Abstract
Background
Escherichia coli is the most predominant pathogen involved in UTIs. Mainly, fimbrial surface appendages are implicated in adherence to urothelium besides non-fimbrial proteins.
Objectives
to determine prevalence of genes encoding fimbrial and non-fimbrial proteins among Uropathogenic Escherichia coli (UPEC). Furthermore, distribution of these genes and biofilm formation capacity were investigated in relation to antimicrobial resistance.
Methods
Antimicrobial susceptibility of 112 UPEC isolates was performed using disc diffusion method. ESBL production was confirmed by double disc synergy test. Genes encoding fimbrial and non-fimbrial proteins were detected using PCR and biofilm formation was investigated using microtitre plate assay.
Results
UPEC isolates exhibited high resistance against doxycyclines (88.39 %), β-lactams (7.14–86.6%), sulphamethoxazole-trimethoprim (53.75%) and fluoro-quinolones (50%). Fifty percent of tested isolates were ESBL producers. PapGII gene was statistically more prevalent among pyelonephritis isolates. SfaS, focG and picU genes were statistically associated with fluoroquinolone (FQs) sensitive isolates and Dr/afaBC gene was statistically associated with ESBL production. Moreover, non-MDR isolates produced sturdier biofilm.
Conclusion
PapGII adhesin variant seems to have a critical role in colonization of upper urinary tract. There is a possible link between antimicrobial resistance and virulence being capable of affecting the distribution of some genes besides its negative impact on biofilm formation.
Keywords: Urinary tract infection, Escherichia coli, UPEC, adhesin genes, ESBL, biofilm
Introduction
Urinary tract infections (UTIs) are one of the most common bacterial infections caused by a wide spectrum of microorganisms, uropathogenic Escherichia coli (UPEC) is the main causative agent of UTIs including community acquired and nosocomial infection1. UTIs can be limited to the bladder (cystitis) with mild localized symptoms or extend to the kidney (pyelonephritis) with more serious symptoms which can be developed into life threating septicemia2.
To establish UTI, the invading UPEC must overcome the repulsive forces present between its surface and the urothelium cells, which is mediated mainly by the mean of chaperon-usher pathway (CUP) fimbrial surface appendages3. Fimbrial appendages are a hetero-polymer protein which ends with adhesin protein subunit recognizing and binding to specific receptor allowing the attachment of invading UPEC to urothelium4. Type -1 fimbriae is the prototype of this family playing a critical role in the colonization of the bladder through recognition of mannosylated uroplakins receptors within superficial umbrella cell layer of bladder urothelium5. UPEC possesses other fimbrial appendages including P-fimbriae, S-fimbriae and F1C fimbriae which recognize different receptors along urinary tract 4, 6. Dr-adhesin family is another member of CUP appendages which is essential for establishment of chronic and recurrent infections7.
Moreover, several non fimbrial surface proteins are involved in the colonization of urinary tract such as iron-regulated gene A homologue adhesin (Iha) and ompT protein which are integrated outer membrane proteins8,9. Autotransport (AT) proteins are another class of non-fimbrial proteins which can be either surface localized or secreted into the surrounding environment. Ag43 is a surface localized AT protein involved in long term colonization and formation of biofilm on abiotic surface as well as intracellular bacterial communities10, 11. PicU is a secreted AT protein facilating the adherence of the invading UPEC by breaking down mucin layer lining the apical surface of the urothelium12.
Antimicrobial resistance is mainly due target genes mutation13, or acquisition of resistance genes via mobile genetic elements such as plasmid and integrons which could provide co-resistance to different antimicrobial agents14. Moreover, biofilm formation provides an additional protective approach by which the encased bacterial cells can avoid the destructive effect of antimicrobial agents as well as drastic environmental conditions15. The fact that virulence genes as well as antimicrobial resistance genes could be transferred together by the mean of plasmid or other transferable genetic element besides the capability of the acquired antimicrobial resistance such as fluoro-quinolones (FQs) resistance to affect gene expression among resistant isolates16 indicates a possible relationship between the acquired antimicrobial resistance and the virulence. In this study, we determined first the distribution of genes encoding fimbrial and non-fimbrial proteins among cystitis and pyelonephritis UPEC isolates. Then we evaluated if the distribution of these genes as well as the biofilm formation capacity could be affected by the antimicrobial resistance of UPEC isolates.
Materials and methods
Bacterial isolates
A total of 382 clean-catch midstream urine specimens were collected from adult patients (20 – 49 years old) admitted to the outpatient urology clinics of Zagazig university hospitals suffering from symptoms of urinary tract infection in the period from June 2016 to August 2017. Quantitative urine culture was performed using a colony count of 105 CFU / mL as a cut off value for positive urine culture17.
