, Prives C. 1992. Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53. Genes Dev 6:1886–1898. doi: 10.1101/gad.6.10.1886. [DOI] [PubMed] [Google Scholar]
- 18.Jiang D, Srinivasan A, Lozano G, Robbins PD. 1993. SV40 T antigen abrogates p53-mediated transcriptional activity. Oncogene 8:2805–2812. [PubMed] [Google Scholar]
- 19.Lilyestrom W, Klein MG, Zhang R, Joachimiak A, Chen XS. 2006. Crystal structure of SV40 large T-antigen bound to p53: interplay between a viral oncoprotein and a cellular tumor suppressor. Genes Dev 20:2373–2382. doi: 10.1101/gad.1456306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fanning E, Zhao K. 2009. SV40 DNA replication: from the A gene to a nanomachine. Virology 384:352–359. doi: 10.1016/j.virol.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stillman B, Gerard RD, Guggenheimer RA, Gluzman Y. 1985. T antigen and template requirements for SV40 DNA replication in vitro. EMBO J 4:2933–2939. doi: 10.1002/j.1460-2075.1985.tb04026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wold MS, Li JJ, Kelly TJ. 1987. Initiation of simian virus 40 DNA replication in vitro: large-tumor-antigen- and origin-dependent unwinding of the template. Proc Natl Acad Sci U S A 84:3643–3647. doi: 10.1073/pnas.84.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang M, Zhao L, Gamez M, Imperiale MJ. 2012. Roles of ATM and ATR-mediated DNA damage responses during lytic BK polyomavirus infection. PLoS Pathog 8:e1002898. doi: 10.1371/journal.ppat.1002898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verhalen B, Justice JL, Imperiale MJ, Jiang M. 2015. Viral DNA replication-dependent DNA damage response activation during BK polyomavirus infection. J Virol 89:5032–5039. doi: 10.1128/JVI.03650-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Justice JL, Needham JM, Thompson SR. 2019. BK polyomavirus activates the DNA damage response to prolong S phase. J Virol 93:e00130-19. doi: 10.1128/JVI.00130-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruda MC, Alwine JC. 1991. Simian virus 40 (SV40) T-antigen transcriptional activation mediated through the Oct/SPH region of the SV40 late promoter. J Virol 65:3553–3558. doi: 10.1128/JVI.65.7.3553-3558.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller JM, Alwine JC. 1984. Activation of the SV40 late promoter: direct effects of T antigen in the absence of viral DNA replication. Cell 36:381–389. doi: 10.1016/0092-8674(84)90231-9. [DOI] [PubMed] [Google Scholar]
- 28.Low J, Humes HD, Szczypka M, Imperiale M. 2004. BKV and SV40 infection of human kidney tubular epithelial cells in vitro. Virology 323:182–188. doi: 10.1016/j.virol.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 29.An P, Saenz Robles MT, Duray AM, Cantalupo PG, Pipas JM. 2019. Human polyomavirus BKV infection of endothelial cells results in interferon pathway induction and persistence. PLoS Pathog 15:e1007505. doi: 10.1371/journal.ppat.1007505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abend JR, Low JA, Imperiale MJ. 2010. Global effects of BKV infection on gene expression in human primary kidney epithelial cells. Virology 397:73–79. doi: 10.1016/j.virol.2009.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. 2015. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shapiro E, Biezuner T, Linnarsson S. 2013. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet 14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 33.Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C. 2017. Single-cell mRNA quantification and differential analysis with Census. Nat Methods 14:309–315. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C. 2017. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhai W, Tuan JA, Comai L. 1997. SV40 large T antigen binds to the TBP-TAF(I) complex SL1 and coactivates ribosomal RNA transcription. Genes Dev 11:1605–1617. doi: 10.1101/gad.11.12.1605. [DOI] [PubMed] [Google Scholar]
- 37.Damania B, Mital R, Alwine JC. 1998. Simian virus 40 large T antigen interacts with human TFIIB-related factor and small nuclear RNA-activating protein complex for transcriptional activation of TATA-containing polymerase III promoters. Mol Cell Biol 18:1331–1338. doi: 10.1128/mcb.18.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cavender JF, Mummert C, Tevethia MJ. 1999. Transactivation of a ribosomal gene by simian virus 40 large-T antigen requires at least three activities of the protein. J Virol 73:214–224. doi: 10.1128/JVI.73.1.214-224.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Assetta B, De Cecco M, O’Hara B, Atwood WJ. 2016. JC polyomavirus infection of primary human renal epithelial cells is controlled by a type I IFN-induced response. mBio 7:e00903-16. doi: 10.1128/mBio.00903-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saenz Robles MT, Case A, Chong JL, Leone G, Pipas JM. 2011. The retinoblastoma tumor suppressor regulates a xenobiotic detoxification pathway. PLoS One 6:e26019. doi: 10.1371/journal.pone.0026019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sompayrac L. 1997. SV40 and adenovirus may act as cocarcinogens by downregulating glutathione S-transferase expression. Virology 233:130–135. doi: 10.1006/viro.1997.8610. [DOI] [PubMed] [Google Scholar]
- 42.Drayman N, Patel P, Vistain L, Tay S. 2019. HSV-1 single-cell analysis reveals the activation of anti-viral and developmental programs in distinct sub-populations. Elife 8:e46339. doi: 10.7554/eLife.46339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russell AB, Trapnell C, Bloom JD. 2018. Extreme heterogeneity of influenza virus infection in single cells. Elife 7:e32303. doi: 10.7554/eLife.32303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shnayder M, Nachshon A, Krishna B, Poole E, Boshkov A, Binyamin A, Maza I, Sinclair J, Schwartz M, Stern-Ginossar N. 2018. Defining the transcriptional landscape during cytomegalovirus latency with single-cell RNA sequencing. mBio 9:e00013-18. doi: 10.1128/mBio.00013-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zanini F, Pu SY, Bekerman E, Einav S, Quake SR. 2018. Single-cell transcriptional dynamics of flavivirus infection. Elife 7:e32942. doi: 10.7554/eLife.32942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crawford L, Leppard K, Lane D, Harlow E. 1982. Cellular proteins reactive with monoclonal antibodies directed against simian virus 40 T-antigen. J Virol 42:612–620. doi: 10.1128/JVI.42.2.612-620.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zilionis R, Nainys J, Veres A, Savova V, Zemmour D, Klein AM, Mazutis L. 2017. Single-cell barcoding and sequencing using droplet microfluidics. Nat Protoc 12:44–73. doi: 10.1038/nprot.2016.154. [DOI] [PubMed] [Google Scholar]
- 48.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]