Abstract
Since 2015, macroalgae blooms have appeared along the Qinhuangdao coast of the Bohai Sea in China and they have recurred annually during the months of April to September. One of the causal species that results in the macroalgal blooms, Ulva pertusa, has been detrimental to the environment and ecosystem along the coast of the Qinhuangdao, China. In the present study, we sequenced the chloroplast genome of U. pertusa for the first time (GenBank accession number MN853875) and found that the annular genome comprised 104,380 base pairs, including 71 protein-coding genes, 26 tRNAs, and 2 rRNAs. We then constructed a phylogenetic tree of U. pertusa and 17 other species based on core genes, which showed that U. pertusa is the closest sister species of U. fasciata.
Keywords: Macroalgae blooms, chloroplast genome, Ulva pertusa, phylogenetic analysis, Ulva fasciata
Since 2007, the world’s largest macroalgae blooms consisted primarily of Ulva prolifera and recurred on the coast of Qingdao City in Shandong Province of China, which has adversely affected the local environment and tourism (Liu et al. 2009, 2010, 2013; Zhao et al. 2013; Zhang et al. 2017). However, in 2015, a novel macroalgae bloom occurred in the coastal waters of Qinhuangdao City on the western coast of the Bohai Sea making it the second coastal area in China influenced by macroalgae blooms (Han et al. 2019; Song, Han, et al. 2019; Song, Wang, et al. 2019). The bloom’s process had obvious species succession exhibited by three distinct development stages (Song, Han, et al. 2019; Song, Wang, et al. 2019). Ulva pertusa was the dominant species of stage I, which occurred from late April to mid-May (Song, Wang, et al. 2019). Compared with the mitogenome, the chloroplast genome contains more protein-coding genes, which plays an important role in cytoplasmic inheritance, plant phylogeny, DNA barcoding, and genetic diversity (Cai et al. 2017, 2018). Therefore, in this study, we determined the complete chloroplast genome sequence of Ulva pertusa.
Ulva pertusa was collected from the Qinhunangdao coastal area of the Hebei Province (39°49′54.69″N, 119°31′30.50″E). The specimens were preserved at the Marine Ecology Research Center of the First Institute Oceanography, Ministry of Natural Resources in Qingdao (Accession number: KSC06). The shape of genome of U. pertusa is annular with GenBank accession number MN853875. The content of A + T is 74.34%. The complete chloroplast genome sequence is 104,380 bps long. There are 71 protein-coding genes in the genome, including 15 psb genes, 11 rps genes, 10 rpl genes, 6 psa genes, 6 atp genes, 5 pet genes, 5 ycf genes, and 4 rpo genes. In addition, there are some genes that appeared only once in the genome, including acc, ccs, cem, chl, clp, fts, inf, rbcL and tufA genes. Furthermore, there are 26 tRNAs and 2 rRNAs (rrl and rrs) which are non-coding genes in the genome. All the coding genes begin with ATG except for rps19, which begins with AAT. The termination codons for rpoA, rpl2, rpl23, psbT, psaA, rpl20, chlI and atpI are TAG. In addition, rpoA terminates with TGA and the rest of the 63 genes end with TAA.
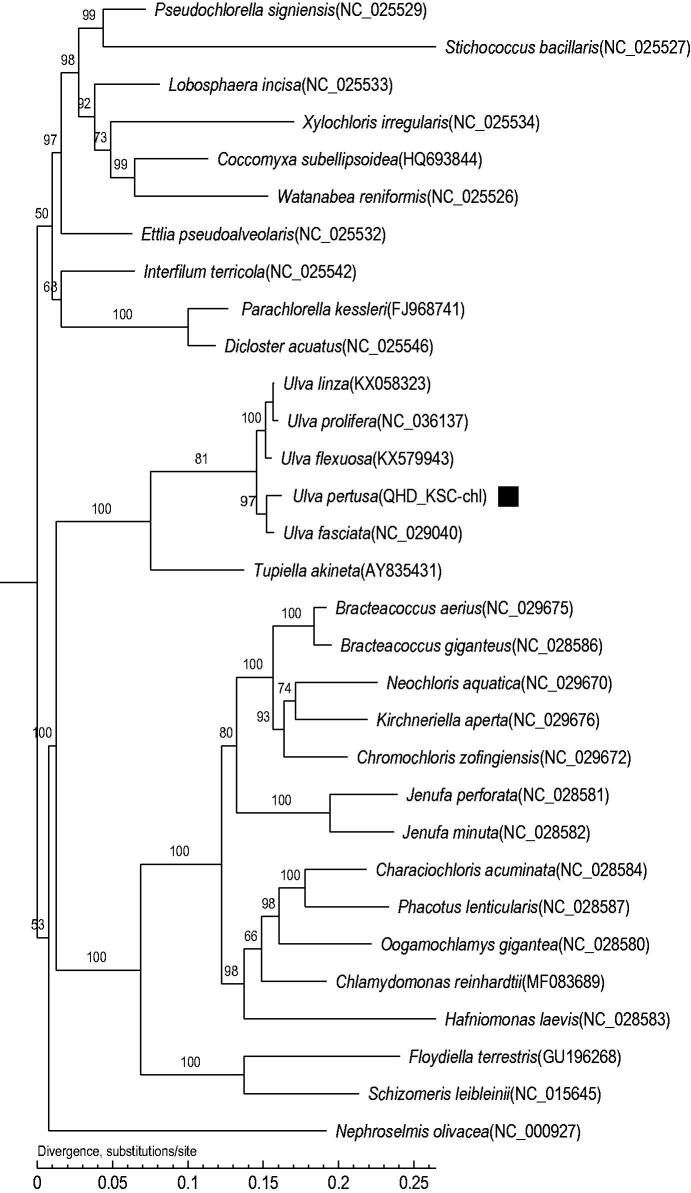
To investigate the phylogenetic status of U. pertusa, a maximum-likelihood (ML) phylogenetic tree including U. pertusa and other 30 species was constructed using the software MEGA-X with 1000 bootstrap replicates (Kumar et al. 2018). As shown in the phylogenetic tree (Figure 1), U. pertusa was most closely related to U. fasciata.