Out of 180 urine specimens showed significant bacteruria, 112 UPEC isolates were recovered including 65 isolates obtained from patients clinically diagnosed with cystitis and the remaining 47 isolates were obtained from patients with pyelonephritis. All isolates were identified by standard microbiological tests18 and stored in Muller Hinton media as 20% glycerol stocks at -80°C.
Antimicrobial susceptibility testing
The antimicrobial susceptibility was performed using standard disc diffusion method19. Tested antimicrobials were belongs to β-lactams, FQs, aminoglycosides, tetracycline, folate pathway inhibitors and nitrofurans classes. Amoxicillin-clavulanate (AMC, 20/10µg), piperacillin-tazobactam (TZP, 100/10 µg), ceftriaxone (CRO, 30 µg), cefpodoxime (CPD, 10 µg), cefuroxime (CXM, 30 µg), aztreonam (ATM, 30 µg), imipenem (IMP, 10 µg), meropenem (MEM, 10 µg), gentamicin (CN, 10 µg), doxycycline (DO, 30µg) and ciprofloxacin (CIP, 5µg) were purchased from Oxoid (Hampshire, UK). The other antimicrobial discs including levofloxacin (LEV, 5 µg), ofloxacin (OFX, 5µg), trimethoprim-sulfamethoxazole (SXT, 1.25/23.75 µg) and nitrofurantoin (F, 300 µg) were obtained from Bioanalysis (Ankra, Turkey). Isolates which were resistant to at least three antimicrobial classes were considered as MDR isolates.
Phenotypic detection of extended spectrum β-lactamases (ESBL) production
Isolates with reduce susceptibility against extended spectrum β lactams were considered as potential ESBL according to CLSI guidelines19. ESBL production in these isolates was confirmed by double disc synergy test (DDST20). This test depends on the detection of synergy between amoxicillin-clavulanate (AMC, 20/10 µg) disc which placed at a distance 20 mm from ceftazidime (CTZ, 30 µg), ceftriaxone (CRO, 30 µg) and cefotaxime (CTX, 30 µg) discs representing third generation cephalosporins along with cefepime (FEB, 30 µg) disc as a fourth generation cephalosporin. Any distortion or enhancement of the inhibition zone of the tested antibiotics toward amoxicillin-clavulanate disc was considered as a positive result for ESBL production20.
Adhesin gene detection using PCR
Bacterial DNA extraction was performed using optimized heat shock method. Briefly, colonies from overnight culture were suspended in 200 µL of sterile water and incubated at 100°C for 10 min and followed by centrifugation where supernatant used as template DNA21.
Primers used in gene amplification were obtained from LGC Biosearch Technologies (Petaluma, CA, USA) and were listed in Table 1. Amplification reaction was performed using Biometra T thermocycler (Analytik Jena, Germany). Each reaction contained 10 µL of MyTaqTM master mix (2x), 1.5 µL forward primer, 1.5 µL reverse primer, 2 µL DNA template and nuclease free water were added to 20 µL, a negative control reaction without DNA was included for each gene amplification. Amplification products along with Quick-Load 100 bp DNA ladder (New England Biolabs, UK) were electrophorised on 1% agarose gel containing ethidium bromide (0.5 µg/mL) and visualized using Cole-Parmer UV-transilluminator (Vernon Hills, USA).
Table 1.
Primers used for amplification of adhesin genes with the corresponding annealing temperature and amplicon size.
| Gene | Primer sequence (5′ to 3′) | Annealing temperature |
Amplicon size (bp) |
Reference |
| PapGII | GGGATGAGCGGGCCTTTGAT, CGGGCCCCCAAGTAACTCG |
62°C | 190 | Tseng et al.21 |
| PapGIII | GGCCTGCAATGGATTTACCTGG, CCACCAAATGACCATGCCAGAC |
65°C | 258 | Tseng et al.21 |
| sfaS | GTGGATACGACGATTACTGTG, CCGCCAGCTTCCCTGTATTC |
63°C | 240 | Johnson and Stell.22 |
| focG | CAGCACAGGCAGTGGATACGA, GAATGTCGCCTGCCCATTGCT |
63°C | 360 | Johnson and Stell.22 |
| Dr/afaBC | GGCAGAGGGCCGGCAACAGGC, CCCGTAACGCGCCAGCATCTC |
69°C | 559 | Johnson and Stell.22 |
| Ag43 | CTGGAAACCGGTCTGCCCTT CCTGAACGCCCAGGGTGATA |
63°C | 433 | Restieri et al.23 |
| picU | ACTGGATCTTAAGGCTCAGGAT GACTTAATGTCACTGTTCAGCG |
60°C | 572 | Restieri et al.23 |
| Iha | CTGGCGGAGGCTCTGAGATCA TCCTTAAGCTCCCGCGGCTGA |
65°C | 827 | Johnson et al.24 |
| ompT | ATCTAGCCGAAGAAGGAGGC CCCGGGTCATAGTGTTCATC |
60°C | 559 | Johnson et al.24 |
In vitro biofilm formation assay
Biofilm forming capacity was performed according to the method of StepanoviC and coworkers25. Briefly, bacterial suspension, from overnight cultures, with a turbidity equivalent to 0.5 McFarland standard was diluted to 1:100 with Tryptone Soya Broth then 100 µL aliquots were dispensed into the wells of 96 wells -microtiter plate where the assay was performed as triplicate including blank wells as a negative control. Microtiter plate incubated for 24 hr at 37°C, non-adherent cells were removed by washing then adherent cells were fixed by adding 150 µL of methanol for 20 min. Formed biofilm was stained with 150 µL 2% crystal violet solution for 15 minutes, excess dye removed by washing and adherent dye was solubilized by adding 150 µL of 33% glacial acetic acid for 30 min
Optical density (OD) values were measured at 570 nm using BioTek synergy HT microtiter plates reader (Vermont, USA) and averaged. The cut -off optical density (ODc) value for each plate was calculated according to following equation:
ODc = average OD of negative control + (3* SD of negative control).
Biofilm forming capacity was interpreted as: non producer when OD ≤ ODc, weak biofilm producer when ODc < OD ≤ 2 ODc, moderate biofilm producer when 2 ODc < OD ≤ 4 ODc and strong biofilm producer when 4 ODc < OD.
Statistical analysis
Comparison of proportions was performed using chisquare test using MedCalc program (version 17.11.5). Results with p ≤ 0.05 or p ≤ 0.001 were marked with single or double asterisks, respectively and considered statistically significant.
Results
Antimicrobial susceptibility testing
UPEC isolates exhibited varied susceptibility rates towards different antimicrobials (Figure 1). The highest resistance was observed against doxycycline, amoxacilin-clavulanic acid, cefuroxime and cefpodoxime (88.39%, 86.8%, 71.4% and 66%, respectively). Intermediate resistance was observed against sulphamethoxazole-trimethoprim (SXT), ceftriaxone, FQs and aztreonam (53.75%, 51%, 50% and 40.17%, respectively).
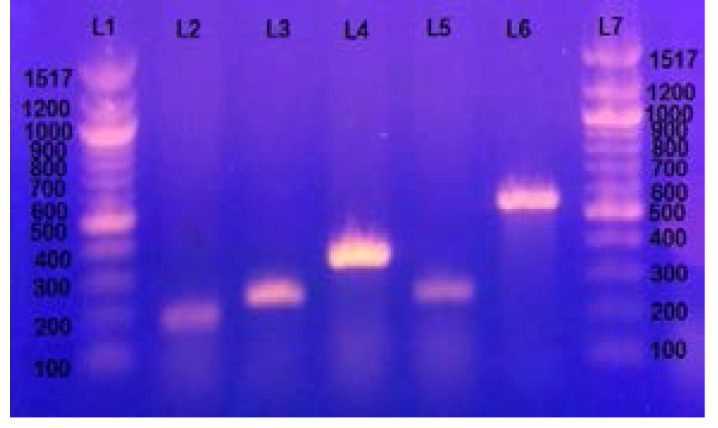
Figure 1.
Gel electrophoresis of genes encoding fimbrial adhesins : L1 and L7 represent 100bp DNA ladder, L2 represents papGII allele band approximately at 190 bp, L3 represents papGIII allele band approximately at 258 bp, L4 represents focG gene approximately at 360 bp, L5 represents sfaS gene approximately at 240 bp and L6 represents Dr/afaBC gene band approximately at 559 bp.
While lower resistance was observed against nitrofurantoin, gentamicin and piperacillin-tazobactam (18.75%, 17.85% and 15.18%, respectively). The lowest resistance was against imipenem and meropenem where 7.41% of tested isolates were resistant for each. Multidrug resistance was observed in 65.2% of the tested isolates.
Extended spectrum β lactamase production
The initial antimicrobial susceptibility testing revealed that 70 UPEC isolates exhibited a reduced susceptibility towards β- lactams. Fifty six isolates were confirmed to be ESBL producers by double disc synergy test representing 50% of the total UPEC isolates.
Prevalence of adhesin genes among UPEC isolates
Detection of genes encoding fimbrial and non-fimbrial proteins was done by PCR and all genes gave a single band at the expected size (Table 1) but with different prevalence.