Figure 1.
Maximum-likelihood (ML) tree based on the complete chloroplast genome sequences of 31 species. The numbers on the branches are bootstrap values.
In this study, we reported and characterized the complete chloroplast genome sequence of U. pertusa, which will be useful for studying genetic diversity and phylogenetic history of U. pertusa and its related species.
Funding Statement
This work was financially supported by the National Key R&D Program of China [2019YFC1407902], National Natural Science Foundation of China [41876140/41606190]. The funders did not play a role in the study design, data collection and analysis, decision to publish, or manuscript preparation.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Cai CE, Wang LK, Jiang T, Zhou LJ, He PM, Jiao BH. 2018. The complete mitochondrial genomes of green tide algae Ulva flexuosa (Ulvophyceae,Chlorophyta). Conserv Genet Resour. 10(3):415–418. [Google Scholar]
- Cai CE, Wang LK, Zhou LJ, He PM, Jiao BH. 2017. Complete chloroplast genome of green tide algae Ulva flexuosa (Ulvophyceae, Chlorophyta) with comparative analysis. PLOS One. 12(9):e0184196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han HB, Song W, Wang ZL, Ding DW, Yuan C, Zhang XL, Li Y. 2019. Distribution of green algae micro-propagules and their function in the formation of the green tides in the coast of Qinhuangdao, the Bohai Sea, China. Acta Oceanol Sin. 8:72–77. [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Keesing J, Dong Z, Zhen Y, Di B, Shi Y, Fearns P, Shi P. 2010. Recurrence of Yellow Sea green tide in June 2009 confirms coastal seaweed aquaculture provides nursery for generation of macroalgal blooms. Mar Pollut Bull. 60(9):1423–1432. [DOI] [PubMed] [Google Scholar]
- Liu D, Keesing JK, Xing Q, Shi P. 2009. World’s largest macroalgal bloom caused by expansion of seaweed aquaculture in China. Mar Pollut Bull. 58(6):888–895. [DOI] [PubMed] [Google Scholar]
- Liu D, Keesing J K, He P, Wang Z, Shi Y, Wang Y. 2013. The world’s largest macroalgal bloom in the Yellow Sea, China: formation and implications. Estuar Coast Shelf Sci. 129:2–10. [Google Scholar]
- Song W, Han HB, Wang ZL, Li Y. 2019. Molecular identification of the macroalgae that cause green tides in the Bohai Sea, China. Molecular identification of the macroalgae that cause green tides in the Bohai Sea. China. Aquat Bot. 156:38–46. [Google Scholar]
- Song W, Wang ZL, Li Y, Han HB, Zhang XL. 2019. Tracking the original source of the green tides in the Bohai Sea, China. Estuar Coast Shelf Sci. 219:354–362. [Google Scholar]
- Zhang JH, Zhao P, Huo YZ, Yu KF, He PM, 2017. The fast expansion of Pyropia aquaculture in Sansha regions should be mainly responsible for the Ulva blooms in Yellow Sea. Estuar Coast Shelf Sci. 189:58–65. [Google Scholar]
- Zhao J, Jiang P, Liu Z, Wei W, Lin HZ, Li FC, Wang JF, Qin S, 2013. The Yellow Sea green tides were dominated by one species, Ulva (Enteromorpha) prolifera, from 2007 to 2011. Chin Sci Bull. 58(19):2298–2302. 5. [Google Scholar]