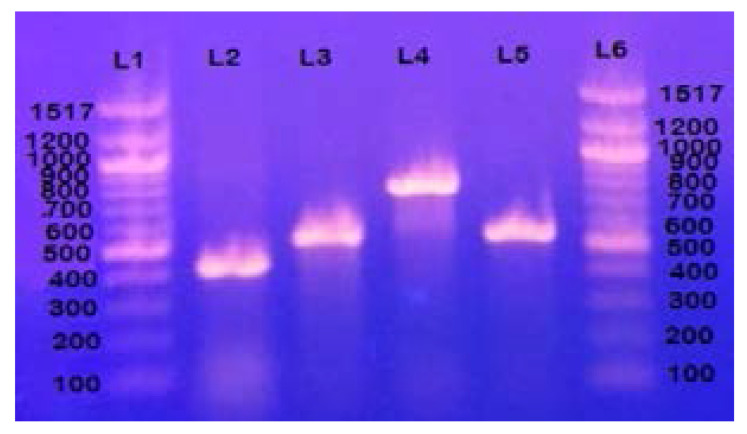
Among the fimbrial adhesin genes (Figure 1 & Table 2), papG gene alleles (papGII and pagGIII) were the most prevalent (49.1 %) followed by focG gene (14.3 %), dr/afaBC gene (9.8%) and sfaS gene (8.9 %). For the genes encoding non fimbrial proteins (Figure 2 & Table 2), ag43 gene was the most prevalent (89.3 %) followed by ompT gene (51.8 %) and iha gene (31.25%), while picU gene was detected only in 9.8% of strains.
Table 2.
prevalence of adhesin genes among cystitis and pyelonephritis isolates
| Gene | Total UPEC isolates n=112 (%) |
Cystitis isolates n =65 (%) |
Pyelonephritis isolates n =47 (%) |
| PapG | 55 (49.1) | 27 (41.5) | 28 (59.5) |
| PapGII | 49 (43.8) | 23 (35.4) | 26 (55.3)* |
| PapGIII | 6 (5.3) | 4 (6.2) | 2 (4.25) |
| FocG | 16 (14.3) | 10 (15.4) | 6 (12.7) |
| SfaS | 10 (8.9) | 6 (9.2) | 4 (8.5) |
| Dr/afa | 11 (9.8) | 6 (9.2) | 5 (10.6) |
| Ag43 | 100 (89.3) | 58 (89.2) | 42 (89.36) |
| Iha | 35 (31.25) | 18 (27.6) | 17 (36.17) |
| PicU | 11 (9.8) | 8 (12.3) | 3 (6.38) |
| OmpT | 58 (51.8) | 33 (50.7) | 25 (53.19) |
p-value ≤ 0.05
p-value ≤0.001
Figure 2.
Gel electrophoresis of genes encoding afimbrial proteins: L1 and L6 represent 100bp DNA ladder, L2 represents gene encoding Ag43 autotransport protein band approximately at 433 bp, L3 represents gene encoding secreted autotransprt protein picU band approximately at 572 bp, L4 gene encoding outer membrane protein Iha band approximately at 827 bp and L5 represents gene encoding outer membrane protein ompt band approximately at 559 bp.
Among papG gene alleles, papGII allele was more prevalent than papGIII allele (43.8 % vs. 5.3%). papGII allele was statistically more prevalent in pyelonephritis isolates than cystitis isolates while the remaining genes showed no significant difference between the two clinical groups (Table 2).
Distribution of the adhesin genes in relation to antimicrobial resistance
Prevalence of genes encoding fimbrial and non fimbrial proteins was assessed in relation to increased resistance rate towards SXT and FQs as well as ESBL production (Table 3).
Table 3.
distribution of genes encoding fimbrial and non fimbrial proteins in relation to FQs, SXT resistance and ESBL production.
| Gene | FQ-sensitive n= 56 (%) |
FQ- Resistant n=56 (%) |
SXT- Sensitive n=52 (%) |
SXT- Resistant n=60 (%) |
ESBL(a) Producer n=56 (%) |
Non-ESBL Producer n=56 (%) |
| PapGII | 21 (37.53) | 28 (50) | 20 (38.46) | 29 (48.3) | 24 (42.8) | 25 (44.6) |
| PapGIII | 4 (7.14) | 2 (3.5) | 4 (7.8) | 2 (3.3) | 2 (3.7) | 4 (7.1) |
| FocG | 14 (25)** | 2 (3.5) | 9 (17.3) | 7 (11.6) | 6 (10.7) | 10 (17.8) |
| SfaS | 9 (16.1)** | 1 (1.8) | 8 (15.4)* | 2 (3.3) | 4 (7.2) | 6 (10.7) |
| Dr/afaBC | 4 (7.14) | 7 (12.5) | 3 (5.7) | 8 (13.3) | 9 (16.1)* | 2 (3.5) |
| Ag43 | 48 (85.7) | 52 (92.9) | 44 (84.6) | 56 (93.3) | 50 (89.2) | 50 (89.2) |
| Iha | 16 (28.6) | 19 (33.9) | 15 (28.8) | 20 (33.3) | 18 (32.1) | 17 (30.35) |
| PicU | 10 (17.9)** | 1 (1.8) | 8 (15.4) | 3 (5) | 5 (8.9) | 6 (10.7) |
| OmpT | 34 (60.7) | 24 (42.8) | 28 (53.8) | 30 (50) | 33 (59) | 25 (44.6) |
Only isolates which were confirmed to be ESBL producer by DDST.
p-value ≤ 0.05
p-value ≤0.001
In relation to FQs resistance; sfaS, focG and picU genes were statistically associated with sensitive strains. ompT gene was more prevalent in sensitive isolates than resistant isolates but it was non-significant (p= 0.059).
sfaS gene was statistically associated with isolates sensitive to SXT than resistant ones. picU gene was more prevalent in sensitive isolates but it was non-significant (p= 0.066).
In relation to ESBL production, dr/afaBC gene was statistically associated with ESBL producing strains while the remaining traits showed a similar prevalence among ESBL producing and non-producing strains.
Biofilm formation capacity in relation to antimicrobial resistance
The biofilm formation capacity of 73 MDR isolates and 39 non MDR isolates was evaluated to determine the impact of multiple drug resistance on biofilm formation capacity (Table 4).
Table 4.
Biofilm formation capacity among MDR and non-MDR isolates.
| Biofilm forming capacity |
Total UPEC isolates n=112 (%) |
MDR isolates n=73 (%) |
Non-MDR isolates n=39 (%) |
| Negative | 23 (20.53) | 17 (23.28) | 6 (15.38) |
| Weak | 39 (34.8) | 31 (42.46)* | 8 (20.51) |
| Moderate | 27 (24.1) | 14 (19.18) | 13 (33.33) |
| Strong | 23 (20.53) | 11 (15) | 12 (30.78)* |
p-value ≤ 0.05
p-value ≤0.001
Non-MDR isolates were statistically more capable of producing strong biofilm than MDR isolates and the percentage of non-MDR isolates that could form moderate biofilm was higher than MDR isolates. While MDR isolates statistically tended to form weak biofilm thanon-MDR isolates and the percentage of MDR isolates that couldn't form biofilms was higher than non MDR isolates.
Discussion
UPEC is the main causative agent involved in UTIs. Adherence to the uroepithelium is a critical step to establish an infection. This step is achieved mainly by fimbrial surface appendages beside other non-fimbrial adhesins. Fimbrial appendages recognize and adhere to certain receptors favoring the tropism of invading pathogen towards a given region within urinary tract6.
UTIs are usually treated empirically especially uncomplicated infections. The selective overuse in addition to the misuse of certain antimicrobial classes, especially those with high propensity for collateral damage, is the main cause of high resistance rates and increased percentages of MDR isolates 26. MDR isolates represents a serious health problem in Egypt27.
In this study, the increased resistance rate among UPEC isolates especially against β-lactams, SXT and FQs limits their role in the treatment of such infections. Fifty percent of tested isolates were ESBL producers coming in agreement with other studies in Egypt 28, 29. Moreover, the increased resistance against SXT and FQs was in consistent with previous studies 30, 31.
The genotypic detection of genes encoding fimbrial and non-fimbrial adhesins revealed that papGII allele was observed to be statistically more prevalent in pyelonephritis isolates than cystitis isolates. This statistical predominance of pyelonephritis UPEC isolates was in agreement with other studies32, 33. This predominance along with the abundance of papGII isoreceptor in kidney tissue indicates the role of papGII adhesin variant in ascending infection4.
The impact of the antimicrobial resistance on the distribution of tested genetic traits was evaluated in the term of resistance towards FQs and SXT, representing the highest resistance rates among non-β lactams, in addition to ESBL enzyme production being the main resistance mechanism for β lactams.
The observed negative impact of FQs resistance acquisition on the prevalence of tested adhesin genes was obvious where three virulence traits including sfaS, focG and picU were statistically more prevalent in sensitive isolates. This statistical prevalence was compatible with other studies34–36. This negative impact could be as a result of acquisition of FQs resistance by strains naturally lacking these virulence factors followed by clonal spreading. This hypothesis was denied by Vila and coworkers37 stated that there was no genetic relationship between these tested isolates using pulsed-field gel electrophoresis. Another possible explanation for this negative impact is related to the mutational efect of fluroquinolone. Exposure to even sub-inhibitory concentrations of FQs not only leads to the acquisition of resistance but also induces SOS response which could lead to partial or total loss of pathogenicity islands within which these virulence genes could be located38,39.
The impact of SXT resistance acquisition on gene prevalence was quite similar to FQs where sfaS trait was statistically more prevalent in sensitive isolates.
However, dr/afaBC gene was statistically more prevalent among ESBL-producing isolates being consistent with Johnson and co-workers demonstrating that dr/afaBC gene was statistically associated with resistance to extended spectrum cephalosporins and cephamycins among UPEC isolates40. This could be related to the co-existence of certain virulence genes along with resistance genes on mobile genetic element like plasmid providing a dual survival advantages to recipient pathogen41.
The negative impact of the acquired antibiotic resistance on the biofilm forming capacity among UPEC was previously reported among Acinetobacter baumannii isolates42. Similarly, Poursina and coworkers43 reported that 69.2% of strong biofilm-producing UPEC isolates were non-MDR isolates while negative and weak biofilm producing isolates were MDR isolates.
Several surface appendages including fimbriae and curli in addition to other non-fimbrial proteins are involved in the biofilm architecture as a supporting scaffold. The acquisition of antimicrobial resistance could affect the expression of these organelles which negatively affect the biofilm formation capacity44. Vila and coworkers37 reported that the expression of type-1 fimbriae among FQ resistant UPEC isolates was statistically lower than sensitive isolates. Similarly, the acquisition of gene encoding ESBL enzymes was previously reported to possess a negative impact on the biofilm formation capacity in E. coli and Pseudomonas aeruginosa45. All of these results confirm that acquisition of antimicrobial resistance could have a negative impact on the biofilm formation capacity among UPEC isolates.
Conclusion
papGII adhesin variant is an important urovirulence factor seeming to have a critical role in ascending UTIs. The acquired antimicrobial resistance could affect the distribution of certain virulence genes. Moreover, there is an inverse relationship between the biofilm forming capacity and the multiple drug resistance among UPEC isolates indicating the role of biofilm formation as an alternative protective approach especially for susceptible isolates as well as a possible link between antimicrobial resistance and virulence.
Conflict of interest
The authors declare that they have no conflict of interest
References
- 1.Da Silva GJ, Mendonça N. Association between antimicrobial resistance and virulence in Escherichia coli. Virulence. 2012;3(1):18–28. doi: 10.4161/viru.3.1.18382. [DOI] [PubMed] [Google Scholar]
- 2.Mahon CR. Textbook of diagnostic microbiology. Elsevier Health sciences. (Fifth edition) 2015;1:884–901. [Google Scholar]
- 3.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nature Reviews Microbiology. 2015;13(5):269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Proft T, Baker EN. Pili in Gram-negative and Gram-positive bacteria, structure, assembly and their role in disease. Cellular and Molecular Life Sciences. 2008;66(4):161–169. doi: 10.1007/s00018-008-8477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spaulding CN, Hultgren SJ. Adhesive pili in UTI pathogenesis and drug development. Pathogens. 2016;5(1):30. doi: 10.3390/pathogens5010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antão EM, Wieler LH, Ewers C. Adhesive threads of extraintestinal pathogenic Escherichia coli. Gut Pathogens. 2009;1(1):22. doi: 10.1186/1757-4749-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nowicki B, Selvarangan R, Nowicki S. Family of Escherichia coli Dr adhesins: decay-accelerating factor receptor recognition and invasiveness. The Journal of Infectious Diseases. 2001;183(1):S24–S27. doi: 10.1086/318846. [DOI] [PubMed] [Google Scholar]
- 8.Johnson JR, Jelacic S, Schoening LM, Clabots C, Shaikh N, Mobley HLT, et al. The IrgA homologue adhesin Iha Is an Escherichia coli virulence factor in murine urinary tract Infection. Infection and Immunity. 2005;73(2):965–971. doi: 10.1128/IAI.73.2.965-971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hritonenko V, Stathopoulos C. Omptin proteins: an expanding family of outer membrane proteases in gram-negative Enterobacteriaceae. Molecular Membrane Biology. 2007;24(5–6):395–406. doi: 10.1080/09687680701443822. [DOI] [PubMed] [Google Scholar]
- 10.Klemm P, Hjerrild L, Gjermansen M, Schembri Mark A. Structure-function analysis of the self-recognizing Antigen 43 autotransporter protein from Escherichia coli. Molecular Microbiology. 2003;51(1):283–296. doi: 10.1046/j.1365-2958.2003.03833.x. [DOI] [PubMed] [Google Scholar]
- 11.Zalewska-Piatek B, Zalewska-Piatek R, Olszewski M, Kur J. Identification of antigen Ag43 in uropathogenic Escherichia coli Dr+ strains and defining its role in the pathogenesis of urinary tract infections. Microbiology. 2015;161(5):1034–1049. doi: 10.1099/mic.0.000072. [DOI] [PubMed] [Google Scholar]
- 12.Heimer SR, Rasko DA, Lockatell CV, Johnson DE, Mobley HLT. Autotransporter genes pic and tsh are associated with Escherichia coli strains that cause acute pyelonephritis and are expressed during urinary tract infection. Infection and Immunity. 2004;72(1):593–597. doi: 10.1128/IAI.72.1.593-597.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shariff VAAR, Shenoy MS, Yadav T, M R. The antibiotic susceptibility patterns of uropathogenic Escherichia coli, with special reference to the fluoroquinolones. Journal of Clinical and Diagnostic Research. 2013;7(6):1027–1030. doi: 10.7860/JCDR/2013/4917.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadry AA, Serry FM, El-Ganiny AM, El-Baz AM. Integron occurrence is linked to reduced biocide susceptibility in multidrug resistant Pseudomonas aeruginosa. British Journal of Biomedical Sciences. 2017;74(2):78–84. doi: 10.1080/09674845.2017.1278884. [DOI] [PubMed] [Google Scholar]
- 15.Stewart PS. Mechanisms of antibiotic resistance in bacterial biofilms. International Journal of Medical Microbiology. 2002;292(2):107–113. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- 16.Yamane T, Enokida H, Hayami H, Kawahara M, Nakagawa M. Genome-wide transcriptome analysis of fluoroquinolone resistance in clinical isolates of Escherichia coli. International Journal of Urology. 2011;19(4):360–368. doi: 10.1111/j.1442-2042.2011.02933.x. [DOI] [PubMed] [Google Scholar]
- 17.Wilson ML, Gaido L. Laboratory diagnosis of Urinary tract Infections in adult patients. Clinical Infectious Diseases. 2004;38(8):1150–1158. doi: 10.1086/383029. [DOI] [PubMed] [Google Scholar]
- 18.Koneman E, Winn WJ. Koneman's Color Atlas and Textbook of Diagnostic Microbiology. 6th ed. London: Lippincott Williams & Wilkins; 2006. [Google Scholar]
- 19.Clinical and Laboratory Standards Institute, author. Performance Standards for Antimicrobial Susceptibility Testing; Twenty 2nd informational Supplement. Wayne, PA, USA: 2012. CLSI document M100S22. [Google Scholar]
- 20.Garrec H, Drieux-Rouzet L, Golmard J-L, Jarlier V, Robert J. Comparison of nine phenotypic methods for detection of Extended-Spectrum β-Lactamase production by Enterobacteriaceae. Journal of Clinical Microbiology. 2011;49(3):1048–1057. doi: 10.1128/JCM.02130-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tseng CC, Huang JJ, Ko WC, Yan JJ, Wu JJ. Decreased predominance of papG class II allele in Escherichia coli strains isolated from adult with acute pyelonephritis and urinary tract abnormalities. The Journal of Urology. 2001;166(5):1643–1646. [PubMed] [Google Scholar]
- 22.Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. The Journal of Infectious Diseases. 2000;181(1):261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 23.Restieri C, Garriss G, Locas MC, Dozois CM. Autotransporter-encoding sequences are phylogenetically distributed among Escherichia coli clinical isolates and reference strains. Applied and Environmental Microbiology. 2007;73(5):1553–1562. doi: 10.1128/AEM.01542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson JR, Russo TA, Tarr PI, Carlino U, Bilge SS, Vary JC, et al. Molecular epidemiological and phylogenetic associations of two novel putative virulence genes, iha and iroN E. coli, among Escherichia coli isolates from Patients with urosepsis. Infection and Immunity. 2000;68(5):3040–3047. doi: 10.1128/iai.68.5.3040-3047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.StepanoviĆ S, VukoviĆ D, Hola V, Bonaventura Giovanni DI, DjukiĆ S, ĆIrkoviĆ I, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by Staphylococci. Journal of pathology, microbiology and immunology. 2007;115(8):891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 26.Gupta K, Hooton TM, Naber KG, Wullt B, Colgan R, Miller LG, et al. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: A 2010 update by the infectious diseases society of America and the European society for microbiology and infectious diseases. Clinical Infectious Diseases. 2011;52(5):103–120. doi: 10.1093/cid/ciq257. [DOI] [PubMed] [Google Scholar]
- 27.Abbas HA, El-Ganiny AM, Kamel HA. Phenotypic and genotypic detection of antibiotic resistance of Pseudomonas aeruginosa isolated from urinary tract infections. African Health Sciences. 2018;18(1):11–21. doi: 10.4314/ahs.v18i1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morsi SS, Tash MR. Virulance determinants among Extended-spectrum B-lactamases producers of uropathogenic Escherichia coli isolates in Zagazig university hospitals, Egypt. Egyptian Journal of Medical Microbiology. 2016;25(2):101–108. [Google Scholar]
- 29.Elsayed TI, Ismail HA, Elgamal SA, Gad AH. The occurrence of multidrug resistant E. coli which produce ESBL and cause urinary tract infections. Journal of Applied Microbiology and Biochemistry. 2017;1:2–8. [Google Scholar]
- 30.Das R, Perrelli E, Towle V, Van Ness PH, Juthani-Mehta M. Antimicrobial susceptibility of bacteria isolated from urine samples obtained from nursing home residents. Infection Control and Hospital Epidemiology. 2009;30(11):1116–1119. doi: 10.1086/647981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neamati F, Firoozeh F, Saffari M, Zibaei M. Virulence genes and antimicrobial resistance pattern in uropathogenic Escherichia coli isolated from hospitalized patients in Kashan, Iran. Jundishapur Journal of Microbiology. 2015;8(2):257–262. doi: 10.5812/jjm.17514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kudinha T, Johnson JR, Andrew SD, Kong F, Anderson P, Gilbert GL. Distribution of phylogenetic groups, sequence type ST131, and virulence-associated traits among Escherichia colii solates from men with pyelonephritis or cystitis and healthy controls. Clinical Microbiology and Infection. 2013;19(4):173–180. doi: 10.1111/1469-0691.12123. [DOI] [PubMed] [Google Scholar]
- 33.Qin X, Hu F, Wu S, Ye X, Zhu D, Zhang Y, et al. Comparison of adhesin genes and antimicrobial susceptibilities between uropathogenic and intestinal commensal Escherichia coli Strains. PLoS One. 2013;8(4):61169. doi: 10.1371/journal.pone.0061169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson JR, O'Bryan TT, Low DA, Ling G, Delavari P, Fasching Cand, et al. Evidence of commonality between canine and human extraintestinal pathogenic Escherichia coli strains that express papG Allele III. Infection and Immunity. 2000;68(6):3327–3336. doi: 10.1128/iai.68.6.3327-3336.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreno E, Prats G, Sabaté M, Pérez T, Johnson JR, Andreu A. Quinolone, fluoroquinolone and trimethoprim/sulfamethoxazole resistance in relation to virulence determinants and phylogenetic background among uropathogenic Escherichia coli. Journal of Antimicrobial Chemotherapy. 2006;57(2):204–211. doi: 10.1093/jac/dki468. [DOI] [PubMed] [Google Scholar]
- 36.Ferjani S, Saidani M, Ennigrou S, Hsairi M, Slim AF, Boutiba Ben Boubaker I. Multidrug resistance and high virulence genotype in uropathogenic Escherichia coli due to diffusion of ST131 clonal group producing CTX-M-15: an emerging problem in a Tunisian hospital. Folia Microbiologica. 2014;59(3):257–262. doi: 10.1007/s12223-013-0292-0. [DOI] [PubMed] [Google Scholar]
- 37.Vila J, Simon K, Ruiz J, Horcajada J P, Velasco M, Barranco M, et al. Are quinolone-resistant uropathogenic Escherichia coli less virulent? The Journal of Infectious Diseases. 2002;186(7):1039–1042. doi: 10.1086/342955. [DOI] [PubMed] [Google Scholar]
- 38.Soto SM, Jimenez de Anta MT, Vila J. Quinolones induce partial or total loss of pathogenicity islands in uropathogenic Escherichia coli by SOS-dependent or -independent pathways, Respectively. Antimicrobial Agents and Chemotherapy. 2006;50(2):649–653. doi: 10.1128/AAC.50.2.649-653.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin T-T, Kang H-Q, Ma P, Li P-P, Huang L-Y, Gu B. SOS response and its regulation on the fluoroquinolone resistance. Annals of Translational Medicine. 2015;3(22):358. doi: 10.3978/j.issn.2305-5839.2015.12.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson JR, Kuskowski MA, Owens K, Gajewski A, Winokur PL. Phylogenetic origin and virulence genotype in relation to resistance to fluoroquinolones and/or Extended-spectrum cephalosporins and cephamycins among Escherichia coli isolates from animals and humans. The Journal of Infectious Diseases. 2003;188(5):759–768. doi: 10.1086/377455. [DOI] [PubMed] [Google Scholar]
- 41.Johnson JR, Kuskowski MA, O'Bryan TT, Colodner R, Raz R. Virulence genotype and phylogenetic origin in relation to antibiotic resistance profile among Escherichia coli urine sample isolates from Israeli women with acute uncomplicated cystitis. Antimicrobial Agents and Chemotherapy. 2005;49(1):26–31. doi: 10.1128/AAC.49.1.26-31.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi L, Li H, Zhang C, Liang B, Li J, Wang L, et al. Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Frontiers in Microbiology. 2016;7:483. doi: 10.3389/fmicb.2016.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poursina F, Sepehrpour S, Mobasherizadeh S. Biofilm formation in non multidrug-resistant Escherichia coli isolated from patients with urinary tract infection in Isfahan, Iran. Advanced Biomedical Research. 2018;7:40. doi: 10.4103/abr.abr_116_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soto SM, Smithson A, Martinez JA, Horcajada JP, Mensa J, Vila J. Biofilm formation in uropathogenic Escherichia coli strains: Relationship with prostatitis, urovirulence factors and antimicrobial resistance. The Journal of Urology. 2007;177(1):365–368. doi: 10.1016/j.juro.2006.08.081. [DOI] [PubMed] [Google Scholar]
- 45.Gallant CV, Daniels C, Leung JM, Ghosh AS, Young KD, Kotra LP, et al. Common β-lactamases inhibit bacterial biofilm formation. Molecular microbiology. 2005;58(4):1012–1024. doi: 10.1111/j.1365-2958.2005.04892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]